Abstract
Purpose
Kinase domain (KD) mutations in the BCR-ABL gene are associated with resistance to imatinib in chronic myeloid leukemia (CML) but their incidence and prognostic significance in chronic phase (CP) patients without resistance are unclear.
Patients and Methods
We analyzed outcome for 319 patients with CML-CP who were treated with imatinib; 171 were in early CP (ECP) and 148 were in late CP (LCP). Patients were screened routinely for mutations using direct sequencing regardless of response status. The 5-year cumulative incidence of mutations was 6.6% for ECP and 17% for LCP patients.
Results
Of the 319 patients, 214 (67%) achieved complete cytogenetic responses (CCyR). The identification of a mutation without other evidence of imatinib resistance was highly predictive for loss of CCyR (RR, 3.8; P = .005) and for progression to advanced phase (RR, 2.3; P = .01), though the intervals from first identification to loss of CCyR and disease progression were relatively long (median, 21 and 16 months, respectively). Mutations in the P-loop (excluding residue 244) were associated with a higher risk of progression than mutations elsewhere.
Conclusion
We conclude that routine mutation screening of patients who appear to be responding to imatinib may identify those at high risk of disease progression.
INTRODUCTION
Imatinib can induce durable responses in the majority of patients with chronic myeloid leukemia (CML) in chronic phase (CP), but some patients either fail to respond (primary resistance) or respond initially and then lose their response (secondary resistance).1-5 This resistance has been ascribed to several possible mechanisms (reviewed by Apperley6) but perhaps the best understood and certainly the most widely documented in clinical practice is the expansion of a Ph-positive clone bearing a mutation in the BCR-ABL kinase domain (KD).7,8 To date, more than 50 different KD mutations have been identified and these confer differing degrees of in vitro resistance to imatinib.6
The presence of KD mutations has been studied mainly in patients in advanced phase and in CP patients when they become resistant to imatinib.6 In both situations KD mutations are frequently present, thus establishing at least a temporal association with advanced phase disease or loss of response, but the prognostic implication of KD mutations for progression-free survival (PFS) has not been established since there are no published studies in which resistant and nonresistant patients have been monitored for mutations systematically. This is particularly important as KD mutations have been identified in patients with stable cytogenetic responses and in patients before starting imatinib therapy.8-13 Here, we describe a series of 319 CP patients who were systematically screened for KD mutations. We detected the presence of mutations independently of response status and were thus able to focus on their role in predicting loss of cytogenetic response and PFS.
PATIENTS AND METHODS
Patient Characteristics and Treatment
Between January 2000 and August 2006, 380 adult patients with BCR-ABL–positive CP CML were treated with imatinib at the Hammersmith Hospital in London. Of the 380 patients, 356 had a follow-up in CP longer than 6 months after starting imatinib and 319 of these had sequential samples available for BCR-ABL KD mutation analysis (see below). These 319 patients were analyzed further (Table 1). Eighty-five of these patients were included in various multicentric phase II3 trials of imatinib and 17 were included in the International Randomized IFN versus STI571(IRIS) study.1 All study protocols were reviewed by the research ethics committee of the Hammersmith Hospital and patients gave written informed consent to participation. CP, complete hematologic responses (CHRs), major cytogenetic responses (MCyR), and complete cytogenetic responses (CCyR) were defined by conventional criteria.1,14 Bone marrow morphology and cytogenetics were assessed at diagnosis and then every 3 months until patients achieved complete cytogenetic response (CCyR). Patients enrolled in the clinical trials had bone marrow examinations performed according to protocol. Patients started treatment with imatinib 400 mg orally daily. The dose was adjusted according to tolerance and response1,2; it was reduced in the presence of grades 3-4 toxicity15 and hematopoietic growth factors were administered with the aim of maintaining imatinib higher than 300 mg/d. Initially the criteria for dose escalation were applied as in the phase II trial or the IRIS study,1,3 but as more evidence accumulated, the criteria evolved and resembled the subsequent recommendations from the European LeukemiaNet.16 Similarly, the criteria for discontinuing imatinib varied according to the availability of the newer tyrosine kinase inhibitors.
Table 1.
Patient Characteristics at Onset of Imatinib Therapy (N = 319)
| Characteristic | No. | % | |
|---|---|---|---|
| Median age, years | 47.2 | ||
| Range | 18-73 | ||
| Sex | |||
| Male | 176 | 55.2 | |
| Female | 143 | 44.8 | |
| Sokal risk group | |||
| Low | 91 | 28.5 | |
| Intermediate | 123 | 38.6 | |
| High | 105 | 32.9 | |
| Interval since diagnosis, months | |||
| Median | 0.9 | ||
| Range | 0-218 | ||
| ≤ 1 year | 196 | 61.4 | |
| Status at the onset of imatinib therapy | |||
| Newly diagnosed chronic phase patients, < 6 months from diagnosis | 171 | 54 | |
| Chronic phase interferon-α failure | 127 | 39 | |
| Late chronic phase never treated with interferon-α | 21 | 7 | |
| Chromosomal abnormalities in addition to the Ph chromosome* | 21 | 6.8 | |
| Transcript type | |||
| e14a2 (b3a2) | 147 | 46 | |
| e13a2 (b2a2) | 131 | 41 | |
| e14a2 and e13a2 | 38 | 12 | |
| Other | 3 | 1 | |
Data missing for 10 patients.
