Abstract
Introduction
Sarcopenia (low muscle mass and strength) causes dysmobility and loss of independence. Sarcopenia is often not directly coded or described in electronic health records (EHR). The objective was to improve sarcopenia detection using structured data from EHR.
Methods
Adults undergoing musculoskeletal testing (December 2017–March 2020) were classified as meeting sarcopenia thresholds for 0 (controls), ≥1 (Sarcopenia-1), or ≥2 (Sarcopenia-2) tests. Electronic health record diagnoses, medications, and laboratory testing were extracted from the Indiana Network for Patient Care. Five machine learning models were applied to EHR data for predicting sarcopenia.
Results
Of 1304 participants, 1055 were controls, 249 met Sarcopenia-1 and 76 met Sarcopenia-2. Sarcopenic participants were older, with higher fat mass, Charlson Comorbidity Index, and more chronic diseases. All models performed better for Sarcopenia-2 than Sarcopenia-1. The top performing models for Sarcopenia-1 were Logistic Regression [area under the curve (AUC) 71.59 (95% confidence interval [CI], 71.51–71.66)] and Multi-Layer Perceptron [AUC 71.48 (95%CI, 71.00–71.97)]. The top performing models for Sarcopenia-2 were Logistic Regression [AUC 91.44 (95%CI, 91.28–91.60)] and Support Vector Machine [AUC 90.81 (95%CI, 88.41–93.20)]. For the best Logistic Regression Model, important sarcopenia predictors included diabetes mellitus, digestive system complaints, signs and symptoms involving the nervous, musculoskeletal and respiratory systems, metabolic disorders, and kidney or urinary tract disorders. Opioids, corticosteroids, and antihyperlipidemic drugs were also more common among sarcopenic participants.
Conclusions
Applying machine learning models, sarcopenia can be predicted from structured data in EHR, which may be developed through future studies to facilitate large-scale early detection and intervention in clinical populations.
Keywords: Sarcopenia, machine learning, health informatics, musculoskeletal
Introduction
Sarcopenia describes a state of low muscle mass and strength, which can occur both with aging,1,2 and in younger and middle-aged adults with chronic diseases.3,4 Sarcopenia is multifactorial in etiology and has features overlapping with cachexia and frailty.1,5,6 Sarcopenia is estimated to affect 10% of older adults across the world7,8 contributing to dysmobility, disability, loss of independence, and hospitalizations with attendant healthcare costs.9–12
There is an unmet clinical need to detect sarcopenia across the life span to implement interventions and improve health outcomes. Current detection methods for sarcopenia with objective measures of muscle mass, strength, and performance, use test thresholds primarily based on studies in older adults who typically exhibit a synchronous decrease in muscle mass and strength. 13 However, sarcopenia also occurs at younger ages and in disease states where loss of muscle mass and strength may not always occur in tandem.14,15 Large electronic health records (EHR) databases provide an opportunity to detect diseases among populations of patients and facilitate clinical care, but have thus far not been used to detect sarcopenia, due to lack of formal assessments in routine clinical reports. For example, lean body mass is rarely quantified or reported when dual-energy x-ray absorptiometry (DXA) is typically performed for assessing bone density. Even when lean body mass or assessments of strength and physical performance are conducted, they may not be easily extractable from the EHR as a specific field or coded variable.
We previously used a combination of ICD codes and text terms to detect 9594 patients with sarcopenia, cachexia and/or frailty from the EHR. 16 Notably the sarcopenia ICD-10 code and the text term sarcopenia were highly specific (>90%) but had poor sensitivity for detecting features suggesting sarcopenia. 16 Additionally, in the absence of an objective gold standard, EHR detection of sarcopenia may be limited by false positive and false negative occurrences.
The purpose of this study was to develop a model to detect sarcopenia by applying machine learning methodologies to structured EHR variables (ICD codes, medications, laboratory tests, etc.) of a large group of adult participants recruited to undergo standardized research measures of muscle mass, strength, and physical function (external to their EHR data) which were used as our standard for the presence of sarcopenia. Machine learning studies have not been applied to sarcopenia studies in EHR in the past. Our study provides a novel approach to identifying risk factors predicting sarcopenia from EHR data. However, EHR-based studies often do not have a true external gold standard, while our study is made possible by utilization of our standard measurements to detect sarcopenia from our research core, which are external to our EHR.
Methods
Study cohort
The study cohort consisted of adult participants (aged 18 years and older) recruited into the Indiana Center for Musculoskeletal Health's Musculoskeletal Function, Imaging and Tissue Resource Core (FIT Core) protocol from December 2017 to March 2020 and having EHR data accessible in the Indiana Network for Patient Care (INPC). The INPC contains EHR data from more than 120 Indiana hospitals and healthcare entities, including Indiana University Health and Eskenazi Health. The FIT Core has Institutional Review Board approval from Indiana University to test any participants who provide written informed consent, broadly including healthy controls and persons with any disease state. The FIT Core informed consent includes access to and analysis of participant EHR data. FIT Core testing includes body composition, bone densitometry, and physical function tests (detailed below). The use of FIT Core data and access to and analysis of EHR data for the current study was also separately approved by the Institutional Review Board of Indiana University.
Data from EHR and the FIT core
The EHR data of the FIT Core participants were extracted for available clinical encounters in INPC between January 2016 and August 2020, with the goal of providing 2 years of EHR data around each participant's FIT Core visit. The EHR data elements included all available demographic data, diagnosis codes, laboratory test results, medication records, and inpatient and outpatient procedures. To ensure adequate EHR data for analysis, the numbers of structured variable occurrences and clinical text notes per participant were determined, then analysis was restricted to those having above the group's 25th percentile for both the total numbers of structured variable occurrences (n ≥ 122) and the number of clinical text notes available per participant (n ≥ 8). Demographic variables were obtained from FIT Core records but were not used for prediction modeling. Body mass index (BMI) was also obtained from FIT Core records.
Participants were categorized as sarcopenic or nonsarcopenic (controls) according to their testing in the FIT Core, using grip strength,17,18 repeat chair stand test, 17 and the Short Physical Performance Battery,19,20 and appendicular skeletal muscle mass adjusted for height in meters2 (ASM/m2) as measured by DXA (see Supplemental Methods). Participants were classified using definitions and thresholds for sarcopenia from the EWGSOP2 guidelines (see Supplemental Methods). 13 For the primary analyses, we categorized persons as controls if meeting none of the criteria thresholds, and as sarcopenic if meeting ≥1 criteria thresholds for sarcopenia (Group Sarcopenia-1) or if meeting ≥2 criteria thresholds for sarcopenia (Group Sarcopenia-2).
Sample size and power
The study used a convenience sample of all adult participants recruited to date in the overall FIT core protocol that had sufficient EHR data as described above, and then the sarcopenia parameters were extracted. Thus, power was calculated based on this available sample size. Given that the resulting cohort was categorized as 1055 Non-sarcopenia, 249 Sarcopenia-1 and 76 Sarcopenia-2, the resulting positive and negative case sample sizes for machine learning analysis has 95% power to gain area under the curve (AUC) of 0.7 or more (with 0.5 as the null hypothesis) using alpha of 0.05.
