Abstract
Radiotherapy is widely used in the management of advanced colorectal cancer (CRC). However, the clinical efficacy is limited by the safe irradiated dose. Sensitizing tumor cells to radiotherapy via interrupting DNA repair is a promising approach to conquering the limitation. The BRCA1–BARD1 complex has been demonstrated to play a critical role in homologous recombination (HR) DSB repair, and its functions may be affected by HERC2 or BAP1. Accumulated evidence illustrates that the ubiquitination–deubiquitination balance is involved in these processes; however, the precise mechanism for the cross-talk among these proteins in HR repair following radiation hasn't been defined. Through activity-based profiling, we identified PT33 as an active entity for HR repair suppression. Subsequently, we revealed that BAP1 serves as a novel molecular target of PT33 via a CRISPR-based deubiquitinase screen. Mechanistically, pharmacological covalent inhibition of BAP1 with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction, interrupting HR repair. Consequently, PT33 treatment can substantially enhance the sensitivity of CRC cells to radiotherapy in vitro and in vivo. Overall, these findings provide a mechanistic basis for PT33-induced HR suppression and may guide an effective strategy to improve therapeutic gain.
Key words: Pharmacological inhibition, BAP1, HERC2 recruitment, BRCA1, BARD1, Competitively dissociation, HR-Mediated DNA repair, CRC radiosensitization
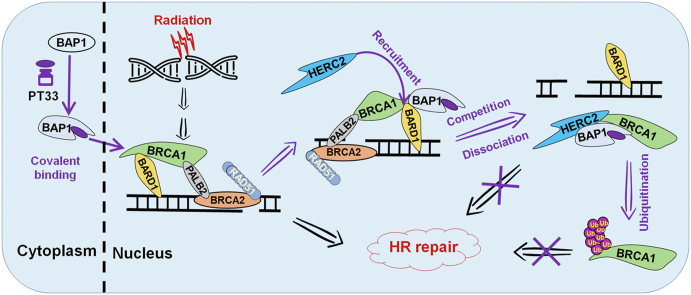
Graphical abstract
Pharmacological covalent inhibition of BAP1 with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction, resulting in the interruption of BRCA1-mediated HR repair.
1. Introduction
Driven by constant technological advances, treatment based on radiotherapy has been the major treatment strategy for advanced colorectal cancer (CRC), especially for locally advanced rectal cancer (LARC)1. As we know, the therapeutic benefit of radiation is enhanced with increasing radiation dose. However, because of a high risk of serious side effects such as radiation enteritis, there is a dose limitation in clinical application2. Thus, an alternative approach to enhance tumor responses and improve clinical outcomes in the standard radiation regimen is to sensitize CRC to radiotherapy. For this to happen, efficient strategies capable of overcoming radioresistance need to be proposed.
Irradiation mainly results in DNA double-strand breaks (DSBs), which are largely responsible for the cellular lethality induced by radiation3. Therefore, the sensitivity of cancer cells to radiation largely depends on the degree of radiation-induced DSBs. In response to DSBs, host of proteins are rapidly recruited to DNA damage sites for repair, which eventually leads to radiotherapy resistance3. In mammalian cells, there are two major pathways for DNA DSB repair: error-free homologous recombination (HR) and error-prone non-homologous end-joining (NHEJ)4, 5, 6. It has been shown that most highly aggressive cancer cells exhibit a higher activity of DSB repair system, which as the common events may render tumors resistant to radiotherapy7. As a consequence, DSB repair mechanisms in response to such radiation must be tightly regulated in order to protect tumors from cell death.
HR-based DSB repair is a multistep process that requires the coordination of multiple repair factors. Thereinto, the BRCA1 DNA repair-associated (BRCA1)–BRCA1-associated RING domain 1 (BARD1) complex plays a critical role in this pathway. Recent investigations have revealed that BARD1 binds to nucleosomes through multivalent interactions, providing the high-affinity recognition of DNA lesions8. And BRCA1 is recruited to DSB sites as a heterodimer with BARD1 to facilitate DNA end resection. Thereby BRCA1 ubiquitinates H2A.X variant histone (H2AX) as an E3 ligase and recruits RAD51 recombinase (RAD51)9, 10, 11. Mutation or deletion of any functional domain on BRCA1 affecting its feature or subcellular location may render cells hypersensitive to DSB damage12, 13, 14. Previous studies implied that BRCA1-associated protein 1 (BAP1) helps to recruit BRCA1 and RAD51 to DSBs in HR repair15,16. However, another study demonstrated that the E3 activity of BRCA1–BARD1 could be inhibited by BAP1, which mediates HR repair and confer radioresistance17. On the other hand, HECT and RLD domain containing E3 ubiquitin-protein ligase 2 (HERC2) is an E3 ligase that targets BRCA1 for ubiquitination and degradation and competes with BARD1 to bind to the RING domain of BRCA118. Nevertheless, neither the regulatory mechanism for the competition of HERC2 and BARD1 binding to BRCA1 nor the precise cross-talk between BAP1 and BRCA1 in HR repair following radiation has been defined.
Given the essential role of ubiquitination in both upstream and downstream of the DSB repair cascade19, it is very likely that deubiquitination, a crucial role in the ubiquitin-mediated regulation of DSB repair, has gradually come to the forefront of mechanism research20, 21, 22. In our preliminary work, we found that the radioresistant CRC cells exhibited a higher activity of intranuclear deubiquitination. We thus speculate that radiation induces more deubiquitinases (DUBs) recruited to DSB sites to facilitate repair. Based on this concept, we applied activity-based profiling to reveal that DUB inhibitors were capable of blocking intranuclear DUB activities elevated by radiation. And identified PT33, a multi-target DUB inhibitor, as an active entity to suppress HR repair following radiation treatment. Subsequently, through a CRISPR-based DUB screen, we identified BAP1 as a novel molecular target of PT33 and found a novel mechanism that pharmacological covalent inhibition of BAP1 with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction, resulting in the interruption of BRCA1-mediated HR repair. Moreover, we showed that PT33 treatment effectively sensitized CRC to radiotherapy in vitro and in vivo. Therefore, our findings provided an optimizing strategy for sensitizing CRC to radiotherapy in clinical application.
2. Materials and methods
2.1. Cell culture and reagents
HCT116, HT-29, HCT-15, SW480, HEK293T and HEK293F cell lines, purchased from American Type Culture Collection (Manassas, VA, USA), were cultured in McCoy's 5A (No. KGM4892N, KeyGen Biotech, Nanjing, China), RPMI-1640 (No. C11875500BT, Thermo Fisher Scientific, Inc. [TFS], Waltham, MA, USA), Leibovitz's L-15 (No. KGM41300N, KeyGen Biotech) and DMEM (No. C11995500, TFS), respectively, supplemented with 10% fetal bovine serum (No. P30-2302, Pan Biotech, Aidenbach, Bavaria, Germany) and 1% penicillin–streptomycin (No. P4333, Sigma–Aldrich, St. Louis, MO, USA). Cells were maintained at 37 °C in 5% CO2. All cell lines were authenticated by short tandem repeat analysis at China Center for Type Culture Collection (Wuhan, China), and the absence of mycoplasma contamination was verified using a PCR detection kit (Shanghai Biothrive Sci. & Tech. Ltd., Shanghai, China). Cells were frozen in liquid nitrogen and used for experiments at passages 3 to 10 after thaw.
The following reagents were obtained from indicated sources: PT33 was synthesized in Prof. Xianzhang Bu's laboratory at School of Pharmaceutical Sciences, Sun Yat-sen University; b-AP15 (No. T1932), IU1 (No. T6107), Spautin1 (No. T1937), P005091 (No. T6925), USP7/USP47i (No. T4438), P22077 (No. T2424), LDN57444 (No. T1924) and ML323 (No. T1757) from Topscience (Shanghai, China); Ub-AMC (No. U-550) from Bostom Biochem (R&D Systems, Inc., Minneapolis, MN, USA).
2.2. Lentiviral transduction and transient transfection
The coding sequences of full-length human BRCA1 tagged with 3 × Flag at the N-terminus, mCherry and ubiquitin (wild-type, K48-, K63-, K6-, K11- or K27-specifity) tagged with HA at the N-terminus were inserted into the pCDH-EF1-MCS-T2A-Puro plasmid respectively. ShRNAs targeting Ku80 (5′-CGCTTTAACAACTTCCTGAAA-3′ in CDS) or RAD51 (5′-GCTAAGACTAACTCAAGATAA-3′ in 3′UTR) were cloned into the pLKO.1 vector. DUBs sgRNAs were cloned into lenti-CRISPR v2 vector. sgRNA information was detailed in Supporting Information Table S1. Lentiviruses were produced by co-transfection of the above constructs, psPAX2 and pMD2. G plasmids into HEK293T cells, and viruses were transduced into CRC cells followed by puromycin selection to generate stable cell lines.
The full length of BAP1, mutated BAP1 (C91A) and truncated HERC2 (3559–4834 aa) were cloned into pcDNA3.1 vector tagged with Myc at the N-terminus. The siRNA of HERC2 (5′-GGCCTTGCCTCCAGAAACA-3′ in CDS) and negative control (5′-GCGACGAUCUGCCUAAGAU-3′) were synthesized. Transient transfection was performed using Lipofectamine 3000 transfection reagent (No. L3000008, Invitrogen) or electroporation (Gene Pulser Xcell, Bio-rad) according to the manufacturer's instructions.
2.3. The activity assays of deubiquitination
Cells, spheroids or xenografts were treated with or without ionizing radiation (IR) (Fig. 1A and B), then were lysed using nuclear and cytoplasmic extraction reagents (No. 78835, Invitrogen) to obtain nuclear and cytoplasmic protein according to manufacturer's instructions. Then, lysates were subject to Ub-AMC hydrolysis assay. In brief, 5 μL of nuclear lysates (NL) or whole cell lysates (WCL) were incubated in assay buffer (50 mmol/L Tris-HCl, 5 nmol/L MgCl2, 1 mmol/L dithiothreitol [DTT], 2 mmol/L adenosine triphosphate [ATP], 250 nmol/L sucrose, pH 7.5) with or without the treatment of inhibitors at indicated concentrations for 30 min at 37 °C in 96-well plates, and then Ub-AMC was added to 400 nmol/L (final reaction volume was 100 μL).
