Background
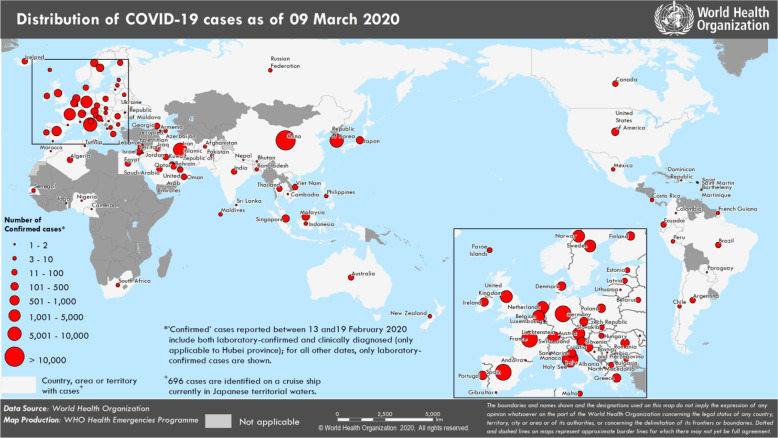
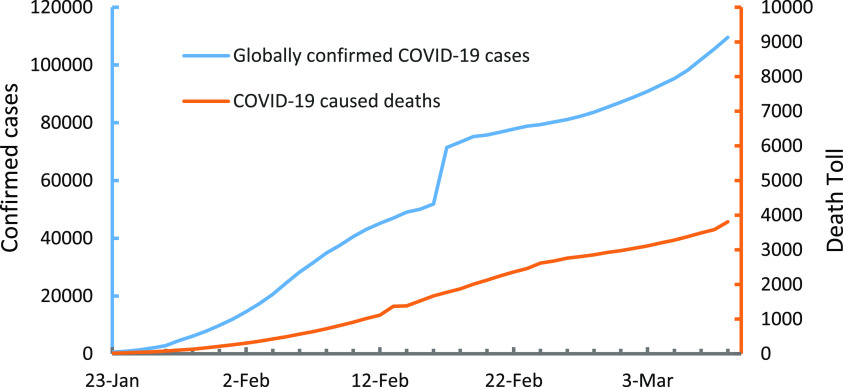
The outbreak of the novel coronavirus disease, COVID-19, caused by the new coronavirus 2019-nCoV that is now officially designated as severe acute respiratory syndrome-related coronavirus SARS-CoV-2, represents a pandemic threat to global public health.1,2 Although the epicenter of the COVID-19 outbreak in December of 2019 was located in Wuhan, China, this disease has spread to more than 100 countries (Figure 1) with over 100 000 confirmed cases and over 3,800 confirmed deaths worldwide (Figure 2) as of March 9, 2020.3 In addition, millions of people’s lives have been affected as a result of mandatory isolations/quarantines. The ripple effect of the COVID-19 outbreak could potentially bring major challenges to worldwide health systems and have far-reaching consequences on the global economy if the spread of the virus is not effectively controlled.1,2,4
Figure 1.
Global distribution of confirmed COVID-19 cases. (Map was reproduced from WHO Coronavirus Disease (COVID-2019) Situation Reports.3 Used with permission from ref (3). Copyright 2020 World Health Organization.)
Figure 2.
Global trend of confirmed COVID-19 cases and associated deaths from January 23 through March 9, 2020. (Data were obtained from WHO Coronavirus Disease (COVID-2019) Situation Reports3).
Coronaviruses (CoVs) are relatively large viruses containing a single-stranded positive-sense RNA genome encapsulated within a membrane envelope. The viral membrane is studded with glycoprotein spikes that give coronaviruses their crown-like appearance (Figure 3). While coronaviruses infect both humans and animals, certain types of animals such as bats that host the largest variety of coronaviruses appear to be immune to coronavirus-induced illness.5 There are four classes of coronaviruses designated as alpha, beta, gamma, and delta. The betacoronavirus class includes severe acute respiratory syndrome (SARS) virus (SARS-CoV), Middle East respiratory syndrome (MERS) virus (MERS-CoV), and the COVID-19 causative agent SARS-CoV-2. Similar to SARS-CoV and MERS-CoV, SARS-CoV-2 attacks the lower respiratory system to cause viral pneumonia, but it may also affect the gastrointestinal system, heart, kidney, liver, and central nervous system leading to multiple organ failure.6,7 Current information indicates that SARS-CoV-2 is more transmissible/contagious than SARS-CoV.8
Figure 3.
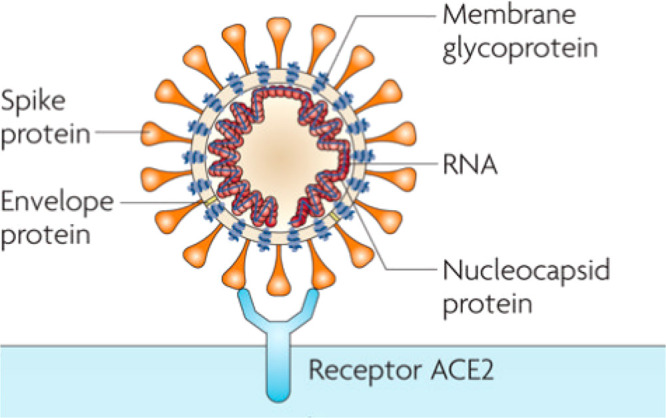
Cartoon illustration of the coronavirus structure and viral receptor ACE2 on the host cell surface. (Image was reproduced with permission from ref (9), Nature Reviews Microbiology 7(3), 226–236. Copyright 2009 Springer Nature.)
The betacoronavirus genome encodes several structural proteins, including the glycosylated spike (S) protein that functions as a major inducer of host immune responses. This S protein mediates host cell invasion by both SARS-CoV and SARS-CoV-2 via binding to a receptor protein called angiotensin-converting enzyme 2 (ACE2) located on the surface membrane of host cells.9−11 A recent study also revealed that this invasion process requires S protein priming which is facilitated by the host cell-produced serine protease TMPRSS211. In addition, the viral genome also encodes several nonstructural proteins including RNA-dependent RNA polymerase (RdRp), coronavirus main protease (3CLpro), and papain-like protease (PLpro).12,13 Upon entrance to the host cells, the viral genome is released as a single-stranded positive RNA. Subsequently, it is translated into viral polyproteins using host cell protein translation machinery, which are then cleaved into effector proteins by viral proteinases 3CLpro and PLpro.12,13 PLpro also behaves as a deubiquitinase that may deubiquinate certain host cell proteins, including interferon factor 3 and NF-κB, resulting in immune suppression.13,14 RdRp synthesizes a full-length negative-strand RNA template to be used by RdRp to make more viral genomic RNA.
The interaction between viral S protein and ACE2 on the host cell surface is of significant interest since it initiates the infection process. Cryo-EM structure analysis has revealed that the binding affinity of SARS-CoV-2 S protein to ACE2 is about 10–20 times higher than that of SARS-CoV S protein.10,15 It is speculated that this may contribute to the reported higher transmissibility and contagiousness of SARS-CoV-2 as compared to SARS-CoV.8
The prospect also exists for discovery of therapeutic agents targeting the highly conserved proteins associated with both SARS-CoV and SARS-CoV-2.15−18 RdRp and 3CLpro protease of SARS-CoV-2 share over 95% of sequence similarity with those of SARS-CoV despite the fact that these two viruses demonstrate only 79% sequence similarity at the genome level.15−18 On the basis of sequence alignment and homology modeling, SARS-CoV and SARS-CoV-2 share a highly conserved receptor-binding domain (RBD), a domain of S protein, and 76% of sequence similarity in their S proteins.15−18 In addition, although the PLpro sequences of SARS-CoV-2 and SARS-CoV are only 83% similar, they share similar active sites.16
To date, there are no SARS-CoV-2-specific antiviral agents. Researchers have been racing to find possible treatments to save lives and produce vaccines for future prevention. To support research and development efforts to discover effective therapeutic and preventive agents for COVID-19, CAS, a division of the American Chemical Society specializing in scientific information solutions, has analyzed scientific data related to the development of therapeutic agents and vaccines for human coronaviruses since 2003. The analyses presented in this report are based on the CAS content collection, a scientist-curated data collection covering published scientific literature and patents from over 60 patent authorities worldwide. For a subset of the analyses, both CAS and MEDLINE data were collectively analyzed.