Variable selection and processing
Supplemental Methods and Tables describe details on processing and groupings applied to the structured data from the EHR, such as diagnosis codes (ICD-10), laboratory test results, and medications, which were included as variables for prediction modeling. Diagnoses were grouped into categories by the third level to the leaf nodes of ICD-10 code hierarchy. 21 The Charlson Comorbidity Index was calculated from ICD-10 codes as a marker of disease burden.22,23 Missing laboratory tests or other data were not imputed.
Analysis
Data were summarized as frequency and proportion or as mean and standard deviation. The Wilcoxon sum-rank test was used to assess the differences for individual variables between the sarcopenic and control groups. P values <0.05 were considered statistically significant. For machine learning model comparisons, the performance of the different models can be compared using one-way ANOVA test. We ran 10-fold cross-validation on the training data to gain the AUC values of each model and applied the one-way ANOVA test to investigate whether there is any significant difference between the model performances to select which models to use on test sets. Since no comparisons had p < 0.05 between models, the numerically best performing models (highest AUC achieved) of each machine learning method were used.
In the study cohort, there were more controls than sarcopenic participants. We split the cohort proportionally into a training (70% of the data), validation (10% of the data), and a test set (20% of the data). The validation set was used to fine-tune the parameters of the models, and the test set was used to evaluate the performance of all models. We developed and compared six different machine models—Support Vector Machine (SVM), 24 Logistic Regression (LR), 25 Random Forest (RF), 26 Gradient Boosting, 27 extreme gradient descent boosting (XGBoost),28,29 and Multilayer Perceptron neural network. 30 Each model has been tuned through hyperparameter search to optimize the performance.
We considered all coded EHR variables for inclusion in machine learning models and then investigated the important variables driving the model decisions. We used SHAP (SHapley Additive exPlanations) 31 to assign an importance value to each variable of the best performing model and explain which variables have greater impact on the output. SHAP has been used to explain machine learning prediction models for many applications, such as prevention of hypoxemia during surgery. 32 We sorted the variables by the sum of SHAP value magnitudes over all samples, drew the variable importance curve and applied the elbow method to select the top variables to interpret the results. The elbow method 33 was originally developed to identify the optimal number of clusters for an analyzed data set. The objective of the elbow method is to find an explicit elbow point in a curve so that before the point, the value rapidly decreases, and after exceeding the point, the decrease plateaus.
Results
From 2209 participants enrolled in the FIT Core as of the cutoff date, 1988 had any structured data and 1981 had any clinical text note data within the EHR, with 1304 meeting the final criteria for amount of EHR data available for analysis. After employing the variable processing steps and groupings, a total of 345 structured EHR variables were available (206 different diagnosis grouping categories, 67 laboratory test variables, 71 medication variables, and the BMI). From the included participants, 1055 met no criteria for sarcopenia (controls) and 249 met ≥1 of the sarcopenia test thresholds (Sarcopenia-1); 76 of Sarcopenia-1 met ≥2 of the sarcopenia test thresholds (Sarcopenia-2). Table 1 shows the demographic and comorbidity characteristics of the study cohort.
Table 1.
Characteristics of the study cohort.
| Total | Sarcopenia-1 | Sarcopenia-2 | Control | p value (1) | p value (2) | |
|---|---|---|---|---|---|---|
| Number of patients | 1304 | 249 | 76 | 1055 | ||
| Age | 53.32 (15.2) | 58.36 (16.1) | 65.53 (14.7) | 52.13 (14.7) | <0.01 | <0.01 |
| Sex | ||||||
| Male | 263 (20.2%) | 72 (28.9%) | 27 (35.5%) | 191 (18.1%) | <0.01 | <0.01 |
| Female | 1041 (79.8%) | 177 (71.1%) | 49 (64.5%) | 864 (81.9%) | <0.01 | <0.01 |
| Race | ||||||
| White | 1098 (84.2%) | 192 (77.1%) | 56 (73.7%) | 906 (85.9%) | <0.01 | <0.01 |
| African American | 111 (8.5%) | 36 (14.5%) | 13 (17.1%) | 75 (7.1%) | <0.01 | <0.01 |
| Asian | 85 (6.5%) | 19 (7.6%) | 6 (7.9%) | 66 (6.3%) | 0.52 | 0.75 |
| Other | 10 (0.8%) | 2 (0.8%) | 1 (1.3%) | 8 (0.8%) | 0.74 | 0.89 |
| BMI, kg/m2 | 28.47 (6.9) | 30.74 (8.54) | 33.14 (10.55) | 27.94 (6.33) | <0.01 | <0.01 |
| Number of different structured EHR variables per person | 73.85 (31.46) | 92.94 (40.61) | 114.98 (45.50) | 69.34 (27) | <0.01 | <0.01 |
| Sarcopenia test variables | ||||||
| Best grip strength (kg) | 27.56 (9.47) | 22.48 (9.21) | 19.38 (7.36) | 28.76 (9.13) | <0.01 | <0.01 |
