Abstract
Lactococcus garvieae (junior synonym Enterococcus seriolicida) is an emerging zoonotic agent isolated from economically important fish (rainbow trout and yellowtail), from cattle, and from humans. Clindamycin susceptibility is the only phenotypic test which can differentiate L. garvieae from Lactococcus lactis, another emerging agent in humans. A PCR assay for the identification of L. garvieae was developed and resulted in an amplified fragment of 1,100 bp in size. The PCR assay was shown to be specific to L. garvieae. The PCR assay was positive for all the L. garvieae strains tested, which originated from three different continents (Asia, Australia, and Europe). The PCR assay was negative for the phenotypically similar L. lactis and for all the other fish pathogens tested, including Streptococcus iniae and Aeromonas salmonicida. The PCR assay was applied to plasma obtained from diseased animals and was found sensitive enough to detect bacteria from 1 μl of plasma. The PCR assay that was developed is the only practical test besides the clindamycin test which can specifically identify the zoonotic agent L. garvieae and which can differentiate it from L. lactis.
Lactococcus garvieae is an emerging zoonotic pathogen which has been isolated from cattle, from various species of fish, and from humans (2, 5, 6, 9, 13). With the development of intensive aquaculture, streptococcal infections of fish have become a major problem worldwide. The symptomatologies of fish infected by the various gram-positive cocci pathogenic for fish are similar and do not allow a rapid identification of the agent responsible for the disease (1). In addition, the isolation and the bacteriological diagnosis of these gram-positive bacteria are not simple and are time-consuming (8, 10, 11). L. garvieae has proven to be one of the major gram-positive coccus pathogens for fish (5, 9). Taxonomic studies based on DNA-DNA hybridization studies (5) and sequence analyses of 16S rRNA (4) indicate that another fish pathogen, Enterococcus seriolicida (12), is in fact synonymous with Lactococcus garvieae and should be classified as L. garvieae (5).
The published identification scheme of L. garvieae based on biochemical and antigenic characteristics (7, 10) can barely differentiate L. garvieae from L. lactis subsp. lactis, which has also been reported as an etiologic agent of infections in humans (6, 13). A clear test is necessary to differentiate the two species. In addition, a rapid bacteriological diagnostic method is necessary to initiate prompt therapeutic and prophylactic measures in order to limit the economic losses generated by these infections in aquaculture. In response to these needs, we have developed a new PCR assay based on primers deduced from the regions carrying the 16S rRNA genes of L. garvieae which specifically identifies L. garvieae, which differentiates L. garvieae from L. lactis, and which is sensitive enough to be applied to clinical specimens.
MATERIALS AND METHODS
Bacterial strains.
The following bacterial strains were used to test the specificity of the PCR assay: L. garvieae ATCC 43921T, E. seriolicida ATCC 49156T, 28 L. garvieae strains isolated from diseased fish (rainbow trout) from different farms in northern Italy during a 4-year period (ITP 1371, ITP 1375, ITP 1457, ITP 1545, ITP 1547, ITP 1815, ITP 1833, ITP 1876, ITP 1877, ITP 1917, ITP 1920, ITP 1921, ITP 1957, ITP 1964, ITP 1991, ITP 1992, ITP 2001, ITP 2224, ITP 2451, ITP 2453, ITP 2454, ITP 2808, ITP 3365, ITP 3433, ITP 3620, ITP 3630, ITP 3631, and ITP 3995), two L. garvieae strains isolated from yellowtail fish in Japan (S1449 and S014), 1 L. garvieae strain isolated from rainbow trout in Australia (88/1400), 2 L. garvieae strains isolated from rainbow trout in Spain (Sp 1 and Sp 2), L. lactis NCFB 604T, Streptococcus iniae ATCC 29178T, Streptococcus difficile CIP 103769T, Lactococcus piscium NCFB 2778T, and Vagococcus salmoninarum NCFB 2777T, as well as Aeromonas salmonicida 3173/86, a gram-negative bacterium pathogenic for fish. Bacteria were grown on blood agar base (Oxoid)–5% sheep blood plates for 24 h at 25°C. All the strains were identified by using either the API Strep system or the API-E system (BioMerieux, Marcy l’Etoile, France) in accordance with the manufacturer’s instructions except that the incubation temperature used was 25°C. Susceptibility to clindamycin was determined for L. garvieae strains and for L. lactis by the Kirby-Bauer test by using 2-μg clindamycin disks (7). DNA-DNA hybridization (5) was used to confirm the speciation of all the L. garvieae strains described above.
