Abstract
In 1996, the dominant (43%) strain of vancomycin-resistant enterococci (VRE; type A) at Massachusetts General Hospital was identified at Brigham and Women’s Hospital (BWH). To characterize the epidemiology of infection with type A isolates of VRE at BWH, we collected demographic and clinical data for all patients from whom VRE were isolated from a clinical specimen through September 1996. The first clinical isolates from all BWH patients from whom VRE were isolated were typed by pulsed-field gel electrophoresis of SmaI digests of chromosomal DNA. Among patients hospitalized after the first patient at BWH infected with a type A isolate of VRE was identified, exposures were compared between patients who acquired type A isolates of VRE and those who acquired other types of VRE. Isolates from 99 patients identified to have acquired VRE were most commonly from blood (n = 27), urine (n = 19), or wounds (n = 19). Three months after the index patient arrived at BWH and at a time when ≥12 types of strains of VRE were present, type A isolates of VRE became dominant; 39 of 75 (52%) of the study cohort had acquired type A isolates of VRE. We found no association between the acquisition of type A isolates of VRE and transfer from another institution or temporal overlap by service, ward, or floor with patients known to have acquired type A isolates of VRE. By multivariate analysis, only residence in the medical intensive care unit (adjusted odds ratio [OR], 3.2; 95% confidence interval [CI], 1.4 to 107) and the receipt of two or more antibiotics per patient-day (adjusted OR, 12.2; 95% CI, 1.2 to 9.0) were associated with the acquisition of strain A. This strain of VRE, dominant at two Boston hospitals, was associated with intensity of antibiotic exposures (i.e., two or more antibiotics per patient-day). We hypothesize that this strain may have unidentified properties providing a mechanism favoring its spread and dominance over other extant isolates, and further studies are needed to define these properties.
Over the past decade, the epidemiology of nosocomial infections caused by enterococci resistant to vancomycin (vancomycin-resistant enterococci [VRE]) has been well studied. Exposures, including the use of cephalosporins, metronidazole, or vancomycin (2, 7, 8, 10, 12, 18, 21), and proximity to other patients (1, 2, 21) infected or colonized with VRE have been determined to be associated with the development of colonization or infection with VRE. Most reports have involved institutions at which VRE had become endemic (2, 13, 18). Although DNA digestion and pulsed-field gel electrophoresis (PFGE) have identified some small clusters of clonal spread among patients at these institutions, most of these studies identified a heterogeneous population of VRE without a predominant clonal strain. These data suggest that increases in the rate of endemicity of VRE in hospitals may result from the introduction of many strains of VRE from multiple sources.
Outbreaks involving a clonal spread from a common source or among a group of patients have been reported (2, 8, 9, 11, 16, 17). Infection control interventions usually control the outbreak and eliminate or reduce the spread of the offending clone. However, there are recent reports of the clonal dissemination of VRE between hospitals in the same geographic area, but the extent of dissemination involved only small numbers of patients at the involved hospitals (3, 6, 12). The establishment of a dominant clonal strain of VRE involving a large number of patients at multiple institutions has not been previously studied. The possibility that some strains of VRE are more prone to dissemination and the establishment of dominance has far-reaching implications for the infection control, long-term-care, and public health communities.
In the latter half of 1994, Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH) integrated many aspects of health care delivery, and attempts were made to coordinate infection control activities between the two hospitals. Isolates of VRE from some BWH patients infected or colonized with isolates of VRE were discovered to have PFGE banding patterns identical to that of the dominant strain of VRE that had rapidly disseminated throughout MGH during 1995 (15). We performed an epidemiologic and laboratory investigation of all BWH patients infected or colonized with isolates of VRE and confirmed our suspicion that a strain of VRE indistinguishable from the dominant strain previously characterized at MGH had established dominance at BWH as well. The observation of clonal spread at BWH and the establishment of dominance among multiple other strains of VRE already present at the institution suggest that some strains of VRE may contain properties that enhance their spread within hospitals.
MATERIALS AND METHODS
Patient selection and definitions.