Detection of BCR-ABL Transcripts
BCR-ABL transcripts were measured in the blood at 6- to 12-week intervals using real time quantitative polymerase chain reaction (RQ-PCR) as described previously.17-20 cDNA was synthesized from nucleated peripheral blood cells and subjected first to multiplex reverse transcriptase PCR to determine the BCR-ABL transcript type; RQ-PCR was then carried out.18 The quantitative results were expressed as percent ratios relative to an ABL internal control and as log10 reductions compared with a standardized median value for the 30 untreated patients used in the IRIS study.17,21 Major molecular response (MMR) was defined as a 3 log reduction in transcript levels21 based on two consecutive molecular studies and complete molecular response (CMR) was defined as two consecutive samples with no detectable transcripts.
BCR-ABL KD Mutations
Samples obtained for RQ-PCR studies from August 2003 were analyzed every 6 months using direct sequencing22 for the presence of mutations in the BCR-ABL KD and more often if resistance to imatinib was suspected. Samples obtained before August 2003 were retrospectively analyzed using the same criteria. Once a mutation was detected the result was confirmed and the level of the mutation in the BCR-ABL-positive cells was quantified using pyrosequencing22; the earlier samples were then analyzed to determine the time at which the mutation first became detectable and the subsequent kinetics of the mutant subclone. The resistance to imatinib of individual KD mutations was classified as high (> 3,000 nmol/L), intermediate (1,000 to 3,000 nmol/L), low (< 1,000 nmol/L), or unknown according to their respective 50% inhibitory concentrations defined in cell-based assays.23-25
Statistical Methods
PFS was defined as survival without evidence of accelerated or blastic phase disease14 and the probabilities of PFS were calculated using the method of Kaplan-Meier. The probabilities of cytogenetic response and cytogenetic relapse were calculated using the cumulative incidence procedure, where cytogenetic response or relapse were the events of interest and death, disease progression and imatinib discontinuation were the competitors. For PFS analyses, patients were censored at the time of stem-cell transplant; for cytogenetic responses and emergence of KD mutations patients were censored at the time of imatinib discontinuation. In the analyses performed to identify prognostic factors for PFS, mutations were considered only when detected for the first time before the loss of CHR. Univariate analyses to identify prognostic factors for PFS, cytogenetic relapse, and emergence of KD mutations were carried out using the log-rank test. Variables found to be significant at the P < .25 level were entered into a proportional hazards regression analysis; a forward stepping procedure was employed to find the best model. The influence of KD mutations on the different outcomes was studied in a time-dependent Cox model. Tests for interactions were carried out but none was found to have statistical significance. The proportional hazards assumption was confirmed by adding a time-dependent covariate for each covariate. P values were two sided and 95% CIs computed.
Patients With No Available Samples for KD Mutation Analysis
Samples for KD mutation analysis were not available for 37 patients. For these patients PFS (P = .4), cumulative incidence of CCyR (P = .2), cumulative incidence of MMR (P = .8), and Sokal risk group distributions (P = .8) were very similar to the 319 patients included in the study.
RESULTS
Responses
The median follow-up for surviving patients on imatinib was 50.5 months (range, 12 to 89.6 months). Three hundred eleven patients (97.5%) achieved CHR; 244 (76%) achieved MCyR (146 ECP and 98 LCP); 214 patients (67%) achieved a CCyR (134 ECP and 80 LCP); and 103 (32%) achieved a MMR (68 ECP and 35 LCP). During follow-up, 53 patients (17%) failed to achieve any degree of cytogenetic response (> 95% Ph-positive metaphases) and were classified as showing primary cytogenetic resistance (12 in ECP and 41 in LCP).
Development of KD Mutations
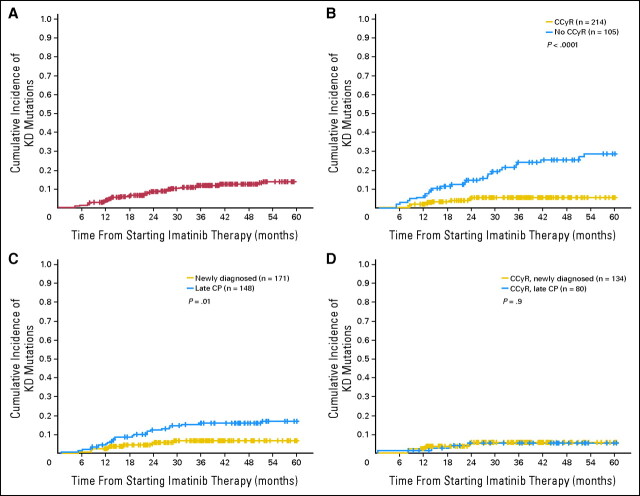
During follow-up, 37 patients (11.6%) developed KD mutations. The median time from starting imatinib to first detection of the mutation was 16.3 months (range, 1 to 52.2). Ten patients developed a mutation before achieving or while in CCyR (median time, 13.5 months; range, 1 to 24.1), an additional 23 while in CHR (median time, 14.1 months; range, 5 to 51.1) and four more before progression to advanced stage (median time, 33.5 months; range, 12.3 to 52.2). The most common mutations were M244V (seven cases; 18.4%), F359V (three cases; 8%), H396R (three cases; 8%), and F486S (three cases; 8%). Appendix Table A1 (online only) shows the different mutations identified and the patient characteristics. The 5-year cumulative incidence of KD mutations was 13.9% (Fig 1).
