Abstract
Despite the remarkable efficacy of COVID-19 vaccines, waning immunity and the emergence of SARS-CoV-2 variants such as Omicron represents a global health challenge. Here, we present data from a study in nonhuman primates demonstrating durable protection against the Omicron BA.1 variant induced by a subunit SARS-CoV-2 vaccine comprising the receptor binding domain of the ancestral strain (RBD-Wu) on the I53–50 nanoparticle adjuvanted with AS03, which was recently authorized for use in individuals 18 years or older. Vaccination induced neutralizing antibody (nAb) titers that were maintained at high concentrations for at least 1 year after two doses, with a pseudovirus nAb geometric mean titer (GMT) of 1978 and a live virus nAb GMT of 1331 against the ancestral strain but not against the Omicron BA.1 variant. However, a booster dose at 6 to 12 months with RBD-Wu or RBD-β (RBD from the Beta variant) displayed on I53–50 elicited high neutralizing titers against the ancestral and Omicron variants. In addition, we observed persistent neutralization titers against a panel of sarbecoviruses, including SARS-CoV. Furthermore, there were substantial and persistent memory T and B cell responses reactive to Beta and Omicron variants. Vaccination resulted in protection against Omicron infection in the lung and suppression of viral burden in the nares at 6 weeks after the final booster immunization. Even at 6 months after vaccination, we observed protection in the lung and rapid control of virus in the nares. These results highlight the durable and cross-protective immunity elicited by the AS03-adjuvanted RBD-I53–50 nanoparticle vaccine.
INTRODUCTION
Waning immunity coupled with the continuing emergence of immune-evasive variants represents a major challenge in controlling the coronavirus disease 2019 (COVID-19) pandemic. The efficacy of the highly effective mRNA vaccines has been shown to decrease 20 to 30% by 6 months after a two-dose vaccine series (1, 2). The efficacy declined more precipitously against Omicron, a variant highly resistant to vaccine-induced antibodies (3–5), reaching 8.8% after the two-dose Pfizer-BioNTech mRNA vaccination. The waning efficacy thus mandates a booster vaccination despite the fact that about 40% of the world’s population are yet to receive full vaccination.
We recently reported the results of a study in nonhuman primates (NHPs) in which we compared the immunogenicity and protective efficacy of the receptor binding domain (RBD)–I53–50 nanoparticle immunogen formulated with five different adjuvants (6). Administering the RBD-I53–50 vaccination with AS03, an oil-in-water emulsion containing α-tocopherol, elicited the most potent and broad neutralizing antibody (nAb) response, as well as substantial T cell responses, and conferred protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge in the upper and lower airways. Furthermore, vaccine-induced nAbs persisted for at least 6 months after vaccination. In addition, AS03-adjuvanted RBD-I53–50 vaccination elicited potent nAb response in humans in a phase 1/2 clinical trial (7). Recently, the AS03-adjuvanted RBD-I53–50 met its primary end point in the phase 3 clinical trial and was approved by the Korean Ministry of Food and Drug Safety for use in individuals 18 years or older. In the present study, we evaluated the durability of immune protection after a booster immunization with RBD-Wu or RBD-β against the immune-evasive Omicron BA.1 variant.
RESULTS
AS03-adjuvanted RBD-I53–50 vaccination elicits robust and durable antibody responses
The study involved four groups of male rhesus macaques. The first group of five animals were immunized thrice with RBD-Wu + AS03, at days 0 and 21, followed by a final booster about 6 months later (group RBD-Wu/RBD-Wu/RBD-Wu; Fig. 1A). The second and third groups were from our previous study (6) in which one group of five animals received two doses of RBD-Wu, and the other group comprising six animals received two doses of HexaPro (HexaPro Spike protein of the ancestral Wu strain displayed on I53–50 nanoparticle). Both immunogens were administered with the AS03 adjuvant on days 0 and 21 using a prime-boost regimen during the previous study. In the current phase of the study, all 11 animals from both groups were boosted with an I53–50 nanoparticle immunogen displaying RBD-β stabilized with the Rpk9 mutations (8) about a year after the first immunization series (groups RBD-Wu/RBD-Wu/ RBD-β and HexaPro/HexaPro/RBD-β; Fig. 1A). The Beta variant was selected because it was the prevalent SARS-CoV-2 strain at the time. Thus, after a prolonged interval after immunization with RBD-β, we had planned to challenge these animals with the Beta variant to assess the durability of protection against this variant. However, the emergence of Omicron as the dominant strain while the study was ongoing prompted us to challenge the animals with Omicron instead to assess heterologous protection against this variant. The fourth group of five animals were unvaccinated controls. To assess the immunogenicity, the antibody responses in the RBD-Wu/RBD-Wu/ RBD-Wu animals were followed longitudinally from the day of the first immunization, whereas the two groups from the previous study were followed from the day of the final booster (Fig. 1A). To assess protective efficacy, we challenged the animals with 2 × 106 plaque-forming units (PFU) of SARS-CoV-2 Omicron BA.1 virus. We challenged the first group at 6 weeks after the final booster and the other two groups at about 6 months after the final booster to assess protection at the peak of immune responses versus when immune responses were waning.
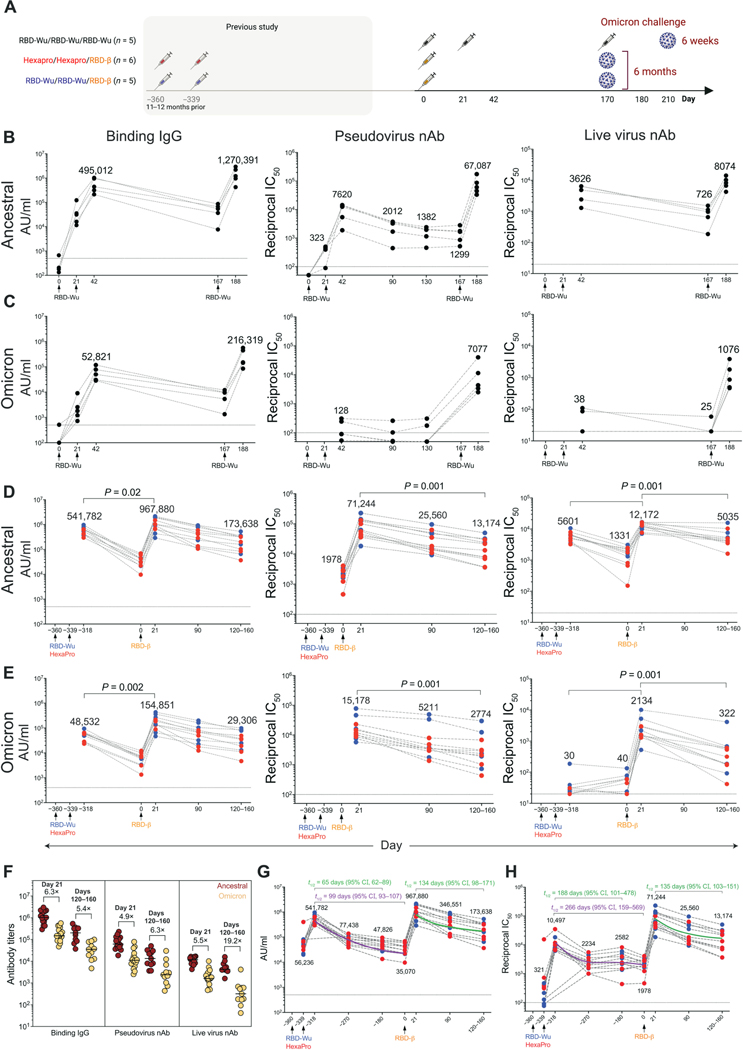
Fig. 1. AS03-adjuvanted RBD-I53–50 immunization elicits potent and durable serum antibody responses.
(A) Schematic of the study design is shown. (B and C) Anti–Spike protein binding IgG, pseudovirus, and live virus nAb titers were measured against ancestral (B), and Omicron (C) strains in the RBD-Wu/RBD-Wu/RBD-Wu group (n = 5). (D and E) Binding IgG, pseudovirus, and live virus nAb titers were measured against ancestral (D) and Omicron (E) strains in the RBD-Wu/RBD-Wu/RBD-β (blue, n = 5) and HexaPro/HexaPro/RBD-β (red, n = 6) groups. The numbers within the graphs show GMTs. (F) Antibody titers against ancestral and Omicron strains are shown at the time points (n = 16 and 11 on day 21 and days 120 to 160, respectively) indicated on top. The numbers indicate fold change between ancestral and Omicron titers. Horizontal bars indicate median. (G and H) Binding (G) and pseudovirus nAb (H) titers were measured against the ancestral strain in serum collected at indicated time points (n = 11; n = 9 or 10 on day −318, 21 days after the second vaccination). The purple (calculated using four data points) and green (calculated using three data points) lines show the fit using the power law model to calculate decay rates. The data in (F) to (H) contain a portion of the data from (B) to (E). The statistical differences between time points were determined using Wilcoxon matched-pairs signed rank test. Horizontal dashed lines throughout indicate the lower limit of quantitation.
Vaccination with RBD-Wu + AS03 (RBD-Wu/RBD-Wu/RBD-Wu group) elicited binding immunoglobulin G (IgG) titers against the Spike protein of the ancestral strain, as well as Omicron and Beta variants, that was detectable on day 21. The titers increased greater than 10-fold after the first booster immunization [geometric mean titers (GMT) of 495,012 and 52,821 arbitrary units (AU)/ml against the ancestral and Omicron strains, respectively; Fig. 1, B and C (left), and fig. S1A] and declined to prebooster magnitude by 6 months. The final booster (third) immunization increased the titers by 2.5- and 4-fold against the ancestral (GMT of 1,270,391) and Omicron (GMT of 216,319) strains, respectively, relative to the titers at the peak of the second immunization (Fig. 1, B and C). We also observed detectable nAb response against the ancestral strain after one immunization, which increased 23-fold (GMT of 7620) after the second immunization, and the titer was maintained substantially (GMT of 1299) until the booster dose 6 months later (Fig. 1B, middle). In contrast, there was low nAb response against the Omicron variant (Fig. 1C, middle). The final booster immunization at 6 months increased the titers against the ancestral strain (GMT of 67,087) and against the Omicron (GMT of 7077) and Beta variants [GMT of 38,888; Fig. 1, B and C (middle), and fig. S1B (left)]. Consistent with the pseudovirus nAb response, vaccination also induced live virus nAb titers against the ancestral (GMT of 3626) and Beta (GMT of 377) strains comparable to the responses seen in our previous study (6) but not Omicron after two immunizations [Fig. 1, B and C (right), and fig. S1B (right)]. The titers against ancestral strain decreased about fivefold by 6 months (GMT of 726) when the booster immunization was administered, which enhanced the responses against all three strains reaching peak GMTs of 8074, 1076, and 4527 against the ancestral strain, Omicron variant, and Beta variant, respectively [Fig. 1, B and C (right), and fig. S1B (right)].