We included all patients from BWH from whom vancomycin-resistant Enterococcus faecium was isolated from a clinical specimen from December 1993 (the month that VRE were first identified at BWH) through September 1996. Because the predominant clone of VRE at MGH was E. faecium, we excluded non-E. faecium Enterococcus strains. Isolates from patients from whom VRE were isolated only from rectal-swab surveillance cultures were not included in the epidemiologic study, but additional isolates obtained in surveillance cultures were typed as part of the microbiologic analysis (see below). Isolates of VRE were classified as associated with active infection if the primary service or infectious disease consultant documented infection with VRE in the patient’s record, initiated treatment targeted at the VRE, or did both. We classified acquisition of VRE as “early acquisition” if VRE were isolated ≤48 h into admission. We used the BWH infection control committee definition of a cluster, which was the isolation of VRE from two or more patients on the same ward in any 2-week period.
Epidemiologic investigation.
For each study patient, data collection focused on the hospitalization during which VRE were first detected (i.e., current admission). Demographic and clinical data, including transfers within and to BWH, were obtained from the hospital data retrieval system. If a patient was hospitalized immediately before the current admission (i.e., discharged ≤5 days before the current admission) the data from both admissions were combined for study. Exposures to antibiotics were obtained from the pharmacy computer database and were abstracted as days of antibiotic use (i.e., antibiotic-days) before detection of the clinical isolate of VRE. The average number of antibiotics used per patient-day was determined by dividing the number of total antibiotic-days by the number of patient-days before detection of VRE. Temporal overlap was considered present if the patient was hospitalized concurrently on the same hospital service, ward, or floor as a patient known to be infected or colonized with a type A isolate of VRE.
Microbiologic analysis.
In the BWH microbiology laboratory, enterococci were identified with the automated Vitek Automicrobe System (Vitek Systems, Hazelwood, Mo.). Standard disk diffusion antimicrobial susceptibility tests were performed for all isolates. Isolates with decreased susceptibility to vancomycin by disk diffusion underwent MIC testing with antimicrobial gradient strips (E-test; AB Biodisk, Solna, Sweden).
All clinical isolates were characterized by PFGE with a CHEF-DRII apparatus (BioRad, Hercules, Calif.) after digestion of chromosomal DNA with SmaI (5). The protocol for plug preparation was modified from that of Murray et al. (14). Strains that differed by three or fewer bands by visual inspection were considered to be derived from the same strain (20). Confirmatory PFGE was repeated for these strains by digestion of chromosomal DNA with ApaI. In order to identify additional type A isolates occurring before or shortly after the index clinical isolates, further PFGE was performed for 12 isolates of VRE obtained from all rectal-swab surveillance cultures obtained 6 months before and 3 months after the first type A isolate of VRE was isolated. Rectal-swab cultures were not performed routinely and were performed only in response to clusters of cases of VRE, defined as two nosocomial clinical cases of VRE occurring on the same patient-care unit in a 2-week period.
Statistical analysis.
Clinical and microbiologic data were analyzed with EpiInfo software (version 6.01) (4). To determine risk factors for the acquisition of type A isolates of VRE compared with the risk of acquisition of isolates of VRE of other PFGE types, we limited our analysis to BWH patients infected or colonized with VRE and hospitalized after the first patient infected or colonized with a type A isolate of VRE had been hospitalized (i.e., on or after 5 April 1995). Proportions were compared by the chi-square test or Fisher’s exact test, as appropriate. Continuous variables were compared by the Wilcoxon test. We calculated the risk ratios (RRs) and 95% confidence intervals (CIs) for selected demographic and clinical characteristics. Logistic regression analysis was performed with Stata Statistical Software (release 5.0; Stata Corporation, College Station, Tex.). All exposures or characteristics with P values of <0.20 on univariate analysis were considered for inclusion in the multivariate model, and interaction terms were assessed.
RESULTS
Spectrum of VRE.