Fig 1.
Cumulative incidence of kinase domain (KD) mutations. (A) Five-year cumulative incidence of KD mutations (13.9%). (B) Five-year cumulative incidence of KD mutations comparing patients whose best response was complete cytogenetic responses (CCyR; 5.5%) with patients who failed to achieve CCyR (29.1%; P < .0001). (C) Five-year cumulative incidence of KD mutations comparing early chronic phase (ECP) patients (6.6%) with late chronic phase (LCP) patients (17%, P = .01). (D) Five-year cumulative incidence of KD mutations comparing ECP (5.6%) and LCP patients (5.3%, P = .89) but restricted only to those patients whose best response was a CCyR.
We performed univariate and multivariate analyses to identify pretherapy predictors for the development of KD mutations (Table 2). In multivariate analysis, LCP and high Sokal scores were independent predictors for the development of mutations (RR, 2.7; P = .005 RR, 2.5; P = .03, respectively).
Table 2.
Five-Year Cumulative Incidence of Development of Kinase Domain Mutations, Loss of Complete Cytogenetic Response, and the Probability of Progression-Free Survival
| Variable | Kinase Domain Mutation |
Loss of Complete Cytogenetic Response* |
Progression-Free Survival |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % | P | % | P | % | P | ||||
| Age, years | .04 | .1 | .4 | ||||||
| > 47 | 16.9 | 14.3 | 79.0 | ||||||
| ≤ 47 | 10.2 | 25.4 | 78.3 | ||||||
| Sex | .8 | .3 | .9 | ||||||
| Male | 11.8 | 15.8 | 79.0 | ||||||
| Female | 14.1 | 23.4 | 79.0 | ||||||
| Sokal risk group | .01 | .08 | .009 | ||||||
| Low | 6.2 | 11.5 | 89.1 | ||||||
| High plus intermediate | 15.3 | 23.5 | 75.0 | ||||||
| Interval from diagnosis to start of imatinib, years | .01 | .6 | .4 | ||||||
| ≤ 1 | 8.7 | 21.4 | 79.5 | ||||||
| > 1 | 19.6 | 17.1 | 77.0 | ||||||
| Status at the start of imatinib therapy | .005 | .6 | .06 | ||||||
| Newly diagnosed chronic phase patients | 6.6 | 16.5 | 83.8 | ||||||
| Late chronic phase | 19.5 | 21.6 | 75.2 | ||||||
| Additional cytogenetic abnormalities at onset of imatinib therapy | .002 | .3 | .01 | ||||||
| Yes | 36 | 0 | 55.9 | ||||||
| No | 11.7 | 20.8 | 81.3 | ||||||
| Transcript type | .02 | .02 | .5 | ||||||
| Either e13a2 or e14a2 | 11.6 | 3.6 | 78.9 | ||||||
| Both | 28.5 | 16.7 | 80.9 | ||||||
| Dose intensity of imatinib during the first 6 months of therapy, mg/d | .36 | .7 | .7 | ||||||
| ≥ 400 | 9.3 | 20.7 | 79.6 | ||||||
| < 400 | 14.2 | 15.6 | 78.3 | ||||||
NOTE. Bold font indicates statistical significance.
Includes only the 214 patients who achieved a complete cytogenetic response.
Development of KD Mutations Predicts for Loss of CCyR
Of the 214 patients who achieved CCyR, a KD mutation was detected in 10 (4.7%), of whom four had developed a mutation before they achieved CCyR. At the time when the mutation was first detected in patients in CCyR, the median log reduction in transcript levels was 2.4 (range, 1.9 to 2.7). The median time from the detection of the KD mutation to loss of CCyR was 20.7 months (range, 2.8 to 49.1 months). The median time between the detection of the KD mutation and a subsequent twofold rise in BCR-ABL transcript levels12 was 12 months (range, 3 to 33).
Thirty (14%) of 214 patients who achieved CCyR lost their response during follow-up. We performed univariate and multivariate analysis to identify prognostic factors for loss of CCyR. The development of mutations was highly predictive for loss of CCyR (RR, 3.8; P = .005). We further classified the KD mutation as belonging to the P-loop (n = 0), as M244V (n = 2), or as non P-loop (n = 8). The RR for loss of CCyR of the patients with non-M244V mutations was 7.1 (P < .0001), whereas the M244V mutation did not seem to have any adverse effect. The rest of the variables analyzed in the univariate analysis are presented in Table 2. The development of a KD mutation was the only independent predictor for loss of CCyR in the multivariate analysis.