Scientific Literature and Patents Related to COVID-19, SARS, and MERS
Trend in Scientific Publications Related to COVID-19
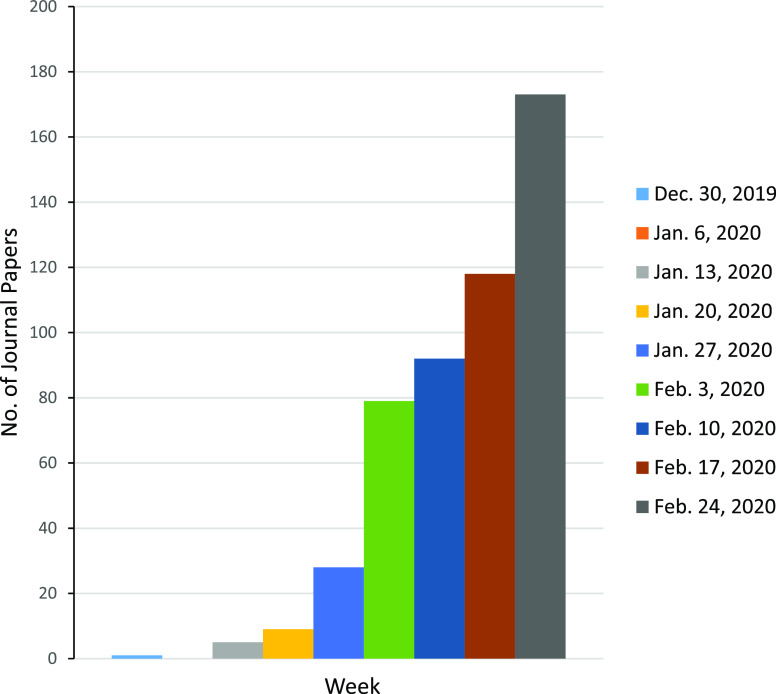
Since the outbreak of COVID-19, this new disease and its causative virus have drawn major global attention. Scientists and physicians worldwide have been conducting a major campaign to understand this new emergent disease and its epidemiology in an effort to uncover possible treatment regimens, discover effective therapeutic agents, and develop vaccines. Figure 4 shows the total number of journal articles related to COVID-19 or SARS-CoV-2 published each week from the last week of 2019 through the week of February 24, 2020. Over 500 journal articles were published electronically or in print during this period, and the number of published articles has increased each week since the week of January 13, 2020. Although a large portion of these articles are about clinical manifestations and treatment options, an increasing number of studies are focused on elucidation of virus structure, virus transmission mechanisms/dynamics, as well as identification of antiviral agents and accurate diagnostics for virus detection. These trends reflect immense interest and desire from the scientific community, including both academic and industrial organizations as well as clinicians, to identify new methods to halt the progression of this epidemic disease and to prevent infection and transmission in the future.
Figure 4.
Number of journal articles related to COVID-19 published each week.
Notable Journal Articles Related to COVID-19 and SARS-CoV-2
Table 1 lists some journal articles published from December 30, 2019 through February 23, 2020. These articles were selected based on collective use of factors such as journal impact factor, citation, and type of study. For example, the No. 8 article listed about the characterization of the SARS-CoV-2 genome has greatly facilitated the global effort to develop a vaccine for prevention of COVID-19. Also shown in this table are journal articles pertaining to potential antiviral drug candidates such as remdesivir, baricitinib, and chloroquine for the treatment of this disease.
Table 1. Notable Journal Articles on COVID-19 and/or SARS-CoV-2 Published as of February 23, 2020a.
| no. | journal | paper title | publication date | organization |
|---|---|---|---|---|
| 1 | The New England Journal of Medicine | A novel coronavirus from patients with pneumonia in China, 2019 | January 24, 2020 | NHC Key Laboratory of Biosafety, China, and National Institute for Viral Disease Control, Chinese Center for Disease Control and Prevention, Beijing, Chinab |
| 2 | Lancet | Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China | January 24, 2020 | Department of Pulmonary and Critical Care Medicine, China-Japan Friendship Hospital, Beijing, China; NHC Key Laboratory of Systems Biology of Pathogens and Christophe Merieux Laboratory, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, Chinab |
| 3 | The New England Journal of Medicine | Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia | January 29, 2020 | Chinese Center for Disease Control and Prevention, Beijing, China; School of Public Health, University of Hong Kong, Hong Kong; Hubei Center for Disease Control and Prevention, Wuhan, Hubei, Chinab |
| 5 | Journal of Virology | Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS | January 29, 2020 | Department of Epidemiology, University of North Carolina, Chapel Hill, NC, USA |
| 6 | Lancet | Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study | January 30, 2020 | Tuberculosis and Respiratory Department, Wuhan Jinyintan Hospital, Wuhan, China |
| 7 | The New England Journal of Medicine | First case of 2019 novel coronavirus in the United States | January 31, 2020 | The Washington State Department of Health Public Health Laboratories, WA, USAb |
| 8 | Lancet | Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding | January 30, 2020 | NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China, Central Theater, People’s Liberation Army General Hospital, Wuhan, China, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Beijing, Chinab |
| 9 | Lancet | Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study | January 31, 2020 | School of Public Health, Li Ka Shing Faculty of Medicine, University of Hong Kong, Hong Kong, Chinab |
| 10 | Nature | A new coronavirus associated with human respiratory disease in China | February 3, 2020 | Shanghai Public Health Clinical Center & School of Public Health, Fudan University, Shanghai, Chinab |
| 11 | Nature | A pneumonia outbreak associated with a new coronavirus of probable bat origin | February 3, 2020 | Key Laboratory of Special Pathogens, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan, Chinab |
| 12 | Lancet | Baricitinib as potential treatment for 2019-nCoV acute respiratory disease | February 4, 2020 | BenevolentAI, London, UK and Department of Surgery and Cancer, Imperial College London, UK |
| 13 | Cell Research | Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro | February 4, 2020 | State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan, China, and National Engineering Research Center for the Emergency Drug, Beijing Institute of Pharmacology and Toxicology, Beijing, Chinab |
| 14 | Emerging Microbes & Infections | RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak | February 5, 2020 | State Key Laboratory of Virology, Modern Virology Research Center, College of Life Sciences, Wuhan University, Wuhan, Chinab |
| 15 | The Journal of the American Medical Association | Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China | February 7, 2020 | Department of Critical Care Medicine, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China |
| 16 | Cell Host & Microbe | Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China | February 7, 2020 | National Institute for Viral Disease Control and Prevention, China CDC, Beijing, China; Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, USA; Center for Systems Medicine, Institute of Basic Medical Sciences & Peking Union Medical College, Beijing, Chinab |
| 17 | Cellular & Molecular Immunology | Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein | February 11, 2020 | Key Laboratory of Medical Molecular Virology, School of Basic Medical Sciences, Fudan-Jinbo Joint Research Center, Fudan University, Shanghai, China |
Note: The publication date is the date for electronic publication.
Only corresponding organization(s) is/are listed for papers published by multiple organizations.
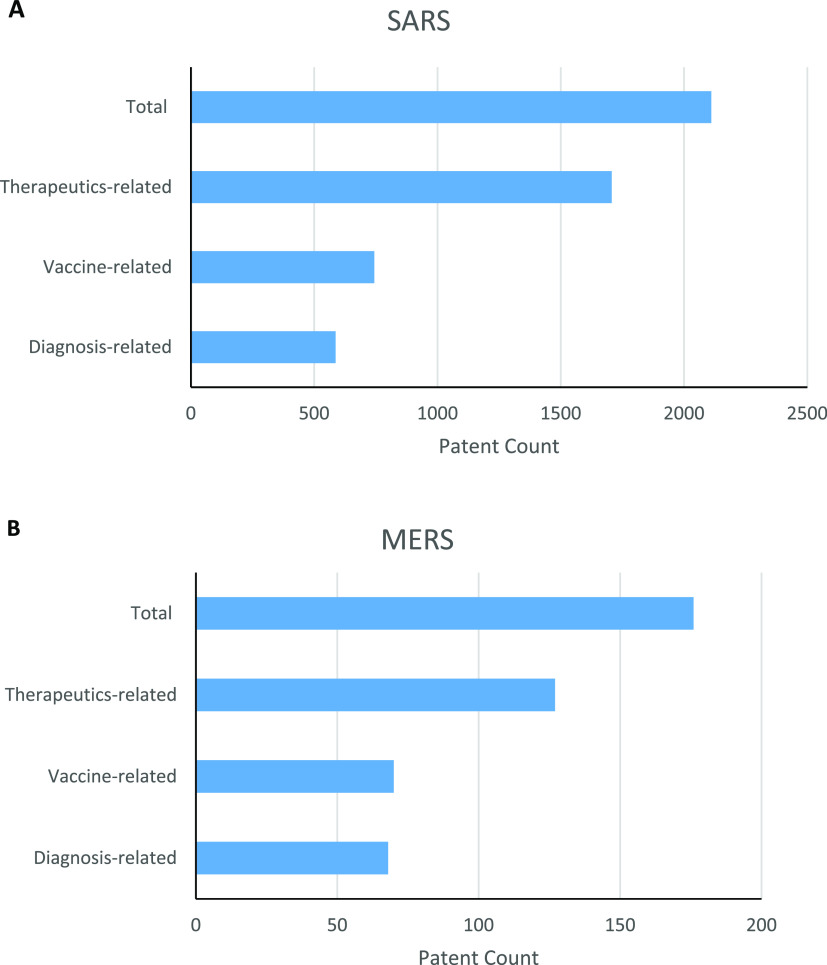
Distribution of patents related to SARS and MERS
As mentioned earlier, COVID-19 is caused by SARS-CoV-2, a new type of coronavirus in the same genus as SARS-CoV and MERS-CoV. Viral proteins responsible for SARS-CoV-2 entry into host cells and replication are structurally similar to those associated with SARS-CoV. Thus, research and development on SARS and MERS may offer insights that would be beneficial to the development of therapeutic and preventive agents for COVID-19. This report identified pertinent data from patents related to these two coronaviruses. Figure 5 shows the distribution of patents in the CAS content collection related to SARS (A) and MERS (B). The number of patents related to SARS is almost 12 times the number related to MERS, probably because the SARS outbreak occurred about 10 years before the MERS outbreak. Among SARS patents, about 80% are related to the development of therapeutics, 35% are related to vaccines, and 28% are related to diagnostic agents or methods. Because an individual patent may cover any two or more areas, the sum of percentage values is greater than 100%. A similar distribution pattern was also observed for patents related to MERS. Thus, for both diseases, more patents have been devoted to the development of therapeutic agents as opposed to diagnostic methods and vaccines.