Clinical specimens.
Blood specimens were obtained from trout farmed in northern Italy. Ten rainbow trout (Oncorhynchus mykiss Walbaum) from a pond with a high level of mortality due to L. garvieae as well as 10 antibiotic-treated rainbow trout from the same farm but another raceway were sampled randomly. In addition, 10 rainbow trout from a farm without disease where the fish were vaccinated against L. garvieae and 10 rainbow trout from a different farm with no reported disease or mortality but where the disease had occurred a previous year were also sampled randomly. Half a milliliter of blood was taken from each of the anesthetized rainbow trout. Heparin at a final concentration of 10 U/ml (Kamada, Beit Kama, Israel) was added. The blood specimens were centrifuged at 2,000 × g for 10 min to obtain the plasma. Then, 1 μl of the plasma was added to a PCR tube and the DNA was extracted by adding 20 μl of Genereleaser (Bioventures, Murfreesboro, Tenn.) in accordance with the manufacturer’s protocol. PCR mix (see below) was added to a final volume of 100 μl. The PCR assay was done under the conditions described below.
Primers.
Specific primers to identify L. garvieae, pLG-1 (5′-CATAACAATGAGAATCGC-3′) and pLG-2 (5′-GCACCCTCGCGGGTTG-3′), were selected by comparing the 16S rDNA sequence of L. garvieae (EMBL accession no. X54262) (3) to other bacterial sequences by using the GCG (version 9.0-UNIX) FastA program.
PCR sensitivity was evaluated by testing 10-fold dilutions of L. garvieae suspensions in saline that were simultaneously plated to determine the number of CFU.
Amplification by PCR.
Typical PCRs were carried out in a final volume of 50 or 100 μl containing 1 U of Vent DNA polymerase (New England Biolabs) per 50 μl, 1× buffer (New England Biolabs), nucleotides (0.25 mmol/liter, final concentration for each nucleotide), bovine serum albumin (0.1 mg/μl), and primers (8 ng/μl each). Bacterial DNA was obtained from cells by touching colonies grown on blood-agar medium with a sterile syringe needle and then dipping the needle directly into the PCR mix. Typical cycling parameters were 1 min of denaturation (94°C), 1 min of annealing (55°C), and 1.5 min of extension (72°C), for 35 cycles. The reaction was started by a denaturation step (3 min at 94°C) and ended by a 10-min extension step at 72°C. The PCR-amplified samples were subjected to electrophoresis (90 min, 90 V) in a 2% agarose gel (FMC) with 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8]) and stained with ethidium bromide after the run. The DNA molecular weight marker VI (Boehringer Mannheim) was used to estimate the size of the amplified fragment.
RESULTS AND DISCUSSION
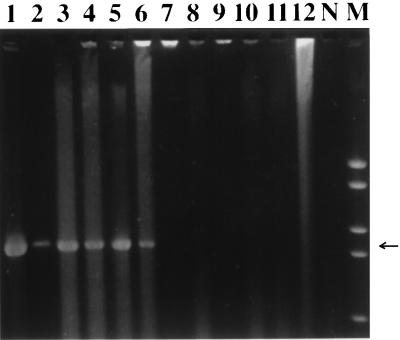
The PCR assay resulted in the amplification of a band of 1,100 bp in size that was detected for all 35 L. garvieae strains tested, including the L. garvieae ATCC 43921T and the E. seriolicida ATCC 49156T reference strains. Examples of the amplified band can be seen in Fig. 1 (lanes 1 to 6). The L. garvieae strains tested by PCR originated from diseased fish from three different continents (Asia, Australia, and Europe), proving the universality of the primers selected for the PCR assay. The specificity of the PCR assay was demonstrated by the fact that no specific band was amplified when L. lactis or any other fish pathogen was used as the DNA template (Fig. 1, lanes 7 to 12). The amplification of the specific 1,100-bp band from both L. garvieae and E. seriolicida reference strains (Fig. 1, lanes 1 and 2) confirms once more the genetic identity of these two bacterial species.
FIG. 1.