Development of KD Mutations Predicts for Progression to Advanced Phase
During follow-up, 49 patients (15%) progressed to advanced phase. A KD mutation was detected in 17 of these patients before progression (median time between detection of mutation and progression was 16.3 months; range, 2.4 to 51.3); in 14 cases the mutation was detected before the loss of CHR. The median time between detection of mutation and loss of CHR was 13.6 months (range 2.3 to 49.1). A KD mutation was detected in 20 patients who did not progress (19 while in CHR); the median follow-up after the detection of the mutation was 60.7 months (range, 29.7 to 88.7).
We performed univariate and multivariate analysis to identify prognostic factors (including the emergence of KD mutations in patients in CHR) for PFS in patients treated with imatinib in CP (Table 2). Patients developing KD mutations had a higher risk of progression (RR, 3.7; P < .0001). The other major on-therapy prognostic factor for risk of progression was the achievement of CCyR (RR, 0.13; P < .0001). In the multivariate analysis, the only independent prognostic factors for risk of progression were the achievement of CCyR (RR, 0.15; P < .0001) and development of KD mutations (RR, 2.3; P = .01).
When the mutations detected in patients before loss of CHR were subclassified according to their level of resistance to imatinib in vitro, we found that the six patients with highly resistant mutations had a significantly worse PFS than the 27 patients with low, intermediate, or unknown levels considered together (RR for progression, 8.6; P = .001). Similarly, when patients were classified according to the position of the KD mutation, the seven patients with P-loop mutations had a significantly worse prognosis than the patients with mutations elsewhere (RR for progression, 5.4; P = .001); patients with the M244V mutation (n = 6) had prognoses similar to that of patients with no mutations (P = .97).
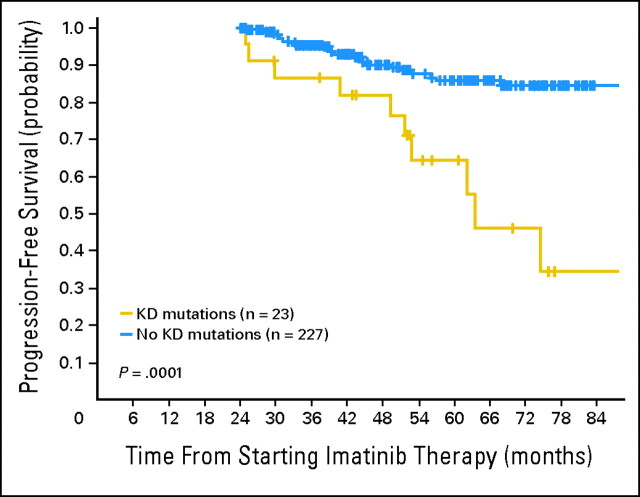
Although KD mutations may develop at any time during therapy, we chose to assess the impact of KD mutations according to response and mutation status at a given time point. At 2 years, 250 patients were still in CHR and formed the cohort for this subanalysis; 23 of these had developed a KD mutation. In this landmark analysis, the 143 patients who had achieved a CCyR by 2 years had a significantly better 5-year PFS than those who had not achieved a CCyR (95.5 v 71.3; P < .0001), as did patients who had not developed a mutation compared to those with who had (85.8% v 64.6%; P = .0001; Fig 2). The Sokal risk score, initial therapy with imatinib, and the presence of additional cytogenetic abnormalities at start of imatinib were also significant predictors for PFS (data not shown). In the multivariate analysis, only the presence of a mutation and the cytogenetic response were independent prognostic factors for risk of progression (RR, 3.0; P = .004; RR, 0.18; P = .0001, respectively). Figure 3 shows the PFS according to presence or absence of KD mutations taking into account the degree of imatinib resistance and the position of the nucleotide substitution.
Fig 2.
Progression-free survival according to the presence of kinase domain (KD) mutations at 2 years: a landmark analysis.
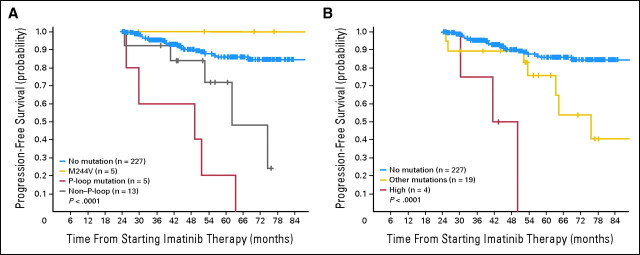
Fig 3.
Progression-free survival (PFS) in a 2-year landmark analysis according to the position of the amino acid substitution and the level of in vitro resistance of the kinase domain (KD) mutation. (A) PFS in the 2-year landmark analysis according to the type of mutation. The 5-year PFS was 86% for patients with no mutation, 100% for patients with the mutation M244V, 20% for patients with P-loop mutations, and 47% for patients with non-P-loop mutations (P < .0001). Patients harboring the M244V mutation had a PFS no different from that of patients with no mutation (P = .9), but patients with a P-loop mutation and patients harboring non-P-loop mutations had a significantly different PFS when compared with patients with no mutation (P = .01 and P < .0001, respectively). The difference in PFS between patients harboring a P-loop mutation and non-P-loop mutations was also significant (P = .02). (B) PFS in the 2-year landmark analysis according to the degree of in vitro resistance of the KD mutation. The 5-year PFS was 86% for patients with no mutation, 0% for patients highly resistant mutations, and 64.7% for patients with other mutations (P < .0001). Patients with a highly resistant mutations and patients harboring other mutations had a significantly different PFS when compared with patients with no mutations (P = .008 and P < .0001, respectively). The difference in PFS for patients harboring a highly resistant mutation and patients with other mutations was also significant (P = .03).