Figure 5.
Distribution of patents related to SARS (A) and MERS (B) based on application purpose.
RESEARCH AND DEVELOPMENT IN SMALL MOLECULE ANTIVIRAL AGENTS FOR COVID-19 AND RELATED CORONAVIRUS DISEASES
Key Proteins and Their Roles in Viral Infection
Identification of targets is important for identifying drugs with high target specificity and/or uncovering existing drugs that could be repurposed to treat SARS-CoV-2 infection. Table 2 lists potential targets, their roles in viral infection, and representative existing drugs or drug candidates that reportedly act on the corresponding targets in similar viruses and thus are to be assessed for their effects on SARS-CoV-2 infection. 3CLpro and PLpro are two viral proteases responsible for the cleavage of viral peptides into functional units for virus replication and packaging within the host cells. Thus, drugs that target these proteases in other viruses such as HIV drugs, lopinavir and ritonavir, have been explored.19 RdRp is the RNA polymerase responsible for viral RNA synthesis that may be blocked by existing antiviral drugs or drug candidates, such as remdesivir.19 Conceivably, the interaction of viral S protein with its receptor ACE2 on host cells, and subsequent viral endocytosis into the cells, may also be a viable drug target. For example, the broad-spectrum antiviral drug Arbidol, which functions as a virus-host cell fusion inhibitor to prevent viral entry into host cells against influenza virus,20 has entered into a clinical trial for treatment of SARS-CoV-2.21,22 The protease TMPRSS2 produced by the host cells plays an important role in proteolytic processing of S protein priming to the receptor ACE2 binding in human cells.11 It has been shown that camostat mesylate, a clinically approved TMPRSS2 inhibitor, was able to block SARS-CoV-2 entry to human cells, indicating its potential as a drug for COVID-19.11
Table 2. Key Proteins and Their Roles during the Viral Infection Process.
| target candidate | full name | role during viral infection | drug candidate |
|---|---|---|---|
| 3CLpro | coronavirus main protease 3CLpro | a protease for the proteolysis of viral polyprotein into functional units | lopinavir19,30 |
| PLpro | papain-like protease PLpro | a protease for the proteolysis of viral polyprotein into functional units | lopinavir19,30 |
| RdRp | RNA-dependent RNA polymerase | an RNA-dependent RNA polymerase for replicating viral genome | remdesivir,19,29,32 ribavirin16,29,31 |
| S protein | viral spike glycoprotein | a viral surface protein for binding to host cell receptor ACE2 | Arbidol20,22,33a |
| TMPRSS2 | transmembrane protease, serine 2 | a host cell-produced protease that primes S protein to facilitate its binding to ACE2 | camostat mesylate11 |
| ACE2 | angiotensin-converting enzyme 2 | a viral receptor protein on the host cells which binds to viral S protein | Arbidol20,22,33a |
| AT2 | angiotensin AT2 receptor | an important effector involved in the regulation of blood pressure and volume of the cardiovascular system | L-16349128 |
An inhibitor of viral entry to host cells. Its direct action on S protein and ACE2 is yet to be confirmed.
ACE2 involvement with coronavirus infection is of further interest since ACE2 is a potent negative regulator restraining overactivation of the renin-angiotensin system (RAS) that may be involved in elicitation of inflammatory lung disease in addition to its well-known role in regulation of blood pressure and balance of body fluid and electrolytes.23,24 It catalyzes degradation of angiotensin II to angiotensin (1–7). The balance between angiotensin II and angiotensin (1–7) is critical since angiotensin II binds to angiotensin AT1 receptor to cause vasoconstriction, whereas angiotensin (1–7) elicits vasodilation mediated by AT2.25−27 Although the notion that ACE2 mediates coronavirus invasion is largely accepted, it remains unclear how the levels or activities of ACE2, AT1 receptors, and AT2 receptors are altered in coronavirus-induced diseases due to the limited number of studies.23,24 Therefore, it is yet to be determined whether some drugs or compounds that target any of these proteins (e.g., L-163491 as a partial antagonist of AT1 receptor and partial agonist of AT2 receptor) may alleviate coronavirus-induced lung injury.28
Patents and Potential Drug Candidates Related to Key Protein Targets
The CAS content collection contains patents related to coronavirus key proteins listed above. Table 3 lists the number of patents related to each protein target and associated therapeutic compounds with a CAS Registry Number (CAS RN) reported in these patents. CAS data show that targets 3CLpro and RdRp attracted more attention than other targets, and more compounds with therapeutic potential were identified for these targets, probably due to the work done for SARS-CoV which also contains 3CLpro and RdRp.
Table 3. Key Protein Targets and Related Patents in the CAS Content Collection and Potential Drug Candidates in CAS REGISTRY of Chemical Substances.
| target | no. of patents | no. of potential drug candidates |
|---|---|---|
| 3CLpro | 49 | 2178 |
| PLpro | 4 | 189 |
| RdRp | 26 | 570 |
| S protein | 46 | 333 |
| ACE2 | 5 | 97 |
| AT2 | 2 | 38 |
Existing Drugs with Potential Therapeutic Applications for COVID-19
Since SARS-CoV-2 is a newly discovered pathogen, no specific drugs have been identified or are currently available. An economic and efficient therapeutic strategy is to repurpose existing drugs. On the basis of genomic sequence information coupled with protein structure modeling, the scientific community has been able to rapidly respond with a suggested list of existing drugs with therapeutic potential for COVID-19. Table 4 provides a summary of such drugs together with potential mechanisms of actions for their activities. Barcitinib was proposed because of its anti-inflammatory effect and possible ability to reduce viral entry.35 A fixed dose of the anti-HIV combination, lopinavir–ritonavir, is currently in clinical trials with Arbidol or ribavirin.22 Remdesivir, developed by Gilead Sciences Inc., was previously tested in humans with Ebola virus disease and has shown promise in animal models for MERS and SARS. The drug is currently being studied in phase III clinical trials in both China and the USA. Favipiravir, a purine nucleoside leading to inaccurate viral RNA synthesis,36 was originally developed by Toyama Chemical of Japan, and has recently been approved for a clinical trial as a drug to treat COVID-19.30 Chloroquine, an antimalarial drug, has proven effective in treating coronavirus in China.32 In addition to the above-mentioned, many other antiviral drugs are also listed.
Table 4. Existing Drugs with Therapeutic Potentials for COVID-19 (Drug Repurposing).
| drug candidate | CAS RN | target | possible mechanism of action on COVID-19 | disease indication |
|---|---|---|---|---|
| baricitinib35 | 1187594-09-7 | JAK kinase | a JAK inhibitor that may interfere with the inflammatory processes | approved drug for rheumatoid arthritis |
| lopinavir19a | 192725-17-0 | viral proteases: 3CLpro or PLpro | protease inhibitors that may inhibit the viral proteases: 3CLpro or PLpro | lopinavir and ritonavir are approved drug combination for HIV infection |
| ritonavir19,37c | 155213-67-5 | |||
| darunavir33 | 206361-99-1 | approved drug for HIV infection | ||
| favipiravir (favilavir)29,36 | 259793-96-9 | RdRp | a purine nucleoside that acts as an alternate substrate leading to inaccurate viral RNA synthesis | viral infections |
| remdesivir19,29,32a | 1809249-37-3 | a nucleotide analogue that may block viral nucleotide synthesis to stop viral replication | Ebola virus infection | |
| ribavirin16,29−31a | 36791-04-5 | RSV infection, hepatitis C, some viral hemorrhagic fevers | ||
| galidesivir34b | 249503-25-1 | hepatitis C, Ebola virus, Marburg virus | ||
| BCX-4430 (salt form of galidesivir)34b | 222631-44-9 | hepatitis C, Ebola virus, Marburg virus | ||
| Arbidol22,33a | 131707-23-8 | S protein/ACE2d | an inhibitor that may disrupt the binding of viral envelope protein to host cells and prevent viral entry to the target cell | influenza antiviral drug |
| chloroquine29,32 | 54-05-7 | endosome/ACE2 | a drug that can elevate endosomal pH and interfere with ACE2 glycosylation | malarial parasite infection |
| nitazoxanide29 | 55981-09-4 | N/A | a drug that may inhibit viral protein expression | various helminthic, protozoal, and viral infection-caused diarrhea |
Drugs under clinical trials for treating COVID-19 (repurposing).