| Best grip strength—females (kg) | 24.85 (6.79) | 19.64 (7.21) | 16.74 (5.93) | 25.92 (6.18) | <0.01 | <0.01 |
| Best grip strength—males (kg) | 38.28 (10.91) | 29.44 (9.91) | 24.18 (7.35) | 41.60 (9.32) | <0.01 | <0.01 |
| Repeat chair stand time (for 5 chair stands) (s) | 9.49 (3.4) | 10.58 (5.97) | 9.71 (8.69) | 9.23 (2.36) | <0.01 | 0.065 |
| Appendicular skeletal mass/ht2 (kg/m2) | 8.26 (2.31) | 6.86 (3.63) | 7.17 (3.92) | 8.59 (1.71) | <0.01 | <0.01 |
| Appendicular skeletal mass/ht2—female (kg/m2) | 8.05 (2.16) | 6.73 (3.56) | 7.35 (3.77) | 8.32 (1.61) | <0.01 | <0.05 |
| Appendicular skeletal mass/ht2—male (kg/m2) | 9.10 (2.67) | 7.16 (3.79) | 6.84 (4.23) | 9.83 (1.59) | <0.01 | <0.01 |
| Usual gait speed (m/s) | 1.34 (0.29) | 1.15 (0.33) | 0.90 (0.34) | 1.39 (0.25) | <0.01 | <0.01 |
| Total SPPB score | 11.24 (1.91) | 9.60 (3.02) | 7.20 (2.93) | 11.63 (1.26) | <0.01 | <0.01 |
| Bone Density: | ||||||
| Total Body BMD T-Score | 0.2227 (1.23) | 0.1350 (1.52) | 0.2172 (1.47) | 0.2434 (1.15) | <0.01 | 0.20 |
| Total Body BMD Z-score | 0.7280 (1.09) | 0.5941 (1.38) | 0.7076 (1.52) | 0.7596 (1.01) | <0.01 | <0.05 |
| Spine BMD T-Score | −0.1343 (1.26) | −0.0716 (1.19) | 0.0596 (1.30) | −0.1491 (1.27) | 0.10 | 0.06 |
| Spine BMD Z-Score | −0.3336 (1.24) | 0.3802 (1.22) | 0.6353 (1.42) | 0.3226 (1.24) | 0.41 | 0.09 |
| Total Hip BMD T-score | −0.9157 (1.30) | −0.9992 (1.34) | −1.1638 (1.38) | −0.8982 (1.28) | 0.40 | 0.11 |
| Total Hip BMD Z-score | 0.1506 (1.01) | 0.0369 (0.91) | 0.0766 (1.02) | 0.1774 (1.03) | <0.05 | 0.09 |
| Femur Neck BMD T-score | −0.4280 (1.19) | −0.6575 (1.24) | −0.8616 (1.26) | −0.3738 (1.17) | <0.05 | <0.01 |
| Femur Neck BMD Z-score | 0.2472 (0.99) | 0.0546 (0.95) | 0.0521 (1.02) | 0.2926 (0.99) | <0.01 | <0.01 |
| Percent Total Body Fat Mass | 37.92 (10.73) | 40.46 (11.02) | 41.12 (10.06) | 37.41 (10.6) | <0.01 | <0.01 |
| Charlson Comorbidity Index | 1.1702 (2.20) | 2.49 (3.17) | 4.0658 (3.59) | 0.8585 (1.75) | <0.01 | <0.01 |
| Charlson Comorbidity Index = 0 or 1 | 996 (76.38%) | 141 (56.63%) | 24 (31.58%) | 855 (81.04%) | <0.01 | <0.01 |
| Charlson Comorbidity Index ≥2 | 308 (23.62%) | 108 (43.37%) | 52 (68.42%) | 200 (18.96%) | <0.01 | <0.01 |
| Specific Diagnoses | ||||||
| Diabetes | 180 (13.8%) | 71 (28.51%) | 42 (55.26%) | 109 (10.33%) | <0.01 | <0.01 |
| Dementia | 7 (0.54%) | 5 (2.01%) | 4 (5.26%) | 2 (0.19%) | <0.01 | <0.01 |
| Cerebrovascular disease | 52 (3.99%) | 19 (7.63%) | 5 (6.58%) | 33 (3.13%) | <0.01 | 0.20 |
| Hemiplegia or paraplegia | 6 (0.46%) | 4 (1.61%) | 1 (1.32%) | 2 (0.19%) | <0.05 | 0.49 |
| Myocardial infarction | 22 (1.69%) | 10 (4.02%) | 3 (3.95%) | 12 (1.14%) | <0.01 | 0.12 |
| Congestive heart failure | 56 (4.29%) | 30 (12.05%) | 16 (21.05%) | 26 (2.46%) | <0.01 | <0.01 |
| Peripheral vascular disease | 41 (3.14%) | 20 (8.03%) | 13 (17.11%) | 21 (1.99%) | <0.01 | <0.01 |
| Chronic pulmonary disease | 210 (16.10%) | 61 (24.50%) | 29 (38.16%) | 149 (14.12%) | <0.01 | <0.01 |
| Peptic ulcer disease | 12 (0.92%) | 6 (2.41%) | 4 (5.26%) | 6 (0.57%) | <0.05 | <0.01 |
| Liver disease | 78 (5.98%) | 28 (11.74%) | 12 (15.79%) | 50 (4.74%) | <0.01 | <0.01 |
| Rheumatic disease | 43 (3.30%) | 14 (5.62%) | 4 (5.26%) | 29 (2.75%) | <0.05 | 0.37 |
| Renal disease | 78 (5.98%) | 39 (15.66%) | 22 (28.95%) | 39 (3.70%) | <0.01 | <0.01 |
| AIDS/HIV | 5 (0.38%) | 3 (1.20%) | 0 (0%) | 2 (0.19%) | 0.078 | 0.30 |
| Any malignancy | 150 (11.50%) | 45 (18.07%) | 22 (28.95%) | 105 (9.95%) | <0.01 | <0.01 |
| Metastatic solid tumor | 35 (2.68%) | 14 (5.62%) | 5 (6.58%) | 21 (1.99%) | <0.01 | <0.05 |
| Osteoporosis | 135 (10.35%) | 30 (12.05%) | 14 (18.42%) | 105 (9.95%) | 0.39 | <0.05 |
| Fractures | 92 (7.06%) | 25 (10.04%) | 9 (11.84%) | 67 (6.35%) | 0.057 | 0.108 |
Continuous variables are shown as mean (SD), and frequencies as n (%). P value (1) indicates comparison with patients meeting ≥1 criteria for sarcopenia vs. controls. P value (2) indicates comparison with patients meeting ≥2 criteria for sarcopenia vs. controls.
The average age of the sarcopenic groups was higher than the control group. The sarcopenia groups had a higher percentage of African Americans, more males, higher BMI, and total body fat mass (p < 0.01). As expected by the sarcopenia threshold definitions, the mean sarcopenia test results differed between groups with sarcopenia and controls. Sarcopenia-1 had a lower mean BMD Z-score than controls at the Total Body, Total Hip, and Femur Neck, but not at the Spine. Sarcopenia-2 had lower mean BMD Z-score than controls at the Total Body and Femur Neck, but not at the Total Hip or Spine.
Multiple chronic diseases including those often associated with sarcopenia were present in higher proportions in the sarcopenia groups than in controls (Table 1). Additionally, the sarcopenia groups had higher Charlson Comorbidity Index and a higher proportion of patients with Charlson Comorbidity Index ≥2 (p < 0.01). Participants with sarcopenia also had more structured EHR variables.