Adverse Effect of KD Mutations on PFS Was Restricted to Patients With Secondary Cytogenetic Resistance to Imatinib
Two hundred sixty-six patients achieved at least a MiCyR, of whom 23 (8.6%) developed a KD mutation, whereas of the 53 patients with primary cytogenetic resistance, 14 (26.4%) developed a KD mutation (P = .001). There was no significant difference in the characteristics of the KD mutations between the two groups, since the median time to first detection of mutation was 18.2 months for the cytogenetic refractory and 16.3 months in the responding group (P = .4). A M244V mutation occurred in three patients (21%) with primary cytogenetic resistance and in four responders (17%; P = .8).
We found that the adverse effect of KD mutations described earlier was limited to patients with secondary resistance to imatinib. In the patients with primary cytogenetic resistance and mutations, the adjusted RR for progression was 0.54 (P = .35), while in patients with secondary cytogenetic resistance and mutations the RR for progression was 6.0 (P < .0001).
DISCUSSION
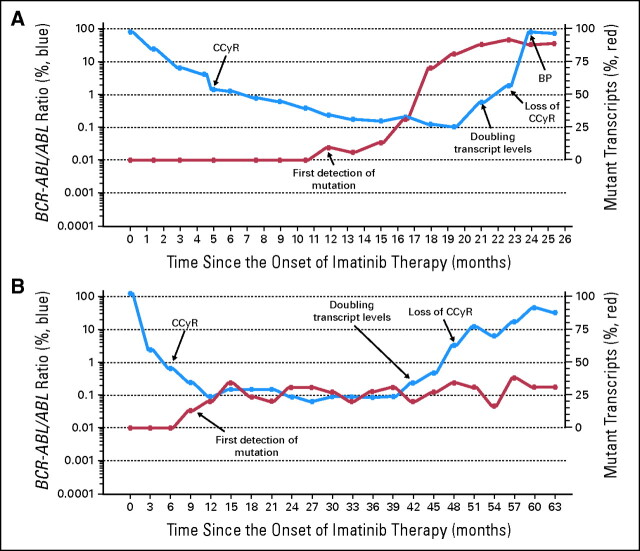
The emergence of mutations in the KD of BCR-ABL is the most commonly identified mechanism of clinical resistance to imatinib.6 However, because the majority of studies on KD mutations have involved patients with acquired resistance to imatinib or patients who have progressed to advanced phase the prognostic implication of KD mutations in CP patients with continuing hematologic or cytogenetic responses has not yet been established. We studied the relationship of KD mutations to loss of CCyR and to PFS by systematically screening our population of CP patients for the presence of mutations irrespective of their response status. We identified KD mutations in 37 patients; 17 of these progressed to advanced phase and the KD mutation was always detected before progression (median time, 16 months). Figure 4 shows the evolution of two representative patients with KD mutations.
Fig 4.
Evolution of total BCR-ABL transcript levels and percentages of the mutated transcripts in two selected patients. The figure shows the evolution of the total transcript levels the percentages of mutated subclones and in two representative patients. In both cases the detection of the kinase domain (KD) mutation clearly antedated any noticeable change in the transcript levels (see text). In 16 patients, exemplified by the patient in (A), the proportion of the mutant BCR-ABL clone (E459K) increased progressively until it became the predominant clone (more than 50%). In 14 of these patients the dose of imatinib was increased higher than 400 mg; in the patient represented in (A) the dose was increased to 800 mg at the moment of losing the complete cytogenetic response (CCyR). Seven of these 16 patients progressed to advanced phase. In the remaining 21 patients, exemplified by the patient in (B), the proportion of mutant transcripts (M351T) remained at or below 50% during follow-up (10 cases) or transiently increased higher than 50% (11 cases). In 20 patients, the dose of imatinib was increased above 400 mg. (In the patient represented in (B) the dose was increased to 600 mg at the moment of losing the CCyR and later changed to dasatinib). Ten of 21 patients progressed to advanced phase. BP, blastic phase.
In order to study the prognostic value of KD mutations detected in patients in CHR, we performed two different analyses. In one analysis, we included all 319 patients and studied the prognostic significance of mutations (and also cytogenetic responses) occurring at any time during follow-up. In this analysis the emergence of a mutation and the achievement of CCyR were the only independent prognostic factors for risk of progression (RR, 2.3; P = .01; RR, 0.15; P < .0001, respectively). In the second analysis, we considered only the 250 patients who still were in CHR at 2 years, of whom 23 had developed a KD mutation and 143 had achieved CCyR. Again the presence of KD mutations and the achievement of CCyR were the only two independent prognostic factors for PFS (Fig 2).
