Abstract
CHROMagar Orientation, a new chromogenic medium, was evaluated for the detection and differentiation of gram-positive and gram-negative pathogenic microorganisms in 900 urine samples from hospitalized patients. Performance characteristics of the medium were evaluated in comparison to those of 5% sheep blood and MacConkey agars by direct inoculation of the urine samples on the three media. Four gram-negative and two gram-positive strains as well as one yeast control strain from the American Type Culture Collection were used to ensure quality control. CHROMagar Orientation succeeded in detecting all the urine pathogens that were detected by the reference media, including gram-negative bacilli, staphylococci, streptococci, and yeasts. Colony color and morphology on CHROMagar Orientation accurately differentiated Escherichia coli, Proteus mirabilis, Proteus vulgaris, Pseudomonas aeruginosa, and Acinetobacter spp. Owing to the similarity in the pigmentation produced by Klebsiella, Enterobacter, and Citrobacter isolates, the medium failed to distinguish among them; however, these isolates were easily recognized as coliforms because of their metallic blue coloration. Staphylococci were clearly perceptible: S. aureus and S. epidermidis grow in regular-size colonies that range from opaque white to yellowish, and S. saprophyticus produces opaque pink colonies. All streptococcus strains, including those from groups B and C, were detected. They grow as undifferentiated flat dry diffused colonies, and additional tests were required for identification. Enterococci were easily discriminated by their strong turquoise pigmentation and their typical growth on the agar’s surface. Yeast grow in typical creamy wet convex colonies. The accuracy of antibiotic susceptibility determinations according to standard methods was also tested by picking isolates directly from CHROMagar Orientation. The results showed excellent correlation with those obtained with microorganisms picked from reference media. Owing to the ease in differentiating mixed flora on CHROMagar Orientation, antimicrobic susceptibility tests were performed directly from primary isolates in all cases without the need for subcultures.
Urinary tract infections (UTI) continue to be a common problem (13). The increase in resistance of microorganisms to antimicrobic agents, especially in hospitalized patients, demands rapid identification of the pathogen (1, 2, 10, 16). Early information enables the selection of the appropriate antibiotic prior to the results of susceptibility tests and may thereby prevent outbreaks (15). For many years blood, cystine lactose electrolyte-deficient, and MacConkey agars have been used for the detection of urinary tract pathogens, as well as for the differentiation of a few of them. In the last few years several chromogenic media have been developed and commercialized, allowing for more specific direct differentiation of microorganisms on primary plates (9, 14). A new one, CHROMagar Orientation, offers simultaneous presumptive identification of gram-positive and gram-negative bacteria and yeasts on a single medium by means of distinct colony colors produced by reactions of genus- or species-specific enzymes with a suitable chromogenic substrate (11).
The aim of this study was to evaluate the sensitivity of this medium and its ability to differentiate urinary pathogens. The accuracy of antimicrobic susceptibility testing by standard methods was also tested by picking isolates directly from CHROMagar Orientation.
MATERIALS AND METHODS
Study population and specimens.
Patients hospitalized at Rabin Medical Center, Petah Tiqva, Israel, were included in the survey. Rabin Medical Center is a 1,000-bed university hospital in central Israel serving a mainly urban population, and it is a reference hospital for other hospitals in the area. Nine hundred urine samples from patients in different departments were tested in this study.
Media. (i) CHROMagar Orientation.
The principle of this medium is the use of chromogenic substrates revealing metabolic enzymes. Dehydrated powder was provided by the CHROMagar Company, Paris, France. The medium is composed of 16 g each of peptone, meat, and yeast extracts and 15 g of agar per liter and a special chromogenic mixture. The medium was prepared by Hy-Laboratories, Rehovot, Israel, according to the manufacturer’s instructions and Hy-Laboratories’ good-manufacturing procedures. The powder was introduced into an automatic preparator, and the sterilization process was performed at 120°C for 15 min. The medium was poured into 90-mm-diameter petri dishes, stored at 4 to 6°C, protected from light, and used within 10 weeks.
(ii) Standard reference media.
Standard reference media consisted of tryptic soy agar no. 2 (TSA) with 5% defibrinated sheep blood and MacConkey and Mueller-Hinton agars. All were from Difco, and all were prepared according to the manufacturers’ directions and dispensed into 90-mm-diameter petri dishes.