Model performance
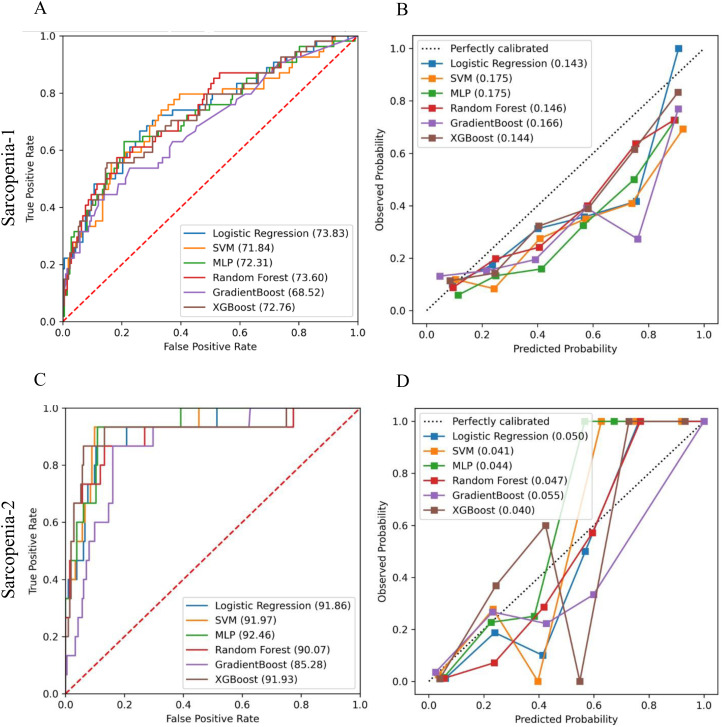
We built predictive models using machine learning separately for Sarcopenia-1 and Sarcopenia-2 (versus controls). Table 2 shows the AUC and Brier scores with confidence intervals (CIs) generated based on both the training and test datasets when different predictive machine learning models were applied to each cohort. Figure 1 shows the AUC-ROC curves and calibration slopes for the test dataset for Sarcopenia-1 and Sarcopenia 2. The mean AUC differences between best results on the different machine learning model methods were quite small in magnitude (6.3% or smaller), and not significantly different based on ANOVA (p > 0.05). None of the machine learning methods performed significantly better than the others on either Sarcopenia-1 or Sarcopenia-2. All models achieved better performance for predicting Sarcopenia-2 versus controls, than for predicting Sarcopenia-1 versus controls. The top performing models on the test data set for Sarcopenia-1 were LR and Multi-Layer Perceptron which gained AUC 71.59 (95% CI, 71.51–71.66) and AUC 71.48 (95%CI, 71.00–71.97), respectively. The top performing model on the test data set for Sarcopenia-2 was LR and SVM which gained AUC 91.44 (95%CI, 91.28–91.60) and AUC 90.81 (95%CI, 88.41–93.20), respectively. The best Brier score gained on test data of Sarcopenia-1 were 0.148 (95%CI, 0.146–0.150) and 0.154 (95%CI, 0.139–0.169) when XGBoost and LR were used, respectively. The best Brier score gained on test data of Sarcopenia-2 were 0.042 (95%CI, 0.037–0.047) and 0.043 (95%CI, 0.041–0.044) when SVM and RF were applied, respectively. With both methods of assessing the models, the machine learning model accuracy of classification was greater for Sarcopenia-2 than for Sarcopenia 1.
Table 2.
Comparison of ROC AUC and Brier scores.
| Cohorts | Predictive models | Training AUC (95% CI) | Test AUC (95% CI) | Training Brier (95% CI) | Test Brier (95% CI) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sarcopenia-1 | Logistic Regression | 76.61 | (74.42, 78.80) | 71.59 | (71.51, 71.66) | 0.131 | (0.118, 0.144) | 0.154 | (0.139, 0.169) |
| Support Vector Machine | 79.92 | (74.65, 85.19) | 71.17 | (69.28, 73.07) | 0.135 | (0.123, 0.146) | 0.169 | (0.160, 0.178) | |
| Multi-Layer Perceptron | 75.20 | (67.42, 82.99) | 71.48 | (71.00, 71.97) | 0.134 | (0.125, 0.143) | 0.160 | (0.149, 0.171) | |
| Random Forest | 74.06 | (72.65, 75.46) | 69.09 | (67.97, 70.22) | 0.132 | (0.129, 0.136) | 0.156 | (0.149, 0.162) | |
| Gradient Boosting | 77.63 | (76.67, 78.59) | 69.56 | (69.28, 69.83) | 0.165 | (0.163, 0.167) | 0.203 | (0.200, 0.206) | |
| XGBoost | 73.68 | (72.18, 75.18) | 70.19 | (69.18, 71.19) | 0.130 | (0.130, 0.131) | 0.148 | (0.146, 0.150) | |
| Sarcopenia-2 | Logistic Regression | 96.46 | (95.13, 97.79) | 91.44 | (91.28, 91.60) | 0.034 | (0.018, 0.051) | 0.047 | (0.028, 0.067) |
| Support Vector Machine | 95.56 | (92.18, 98.95) | 90.81 | (88.41, 93.20) | 0.029 | (0.022, 0.036) | 0.042 | (0.037, 0.047) | |
| Multi-Layer Perceptron | 94.20 | (89.94, 98.46) | 88.50 | (85.83, 91.18) | 0.035 | (0.021, 0.050) | 0.047 | (0.039, 0.056) | |
| Random Forest | 96.18 | (95.92, 96.44) | 90.04 | (88.16, 91.92) | 0.031 | (0.030, 0.032) | 0.043 | (0.041, 0.044) | |
| Gradient Boosting | 94.63 | (89.67, 99.60) | 86.18 | (79.56, 92.80) | 0.029 | (0.018, 0.040) | 0.048 | (0.035, 0.060) | |
| XGBoost | 92.89 | (90.94, 94.84) | 89.56 | (82.45, 96.68) | 0.033 | (0.027, 0.040) | 0.045 | (0.035, 0.056) | |
For Brier score, lower is better and <0.11 would be excellent. (The columns marked Test area under the curve [AUC] and Test Brier correspond to the graphs in Figure 1.)
Figure 1.
Performance of machine learning models in the test set for sarcopenia-1 (≥1 sarcopenia criteria) vs. Control (A) area under the curve (AUC) of ROC and (B) Brier Score Calibration Curves; and for Sarcopenia-2 (≥2 sarcopenia criteria) vs. Controls (C) AUC of ROC and (D) Brier Score Calibration Curves.
Prediction model interpretation
We applied SHAP to interpret the important variables driving the prediction of the best performing model (LR model with the highest AUC on the test set for Sarcopenia-2). Applying the elbow method (Supplemental Figure 1) to determine the most impactful variables, the top 31 variables with the highest SHAP values were selected (Table 3). The selected variables show statistically significant differences between the Sarcopenic-2 and control groups (with differences for all but one variable p < 0.01). Several chronic condition categories were more represented in the sarcopenia group including: diabetes mellitus, upper gastrointestinal disorders and other digestive system complaints, signs and symptoms involving general systems, the nervous and musculoskeletal systems, the respiratory system, metabolic disorders, and kidney and other urinary tract disorders.
Table 3.