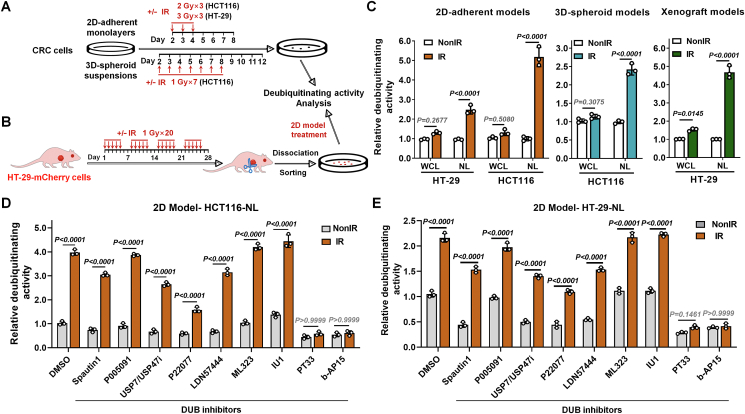
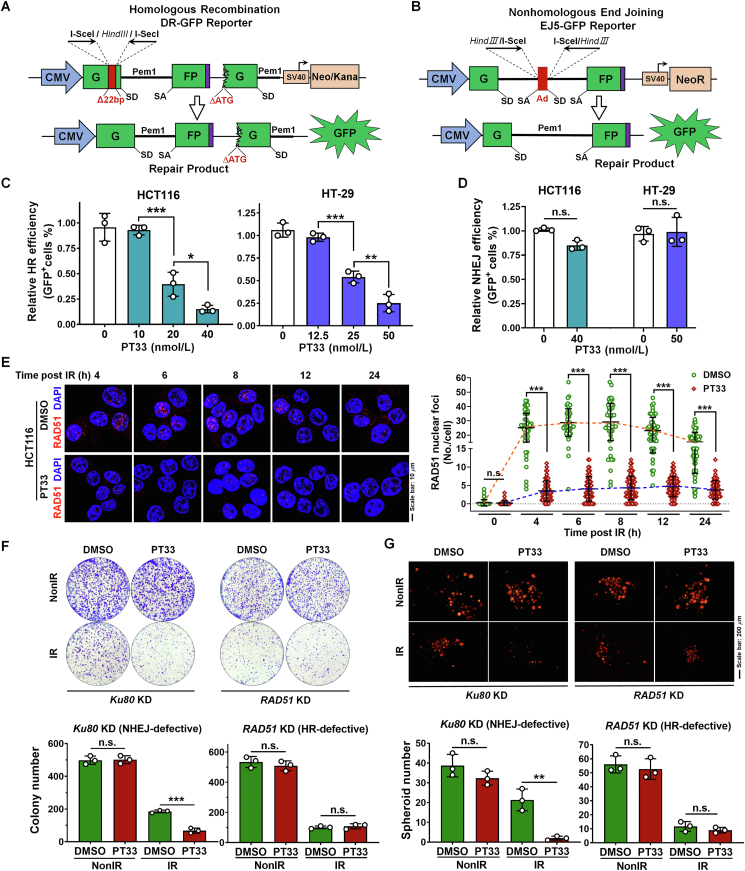
Figure 1.
PT33 inhibited nuclear deubiquitinase activity which increased after IR in CRC cells. (A, B) Treatment schemes of 2D-adherent, 3D-spheroid (A) and xenograft (B) models. IR, ionizing radiation. (C) Relative deubiquitinating activity of whole cell lysates (WCL) or nuclear lysates (NL) from CRC cells. The DUB activity was calculated as the AMC fluorescence increment (folds) per minute and normalized to the corresponding control group, similarly hereinafter. (D, E) The relative deubiquitinating activity of NL from HCT116 (D) and HT-29 (E) was treated in 2D-adherent model as in (A). After the treatment and culture, cells were collected and NL was extracted. NL was treated with various DUB inhibitors (2 μmol/L) for 1 h (DMSO as a vehicle control), and the DUB activities were detected. Data are represented as mean ± SD (n = 3); P values were determined by Student's t test.
For the deubiquitinating activity assays of BAP1, recombinant BAP1 protein (No. ab268359, Abcam, Waltham, USA) or Flag-BAP1 purified from cell lysates using Anti-Flag Affinity Gel (No. P2271, Beyotime, Haimen, China) according to manufacturer's instructions was used. In brief, enzymes (final concentration, 2 nmol/L) were incubated in assay buffer (50 mmol/L Tris-HCl, 1 mmol/L ethylene diamine tetraacetic acid [EDTA], 5 nmol/L MgCl2, 1 mmol/L DTT, 2 mmol/L ATP, 250 nmol/L sucrose, and 1 mg/mL ovalbumin, pH 7.5) with or without inhibitors of indicated concentrations for 30 min at 37 °C in 96-well plates, and then Ub-AMC was added to 400 nmol/L (final reaction volume was 100 μL)23.
The plates were subjected to measure fluorescence density on a BioTek Epoch Multi-Mode Microplate Reader at 380 nm of excitation wavelength and 460 nm of emission wavelength every 4 min for 2 h. The deubiquitinating activities were calculated as the fluorescence increment (folds) per minute, indicating the deubiquitination rate of substrate Ub-AMC.
2.4. Determine the kinetic parameters of covalent binding of PT33 towards BAP1
In a covalent inhibition, the total enzyme concentration is calculated according to Eq. (1):
| (1) |
whereas [E0] is the total concentration of BAP1 which is not covalently coupled with PT33, which is active and releases AMC from its substrate (Ub-AMC) to produce fluorescence, and [E–I] is the concentration of BAP covalently coupled with PT33. The fluorescence intensity observed is caused by the un-covalently coupled BAP1 (E0). According to the Michaelis–Mentent equation without inhibitor, the generate rate of AMC released by BAP1 from Ub-AMC (vAMC) equals to:
| (2) |
which exhibits a linear correlation with the [E0]. Therefore a standard curve can be obtained:
| (3) |
By recording the time course responses of various concentrations of BAP1 in the presence of 400 nmol/L Ub-AMC, which could be used to determine the concentration of un-covalently coupled BAP1 ([E0]) in the presence of PT33, and the covalently coupled BAP1 ([E–I]) can be calculated by [Etotal] – [E0], and the generation rate of BAP1–PT33 covalent complex E–I (vcc) can be calculated by dividing the [Etotal] with the incubation time of PT33 with BAP1 (30 min). Therefore, a curve can be obtained:
| (4) |
By recording the response of BAP1 (2 nmol/L)/Ub-AMC (400 nmol/L) reaction in the presence of various concentrations of PT33, whereas Ki is the reversible binding affinity of PT33 towards BAP1, kinact is covalent inactivation rate constant, both of which were obtained by fitting the curve.
2.5. Comet assay
DNA damage was analyzed by comet assay as described previously24,25. Briefly, CRC cells were treated with drugs for 12 h, then exposed to X-rays. Six hours after that, comet assay was performed using a comet assay kit (No. 4250-050-K, Trevigen, Gaithersburg, MD, USA) according to the manufacturer's instructions. Comet images were captured by fluorescence microscopy. The average comet tail moment (percentage of DNA in tail × tail length) was scored in three fields for each well using Comet Assay IV software (Perceptive Instruments, Bury St. Edmunds, UK).
2.6. HR and NHEJ reporter assays
The reporter constructs for HR and NHEJ, and the I-SceI expression plasmid pCMV-NLS-I-SceI were gifts from Dr. Lin Feng, Sun Yat-sen University Cancer Center (SYSUCC, Guangzhou, China). Briefly, parent HCT116 or HT-29 cells were transfected with the DR-GFP (HR) or EJ5-GFP (NHEJ) reporter plasmid and pCMV-NLS-I-SceI plasmid; BAP1-WT or BAP1-KO HCT116 cells were transfected with the DR-GFP (HR) or EJ5-GFP (NHEJ) reporter plasmid, pCMV-NLS-I-SceI plasmid, pCMV-DsRed plasmids (as an internal reference to monitor transfection efficiency) and corresponding siRNA. Meanwhile, the transfection system in absence of pCMV-NLS-I-SceI plasmid was set as the negative control. Transfection was accomplished via electroporation with a DT-130 program using the kit (V4XP-2032) on a Lonza 4D machine (Cologne, Germany). Thirty-six hours after transfection, cells were harvested and replanted. And post 12 h, cells were treated with drugs at the indicated concentration for 12 h and then collected for analysis of DNA repair efficiency (presented as GFP+ rate for parent HCT116 or HT-29 cells or relative ratio of GFP+/DsRed + cells for BAP1-WT or BAP1-KO HCT116 cells on a CytoFLEX flow cytometer.
2.7. Colony formation assay
The colony formation assay was performed in a 2D culture model as described previously24,26. Briefly, CRC cells were seeded into 6-well plates at a density of 1000 cells/well. After 24 h, the cells were treated with X-rays or inhibitors and cultured for 10–14 days, and then followed by staining with 0.5% crystal violet fixed in methanol. Colonies containing more than 50 cells were counted.
2.8. The DUBs CRISPR library generation and screen
Human DUBs CRISPR Knockout Pooled Library was generated. sgRNAs targeting 87 DUB genes (2 sgRNAs per target gene, Supporting Information Table S1) were cloned into the lenti-CRISPR v2 vector. HCT116 cells were transduced with lentivirus particles containing the DUBs sgRNA library with no greater than one sgRNA per cell. The Cas9-sgRNA-HCT116 cells were selected with 2 μg/mL puromycin for two weeks to achieve >95% gene knockdown. These Cas9/sgRNA-HCT116 cells were then treated as Fig. 4A model, and the surviving resistant population was expanded and subjected to deep sequencing analysis for candidate genes27.
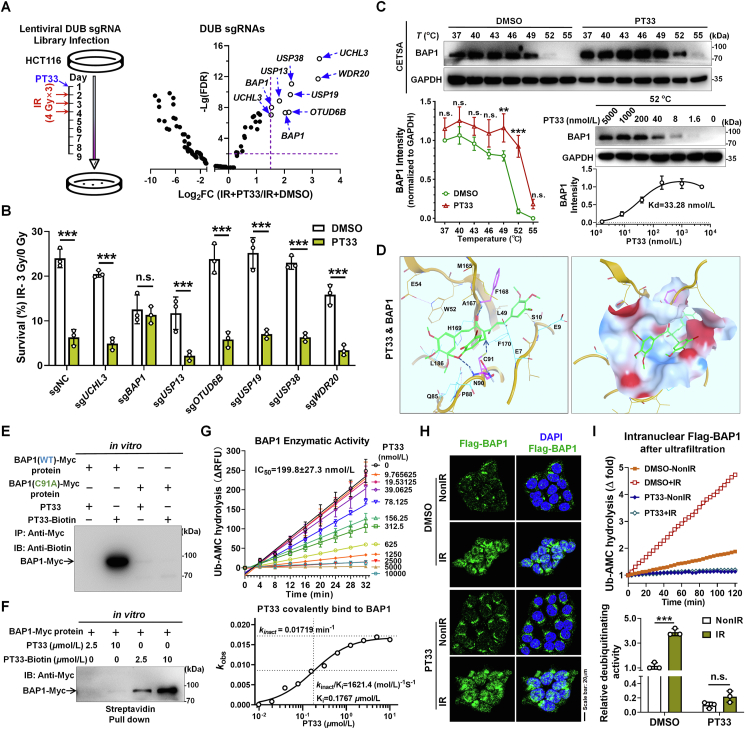
Figure 4.