Quality control.
Each batch of medium used in this trial was tested for sterility, culture response to a minimum inoculum, and biochemical and chromogenic reactions with American Type Culture Collection (ATCC) strains according to directions from the Hy-Laboratories quality control manual. Quality control testing of CHROMagar Orientation against the reference medium TSA plus 5% sheep blood was performed by comparing bacterial counts from an estimated inoculum of 102 CFU per ml.
ATCC control strains.
Staphylococcus aureus ATCC 25923, Enterococcus faecalis ATCC 19433, Escherichia coli ATCC 25922, Proteus mirabilis ATCC 4630, Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumoniae 13883, and Candida albicans ATCC 10231 were used for quality control. The microorganisms were inoculated into tryptic soy broth (Difco) and incubated overnight at 35 ± 2°C. Serial 10-fold dilutions in sterile saline were performed to reduce the microbial count to the desired inoculum concentration. The number of viable CFU per milliliter in each suspension was monitored in duplicate by the plate count method on TSA.
Bacteriological procedures.
CHROMagar Orientation was evaluated in comparison to standard reference media: TSA plus 5% sheep blood and MacConkey agar plates. The urine samples were inoculated at the same time on the three agars with a calibrated 10-μl loop and were incubated aerobically at 35 ± 2°C overnight or for 48 h on weekends. The antimicrobic susceptibilities of the isolates were tested by the disk diffusion technique according to National Committee for Clinical Laboratory Standards (NCCLS) recommendations (3, 12). The accuracy of antimicrobic susceptibility testing was evaluated by picking isolates directly from CHROMagar Orientation to Mueller-Hinton agar and comparing the results with those from parallel tests of isolates picked from reference media; the gram-positive bacteria were picked from 5% sheep blood agar, and the gram-negative bacteria were picked from MacConkey agar.
Microorganism identification.
Enterobacteriaceae isolates were identified by the following biochemical reactions: motility, indole production, o-nitrophenyl-β-d-galactopyranoside hydrolysis, glucose fermentation with or without CO2 production, hydrogen sulfide production, urea hydrolysis, and lysine and ornithine decarboxylase and sodium citrate utilization. Gram-negative microorganisms other than Enterobacteriaceae were also tested for colony morphology and pigmentation as well as for additional biochemical reactions: gelatin, catalase, and oxidase utilization.
The identification of streptococci was confirmed by hemolysis on 5% sheep blood agar, hydrolysis of l-pyrrolindonyl-β-naphthylamide substrate by PYRase, aesculin hydrolysis, and agglutination tests. Isolates suspected to be S. aureus (lack of growth on MacConkey agar, growth of beta-hemolytic colonies on blood agar, and white-to-yellowish colonies on CHROMagar Orientation) were Gram stained and checked by the slide coagulase test for final identification.
Candida isolates were subcultured on CHROMagar Candida (CHROMagar), a medium allowing the identification of C. albicans, C. tropicalis, and C. krusei by their different colony colors.
RESULTS
Quality control assay results of bacterial counting and color and colony morphology determinations for ATCC strains on CHROMagar Orientation are given in Table 1. Colony count results on CHROMagar Orientation and on blood agar showed excellent correlation. No significant difference between the results of any CHROMagar Orientation batch and those of reference media were found for any bacteria (P > 0.05; paired t test). The colony morphology and pigmentation results for microorganisms were consistent among all seven batches used in this trial. Of the 900 urine samples assayed, 190 were found to be positive. A single species was isolated in 176 samples; two species were isolated in 12 samples, and three species were isolated in 2 samples.
TABLE 1.
Quality control testing of CHROMagar Orientation used for the evaluation
| Test microorganism | Mean colony count (CFU) ± SD with:
|
Pa | |
|---|---|---|---|
| Reference mediumb | CHROMagar Orientationc | ||
| E. coli ATCC 25922 | 174 ± 98 | 166 ± 47 | 0.511 |
| K. pneumoniae ATCC 13883 | 210 ± 77 | 230 ± 99 | 0.747 |
| P. aeruginosa ATCC 27853 | 138 ± 15 | 128 ± 11 | 0.084 |
| P. mirabilis ATCC 4630 | 110 ± 34 | 128 ± 43 | 0.132 |
| S. aureus ATCC 25923 | 129 ± 69 | 132 ± 68 | 0.219 |
| E. faecalis ATCC 19433 | 168 ± 69 | 174 ± 75 | 0.468 |
No significant differences were found between CHROMagar Orientation and the reference medium (P > 0.05 [paired t test]).