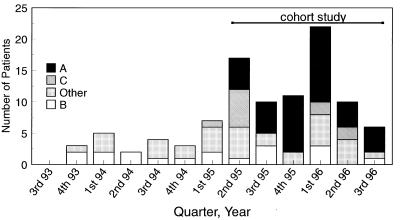
Ninety-nine patients had acquired at least one clinical isolate of E. faecium resistant to vancomycin. Of the 99 vancomycin-resistant E. faecium isolates, 27 were from blood, 19 were from urine, 23 were from drainage or other normally sterile sites, 19 were from wounds, and 11 were from other nonsterile sites (respiratory tract, n = 4; decubitus ulcers, n = 4; femoral catheter exit site, n = 2; J-tube exit site, n = 1). The frequency of isolation of VRE after the identification on 14 December 1993 of the first patient who had acquired VRE remained relatively stable until the second quarter of 1995 (Fig. 1). The rate of acquisition of VRE from clinical isolates increased from 0.2/1,000 patient-days in 1994 to 0.6/1,000 patient-days in the first three quarters of 1996. VRE were isolated from patients on 32 of 40 inpatient, adult, nonobstetric patient-care wards.
FIG. 1.
Frequency of isolation of VRE from cultures of clinical samples from patients at BWH by strain type over time (September 1993 to September 1996). Data include only the date that an isolate of VRE was first isolated from each patient.
Classification of VRE by PFGE type.
PFGE analysis of the 99 isolates of VRE revealed at least 21 distinct PFGE strain types. However, most (68 of 99) isolates had zero to three band differences from one of three unique strain types: 39 classified as type A, 18 classified as type B, and 11 classified as type C (Fig. 2). There were 18 distinct strain types among the remaining 31 VRE, none of which was common to >4 isolates. The first isolate of VRE collected at BWH, in December 1993, was PFGE type B. This strain type continued to occur throughout the study period (Fig. 1). Type C isolates of VRE appeared in March 1995 and represented the majority of the PFGE strain types over the next quarter (Fig. 1), during which most of the type C isolates of VRE (4 of 7 [57%]) were collected from patients on two surgical wards. A clinical type A isolate of VRE was first detected on 5 April 1995 in a surgical intensive care unit patient (i.e., the index patient). This day was the first day of admission to BWH for this patient, in whom VRE had been identified in a surgical wound 1 week earlier at the transferring institution, which was not MGH.
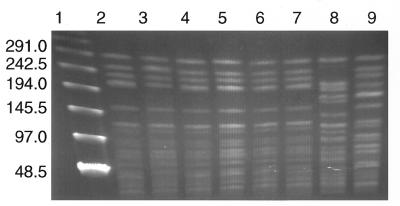
FIG. 2.
PFGE banding patterns of chromosomal DNAs from enterococcal strains from eight patients infected with E. faecium. Strains were selected from the index patient from October 1994 at MGH (lane 2), the index patient from April 1995 at BWH (lane 3), patients who acquired VRE ≥48 h into their stay at BWH (October 1995, lane 4; April 1996, lane 5), patients who acquired VRE <48 h into their stay at BWH (November 1995, lane 6; October 1995, lane 7), and patients at BWH who acquired type B strains of VRE (December 1993; lane 8) or type C strains of VRE (March 1996; lane 9) were subjected to electrophoresis with the CHEF-DRII system by using pulse intervals of 1 to 28 s at 180 V for 20 h. Lane 1, bacteriophage lambda ladder molecular size standards. Numbers to the left of the figure indicate DNA fragment sizes (in kilobases).
Over the next 4 months, type A isolates of VRE were identified in five additional patients, three of whom received care in the medical intensive care unit (MICU). These patients preceded a cluster of type A isolates of VRE isolated from five MICU patients between 13 September and 13 October 1995. After this cluster, VRE having 11 distinct PFGE patterns were identified in 52 patients at BWH, including 14 of type B (27%), 11 of type A (21%), and 7 of type C (14%). Furthermore, the PFGE patterns of 12 VRE isolated from surveillance cultures of rectal swabs obtained during the 6 months before and 3 months after the index patient was identified revealed no PFGE pattern similar to those of type A isolates of VRE. Thus, an earlier index case of infection or colonization with a type A isolate of VRE was not identified by rectal-swab surveillance cultures. Despite this heterogeneous population, type A isolates of VRE quickly became established as the dominant type of VRE, being identified in 39 (52%) of the 75 patients infected or colonized with VRE and hospitalized after the identification of the first type A isolate of VRE at BWH (Fig. 1). Type A isolates of VRE were indistinguishable (i.e., zero band differences) from the VRE previously shown to be dominant (43% of VRE in 1995) at MGH (15) (Fig. 2).