CRISPR-Cas9 screening identified BAP1 as a potential target of PT33. (A) Left: treatment scheme of DUBs sgRNA library screening experiment; right: relative abundance of individual genes from CRISPR-Cas9 screen. Genes with Log2FC > 1.5 and –Lg(FDR) > 2 were considered as potential target of PT33 to sensitize CRC cells to IR. (B) Colony formation assay detecting survival ratio of HCT116 cells knocking out corresponding genes treated with PT33 and IR. (C) PT33 binding to BAP1 was evaluated by cellular thermal shift assay. Upper and left below: HCT116 cells were treated with PT33 (5 μmol/L) for 1 h incubated in indicated temperature for 3 min; right below, HCT116 cells were treated with indicating concentrations of PT33 for 1 h and incubated in 52 centigrade for 3 min. Immunoblot detecting BAP1 intensity, GAPDH as loading control. (D) 3D presentation of the predicted binding mode of PT33 with BAP1 by molecular docking. Hydrophobic and hydrophilic residues of BAP1 were labeled by dashed lines (upper); and the surface is shown in below. PT33 is shown in green stick. (E, F) In vitro pull-down assays detecting PT33 and BAP1 covalent interaction. Purified myc-BAP1 was incubated with PT33 or PT33–Biotin. (E) BAP1 was immunoprecipitated and immunoblotted by biotin antibody. (F) Streptavidin pull down of biotin and immunoblotted by Myc antibody. (G) In vitro BAP1 deubiquitinating activity of BAP1 measured by UB-AMC hydrolysis assay. Upper: fluorescence-time curve; Below: the curve of covalent binding Kobs to determinate the covalent inactivation rate (kinact) and reversible binding affinity Ki for PT33 against BAP1. (H) HCT116 cells were treated with PT33 (125 nmol/L) for 12 h followed by 6 Gy IR, 6 h later, Flag-BAP1 was detected by IF assay. (H) HCT116 cells were treated as indicated and nuclear Flag-BAP1 was enriched and ultrafiltration, followed by Ub-AMC hydrolysis assay. Upper: representative fluorescence-time curve; below: statistical analysis of relative DUB activity. (B, C, G and I) Data are shown as mean ± SD (n = 3). Statistical significance was determined by (B, I) Student's t test, (C) two-way ANOVA (n.s., not significant; ∗∗∗P < 0.001).
2.9. Cellular thermal shift assay (CETSA)
The CETSA assay was performed as previously described28. Briefly, HCT116 cells were treated with PT33 for 1 h and followed by incubation at different temperatures (37–55 °C) for 3 min. For dose–effect evaluation, HCT116 cells were treated with different doses of PT33 for 1 h and then incubated at 52 °C for 3 min. The cell lysate was prepared as described below and subject to Western blot assay.
2.10. Molecular docking
The complex model of BAP1 with PT33 was generated by the Molecular Operating Environment (MOE) software. The predicted structure of BAP1 by AlphaFold (https://alphafold.ebi.ac.uk, AF-Q92560-F1) was used, and the complex structure of BAP1 with PT33 was first generated by the docking module of MOE package following the same protocol as in previous study29. The best rank of the docked structure was relaxed and optimized by the dynamic module of MOE package.
2.11. In vitro pull-down assay
The expression of Myc-BAP1 was conducted referring to the previous protocol30. Briefly, the full length of BAP1 was cloned into pcDNA3.1 and transfected into HEK293F. Myc-BAP1 was purified with anti-Myc magnetic beads from cell lysates. Biotin-labeled PT33 was synthesized as detailed in Supporting Information (Supporting methods) and Fig. S7C. Myc-BAP1 protein and PT33-Biotin were mixed and incubated in buffer (50 mmol/L Tris-HCl, 5 nmol/L MgCl2, 1 mmol/L DTT, 2 mmol/L ATP, 250 nmol/L sucrose, pH 7.5) at 37 °C for 1 h. Then, the mixture was subject to immunoprecipitation as described above to harvest Myc-BAP1 or streptavidin pull down to harvest PT33-Biotin. The enriched protein was boiled and subject to Western blot assay.
2.12. Chromatin fraction
The preparation of chromatin fractions was as described previously. Briefly, 6 h after treatment with PT33 or IR, cells were collected and washed once with phosphate-buffered saline (PBS). Cell pellets were subsequently resuspended in low salt permeabilization buffer (10 mmol/L 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid [HEPES] at pH 7.4, 10 mmol/L KCl, 0.05% Nonidet P-40 [NP-40] and protease inhibitors) and incubated on ice for 20 min. Nuclei were then recovered and resuspended in HCl (0.2 mol/L). The soluble fraction was neutralized with Tris-HCl (1 mol/L at pH 8.0) for further analysis. For micrococcal nuclease (MNase) treatment, nuclei recovered after low salt extraction was washed and resuspended in nuclease reaction buffer (10 mmol/L HEPES at pH 7.4, 10 mmol/L KCl, 0.5 mmol/L MgCl2 and 2 mmol/L CaCl2). Nuclease (20 U) was added and nuclei were incubated for 30 min on ice. The insoluble fraction was treated as above for isolating the chromatin-bound proteins.
2.13. Co-immunoprecipitation (co-IP) and Western blot (WB) assays
Cells were lysed in lysis buffer (No. 9803, CST) supplemented with protease and phosphatase inhibitors cocktail (No. 78442, TFS), and then protein concentration was quantified using a BCA protein assay kit (No. KGP903, KeyGen Biotech). For co-IP, cell lysates were incubated with antibody beads overnight at 4 °C or primary antibodies overnight at 4 °C plus protein G agarose beads (No. sc-2003, SCBT) for 2 h at 4 °C. After washing, the pulldown products were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For WB assay, protein bands separated by SDS-PAGE were transferred onto polyvinylidene fluoride membrane (No. 03010040001, Sigma–Aldrich) and the membrane was blocked with 5% milk in tris buffer saline (TBS) and incubated overnight at 4 °C with specific primary antibodies, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. Signals were visualized using an enhanced chemiluminescence reagent (No. WP20005, TFS) and captured by ChemiDoc IRS system (Bio-Rad), and quantified using Image J.
2.14. Immunofluorescence (IF) assay
CRC cells on confocal dishes were fixed with 4% paraformaldehyde for 20 min at room temperature, then permeabilized using 0.5% Triton X-100 in PBS and blocked with 3% goat serum for 1 h at 37 °C. The cells were then incubated with primary antibodies at 4 °C overnight followed by incubation with fluorogenic secondary antibodies at room temperature for 1 h. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 2 min. Images were acquired using a LSM880 with Airyscan FAST Confocal microscopy (ZEISS, Oberkochen, Germany).
2.15. Cell viability assay
CRC cells (2500/well) were cultured in 96-well plates for 24 h and then treated with inhibitors or IR. After 72 h, cell viability was determined by cell counting kit-8 (CCK-8, No. HY-K0301, MCE, Monmouth Junction, NJ, USA) according to the manufacturer's instructions.
2.16. Spheroid formation assay
Anchorage-independent growth and spheroid formation of CRC cells was investigated in a 3D spheroid model. Briefly, cells (1000–4000 viable cells/well) were seeded into 6-well plates with ultra-low attachment surface (No. CLS3471, Corning Inc., Corning, NY, USA) in 2 mL of serum-free medium supplemented with epidermal growth factor (EGF) (100 ng/μL, No. AF-100-15), vascular endothelial growth factor (VEGF) (10 ng/μL, No. AF-100-20A) (PeproTech, Cranbury, NJ, USA) and 0.5% methylcellulose (No. M7027, Sigma–Aldrich). The cell spheres were allowed to grow for 10–15 days and imaged under an inverted fluorescence microscope. The number of spheres was counted and sphere size was assessed using ImageJ.
2.17. Nude mouse xenograft model and experimental therapy
Animal experiments were approved by the SYSUCC Institutional Animal Care and Usage Committee following the Animal Welfare and Rights in China (Approval number: L102012018005X). Female BALB/c nude mice (4–5 weeks, 15–18 g; SLRC Laboratory Animal Co., Shanghai, China) were used in animal experiments. For protogenetic HCT116 cells, 3 × 106 cells were subcutaneously injected into right flank to establish primary xenografts. Then, primary xenografts were cut into sections (approximately 5 mm3) and transplanted into new mice. Once the xenograft volume reached approximately 100 mm3, mice were treated alternately with PT33 and X-rays. For BAP1-WT and BAP1-KO HCT116 cells, 3 × 106 cells were subcutaneously injected into right flank to generate xenografts directly. Once the xenograft volume reached approximately 250 mm3, mice were treated alternately with PT33 and X-rays. The tumor volume was calculated using the Eq. (5) 24,31:
| (5) |
Mice were sacrificed 1–3 weeks after the last treatment, and then xenografts were removed, weighed and subjected to pathological analysis. In the case of survival analysis, mice were observed for additional 4 weeks.
2.18. Antibodies
Primary antibodies to γ-H2AX (No. 2577; WB 1:1000; IF 1:200), Lamin A/C (No. 4777; WB 1:1000), BRCA1 (No. 9010; WB 1:1000; IF 1:200), K48-linkage specific polyubiquitin (No. 8081; WB 1:1000), FLAG-tag (No. 8146; WB 1:1000; IF 1:200), Myc-tag (No. 2276; WB 1:1000), GAPDH (No. 2118; WB 1:1000), Ku80 (No. 2753, WB 1:1000), ORC2 (No. 4736, WB 1:1000), Actin (No. 3700, WB 1:1000) and HRP-linked secondary antibody to rabbit IgG (No. 7074; WB 1:1000), mouse IgG (No. 7076; WB 1:1000) from Cell Signal Technology (CST, Danvers, MA, USA); antibody to β-tubulin (No. T0023; WB 1:1000) from Affinity Biologicals; antibody to HA (No. ab18181; WB 1:1000; IP 1:1000), RAD51 (No. ab63801; WB 1:1000; IF 1:200) from Abcam; BAP1 (No. sc-28283; WB 1:500) from Santa Cruz Biotechnology; primary antibody to Biotin (No. bs-0311R; WB 1:1000) from Bioss (Beijing, China); primary antibody to BARD1 (No. PA5-85707; WB 1:1000) and fluorogenic secondary antibody goat anti-rabbit Alexa Fluro-488, anti-rabbit Alexa Fluro-594, goat anti-mouse Alexa Fluro-488, anti-mouse Alexa Fluro-594 (No. A11034, A11037, A32723, A11032, respectively; IF 1:1000) from Invitrogen (Carlsbad, CA, USA); M2 anti-Flag agarose (No. A2220) from Sigma–Aldrich.
2.19. Statistical analysis
All in vitro experiments were repeated at least three times, and the animal experiments were repeated twice, except those specified in the figure legends. Data are presented as mean ± standard deviation (SD). The data were analyzed using GraphPad Prism 8.0 or SPSS 20.0 software. Differences in the average between two groups with one variate were determined by Student's t test. Differences in the average between more than two groups with one variate were determined by one-way analysis of variance (ANOVA). Differences between groups with dose factor or with more than one variate were determined by two-way ANOVA. Survival was analyzed using the log-rank test. P value < 0.05 indicates a significant difference.
3. Results
3.1. Identification of intranuclear deubiquitylation inhibitors in CRC
Recent observations highlighted the balance of ubiquitylation and deubiquitylation in orchestrating the chemo-/radio-resistance, providing a dynamic cellular regulatory circuit to repair DNA and guarantee nuclear homeostasis. To investigate if deubiquitylation can be involved in the resistance of radiotherapy in colorectal cancer (CRC), we generated two-dimensional (2D) adherent monolayers, three-dimensional (3D) spheroid suspensions and nude mouse xenograft models in CRC cells (Fig. 1A and B). Then they are used for the assays of deubiquitinating activity with substrates ubiquitin-7-amido-4-methyl coumarin (Ub-AMC). The results show that deubiquitinating activities were significantly higher in nuclear lysates (NL) of radiation-treated CRC cells than in NL of untreated cells (all P < 0.0001). But there are no obvious differences in whole cell lysates (WCL) (Fig. 1C, Supporting Information Fig. S1A–S1D). These data suggest that radiation can induce a significant elevation of intranuclear deubiquitinating activity, which is likely an important reason for the enhancement of radioresistance.