In the two groups that were boosted with RBD-β at about a year after the two-dose primary immunization series (Fig. 1A), the booster vaccination elicited binding IgG responses against ancestral, Omicron, and Beta strains comparable to the titers in the RBD-Wu/ RBD-Wu/RBD-Wu group. The responses were maintained durably through the 5-month follow-up period [Fig. 1, D and E (left), and fig. S1C]. Because we observed no difference in antibody titers between the RBD-Wu and HexaPro groups before or after the booster immunization, the GMTs indicated in the text below and the figures represent the geometric mean of all 11 animals combined from both groups. Consistent with the binding IgG response, the pseudovirus (GMT of 1978) and live virus (GMT of 1331) nAb titers against the ancestral strain, after the two-dose primary vaccine series, were still detectable about 1 year later before the final booster immunization (Fig. 1D, middle and right). The RBD-β booster immunization enhanced the titers against the ancestral strain, Omicron, and Beta variants. The GMTs against the ancestral strain reached as high as 71,244 and 12,172 in the pseudovirus and live virus neutralization assays, respectively. Although the pseudovirus nAb titers decreased only by about fivefold against all three strains, the live virus nAb titers decreased by 2.5- and 7-fold against the ancestral and Omicron strains, respectively, over the 5-month follow-up period [Fig. 1, D and E (middle and right), and fig. S1D]. Boosting with RBD-Wu or RBD-β elicited comparable titers in both groups against each of the three viral strains measured (fig. S1E). In addition, we assessed binding and live virus nAb titers in serum samples collected on day 21 after the second vaccination (represented as day −318 in the figures, relative to the final booster dose) to compare the magnitude and durability after the second and third doses. Although the nAb response against Omicron was low to undetectable after the second dose (consistent with the responses in the RBD-Wu/RBD-Wu/ RBD-Wu group), the live virus nAb titers against the ancestral strain reduced less than 10-fold within the 1 year before the final booster vaccination (Fig. 1D, right). Last, we examined the reduction in nAb titers against Omicron in comparison with the ancestral strain. Consistent with several recent studies (5, 9–17), the antibody titers were about fivefold lower to the Omicron variant than was elicited against the ancestral strain (Fig. 1F). Although the magnitude of nAbs measured using the pseudovirus assay was 10-fold higher than the live virus neutralization assays, the titers correlated with each other and with binding antibodies (fig. S1F). Last, we also observed pseudovirus nAb titers against the Omicron BA.2 subvariant in all three groups, four- to fivefold lower than that of the ancestral strain (fig. S1G).
Next, we estimated the half-life of binding and nAb titers using exponential decay and power law decay models. The exponential decay model assumes a constant decay rate over time, whereas the power law decay model assumes that decay rates decrease over time and may more accurately reflect the true nature of the kinetics of antibody response in vivo. Consistent with this hypothesis, the power law model fitted the data better than the exponential decay model after the second vaccine dose for both binding (ΔBICc = 41.2) and neutralizing (ΔBICc = 15.17) antibody titers. Therefore, we used the power law model to calculate half-lives after the second and third were 99 days [95% Confidence Interval (CI), 93 to 107] and 266 days (95% CI, 159 to 569), respectively, after the second immunization (Fig. 1, G and H). After the final booster immunization, the estimated half-lives were 134 days (95% CI, 98 to 171) and 135 days (95% CI, 103 to 151) for binding and nAb titers, respectively (Fig. 1, G and H). Comparison of half-lives using data from three equivalent time points after the second (days 21, 98, and 180) and third (days 21, 90, and 120 to 160) immunizations showed that the half-life of binding titers increased twofold, whereas the half-life of neutralizing titers decreased from 188 to 135 days (indicated in green font in Fig. 1, G and H). Collectively, these data demonstrate that the RBD-I53–50 immunogen adjuvanted with AS03 stimulates robust and durable nAb response against multiple SARS-CoV-2 variants, including Omicron BA.1 and BA.2 subvariants. In addition, there was no difference in the antibody responses elicited by RBD-Wu or RBD-β booster immunizations, consistent with recent studies assessing variant booster vaccination (18, 19).
Booster immunization 6 to 12 months after a two-dose primary series promotes broadly neutralizing antibodies against sarbecoviruses
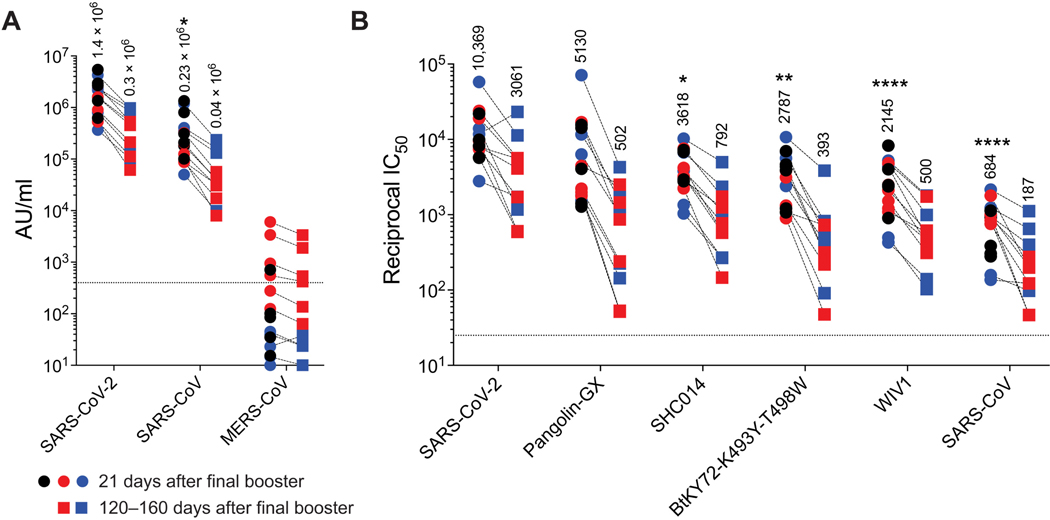
The increased half-life of nAbs in comparison to the binding antibodies after the second vaccination suggested continuing evolution of the memory B cell repertoire, which can result in broadly cross-reactive nAb response after the final booster (20). This prompted us to examine whether the final booster vaccination elicited antibodies that neutralize other sarbecoviruses and the Merbecovirus, MERS-CoV (Middle East respiratory syndrome coronavirus). Binding antibody responses were detected in all three groups against SARS-CoV but not MERS-CoV Spike protein, consistent with the knowledge that the RBD region of MERS-CoV is structurally distinct from that of SARS-CoV and SARS-CoV-2 (Fig. 2A). Furthermore, the final booster notably increased the magnitude of nAb response to a pseudoviral panel of sarbecoviruses including SARS-CoV (Fig. 2B), but there was no neutralization capacity against MERS-CoV in a live virus neutralization assay (fig. S2). The magnitude of binding and nAb responses did not differ between groups and was highest against SARS-CoV-2 followed by Pangolin-GX, SHC014, BtKY72, WIV1, and SARS-CoV. Although the nAb titers were 15-fold lower against SARS-CoV in comparison with SARS-CoV-2, the half maximal inhibitory concentration (IC50) GMT was 684. Thus, the AS03-adjuvanted RBD-I53–50 platform elicits potent nAb response not only against SARS-CoV-2 variants but also against other sarbecoviruses.
Fig. 2. A second booster immunization with RBD-Wu + AS03 or RBD-β + AS03 promotes broadly nAb responses.
(A) Binding IgG responses are shown against Spike protein from viruses indicated on the x axis. (B) Pseudovirus nAb responses against viruses indicated on X-axis are shown. Each symbol represents an animal [RBD-Wu/RBD-Wu/RBD-Wu (black; n = 5), RBD-Wu/RBD-Wu/RBD-β (blue; n = 5), and HexaPro/HexaPro/RBD-β (red; n = 6)], and paired samples are connected with a dashed line. The numbers within the graphs show GMTs. The statistical differences between viruses in were calculated using Friedman test for paired samples (*P < 0.05, **P < 0.01, and ****P < 0.0001). The horizontal dashed lines indicate the lower limit of quantitation.
AS03-adjuvanted RBD-I53–50 vaccination elicits mucosal antibody responses
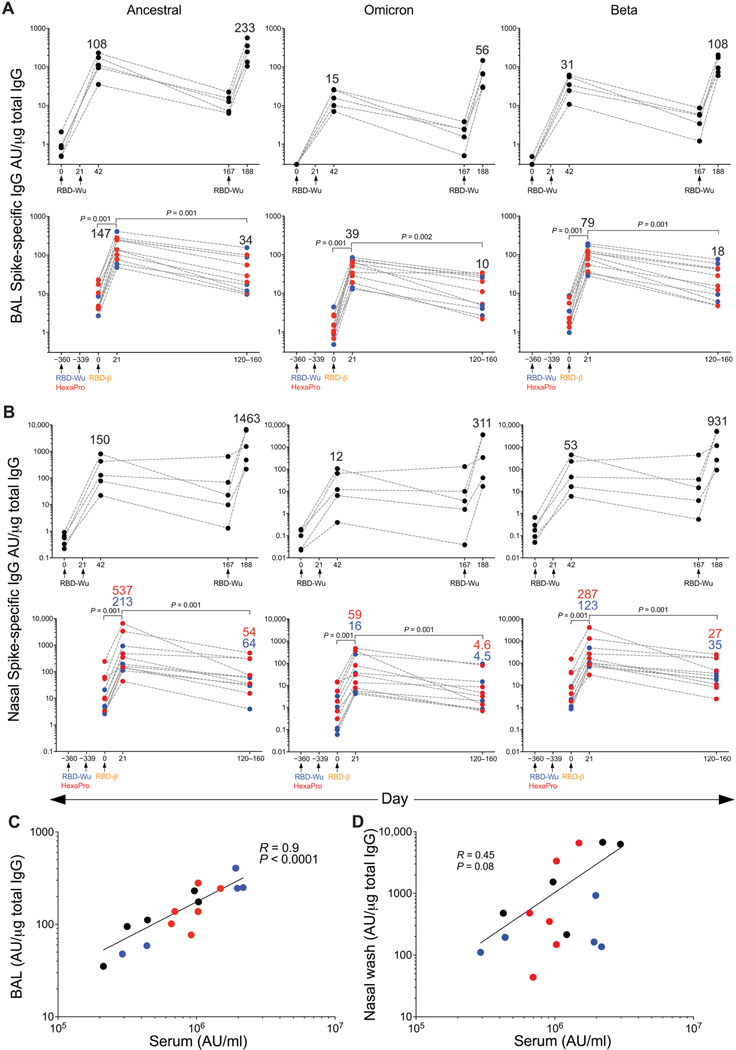
Antibody responses at the mucosa are critical to developing protective immunity against SARS-CoV-2. To assess whether this vaccine regimen elicits antibody responses at the mucosa, we measured binding antibody titers in the bronchoalveolar lavage (BAL) fluid and nasal swabs collected longitudinally from vaccinated animals using the Meso Scale platform. Consistent with the serum antibody responses, RBD-Wu + AS03 immunization (RBD-Wu/RBD-Wu/ RBD-Wu group) resulted in induction of antibodies that bound to the ancestral (GMT of 108 and 150 AU/μg total IgG in BAL fluid and nasal swab, respectively) and Omicron (GMT of 15 and 12 AU/μg in BAL fluid and nasal swab, respectively) Spike proteins after two doses (Fig. 3, A and B, left and middle). The final dose boosted the titers to 233 and 1463 AU/μg against the ancestral strain in BAL fluid and nasal swab, respectively. There was also a substantial increase in Omicron- and Beta-binding antibodies after the third dose (Fig. 3, A and B, middle and right). Similarly, the animals in the other two groups that were boosted with RBD-β elicited titers comparable to that of the RBD-Wu/RBD-Wu/RBD-Wu group and maintained durably up to 4 to 5 months after vaccination with only a fourfold reduction in titers (Fig. 3, A and B). The binding antibody titers in BAL fluid and nasal swab correlated with serum binding antibody titers (Fig. 3, C and D).
Fig. 3. Mucosal antibody responses are induced by AS03-adjuvanted RBD- nanoparticle vaccination.
(A and B) Spike protein–binding IgG titers against the ancestral strain (left panel), Omicron (middle panel) and Beta (right panel) variants were measured in the BAL fluid (A) and nasal swabs (B) of animals in the RBD-Wu/RBD-Wu/RBD-Wu (top, n = 5) group, as well as the RBD-Wu/ RBD-Wu/RBD-β (blue, n = 5) and HexaPro/HexaPro/RBD-β (red, n = 6) groups (bottom). The numbers within the graphs show GMTs. The statistical differences between time points were determined using Wilcoxon matched-pairs signed rank test combining 11 animals from the RBD-Wu/RBD-Wu/RBD-β and HexaPro/HexaPro/RBD-β groups. (C and D) Spearman’s correlation between binding IgG measured at 21 days after final booster in serum versus BAL fluid (C) and nasal swabs (D) is shown.