Antibiotic susceptibilities.
Seventy-five patients whose first clinical sample that grew VRE was obtained on or after 5 April 1995 (i.e., the date that a type A isolate of VRE was first isolated at BWH) constituted the study cohort. The vancomycin MIC was ≥256 μg/ml for all VRE isolated from these patients. Routine antibiotic susceptibility testing by disc diffusion of the patients’ isolates revealed that almost all isolates were susceptible to chloramphenicol but were resistant to most other antibiotics tested, with few exceptions (e.g., one non-type A isolate of VRE was resistant to chloramphenicol and three non-type A isolates of VRE were susceptible to gentamicin). However, while only one type A isolate of VRE was susceptible to tetracycline, 20 (55%) of 36 nontype A isolates of VRE were susceptible to this drug (P < 0.01). No isolate of VRE was resistant to clindamycin by disk diffusion testing.
Source of VRE.
Among the study cohort, most patients appeared to have acquired VRE at BWH. For only 17 (23%) of the 75 study patients was their first sample for culture that was positive for VRE obtained <48 h into their admission (i.e., early acquisition), suggesting possible acquisition before admission to BWH. However, patients with recent admission to BWH were significantly more likely to have early acquisition of VRE; 10 (50%) of 20 patients admitted to BWH <1 month previously had early acquisition, whereas 4 (11%) of 34 patients with previous admission to BWH >6 months previously or never admitted to BWH had early acquisition (RR, 4.3; 95% CI, 1.5 to 11), suggesting possible prior acquisition at BWH. Although the rate of early acquisition among those transferred from other health care institutions was slightly higher than those admitted from home (9 of 28 [32%] versus 8 of 47 [17%]; RR, 1.9; 95% CI, 0.8 to 4.3), the difference was not statistically significant.
Risk factors for acquisition of type A isolates of VRE.
We determined the risk of acquiring type A isolates of VRE among all patients in the study cohort hospitalized during the same time as another patient who was known to have been infected or colonized with a type A isolate of VRE (n = 75). Although there tended to be a higher risk of acquiring a type A isolate of VRE with a longer duration of temporal overlap by floor or ward, the differences did not reach statistical significance (Table 1). The proportion of patients who acquired type A isolates of VRE was similar among patients who had been associated with a cluster and patients who had not been associated with a cluster (6 of 14 [43%] versus 33 of 61 [54%]; RR, 0.8; 95% CI, 0.4 to 1.5).
TABLE 1.
Risk of acquiring type A isolates of VRE among study patients by temporal overlap with patients from whom type A isolates of VRE were previously isolated, BWH, April 1995 to September 1996
| Location (time) of overlap with patients known to be infected or colonized with type A isolate of VRE | No. of patients who acquired type A isolate of VRE/total no. of patients who acquired any isolate of VRE (%) | RR | 95% CI |
|---|---|---|---|
| Same ward (days) | |||
| 0 | 27/55 (49) | Referent | |
| 1–6 | 9/15 (60) | 1.2 | 0.8–2.0 |
| >6 | 3/5 (60) | 1.2 | 0.6–2.6 |
| Same floor (days) | |||
| 0 | 15/35 (43) | Referent | |
| 1–6 | 16/27 (59) | 1.4 | 0.8–2.3 |
| Same service (day) | |||
| 0 | 26/51 (51) | Referent | |
| 1–6 | 9/16 (56) | 1.1 | 0.7–1.8 |
| >6 | 4/8 (50) | 1 | 0.5–2.0 |
The proportion of patients who acquired type A isolates of VRE was significantly higher among patients admitted to the Burn/Trauma Service (5 of 6; 83%) or the Pulmonary Service (7 of 8; 87%) services and among those receiving treatment in the MICU (15 of 19; 79%) than among patients not admitted to these services or wards (Table 2). In contrast, no other wards or services were associated with a significantly different risk of acquiring a type A isolate of VRE (data not shown). Patients who acquired type A isolates of VRE were significantly younger than patients who acquired non-type A isolates of VRE (median ages, 54 and 66 years, respectively; P = 0.01). Gender, compromised immune status, previous infection with Clostridium difficile or methicillin-resistant Staphylococcus aureus, use of mechanical ventilation, use of total parenteral nutrition, or the presence of a gastrostomy or percutaneous enterogastric tube did not have an effect on the proportions of patients who acquired type A isolates of VRE.