Statistical summary of the selected 31 variables with the highest SHAP values from the logistic regression analysis of participants meeting ≥2 sarcopenia criteria.
| Variables | SHAP score | Total (1131) a | Sarcopenic-2 (76) | Control (1055) | p value |
|---|---|---|---|---|---|
| Diagnosis | |||||
| Diabetes mellitus (E08-E13) | 11.0021 | 157 (13.88%) | 43 (56.58%) | 114 (10.81%) | <0.01 |
| Diseases of esophagus, stomach, and duodenum (K20-K31) | 10.2377 | 228 (20.16%) | 44 (57.89%) | 184 (17.44%) | <0.01 |
| Symptoms and signs involving the nervous and musculoskeletal systems (R25-R29) | 7.5986 | 120 (10.61%) | 34 (44.74%) | 86 (8.15%) | <0.01 |
| Disorders of thyroid gland (E00-E07) | 7.3267 | 267 (23.61%) | 35 (46.05%) | 232 (21.99%) | <0.01 |
| General symptoms and signs (R50-R69) | 6.7480 | 451 (39.88%) | 52 (68.42%) | 399 (37.82%) | <0.01 |
| Aplastic and other anemias and other bone marrow failure syndromes (D60-D64) | 5.5454 | 131 (11.58%) | 34 (44.74%) | 97 (9.19%) | <0.01 |
| Other disorders of kidney and ureter (N25-N29) | 4.4251 | 69 (6.10%) | 19 (25.00%) | 50 (4.74%) | <0.01 |
| Symptoms and signs involving the digestive system and abdomen (R10-R19) | 4.1636 | 435 (38.46%) | 40 (52.63%) | 395 (37.44%) | <0.01 |
| Noninflammatory disorders of female genital tract (N80-N98) | 4.1513 | 336 (29.71%) | 11 (14.47%) | 325 (30.81%) | <0.01 |
| Other diseases of the urinary system (N30-N39) | 4.0818 | 202 (17.86%) | 29 (38.16%) | 173 (16.40%) | <0.01 |
| Symptoms and signs involving the circulatory and respiratory systems (R00-R09) | 3.7241 | 485 (42.88%) | 48 (63.16%) | 437 (41.42%) | <0.01 |
| Symptoms and signs involving cognition, perception, emotional state and behavior (R40-R46) | 3.5150 | 156 (13.79%) | 28 (36.84%) | 128 (12.13%) | <0.01 |
| Acute kidney failure and chronic kidney disease (N17-N19) | 2.9535 | 72 (6.37%) | 28 (36.84%) | 44 (4.17%) | <0.01 |
| Other dorsopathies (M50-M54) | 2.8545 | 378 (33.42%) | 42 (55.26%) | 336 (31.85%) | <0.01 |
| Metabolic disorders (E70-E88) | 2.3787 | 446 (39.43%) | 59 (77.63%) | 387 (36.68%) | <0.01 |
| Other disorders of the skin and subcutaneous tissue (L80-L99) | 2.3779 | 210 (18.57%) | 20 (26.32%) | 190 (18.01%) | <0.05 |
| Mental and behavioral disorders due to psychoactive substance use (F10-F19) | 2.2387 | 76 (6.72%) | 16 (21.05%) | 60 (5.69%) | <0.01 |
| Slipping, tripping, stumbling, and falls (W00-W19) | 2.2203 | 82 (7.25%) | 19 (25.00%) | 63 (5.97%) | <0.01 |
| Other forms of heart disease (I30-I5A) | 2.1712 | 166 (14.68%) | 33 (43.42%) | 133 (12.61%) | <0.01 |
| Medication | |||||
| Analgesics—Opioid | 7.9003 | 373 (32.98%) | 47 (61.84%) | 326 (30.90%) | <0.01 |
| Corticosteroids | 6.1217 | 344 (30.42%) | 33 (43.42%) | 311 (29.48%) | <0.01 |
| Antihyperlipidemics | 4.4709 | 268 (23.70%) | 39 (51.32%) | 229 (21.71%) | <0.01 |
| Antiasthmatic and bronchodilator agents | 3.2897 | 225 (19.89%) | 29 (38.16%) | 196 (18.58%) | <0.01 |
| Endocrine and metabolic agents—miscellaneous. | 3.0673 | 91 (8.05%) | 19 (25.00%) | 72 (6.82%) | <0.01 |
| Antiemetics | 2.9599 | 182 (16.09%) | 24 (31.58%) | 158 (14.98%) | <0.01 |
| Thyroid Agents | 2.7711 | 199 (17.60%) | 24 (31.58%) | 175 (16.59%) | <0.01 |
| Clinical measurements | |||||
| Glucose | 13.7524 | ||||
| Low | 19 (1.68%) | 0 (0.0%) | 19 (1.80%) | 0.12 | |
| Normal | 720 (63.66%) | 21 (27.63%) | 699 (66.26%) | <0.01 | |
| High | 392 (34.66%) | 55 (72.37%) | 337 (31.94%) | <0.01 | |
| LDL | 5.6637 | ||||
| Low | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1.00 | |
| Normal | 651 (57.56%) | 58 (76.32%) | 593 (56.21%) | <0.01 | |
| High | 480 (42.44%) | 18 (23.68%) | 462 (43.79%) | <0.01 | |
| HDL | 4.8129 | ||||
| Low | 96 (8.49%) | 19 (25.00%) | 77 (7.30%) | <0.01 | |
| Normal | 1035 (91.51%) | 57 (75.00%) | 978 (92.70%) | <0.01 | |
| High | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1.00 | |
| Hemoglobin | 3.5917 | ||||
| Low | 154 (13.62%) | 30 (39.47%) | 124 (11.75%) | <0.01 | |
| Normal | 932 (82.40%) | 43 (56.58%) | 889 (84.27%) | <0.01 | |
| High | 45 (3.98%) | 3 (3.95%) | 42 (3.98%) | 0.49 | |
| QRS duration | 2.4131 | ||||
| Low | 72 (6.37%) | 11 (14.47%) | 61 (5.78%) | <0.01 | |
| Normal | 1018 (90.01%) | 53 (69.74%) | 965 (91.47%) | <0.01 | |
| High | 41 (3.63%) | 12 (15.79%) | 29 (2.75%) | <0.01 | |
Total for this table is only those in the Sarcopenia-2 and control groups.
Variables are listed in order of importance within each of the categories (diagnosis, medication groupings, and clinical measurements).
Regarding medication categories, participants with sarcopenia criteria were also more likely to be prescribed opioid analgesics, corticosteroids, antihyperlipidemic drugs, as well as anti-asthmatic/bronchodilators, antiemetics, and miscellaneous endocrine/metabolic agents (each p < 0.01). The drugs listed under the NDC classification “endocrine and metabolic agents (miscellaneous),” consisted mostly of hormone receptor modulators and bone density medications. The main hormone receptor modulators were selective estrogen receptor modulators which were present in none of the sarcopenic group and 17 (1.61%) of the control group (p = 0.1327). Bone density regulators were used by 16 (21.05%) in the sarcopenic group and 49 (4.64%) of the control group (p < 0.01), most of which were bisphosphonates [sarcopenia group 15 (19.74%) and control group 41 (3.89%), p < 0.01). Parathyroid hormone agonists were administered to none of the sarcopenia group during this period and only 6 (0.57%) of the control group.