KD mutations were found in four patients before they achieved CCyR and in six patients already in CCyR (Table A1). As shown previously,26 KD mutations were the only significant predictor for loss of CCyR (RR, 3.8; P = .005), but there was a long delay between the identification of the mutation and this loss of response (median time, 21 months). These six mutations were all detected before any discernible change in the transcript levels (median time to doubling in transcript levels was 12 months).
Our data suggest that KD mutations should be regarded as an event clinically analogous to the observation of a new cytogenetic abnormality (so-called clonal evolution). Thus, theoretically the identification of a KD mutation before any discernible increase in BCR-ABL transcripts levels might be an indication for changing therapy,26,27 but in practice to systematically screen all CCyR patients for the relatively rare occurrence of a KD mutation might not be cost-effective. A reasonable compromise could be to screen patients with incomplete cytogenetic responses and patients in CCyR with relatively modest reductions in transcript levels (eg, < 2.5 logs) twice a year, especially if they have high Sokal risk scores.
The precise prognostic significance of P-loop mutations is still contentious. Some investigators have reported that P-loop mutations were associated with worse prognosis,28,29,30 while others31 were unable to confirm this finding. A possible explanation for this discrepancy might relate to the prognostic significance of the M244V mutation, which may be relatively innocuous. Thus, this mutation was predominant in the Houston31 study but was excluded from definition of the P-loop in the Adelaide, Bologna, and French intergroup studies.28,29,30 We classified mutations in three categories: M244V (our most frequent mutation), mutations involving residues 245 to 255 (P-loop), and other mutations. The mutation M244V did not have any adverse effect on PFS, while the remaining mutations did confer a worse prognosis. Patients with P-loop mutations (excluding M244V) had a significantly worse prognosis than patients with other mutations (RR for progression, 5.4; P = .001; Fig 3). The reason why patients with a P-loop mutation fare worse remains unclear.
Patients with primary cytogenetic resistance to imatinib were more likely to develop KD mutations after the resistance was already identified, indicating that the mechanism of resistance was in operation since the beginning of therapy, which in turn suggests that the emergence of mutations merely reflects a higher level of genomic instability.32 Moreover, the fact that mutations were more frequent in patients in the high Sokal risk group supports the hypothesis that the probability of developing a mutation is related to the basic biology of the disease rather than being merely a random event.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: David Marin, Novartis (C), Bristol-Myers Squibb (C) Stock Ownership: None Honoraria: None Research Funding: David Marin, Novartis Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Jamshid S. Khorashad, Jaspal Kaeda, John M. Goldman, David Marin
Provision of study materials or patients: Jamshid S. Khorashad, Hugues de Lavallade, Jane F. Apperley, Dragana Milojkovic, Alistair G. Reid, Eduardo Olavarria, John M. Goldman, David Marin
Collection and assembly of data: Jamshid S. Khorashad, Hugues de Lavallade, Jane F. Apperley, Dragana Milojkovic, Alistair G. Reid, Marco Bua, Jaspal Kaeda, David Marin
Data analysis and interpretation: Jane F. Apperley, Marco Bua, Richard Szydlo, David Marin
Manuscript writing: Hugues de Lavallade, John M. Goldman, David Marin
Final approval of manuscript: Jamshid S. Khorashad, Hugues de Lavallade, Jane F. Apperley, Dragana Milojkovic, Alistair G. Reid, Marco Bua, Richard Szydlo, Eduardo Olavarria, Jaspal Kaeda, John M. Goldman, David Marin
Appendix
Table A1.