RBD-I53–50 immunization with AS03 elicits CD4 T cell responses
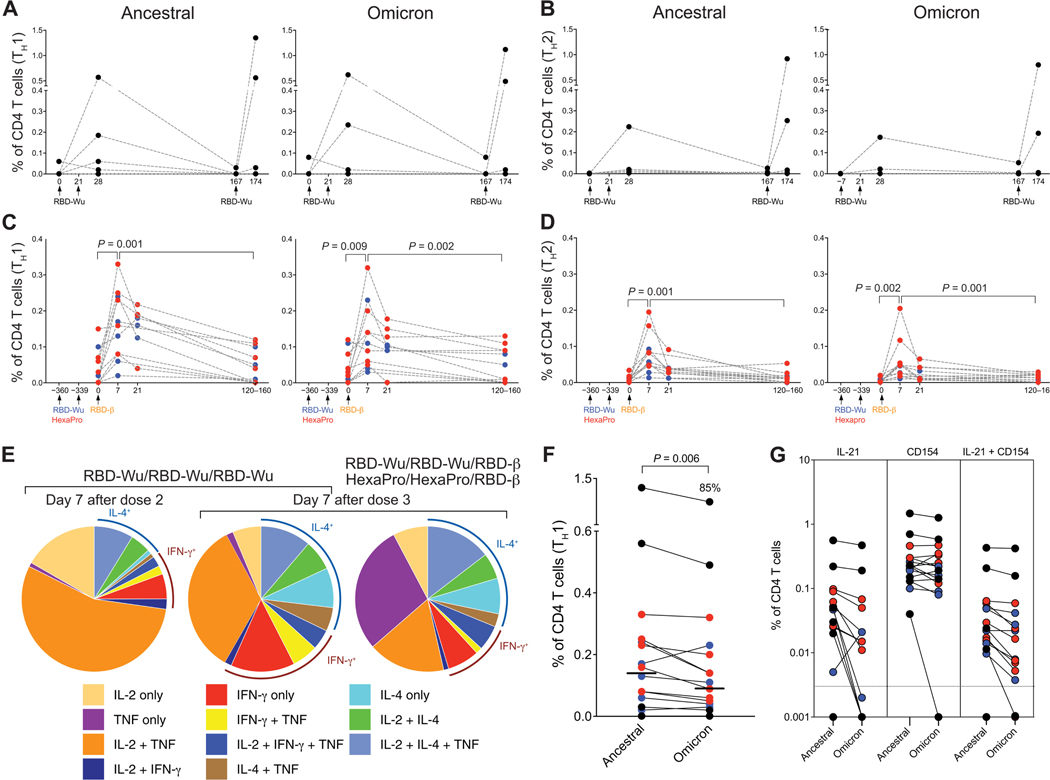
We measured T cell responses using intracellular cytokine staining (ICS) assay after a 6-hour stimulation of peripheral blood mononuclear cells (PBMCs) with overlapping peptide pools spanning the Spike proteins of the ancestral, Omicron, and Beta variants (21). Consistent with our previous study (6), RBD-Wu + AS03 vaccination (RBD-Wu/RBD-Wu/RBD-Wu group) elicited CD4 T cell responses, both T helper 1 (TH1) and TH2 types, after two doses (Fig. 4, A and B). The responses subsided to near baseline frequencies by 6 months, which were boosted by the final booster immunization (Fig. 4, A and B, and fig. S3A). Similarly, the final booster immunization with RBD-β in the other two groups also increased antigen-specific CD4 T cell responses (TH1 and TH2; Fig. 4, C and D, and fig. S3, B and C). The responses decreased considerably by 4 to 5 months after final booster immunization but were still detectable. Antigen-specific CD8 T cell responses could not be detected after two or three vaccinations (fig. S3D). The polyfunctional profile of Spike protein–specific CD4 T cells after two doses was comparable to our previous study (6), with the majority (about 70%) of the cells expressing interleukin-2 (IL-2) with or without tumor necrosis factor (TNF) and a balanced TH1/TH2 profile expressing interferon-γ (IFN-γ) or IL-4 (Fig. 4E). The profile after the third dose was similar; however, the cells expressing IL-2 or TNF without IFN-γ or IL-4 were reduced marginally (40 to 60% in contrast to 70%; Fig. 4E). The CD4 T cell response to Omicron was reduced only modestly (15%), albeit significantly (P = 0.006), in comparison to responses to the ancestral strain (Fig. 4F), consistent with studies in humans (21–23). In addition to TH1 and TH2 cytokines, vaccination also promoted follicular T helper responses, with low but detectable IL-21 and high CD154-expressing CD4 T cell frequencies in response to vaccination (Fig. 4G). In summary, the booster immunization with RBD-Wu or RBD-β elicited considerable CD4 T cell responses with only a marginal reduction in Omicron- or Beta-specific cellular immunity.
Fig. 4. AS03-adjuvanted RBD-I53–50 immunization elicits Spike protein–specific CD4 T cell responses.
(A and B) Frequency of Spike protein–specific CD4 T cells against ancestral (left) and Omicron (right) strains are shown for PBMC samples from the RBD-Wu/RBD-Wu/RBD-Wu group. (C and D) Frequency of Spike protein–specific CD4 T cells against ancestral (left) and Omicron (right) strains is shown for PBMC samples from the RBD-Wu/RBD-Wu/RBD-β (blue) and HexaPro/HexaPro/RBD-β (red) groups. CD4 T cells secreting IL-2, IFN-γ, or TNF are plotted as TH1-type responses (A and C), and IL-4–producing CD4 T cells are shown as TH2-type responses (B and D). The statistical differences between time points were determined using Wilcoxon matched-pairs signed rank test. (E) Pie charts representing the proportions of Spike protein–specific CD4 T cells expressing one, two, or three cytokines are shown. (F) Comparison of CD4 T cell frequencies between ancestral and Omicron viral strains measured on day 7 after final booster immunization is shown. The statistical difference was determined using Wilcoxon matched-pairs signed rank test. The percent value on top of Omicron represents the proportion of Omicron-specific responses relative to responses against the ancestral strain. The horizontal bars indicate median. (G) Spike protein–specific IL-21+, CD154+, and CD154+IL-21+ CD4 T cell responses were measured in samples collected on day 7 after final immunization. The horizontal dashed line indicates the lower limit of detection. In all plots, each circle represents an animal. In (F) and (G), black, blue, and red colors indicate RBD-Wu/RBD-Wu/RBD-Wu, RBD-Wu/RBD-Wu/RBD-β, and HexaPro/HexaPro/RBD-β groups, respectively.
AS03-adjuvanted RBD-I53–50 immunization elicits polyreactive memory B cell responses
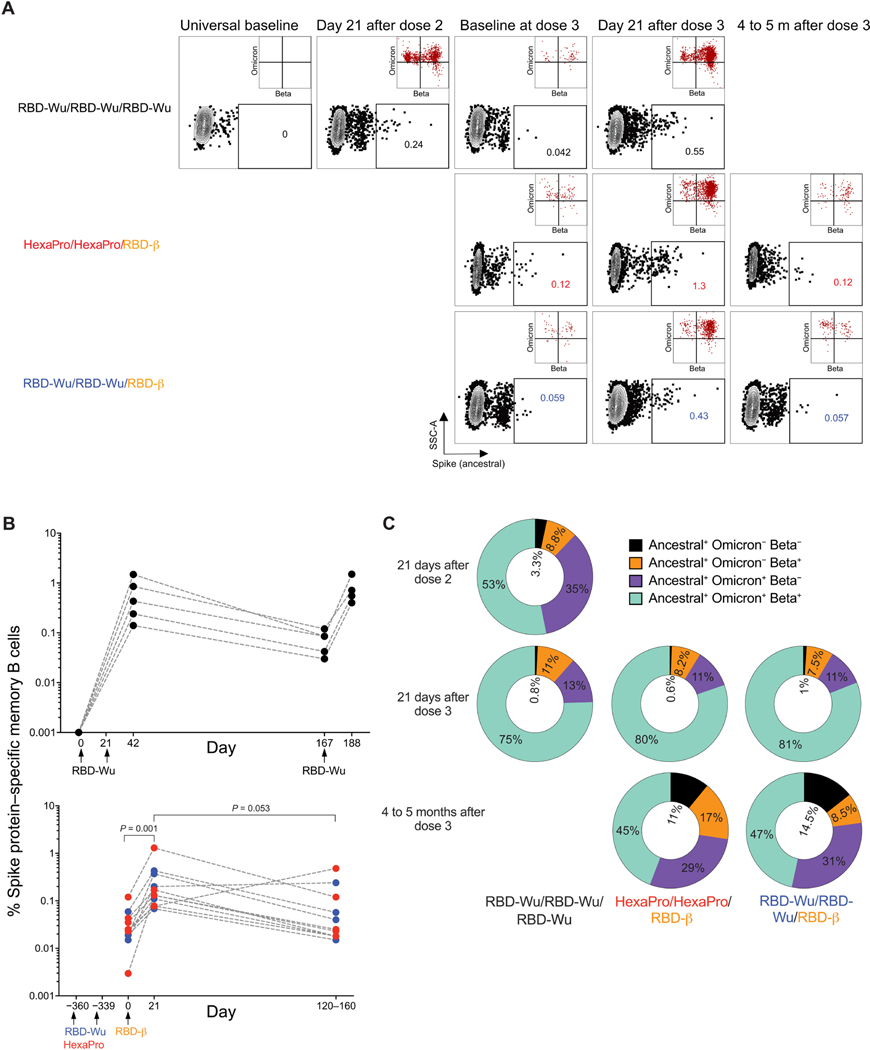
Next, we assessed Spike protein– specific memory B cells by flow cytometry analysis of PBMCs labeled with fluorescent-tagged Spike protein of the ancestral, Omicron, and Beta variants (Fig. 5A and fig. S4A). Immunization with RBD-Wu + AS03 elicited robust Spike protein–specific memory B cells (up to 1%) 21 days after the second immunization, more than 50% of which bound to all three probes (Fig. 5, B and C). The memory B cells consisted of 0.021 and 0.011% of all CD20+ B cells 6 months and 1 year after two doses, respectively (Fig. 5B). The third dose, either with RBD-Wu or RBD-β, boosted the memory B cell frequencies by 10-fold with a phenotype transition from resting (CD21+/CD27+) to activated (CD21−/CD27+) memory (Fig. 5, A and B, and fig. S4B) and an increased proportion of memory B cells binding to all three probes (Fig. 5C). The frequency of total Spike protein–specific memory B cells decreased gradually and reached a magnitude close to the prebooster time point by 4 to 5 months (Fig. 5B). Together, these data show that two doses of immunization elicit a durable memory B cell response, and the booster immunization with RBD-Wu or RBD-β further broadens the cross-reactivity of memory B cells to SARS-CoV-2.
Fig. 5. Memory B cell responses are induced by AS03-adjuvanted RBD-nanoparticle vaccination.
(A) Representative flow cytometry profile shows ancestral Spike protein–specific B cell frequencies gated as live, CD20+ IgD− IgM− Spike+ cells. The insets show proportion of Wu Spike+ cells that bind to Omicron versus Beta probes. A higher number of events is shown in the insets to improve visibility. SSC-A, Side Scatter-Area. (B) Frequency of Spike protein–specific memory B cells relative to CD20+ B cells is shown for samples from the RBD-Wu/RBD-Wu/RBD-Wu (top), RBD-Wu/RBD-Wu/RBD-β (blue, bottom), and HexaPro/HexaPro/RBD-β groups (red, bottom) groups. (C) Donut charts showing proportion of Spike protein–specific memory B cells bound to ancestral (WT), Omicron, and Beta probes, as indicated in the legend.
AS03-adjuvanted RBD-I53–50 immunization results in durable protection against Omicron BA.1 infection
Omicron is characterized by high resistance to vaccine-induced and therapeutic monoclonal nAbs (5, 24), which results in a notable decline in vaccine efficacy (25). Booster vaccination has been recommended to improve effectiveness (26); however, the efficacy against symptomatic infection remains at about 65% at 2 to 4 weeks after a booster vaccination with BNT162b2 or mRNA-1273 (25, 27). These findings prompted us to examine protection at the peak time point in our study in addition to evaluating protection at 6 months when the vaccine-induced immunity is waning. Therefore, we challenged the animals receiving RBD-Wu/RBD-Wu/RBD-Wu at 6 weeks after final booster with the Omicron BA.1 variant. Given that the immune responses in the animals belonging to the other two groups (RBD-Wu/RBD-Wu/RBD-β and HexaPro/HexaPro/RBD-β) were comparable, we challenged all 11 animals at 6 months after final booster (Fig. 1A). All the animals were challenged with 2 × 106 PFU by the intranasal (1 × 106 PFU) and intratracheal (1 × 106 PFU) routes.