Therefore, we further evaluated the effects of 9 small-molecule inhibitors of DUBs (Supporting Information Fig. S2A) on intranuclear deubiquitinating activity by Ub-AMC assay. And then PT33 or b-AP15 was identified as a really effective nuclear deubiquitinating inhibitor in CRC cells (Fig. 1D, E and Fig. S2B–S2D). As an active-site-directed thiol protease inhibitor, PT33 or b-AP15 was reported to target 19S regulatory particle-associated DUBs32,33. Overall, these data indicate that radiation-mediated elevation of intranuclear DUB activity can be arrested by PT33 or b-AP15.
3.2. PT33 impairs HR-mediated DNA DSB repair
Since DNA double-strand breaks (DSBs) are considered the most severe lesions produced by radiation to generate main antitumor effects, we investigated whether the abrogation of intranuclear deubiquitination influenced the DNA DSB damage or repair. Firstly, we measured DNA damage levels by the comet assay in CRC cells after IR. The results show that there were significantly longer comet tails in PT33-pretreated HCT116 and HT-29 cells at 6 h after IR than in cells without PT33 pretreatment, whereas no obvious comet tails were observed in cells with treatment of PT33 alone (Fig. 2A). However, there was no statistical significance of comet tails between b-AP15-pretreated and control CRC cells after IR treatment (Supporting Information Fig. S3A). In addition, PT33 was reported to inhibit proteasome-associated DUBs. We then asked whether PT33 acts as a proteasome inhibitor that may facilitate DSB damage. For this, we carried out comet assays to compare the effect between PT33 and proteasome 20S core particle inhibitor—bortezomib (BTZ). Our results show BTZ or b-AP15 had no apparent effect on the length of comet tails after IR. Meanwhile, the addition of PT33 made the tail moments longer (Fig. S3B). Together, these results demonstrate that PT33 leads to an enhanced degree of DSB damage, but not b-AP15 or BTZ, suggesting that PT33 is a promoter of DNA damage after IR treatment.
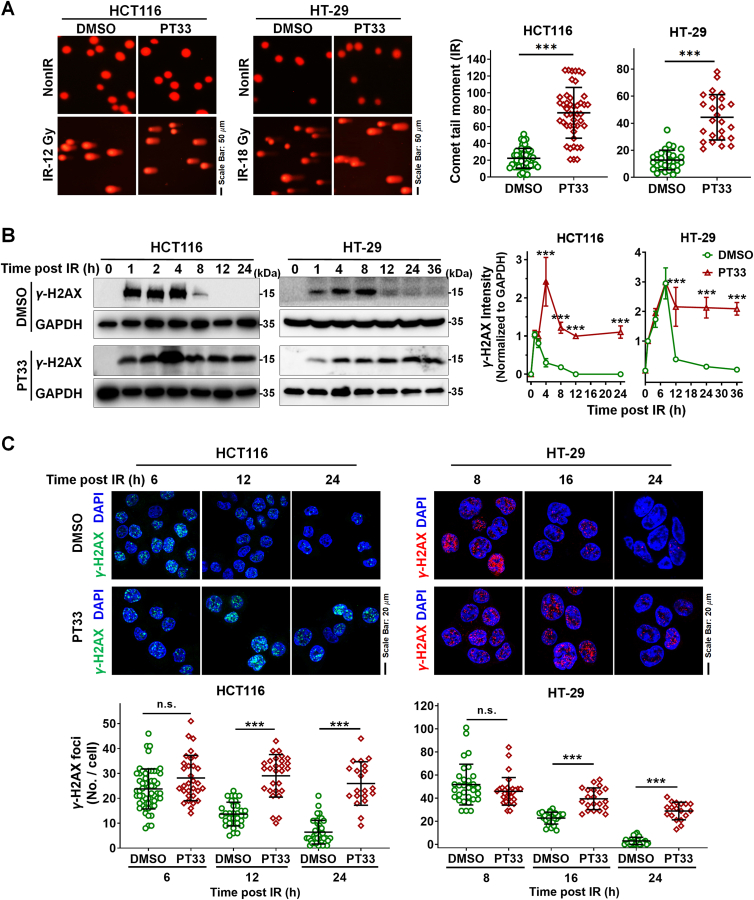
Figure 2.
PT33 retarded repairment of DNA damage induced by IR. CRC cells were treated with PT33 (HCT116: 20 nmol/L; HT-29: 25 nmol/L) 12 h ahead of ionizing radiation (IR). (A) Comet assays for DNA damage at 6 h after IR. Left, representative images; right, spot charts indicating the average tail moment per cell. The results are represented as cells in all views from 3 biological replicates. (B) Immunoblot assay detecting γ-H2AX at the indicated time after IR. Left, representative images; right, the relative γ-H2AX levels, shown as the ratio of γ-H2AX intensity (grayscale normalized to corresponding GAPDH) at the indicated time to that at 1 h after IR. Data are represented as mean ± SD (n = 3). (C) Immunofluorescence (IF) assay for γ-H2AX foci at the indicated time after IR. Upper, representative images; below, quantification of γ-H2AX foci. The results are represented as cells in all views from 3 biological replicates. Statistical significance was determined by (A) Student's t test, (B, C) two-way ANOVA (n.s., not significant; ∗∗∗P < 0.001).
As is known that the level of histone H2AX phosphorylation at serine 139 (γ-H2AX) subsequently declines as DNA DSBs damage is repaired. Next, we compared the kinetics of γ-H2AX induced by IR between PT33-pretreated and control CRC cells. Data from these different cell lines consistently show that the γ-H2AX signal disappeared quickly after IR in control CRC cells (<12 h), but persisted to 24 h in PT33-treated cells using WB assays (Fig. 2B). Immunofluorescence assays showed that at 6 h after IR, there was no significance of the number of γ-H2AX foci between PT33-pretreated and control CRC cells. After that, the number of γ-H2AX foci gradually decreased and almost reached the basal level at 24 h in control CRC cells. By contrast, PT33-pretreated cells contained more numbers of γ-H2AX foci and the difference remained significant over 24 h (Fig. 2C), suggesting that PT33-pretreated cells were unable to efficiently repair DSBs. These data indicate that PT33 treatment leads to the inhibition of DSB damage repair.
Since DNA DSBs are repaired by two major pathways: HR and NHEJ24,34, we further investigate which of these two DSB repair pathways might be interrupted by PT33. For this purpose, we used the DR-GFP or EJ5-GFP reporter assay to quantify the activity of HR or NHEJ respectively35,36 (Fig. 3A and B). We found PT33 caused a dose-dependent inhibition effect on HR repair in CRC cells (Fig. 3C and Supporting Information Fig. S4A and S4B), whereas the concentrations of PT33 almost had no killing effect on cells with 24 h treatment. Notably, PT33 at the above concentrations had failed to affect NHEJ repair in CRC cells (Fig. 3D and Fig. S4C and S4D). Additionally, BTZ or b-AP15 alone had no apparent effect on HR repair, but the addition of PT33 exhibited a significantly inhibitory effect on HR repair (Fig. S4E). Considering that the formation of RAD51 foci in nucleus is an essential step in HR-mediated repair, we next examined whether PT33 may affect the foci formation. Our results show that compared to control CRC cells where the number of RAD51 foci peaked at 4–6 h and gradually decreased, the RAD51 foci number was attenuated in cells pretreated with PT33 (Fig. 3E). These results reveal that PT33 prevents DNA repair through inhibition of the HR repair. This conclusion was further confirmed that PT33 enhanced the inhibitory effect of IR on 2D adherent colony or 3D spheroid formation in NHEJ-defective (Ku80 knockdown) HCT116 cells, but not in HR-defective (RAD51 knockdown) HCT116 cells (Fig. 3F and G and Supporting Information Fig. S5). Because HR repair is related to the cell cycle, we further determined whether the inhibitory effect of PT33 on HR repair is dependent on the regulation of cell cycle. Cell cycle profiling showed that IR treatment was associated with the increase of cell population in the G2–M phase, but not G2–M phase arrest in CRC cells with PT33 treatment (Supporting Information Fig. S6), indicating that PT33 did not affect cell–cycle progression. Taken together, these results suggest that PT33 inhibits HR-mediated DSB repair probably through interrupting the function of one or more core regulators.
Figure 3.
PT33 restrained HR-mediated DSB repair. (A, B) The HR (A) or NHEJ (B) reporter construct for the analysis of DNA DSB repair. SD, splice donor; SA, splice acceptor. (C, D) Quantification of HR (C) and NHEJ (D) reporter assay by flow cytometry in CRC cells treated with PT33 of indicated doses. (E) HCT116 cells were treated with PT33 (125 nmol/L) ahead of IR. RAD51 foci were detected at indicated time points after IR by IF assay. Left, representative images; right, quantification of RAD51 foci. The results are represented as cells in all views from 3 biological replicates. (F) Colony formation assay for CRC cells treated with PT33 (25 nmol/L) plus IR in NHEJ- or HR-defective HCT116 cells (scheme as in Fig. S5C). Upper: representative images; below: quantification of colony number. (G) 3D spheroid formation assay for CRC cells treated with PT33 plus IR in NHEJ- or HR-defective HCT116 cells (scheme as in Fig. S5C). Upper: representative images; below: quantification of spheroid number. (C–F and G) Data are represented as mean ± SD (n = 3). Statistical significance was determined by (C) one-way ANOVA, (D, F and G) Student's t test, (E) two-way ANOVA (n.s., not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
3.3. The CRISPR-based DUB screen identifies BAP1 as a novel target of PT33 that confers radiosensitivity
To explore the mechanisms of PT33 on HR suppression, we generated Cas9/sgRNA-expressing HCT116 cells using a DUBs CRISPR library to identify potential DUB targets of PT33. The cells were then treated with PT33 followed by continuous radiation at 4 Gy/dose × 3 times, which can kill all wild-type (WT) cells. Meanwhile, as a control, the cells were treated with DMSO followed by continuous radiation, whereas which slightly killed WT cells (Fig. 4A-left). A small fraction of cells transduced with the CRISPR library survived in PT33 + IR group and these surviving cells were subjected to secondary sequencing to analyze DUB sgRNAs. A complete list of identified DUBs are provided in Supporting Information Table S1, the sgRNAs of the top 9 candidate DUBs were ranked based on the read number ratio of IR + PT33 group to IR + DMSO group (Log2 FC > 1.5; and FDR <0.01) and listed in Fig. 4A-right. We further generated HCT116 cell clones with stable expression of Cas9 and sgRNAs targeting 9 candidate DUBs. An in vitro colony formation assay showed that only BAP1 knockout (KO) eliminated the inhibitory effect of PT33 on post-radiation cell survival (Fig. 4B), suggesting that BAP1 may be a potential target of PT33 to enhance radiation sensitivity. To confirm this, the cellular thermal shift assay (CETSA) was employed to validate the interaction of PT33 with BAP1. We observed that PT33 resulted in the increase of the thermal stability of BAP1, thus confirming that PT33 can bind to BAP1. Based on the curves, 52 °C was selected for isothermal dose–response experiments in which PT33 showed dose-dependent stabilization of BAP1 with a binding constant (Kd) value of 33.28 nmol/L (Fig. 4C).