The reference medium was TSA plus 5% defibrinated sheep blood. The inoculum was diluted to an estimated concentration of 102 CFU/ml. Four batches were tested.
The inoculum was diluted to a concentration of 102 CFU/ml. Seven batches were tested.
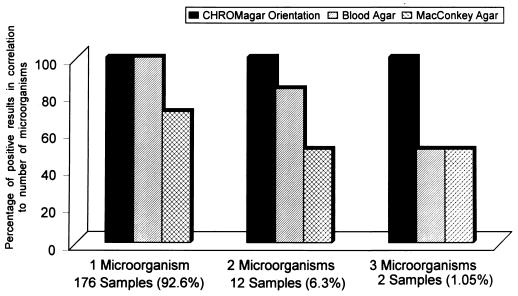
The colony characteristics of the different microorganisms detected in the trial are described in Table 2 and are shown in Fig. 1. E. coli, Proteus spp., and enterococci grow on this medium in typical differentiated colonies. Acinetobacter spp. were also easily differentiated and distinguished from Pseudomonas isolates. The similarity of colors produced by Klebsiella, Enterobacter, and Citrobacter spp. prevents differentiation among them, and additional biochemical tests were required for final identification. The distribution of the different urine pathogens among the 190 positive urine samples is given in Table 3. The ability of the media to detect single or multiple species is given in Fig. 2. The results showed that overnight incubation is optimal for the growth response of microorganisms on CHROMagar Orientation (data not shown). Longer incubation of up to 72 h confirmed the results and deepened the colony colors.
TABLE 2.
Urine isolates presumptively identified on CHROMagar Orientation according to pigment reaction
| Organism | Morphology and/or color (18–24-h incubation) |
|---|---|
| E. coli | Small, pink-red |
| K. pneumoniae | Mucoid,a metallic blue |
| Citrobacter freundii | Metallic blueb |
| Enterobacter sp. | Metallic blue |
| P. mirabilis | Clear diffusible beige on beige background |
| M. morganii | Clear diffusible beige on beige background |
| P. aeruginosa | Transparent, yellow serrated edges, diffusec |
| Acinetobacter sp. | Nontransparent, entire edges, white |
| Enterococcus sp. | Dry, turquoise |
| Streptococcus sp. | Small, translucent; diffuse light blue within agar |
| Staphylococcus sp. | Opaque, white to yellowish |
| S. saprophyticus | Pink, opaque |
| Lactobacillus sp. | Scanty, light blue within agar |
| Corynebacterium sp. | Colorless, small, undifferentiated |
| Candida sp. | Creamy, wet convex |
| Torulopsis glabrata | Creamy, very small, indistinct |
Slight pink halo around the periphery after 24 to 36 h.
Strong purple-pink halo (diffuse) after 24 to 36 h.
Green after 24 to 36 h.
FIG. 1.
Specific color reactions of microorganisms on CHROMagar Orientation. 1, P. mirabilis; 2, E. faecalis; 3, K. pneumoniae; 4, P. aeruginosa; 5, E. coli; 6, S. aureus.
TABLE 3.
Distribution of isolates among positive urine specimens
| Microorganism | No. of isolates on:
|
|||||
|---|---|---|---|---|---|---|
| CHROMagar Orientationd
|
Blood agard
|
MacConkey agar
|
||||
| Total | Pure | Total | Pure | Total | Pure | |
| E. coli | 87 | 73 | 85 | 73 | 87 | 73 |
| Enterococcus sp. | 30 | 28 | 30a | 28a | ||
| Proteus sp. | 13 | 8 | 13 | 8 | 13 | 8 |
| M. morganii | 4 | 2 | 4 | 2 | 4 | 2 |
| K. pneumoniaeb | 16 | 15 | 16 | 15 | 14 | 13 |
| Enterobacter sp.b | 5 | 2 | 5 | 2 | 2 | 2 |
| P. aeruginosac | 20 | 20 | 20 | 20 | 20 | 20 |
| Citrobacter sp.b | 3 | 2 | 2 | 2 | 3 | 2 |
| Acinetobacter sp. | 4 | 4 | 4 | 4 | 4 | 4 |
| Staphylococcus sp. | 11 | 9 | 11 | 9 | ||
| Streptococcus group B sp. | 5 | 5 | 5 | 5 | ||
| Streptococcus group C sp. | 1 | 1 | 1 | 1 | ||
| Candida sp. | 7 | 7 | 7 | 7 | ||
| Total | 206 | 176 | 203 | 176 | 147 | 124 |
For 8 of the 30 isolates, additional tests were necessary to confirm the identification.