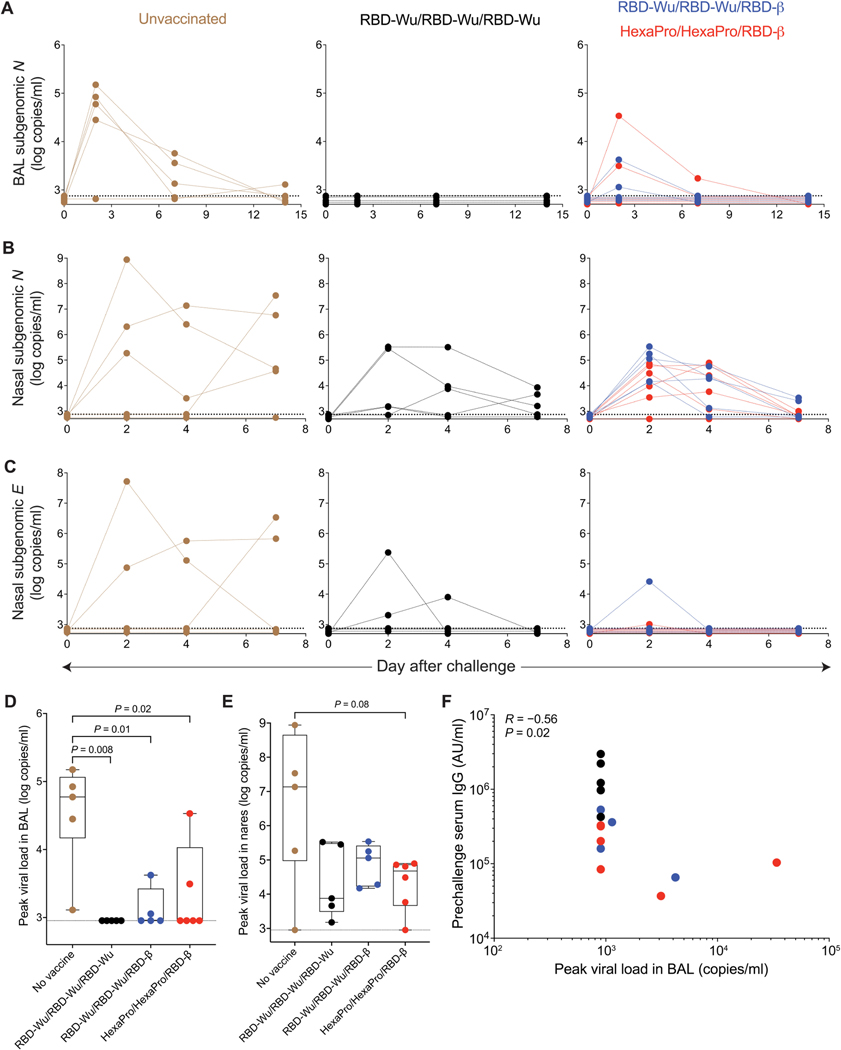
Two days after challenge, four of the five unvaccinated animals had a subgenomic viral load of 28,000 to 150,000 copies/ml (N gene) in the BAL fluid. The viral load persisted until day 7 and reduced to detection limit by day 14. In contrast, the animals challenged at the peak of immune responses (RBD-Wu/RBD-Wu/RBD-Wu group) demonstrated undetectable viral load in the lung, with none of the five animals showing a detectable viral load at any time point in the BAL fluid (Fig. 6A). Of the 11 animals challenged at 6 months after final booster vaccination, there was incomplete but significant (P < 0.05) protection in the lung compartment. Seven of the 11 animals showed an undetectable viral load, whereas the remaining 4 animals (2 animals from each vaccination group consistent with the equivalent immunogenicity in these groups) had a peak viral load of 1135, 3114, 4195, and 33845 copies/ml, in contrast to a median viral load of 59,260 copies/ml in the unvaccinated controls. Of the four animals showing a viral load in BAL fluid, there was rapid viral control, with only one animal showing detectable viral load by day 7 (Fig. 6A).
Fig. 6. AS03-adjuvanted RBD-nanoparticle vaccination confers protection against SARS-CoV-2 Omicron challenge.
(A) SARS-CoV-2 Omicron viral load in BAL fluid was measured using N gene subgenomic PCR. (B and C) SARS-CoV-2 Omicron viral load in nasal swabs was measured using N (B) and E (C) gene subgenomic PCR. (D and E) Peak N gene viral load in the BAL fluid (D) and nasal swabs (E) was measured. Data are presented as box plots in which the boxes show median and 25th to 75th percentiles and the whiskers show the range. The statistical differences between groups were determined using Mann-Whitney test. The dashed horizontal lines indicate the lower limit of quantitation. (F) Spearman’s correlation between serum IgG response measured before challenge (the time point closest to the challenge) and peak BAL fluid viral load is shown.
To assess protection in the upper airways, we assessed viral loads in nasal swabs using N and E gene subgenomic polymerase chain reaction (PCR). Four of the five control animals showed detectable virus in nares. It was notable that the animal with undetectable viral load in nares was productively infected with a peak viral load of 28,063 copies in the BAL fluid. The median peak viral load was 1.36 × 107 copies/ml of the N gene, which was maintained up to day 7 in all four animals [Fig. 6, B and C (left)]. In contrast, although all vaccinated animals showed detectable viral loads irrespective of whether they were challenged at peak or 6 months later, the viral burden was rapidly suppressed. The peak median viral loads were 7567 and 64,366 copies/ml in the animals challenged at peak and 6 months, respectively, and viral loads were reduced to detection limit by day 7 (Fig. 6, B and C). Comparing peak viral loads revealed a statistically significant (P ≤ 0.02) difference in the BAL fluid between each of the three vaccinated groups compared to unvaccinated controls, and viral loads in the nasal compartment were substantially reduced (Fig. 6, D and E).
The magnitude of serum IgG titers correlated negatively with peak viral load in the BAL fluid (Fig. 6F) and the nasal IgG response correlated with peak viral load in the nasal swab (fig. S5A). Concurrent with the infection of control animals, there was an induction of Omicron nAbs in serum (fig. S5B) and an inflammatory cytokine response in the BAL fluid (fig. S5C) but no clinical symptoms of severity (fig. S6), as has been seen in the macaque model. Collectively, these data demonstrate that the RBD-I53–50 nanoparticle vaccine adjuvanted with AS03 confers protection against Omicron even at 6 months after the final booster immunization.
DISCUSSION
Waning immunity, especially against Omicron, represents a major challenge in managing the ongoing pandemic. The efficacy against symptomatic infection of the Pfizer-BioNTech mRNA vaccine, one of the most efficacious vaccines against SARS-CoV-2, declined from greater than 90% to 67% against Omicron 2 to 4 weeks after the second booster (third inoculation) vaccination. The efficacy further declined to 45% in the following 10 weeks, demonstrating the effect of rapidly waning immunity (25). We show here that the AS03-adjuvanted RBD-I53–50 nanoparticle vaccine confers protection against Omicron infection in the lung at 6 weeks after final booster and about 65% protection even at 6 months after final booster immunization. Recent studies in NHPs vaccinated with the Moderna mRNA platform (28) or a homologous or a heterologous prime-boost regimen with the Ad26 and BNT162b2 vaccine platforms (29) evaluated protection only at the peak (1 month) of the vaccine- induced immunity. We also observed protection in the lung and rapid control of virus in the nares. Our study provides data demonstrating protection at 6 months after final booster. Whether similar protection is observed in humans remains to be seen.
A second key finding of our study is the durability of binding and nAb titers elicited by this vaccine platform. After an initial decline in the first 2 months (10,497 at peak and 2234 at about 2 months), the neutralizing titers remained high over the 10-month period until the day of the final booster, with an estimated half-life of 266 days, which is among the highest of all half-life estimates reported for COVID-19 vaccines in humans (30, 31). Furthermore, comparison of live virus nAb titers measured using the same assay by the same laboratory (13, 32) shows that the peak response after two doses of mRNA-1273 [live virus nAb titer GMT: 5560 (28)] and the RBD-I53–50 + AS03 (GMT of 6697) vaccines were comparable; however, the live virus neutralizing GMTs were 330 (28) and 1964 at the time of the final booster at 10 to 11 months after the second dose of mRNA-1273 and RBD-I53–50 + AS03, respectively, suggesting that the nAb titers were relatively more durable after vaccination with this adjuvanted nanoparticle platform. However, the assays were not performed simultaneously to allow for an ideal head-to-head comparison.
In addition to the durability of the antibody responses, our data show that the final booster immunization elicits a high magnitude of binding and nAb responses to Omicron BA.1 and BA.2 subvariants, as well as other sarbecoviruses, including SARS-CoV against which there was no response after the primary two-immunization series as reported in Fig. 1 (for Omicron) and our previous work (33). These data are consistent with there being continuing somatic hypermutation and affinity maturation of memory B cells long after the second immunization, leading to selection of B cell clones producing broad and qualitatively superior nAbs. Moreover, the frequencies of antigen-specific memory B cells with polyreactive specificities against the ancestral, Beta, or Omicron Spike protein variants were similar in animals boosted with RBD-Wu or RBD-β, suggesting that there are conserved epitopes targeted by B cells that elicit potent and broad neutralization.
The RBD-I53–50 + AS03 vaccination results in substantial memory T and B cell responses. Vaccination stimulates substantial TH1- and TH2-type CD4 T cell responses but little CD8 T cell responses. We observed no evidence of vaccine-associated enhanced respiratory disease in vaccinated animals as measured by positron emission tomography with computed tomography in our previous study (6) and the induction of inflammatory cytokines in BAL fluid in both studies. Despite the lack of CD8 T cell immunity, the protection observed in these animals was comparable to vaccine modalities that elicit CD8 T cells, such as the Ad26 platform (29).
The landscape of the COVID-19 pandemic continues to evolve rapidly. The animals challenged at the peak time point received the final booster vaccination with the ancestral vaccine strain, but the animals challenged 6 months after the final booster vaccination were boosted with the Beta variant and not the ancestral strain. Although we demonstrated that the immune responses did not differ between boosting with the ancestral or the Beta strain, we could not examine whether the durable protection was also true after boosting with the ancestral vaccine, which is the clinical vaccine. Since the completion of the study, the landscape further changed and Omicron BA.1 was replaced by BA.2, which was then replaced by BA.4 and BA.5. The Omicron subvariant BA.5 is the dominant circulating virus as we write this paper currently. Our challenge data have been with the Omicron BA.1 variant. It is conceivable that BA.2, BA.4, BA.5, and other emerging subvariants may differ in their sensitivity to vaccine-elicited immune responses, although our data currently indicate that it is not the case (16, 17). Last, we acknowledge that the magnitude of immune responses, particularly nAb responses, is generally higher in NHPs than in humans. The magnitude, durability, and protective capacity of this vaccine platform remain to be tested in humans. In summary, our study demonstrates potent, broad, and durable immunity elicited by the AS03-adjuvanted RBD-I53–50 vaccine platform, which confers protection against Omicron at least until 6 months after vaccination. These results have important implications for the vaccine, which recently met the study end points in a phase 3 clinical trials.