TABLE 2.
Risk of acquiring a type A isolate of VRE among study patients at BWH infected or colonized with VRE, by exposure or characteristic, from April 1995 to September 1996
| Characteristic or exposure | No. of patients who acquired type A/total no. of patients infected or colonized with any isolate of VRE (%)
|
RR (95% CI) | |
|---|---|---|---|
| With characteristic or exposure | Without characteristic or exposure | ||
| Male gender | 20/35 (57) | 19/40 (48) | 1.2 (0.8–1.9) |
| Isolate considered an infection | 16/28 (60) | 23/47 (49) | 1.2 (0.9–1.8) |
| Isolate obtained <48 h into admission | 8/17 (47) | 31/58 (53) | 0.9 (0.5–1.5) |
| Admitted from another institution | 12/28 (43) | 27/47 (57) | 0.8 (0.5–1.2) |
| Infection or colonization associated with cluster of VRE | 6/14 (43) | 33/61 (54) | 0.8 (0.4–1.5) |
| Died during current admission | 12/20 (60) | 27/55 (50) | 1.2 (0.8–1.9) |
| Initially admitted to the following service: | |||
| Hematology or oncology | 7/10 (70) | 32/65 (49) | 1.4 (0.9–2.3) |
| Burn/Trauma | 5/6 (83) | 34/69 (49) | 1.7 (1.1–2.6) |
| Pulmonary | 7/8 (87) | 32/67 (47) | 1.8 (1.3–2.6) |
| Received care at any time in MICU | 12/19 (79) | 24/56 (43) | 1.8 (1.3–2.7) |
| Before isolation of VRE the patient received the following: | |||
| Vancomycin | 24/43 (56) | 15/32 (47) | 1.2 (0.8–1.9) |
| Metronidazole | 16/32 (50) | 23/43 (54) | 0.9 (0.6–1.5) |
| Fluoroquinolones | 22/39 (56) | 17/36 (49) | 1.2 (0.8–1.9) |
| Expanded-spectrum cephalosporins | 23/39 (59) | 16/36 (44) | 1.3 (0.9–2.1) |
| Broad-spectrum cephalosporins | 15/29 (52) | 24/46 (52) | 1 (0.6–1.6) |
| Aminoglycosides | 16/27 (59) | 23/48 (47) | 1.2 (0.8–1.9) |
| Clindamycin | 8/10 (80) | 31/65 (48) | 1.7 (1.1–2.51) |
| Receipt of an average of two or more antibiotics per patient-day | 28/45 (62) | 11/30 (37) | 1.7 (1.1–2.86) |
| Receipt of four or more antibiotics per patient-day | 26/40 (65) | 13/35 (37) | 1.8 (1.1–2.85) |
In contrast to the risk of acquiring a type A isolate of VRE associated with the use of most antimicrobial agents evaluated, the risk of acquiring a type A isolate of VRE was significantly higher among patients receiving clindamycin than among those not receiving this antibiotic (RR, 1.68; 95% CI, 1.12 to 2.51) (Table 2). Also, the risk of acquisition was significantly higher among patients receiving higher numbers of antibiotics (i.e., above the median for the study cohort) than among those receiving fewer antibiotics, as measured by either the number of antibiotics per patient-day (median, two antibiotics/patient-day; RR, 1.7; 95% CI, 1.01 to 2.86) or the total number of antibiotics (median, four antibiotics/patient-day; RR, 1.75; 95% CI, 1.07 to 2.85).