In SHAP analysis, some laboratory measures contributed among the top variables in the model. Participants with sarcopenia had more hyperglycemia, but less documented hypoglycemia on laboratory testing. Differences were also seen in the proportion with low hemoglobin (more in sarcopenia), low HDL (more in sarcopenia) and high LDL (less in sarcopenia). Triglyceride measurements were not among the top 31 variables in SHAP analysis (it ranked number 45). There was no difference between groups in the proportions having high or low triglycerides. However, a higher proportion of the sarcopenia group had low HDL [19 (25%)] than in the control group [(77 (7.3%)], and patients with sarcopenia were more likely to have the combination of high triglycerides with low HDL [19 (25.00%)] than the control group [64 (6.06%)] (p < 0.01). Participants with sarcopenia were more likely than controls to have recorded either high or low QRS interval.
Discussion
In this study of 1304 individuals undergoing measurements of muscle function and lean mass, 19.1% met at least one criterion for sarcopenia and 5.8% met at least two criteria. Using only structured data from their EHR and machine learning approaches, we were able to identify patterns of structured variables that were highly predictive of sarcopenia. Models with mean AUCs over 90% were achieved across test data using multiple approaches, with similar overall AUC between models. Each of the machine learning methods performed better at predicting Sarcopenia-2 than Sarcopenia-1 vs. controls. Sarcopenia is often undercoded and undetected; indeed, only one participant had the sarcopenia ICD code, and none had the related codes cachexia or frailty.
The mean age of sarcopenic patients was older than that of controls but was still relatively young (58 years). This study highlights the importance of chronic diseases as risks for sarcopenia among the non-elderly population3,4 and the importance of recognizing early signs of sarcopenia at younger ages to support preventative interventions to preserve mobility.
As expected, the Charlson Comorbidity Index was higher and several chronic conditions were more common in the sarcopenia groups than in controls. These included diabetes, renal diseases, malignancy, osteoporosis, and pulmonary, cardiac, and liver diseases. While all diagnosis groups were considered for models, in the highest performing models most of these were not among the top importance variables by SHAP for predicting sarcopenia. The most important diagnosis group variables were diabetes, upper gastrointestinal diseases, signs and symptoms for the nervous and musculoskeletal systems, thyroid disorders, and kidney disease along with circulatory and respiratory symptoms and disorders. Malignancy was more common among our sarcopenia group than controls and also is commonly clinically associated with sarcopenia and cachexia, but was not one of the top model variables detected by SHAP. Malignancies, with SHAP of 0.7228 (ranked 83 among all variables) were far below the cutoff from the elbow method. One possible reason for this is the overall small proportion of participants in this study with any malignancy.
Sarcopenia can be present even among obese patients, 34 where it is often missed clinically. Sarcopenia, especially in combination with obesity, has also been associated with the metabolic syndrome. 35 In our population, those with sarcopenia had higher BMI, and higher total body fat mass, despite lower height-adjusted appendicular lean mass. More participants with sarcopenia also had general metabolic abnormalities as indicated by diabetes, kidney disease, and ICD codes for metabolic disorders. Indeed, glucose value and diabetes diagnosis had the highest overall SHAP values. Sarcopenic participants were more likely to have low HDL in combination with high triglycerides, features of the metabolic syndrome often present with diabetes. Similarly, a study of Korean adults also indicated those with sarcopenia had more metabolic syndrome, higher triglycerides, and lower HDL than nonsarcopenic controls. 35 In contrast, in the West China Health and Aging Trend study, sarcopenic patients had lower triglycerides but higher HDL compared to controls. 36 In our study, we also note that fewer participants with sarcopenia than controls had high LDL on laboratory measurements, which might have been influenced by the greater proportion of sarcopenic participants already receiving antihyperlipidemic drugs.
The presence of low hemoglobin level was also among the top variables in our model. More patients with sarcopenia had low hemoglobin levels, which can be a marker of nutritional status, similar to albumin. This is consistent with studies that have shown associations between anemia and sarcopenia, slow gait speed and low muscle strength in older adults,37,38 as well as post-stroke 39 or with malignancies. 40
Machine learning models are data-driven approaches and depend on the information available within the EHR. Additional variables from unstructured text notes describing patients’ mobility may be useful to further improve performance. We used EHR data available from a multi-institution health data exchange, but different variables may be more important for models built on different individual EHR systems.
Strengths of our study and approach include standardized measurements of muscle strength and function, inclusion of a broad age range and >1000 adults, and combining “gold-standard” sarcopenia assessments with robust multi-health system EHR data (rather than relying on a single health system EHR). Our approach to exclude subjects who did not have sufficient EHR data ensured that robust structured EHR data were available for both cases and controls. This was likely to exclude some healthy control participants, who may have less interaction with the healthcare system. An additional strength was the use of multiple robust and unbiased machine learning models, all of which demonstrated consistent AUC within a narrow range, in support of the method. This allowed the detection of the most important predictors from the large structured EHR data.
Some weaknesses of our study include that a limited total number of patients met criteria for sarcopenia. Because of this, we performed analysis based on participants having either 1 or more criteria for sarcopenia or 2 or more criteria for sarcopenia. Thus, some of our patients were perhaps more mildly affected, which might have decreased the differences between groups. Additionally, it should be noted that factors beyond muscle influence performance on standardized muscle tests, including pain, joint dysfunction, neurologic, cardiac, and pulmonary function. In this study, our sample size was not large enough to evaluate the performance in different demographic, racial/ethnic, or disease-based populations. Finally, while we have demonstrated internal validation by dividing the cohort into training and test sets, we did not have an external cohort with which to validate the findings. External validation will be required in future studies of external datasets and different EHR systems to confirm the ability to accurately predict sarcopenia and demonstrate utility for screening purposes in various populations.
Sarcopenia and frailty are likely to be related in an individual, as both increase with aging, and many of the tests used in sarcopenia represent components of frailty. We designed this analysis specifically around sarcopenia or a loss of skeletal muscle mass, strength, and function, whereas frailty may more broadly include states of declining energy, disability, symptoms, and comorbidities resulting in overall decompensation of the individual.41–43 However, there is overlap between these conditions, as also noted in our prior publication evaluating EHR-coded diagnoses and simple text terms for sarcopenia, frailty, and cachexia. 16 While to our knowledge, there have not been prior electronic sarcopenia instruments to identify sarcopenia from EHR, other authors have reported EHR-based electronic frailty indexes, focused on either collections of preconceived concepts for contributors to frailty or on the accumulations of deficits in relation to aging, defined by ICD codes, and laboratory tests or on focused interview and examination results.42–44 Using this accumulated deficit concept to address frailty, electronic frailty indices have been developed to predict various outcomes including fracture, hospitalization, mortality, and other healthcare utilization.42,44 Some frailty models required specific history to be obtained regarding activities of daily living and various symptoms, which also may not be consistently obtainable from the structured data within the EHR.41–43
However, these previous electronic frailty index tools were not devised based on actual gold standard outcome measures of body composition or muscle function tests as the outcome of interest. This is not surprising as the necessary measurements are often not conducted clinically, and even when performed, the results are recorded usually in a manner not readily extracted from most EHR systems. In contrast, our models were developed based on all participants having undergone standardized measures through a research protocol, allowing us to develop EHR-based models for these outcomes.