MATERIALS AND METHODS
Study design
The study involved four groups of male rhesus macaques (Macaca mulatta). The first group of five animals were immunized thrice with RBD-Wu + AS03, at days 0 and 21, followed by a final booster about 6 months later. The second and third groups were from our previous study (6) in which one group of the five animals received two doses of RBD-Wu and the other group comprising six animals received two doses of HexaPro (HexaPro Spike protein from the ancestral strain displayed on I53–50 nanoparticles). Both immunogens were administered with the AS03 adjuvant on days 0 and 21 using a prime-boost regimen. In the current phase of the study, all 11 animals from both groups were boosted with an I53–50 nanoparticle immunogen displaying RBD-β stabilized with the Rpk9 mutations (8) about a year after the first immunization series. The fourth group of five animals were unvaccinated controls. The number of animals in each group was determined to identify large differences between groups based on our previous experience. We did not do a power calculation to determine the sample size. The animals were randomly distributed between groups, considering body weight and age as the critical variables. The experiments were performed in an unblinded fashion through the immunogenicity phase of the study at New Iberia Research Center (NIRC). The baseline samples (indicated as day 0 in the figures for clarity) at primary and tertiary vaccinations were collected 7 days before vaccination. The animals were transported to Tulane National Primate Research Center (TNPRC) for the challenge phase when the investigators were blinded to the experimental groups. To assess immunogenicity, the RBD-Wu/RBD-Wu/RBD-Wu animals were followed longitudinally from the day of the first immunization, whereas the two groups from the previous study were followed from the day of the final booster. To assess protective efficacy, we challenged the animals with 2 × 106 PFU of SARS-CoV-2 Omicron BA.1 virus at 6 weeks or 6 months after the final booster vaccination.
Animal subjects
Twenty-one male rhesus macaques of Indian origin, aged 5 to 15 years, including 11 animals from the previous study (6) were housed and maintained as per National Institutes of Health (NIH) guidelines at the NIRC of the University of Louisiana at Lafayette in accordance with the rules and regulations of the Committee on the Care and Use of Laboratory Animal Resources. The entire study (protocol 2020–8808-015) was reviewed and approved by the University of Louisiana at Lafayette Institutional Animal Care and Use Committee (IACUC). All animals were negative for simian immunodeficiency virus, simian T cell leukemia virus, and simian retrovirus. For the challenge, the animals were transferred to the regional biosafety level 3 (BSL-3) facility at the TNPRC, where the study was reviewed and approved by the Tulane University IACUC (protocol 3930).
RBD-I53–50 nanoparticle immunogen production, adjuvant formulation, and immunization
Nanoparticle immunogen components and nanoparticles were produced as previously described in detail (34), with the exception that the RBD-Wu was in a buffer containing 50 mM tris (pH 8), 150 mM NaCl, 100 mM l-arginine, and 5% (w/v) sucrose and the RBD-β was in a buffer containing 50 mM tris (pH 8), 150 mM NaCl, 100 mM l-arginine, and 5% (v/v) glycerol. AS03 is an oil-in-water emulsion that contains 11.86 mg of α-tocopherol, 10.69 mg of squalene, and 4.86 mg of polysorbate 80 (Tween 80) in phosphate-buffered saline (PBS). For each dose, the nanoparticle immunogens were diluted to 50 μg ml−1 (SARS-CoV-2 antigen component) in 250 μl of tris-buffered saline and mixed with an equal volume of AS03. The dose of AS03 was 50% (v/v; equivalent of one human dose). All immunizations were administered by the intramuscular route in right forelimbs. The volume of each dose was 0.5 ml.
Extraction of antibodies from nasal swabs
Nasal swabs (two swabs per animal) were collected using Merocel sponges and were stored in a 5-ml polypropylene tube at −80°C. The samples were thawed on ice. One sponge head was cut and inserted into the upper chamber of a QIAshredder (QIAGEN, catalog no. 79656). The samples were centrifuged at 12,000 rpm for 5 min at 4°C. Elution buffer (0.2 ml) was added into the upper chamber, incubated on ice for 15 min, and centrifuged at 12,000 rpm for 10 min at 4°C. The same protocol was then used for the second sponge head. In total, mucosal secretions were extracted in 400 μl of PBS buffer containing 5% (v/v) IGEPAL (Sigma, catalog no. I8896–50ML) and 1× protease inhibitor (Clontech, catalog no. 635673).
Anti–Spike protein electrochemiluminescence binding enzyme-linked immunosorbent assay
Anti–Spike protein IgG titers were measured using V-PLEX COVID-19 panel 23 from Meso Scale Discovery (MSD; catalog no. K15567U) or coronavirus panel 3 (MSD, catalog no. K15399U). The assay was performed as per the manufacturer’s instructions. Briefly, the multispot 96-well plates were blocked in 0.15 ml of blocking solution with shaking at 700 rpm at room temperature. After 30 min of incubation, 50 μl of serum, BAL fluid, or nasal wash samples was diluted in antibody diluent solution, and serially diluted calibrator solution was added to each plate in the designated wells and incubated at room temperature for 2 hours with shaking. After 2 hours of incubation, the plates were washed and 50 μl of Sulfo-tag–conjugated anti-IgG was added, and the plates were incubated at room temperature for 1 hour. After incubation, the plates were washed, and 0.15 ml of MSD GOLD Read buffer was added. The plates were immediately read using the MSD instrument. The unknown concentrations were extrapolated using a standard curve drawn using the calibrators in each plate and presented as relative MSD AU/ml. For BAL fluid and nasal swab samples, the total IgG was measured using the MSD Isotyping panel (NHP/Human kit, catalog no. K15203D) using the manufacturer’s recommendations, and the response was normalized and presented per microgram of total IgG.
Pseudovirus production and neutralization assay
Vesicular stomatitis virus (VSV)–based green fluorescent protein (GFP)/nanoluciferase-expressing SARS-CoV-2 pseudoviruses were produced as described previously (12). VSV-ΔG-GFP/nanoluciferase and plasmids encoding Spike proteins of SARS-CoV-2 Wuhan (SΔ19), Beta (B.1.351), and Omicron (B.1.529) were provided by G. S. Tan (J. Craig Venter Institute). To perform neutralization assay, VeroE6-TMPRSS2-T2A-ACE2 cells [Biodefense and Emerging Infections Research Resources, National Institute of Allergy and Infectious Diseases (NIAID), NR-54970] were seeded at a density of 1 × 104 per well in half-area 96-well black opaque plates (Greiner Bio-One) and were grown overnight at 37°C in a 5% CO2 atmosphere. Serum samples were fivefold serially diluted using the infection medium [Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2% fetal bovine serum (FBS) and penicillin-streptomycin (100 U/ml)] in duplicates. Diluted serum samples were then mixed with an equal volume of Wuhan, Beta, or Omicron pseudoviruses and diluted in infection medium at an amount of 200 to 400 focus-forming units/ml per well, followed by incubation at 37°C for 1 hour. Subsequently, immune complexes were added onto the monolayers of PBS-washed VeroE6-TMPRSS2-T2A-ACE2 cells and incubated at 37°C. At 18 hours after incubation, supernatants were removed, cells were washed once with PBS, and nanoluciferase enzymatic activities were measured using the Nano-Glo Luciferase Assay System (Promega, #N1120) and a SpectraMax iD3 multimode microplate reader. Percent inhibition values were calculated by subtracting the percent infection from 100. Nonlinear curves and IC50 values were determined using GraphPad Prism.
Viruses and cells for focus reduction neutralization test
VeroE6-TMPRSS2 cells were described previously and cultured in complete DMEM (DMEM with 10% FBS + penicillin-streptomycin) in the presence of Gibco puromycin (10 mg/ml; #A11138–03). nCoV/USA_WA1/2020 (WA/1) was propagated from an infectious SARS-CoV-2 clone as previously described (35). icSARS-CoV-2 was passaged once to generate a working stock. The hCoV-19/USA/ MD- HP01542/2021 (B.1.351) was provided by A. Pekosz (Johns Hopkins University) and was propagated in Vero-TMPRSS2 cells (36). hCoV19/EHC_C19_2811C (referred to as the B.1.1.529 variant) was derived from a mid-turbinate nasal swab collected in December 2021. This SARS-CoV-2 genome is available under Global Initiative on Sharing Avian Influenza Data accession number EPI_ ISL_7171744. All viruses used in the focus reduction neutralization test (FRNT) assay were deep-sequenced and confirmed as previously described (32).
FRNT assays were performed as previously described (13, 32, 37). Briefly, samples were diluted threefold in eight serial dilutions using DMEM in duplicates with an initial dilution of 1:10 in a total volume of 60 μl. Serially diluted samples were incubated with an equal volume of WA1, B.1.351, or B.1.1.529 (100 to 200 foci per well based on the target cell) at 37°C for 45 min in a round-bottomed 96-well culture plate. The antibody-virus mixture was then added to VeroE6-TMPRSS2 cells and incubated at 37°C for 1 hour. After incubation, the antibody-virus mixture was removed, and 100 μl of prewarmed 0.85% methylcellulose (Sigma-Aldrich, #M0512–250G) overlay was added to each well. Plates were incubated at 37°C for either 18 hours (WA1 or B.1.351) or 40 hours (B.1.1.529), and the methylcellulose overlay was removed and washed six times with PBS. Cells were fixed with 2% paraformaldehyde in PBS for 30 min. After fixation, plates were washed twice with PBS and permeabilization buffer (0.1% bovine serum albumin and 0.1% saponin in PBS) was added to permeabilized cells for at least 20 min. Cells were incubated with an anti–SARS-CoV Spike protein primary antibody directly conjugated to Alexa Fluor 647 (CR3022-AF647) for 4 hours at room temperature or overnight at 4°C. Cells were washed three times in PBS, and foci were visualized on an ELISpot reader. Antibody neutralization was quantified by counting the number of foci for each sample using the Viridot program (38). The neutralization titers were calculated as follows: 1 − (ratio of the mean number of foci in the presence of serum and foci at the highest dilution of respective serum sample). Each specimen was tested in duplicate. The FRNT-50 titers were interpolated using a four-parameter nonlinear regression in GraphPad Prism 9.2.0. Samples that do not neutralize at the limit of detection at 50% are plotted at 10 and were used for geometric mean and fold change calculations.
Antibody half-life calculations
Mixed-effects models implemented in MonolixSuite 2021R1 (Lixoft) were used to estimate the corresponding half-lives of antigen-specific antibodies. The equations dAb/dt = −k × Ab (for the exponential decay model) and dAb/dt = −k/t × Ab (for the power law decay model) were fitted to the longitudinal data starting from day 21 after the second or third vaccine doses, where Ab is the antibody concentration and k is the exponential or power law decay rates, respectively. The corresponding half-lives were calculated as t1/2 = ln(2)/k for the exponential decay model or t1/2 (estimated from a given time T) = T(21/k − 1) for the power law decay model. For the power law model, the half-lives were estimated at day T = 100 days after the second or third vaccine doses. Longitudinal data from the comparable three time points were used to estimate the decay rates after the second dose (days 21, 98, and 180) and the third dose (days 21, 90, and 120 to 160). The individual-level parameters were lognormally distributed for the initial antibody concentration (at day 21) and normally distributed for the decay rate k with an assumption of no correlations between the random effects. We assumed multiplicative independent lognormal observation error. The estimation of the population parameters was performed using the stochastic approximation expectation-maximization algorithm.
Pseudovirus neutralization assay for breadth
The pseudovirus neutralizing titers against ancestral (D614G Spike protein), Omicron BA.2 Spike protein, Pangolin-Guangxi Spike protein (QIA48623.1), SARS-CoV Spike protein (YP 009825051.1), WIV1 Spike protein, SHC-014 spike protein, and BtKY72-K493Y-T498W Spike protein (39) were measured using a protocol adapted from our previous study (15, 24). Briefly, human embryonic kidney (HEK) 293T cells in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin seeded in 10-cm dishes were transfected with the plasmid encoding for the corresponding Spike glycoprotein using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. One day after transfection, cells were infected with VSV (G*ΔG-luciferase) and, after 2 hours, were washed five times with DMEM before adding medium supplemented with anti–VSV-G antibody (I1-mouse hybridoma supernatant; CRL- 2700, American Type Culture Collection). Virus pseudotypes were harvested 18 to 24 hours after inoculation, clarified by centrifugation at 2500g for 5 min, filtered through a 0.45-μm cutoff membrane, concentrated 10 times with a 30-kDa cutoff membrane, aliquoted, and stored at −80°C.