In a multivariate model including data for all 75 patients in the study cohort, independent predictors of the risk of acquiring a type A isolate of VRE were residence in the MICU (adjusted odds ratio, 12.2; 95% CI, 1.4 to 107) or the receipt of two or more antibiotics per patient-day (adjusted odds ratio, 3.2; 95% CI, 1.2 to 9.0). When we evaluated only those patients who acquired VRE >48 h into their hospitalization at BWH, only two or more antibiotics per patient-day remained significant (adjusted odds ratio, 3.5; 95% CI, 1.2 to 10.3), controlling for MICU residence.
DISCUSSION
Since VRE were first identified at BWH in December 1993, the rate of isolation of VRE from cultures of patients’ clinical samples has reached levels consistent with a pattern of endemicity. However, unlike in most tertiary-care centers, our molecular biology-based studies provide data suggesting that even though a heterogeneous population of VRE exists at BWH, one particular clone established itself as the dominant strain at BWH and persisted as such for more than a year. Although we document a cluster of type A isolates of VRE among MICU patients within 4 months of this strain’s first appearance at BWH, this cluster involved only five patients and does not explain the strain’s rapid dissemination throughout BWH. Furthermore, we evaluated the possibility that type A isolates of VRE had a large hidden reservoir among unidentified colonized patients at BWH around the time that a type A isolate of VRE was first identified by culturing a clinical sample; however, no surveillance rectal swabs from this period contained type A isolates of VRE.
Most studies of VRE address the risk factors for the acquisition of any strain of VRE. In contrast, our study evaluated the risk of acquiring a particular strain of VRE among all patients who acquired VRE that were first isolated from cultures of clinical samples. This is the second study describing the hospital-wide dissemination of a single clone, with the first such study describing a single clone at MGH (15). Most striking is the fact that our type A isolate of VRE has a PFGE pattern indistinguishable from that of the strain of VRE that established dominance at MGH. Unlike the previous study, our study evaluated in detail the possible contribution of type A isolates of VRE from the community or other hospitals, including MGH. Although some patients probably arrived at BWH already infected or colonized with VRE (including the index patient), we found that the likelihood of isolation of VRE ≤48 h after the beginning of the hospitalization was related to the previous time spent in BWH. The risk of such early acquisition was inversely related to the number of months since prior admission to BWH. These data suggest that most of the VRE identified at BWH were acquired at BWH.
Most (77%) isolates of VRE from the cohort study were collected from patients hospitalized >48 h and may represent cross-transmission from patient to patient. Among the cohort, type A isolates of VRE comprised about half of the isolates. We found no evidence, however, that type A isolates of VRE established dominance because of the proximity of patients to one another. Despite a detailed evaluation of all patients’ transfer activities, we were unable to show an increased risk of acquiring type A isolates of VRE among study patients when the patients were stratified by duration of time (in days) that the patients shared a ward, floor, or service with a patient known to have acquired a type A isolate of VRE. Furthermore, we found that patients who acquired type A isolates of VRE were no more likely than patients who acquired non-type A isolates of VRE to have been associated with a cluster of cases. These data suggest that the acquisition of a type A isolate of VRE is not explained by receipt of care near other patients who had acquired type A isolates of VRE or from health care providers who cared for other patients who were infected or colonized with a type A isolate of VRE. It is important, however, that without a comprehensive, prospective surveillance study with perirectal swab cultures we underestimate the numbers of colonized patients who may have served as a reservoir of type A strains of VRE (or other types of VRE) for patient-to-patient transmission.
Our data and those from another Boston hospital (15) suggest that type A isolates of VRE may have certain microbiologic properties distinct from those of other heterogeneous VRE found at BWH. The presence of such properties may explain why we were unable to identify proximity to other patients who were infected or colonized with type A isolates of VRE as a risk factor for the acquisition of this organism. More likely, the clonal dissemination of type A isolates of VRE may be related to host-bacteria interactions. Such a hypothesis is supported by our analysis of host risk factors for the acquisition of type A isolates of VRE.