Specificity of the L. garvieae PCR assay. Agarose gel (2%)-resolved ethidium bromide-stained PCR products from template DNA of various needle-touched bacterial colonies are shown. Lanes: 1, L. garvieae ATCC 43921T; 2, E. seriolicida ATCC 49156T; 3, L. garvieae ITP 2001 (Italy); 4, L. garvieae S1449 (Japan); 5, L. garvieae 88/1400 (Australia); 6, L. garvieae Sp 1 (Spain); 7, L. lactis NCFB 604T; 8, L. piscium NCFB 2778T; 9, V. salmoninarum NCFB 2777T; 10, S. iniae ATCC 29178T; 11, S. difficile CIP 103769T; 12, A. salmonicida 3173/86; N, negative control (no DNA); M, DNA molecular weight marker VI. The arrow indicates the band of the expected length of 1,100 bp.
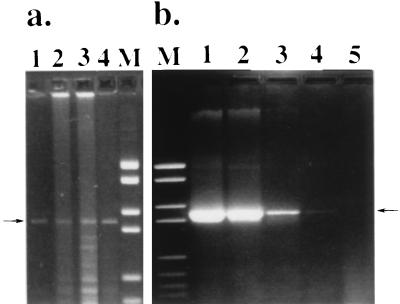
The PCR assay was applied to 10-fold dilutions of L. garvieae ITP 1992. The PCR assay was positive (presence of a specific 1,100-bp band) down to the dilution corresponding to 4 bacteria (4 CFU) per reaction tube (Fig. 2b, lane 4) but not for the next dilution (0.4 CFU) (Fig. 2b, lane 5). These data showed that the primers that were selected allowed a highly sensitive detection of L. garvieae cells suspended in saline. In fact, we have also tested the sensitivity of the PCR assay for L. garvieae cells suspended in trout plasma and found a similar level of detection (data not shown).
FIG. 2.
Field application and sensitivity of the L. garvieae PCR assay. (a) PCR product from 1 μl of fish plasma. Lanes: 1 to 3, diseased fish; 4, antibiotic-treated fish. (b) PCR assay sensitivity evaluated with a serially diluted bacterial suspension. Lanes: 1, 3,600 CFU; 2, 360 CFU; 3, 36 CFU; 4, 4 CFU; 5, 0.4 CFU. The arrows indicate the band of the expected length of 1,100 bp. Lane M contains DNA molecular weight marker VI.
Seven of the 10 plasma specimens obtained from trout sampled from a pond with active L. garvieae disease as well as 1 of 10 of the antibiotic-treated trout originating from the same fish farm with active clinical L. garvieae disease were found positive by PCR (Fig. 2a). More animals might have been found PCR positive in the pond stricken by L. garvieae mortalities if a larger volume of plasma from each fish had been used or more than one PCR had been performed. The PCR assay was negative for the plasma samples obtained from 10 vaccinated trout originating from a farm without active disease. However, the PCR assay was positive for 3 of 10 plasma samples from nonvaccinated trout reared at another farm without current active disease but where the disease was active the previous year. This observation may reflect the presence of L. garvieae carriers among fish that will develop the disease only under detrimental environmental conditions, as already suggested (1). The data presented above show that the PCR assay performed on a very limited amount of plasma (1 μl) is sensitive enough to detect specifically L. garvieae in fish originating from a pond where an active epizootic is occurring (Fig. 2a, lanes 1 to 4). In addition, the PCR assay could detect carriers of L. garvieae in a pond where the disease had occurred a previous year (Fig. 2a, lane 4). From these data, it can be concluded that the proposed PCR assay can be useful not only for diagnostic but also for epidemiological surveys and could be an efficient tool to establish preventive measures on time (antibiotic prophylaxis, vaccination).
Finally, all the L. garvieae strains, including the E. seriolicida ATCC 49156T strain, were contact resistant to clindamycin, whereas the L. lactis strain presented a 22-mm-diameter inhibition zone around the clindamycin disk. Our results confirm those of Elliot and Facklam (7) in that all the L. garvieae strains tested were resistant to clindamycin whereas the L. lactis strain tested was susceptible to clindamycin.