Since PT33 bears an α,β-unsaturated motif as a Michael receptor, therefore, we wondered whether PT33 can covalently bind to the Cys91 in the catalytic active pocket of BAP1. We then performed molecular docking to predict the binding mode and investigate the PT33–BAP1 interaction interface. As illustrated in Fig. 4D and Supporting Information Fig. S7A and S7B, PT33 binds to BAP1 catalytic active pocket through several hydrogen bonds. The inserted methoxyl and carbonly groups of PT33 form hydrogen bonds with Asn90 and Phe168, and sulfhydryl group of Cys91 point to PT33 facilitating the formation of a covalent bond by nucleophilic addition. To investigate the possibility of PT33 covalently binding to BAP1, we synthesized a biotinylated affinity probe-PT33–Biotin by modifying the hydroxy of PT33 with biotin (Supporting Information Figs. S7C and S8), and purified recombinant the WT and mutant (C91A) BAP1-myc tagged protein. We observed the presence of PT33–Biotin in the wild-type protein by WB assay, but not in the C91A mutant BAP1 protein (Fig. 4E). Meanwhile, the streptavidin pull-down experiments were conducted to further reveal that a dose-depended covalently interaction of PT33–Biotin with BAP1 (Fig. 4F). Moreover, we performed Ub-AMC hydrolysis assays to evaluate the kinetic mechanism of PT33-mediated covalent inhibition on BAP1, our in vitro kinetic results showed that the IC50 value of PT33 is 199.8 ± 27.3 nmol/L (Fig. 4G and Supporting Information Fig. S9A), with the value of covalent inactivation rates (Kinact) is 0.01719 min−1 and reversible binding affinity (Ki) is 0.1767 μmol/L (Fig. 4G). By contrast, the IC50 value of b-AP15 (>10,000 nmol/L, Fig. S9B) is far higher than that of PT33, indicating that b-AP15 unlike PT33, failed to efficiently inhibit BAP1 activity. Collectively, these results prove that PT33 is a covalent inhibitor of BAP1.
On the basis of these findings, we next examined whether the covalently interaction of BAP1 with PT33 may alter BAP1 sub-cellular location, and found that after IR treatment, PT33 had no apparent effect on the nuclear translocation of BAP1 in HCT116 cells (Fig. 4H). Therefore, we then asked whether PT33 inhibits nuclear BAP1 activity after IR. For this, we carried out Ub-AMC hydrolysis assay and found that there was indeed significantly more purified BAP1 in NL of radiation-treated CRC cells than in NL of non-treated cells. And PT33 markedly reduced the deubiquitinating activity of intranuclear BAP1 in both irradiated and non-irradiated tumor cells (Fig. 4I). Taken together, these results suggest that the function of PT33 in HR is related to its covalently binding ability to the active site of BAP1.
3.4. Targeting BAP1 with PT33 facilitates the K48-linked polyubiquitination of BRCA1
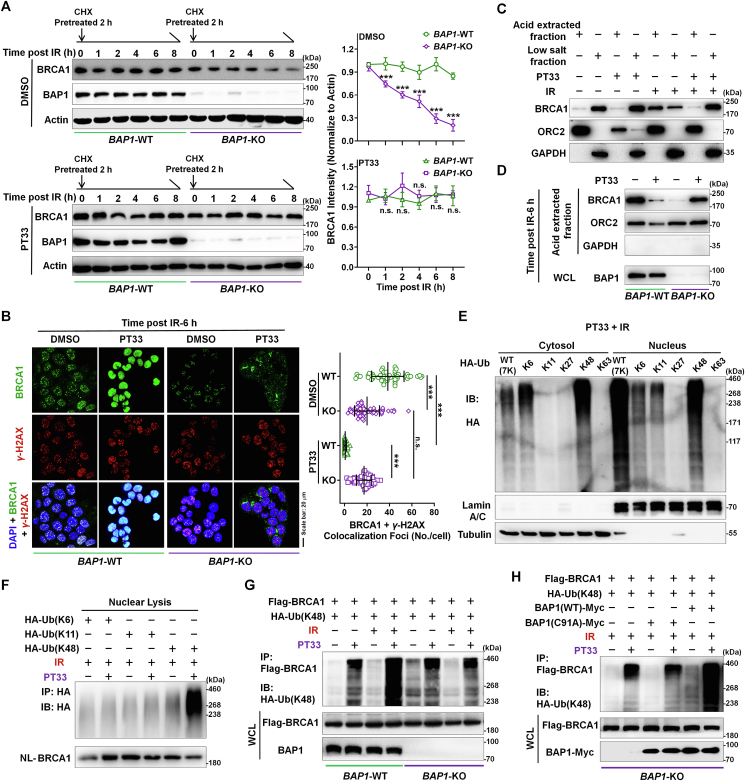
Since BAP1 as a BRCA1-associated protein was revealed to bind the BRCA1–BARD1 complex and modulate its function in HR pathway15,16, we thus wanted to understand whether BAP1 as a DUB enzyme may alter BRCA1 ubiquitin-dependent turnover to maintain its stability after IR treatment. A measurement of the rate of BRCA1 protein decay in cycloheximide-treated cells after IR treatment showed that the half-life of endogenous BRCA1 protein was markedly decreased upon depletion of BAP1 (Fig. 5A). However, IR caused BRCA1 degradation was attenuated by PT33 treatment in BAP1 deficient cells, which may be caused by PT33 inducible proteasome inhibition32 (Fig. 5A). These results indicate that PT33 treatment had no apparent impact on the protein level of BRCA1 in presence or absence of BAP1, and suggest that BAP1 inhibition with PT33 (or PT33-bound BAP1) is radically different from BAP1 depletion in HR suppression. To further confirm this, we next compared the distribution of BRCA1 with or without BAP1, with or without PT33, respectively. Our results show that IR-induced BRCA1 foci colocalizing with γ-H2AX foci, indicating that BRCA1 located on DSB sites to involve in DNA repair (Fig. 5B), accordingly, the observed diffusion of BRCA1 along with the loss of γ-H2AX may suggest that repair was almost completed at 48 h after IR in control cells (Supporting Information Fig. S10). But BAP1 deficiency markedly decreased the BRCA1 foci after IR (Fig. 5B and Fig. S10). Of note, in control cells, PT33 pretreatment failed to produce IR-induced BRCA1 foci formation and BRCA1 signals were distributed diffusely, not as foci colocalizing with γ-H2AX foci in nucleus (Fig. 5B). However, after IR, we still observed the existence of BRCA1 foci in nucleus until DNA repair completed in BAP1-KO cells pretreated with PT33 (Fig. 5B and Fig. S10). In addition, given that the binding of repair proteins to chromatin in the vicinity of DSB sites results in damage or repair-induced foci, we then asked whether PT33 may affect the chromatin location of BRCA1 through binding to BAP1. In a fractionation experiment, we observed that a marked portion of BRCA1 shifted from the acid-extracted fraction (chromatin fraction) to the low salt-extracted fraction (soluble fraction) after IR following PT33 treatment (Fig. 5C). Moreover, the chromatin fraction of BRCA1 can be released by PT33 treatment in presence of BAP1, but not in absence of BAP1 (Fig. 5D). Together, these data suggest that PT33 caused the diffuse distribution of BRCA1 is depended on the present of BAP1.
Figure 5.
Targeting to BAP1 with PT33 facilitates the K48-linked polyubiquitination of BRCA1. (A) BAP1-WT or BAP1-KO HCT116 cells were treated with PT33 (125 nmol/L) for 12 h and cycloheximide (CHX) for 2 h ahead of exposed to IR (6 Gy). BRCA1 levels at indicating time points after IR were detected by WB assay. Left: representative images; right: the relative BRCA1 levels, shown as the ratio of BRCA1 gray scale to Actin at indicated time normalized to that of 0 h. (B) BAP1-WT and BAP1-KO HCT116 cells were treated with PT33 for 12 h ahead of IR. IF staining for BRCA1 and γ-H2AX at the indicated time point. Left: representative images; right: quantification of colocalized foci. (C) HCT116 cells were treated with PT33 for 12 h and followed by IR. After 6 h, chromatin fractions were isolated and subject to immunoblotting. (D) BAP1-WT and BAP1-KO HCT116 cells were treated with PT33 for 12 h and followed by IR. After 6 h, the acid extracted fraction was isolated and subject to immunoblotting. (E) HCT116 cells expressing HA-Ubiquitin (WT, K6/K11/K27/K48/K63-specific) respectively were treated with PT33 for 12 h followed by IR (6 Gy); 6 h later, these cells were collected. WB assays for various HA-ubiquitin in cytoplasmic and nuclear fractions. (F) HCT116 cells expressing HA-Ubiquitin K6, K11 or K48 were treated with PT33 for 12 h followed by IR (6 Gy); 6 h later, these cells were collected. The nuclear lysate was subject to immunoprecipitation (IP) and WB assays. (G, H) IP and WB assays detecting the K48-ubiquitinated level of BRCA1. (G) Flag-BRCA1 and HA-Ubiquitin (K48) plasmids were transferred into BAP1-WT or BAP1-KO HCT116 cells following treatment with PT33 and then IR; (H) Myc-BAP1 (WT) or Myc-BAP1(C91A) plasmid was transfected into HCT116 and treated with PT33 and then IR. DMSO or NonIR as control; PT33, 125 nmol/L; IR, 6 Gy. (A, B) The results are represented as cells in all views from 3 biological replicates. Quantitative data are shown as mean ± SD (n = 3). Statistical significance was determined by two-way ANOVA (n.s., not significant; ∗∗∗P < 0.001).
We investigated if PT33 caused the diffuse BRCA1 signals by facilitating the accumulation of polyubiquitinated BRCA1(Ubn-BRCA1). To test this possibility, we performed WB/co-IP assays to confirm that PT33 mainly triggered the internuclear accumulation of K48-linked Ubn-BRCA1, but not K6- or K11-linked Ubn-BRCA1 (Fig. 5E and F). To determine whether the internuclear accumulation of K48-linked Ubn-BRCA1 caused specifically by PT33 might be influenced by BAP1, co-IP/WB assays were performed. The results show that when BAP1 is present, PT33 promoted sharply the accumulation of K48-linked Ubn-BRCA1 (full-length or a fragment of 1–342 aa containing a degron domain) after IR treatment (Fig. 5G and Supporting Information Fig. S11). On the contrary, when BAP1 is absent, PT33 had no capability to further enhance the level of K48-linked Ubn-BRCA1 after IR (Fig. 5G). To further confirm this, we carried out rescue experiments using BAP1-KO cells. Our results show that re-expression of BAP1-WT markedly increased the accumulation of K48-linked Ubn-BRCA1 in BAP1-KO cells with PT33 treatment during the post-IR recovery phase. In contrast, re-expression of mutant BAP1 (C91A) had no alteration of the level of K48-linked Ubn-BRCA1 (Fig. 5H), suggesting that PT33 inducible enhancement of the accumulation of K48-linked Ubn-BRCA1 is dependent on binding to BAP1. In brief, these results indicate that PT33 suppresses HR-mediated DSB repair by targeting BAP1 to trigger the accumulation of K48-linked polyubiquitinated BRCA1, which interrupts HR process.