Drugs under clinical trials for other virus-induced diseases.
Ritonavir is a pharmacokinetic profile enhancer that may potentiate the effects of other protease inhibitors due to its ability to attenuate the degradation of those drugs by the liver enzyme CYP3A4 and thus is used in combination with antivirial Lopinavir.37
An inhibitor of viral entry to host cells. Its direct action on S protein and ACE2 is yet to be confirmed.
Selected Patents Related to Promising Small Molecule Drug Candidates
Table 5 shows selected patents associated with the aforementioned potential drugs, together with patents disclosing small molecules for treatment of SARS or MERS. The selection was based on the presence of important terms in CAS-indexed patents as well as the presence of the synthetic preparation role assigned by CAS scientists during document indexing. Patent applications WO2009114512 and WO2014028756 disclose preparation of compounds active as JAK inhibitors, one of which was later named as baricitinib and developed for reducing inflammation in rheumatoid arthritis. Patent application JP5971830 discloses preparation of polycyclic pyridone compounds and their use as endonuclease inhibitors. Patent applications US20160122374 and US20170071964 disclose preparation of the nucleotide analog drug remdesivir that was later developed as a therapeutic agent for Ebola and Marburg virus infections (Patent US20170071964). Because of its promising results in at least two COVID-19 patients, remdesivir has now entered into phase III clinical trials.
Table 5. Selected Patents Associated with Potential Drugs (Repurposing) for COVID-19 or Small Molecules for Treatment of SARS or MERS.
| patent no. | priority date | title | organization |
|---|---|---|---|
| WO2009114512 | 20080311 | Preparation of azetidine and cyclobutane derivatives as JAK inhibitors | Incyte Corporation, USA |
| WO2014028756 | 20140220 | Deuterated baricitinib | Concert Pharmaceuticals, Inc., USA |
| JP5971830 | 20150428 | Preparation of polycyclic pyridone derivatives as cap-dependent endonuclease (CEN) inhibitors and prodrugs thereof | Shionogi and Co., Ltd., Japan |
| US20160122374 | 20141029 | Preparation of nucleosides and methods for treating Filoviridae virus infections | Gilead Sciences, Inc., USA |
| US20170071964 | 20160916 | Preparation of amino acid-containing nucleotides and methods for treating arenaviridae and coronaviridae virus infections | Gilead Sciences, Inc., USA |
| WO2007075145 | 20070704 | Preparation of benzopyranone derivatives as anti-coronaviral agents | Singapore Polytechnic, Singapore; Shanghai Institute of Materia Medica Chinese Academy of Sciences, China |
| WO2005021518 | 20050310 | Preparation of 3,4-dihydro-2H-1,4-benzoxazine-2-carboxylic acid derivatives as cysLT2 receptor antagonists for treatment of respiratory diseases | Ono Pharmaceutical Co., Ltd., Japan |
| WO2007120160 | 20071025 | Preparation of N-heterocyclic acetamides useful for viral inhibition | Novartis AG, USA |
| WO2009119167 | 20091001 | Aniline derivative having anti-RNA viral activity | KinoPharma, Inc., Japan |
| WO2013049382 | 20130404 | Broad-spectrum antivirals against 3c or 3c-like proteases of picornavirus-like supercluster: picornaviruses, caliciviruses and coronaviruses | Kansas State University Research Foundation; The Ohio State University; Wichita State University - all in USA |
| WO2018042343 | 20180308 | Preparation of peptides that inhibit 3C and 3CL proteases and methods of use thereof | GlaxoSmithKline, UK |
| WO2007067515 | 20070614 | Five-membered iminocyclitol derivatives as selective and potent glycosidase inhibitors: new structures for antivirals and osteoarthritis therapeutics | Academia Sinica, Taiwan |
Patent application WO2013049382 discloses both structures and syntheses of compounds from various structure classes (peptidyl aldehydes, peptidyl α-ketoamides, peptidyl bisulfite salts, and peptidyl heterocycles), as well as certain formulation compositions, developed to inhibit viral 3C protease or 3C-like protease (i.e., 3CLpro).
Patent application WO2018042343 presents both preparation methods and biological assay results for compounds capable of inhibiting the SARS virus proteases. These compounds appeared to exhibit good enzyme-inhibiting activity (pIC50 ≈ 7 or IC50 ≈ 0.1 μM) and antiviral activity, which was assessed by host cell viability using cultured human lung fibroblast MRC-5 cells infected with a specified virus (e.g., MERS virus) expressing the viral S protein. Drug administration routes were also mentioned in this patent.
Small Molecule Compounds in Research and Development with Potential Effects on Key Protein Targets for Human Coronavirus-Induced Diseases
Besides various commercialized antiviral drugs, there are also small molecule compounds currently in research and development that have shown significant inhibitory effects on many key proteins from similar coronaviruses such as SARS-CoV and MERS-CoV (Table 6). These drug candidates mostly inhibit viral enzymes including proteases and components for RdRp. Since 3CLpro protease has a high level of sequence homology between SARS-CoV and SARS-CoV-2, inhibitors against 3CLpro of SARS-CoV may also be applicable to SARS-CoV-2. Compounds, including benzopurpurin B, C-467929, C-473872, NSC-306711 and N-65828, which may inhibit the activity of viral NSP15, poly(U)-specific endoribonuclease, were tested for reduced SARS-CoV infectivity in cultured cells with IC50 of 0.2–40 μM.38 Compound C-21 and CGP-42112A are two AT2 agonists, whereas L-163491 has dual functions as a partial agonist for AT2 receptor and a partial antagonist of AT1 receptor. Since AT1 and AT2 are important effectors in the RAS system to which ACE2 belongs, it has been speculated that these compounds may be used to adjust the balance between AT1 and AT2, which may be affected by coronavirus infection and to alleviate viral-induced lung injury during the infection.24
Table 6. Small Molecule Compounds in Research and Development with Therapeutic Potential for COVID-19.
| CAS RN | small molecule compound | target | possible mechanism of action on COVID-19 |
|---|---|---|---|
| 4431-00-9 | aurine tricarboxylic acid | RNA-dependent RNA polymerase (RdRp) | an inhibitor that may bind to viral RdRp, as tested against SARS-CoV in cell culture16 |
| 502960-90-9 | 4-methyl-N-[(1S)-2-oxo-2 [[(1S,2E)-1-(2-phenylethyl)-3-(phenylsulfonyl)-2-propen-1-yl]amino]-1-(phenylmethyl)ethyl]- 1-piperazinecarboxamide | viral proteases: 3CLpro and PLpro | an inhibitor that may disrupt the function of 3CLpro and PLpro, which was tested against SARS-CoV16,39,40 |
| 1851279-09-8 | 4-(1,1-dimethylethyl)-N-[(1S)-2-oxo-2-[[(1S,2E)-1-(2-phenylethyl)-3-(phenylsulfonyl)-2-propen-1-yl]amino]-1-(phenylmethyl)ethyl]- 1-piperazinecarboxamide | ||
| 1851280-00-6 | 4-(2-methoxyethyl)-N-[(1S)-2-oxo-2-[[(1S,2E)-1-(2-phenylethyl)-3-(phenylsulfonyl)-2-propen-1-yl]amino]-1-(phenylmethyl)ethyl]- 1-piperazinecarboxamide | ||
| 223537-30-2 | rupintrivir | a cysteine protease inhibitor that may disrupt the function of 3CLpro and PLpro41 | |
| 2409054-43-7 | (αR)-α-[[3-(4-chloro-2-fluorophenyl)-1-oxo-2-propen-1-yl]amino]-N-[(1R)-1-methyl-2-(2-oxo-3-pyrrolidinyl)ethyl]- benzenepropanamide | viral proteases: 3CLpro or PLpro | an inhibitor that may disrupt the function of 3CLpro or PLpro, as tested against SARS-CoV or MERS-CoV39,40 |
| 452088-38-9 | 5-[(4-methyl-1-piperidinyl)sulfonyl]-1H-indole-2,3-dione | ||
| 2409054-44-8 | 3-hydroperoxy-4-[2-hydroxy-3-[3-(4-hydroxyphenyl)-1-oxo-2-propen-1-yl]-6-methoxyphenyl]-2-butanone | ||
| 41137-87-5 | hirsutenone | ||
| 992-59-6 | benzopurpurin B | NSP15 (poly(U)-specific endoribonuclease) | chemical inhibitors that may suppress viral infectivity by inhibiting endoribonuclease NSP15, as tested against SARS-CoV in cultured cells38 |
| 351891-58-2 | C-467929 | ||
| 331675-78-6 | C-473872 | ||
| 813419-93-1 | NSC-306711 | ||
| 501444-06-0 | N-65828 | ||
| 477775-14-7 | C-21 | AT2 | an angiotensin AT2 receptor agonist that may alleviate the virus-induced lung injury24 |
| 127060-75-7 | CGP-42112A | ||
| 170969-73-0 | L-163491 | a dual-property molecule that functions as angiotensin AT1 partial antagonist and AT2 agonist which may alleviate the virus-induced lung injury24 |
Small Molecules Identified by Structure Similarity, Lipinski’s Rule of 5, and CAS-Indexed Pharmacological Activity and/or Therapeutic Usage
Besides the aforementioned antiviral drugs, there may be additional small molecule compounds with therapeutic or pharmacological potential against viruses such as SARS-CoV and MERS-CoV. Compounds listed in Tables 4 and 6 were subjected to a Tanimoto similarity search in CAS REGISTRY using CAS proprietary fingerprints.a Those substances with at least 60% structural similarity match and meeting Lipinski’s rule of 5 were identified. Table 7 lists selected compounds that were also identified to have a pharmacological activity or therapeutic usage role. Compound name and CAS RN are provided for each compound. The second column lists the number of compounds that met the structure similarity and Lipinski’s rule criteria. Although more work remains to be done in this regard, the methodology and results mentioned here point to a strategy that may help streamline the process of drug discovery for COVID-19.