Additional tests were required for final identification.
For 1 of the 20 samples, only a few colonies were detected.
No significant difference (McNemar test; P = 1).
FIG. 2.
Abilities of different media to detect microorganisms in positive urine samples (24-h incubation).
Table 3 and Fig. 2 show that the ability of CHROMagar Orientation to detect urine pathogens is equal to that of the combination of the two reference media (TSA plus 5% sheep blood and MacConkey agar), according to the McNemar test (P = 1).
Due to its specificity with respect to color and colony morphology, CHROMagar Orientation made the differentiation of bacterial colonies than did the reference media. This fact, together with the ability of the medium to limit the spread of bacteria, allowed the presumptive identification of several microorganisms directly from the primary plates, as well as allowing the performance of antimicrobic susceptibility tests without the need for subculturing (Table 3), even when one of the isolates was a Proteus sp. Equivalent susceptibilities were obtained in all the cases, and very few differences between zone diameters of 1 to 2 mm were detected randomly. Note that none of these differences were out of the range specified by NCCLS criteria for the disk diffusion method of susceptibility testing (Tables 4 and 5). The numbers of the susceptible isolates picked from CHROMagar Orientation were exactly the same as the numbers of those picked from the reference media. No significant differences (paired t test) between the numbers of intermediate (P = 0.7725) and resistant (P = 0.7168) gram-negative isolates were observed (Table 4). For gram-positive bacteria P = 0.3306 for both intermediate and resistant isolates (Table 5).
TABLE 4.
Susceptibility results for 152 gram-negative isolates according to the NCCLS criteria
| Antimicrobic agent (disk content [μg]) | No. of isolates with indicated resulta picked from:
|
|||||
|---|---|---|---|---|---|---|
| CHROMagar Orientation
|
MacConkey agar
|
|||||
| S | I | R | S | I | R | |
| Ampicillin (10) | 41 | 1 | 110 | 41 | 2 | 109 |
| Amoxicillin (20) + clavulanic acid (10) | 98 | 3 | 51 | 98 | 4 | 50 |
| Cefotaxime (30) | 108 | 3 | 41 | 108 | 2 | 42 |
| Ceftriaxone (30) | 107 | 3 | 42 | 107 | 3 | 42 |
| Cefuroxime (30) | 105 | 1 | 46 | 105 | 1 | 46 |
| Ceftazidime (30) | 137 | 2 | 13 | 137 | 1 | 14 |
| Cefonicid (30) | 99 | 0 | 53 | 99 | 0 | 53 |
| Gentamicin (10) | 119 | 2 | 31 | 119 | 2 | 31 |
| Amikacin (30) | 138 | 1 | 13 | 138 | 2 | 12 |
| Ofloxacin (10) | 129 | 0 | 23 | 129 | 0 | 23 |
| Ciprofloxacin (5) | 125 | 0 | 27 | 125 | 0 | 27 |
| Imipenem (10) | 149 | 2 | 1 | 149 | 2 | 1 |
| Aztreonam (30) | 134 | 1 | 17 | 134 | 1 | 17 |
| Minocycline (30) | 83 | 10 | 59 | 83 | 11 | 58 |
| Piperacillin (100) | 82 | 15 | 55 | 82 | 14 | 56 |
| Nitrofurantoin (30) | 75 | 24 | 53 | 75 | 25 | 52 |
| Nalidixic acid (30) | 90 | 0 | 62 | 90 | 0 | 62 |
| Sulfamethoxazole (23.75) + trimethoprim (1.25) | 78 | 0 | 74 | 78 | 0 | 74 |
S, susceptible; I, intermediate; R, resistant. Paired t test results: for intermediate isolates, P = 0.7725; for resistant isolates, P = 0.7168.
TABLE 5.