3.5. BAP1 binding with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction
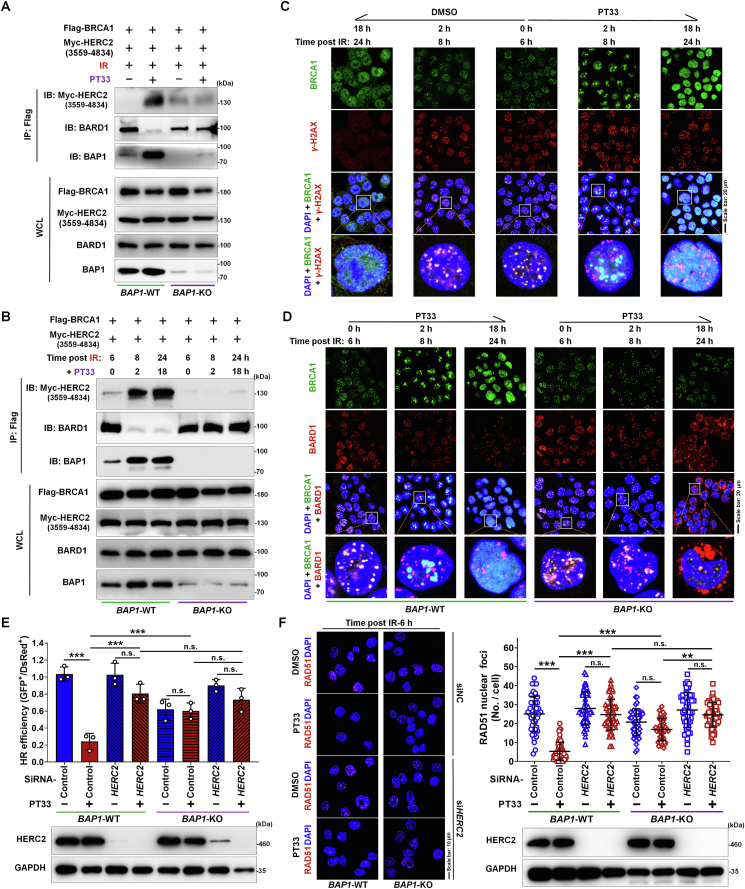
According to the results above, the accumulation of K48-linked Ubn-BRCA1 caused by PT33 is dependent on binding to BAP1. As is known that HERC2 is an E3 ligase targeting BARD1-uncoupled BRCA1, which catalyzes the K48-linked ubiquitination of BRCA1. Therefore, we carried out co-IP/WB assays with ectopic expression of Myc-HERC2 (3559–4834 aa18), and found that after IR, PT33-bound BAP1 showed markedly stronger interaction with BRCA1 than uncombined BAP1; only PT33-bound BAP1 could recruit HERC2 to target BRCA1, but not uncombined BAP1. Concordantly, the BRCA1–BARD1 interaction was substituted by the BRCA1–HERC2 interaction upon PT33 bond to BAP1 (Fig. 6A). By contrast, recruitment of HERC1 to BRCA1 could not occurred in cells by depletion of BAP1, either with or without PT33 treatment (Fig. 6A). To further confirm this, we treated the cells with PT33 at 6 h post-IR. At this time, numerous BRCA1–BARD1 complexes have already existed. Our co-IP/WB assays reveal that the BRCA1–BARD1 couples were rapidly substituted by enhanced interaction of HERC2 with BRCA1 after PT33 treatment in presence of BAP1 (WT). However, PT33 did not affect the coupling of BRCA1 and BARD1 in absence of BAP1 (Fig. 6B).
Figure 6.
BAP1 binding with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction. (A) IP and WB assays were used to detect the interaction between Flag-BRCA1 and Myc-HERC2, BARD1 or BAP1. Flag-BRCA1 and Myc-HERC2 (3559–4834 aa) plasmids were transfected into BAP1-WT or BAP1-KO HCT116 cells following treatment with PT33 (125 nmol/L) for 12 h followed by IR (6 Gy), 6 h later, these cells were collected. (B) HCT116 cells with Flag-BRCA1 and Myc-HERC2 (3559–4834 aa) expression were exposed to IR; after 6 h, the cells were treated with PT33. At the indicated times, IP and WB assays were used to detect these interactions. (C) HCT116 cells were exposed to IR; after 6 h, the cells were treated with PT33. At the indicated times, IF assays for observing the distribution of BRCA1 and γ-H2AX. (D) BAP1-WT and BAP1-KO HCT116 cells were exposed to IR; after 6 h, the cells were treated with PT33. At the indicated times, IF assays for the distribution of BRCA1 and BARD1. (E, F) HERC2 was knocked down in BAP1-WT and BAP1-KO HCT116 cells with siRNA. (E) Cells were treated with PT33 and subject to HR reporter assay. (F) IF assay for distribution of RAD51. Cells were treated with PT33 (125 nmol/L) ahead of IR. At 6 h after IR, Nuclear foci were counted and the results are represented as cells in all views from 3 biological replicates. (E, F) Quantitative data are shown as mean ± SD (n = 3). Statistical significance was determined by two-way ANOVA (n.s., not significant; ∗∗∗P < 0.001).
Immunofluorescence assay demonstrated that obvious BRCA1 foci were formed in DSB sites at 6 h post-IR, but the BRCA1 foci rapidly estranged from DSB sites, and were transformed into a diffuse distribution in nucleus following by PT33 treatment (Fig. 6C and Supporting Information Fig. S12). Furthermore, to observe whether BRCA1–BARD1 complex altered in the presence or absence of BAP1, with or without PT33 treatment, respectively, we examined their assembly or dissociation using IF assays. Consistent with our co-IP/WB results, in control cells, the BRCA1–BARD1 colocation foci were noticeably observed at 6 h post-IR, but the addition of PT33 rapidly caused the loss of colocation foci of BRCA1 and BARD1. And BRCA1 was gradually transformed into a diffuse distribution, but BARD1 foci were persist existed in nucleus (Fig. 6D). However, in contrast to BAP1-WT counterpart, after adding PT33, BRCA1 localization to BARD1 foci was not noticeably affected in cells with BAP1 deficiency until repair almost completed (Fig. 6D), suggesting that PT33 requires BAP1 for dissociation of BRCA1–BARD1 complex. Together, these results demonstrate that PT33-bound BAP1 recruits HERC2 and competes with BARD1 for BRCA1 interaction, suppressing BRCA1-mediated DNA repair.
Based on these observations, we noticed that PT33 functions may require cooperation with BAP1 or HERC2. Therefore, we sought to determine if BAP1 or HERC2 was sufficient to influence the function of PT33 in HR repair. To this end, we performed a DR-GFP reporter assay for HR repair efficiency, the results show that the inhibitory efficacy on HR repair of PT33 was eliminated by depletion of BAP1 or HERC2 (Fig. 6E and Supporting Information Fig. S13). Consistently, the decrease of RAD51 nuclear foci number caused by PT33 was substantially rescued in cells with BAP1 or HERC2 depletion (Fig. 6F). These results demonstrate that the function of PT33 in HR is related to the existence of BAP1 and HERC2, both are necessary.
3.6. PT33 and IR in combination synergistically kill CRC cells, which is depended on BAP1
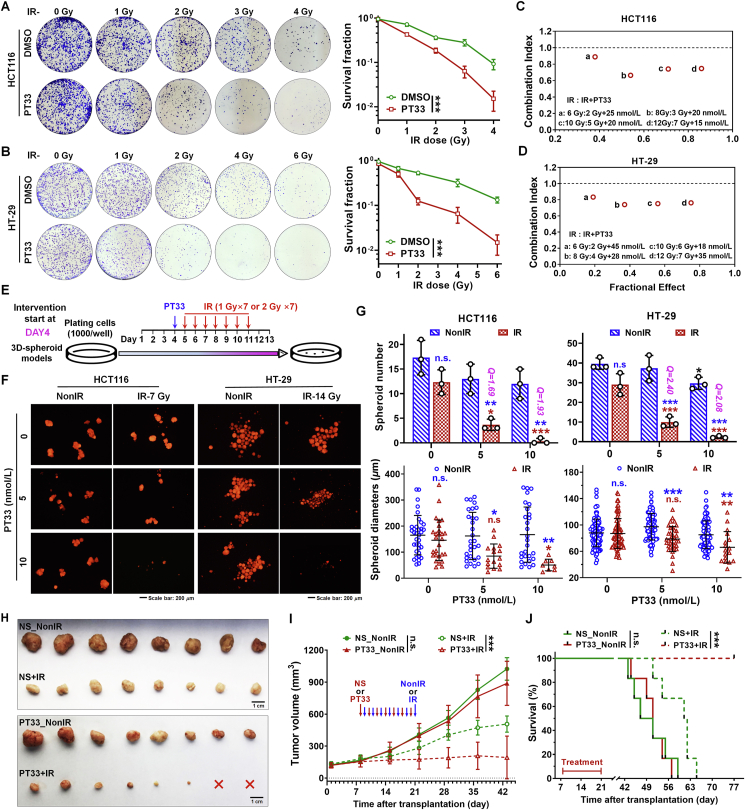
Radiotherapy is a standard therapy for CRC patients. Because PT33 inhibits HR-based DNA repair, we thus postulated that PT33 might facilitate the sensitivity of CRC cells to radiotherapy. To investigate this possibility, we first performed colony formation assays to reveal that PT33 treatment significantly weakened the ability of colony formation in CRC cells (HCT116, HT-29, SW480, and HCT-15) after IR, while PT33 alone did not impact colony formation at those indicated doses (Fig. 7A and B and Supporting Information Fig. S14A and S14B). Consistently, cell viability assays revealed that irradiation and PT33 exhibited a significantly synergistic antitumor effect, defined as Combination Index (CI) < 1 by a fraction affected (Fa)-CI plot analysis (Fig. 7C, D, Fig. S14C and S14D). Then we investigated the impacts of PT33 on radiation sensitivity in 3D spheroid models, growing in an anchorage-independent manner. We engineered two treatment windows starting on Day 1 or Day 4 after planting CRC cells expressing mCherry (Fig. 7E and Supporting Information Fig. S15A), representing the intervention before and during spheroid formation respectively. When intervention started during spheroid formation (on Day 4), PT33 at higher doses (10 nmol/L) but not at lower doses (5 nmol/L) significantly inhibited the 3D spheroid formation of HCT116 and HT-29 cells, both doses of PT33 synergistically enhanced the inhibitory effects of IR on the spheroid formation (all Q > 1.15; Fig. 7F). Also, the spheroid sizes were markedly smaller in the combined treatment groups than in relevant groups treated with IR or PT33 alone (Fig. 7G). Meanwhile, when intervention started before spheroid formation (on Day 1), PT33 similarly enhanced the inhibitory effects of IR, even though IR or PT33 alone already exhibited obvious inhibitory effects (Fig. S15B–S15D).
Figure 7.