Long-term studies of the natural history of sarcopenia have been hampered by inconsistent definitions, which led to relatively recent alterations of consensus definitions. 45 However, given that the sarcopenia prevalence increases with advancing age, and that some sarcopenia feature measures predict the development of other sarcopenia features, it is likely that the natural history of sarcopenia is to worsen over time. Multiple studies have indicated that resistance training is likely beneficial in treatment of sarcopenia,46,47 though with heterogeneity in the duration and delivery of the intervention as well as the magnitude of results. Additionally, multiple studies of various nutrition supplementation (leucine, whey, casein, omega 3 fatty acids, collagen peptides, and others) also show benefit in small trials, especially when combined with resistance training.48–51 The development of EHR-based tools to identify and recruit sarcopenic patients early may be useful to support targeted large-scale interventions in the future.
Future directions include testing of our models in external EHR cohorts having similar standardized muscle function measures, incorporation of unstructured text notes to further improve detectability of sarcopenia in special patient populations and to use machine learning to generate algorithms that could then be applied to EHR data to prospectively identify sarcopenia patients for selection for targeted interventions.
Conclusion
Sarcopenia can occur in individuals throughout their life span. While standardized muscular testing is needed to define sarcopenia, machine learning can effectively detect combinations of variables from within the EHR that are predictive of sarcopenia. This method may provide opportunities in the future for targeted selection of individual patients for more detailed sarcopenia testing and clinical intervention.
Contributorship: EAI and XL had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The study was conceptualized and designed by XL, SJW, AB, RNM, and EAI; funding was obtained by XL, RNM, and EAI. Data collection and cleaning was by AB, HD, SJW, RNM, EAI, and XL. Analysis was by XL, HD, RNM, and EAI. Interpretation was by XL, RNM, SJW, and EAI. The first draft was completed by XL, RNM, and EAI, and subsequent versions edited and approved by all authors.
Data sharing statement: De-identified data used in this study may be shared with investigators with formal request. Our own data use agreement with the institutional source of electronic health records data requires that a separate data use agreement be put in place and approved institutionally for every reuse case requested. Requests can be sent to eimel@iu.edu.
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: This cohort study was approved by the Institutional Review Board of Indiana University, under two protocols (#2004191295 and #1707550885). All participants provided written informed consent.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by funding from the NIH by NIAMS (P30AR072581 and R01AR077273) and by NIDDK (K23DK102824) and by NCATS (UL1TR002529). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The sponsor was not involved in the conduct of the study or writing of the manuscript.
Guarantor: EAI
Supplemental Material
Supplemental material, sj-docx-1-dhj-10.1177_20552076231197098 for Using machine learning to detect sarcopenia from electronic health records by Xiao Luo, Haoran Ding, Andrea Broyles, Stuart J Warden and Ranjani N Moorthi, Erik A Imel in DIGITAL HEALTH
Supplemental material, sj-docx-2-dhj-10.1177_20552076231197098 for Using machine learning to detect sarcopenia from electronic health records by Xiao Luo, Haoran Ding, Andrea Broyles, Stuart J Warden and Ranjani N Moorthi, Erik A Imel in DIGITAL HEALTH
Footnotes
ORCID iD: Erik A Imel https://orcid.org/0000-0002-7284-3467
Supplemental material: Supplemental material for this article is available online.
References
- 1.Cruz-Jentoft AJ, Bahat G, Bauer J, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019; 48: 16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Studenski SA, Peters KW, Alley DE, et al. The FNIH sarcopenia project: rationale, study description, conference recommendations, and final estimates. J Gerontol A Biol Sci Med Sci 2014; 69: 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandberg C, Johansson K, Christersson C, et al. Sarcopenia is common in adults with complex congenital heart disease. Int J Cardiol 2019; 296: 57–62. [DOI] [PubMed] [Google Scholar]
- 4.Silva TLD, Mulder AP. Sarcopenia and poor muscle quality associated with severe obesity in young adults and middle-aged adults. Clin Nutr ESPEN 2021; 45: 299–305. [DOI] [PubMed] [Google Scholar]
- 5.Rolland Y, Abellan van Kan G, Gillette-Guyonnet Set al. et al. Cachexia versus sarcopenia. Curr Opin Clin Nutr Metab Care 2011; 14: 15–21. [DOI] [PubMed] [Google Scholar]
- 6.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001; 56: M146–M156. [DOI] [PubMed] [Google Scholar]
- 7.Shafiee G, Keshtkar A, Soltani A, et al. Prevalence of sarcopenia in the world: a systematic review and meta- analysis of general population studies. J Diabetes Metab Disord 2017; 16: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayhew AJ, Amog K, Phillips S, et al. The prevalence of sarcopenia in community-dwelling older adults, an exploration of differences between studies and within definitions: a systematic review and meta-analyses. Age Ageing 2019; 48: 48–56. [DOI] [PubMed] [Google Scholar]
- 9.Steffl M, Sima J, Shiells Ket al. et al. The increase in health care costs associated with muscle weakness in older people without long-term illnesses in the Czech Republic: results from the survey of health, ageing and retirement in Europe (SHARE). Clin Interv Aging 2017; 12: 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes AC, Araujo DA, Verissimo MTet al. et al. Sarcopenia and hospitalisation costs in older adults: a cross-sectional study. Nutr Diet 2017; 74: 46–50. [DOI] [PubMed] [Google Scholar]
- 11.Landi F, Cruz-Jentoft AJ, Liperoti R, et al. Sarcopenia and mortality risk in frail older persons aged 80 years and older: results from ilSIRENTE study. Age Ageing 2013; 42: 203–209. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Zhang W, Wang C, et al. Sarcopenia as a predictor of hospitalization among older people: a systematic review and meta-analysis. BMC Geriatr 2018; 18: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz-Jentoft AJ, Bahat G, Bauer J, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019; 48: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souza RMP, Cardim AB, Maia TO, et al. Inspiratory muscle strength, diaphragmatic mobility, and body composition in chronic obstructive pulmonary disease. Physiother Res Int 2019; 24: e1766. [DOI] [PubMed] [Google Scholar]
- 15.Carrero JJ, Johansen KL, Lindholm B, et al. Screening for muscle wasting and dysfunction in patients with chronic kidney disease. Kidney Int 2016; 90: 53–66. [DOI] [PubMed] [Google Scholar]
- 16.Moorthi RN, Liu Z, El-Azab SA, et al. Sarcopenia, frailty and cachexia patients detected in a multisystem electronic health record database. BMC Musculoskelet Disord 2020; 21: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warden SJ, Liu Z, Moe SM. Sex- and age-specific centile curves and downloadable calculator for clinical muscle strength tests to identify probable sarcopenia. Phys Ther 2022; 102. DOI: 10.1093/ptj/pzab299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts HC, Denison HJ, Martin HJ, et al. A review of the measurement of grip strength in clinical and epidemiological studies: towards a standardised approach. Age ageing 2011; 40: 423–9. [DOI] [PubMed] [Google Scholar]
- 19.Guralnik JM, Simonsick EM, Ferrucci L, et al. A Short Physical Performance Battery assessing lower extremity function: association with self-reported disability and prediction of mortality and nursing home admission. J Gerontol 1994; 49: M85–M94. [DOI] [PubMed] [Google Scholar]
- 20.Warden SJ, Kemp AC, Liu Zet al. et al. Tester and testing procedure influence clinically determined gait speed. Gait Posture 2019; 74: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YJ, Do Shin S, Park HS, et al. International Classification of Diseases 10th edition-based disability adjusted life years for measuring of burden of specific injury. Clin Exp Emerg Med 2016; 3: 219–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol 1992; 45: 613–619. [DOI] [PubMed] [Google Scholar]
- 23.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005; 43: 1130–1139. [DOI] [PubMed] [Google Scholar]
- 24.Noble WS. What is a support vector machine? Nat Biotechnol 2006; 24: 1565–1567. [DOI] [PubMed] [Google Scholar]
- 25.Menard S. Applied logistic regression analysis. In Sage University Papers Series on Quantitative Applications in the Social Sciences, Thousand Oaks, California, USA, Vol. 07-106. Sage, 2002. [Google Scholar]
- 26.Qi Y. Random forest for bioinformatics. In: Ensemble machine learning. Editors Zhang and Ma, Springer, New York, NY, USA, 2012, pp.307–323. [Google Scholar]
- 27.Zhang Z, Zhao Y, Canes A, et al. Predictive analytics with gradient boosting in clinical medicine. Ann Transl Med 2019; 7(7): 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanwal F, Taylor TJ, Kramer JR, et al. Development, validation, and evaluation of a simple machine learning model to predict cirrhosis mortality. JAMA Netw Open 2020; 3: e2023780–e2023780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. 2016: 785–794.