Susceptibility results for 47 gram-positive isolates according to the NCCLS criteria
| Antimicrobic agent (disk content) | No. of isolates with indicated resulta picked from:
|
|||||
|---|---|---|---|---|---|---|
| CHROMagar Orientation
|
Blood agar
|
|||||
| S | I | R | S | I | R | |
| Penicillin G (10 IU) | 20 | 2 | 25 | 20 | 1 | 26 |
| Ampicillin (10 μg) | 39 | 0 | 8 | 39 | 0 | 8 |
| Amoxicillin (20 μg) + clavulanic acid (10 μg) | 44 | 0 | 3 | 44 | 0 | 3 |
| Methicillin (5 μg) | 10 | 0 | 37 | 10 | 0 | 37 |
| Cefotaxime (30 μg) | 10 | 2 | 35 | 10 | 3 | 34 |
| Ceftriaxone (30 μg) | 10 | 2 | 35 | 10 | 2 | 35 |
| Cefuroxime (30 μg) | 10 | 4 | 33 | 10 | 4 | 33 |
| Cephalothin (30 μg) | 14 | 2 | 29 | 14 | 3 | 30 |
| Amikacin (30 μg) | 12 | 0 | 35 | 12 | 0 | 35 |
| Gentamicin (10 μg) | 6 | 0 | 41 | 6 | 0 | 41 |
| Erythromycin (15 μg) | 18 | 0 | 29 | 18 | 0 | 29 |
| Vancomycin (30 μg) | 45 | 0 | 2 | 45 | 0 | 2 |
| Clindamycin (2 μg) | 14 | 0 | 33 | 14 | 0 | 33 |
| Rifampin (2 μg) | 20 | 2 | 25 | 20 | 2 | 25 |
| Ofloxacin (10 μg) | 14 | 0 | 33 | 14 | 0 | 33 |
| Nitrofurantoin (30 μg) | 41 | 0 | 6 | 41 | 0 | 6 |
| Fusidic acid (10 μg) | 13 | 0 | 34 | 13 | 0 | 34 |
| Chloramphenicol (30 μg) | 26 | 6 | 15 | 26 | 6 | 15 |
| Sulfamethoxazole (23.75 μg) + trimethoprim (1.25 μg) | 14 | 0 | 33 | 14 | 0 | 33 |
S, susceptible; I, intermediate; R, resistant. Paired t test result: P = 0.3306 for intermediate and resistant isolates.
DISCUSSION
In the present study, CHROMagar Orientation was evaluated for the first time as a direct isolation medium for clinical specimens. Nine hundred urine samples were tested by parallel inoculation on CHROMagar Orientation and on two reference media, TSA plus 5% sheep blood and MacConkey agar.
CHROMagar Orientation showed the same ability to detect urine pathogens as the combination of the two reference media. The results show that the growth factors included in the formula supported the growth of all UTI pathogens, even those nutritionally dependent, which required routine enriched media (blood agar). CHROMagar Orientation offered the advantage of limiting the spread of some isolates, such as Proteus spp., K. pneumoniae, and E. coli mucoid strains, which may yield confluent growth on plates. This increased the ability of the medium to detect urinary tract pathogens when mixed flora were present (11).
Color and morphology characteristics on CHROMagar Orientation allowed for easy differentiation of the bacterial colonies.
The results of this trial showed that, among the cases studied, about 70% of the UTI were caused by gram-negative pathogens, 26% were caused by gram-positive pathogens, and the remaining 4% were caused by fungi. These results correlate with other data reported recently (8). Among the gram-negative isolates, E. coli was the predominant species (65%). All these isolates grew on CHROMagar Orientation in reddish colonies and were very easy to distinguish. Since E. coli is responsible for many of the UTI in nosocomial patients (6, 7), CHROMagar Orientation seems to be very suitable as a differential medium for direct isolation of urine samples.
The medium failed to differentiate among Klebsiella, Enterobacter, and Citrobacter owing to the similarity of color produced. However, these isolates were distinguished as coliforms among other gram-negative bacteria. Their final identification required additional biochemical tests (11).
Proteus spp. were also easily distinguished on the primary plates because of their characteristic brown colonies on a diffuse beige background. Proteus spp. are important pathogens in patients with indwelling urinary catheters (4, 5). The differentiation between P. mirabilis and Morganella morganii was possible only after the indole test was performed.