VeroE6-TMPRSS cells (40) used for ancestral and Omicron BA.2 (fig. S1G) neutralization were cultured in DMEM with 10% FBS (HyClone), 1% penicillin-streptomycin, and puromycin (8 μg/ml; to ensure retention of TMPRSS2) with 5% CO2 in a 37°C incubator (Thermo Fisher Scientific). HEK-ACE2 cells were used for the sarbecovirus panel (G614, SARS-CoV-1, WIV1, Pangolin-GX, SHC014, and BtKY72 in Fig. 2) and were cultured in DMEM with 10% FBS (HyClone) and 1% penicillin-streptomycin. Cells were trypsinized using 0.05% trypsin and plated to 40,000 cells per well in a white 96-well plate. The following day, cells were checked to be at 80% confluence. In an empty half-area 96-well plate, a 1:3 serial dilution of serum was made in DMEM and diluted pseudovirus was then added and incubated at room temperature for 30 to 60 min before addition of the serum-virus mixture to the cells at 37°C. Two hours later, 40 μl of DMEM containing 20% FBS and 2% penicillin-streptomycin was added to each well. After 17 to 20 hours, 40 μl per well of ONE-Glo EX substrate (Promega) was added to the cells and incubated in the dark for 5 to 10 min before reading on a BioTek plate reader. Measurements were done at least in duplicate using distinct batches of pseudoviruses, and one representative experiment is shown. Relative luciferase units were plotted and normalized in Prism (GraphPad). The nonlinear regression of log (inhibitor) versus normalized response was used to determine IC50 values from curve fits.
MERS-CoV neutralization assay
NHP serum samples were diluted 1:80 in VeroE6 cell growth media and further diluted in a twofold series for 12 wells. MERS-CoV-Jordan was incubated with serum for 30 min at room temperature. Mock-infected NHP serum and MERS-CoV-Jordan VeroE6 medium served as negative and positive controls, respectively. The inhibitory capacity of each serum dilution was assessed for cytopathic effect, and the serum dilution at which MERS-CoV-Jordan was not inhibited was recorded as the Neut99.
ICS assay
Antigen-specific T cell responses were measured using the ICS assay. Live frozen PBMCs were revived, counted, and resuspended at a density of 2 × 106 live cells/ml in complete RPMI 1640 (RPMI 1640 supplemented with 10% FBS and penicillin-streptomycin). The cells were rested overnight at 37°C in a CO2 incubator. The next morning, the cells were counted again and resuspended at a density of 12 × 106/ml in complete RPMI 1640, and 100 μl of cell suspension containing 1.2 × 106 cells was added to each well of a 96-well round- bottomed tissue culture plate. Each sample was treated with three or four conditions depending on cell numbers: no stimulation or a peptide pool spanning the Spike protein of the ancestral Wu strain, Omicron BA.1 variant, or Beta variant (where cell numbers permitted) in the presence of anti-CD28 (1 μg ml−1; clone CD28.2, BD Biosciences) and anti-CD49d (clone 9F10, BD Biosciences), as well as anti-CXCR3 and anti-CXCR5. The details of peptide synthesis and purity are described previously (21). Briefly, the peptide pools were 15-nucleotide oligomer peptides with 10-nucleotide oligomer overlaps spanning the entire Spike protein sequence of each variant. The amino acids in the variant peptide pools that vary from the ancestral Spike protein sequence are provided in table S3 of (21). Each peptide was dissolved at a concentration of 20 mg/ml in dimethyl sulfoxide (DMSO), and individual peptides were pooled to prepare each variant-specific peptide pool after sequential lyophilization, as previously reported (21). Each peptide pool contained 253 peptides and was resuspended in DMSO at a concentration of 1 mg/ml. PBMCs were stimulated at a final concentration of 1 μg/ml of each peptide in the final reaction with an equimolar amount of DMSO [0.5% (v/v) in 0.2-ml total reaction volume] as negative control. The samples were incubated at 37°C in CO2 incubators for 2 hours before addition of brefeldin A (10 μg ml−1). The cells were incubated for an additional 4 hours. The cells were washed with PBS and stained with Zombie ultraviolet (UV) fixable viability dye (BioLegend). The cells were washed with PBS containing 5% FBS before the addition of surface antibody cocktail (table S1). The cells were stained for 20 min at 4°C in 100-μl volume. Subsequently, the cells were washed, fixed, and permeabilized with Cytofix/Cytoperm buffer (BD Biosciences, #555028) for 20 min. The permeabilized cells were stained with ICS antibodies for 20 min at room temperature in 1× Perm/Wash buffer (BD Biosciences, #555028). The details of antibodies used in the assay are provided in table S1. Cells were then washed twice with Perm/Wash buffer and once with staining buffer before acquisition using the BD FACSymphony flow cytometer and the associated BD FACSDiva software. All flow cytometry data were analyzed using FlowJo software v10 (TreeStar Inc.).
Spike protein–specific memory B cell staining
Cryopreserved PBMCs were thawed and washed twice with 10 ml of fluorescence-activated cell sorting (FACS) buffer (1× PBS containing 2% FBS and 1 mM EDTA) and resuspended in 100 μl of PBS containing Zombie UV LIVE/DEAD dye at 1:200 dilution (BioLegend, #423108) and incubated at room temperature for 15 min. After washing, cells were incubated with an antibody cocktail for 1 hour protected from light on ice. The following antibodies were used: IgD phycoerythrin (PE; Southern Biotech, #2030–09), IgM peridinin chlorophyll protein–Cy5.5 (BioLegend, #314512), CD20 allophycocyanin-H7 (BD Biosciences, #560734), CD27 PE-Cy7 (BioLegend, #302838), CD14 brilliant violet (BV) 650 (BioLegend, #301836), CD16 BV650 (BioLegend, #302042), IgG brilliant UV 395 (BD Biosciences, #564229), CD3 BV650 (BD Biosciences, #563916), CD21 PE-CF594 (BD Biosciences, #563474), Alexa Fluor 488–labeled Beta Spike protein (antibodies-online, #ABIN6963740), Alexa Fluor 647– labeled Omicron Spike protein (Sino Biological, #40589-V08H26), and BV421-l abeled Wuhan Spike protein (Sino Biological, #40589-V27B-B). All antibodies were used as per the manufacturer’s instruction, and the final concentration of each probe was 0.1 μg/ml. Cells were washed twice in FACS buffer and immediately acquired on a BD FACSAria III. FlowJo software v10 (TreeStar Inc.) was used for data analysis.
Viral challenge
The animals were challenged in three cohorts. Cohorts 1, 2, and 3 contained 5, 10, and 6 animals, respectively. Each cohort contained unvaccinated and vaccinated animals. All the animals were housed at the NIRC for the vaccination phase and moved to TNPRC for challenge at least 3 weeks before challenge. The animals were housed in the BSL-3 facility for 7 to 10 days before challenge for acclimatization. The animals were inoculated by the intratracheal and intranasal routes with a total of 2 × 106 PFU of SARS-CoV-2 Omicron. The inoculum was divided into two equal parts, and 1 × 106 PFU was inoculated intratracheally and 0.5 × 106 PFUs was inoculated into each nostril in 0.5-ml volume.
The details of the Omicron challenge stock and the sequencing confirmation have been described previously (13, 28). We used the same challenge stock. Briefly, the Omicron BA.1 variant virus (B.1.1.529) was derived from a mid-turbinate nasal swab collected in December 2021. The virus was plaque-purified using VeroE6-TMPRSS cells from the nasal swab and propagated once to expand, and a working stock was generated. The virus was deep-sequenced and confirmed.
Sampling of nares
The monkeys were anesthetized using Telazol and placed in dorsal recumbency or a chair designed to maintain an upright posture. Sterile swabs were gently inserted into the nares. Once inserted, the sponge or swab was rotated several times within the cavity or region and immediately withdrawn.
BAL fluid collection and processing after challenge
The animals were anesthetized using Telazol and placed in a chair designed specifically for the proper positioning for BAL procedures. A local anesthetic (2% lidocaine) was applied to the larynx at the discretion of the veterinarian. A laryngoscope was used to visualize the epiglottis and larynx. A feeding tube was carefully introduced into the trachea after which the stylet was removed. The tube was advanced further into the trachea until slight resistance was encountered. The tube was slightly retracted, and the syringe was attached. Aliquots of warmed normal saline were instilled into the bronchus. The saline was aspirated between each lavage before a new aliquot was instilled. When the procedure was complete, the monkey was placed in right lateral recumbency. The monkey was carefully monitored, with observation of the heart rate, respiratory rate and effort, and mucous membrane color. An oxygen facemask may be used after the procedure at the discretion of the veterinarian. The monkey was returned to its cage, positioned on the cage floor in right lateral recumbency, and monitored closely until recovery was complete. The BAL fluid samples were filtered twice using 100-μl strainers and collected in 50-ml centrifuge tubes. The samples were centrifuged at 300g for 10 min at 4°C. The supernatant was transferred into new tubes, aliquoted, and stored at −80°C until downstream processing.
Viral load
Quantitative reverse transcription PCR (qRT-PCR) for the subgenomic RNA encoding the N protein was performed using the primers, probes, and conditions described recently (28). The E gene subgenomic PCR was performed as described previously (6, 41). Primers and probes for the N subgenomic qRT-PCR were as follows: forward, 5′-CGATCTCTTGTAGATCTGTTCTC-3′; reverse, 5′-GGTGAAC-CAAGACGCAGTAT-3′; and probe, 5′-FAM-CGATCAAAAC-AACGTCGGCCCC-BHQ1–3′. Both PCRs were run in a 20-μl volume containing 5 μl of sample, 900 nM primers, 250 nM probe with TaqPath 1-Step RT-qPCR Master Mix, CG (Thermo Fisher Scientific, #A15300). The PCR conditions were 2 min at 25°C for uracil N-glycosylase incubation, 15 min at 50°C for RT, and 2 min at 95°C (Taq activation), followed by 40 cycles of 95°C for 3 s (denaturation) and 60°C for 30 s (annealing and elongation).
Plaque reduction microneutralization test after challenge
Plaque reduction microneutralization test assays were conducted essentially as described (42), with modifications. Briefly, serially diluted serum (starting with 1:10 and followed by threefold dilutions) were mixed 1:1 with a 10,000 tissue culture infectious dose (TCID50)/ ml solution of SARS-CoV-2 B.1.1.529 (Omicron), and 100 μl of the mixture was overlaid on Vero-TMPRSS2 cells and incubated for 48 hours in DMEM with 2% FBS. Cells were fixed and labeled using a mouse anti–SARS-CoV nucleoprotein monoclonal antibody (clone MM08, Sino Biological) followed by a horseradish peroxidase– conjugated goat anti-mouse secondary antibody. Infection was quantified by measuring the absorbance at 490 nm. Neutralization titer was calculated as the reciprocal of the serum dilution that caused a 50% reduction of infection.
Meso Scale assessment of cytokine response in BAL fluid
V-PLEX MSD Multi-Spot Assay System kits were used to measure concentrations of various protein targets: chemokine panel 1 [eotaxin, macrophage inflammatory protein 1β (MIP-1β), eotaxin-3, thymus and activation-regulated chemokine, IFN-γ–induced protein-10, MIP-1α, IL-8, monocyte chemoattractant protein–1 (MCP-1), macrophage-derived chemokine, and MCP-4], cytokine panel 1 (granulocyte-macrophage colony-stimulating actor, IL-1α, IL-5, IL-7, IL-12/IL-23p40, IL-15, IL-16, IL-17A, TNF-β, and vascular endothelial growth factor–A), and proinflammatory panel 1 (IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, and TNF) (Meso Scale Diagnostics). Assays were performed on frozen BAL supernatants following the manufacturer’s instructions with an extended incubation of samples overnight at 4°C to improve sensitivity. Kits were brought to room temperature, and samples were thawed in a room temperature water bath. Plates were washed three times, followed by the addition of prepared samples and calibrator standards. Plates were then sealed and incubated at room temperature on a shaker for 2 hours. Plates were immediately transferred to 4°C overnight. The next day, detection antibody cocktails were added after washing the plates three times. The plates were then sealed and incubated at room temperature on a shaker for 2 hours. Last, the plates were washed three times, and MSD read buffer T was added and immediately read on an MESO QuickPlex SQ 120MM instrument (Meso Scale Diagnostics). The concentration of each analyte was determined on the basis of the standard curve plotted between the known concentrations of calibrators and their respective signals.