By univariate analysis, we found that receiving care on the Burn/Trauma Service, the Pulmonary Service, or the MICU was associated with a higher risk of acquiring a type A isolate of VRE. Although this may suggest some clustering not detected by our previous analysis, combining this finding with the lack of evidence supporting proximity as a risk factor suggests that the patient mixture on these services or units rather than only proximity to other patients infected or colonized with type A isolates of VRE may be a risk factor. Furthermore, by univariate analysis we found that receipt of clindamycin or receipt of more antibiotics significantly increased the risk of acquiring a type A isolate of VRE. These findings are strikingly similar to those found among patients at MGH who acquired VRE (15), for whom receipt of clindamycin, duration of hospitalization, and care on one medical ward were independent predictors of acquiring a type A isolate of VRE.
We attempted to evaluate antibiotic exposure in several ways, including measurement of the total number of antibiotics received, the number of patient-days on antibiotics, and the numbers of antibiotics per patient-day. As a group, the study cohort was exposed to a large number and large amounts of antibiotics, with a median of two antibiotics per patient-day. This finding indicates that at least half of the patients in the study cohort were exposed, on average, to two antibiotics each day that they were at BWH before acquiring VRE. By multivariate analysis, we found that receipt of an average of at least two antibiotics per patient-day increased the patient’s risk of acquiring a type A isolate of VRE compared with the risk of acquiring a non-type A isolate of VRE, independent of admission to the MICU. These data, together with the lack of evidence that proximity is a risk factor for acquiring a type A isolate of VRE, suggest that characteristics of antimicrobial exposure in the host may play an important role in the selection of strains of VRE.
Several other studies have documented the spread of a clone of VRE between hospitals (3, 6, 12). An occurrence in the San Antonio, Tex., area during 1993 and 1994 involved only one to two patients at five separate hospitals. Likewise, an occurrence in three midwestern states involved only two to four patients from three hospitals (3). Other studies have documented the dissemination of a single clone within a hospital (2, 8). Such occurrences, unfortunately, not uncommon now, are usually related to an outbreak setting, and progress to involve a large heterogeneous population of VRE. Our study documents a dominant strain of VRE involving a large number of patients infected or colonized with a strain of VRE indistinguishable from the dominant strain of VRE found at another institution in the same city. In addition, a detailed epidemiologic study found a risk factor for acquisition similar to that found in the MGH study: increased antibiotic exposure, particularly to clindamycin, appears to provide a favorable host environment for type A isolates of VRE.
The separate emergence at BWH and MGH of the same predominant strain type among multiple competing strains of VRE at both institutions strongly suggests that bacterial factors play a role in dissemination throughout a hospital. The nature of such bacterial factors has not yet been defined, but elucidation of these factors may provide insight into the molecular basis of the different levels of virulence that are operating to enhance spread rather than cause disease in an individual patient. Preventing the interhospital spread of antimicrobial agent-resistant pathogens is integral to reducing the threat of increasing antimicrobial resistance among human pathogens (19); further study of strains of VRE that tend to establish dominance at institutions must be done to obtain an understanding of and prevent such spread.
ACKNOWLEDGMENTS
We thank Richard Platt of BWH, Harvard Medical School, for assistance in the investigation and review of the manuscript.
REFERENCES
- 1.Bonten M J M, Hayden M K, Nathan C, Van Voorhis J, Matushek M, Slaughter S, Rice T, Weinstein R A. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet. 1996;348:1615–1619. doi: 10.1016/S0140-6736(96)02331-8. [DOI] [PubMed] [Google Scholar]
- 2.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow J W, Kuritza A, Shlaes D M, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean A G, Dean J A, Burton J A, Dicker R C. EpiInfo, version 6: a wordprocessing, database, and statistics program for epidemiology for public health on IBM-compatible microcomputers. Atlanta, Ga: Centers for Disease Control and Prevention; 1994. [Google Scholar]