PT33 sensitizes CRC cells to irradiation therapy in vitro and in vivo. (A, B) Representative images and statistical analysis of colony formation assays in HCT116 (A) and HT-29 (B) cells treated with PT33 (25 nmol/L) and irradiation at the indicated dose. Data are shown as mean ± SD (n = 3, two-way ANOVA, ∗∗∗P < 0.001). (C, D) The combination index (CI) of IR and PT33 on HCT116 (C) and HT-29 (D) cells based on cell viability assays using CCK-8. CI for x% inhibition = C/Cx + D/Dx + C × D/(Cx × Dx) (C and D are doses of IR and PT33 for x% inhibition by combined treatment; Cx or Dx is the dose of IR or PT33 for x% inhibition by treatment with PT33 or IR alone, respectively). CI > 1, CI = 1 or CI < 1 indicated an antagonist, additive or synergistic effect respectively. (E) Treatment scheme of 3D-spheroid formation assay. (F, G) Representative images (F) and statistical analysis (G) of 3D-spheroid formation assays for CRC cells treated as in (E). Upper: Q value = EAB/[EA + EB × (1–EA)] (EA, EB and EAB indicated the inhibitory rates of PT33, IR and PT33 + IR), antagonist (Q < 0.85), additive (0.85 ≤ Q ≤ 1.15) and synergistic (Q > 1.15) effect were indicated. Statistical significance symbols in blue, compared to corresponding NonIR group; symbols in red, compared to PT33 0 nmol/L + IR group. Data are shown as mean ± SD (n = 3; two-way ANOVA; n. s., not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001), (G) Below: the statistical data from all spheroid diameters of 3 independent biological replicates. (H–J) HCT116 xenografts formed in the right flank of BALB/c nude mice were treated alternately with PT33 (1 mg/kg/day for 7 times, normal saline (NS) as control) and IR (1 Gy/day for 7 times). Tumor images (H); and tumor volume (I) are shown (n = 8/group, two-way ANOVA; n. s., not significant; ∗∗∗P < 0.001). (J) Kaplan–Meier survival analysis of mice bearing xenografts (n = 6/group; log-rank test; n. s., not significant; ∗∗∗P < 0.001).
To further investigate the potential role of PT33 on radiosensitivity in vivo, we performed an experimental therapy for nude mice bearing subcutaneous HCT116 xenografts. As shown in Fig. 7H and I, PT33 administration markedly increased the sensitivity of radiotherapy, whereas PT33 alone treatment was almost noneffective. Notably, PT33 addition improved the tumor inhibition ratio of IR from 61.8% (vehicle + IR) to 93.9% (PT33 + IR), and the combination treatment exhibited a conspicuously synergistic effect (Q = 1.33; Supporting Information Fig. S16). No body weight loss or overt signs of general health deficits were observed in mice treated with the combination of PT33 and IR (data not shown). Furthermore, the combination treatment with IR and PT33 significantly improved survival rates and prolonged survival times compared to the treatment with IR or PT33 alone (Fig. 7J). All in all, these findings indicate that PT33 treatment effectively sensitized CRC to radiotherapy in vitro and in vivo.
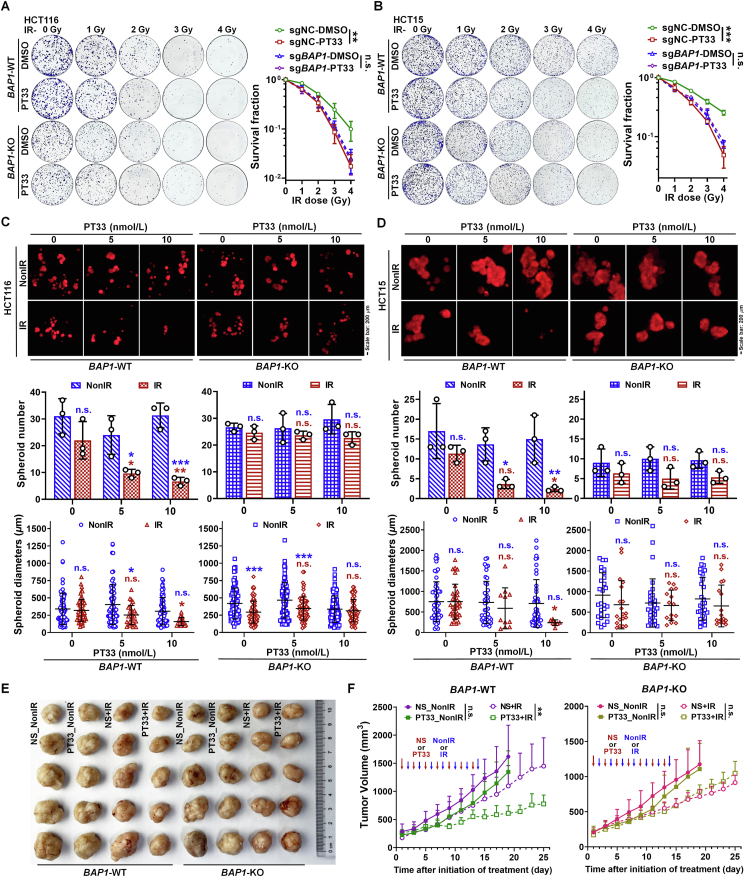
To investigate the important role of BAP1 in the sensitivity of CRC cells to radiotherapy induced by PT33, we exposed BAP1-WT and -KO CRC cells to IR followed by treatment of PT33 in vitro and in vivo. Using colony formation assays, we found that PT33 only rendered BAP1-WT cells (HCT116 and HCT15) sensitive to IR, but not BAP1-KO cells (Fig. 8A and B). In addition, when intervention started during spheroid formation (on Day 4), our results show that PT33 only enhanced the inhibitory effects of IR on the spheroid formation in BAP1-WT cells (HCT116 and HT15), consistently, but not in BAP1-KO cells (Fig. 8C and D). Furthermore, we performed in vivo experiments by subcutaneously implanting corresponding cells into nude mice, and treated the mice with PT33, IR, and their combination. In keeping with the in vitro results observed in cells, the combination of PT33 and radiotherapy showed a stronger effect on tumor suppression only in tumors derived from BAP1-WT cells, but not from BAP1-KO cells (Fig. 8E and F, and Supporting Information Fig. S17). Taken together, these results demonstrate that, when PT33 was used in combination with IR, antitumor effect is substantially enhanced in CRC cells with BAP1 expression.
Figure 8.
BAP1 deletion impairs PT33 sensitizing CRC to radiotherapy. (A, B) Representative images and statistical analysis of colony formation assays in BAP1-WT and BAP1-KO cells treated with PT33 (25 nmol/L) and IR at the indicated dose. Data are shown as mean ± SD (n = 3; two-way ANOVA; n. s., not significant; ∗∗P < 0.01 ∗∗∗P < 0.001). (C, D) Representative images (upper) and quantitative analysis (below) of 3D-spheroid formation assays for BAP1-WT and BAP1-KO HCT116 (C) and HCT15 (D) cells treated as in Fig. 7E. For spheroid number, data are mean ± SD (n = 3); for spheroid diameter, data are all spheroids in each well from 3 independent biological replicates and shown as mean ± SD. Statistical significance was determined by two-way ANOVA (n.s, not significant; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; blue symbols: compared to corresponding NonIR group; red symbols: compared to PT33 0 nmol/L + IR group). (E, F) BAP1-WT and BAP1-KO HCT116 xenografts were treated alternately with PT33 (1 mg/kg/day for 7 times, normal saline (NS) as control) and IR (1 Gy/day for 7 times). Representative images of tumors (E) and quantitative analysis of tumor volume (F). Data are mean ± SD (n = 5/group). Statistical significance was determined by two-way ANOVA (n.s., not significant; ∗∗P < 0.01).
4. Discussion
Radiation-induced DSBs are often repaired through either error-prone NHEJ or error-free HR in tumor cells37. Activation of the two pathways is accompanied by the recruitment and accumulation of numerous factors to the chromatin regions surrounding DSBs to protect and sequester the broken DNA ends and modulate DNA repair. Tumors are dependent on such mechanisms regulated tightly to effectively complete DSB repair for survival and progression5,38. An important way to regulate DSB repair is ubiquitination, which controls the localization, bioactivity and stability of repair factors. Deubiquitylation, a reversal of ubiquitination, has gradually come to the forefront of mechanism research as a crucial role in the ubiquitin-mediated regulation of DSB repair20, 21, 22. As such, DUBs have emerged as a key factor in HR or NHEJ repair39, 40, 41, 42. Consistent with our findings, the radioresistant CRC cells exhibited a higher activity of intranuclear deubiquitination for DSB repair. Thus, we speculate that radiation induces more DUBs recruited to DSB sites to facilitate DNA repair, which may result in radioresistance.
The human genome encodes approximately 100 DUBs. The major subgroups of DUBs are cysteine proteases, including ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs) and ovarian tumor proteases (OTUs)43. Unlike E3 ligases, DUBs are more likely to be targeted by small molecular inhibitors due to their well-defined catalytic residues, which provide a promising avenue for developing novel cancer therapy. Normally, because of the therapeutic value of deubiquitinating inhibition, DUB inhibitors could be used to clarify the role of ubiquitin-conjugation machinery in DNA repair16,44, 45, 46, 47. Based on these, we identified PT33 as an active entity capable of abrogating intranuclear DUB activities, which selectively interrupts HR-based DSB repair. PT33, a multitargeting active-site-directed thiol protease inhibitor for DUBs, had been reported to target 19S regulatory particles associated with deubiquitinases-USP14 and UCHL5 in our previous study32. We believe that as a DUB inhibitor, PT33 carries out distinct functions in interrupting HR repair process. But the functions of USP14, UCHL5 or other unidentified DUBs targeted by PT33 have not been characterized in HR, and the downstream of hydrolytic substrates remain unknown.
According to our results, BAP1 is identified as a novel molecular target of PT33 and confers the radiosensitization of PT33. In brief, pharmacological covalent inhibition of BAP1 with PT33 was involved in the competition of HERC2 and BARD1 for the BRCA1 interaction, which is similar to mutation or deletion in RING domain of BRCA1, ultimately leading to dissociation the BRCA1–BARD1 complex and then suppress BRCA1-mediated HR repair12, 13, 14.
One study reported that BAP1 interferes with the BRCA1/BARD1 association via interacting with BARD1, which perturbs the autoubiquitination of BRCA1 and the BRCA1–BARD1 complex mediated ubiquitination, conferring radioresistance in HeLa cells17. By contrast, several other reports demonstrated that autoubiquitination of BRCA1 contributes significantly to the stability of the BRCA1–BARD1 complex and enhances the affinity of BRCA1 for DNA48,49. And BRCA1 autoubiquitination occurs principally by conjugation with K6-linked polymers, which is the premise of BRCA1–BARD1 heterodimer functioning as an E3 ligase involved in repair50. Apparently, it has been a subject of debate that how cross-talk between BAP1 and BRCA1 functions in HR repair. Here, we described one example of such a mechanism in which PT33 covalently bound BAP1 functions as a molecular linker between BRCA1 and its E3 ligase-HERC2. We showed that HERC2 can be recruited by the PT33-bound BAP1 and competes with BARD1 to bind to the RING domain of BRCA1, ultimately leading to BRCA1 estrangement from DSB sites. Of note, the function of PT33 in HR is related to the existence of BAP1 and HERC2, both are necessary.