Kinase Domain Mutations and Patient Characteristics
| Patient | Best Response to Imatinib | Status at Last Follow-Up | Aa Substitution | Time in Months From Start of Imatinib to Best Response | Time in Months From Start of Imatinib to Detection of Mutation | Time in Months From Detection of Mutation to Loss of CCyR | Time in Months From Detection of Mutation to CHR* | Time From First Detection of a Mutation to Progression or Last Follow-Up | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Pri Hem Res | UCP | F359V | NA | 12 | NA | 25 | ||
| 2 | Pri Hem Res | UCP | M351T | NA | 28 | NA | 57 | ||
| 3 | Pri Cy Res | BP | Y253F | 1 | 26 | 4 | 4 | ||
| 4 | Pri Cy Res | UCP | F486S | 1 | 10 | 41 | 45 | ||
| 5 | Pri Cy Res | BP | M244V | 1.5 | 60 | 19 | 13 | ||
| 6 | Pri Cy Res | AP | G250E | 1 | 12 | 10 | 51 | ||
| 7 | Pri Cy Res | UCP | F486S | 1 | 31 | 2 | 48 | ||
| 8 | Pri Cy Res | BP | L248V | 2 | 8 | 17 | 17 | ||
| 9 | Pri Cy Res | BP | F359V | 1.1 | 49 | −2 | 4 | ||
| 10 | Pri Cy Res | AP | E453K | 1 | 14 | 9 | 48 | ||
| 11 | Pri Cy Res | BP | E255V | 1.1 | 14 | 16 | 16 | ||
| 12 | Pri Cy Res | UCP | M244V | 1.7 | 5 | 4 | 25 | ||
| 13 | Pri Cy Res | AP | F359V | 1 | 82 | −29 | 16 | ||
| 14 | Pri Cy Res | UCP | M244V | 1 | 22 | 24 | 55 | ||
| 15 | MiCyR | CHR | E453V | 13 | 29 | NA | 32 | ||
| 16 | MiCyR | AP | Y253H | 6 | 5 | 8 | 8 | ||
| 17 | MiCyR | UCP | F486S | 15 | 70 | 0 | 34 | ||
| 18 | MCyR | CHR | E453K | 14 | 51 | NA | 18 | ||
| 19 | MCyR | AP | G250E | 6 | 6 | 46 | 46 | ||
| 20 | MCyR | UCP | M244V | 6 | 8 | 20 | 44 | ||
| 21 | MCyR | AP | Y253H | 12 | 13 | 22 | 36 | ||
| 22 | MCyR | BP | H396R | 20 | 23 | 2 | 2 | ||
| 23 | MCyR | AP | T315I | 12 | 12 | 24 | 29 | ||
| 24 | MCyR | CHR | M244V | 16 | 35 | NA | 43 | ||
| 25 | MCyR | BP | F311I | 8 | 16 | 22 | 36 | ||
| 26 | MCyR | UCP | H396R | 19 | 19 | 21 | 37 | ||
| 27 | MCyR | AP | L384M | 17 | 28 | 11 | 11 | ||
| 28 | CCyR | AP | E459K | 3 | 13 | 3 | 3 | 3 | |
| 29 | CCyR | CHR | M351T | 6 | 9 | 38 | NA | 52 | |
| 30 | CCyR | MCyR | T315I | 4 | 8 | 21 | NA | 35 | |
| 31 | CCyR† | CCyR | M244V | 18 | 14 | NA | NA | 56 | |
| 32 | CCyR | MCyR | L387M | 20 | 24 | 20 | NA | 21 | |
| 33 | CCyR | MCyR | S417F | 12 | 24 | 12 | NA | 29 | |
| 34 | CCyR | BP | E459K | 5 | 12 | 23 | 25 | 12 | |
| 35 | CCyR† | CCyR | M244V | 35 | 19 | NA | NA | 70 | |
| 36 | MMR† | MMR | F317L, H396R | 30 | 1 | NA | NA | 42 | |
| 37 | CMR† | CMR | S438C | 57 | 8 | NA | NA | 67 | |
NOTE. Bold font indicates early CP patients.
Abbreviations: CCyR, complete cytogenetic responses; CHR, complete hematologic response; NA, not applicable; Pri Hem Res, primary hematologic resistance; Pri Cy Res, primary cytogenetic resistance; CP, chronic phase; UCP, uncontrolled chronic phase; AP, accelerated phase; BP, blastic phase; MiCyR, minor cytogenetic responses; MCyR, major cytogenetic responses; MMR, major molecular response; CMR, complete molecular response.
Negative values indicate that a mutation was detected after the loss of CHR.
Mutation detected before CCyR.
We thank the many members of the medical, nursing, and technical staff who contributed to patient care and production of laboratory data.
Footnotes
published online ahead of print at www.jco.org on July 21, 2008
Supported by the NIHR Biomedical Research Centre Funding Scheme; and by a grant from the Fondation de France (Paris, France; H.d.L.).
J.S.K. and H.d.L. contributed equally to this article.
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.O'Brien SG, Guilhot F, Larson RA, et al: Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 348::994,2003-1004, [DOI] [PubMed] [Google Scholar]
- 2.Druker B, Guilhot F, O'Brien S, et al: Five-year follow-up of imatinib therapy for newly diagnosed chronic myelogenous leukemia in chronic-phase shows sustained responses and high overall survival. N Engl J Med 355::2408,2006-2417, 17151364 [Google Scholar]
- 3.Kantarjian H, Sawyers C, Hochhaus A, et al: Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med 346::645,2002-652, [DOI] [PubMed] [Google Scholar]
- 4.Kantarjian HM, Cortes JE, O'Brien S, et al: Long-term survival benefit and improved complete cytogenetic and molecular response rates with imatinib mesylate in Philadelphia chromosome-positive chronic-phase chronic myeloid leukemia after failure of interferon-alpha. Blood 104::1979,2004-1988, [DOI] [PubMed] [Google Scholar]
- 5.Hochhaus A, Druker B, Sawyers C, et al: Favorable long-term follow-up results over six years for response, survival and safety with imatinib mesylate therapy in chronic phase chronic myeloid leukemia post failure of interferon-alpha treatment. Blood 111::1039,2008-1043, [DOI] [PubMed] [Google Scholar]
- 6.Apperley JF: Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol 8::1018,2007-1029, [DOI] [PubMed] [Google Scholar]
- 7.Gorre ME, Mohammed M, Ellwood K, et al: Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293::876,2001-880, [DOI] [PubMed] [Google Scholar]