Table 7. Examples of Similar Molecules with Possible Therapeutic Effects Identified by Structural Similarity, Lipinski’s Rule of 5, and Pharmacology/Therapeutic Role Assigned by CAS Scientists during Document Indexing.
| query substance name (CAS RN) | no. of substances with >60% similarity | example of selected similar substance | Registry Number of selected similar substance |
|---|---|---|---|
| ribavirin (36791-04-5) | 1520 | viramidine | 119567-79-2 |
| galidesivir (249503-25-1) | 502 | (2R,3S,5R)-5-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-3-hydroxy-2-pyrrolidinemethanol | 1610426-50-0 |
| (2S,4R,5S)-5-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-4-hydroxy-2-pyrrolidinemethanol | 872534-76-4 | ||
| (2R,3R,4S,5S)-5-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-3-hydroxy-4-methoxy-2-pyrrolidinemethanol | 1610426-51-1 | ||
| chloroquine (54-05-7) | 21176 | hydroxychloroquine | 118-42-3 |
| (±)-chloroquine diphosphate | 50-63-5 | ||
| chloroquine hydrochloride | 3545-67-3 | ||
| chloroquine sulfate | 132-73-0 | ||
| favipiravir (259793-96-9) | 309 | 6-bromo-3,4-dihydro-3-oxo-2-pyrazine-5-d-carboxamide | 1476773-04-2 |
| 6-fluoro-3,4-dihydro-3-oxo-2-pyrazine-5-d-carboxamide | 1492021-26-7 | ||
| 2-butanone, 3-hydroperoxy-4-[2-hydroxy-3-[3-(4-hydroxyphenyl)-1-oxo-2-propen-1-yl]-6-methoxyphenyl] (2409054-44-8) | 63195 | xanthoangelol D | 132998-83-5 |
BIOLOGICS FOR CORONAVIRUS-ASSOCIATED DISEASES
Distribution of Biologics Patents Related to SARS and MERS
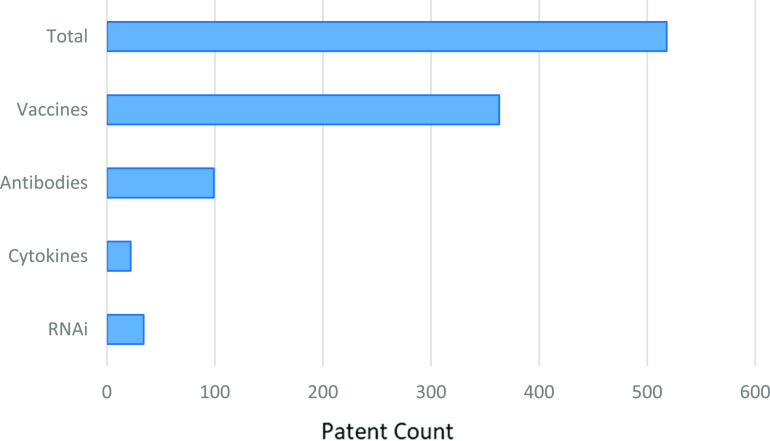
The new coronavirus SARS-CoV-2 related to SARS and MERS viruses is causing serious and ongoing epidemiological problems around the world. Since there is limited clinical and basic research information at this time, treatment options for COVID-19 currently comprise investigational drugs and management of symptoms. As biologics have the potential to broaden the spectrum of the treatment options for coronavirus-induced diseases, leveraging prior knowledge and practices used to address the SARS and MERS outbreaks provides a practical strategy for developing new target-specific therapeutic agents for SARS-CoV-2. To this end, an analysis of biologics from patents contained in the CAS content collection was performed. The patent analysis included information related to therapeutic antibodies, cytokines, interfering and other therapeutic RNAs, and vaccines for potential treatment and/or prevention of SARS-related diseases from patents published from 2003 to the present. Figure 6 shows more than 500 patents that disclose the use of these four biologics classes to treat and prevent SARS and MERS. Of these patents, vaccine development was the largest class (363), followed by therapeutic antibodies (99), interfering RNAs (35), and cytokines (22). Given the indispensable role of vaccines in viral disease prevention, detailed analysis of vaccines will be presented in a later section.
Figure 6.
Distribution of biologics patents related to SARS and MERS.
Antibodies
Ninety-nine patents containing information about antibodies with therapeutic and/or diagnostic potential for SARS and MERS were identified. Of these, 61 patents claimed preparation of SARS-specific antibodies (23), MERS-specific antibodies (17), or antibodies with diagnostic application (21). Similar to SARS-CoV, the receptor-binding domain (RBD) in the S protein of SARS-CoV-2 binds to human ACE2 receptor in order to gain access into host cells.42 In viral infection, the S protein, but not the other structural proteins, M, E, and N in SARS-CoV, elicits an immune response.43Table 8 shows the target analysis of patents related to development of therapeutic antibodies for SARS. Over 90% of these antibodies are directed against S protein including its RBD. The data indicate that the S protein is a putative target for SARS-CoV-2 antibody development.
Table 8. Target Analysis of Patents on Developing Therapeutic Antibodies for SARS.