Besides determination of clindamycin susceptibility, only comparisons of whole-cell protein patterns (6) and DNA, RNA, and restriction fragment length polymorphism data (5, 7) can distinguish between L. lactis and L. garvieae, which are phenotypically similar. These techniques are useless for routine identification. The clindamycin susceptibility test has been the only practical phenotypic test which differentiates L. garvieae from L. lactis. However, the occurrence of clindamycin-resistant L. lactis strains would result in an erroneous diagnosis. The data presented here show that L. garvieae and L. lactis subsp. lactis, which are both recently recognized pathogens, can be easily differentiated by the proposed PCR assay. The PCR assay that we have developed is a convenient, fast, and simple technique that can be accomplished in 5 h and that can be applied not only to the specific identification of L. garvieae but also to the surveillance of L. garvieae infections in aquaculture.
ACKNOWLEDGMENTS
We thank E. Bozetta, M. Goria, and M. Prearo for their technical assistance.
This work was supported in part by a joint American-Israeli grant (BARD IS-2307-93) and by grant CA13-O18, US-Israel CDR, Human Capacity Development, USAID.
REFERENCES
- 1.Bercovier H, Ghittino C, Eldar A. Immunization with bacterial antigens: infections with streptococci and related organisms. Dev Biol Stand. 1997;90:153–160. [PubMed] [Google Scholar]
- 2.Carson J, Gudkovs N, Austin B. Characteristics of an Enterococcus-like bacterium from Australia and South Africa, pathogenic for rainbow trout (Oncorhynchus mykiss Walbaum) J Fish Dis. 1993;16:381–388. [Google Scholar]
- 3.Collins M D, Ash C, Farrow J A E, Wallbanks S, Williams A M. 16S ribosomal ribonucleic acid sequence analyses of lactococci and related taxa. Description of Vagococcus fluvialis gen. nov., sp. nov. J Appl Bacteriol. 1989;67:453–460. doi: 10.1111/j.1365-2672.1989.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 4.Domenech A, Prieta J, Fernandez-Garayzabal J F, Collins M D, Jones D, Dominguez L. Phenotypic and phylogenetic evidence for a close relationship between Lactococcus garvieae and Enterococcus seriolicida. Microbiologica. 1993;9:63–68. [PubMed] [Google Scholar]
- 5.Eldar A, Ghittino C, Asanta L, Bozzetta E, Goria M, Prearo M, Bercovier H. Enterococcus seriolicida is a junior synonym of Lactococcus garvieae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol. 1996;32:85–88. doi: 10.1007/s002849900015. [DOI] [PubMed] [Google Scholar]
- 6.Elliott J A, Collins M D, Pigott N E, Facklam R R. Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole-cell protein patterns. J Clin Microbiol. 1991;29:2731–2734. doi: 10.1128/jcm.29.12.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliot J A, Facklam R R. Antimicrobial susceptibilities of Lactococcus lactis and Lactococcus garvieae and a proposed method to discriminate between them. J Clin Microbiol. 1996;34:1296–1298. doi: 10.1128/jcm.34.5.1296-1298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facklam R R, Hollis D, Collins M D. Identification of gram-positive coccal and coccobacillary vancomycin-resistant bacteria. J Clin Microbiol. 1989;27:724–730. doi: 10.1128/jcm.27.4.724-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghittino C, Prearo M. Comparison of some strains isolated from rainbow trout affected by streptococcosis. Boll Soc Ital Patol Ittica. 1993;11:30–43. [Google Scholar]
- 10.Holt J G, Krieg N R, Sneath P H A, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. pp. 527–558. [Google Scholar]
- 11.Kitao T. Streptococcal infections. In: Inglis V, Roberts R J, Bromage N R, editors. Bacterial diseases of fish. Oxford, United Kingdom: Blackwell Scientific Publications; 1993. pp. 196–210. [Google Scholar]
- 12.Kusuda K, Kawai K, Salati F, Banner C R, Freyer J L. Enterococcus seriolicida sp. nov., a fish pathogen. Int J Syst Bacteriol. 1991;41:406–409. doi: 10.1099/00207713-41-3-406. [DOI] [PubMed] [Google Scholar]
- 13.Mannion P T, Rothburn M M. Diagnosis of bacterial endocarditis caused by Streptococcus lactis and assisted by immunoblotting of serum antibodies. J Infect. 1990;21:317–318. doi: 10.1016/0163-4453(90)94149-t. [DOI] [PubMed] [Google Scholar]