- 8.Shah N, Nicoll J, Nagar B, et al: Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2::117,2002-223, [DOI] [PubMed] [Google Scholar]
- 9.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al: Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood 100::1014,2002-1018, [DOI] [PubMed] [Google Scholar]
- 10.Willis S, Lange T, Demehri S, et al: High sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: Correlation with clonal cytogenetic evolution but not response to therapy. Blood 106::2128,2005-2136, [DOI] [PubMed] [Google Scholar]
- 11.Chu S, Xu H, Shah NP, et al: Detection of BCR-ABL kinase mutations in CD34+ cells from chronic myelogenous leukemia patients in complete cytogenetic remission on imatinib mesylate treatment. Blood 105::2093,2005-2098, [DOI] [PubMed] [Google Scholar]
- 12.Branford S, Rudzki Z, Parkinson I, et al: Real-time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood 104::2926,2004-2932, [DOI] [PubMed] [Google Scholar]
- 13.Sherbenou DW, Wong MJ, Humayun A, et al: Mutations of the BCR-ABL-kinase domain occur in a minority of patients with stable complete cytogenetic response to imatinib. Leukemia 21::489,2007-493, [DOI] [PubMed] [Google Scholar]
- 14.Kantarjian HM, Dixon D, Keating MJ, et al: Characteristics of accelerated disease in chronic myelogenous leukemia. Cancer 61::1441,1988-1446, [DOI] [PubMed] [Google Scholar]
- 15.Cancer Therapy Evaluation Program: Common Toxicity Criteria. In: National Cancer Institute, ed. 1998
- 16.Baccarani M, Saglio G, Goldman J, et al: Evolving concepts in the management of chronic myeloid leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 1809,2006-1820, [DOI] [PubMed]
- 17.Kaeda J, Chase A, Goldman JM: Cytogenetic and molecular monitoring of residual disease in chronic myeloid leukaemia. Acta Haematol 107::64,2002-75, [DOI] [PubMed] [Google Scholar]
- 18.Marin D, Kaeda J, Szydlo R, et al: Monitoring patients in complete cytogenetic remission after treatment of CML in chronic phase with imatinib: Patterns of residual leukaemia and prognostic factors for cytogenetic relapse. Leukemia 19::507,2005-512, [DOI] [PubMed] [Google Scholar]
- 19.Kaeda J, O'Shea D, Szydlo RM, et al: Serial measurement of BCR-ABL transcripts in the peripheral blood after allogeneic stem cell transplantation for chronic myeloid leukemia: An attempt to define patients who may not require further therapy. Blood 107::4171,2006-4176, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes T, Deininger M, Hochhaus A, et al: Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 108::28,2006-37, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes TP, Kaeda J, Branford S, et al: Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med 349::1423,2003-1432, [DOI] [PubMed] [Google Scholar]
- 22.Khorashad JS, Anand M, Marin D, et al: The presence of a BCR-ABL mutant allele in CML does not always explain clinical resistance to imatinib. Leukemia 20::658,2006-663, [DOI] [PubMed] [Google Scholar]
- 23.Martinelli G, Soverini S, Rosti G, et al: Dual tyrosine kinase inhibitors in chronic myeloid leukemia. Leukemia 19::1872,2005-1879, [DOI] [PubMed] [Google Scholar]
- 24.Burgess MR, Skaggs BJ, Shah NP, et al: Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A 102::3395,2005-3400, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Hare T, Pollock R, Stoffregen EP, et al: Inhibition of wild-type and mutant Bcr-Abl by AP23464, a potent ATP-based oncogenic protein kinase inhibitor: Implications for CML. Blood 104::2532,2004-2539, [DOI] [PubMed] [Google Scholar]
- 26.de Lavallade H, Apperley JF, Khorashad JS, et al: Imatinib for newly diagnosed patients with chronic myeloid leukaemia: Incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol doi: [epub ahead of print on June 2, 2008] 10.1200/JCO.2007.15.8154 [DOI] [PubMed]
- 27.Kantarjian H, Quintas-Cardama A, O'Brien S, et al: Importance of early intervention with dasatinib at cytogenetic rather than hematologic resistance to imatinib. Blood 110::314a,2007, [Google Scholar]
- 28.Branford S, Rudzki Z, Walsh S, et al: Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 102::276,2003-283, [DOI] [PubMed] [Google Scholar]
- 29.Soverini S, Martinelli G, Rosti G, et al: ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: A study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J Clin Oncol 23::4100,2005-4109, [DOI] [PubMed] [Google Scholar]
- 30.Nicolini FE, Corm S, Le QH, et al: Mutation status and clinical outcome of 89 imatinib mesylate-resistant chronic myelogenous leukemia patients: A retrospective analysis from the French intergroup of CML (Fi(phi)-LMC GROUP). Leukemia 20::1061,2006-1066, [DOI] [PubMed] [Google Scholar]
- 31.Jabbour E, Kantarjian H, Jones D, et al: Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia 20::1767,2006-1773, [DOI] [PubMed] [Google Scholar]
- 32.Jiang X, Saw KM, Eaves A, Eaves C: Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst 99::680,2007-693, [DOI] [PubMed] [Google Scholar]