Statistical analysis
Individual-level data for all the figures are presented in data file S1. The difference between any two groups at a time point was measured using a two-tailed nonparametric Mann-Whitney unpaired rank-sum test. The difference between time points within a group was measured using a Wilcoxon matched-pairs signed rank test. All correlations were Spearman’s correlations based on ranks. All statistical analyses were performed using GraphPad Prism v.9.0.0 or R version 3.6.1. All the figures were made in GraphPad Prism or R and organized in Adobe Illustrator.
Supplementary Material
Acknowledgments:
We thank the staff at the NIRC, UL Lafayette, and the TNPRC for conducting the animal studies; R. Seder and D. Douek for the N gene probe; L. Stuart at the Bill and Melinda Gates Foundation for inputs and insights throughout the study; and all the members of GlaxoSmithKline (GSK) for critical reading of the manuscript. The schematic was made using the BioRender.
Funding:
This study was supported by the Bill and Melinda Gates Foundation INV- 018675 (to B.P.); NIH grants U19AI057266 [to B.P. and principal investigator R. Ahmed (Emory University)]; U19 AI111825 (to T.T.W.), U54 CA260517 (to T.T.W.); and P51OD011104 (to J.R.); NIAID, National Institutes of Health, Department of Health, and Human Services contract nos. 75N93021C00016 and 75N93019C00065 (to A.S.).; NIAID grants DP1AI158186 and HHSN272201700059C (to D.V.); Open Philanthropy (to B.P.); the Soffer endowment (to B.P.); OPP1156262 (to D.V. and N.P.K.); a Pew Biomedical Scholars Award (to D.V.); an Investigators in the Pathogenesis of Infectious Disease awards from the Burroughs Wellcome Fund (to D.V.); fast grants (to D.V.); a gift from the Audacious Project (to N.P.K.); a gift from J. Green and M. Halperin (to N.P.K.); a gift from the Hanauer family (to N.P.K.); the Defense Threat Reduction Agency HDTRA1–18-1–0001 (to N.P.K.); and fast grants (to T.T.W.). D.V. is an investigator of the Howard Hughes Medical Institute.
Competing interests:
B.P. serves on the External Immunology Board of GSK and on the Scientific Advisory Board of Sanofi, Medicago, CircBio, Boehringer-Ingelheim, Icosavax, and EdJen. D.T.O., R.V.D.M., and R. Rappuoli are employees of GSK group of companies. H.K. is an employee of Bill and Melinda Gates Foundation. A.S. is a consultant for Gritstone bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, Cell Carta, Avalia, Moderna, Fortress, and Repertoire. La Jolla Institute for Immunology has filed for patent protection for various aspects of T cell epitope and vaccine design work. M.S.S. serves in a consulting role for Moderna and Ocugen. N.P.K. is a cofounder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax Inc. N.K.’s laboratory has received unrelated sponsored research agreements from Pfizer and GSK. D.V.’s laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. A.C.W., B.F., R. Ravichandran., D.E., L.C, D.V., and N.P.K. are inventors on patent applications 14/930,792, PCT/US2019/020029, 63/132,863, and PCT/US2021/017799 held/submitted by the University of Washington that covers nanoparticle scaffolds and nanoparticle immunogens related to the materials reported here. B.F. is an employee and shareholder of Icosavax Inc. M.F. received funding from the NIAID Biomedical Advanced Research and Development Authority (BARDA), Defense Advanced Research Projects Agency (DARPA), Gates Foundation, Aikido Pharma, Emergent, AstraZeneca, Novavax, Regeneron, and the CDC, outside the submitted work. M. F. received royalties/licenses from Aikido Pharma for antiviral drug patent licensing, consulting fees from Aikido Pharma, Observatory Group, for consulting for COVID-19 and participation on scientific advisory board for Aikido Pharma, outside the submitted work.
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. AS03, RBD-NP, and Spike protein HexaPro are respectively available from GSK; Institute for Protein Design, University of Washington; and the University of Texas Austin under a material transfer agreement with the university or institution. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
REFERENCES AND NOTES
- 1.Feikin DR, Higdon MM, Abu-Raddad LJ, Andrews N, Araos R, Goldberg Y, Groome MJ, Huppert A, O’Brien KL, Smith PG, Wilder-Smith A, Zeger S, Deloria Knoll M, Patel MK, Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 399, 924–944 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg Y, Mandel M, Bar-On YM, Bodenheimer O, Freedman L, Haas EJ, Milo R, Alroy-Preis S, Ash N, Huppert A, Waning immunity after the BNT162b2 vaccine in israel. N. Engl. J. Med. 385, e85 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer A-S, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck H-M, Behrens GMN, Pöhlmann S, The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Clemens SAC, Naveca FG, Nascimento V, Nascimento F, da Costa CF, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Silva KD, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, Townsend AR, Huang K-YA, Tan TK, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Constantinides B, Webster H, Crook D, Pollard AJ, Lambe T; OPTIC Consortium; ISARIC4C Consortium, Paterson NG, Williams MA, Hall DR, Fry EE, Mongkolsapaya J, Ren J, Schreiber G, Stuart DI, Screaton GR, SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185, 467–484.e15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, de Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini A Ceschi, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, Telenti A, Virgin HW, Lanzavecchia A, Snell G, Veesler D, Corti D, Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, Aye P, Navarro MJ, Lai L, Edara VV, Röltgen K, Rogers K, Shirreff L, Ferrell DE, Wrenn S, Pettie D, Kraft JC, Miranda MC, Kepl E, Sydeman C, Brunette N, Murphy M, Fiala B, Carter L, White AG, Trisal M, Hsieh CL, Russell-Lodrigue K, Monjure C, Dufour J, Spencer S, Doyle-Meyers L, Bohm RP, Maness NJ, Roy C, Plante JA, Plante KS, Zhu A, Gorman MJ, Shin S, Shen X, Fontenot J, Gupta S, O’Hagan DT, van der Most R, Rappuoli R, Coffman RL, Novack D, McLellan JS, Subramaniam S, Montefiori D, Boyd SD, Flynn JAL, Alter G, Villinger F, Kleanthous H, Rappaport J, Suthar MS, King NP, Veesler D, Pulendran B, Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Song JY, Choi WS, Heo JY, Lee JS, Jung DS, Kim S-W, Park K-H, Eom JS, Jeong SJ, Lee J, Kwon KT, Choi HJ, Sohn JW, Kim YK, Noh JY, Kim WJ, Roman F, Ceregido MA, Solmi F, Philippot A, Carter L, Veesler D, King N, Kim H, Ryu JH, Lee SJ, Park YW, Park HK, Cheong HJ, Safety and immunogenicity of a SARS-CoV-2 recombinant protein nanoparticle vaccine (GBP510) adjuvanted with AS03: A phase 1/2, randomized, placebo-controlled, observer-blinded trial. medRxiv 10.1101/2022.03.30.22273143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis D, Brunette N, Crawford KHD, Walls AC, Pham MN, Chen C, Herpoldt KL, Fiala B, Murphy M, Pettie D, Kraft JC, Malone KD, Navarro MJ, Ogohara C, Kepl E, Ravichandran R, Sydeman C, Ahlrichs M, Johnson M, Blackstone A, Carter L, Starr TN, Greaney AJ, Lee KK, Veesler D, Bloom JD, King NP, Stabilization of the SARS-CoV-2 spike receptor-binding domain using deep mutational scanning and structure-based design. Front. Immunol. 12, 710263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt F, Muecksch F, Weisblum Y, da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, Hatziioannou T, Bieniasz PD, Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N. Engl. J. Med. 386, 599–601 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng SMS, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, Santos MDL, Leon P, Breban MI, Billig K, Yildirim I, Pearson C, Downing R, Gagnon E, Muyombwe A, Razeq J, Campbell M, Ko AI, Omer SB, Grubaugh ND, Vermund SH, Iwasaki A, Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 28, 481–485 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Beltran WF, Denis KJS, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB, mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 185, 457–466.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sievers BL, Chakraborty S, Xue Y, Gelbart T, Gonzalez JC, Cassidy AG, Golan Y, Prahl M, Gaw SL, Arunachalam PS, Blish CA, Boyd SD, Davis MM, Jagannathan P, Nadeau KC, Pulendran B, Singh U, Scheuermann RH, Frieman MB, Vashee S, Wang TT, Tan GS, Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 14, eabn7842 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edara VV, Manning KE, Ellis M, Lai L, Moore KM, Foster SL, Floyd K, Davis-Gardner ME, Mantus G, Nyhoff LE, Bechnak S, Alaaeddine G, Naji A, Samaha H, Lee M, Bristow L, Gagne M, Roberts-Torres J, Henry AR, Godbole S, Grakoui A, Saxton M, Piantadosi A, Waggoner JJ, Douek DC, Rouphael N, Wrammert J, Suthar MS, mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 3, 100529 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pajon R, Doria-Rose NA, Shen X, Schmidt SD, O’Dell S, McDanal C, Feng W, Tong J, Eaton A, Maglinao M, Tang H, Manning KE, Edara VV, Lai L, Ellis M, Moore KM, Floyd K, Foster SL, Posavad CM, Atmar RL, Lyke KE, Zhou T, Wang L, Zhang Y, Gaudinski MR, Black WP, Gordon I, Guech M, Ledgerwood JE, Misasi JN, Widge A, Sullivan NJ, Roberts PC, Beigel JH, Korber B, Baden LR, el Sahly H, Chalkias S, Zhou H, Feng J, Girard B, Das R, Aunins A, Edwards DK, Suthar MS, Mascola JR, Montefiori DC, SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. N. Engl. J. Med. 386, 1088–1091 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, Stewart C, Cameroni E, Callum MM, Goecker EA, Degli-Angeli EJ, Logue J, Greninger A, Corti D, Chu HY, Veesler D, SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell 185, 872–880.e3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen JE, Sprouse KR, Walls AC, Mazzitelli IG, Logue JK, Franko NM, Ahmed K, Shariq A, Cameroni E, Gori A, Bandera A, Posavad CM, Dan JM, Zhang Z, Weiskopf D, Sette A, Crotty S, Iqbal NT, Corti D, Geffner J, Grifantini R, Chu HY, Veesler D, Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines. bioRxiv 2022.03.15.484542 (2022). [Google Scholar]