Our data also raises the question of why unbound wild-type BAP1 or mutant BAP1 (C91A) failed to function in the recruitment of HERC2. One possible explanation is that the covalent binding of PT33 might be involved in changing the structure and exposing the recognized domains of BAP1. Subsequently, the combination between the changed BAP1 and BRCA1 has been greatly enhanced, and then recruits HERC2 via its exposed ubiquitin-interacting domains, thereby linking the binding of HERC2 and BRCA1. To investigate the alteration of BAP1 induced by PT33 and the formation of HERC2–BAP1–BRCA1 complex, more work is required to conduct in the future.
5. Conclusions
Our findings reveal a novel mechanism that pharmacological covalent inhibition of BAP1 with PT33 recruits HERC2 to compete with BARD1 for BRCA1 interaction, resulting in the interruption of BRCA1-mediated HR repair. Moreover, PT33 treatment effectively sensitized CRC to radiotherapy. Therefore, our findings provide a strategy that PT33 as a promising drug can be used to sensitize CRC to radiotherapy with potential clinical applicability.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (NSFC) (No. 82272743 to Xin Yue; (82172812) of NSFC to Ran-yi Liu; 81871996 to Ran-yi Liu; 82003218 to Xuecen Wang; 82072029 to Zhenwei Peng and 81973174 to Xianzhang Bu), Guangdong Basic and Applied Basic Research Foundation (No. 2021A1515012496 to Xin Yue and 2022A1515012221 to Xianzhang Bu) and Basic Scientific Research Operation of Sun Yat-sen University (No. 19ykpy192 to Xin Yue). The authenticity of this article was validated by uploading the raw data to the Research Data Deposit public platform (www.researchdata.org.cn) with approval number RDDB2023965515.
Author contributions
Study concept and design: Ran-yi Liu, Zhenwei Peng, Xianzhang Bu and Xin Yue; In vitro functional studies and data analysis: Xin Yue, Tingyu Liu, Xuecen Wang, Jiaxin Wu, Yuan Meng, Liyuan Le, Wenyan Qiu, and Zhenyu Li; Animal experiments and data analysis: Xuecen Wang, Weijian Wu, Yang Yi, Ruirui Wu, and Xiaoyue Zhang; Critical cellular tools: Ziyang Wang, Weixiang Zhan, and Tingyu Liu; Mechanistical studies and data analysis: Xin Yue, Xuecen Wang, Tingyu Liu, Zhirui Cao, and Gesi Wen; Statistical analysis: Xin Yue, Tingyu Liu, and Xuecen Wang; Supervision of data: Guohui Wan and Yong Chen; Analysis and interpretation of data and writing of the manuscript: Ran-yi Liu, Xianzhang Bu, Zhenwei Peng and Xin Yue. All authors have read and approved the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Appendix A. Supporting information
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.05.017.
Contributor Information
Xianzhang Bu, Email: phsbxzh@mail.sysu.edu.cn.
Zhenwei Peng, Email: pzhenw@mail.sysu.edu.cn.
Ran-yi Liu, Email: liury@sysucc.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Häfner M.F., Debus J. Radiotherapy for colorectal cancer: current standards and future perspectives. Visc Med. 2016;32:172–177. doi: 10.1159/000446486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Cutsem E., Cervantes A., Adam R., Sobrero A., Van Krieken J.H., Aderka D., et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 3.Morgan M.A., Lawrence T.S. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin Cancer Res. 2015;21:2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang R.X., Zhou P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct Target Ther. 2020;5:60. doi: 10.1038/s41392-020-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg E.C. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M., Jia K., Wang L., Li W., Chen B., Liu Y., et al. Alterations of DNA damage response pathway: biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm Sin B. 2021;11:2983–2994. doi: 10.1016/j.apsb.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratnaparkhe M., Wong J.K.L., Wei P.C., Hlevnjak M., Kolb T., Simovic M., et al. Defective DNA damage repair leads to frequent catastrophic genomic events in murine and human tumors. Nat Commun. 2018;9:4760. doi: 10.1038/s41467-018-06925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Q., Botuyan M.V., Zhao D., Cui G., Mer E., Mer G. Mechanisms of BRCA1–BARD1 nucleosome recognition and ubiquitylation. Nature. 2021;596:438–443. doi: 10.1038/s41586-021-03716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarsounas M., Sung P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol. 2020;21:284–299. doi: 10.1038/s41580-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao W., Wiese C., Kwon Y., Hromas R., Sung P. The BRCA tumor suppressor network in chromosome damage repair by homologous recombination. Annu Rev Biochem. 2019;88:221–245. doi: 10.1146/annurev-biochem-013118-111058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez J.A., Henderson B.R. Identification of a functional nuclear export sequence in BRCA1. J Biol Chem. 2000;275:38589–38596. doi: 10.1074/jbc.M003851200. [DOI] [PubMed] [Google Scholar]
- 12.Kan C., Zhang J. BRCA1 mutation: a predictive marker for radiation therapy?. Int J Radiat Oncol Biol Phys. 2015;93:281–293. doi: 10.1016/j.ijrobp.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljungman M. Targeting the DNA damage response in cancer. Chem Rev. 2009;109:2929–2950. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]
- 14.Schlich-Bakker K.J., Ausems M.G., Schipper M., ten Kroode H.F., Wárlám-Rodenhuis C.C., van den Bout J. BRCA1/2 mutation testing in breast cancer patients: a prospective study of the long-term psychological impact of approach during adjuvant radiotherapy. Breast Cancer Res Treat. 2008;109:507–514. doi: 10.1007/s10549-007-9680-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismail I.H., Davidson R., Gagné J.P., Xu Z.Z., Poirier G.G., Hendzel M.J. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74:4282–4294. doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 16.Yu H., Pak H., Hammond-Martel I., Ghram M., Rodrigue A., Daou S., et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A. 2014;111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa H., Wu W., Koike A., Kojima R., Gomi H., Fukuda M., et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2008;69:111–119. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 18.Wu W., Sato K., Koike A., Nishikawa H., Koizumi H., Venkitaraman A.R., et al. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010;70:6384–6392. doi: 10.1158/0008-5472.CAN-10-1304. [DOI] [PubMed] [Google Scholar]
- 19.Schwertman P., Bekker-Jensen S., Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 20.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Atanassov B.S., Koutelou E., Dent S.Y. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X.C., Yue X., Zhang R.X., Liu T.Y., Pan Z.Z., Yang M.J., et al. Genome-wide RNAi screening identifies RFC4 as a factor that mediates radioresistance in colorectal cancer by facilitating nonhomologous end joining repair. Clin Cancer Res. 2019;25:4567. doi: 10.1158/1078-0432.CCR-18-3735. [DOI] [PubMed] [Google Scholar]
- 25.Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 26.Liao J., Yi Y., Yue X., Wu X., Zhu M., Chen Y., et al. Methyltransferase 1 is required for nonhomologous end-joining repair and renders hepatocellular carcinoma resistant to radiotherapy. Hepatology. 2023;77:1896–1910. doi: 10.1002/hep.32615. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y., Jiang T., Zhang H., Gou X., Han C., Wang J., et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat Cell Biol. 2020;22:1276–1285. doi: 10.1038/s41556-020-00586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundback T., Nordlund P., et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 29.Sun H., Chen D., Zhan S., Wu W., Xu H., Luo C., et al. Design and discovery of natural cyclopeptide skeleton based programmed death ligand 1 inhibitor as immune modulator for cancer therapy. J Med Chem. 2020;63:11286–11301. doi: 10.1021/acs.jmedchem.0c01262. [DOI] [PubMed] [Google Scholar]
- 30.Portolano N., Watson P.J., Fairall L., Millard C.J., Milano C.P., Song Y., et al. Recombinant protein expression for structural biology in HEK 293F suspension cells: a novel and accessible approach. J Vis Exp. 2014;(92) doi: 10.3791/51897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L., Liu R.Y., Huang J.L., Liu Q.C., Li Y., Wu P.H., et al. Adenovirus-mediated intra-tumoral delivery of the human endostatin gene inhibits tumor growth in nasopharyngeal carcinoma. Int J Cancer. 2006;118:2064–2071. doi: 10.1002/ijc.21585. [DOI] [PubMed] [Google Scholar]
- 32.Yue X., Zuo Y., Ke H., Luo J., Lou L., Qin W., et al. Identification of 4-arylidene curcumin analogues as novel proteasome inhibitors for potential anticancer agents targeting 19S regulatory particle associated deubiquitinase. Biochem Pharmacol. 2017;137:29–50. doi: 10.1016/j.bcp.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 33.D'Arcy P., Brnjic S., Olofsson M.H., Fryknäs M., Lindsten K., De Cesare M., et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1630–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 34.Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 35.Seluanov A., Mittelman D., Pereira-Smith O.M., Wilson J.H., Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Z., Seluanov A., Jiang Y., Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 2007;104:13068–13073. doi: 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoeijmakers J.H.J. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y., Gao J., Mu G., Zhang Y., Huang F., Zhang W., et al. Selectively enhancing radiosensitivity of cancer cells via in situ enzyme-instructed peptide self-assembly. Acta Pharm Sin B. 2020;10:2374–2383. doi: 10.1016/j.apsb.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma A., Alswillah T., Kapoor I., Debjani P., Willard B., Summers M.K., et al. USP14 is a deubiquitinase for Ku70 and critical determinant of non-homologous end joining repair in autophagy and PTEN-deficient cells. Nucleic Acids Res. 2020;48:736–747. doi: 10.1093/nar/gkz1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cukras S., Lee E., Palumbo E., Benavidez P., Moldovan G.L., Kee Y. The USP1–UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair. Cell Cycle. 2016;15:2636–2646. doi: 10.1080/15384101.2016.1209613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo K., Li L., Li Y., Wu C., Yin Y., Chen Y., et al. A phosphorylation–deubiquitination cascade regulates the BRCA2–RAD51 axis in homologous recombination. Genes Dev. 2016;30:2581–2595. doi: 10.1101/gad.289439.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nardi I.K., Stark J.M., Larsen A., Salgia R., Raz D.J. USP22 interacts with PALB2 and promotes chemotherapy resistance via homologous recombination of DNA double-strand breaks. Mol Cancer Res. 2020;18:424. doi: 10.1158/1541-7786.MCR-19-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He M., Zhou Z., Shah A.A., Zou H., Tao J., Chen Q., et al. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016;6:62. doi: 10.1186/s13578-016-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Q., Dexheimer T.S., Zhang P., Rosenthal A.S., Villamil M.A., You C., et al. A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato K., Nakajima K., Ui A., Muto-Terao Y., Ogiwara H., Nakada S. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell. 2014;53:617–630. doi: 10.1016/j.molcel.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 46.Wijnhoven P., Konietzny R., Blackford A.N., Travers J., Kessler B.M., Nishi R., et al. USP4 auto-deubiquitylation promotes homologous recombination. Mol Cell. 2015;60:362–373. doi: 10.1016/j.molcel.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sy S.M.H., Jiang J., Ws O., Deng Y., Huen M.S.Y. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013;41:8572–8580. doi: 10.1093/nar/gkt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen D.E., Brzovic P.S., Klevit R.E. E2–BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 49.Simons A.M., Horwitz A.A., Starita L.M., Griffin K., Williams R.S., Glover J.N.M., et al. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–2018. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 50.Wu-Baer F., Lagrazon K., Yuan W., Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278:34743–34746. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.