- 5.Donabedian S M, Chow J W, Boyce J M, McCabe R E, Markowitz S M, Coudron P E, Kuritza A, Pierson C L, Zervos M J. Molecular typing of ampicillin-resistant, non-β-lactamase-producing Enterococcus faecium isolates from diverse geographic areas. J Clin Microbiol. 1992;30:2757–2761. doi: 10.1128/jcm.30.11.2757-2761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunne W M, Jr, Wang W. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J Clin Microbiol. 1997;35:388–392. doi: 10.1128/jcm.35.2.388-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmond M B, Ober J F, Weinbaum D L, Pfaller M A, Hwang T, Sanford M D, Wenzel R P. Vancomycin-resistant Enterococcus faecium bacteremia: risk factors for infection. Clin Infect Dis. 1995;20:1126–1133. doi: 10.1093/clinids/20.5.1126. [DOI] [PubMed] [Google Scholar]
- 8.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh K V, Murray B E, Wolff J, Walters B. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 9.Karanfil L V, Murphy M, Josephson A, Gaynes R, Mandel L, Hill B C, Swenson J M. A cluster of vancomycin-resistant Enterococcus faecium in an intensive care unit. Infect Control Hosp Epidemiol. 1992;13:195–200. doi: 10.1086/646509. [DOI] [PubMed] [Google Scholar]
- 10.Livornese L L, Jr, Dias S, Samel C, Romanowski B, Taylor S, May P, Pitsakis P, Woods G, Kaye D, Levison M E, et al. Hospital-acquired infection with vancomycin-resistant Enterococcus faecium transmitted by electronic thermometers. Ann Intern Med. 1992;117:112–116. doi: 10.7326/0003-4819-117-2-112. [DOI] [PubMed] [Google Scholar]
- 11.Montecalvo M A, Horowitz H, Gedris C, Carbonaro C, Tenover F C, Issah A, Cook P, Wormser G P. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother. 1994;38:1363–1367. doi: 10.1128/aac.38.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno F, Grota P, Crisp C, Magnon K, Melcher G P, Jorgensen J H, Patterson J E. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin Infect Dis. 1995;21:1234–1237. doi: 10.1093/clinids/21.5.1234. [DOI] [PubMed] [Google Scholar]
- 13.Morris J G, Jr, Shay D K, Hebden J N, McCarter R J, Jr, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Establishment of endemicity in a university medical center. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 14.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pegues D A, Pegues C F, Hibberd P L, Ford D S, Hooper D C. Emergence and dissemination of a highly vancomycin-resistant vanA strain of Enterococcus faecium at a large teaching hospital. J Clin Microbiol. 1997;35:1565–1570. doi: 10.1128/jcm.35.6.1565-1570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhinehart E, Smith N E, Wennersten C, Gorss E, Freeman J, Eliopoulos G M, Moellering R C, Jr, Goldmann D A. Rapid dissemination of beta-lactamase-producing, aminoglycoside-resistant Enterococcus faecalis among patients and staff on an infant-toddler surgical ward. N Engl J Med. 1990;323:1814–1818. doi: 10.1056/NEJM199012273232606. [DOI] [PubMed] [Google Scholar]
- 17.Rubin L G, Tucci V, Cercenado E, Eliopoulos G, Isenberg H D. Vancomycin-resistant Enterococcus faecium in hospitalized children. Infect Control Hosp Epidemiol. 1992;13:700–705. doi: 10.1086/648342. [DOI] [PubMed] [Google Scholar]
- 18.Shay D K, Maloney S A, Montecalvo M, Banerjee S, Wormser G P, Arduino M J, Bland L A, Jarvis W R. Epidemiology and mortality risk of vancomycin-resistant enterococcal bloodstream infections. J Infect Dis. 1995;172:993–1000. doi: 10.1093/infdis/172.4.993. [DOI] [PubMed] [Google Scholar]
- 19.Shlaes D M, Gerding D N, John J F, Jr, Craig W A, Bornstein D L, Duncan R A, Eckman M R, Farrer W E, Greene W H, Lorian V, Levy S, McGowan J E, Jr, Paul S M, Ruskin J, Tenover F C, Watanakunakorn C. Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the Prevention of Antimicrobial Resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Infect Control Hosp Epidemiol. 1997;18:275–291. doi: 10.1086/647610. [DOI] [PubMed] [Google Scholar]
- 20.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstein J W, Roe M, Towns M, Sanders L, Thorpe J J, Corey G R, Sexton D J. Resistant enterococci: a prospective study of prevalence, incidence, and factors associated with colonization in a university hospital. Infect Control Hosp Epidemiol. 1996;17:36–41. doi: 10.1086/647186. [DOI] [PubMed] [Google Scholar]