| patent number | antigen of SARS antibody | patent title | organization | priority date |
|---|---|---|---|---|
| EP2112164 | lipid attachment signals or GPI | Antiviral peptides linked to a lipid attachment signals or GPI against enveloped virus such as HIV, avian flu, SARS or Ebola virus | Institute Pasteur of Shanghai | 20080229 |
| WO2009128963 | spike protein | Cross-neutralizing human monoclonal antibodies to SARS-CoV and methods of use thereof | Institute for Research In Biomedicine | 20080117 |
| WO2009128963 | spike protein | Cross-neutralizing human monoclonal antibodies to spike protein of SARS coronavirus and methods of use thereof | Humab, LLC | 20080117 |
| WO2008035894 | viral infection | Preparation of antiviral antibody 3D8 fragments and their use in treatment of viral infection | Sung Kyun Kwan University; Ajou University; Invitroplant Co., Ltd. | 20060919 |
| WO2008060331 | spike protein | Antibodies to SARS coronavirus | Amgen Inc. | 20060519 |
| WO2007044695 | spike protein | Neutralizing monoclonal anti-spike protein antibodies for diagnosis and treatment of SARS-coronavirus-associated disease and screening of vaccine or anti-SARS agent | Dana-Farber Cancer Institute | 20051007 |
| CN1911963 | RBD of S protein | Method for preparing neutralizing monoclonal antibody against severe acute respiratory syndrome coronavirus and its application | Chinese Academy of Sciences | 20050810 |
| CN1903878 | spike protein | Fab fragment of human antibody IgG against SARS coronavirus | Fudan University | 20050726 |
| WO2006095180 | S2 protein | Human monoclonal antibodies against SARS-associated coronavirus and treatment of patients with SARS | Ultra Biotech Ltd.; University of California | 20050310 |
| WO2006086561 | spike protein | Neutralizing monoclonal antibodies against severe acute respiratory syndrome-associated coronavirus | New York Blood Center, Inc. | 20050208 |
| CN1664100 | spike protein | Preparation of heavy chain and light chain variable regions of anti-SARS coronavirus antigen antibodies and their diagnostic and therapeutic uses thereof | Chen Zhinan | 20050204 |
| CN1660912 | IL-8 | Sequences of monoclonal antibodies against human interleukin 8 and therapeutic use | Ye Qingwei | 20041208 |
| WO2006051091 | spike protein | Compositions against SARS-coronavirus and uses thereof | Crucell Holland BV | 20041111 |
| WO2006051091 | spike protein | Compositions against SARS-coronavirus comprising at least two immunoglobulins reacting with non-competing epitopes, and therapeutic and diagnostic uses thereof | Crucell Holland BV | 20041111 |
| CN1673231 | spike protein | Monoclonal antibody of SARS coronavirus N protein and its use in treatment of SARS virus infections | Chinese Academy of Sciences | 20040715 |
| US20060240551 | spike protein | Neutralizing monoclonal antibodies against severe acute respiratory syndrome-associated coronavirus | New York Blood Center, Inc. | 20040602 |
| WO2005054469 | spike protein | Anti-SARS-coronavirus monoclonal antibodies, and diagnostic, therapeutic and vaccine preparation uses | Health Canada | 20031205 |
| WO2005060520 | spike protein | Antibodies specific to SARS-CoV spike protein for diagnosis and therapy of SARS and for screening of epitopic vaccines or anti-SARS therapeutics | Dana-Farber Cancer Institute, Inc. | 20031125 |
| US20050106563 | spike protein | Epitope profiles of SARS coronavirus for use in antigen detection, antibody production, and defense against infection | Genesis Biotech Inc. | 20030908 |
| US20050069869 | spike protein | SARS coronavirus codon-optimized sequences for spike (S) protein expression, anti-S human monoclonal antibodies, and therapeutic and diagnostic uses thereof | University of Massachusetts | 20030804 |
| WO2005012360 | S and N proteins | Antibody binding molecules specific for SARS coronavirus | Crucell Holland BV | 20030722 |
| CN1566155 | S, N, and M proteins | Antibody library-derived human monoclonal anti-SARS virus antibodies for treating severe acute respiratory syndrome | Igcon Therapeutics Co., Ltd.; Genetastix Corporation | 20030710 |
| WO2005007671 | spike protein | Compositions and methods for treating SARS using peptides derived from SARS virus E2 N-terminal-alpha helix or C-terminal-alpha helix and related monoclonal antibody | Epitomics, Inc. | 20030429 |
An additional 38 patents contained information pertaining to other types of antiviral antibodies that were useful for SARS and MERS therapies. These included neutralizing antibodies or antibodies designed to target proteins such as IL-6/IL-6R, TLR3 (Toll-like receptor 3), CD16, ITAM (immunoreceptor tyrosine-based activation motif), DC-SIGN (dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin), ICAM-3 (intercellular adhesion molecule 3), or IP-10/CXCL10 (interferon γ-inducible protein 10). Cytokine storm has been reported to correlate with disease severity in SARS-CoV-2 infection. Patients admitted to an ICU had higher concentrations of proinflammatory cytokines and chemokines, particularly G-CSF, IP-10/CXCL10, MCP1 (monocyte chemoattractant protein 1), and TNFα, as well as elevated cytokines from T helper 2 cells such as IL-4 and IL-10.44 Patent application WO2005058815 discloses human anti-IP-10 antibodies, including bispecific molecules and immunoconjugates that bind to IP-10 with high affinity, for treating inflammation, autoimmune disease, neurodegenerative disease, bacterial infection, and viral infection. Patent application WO2017095875 discloses the preparation of human antibodies and immunoconjugates specifically targeting chemokine IP-10, including an anti-IP-10 antibody shown to suppress free serum IP-10 in about 3 days at 0.5 mg/kg and in approximately 10 days at 10 mg/kg in Cynomolgus macaques.
In addition, DC-SIGN/CD209, a type II transmembrane adhesion molecule with C-type lectin function, is mainly expressed on interstitial dendritic cells and lung alveolar macrophages.45 DC-SIGN functions as an entry cofactor in transferring SARS-CoV to susceptible cells such as pneumocytes.46 Patent application WO200505824 claims the production of a humanized anti-DC-SIGN antibody that interfered with the interaction of DC-SIGN with its receptor, ICAM-3. The antibody effectively blocked viral binding, infection, and transmission for viral infections/diseases, including SARS.
Cytokines
Cytokines are low-molecular-weight proteins that act as chemical signals in the immune response to pathogen invasion. The production of various cytokines in response to an invading pathogen such as a virus contributes to the host organism’s ability to eliminate the pathogen. Specific types of cytokines, including chemokines, interferons (IFNs), interleukins, and lymphokines, have been reported and characterized in the literature over the past 40 years. By early 2020, the CAS Lexicon contained over 700 terms for specific types of cytokines associated with 76 724 documents, including 11 837 patents.
During a viral infection, the most prominent cytokines produced are IFNs, which interfere with viral replication. IFNs are classified as type I (IFN-α, IFN-β, IFN-δ, IFN-ε, IFN-κ, IFN-ν, IFN-τ, and IFN-ω), type II (IFN-γ), or type III (IFN-λ) based on the receptor complex used for signaling as well as sequence homology.47 Because of their ability to interfere with viral replication, interferons and interferon fusion proteins have been utilized as therapeutic agents for treatment of viral infections for the past 20 years. A few patents disclosing these proteins and their use for treating SARS are described below.
rSIFN-co
Patent applications WO2011072487 and WO2016180335 describe the cloning of a recombinant interferon (rSIFN-co, CAS RN 2043378-94-3) as well as a method for determining its potency that was effective for treating various viral infections/diseases, including SARS. The invention relates that rSIFN-co has an identical amino acid sequence to Infergen (118390-30-0), but it has an altered spatial conformation and different biological potency. The rSIFN-co not only has an antiviral activity that is 20 times stronger than the clinically available interferon, but also has significantly stronger antitumor properties against breast cancer and cervical cancer than other recombinant human α-interferons. The invention further relates that rSIFN has reduced toxic and side effects and can be safely used in large doses (each dose can be >10 million IU), making it possible to successfully treat some viral diseases or tumors that require large doses of interferon.
IFN-ω
Patent application WO2004096852 discloses the amino acid sequence for recombinant human interferon ω (rhIFN-ω) (RN 791910-34-4) that was shown to have an anti-SARS viral activity similar to that of IFN-β. IFN-ω effectively decreased disease severity and inhibited proliferation of coronavirus strain BJ01 in monkeys.
IL-28A (IFN-λ2), IL-28B (IFN-λ3), and IL-29 (IFN-λ1) Variants
Patent application WO2005097165 claims a method for treating SARS viral infection using IL-28A, IL-28B, and IL-29 cysteine variants conjugated to polymers (e.g., polyethylene glycol) and discloses the amino acid and nucleic acid sequences for these cysteine variants. Of these variants, MetIL-29C172S-PEG (RN 867228-40-8) was specifically shown to inhibit viral replication.
Interferon-Human Serum Albumin Fusion Protein
Patents applications US20090053173 and CN101942026 both disclose long-lasting fusion proteins (HSA-IFN) with each of them being composed of an interferon fused with human serum albumin-binding peptide for treatment of a wide range of diseases, including SARS. Specific HSA-IFN fusion proteins were constructed using five different interferons (IFN-α1b, IFN-α2b, IFN-β, IFN-ω, IFN-γ) with corresponding CAS RNs 1122730-20-4, 1122730-23-7, 1122730-25-9, 1122730-27-1, and 1122730-29-3, respectively. These HSA-IFN fusion proteins significantly lengthened the plasma half-life of interferons (e.g., from 10 h to 12 days for HSA-IFN-α2b) due to slower free interferon release into the plasma and thus may prolong the effects of interferon for each injection.
RNA Therapies
RNA interference (RNAi) is a biological process wherein small complementary RNA duplexes target and neutralize specific mRNA molecules, resulting in inhibition of gene expression or genetic translation. Interfering RNAs include microRNAs and small interfering RNAs (siRNAs) that are generally about 21–25 nucleotides in length. Short hairpin RNAs (shRNAs) are artificial synthetic RNA molecules designed to fold into a tight hairpin conformation that allows them to silence their target genes, and can serve as precursors of siRNAs. The expression of shRNAs in cells is typically accomplished by their delivery via plasmids or viral or bacterial vectors.48 Although microRNAs are noncoding and naturally found in plants, animals, and some viruses, synthetic versions are currently being used to silence a variety of genes.49 The ability to chemically synthesize modified analogues of microRNAs as well as siRNAs, which are capable of altering disease-related gene expression or inhibiting pathogen gene expression, has created a host of new therapeutic options.50
In contrast to the microRNAs and siRNAs, antisense RNAs are single-stranded RNAs which are naturally occurring or synthetic and usually around 19–23 nucleotides in length with a sequence complementary to that of a protein coding mRNA, allowing it to hybridize and block protein translation.48
Since the discovery of RNAi in the late 1990s, it has become a well-known method for silencing/suppressing target genes associated with virulence and pathogenesis. Thirty-five patents in the CAS content collection disclose the use of RNAi in treating SARS, with 28 patents using siRNA molecules, three patents using antisense oligonucleotides, two patents using RNA aptamers, one patent using a ribozyme, and one patent using a microRNA inhibitor. Supporting Information Table S1 provides a high-level view of these 35 patents including the specific RNAi targets. A few of these patents are further discussed below.
siRNAs Targeting Coronavirus Proteins M, N, or E
Patent application CN101173275 discloses two double-stranded RNAs (dsRNAs) designed to specifically target two separate regions of the SARS protein M mRNA. The siRNA-M1 sequences targeting the 220–241 region of protein M mRNA correspond to CAS RNs 1023405-01-7 and 1023405-02-8, while siRNA-M2 sequences targeting the 460–480 region correspond to CAS RNs 1023405-03-9 and 1023405-04-0. The interference efficiency of these two siRNAs on SARS M protein gene expression was greater than 70%.