Acinetobacter spp. should be added to the list of gram-negative microorganisms that can be presumptively differentiated directly on CHROMagar Orientation. They grew in nontransparent, white, entire-edge colonies. These strains were very distinct from Pseudomonas isolates, which grew in diffuse, yellow-to-green colonies with serrated edges.
The results of the trial to differentiate the most commonly encountered gram-negative pathogens in UTI on the basis of color and morphology alone were favorable for CHROMagar Orientation compared to MacConkey agar.
CHROMagar Orientation succeeded in detecting all yeasts and gram-positive microorganisms that grew on 5% sheep blood agar, including group B and C streptococci. Both pathogens grew in small, translucent, diffuse light blue colonies within agar.
Enterococci, one of the most commonly encountered gram-positive pathogens in UTI, were easily distinguished on CHROMagar Orientation by their typical growth in turquoise colonies on agar. The ability of the medium to prevent the spread of Proteus spp., the greater differentiation among gram-negative bacteria, and the ease of distinguishing enterococci allowed for the performance of direct biochemical and antimicrobic susceptibility tests without the need for subcultures when multiple probable pathogens were present.
The results of the antimicrobial susceptibility tests of microorganisms picked from CHROMagar Orientation showed an excellent correlation with test results of microorganisms picked from reference media.
In summary, CHROMagar Orientation is recommended as a single medium for direct isolation and presumptive identification of UTI pathogens. The use of this medium for other clinical specimens requires further evaluation.
REFERENCES
- 1.Alon U, Davidai G, Berant M, Merzbach D. Five-year survey of changing patterns of susceptibility of bacterial uropathogens to trimethoprim-sulfamethoxazole and other antimicrobial agents. Antimicrob Agents Chemother. 1987;31:126–128. doi: 10.1128/aac.31.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashkenazi S, Even-Tov S, Samra Z, Dinari G. Uropathogens of various childhood populations and their antibiotic susceptibility. Pediatr Infect Dis J. 1991;10:742–746. doi: 10.1097/00006454-199110000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Bauer A M, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized simple disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Daifuku R, Stamm W E. Association of rectal and urethral colonization with urinary tract infection in patients with indwelling catheters. JAMA. 1984;252:2028–2030. [PubMed] [Google Scholar]
- 5.Damron D J, Warren J W, Chippendale G R, Fenney J H. Do clinical microbiology laboratories report complete bacteriology in urine from patients with long-term urinary catheters? J Clin Microbiol. 1986;24:400–404. doi: 10.1128/jcm.24.3.400-404.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edberg S C, Kontnick C M. Comparison of β-glucoronidase-based substrate systems for identification of Escherichia coli. J Clin Microbiol. 1986;24:368–371. doi: 10.1128/jcm.24.3.368-371.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edberg S C, Trepeta R W. Rapid and economical identification and antimicrobial susceptibility test methodology for urinary tract pathogens. J Clin Microbiol. 1983;18:1287–1291. doi: 10.1128/jcm.18.6.1287-1291.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emori T G, Gaynes R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freydiere A M, Gille Y. Yeast: appearance of colonies on CHROMagar Candida. Presented at IVMS Congress 94, 7th International Congress of Mycology Division, Prague, Czech Republic. 1994. [Google Scholar]
- 10.Marray B E. Problems and mechanisms of antimicrobial resistance. Infect Dis Clin North Am. 1990;3:423–439. [PubMed] [Google Scholar]
- 11.Merlino J, Siarakas S, Robertson G J, Funnell G R, Gottlieb T, Bradbury R. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J Clin Microbiol. 1996;34:1788–1793. doi: 10.1128/jcm.34.7.1788-1793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobic disc susceptibility tests. Approved standard ASM-2. Villanova, PA: National Committee for Clinical Laboratory Standards; 1979. [Google Scholar]
- 13.Neu, C. H. 1985. Infections due to gram negative bacteria: an overview, session VIII. Rev. Infect. Dis. 7(Suppl. 4):S778–S782. [DOI] [PubMed]
- 14.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trend in the microbiology and etiology of nosocomial infections. Am. J. Med. 91(Suppl. 3B):72S–75S. [DOI] [PubMed]
- 16.Slak, R. 1984. Review of bacterial resistance: a challenge to the treatment of urinary infection. J. Antimicrob. Chemother. 13(Suppl. B):1–7. [DOI] [PubMed]