- 17.Park Y, Pinto D, Walls AC, Liu Z, De Marco A, Benigni F, Zatta F, Silacci-Fregni C, Bassi J, Sprouse KR, Addetia A, Bowen JE, Stewart C, Giurdanella M, Saliba C, Guarino B, Schmid MA, Franko N, Logue J, Dang HV, Hauser K, di Iulio J, Rivera W, Schnell G, Lempp FA, Janer J, Abdelnabi R, Maes P, Ferrari P, Ceschi A, Giannini O, de Melo GD, Kergoat L, Bourhy H, Neyts J, Soriaga L, Purcell LA, Snell G, Whelan SPJ, Lanzavecchia A, Virgin HW, Piccoli L, Chu H, Pizzuto MS, Corti D, Veesler D, Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. bioRxiv 2022.05.08.491108 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Choi A, Koch M, Wu K, Chu L, Ma LZ, Hill A, Nunna N, Huang W, Oestreicher J, Colpitts T, Bennett H, Legault H, Paila Y, Nestorova B, Ding B, Montefiori D, Pajon R, Miller JM, Leav B, Carfi A, McPhee R, Edwards DK, Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: An interim analysis. Nat. Med. 27, 2025–2031 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbett KS, Gagne M, Wagner DA, O’ Connell S, Narpala SR, Flebbe DR, Andrew SF, Davis RL, Flynn B, Johnston TS, Stringham CD, Lai L, Valentin D, van Ry A, Flinchbaugh Z, Werner AP, Moliva JI, Sriparna M, O’Dell S, Schmidt SD, Tucker C, Choi A, Koch M, Bock KW, Minai M, Nagata BM, Alvarado GS, Henry AR, Laboune F, Schramm CA, Zhang Y, Yang ES, Wang L, Choe M, Boyoglu-Barnum S, Wei S, Lamb E, Nurmukhambetova ST, Provost SJ, Donaldson MM, Marquez J, Todd JPM, Cook A, Dodson A, Pekosz A, Boritz E, Ploquin A, Doria-Rose N, Pessaint L, Andersen H, Foulds KE, Misasi J, Wu K, Carfi A, Nason MC, Mascola J, Moore IN, Edwards DK, Lewis MG, Suthar MS, Roederer M, McDermott A, Douek DC, Sullivan NJ, Graham BS, Seder RA, Protection against SARS-CoV-2 Beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science 374, 1343–1353 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Kotaki R, Adachi Y, Moriyama S, Onodera T, Fukushi S, Nagakura T, Tonouchi K, Terahara K, Sun L, Takano T, Nishiyama A, Shinkai M, Oba K, Nakamura-Uchiyama F, Shimizu H, Suzuki T, Matsumura T, Isogawa M, Takahashi Y, SARS-CoV-2 Omicron-neutralizing memory B-cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci. Immunol. 7, eabn8590 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, Bloom NI, Goodwin B, Phillips E, Mallal S, Sidney J, Filaci G, Weiskopf D, da Silva Antunes R, Crotty S, Grifoni A, Sette A, SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185, 847–859.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, Khan K, Cele S, Bernstein M, Karim F, Madzorera SV, Moyo-Gwete T, Mennen M, Skelem S, Adriaanse M, Mutithu D, Aremu O, Stek C, du Bruyn E, van der Mescht MA, de Beer Z, de Villiers TR, Bodenstein A, van den Berg G, Mendes A, Strydom A, Venter M, Giandhari J, Naidoo Y, Pillay S, Tegally H, Grifoni A, Weiskopf D, Sette A, Wilkinson RJ, de Oliveira T, Bekker LG, Gray G, Ueckermann V, Rossouw T, Boswell MT, Bhiman JN, Moore PL, Sigal A, Ntusi NAB, Burgers WA, Riou C, T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 603, 488–492 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Cai C, Grifoni A, Müller TR, Niessl J, Olofsson A, Humbert M, Hansson L, Österborg A, Bergman P, Chen P, Olsson A, Sandberg JK, Weiskopf D, Price DA, Ljunggren HG, Karlsson AC, Sette A, Aleman S, Buggert M, Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 28, 472–476 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Walls AC, Hauser K, Joshi A, Stewart C, Dillen JR, Powell AE, Croll TI, Nix J, Virgin HW, Corti D, Snell G, Veesler D, Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375, 864–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, Gower C, Kall M, Groves N, O’Connell A-M, Simons D, Blomquist PB, Zaidi A, Nash S, Aziz NIBA, Thelwall S, Dabrera G, Myers R, Amirthalingam G, Gharbia S, Barrett JC, Elson R, Ladhani SN, Ferguson N, Zambon M, Campbell CNJ, Brown K, Hopkins S, Chand M, Ramsay M, Bernal JL, Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 386, 1532–1546 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omer SB, Malani PN, Booster vaccination to prevent COVID-19 in the Era of Omicron: An effective part of a layered public health approach. JAMA 327, 628–629 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Accorsi EK, Britton A, Fleming-Dutra KE, Smith ZR, Shang N, Derado G, Miller J, Schrag SJ, Verani JR, Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. JAMA 327, 639–651 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagne M, Moliva JI, Foulds KE, Andrew SF, Flynn BJ, Werner AP, Wagner DA, Teng IT, Lin BC, Moore C, Jean-Baptiste N, Carroll R, Foster SL, Patel M, Ellis M, Edara VV, Maldonado NV, Minai M, McCormick L, Honeycutt CC, Nagata BM, Bock KW, Dulan CNM, Cordon J, Flebbe DR, Todd JPM, McCarthy E, Pessaint L, van Ry A, Narvaez B, Valentin D, Cook A, Dodson A, Steingrebe K, Nurmukhambetova ST, Godbole S, Henry AR, Laboune F, Roberts-Torres J, Lorang CG, Amin S, Trost J, Naisan M, Basappa M, Willis J, Wang L, Shi W, Doria-Rose NA, Zhang Y, Yang ES, Leung K, O’Dell S, Schmidt SD, Olia AS, Liu C, Harris DR, Chuang GY, Stewart-Jones G, Renzi I, Lai YT, Malinowski A, Wu K, Mascola JR, Carfi A, Kwong PD, Edwards DK, Lewis MG, Andersen H, Corbett KS, Nason MC, McDermott AB, Suthar MS, Moore IN, Roederer M, Sullivan NJ, Douek DC, Seder RA, mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell 185, 1556–1571.e18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrashekar A, Yu J, McMahan K, Jacob-Dolan C, Liu J, He X, Hope D, Anioke T, Barrett J, Chung B, Hachmann NP, Lifton M, Miller J, Powers O, Sciacca M, Sellers D, Siamatu M, Surve N, VanWyk H, Wan H, Wu C, Pessaint L, Valentin D, van Ry A, Muench J, Boursiquot M, Cook A, Velasco J, Teow E, Boon ACM, Suthar MS, Jain N, Martinot AJ, Lewis MG, Andersen H, Barouch DH, Vaccine protection against the SARS-CoV-2 Omicron Variant in Macaques. Cell 185, 1549–1555.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doria-Rose N, Suthar MS, Makowski M, O’Connell S, McDermott A, Flach B, Ledgerwood JE, Mascola JR, Graham BS, Lin BC, O’Dell S, Schmidt SD, Widge AT, Edara VV, Anderson EJ, Lai L, Floyd K, Rouphael NG, Zarnitsyna V, Roberts PC, Makhene M, Buchanan W, Luke CJ, Beigel JH, Jackson LA, Neuzil KM, Bennett H, Leav B, Albert J, Kunwar P; mRNA-1273 Study Group, Antibody persistence through 6 months after the second dose of mRNA-1273 Vaccine for Covid-19. N. Engl. J. Med. 384, 2259–2261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suthar M, Arunachalam PS, Hu M, Reis N, Trisal M, Raeber O, Chinthrajah S, Davis-Gardner ME, Manning K, Mudvari P, Boritz E, Godbole S, Henry AR, Douek DC, Halfmann P, Kawaoka Y, Boyd SD, Davis MM, Zarnitsyna VI, Nadeau K, Pulendran B, Durability of immune responses to the BNT162b2 mRNA vaccine. Med 3, 25–27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edara VV, Pinsky BA, Suthar MS, Lai L, Davis-Gardner ME, Floyd K, Flowers MW, Wrammert J, Hussaini L, Ciric CR, Bechnak S, Stephens K, Graham BS, Mokhtari EB, Mudvari P, Boritz E, Creanga A, Pegu A, Derrien-Colemyn A, Henry AR, Gagne M, Douek DC, Sahoo MK, Sibai M, Solis D, Webby RJ, Jeevan T, Fabrizio TP, Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 385, 664–666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walls AC, Miranda MC, Pham MN, Schäfer A, Greaney A, Arunachalam PS, Navarro M-J, Tortorici MA, Rogers K, O’Connor MA, Shireff L, Ferrell DE, Brunette N, Kepl E, Bowen J, Zepeda SK, Starr T, Hsieh C-L, Fiala B, Wrenn S, Pettie D, Sydeman C, Johnson M, Blackstone A, Ravichandran R, Ogohara C, Carter L, Tilles SW, Rappuoli R, O’Hagan DT, Van Der Most R, Van Voorhis WC, McLellan JS, Kleanthous H, Sheahan TP, Fuller DH, Villinger F, Bloom J, Pulendran B, Baric R, King N, Veesler D, Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 184, 5432–5447.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walls AC, Fiala B, Schäfer A, Wrenn S, Pham MN, Murphy M, Tse LV, Shehata L, O’Connor MA, Chen C, Navarro MJ, Miranda MC, Pettie D, Ravichandran R, Kraft JC, Ogohara C, Palser A, Chalk S, Lee E-C, Guerriero K, Kepl E, Chow CM, Sydeman C, Hodge EA, Brown B, Fuller JT, Dinnon KH III, Gralinski LE, Leist SR, Gully KL, Lewis TB, Guttman M, Chu HY, Lee KK, Fuller DH, Baric RS, Kellam P, Carter L, Pepper M, Sheahan TP, Veesler D, King NP, Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi P-Y, An infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, Albert J, Anderson E, Bennett H, Corbett KS, Flach B, Jackson L, Leav B, Ledgerwood JE, Luke CJ, Makowski M, Nason MC, Roberts PC, Roederer M, Rebolledo PA, Rostad CA, Rouphael NG, Shi W, Wang L, Widge AT, Yang ES; The mRNA-1273 Study Group§, Beigel JH, Graham BS, Mascola JR, Suthar MS, McDermott AB, Doria-Rose NA, Arega J, Beigel JH, Buchanan W, Elsafy M, Hoang B, Lampley R, Kolhekar A, Koo H, Luke C, Makhene M, Nayak S, Pikaart-Tautges R, Roberts PC, Russell J, Sindall E, Albert J, Kunwar P, Makowski M, Anderson EJ, Bechnak A, Bower M, Camacho-Gonzalez AF, Collins M, Drobeniuc A, Edara VV, Edupuganti S, Floyd K, Gibson T, Ackerley CMG, Johnson B, Kamidani S, Kao C, Kelley C, Lai L, Macenczak H, McCullough MP, Peters E, Phadke VK, Rebolledo PA, Rostad CA, Rouphael N, Scherer E, Sherman A, Stephens K, Suthar MS, Teherani M, Traenkner J, Winston J, Yildirim I, Barr L, Benoit J, Carste B, Choe J, Dunstan M, Erolin R, ffitch J, Fields C, Jackson LA, Kiniry E, Lasicka S, Lee S, Nguyen M, Pimienta S, Suyehira J, Witte M, Bennett H, Altaras NE, Carfi A, Hurley M, Leav B, Pajon R, Sun W, Zaks T, Coler RN, Larsen SE, Neuzil KM, Lindesmith LC, Martinez DR, Munt J, Mallory M, Edwards C, Baric RS, Berkowitz NM, Boritz EA, Carlton K, Corbett KS, Costner P, Creanga A, Doria-Rose NA, Douek DC, Flach B, Gaudinski M, Gordon I, Graham BS, Holman LS, Ledgerwood JE, Leung K, Lin BC, Louder MK, Mascola JR, McDermott AB, Morabito KM, Novik L, O’Connell S, O’Dell S, Padilla M, Pegu A, Schmidt SD, Shi W, Swanson PA II, Talana CA, Wang L, Widge AT, Yang ES, Zhang Y, Chappell JD, Denison MR, Hughes T, Lu X, Pruijssers AJ, Stevens LJ, Posavad CM, Gale M Jr., Menachery V, Shi PY, Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 373, 1372–1377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanderheiden A, Edara VV, Floyd K, Kauffman RC, Mantus G, Anderson E, Rouphael N, Edupuganti S, Shi PY, Menachery VD, Wrammert J, Suthar MS , Development of a rapid focus reduction neutralization test assay for measuring SARS-CoV-2 neutralizing antibodies. Curr. Protoc. Immunol. 131, e116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzelnick LC, Coello Escoto A, McElvany BD, Chávez C, Salje H, Luo W, Rodriguez-Barraquer I, Jarman R, Durbin AP, Diehl SA, Smith DJ, Whitehead SS, Cummings DAT, Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLOS Negl. Trop. Dis. 12, e0006862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starr TN, Zepeda SK, Walls AC, Greaney AJ, Alkhovsky S, Veesler D, Bloom JD, ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature 603, 913–918 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park YJ, Bianchi S, Walls AC, Bowen JE, Zhou J, Kaiser H, Joshi A, Agostini M, Meury M, Dellota E Jr., Jaconi S, Cameroni E, Martinez-Picado J, Vergara-Alert J, Izquierdo-Useros N, Virgin HW, Lanzavecchia A, Veesler D, Purcell LA, Telenti A, Corti D, Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598, 342–347 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C, Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Amanat F, White KM, Miorin L, Strohmeier S, McMahon M, Meade P, Liu WC, Albrecht RA, Simon V, Martinez-Sobrido L, Moran T, García-Sastre A, Krammer F, An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr. Protoc. Microbiol. 58, e108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. AS03, RBD-NP, and Spike protein HexaPro are respectively available from GSK; Institute for Protein Design, University of Washington; and the University of Texas Austin under a material transfer agreement with the university or institution. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.