Table 9. Representative siRNA Data from Chinese Patent CN1648249.
| siRNA | sense strand (CAS RN) | antisense strand (CAS RN) |
|---|---|---|
| No. 8* | 5′-cgucgcagcguguaggcacua-3′ | 5′-cagugccuacacgcugcgacg-3′ |
| (RN 874840-18-3) | (RN 874840-32-1) | |
| No. 51* | 5′-aacgguuuacgucuacucgca-3′ | 5′-cgcgaguagacguaaaccguu-3′ |
| (RN 874840-19-4) | (RN 874840-47-8) | |
| No. 56* | 5′-aacguacugccacaaaacagc-3′ | 5′-acuguuuuguggcaguacguu-3′ |
| (RN 874840-20-7) | (RN 874840-46-7) |
siRNAs Targeting Replicase and RNA Polymerase Region
Table 10. Representative siRNA Data from US20050004063.
| siRNA | sense strand | CAS RN | target region or gene |
|---|---|---|---|
| SARSi-1 | 5′-gugaacucacucgugagcuctt-3′ | 821121-35-1 | 512–531 bp of replicase A1 region |
| SARSi-2 | 5′-guacccucuugauugcauctt-3′ | 821121-36-2 | 586–604 bp of replicase A1 region |
| SARSi-3 | 5′-gagucgaagagaggugucutt-3′ | 821121-37-3 | 916–934 bp of replicase A1 region |
| SARSi-4 | 5′-gcacuugucuaccuugaugtt-3′ | 821121-38-4 | 1194–1213 of replicase A1 region |
| SARSi-5 | 5′-ccuccagaugaggaagaagtt-3′ | 821121-39-5 | 3028–3046 bp of replicase A region |
| SARSi-6 | 5′-gguguuuccauuccaugugtt-3′ | 821121-40-8 | 5024–5042 bp of replicase A region |
| SARSi-7 | 5′-cacugauuccguucgagauctt-3′ | 821121-41-9 | S gene |
| SARSi-8 | 5′-cguuucggaagaaacagguactt-3′ | 821121-42-0 | E gene |
| SARSi-9 | 5′-caagccucuucucgcuccuctt-3′ | 821121-43-1 | N gene |
| SARSi-10 | 5′-guggcuuagcuacuucguugtt-3′ | 821121-44-2 | M gene |
| SARSi-11 | 5′-ugcuugcugcugucuacagtt-3′ | 821121-45-3 | M gene |
The authors demonstrated that SARSi-2, SARSi-3, SARSi-4, and SARSi-7-11 inhibited coronavirus infection and replication in FRhk-4 cells. SARSi-4 was the most effective with nearly complete inhibition, followed by SARSi-2 and SARSi-3.
Patent application CN1569233 discloses siRNAs, shown in Table 11, that target SARS genes encoding RNA-dependent RNA polymerase, helicase, nucleoprotein N, and proteolytic enzymes. These siRNAs were able to inhibit or kill 50–90% of the SARS virus BJ01 strain, with the proteolytic enzyme-targeting siRNAs being the most effective.
Table 11. Representative siRNA Data from CN1569233.
| sequence | CAS RN | gene target | % inhibition of SARS virus |
|---|---|---|---|
| 5′-caucauccggugaugcuac-3′ | 872062-80-1 | RNA-dependent RNA polymerase | ∼50 |
| 5′-uaguguauacggcaugcuc-3′ | 872062-81-2 | helicase | ∼70 |
| 5′-gugcgugcagacgguucgu-3′ | 872062-82-3 | nucleoprotein N | ∼95 |
| 5′-cguagucgcgguaauucaa-3′ | 872067-98-6 | proteolytic enzyme | ∼90 |
RNA Aptamers
Two Korean patents describe the use of RNA aptamers for inhibition of SARS viruses. Patent application KR2009128837 identifies RNA aptamers as anti-SARS agents capable of binding to and inhibiting the double-stranded DNA unwinding of the SARS virus helicase. Related patent application KR 2012139512 describes RNA aptamers with distinct affinity for the nucleocapsid of SARS-CoV for potential pharmaceutical use.
Ribozymes
Patent application JP2007043942 describes a therapeutic RNA/DNA chimeric ribozyme designed to recognize and cleave conserved common regions and regions with loop structures in the genes of coronaviruses, including SARS. This ribozyme specifically recognizes the GUC in viral genes with loop conformations.
Antisense Oligonucleotides
Antisense oligonucleotides have also been developed to reduce the severity of SARS virus infections and to prevent or treat SARS virus-associated disease, to detect the virus in human samples, and to diagnose SARS virus-associated diseases. Patent application WO2005023083 published by Ionis Pharmaceuticals describes hybrid DNA/RNA antisense oligonucleotides designed to disrupt the pseudoknot in the frameshift site of the SARS coronavirus RNA. In addition to directly targeting the virus, antisense oligonucleotides may be used to target disease-related proteins involved in the inflammatory process.
Vaccines
It is crucial to develop safe and effective vaccines to control the COVID-19 pandemic, eliminate its spread, and ultimately prevent its future recurrence. Since the SARS-CoV-2 virus shares significant sequence homology with two other lethal coronaviruses, SARS and MERS, the vaccines identified in these patents related to SARS and MERS viruses could potentially facilitate the design of anti-SARS-CoV-2 vaccines.
Distribution of Patents Related to SARS and MERS among Vaccine Types
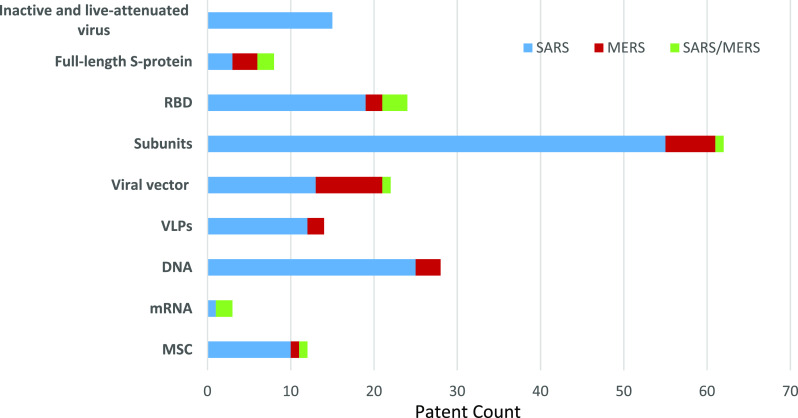
Antiviral vaccines generally fall into one of the following types: inactive or live-attenuated viruses, virus-like particle (VLP), viral vectors, protein-based, DNA-based, and mRNA-based vaccines. There are 363 patents in the CAS content collection related to vaccine development to prevent viral disorders/diseases, including SARS and MERS. Of these, 175 patents disclose vaccines for non-coronaviruses that may have relevance to SARS and MERS, while 188 patents are directly associated with anti-SARS and anti-MERS vaccines with a demonstrated immune response. Supporting Information Table S2 contains additional information on these SARS/MERS vaccine-related patents.
Figure 7 reveals the distribution of patents among these vaccine types related to SARS and MERS. As can be seen, 15 patents disclose information about inactive and live-attenuated virus vaccines, 28 patents describe DNA vaccines, 21 patents disclose information on viral vector vaccines, 13 patents disclose information on VLP vaccines, and three patents are focused on mRNA vaccines.
Figure 7.
Distribution of vaccine-related patent associated to SARS and MERS.
It was reported that viral S protein subunit vaccines produced higher neutralizing antibody titers and more complete protection than live-attenuated SARS-CoV, full-length S protein, and DNA-based S protein vaccines.51 Unsurprisingly, about half of the patents focused on protein vaccines comprising the S protein subunit vaccine and vaccines specifically targeting the receptor binding domain (RBD) of the S1 subunit of the viral S protein. Collectively, S protein/gene is the preferred target site in SARS/MERS vaccine development, and the same strategy can be potentially useful in developing SARS-CoV-2 vaccines. A condensed report on several patents that describe vaccines for generating immunity to SARS and MERS follows.