- 30.Abdar M, Yen NY, Hung JC-S. Improving the diagnosis of liver disease using multilayer perceptron neural network and boosted decision trees. J Med Biol Eng 2018; 38: 953–965. [Google Scholar]
- 31.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Proceedings of the 31st international conference on neural information processing systems. 2017: 4768–4777.
- 32.Lundberg SM, Nair B, Vavilala MS, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng 2018; 2: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ketchen DJ, Shook CL. The application of cluster analysis in strategic management research: an analysis and critique. Strateg Manage J 1996; 17: 441–458. [Google Scholar]
- 34.Donini LM, Busetto L, Bischoff SC, et al. Definition and diagnostic criteria for sarcopenic obesity: ESPEN and EASO consensus statement. Clin Nutr 2022; 41: 990–1000. [DOI] [PubMed] [Google Scholar]
- 35.Lee DY, Shin S. Sarcopenia is associated with metabolic syndrome in Korean adults aged over 50 years: a cross-sectional study. Int J Environ Res Public Health 2022; 19: 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin M, Zhang H, Liu Q, et al. Diagnostic performance of clinical laboratory indicators with sarcopenia: results from the West China health and aging trend study. Front Endocrinol (Lausanne) 2021; 12: 785045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng SH, Lee WJ, Peng LN, et al. Associations between hemoglobin levels and sarcopenia and its components: results from the I-Lan longitudinal study. Exp Gerontol 2021; 150: 111379. [DOI] [PubMed] [Google Scholar]
- 38.Bani Hassan E, Vogrin S, Hernandez Vina I, et al. Hemoglobin levels are low in sarcopenic and osteosarcopenic older persons. Calcif Tissue Int 2020; 107: 135–142. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimura Y, Wakabayashi H, Nagano F, et al. Low hemoglobin levels are associated with sarcopenia, dysphagia, and adverse rehabilitation outcomes after stroke. J Stroke Cerebrovasc Dis 2020; 29: 105405. [DOI] [PubMed] [Google Scholar]
- 40.Di Sebastiano KM, Yang L, Zbuk K, et al. Accelerated muscle and adipose tissue loss may predict survival in pancreatic cancer patients: the relationship with diabetes and anaemia. Br J Nutr 2013; 109: 302–312. [DOI] [PubMed] [Google Scholar]
- 41.Anzaldi LJ, Davison A, Boyd CM, et al. Comparing clinician descriptions of frailty and geriatric syndromes using electronic health records: a retrospective cohort study. BMC Geriatr 2017; 17: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pajewski NM, Lenoir K, Wells BJ, et al. Frailty screening using the electronic health record within a medicare accountable care organization. J Gerontol A Biol Sci Med Sci 2019; 74: 1771–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lekan DA, Wallace DC, McCoy TP, et al. Frailty assessment in hospitalized older adults using the electronic health record. Biol Res Nurs 2017; 19: 213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clegg A, Bates C, Young J, et al. Development and validation of an electronic frailty index using routine primary care electronic health record data. Age Ageing 2016; 45: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cruz-Jentoft AJ, Baeyens JP, Bauer JM, et al. Sarcopenia: European consensus on definition and diagnosis: report of the European Working Group on sarcopenia in older people. Age Ageing 2010; 39: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lozano-Montoya I, Correa-Pérez A, Abraha I, et al. Nonpharmacological interventions to treat physical frailty and sarcopenia in older patients: a systematic overview—the SENATOR project ONTOP series. Clin Interv Aging 2017; 12: 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao H, Cheng R, Song G, et al. The effect of resistance training on the rehabilitation of elderly patients with sarcopenia: a meta-analysis. Int J Environ Res Public Health 2022; 19: 15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cruz-Jentoft AJ, Landi F, Schneider SM, et al. Prevalence of and interventions for sarcopenia in ageing adults: a systematic review. Report of the International Sarcopenia Initiative (EWGSOP and IWGS). Age Ageing 2014; 43: 748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilsson MI, Mikhail A, Lan L, et al. A five-ingredient nutritional supplement and home-based resistance exercise improve lean mass and strength in free-living elderly. Nutrients 2020; 12: 2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zdzieblik D, Oesser S, Baumstark MW, et al. Collagen peptide supplementation in combination with resistance training improves body composition and increases muscle strength in elderly sarcopenic men: a randomised controlled trial. Br J Nutr 2015; 114: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori H, Tokuda Y. De-training effects following leucine-enriched whey protein supplementation and resistance training in older adults with sarcopenia: a randomized controlled trial with 24 weeks of follow-up. J Nutr Health Aging 2022; 26: 994–1002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-dhj-10.1177_20552076231197098 for Using machine learning to detect sarcopenia from electronic health records by Xiao Luo, Haoran Ding, Andrea Broyles, Stuart J Warden and Ranjani N Moorthi, Erik A Imel in DIGITAL HEALTH
Supplemental material, sj-docx-2-dhj-10.1177_20552076231197098 for Using machine learning to detect sarcopenia from electronic health records by Xiao Luo, Haoran Ding, Andrea Broyles, Stuart J Warden and Ranjani N Moorthi, Erik A Imel in DIGITAL HEALTH