Attenuated Virus Vaccines
Patent application US20060039926 discloses live attenuated coronavirus or torovirus vaccines. Introduction of a mutation (Y6398H) into the Orf1a/b polyprotein (p59/nsp14/ExoN) was shown to completely attenuate virulence of mouse coronavirus (MHV-A59). The attenuated MHV virus exhibited reduced replication in mice at day five following intracerebral inoculation.
DNA-Based Vaccines
Patent application WO2005081716 discloses compositions and methods for inducing/enhancing immune responses, particularly antigen-specific CD8+ T cell-mediated responses, against antigens of the SARS coronavirus. An enhancement of the immune response involving particularly cytotoxic T cell immune responses is induced in vivo by chimeric nucleic acids that encode an endoplasmic reticulum chaperone polypeptide (e.g., calreticulin) linked to at least one antigenic polypeptide or peptide from SARS-CoV. Using gene gun delivery of DNA-coated gold particles, vaccination of mice against a calreticulin–nucleocapsid fusion protein resulted in potent nucleocapsid-specific humoral and T cell-mediated immune responses. Vaccinated animals were capable of significantly reducing the titer of a challenging vaccinia vector expressing the N protein of the SARS virus.
Patent application WO2015081155 discloses immunogens, which comprise consensus proteins derived from the MERS-CoV spike protein, for use in DNA-based vaccines targeting MERS-CoV. The consensus spike protein significantly induced both humoral and cellular immune responses, including increased titers of IgG and neutralizing antibodies. The induced cellular immune response involved increased CD3+CD4+ and CD3+CD8+ T cell responses that produced IFN-γ, TNF-α, IL-2, or both IFN-γ and TNF-α. On March 3, 2020, Inovio Pharmaceutical, Inc. announced they had designed the DNA vaccine called INO-4800 to be planned for human trials in the United States in April.57
Protein-Based Vaccines
Patent application WO2010063685 by GlaxoSmithKline (GSK) discloses a vaccine capable of provoking a protective immune response against SARS. The vaccine comprises an S protein immunogen and an oil-in-water emulsion adjuvant. An engineered ectodomain immunogen (soluble S protein), in combination with the emulsion adjuvant, GSK2, induced high levels of anti-SARS-CoV IgG2a or IgG2b antibody responses and neutralizing antibody responses in animal models. In late February 2020, GSK announced a collaboration with Chinese firm Clover Biopharmaceuticals to assess a coronavirus (COVID-19) vaccine candidate.52 This collaboration will involve the use of Clover’s protein-based coronavirus vaccine candidate (COVID-19 S-Trimer) with GSK’s adjuvant system. By applying their Trimer-Tag technology, Clover has manufactured an S-Trimer subunit vaccine using a rapid mammalian cell culture-based expression system. The Trimer-Tag is an advanced drug development platform, which enables the production of novel, covalently trimerized fusion proteins that can better target previous undruggable pathways.
Patent application US20070003577 discloses immunogenic compositions and vaccines associated with the S protein of SARS coronavirus. A TriSpike SARS coronavirus vaccine was prepared from a recombinant full-length trimeric S protein. The recombinant protein was shown to (1) exhibit native antigenicity as shown by reactivity with convalescent SARS patient sera; (2) exhibit specific binding to soluble ACE2 receptor; (3) promote antibody-dependent viral entry in otherwise refractory human Raji B cells; and (4) elicit protection against a challenge infection in an animal model.
Patent application US20060002947 (Antigen Express, Inc., a subsidiary of Generex) discloses the preparation of hybrid peptides composed of three elements, including (a) an invariant chain (Ii) key peptide for antigen presentation enhancing activity, (b) a chemical structure linking the Ii to the antigenic epitope, and (c) an antigenic epitope that binds to a MHC class II molecule. The methodology was used to create Ii-Key/MHC II SARS hybrids. Recently, Generex announced that it is developing a COVID-19 vaccine following a contractual agreement with a Chinese consortium comprised of China Technology Exchange, Beijing Zhonghua Investment Fund Management, Biology Institute of Shandong Academy of Sciences, and Sinotek-Advocates International Industry Development. The company will utilize its Ii-Key immune system activation technology to produce a COVID-19 viral peptide for human clinical trials.53
Virus-like Particle Vaccines
In 2015, patent application WO2015042373 by Novavax disclosed an immunogenic composition composed of MERS-CoV nanoparticle VLPs containing at least one trimer of a S protein, produced by baculovirus overexpression in Sf9 cells. This VLP preparation induced a neutralizing antibody response in mice and transgenic cattle, when administered along with their proprietary adjuvant Matrix M (RN 1235341-17-9). In addition, preparations of sera from vaccinated cattle (SAB-300 or SAB-301) were injected into Ad5-hDPP4 transduced BALB/c mice prior to challenge with MERS-CoV. Both SAB-300 and SAB-301 were able to protect these mice from MERS-CoV infection with a single prophylactic injection. Novavax announced on February 26, 202054 that it was beginning animal testing on potential COVID-19 vaccine candidates due to their previous experiences working with other coronaviruses, including both MERS and SARS. Their COVID-19 candidate vaccines targeting the S protein of SARS-CoV-2 were developed using their recombinant nanoparticle vaccine technology along with their proprietary adjuvant Matrix-M.
mRNA-Based Vaccines
The potential advantages of an mRNA approach to prophylactic vaccines include the ability to mimic natural infection to stimulate a more potent immune response as well as the ability to combine multiple mRNAs into a single vaccine. Patent application WO2017070626 by Moderna discloses mRNA vaccines composed of mRNAs encoding antigenic viral full-length S, S1, or S2 proteins from SARS-CoV and MERS-CoV virus, formulated in cationic lipid nanoparticles. They show that mice vaccinated with mRNA encoding coronavirus full-length S protein generated much higher neutralizing antibody titers compared to mRNA encoding the S protein S2 subunit. New Zealand white rabbits immunized with MERS-CoV mRNA vaccine encoding the full-length S protein reduced more than 90% of the viral load in the lungs of the rabbits and induced a significant amount of neutralizing antibody against MERS-CoV. Moderna announced on February 24, 202055 that it has released the first batch of mRNA-1273 against SARS-CoV-2 for use in humans, prepared using methods and strategies outlined in their previous patents. Vials of mRNA-1273 have been shipped to the National Institute of Allergy and Infectious Diseases (NIAID), a division of the National Institutes of Health (NIH), to be used in the planned Phase 1 study in the United States. Moderna reports that mRNA-1273 is an mRNA vaccine targeting a prefusion-stabilized form of the S protein associated with SARS-CoV-2, which was selected by Moderna in collaboration with investigators at the NIAID Vaccine Research Center. Manufacture of this batch was funded by the Coalition for Epidemic Preparedness Innovations.
Patent application WO2018115527 describes vaccines comprising mRNA encoding at least one antigen of a MERS coronavirus, preferably a S protein or a S protein fragment (S1), an envelope protein (E), a membrane protein (M), or a nucleocapsid protein (N), all of which were effective in inducing an antigen-specific immune response. Intradermal administration into mice of a lipid nanoparticle (LNP)-encapsulated mRNA mixture encoding MERS-CoV S proteins was shown to result in translation in vivo and induction of humoral immune responses.
SUMMARY AND PERSPECTIVES
This report provides an overview of published information on global research and development of coronavirus-related therapeutic agents and preventive vaccines based on the extensive CAS content collection, with a focus on patents. It includes an overview of coronavirus morphology, biology, and pathogenesis with a particular focus on antiviral strategies involving small molecule drugs, as well as biologics targeting complex molecular interactions involved in coronavirus infection and replication. The drug-repurposing effort summarized in this report is focused primarily on agents currently known to be effective against other RNA viruses including SARS-CoV, MERS-CoV, influenza, HCV, and Ebola as well as anti-inflammatory drugs. The potential impact of biologics for treatment of coronavirus infections is promising and includes a wide variety of options including bioengineered and vectored antibodies, cytokines, and nucleic acid-based therapies targeting virus gene expression as well as various types of vaccines.
The information provided in this report provides a strong intellectual groundwork for support of ongoing research and development for discovery and development of therapeutic agents and vaccines for treatment of COVID-19 and coronavirus-related diseases. Because of limited space, this report devotes minimal attention to current efforts involved in advancing more efficient and accurate COVID-19 diagnosis methods and products.
Novel infectious diseases resulting from RNA viruses subject to mutation and genetic recombination, as well as cross-species transmission, will continue to present a serious global health threat, as exemplified by COVID-19. Despite two former major outbreaks of coronavirus infections causing the SARS and MERS respiratory illnesses, the world remains underprepared to effectively manage the current COVID-19 outbreak, as evidenced by the fact that COVID-19 has resulted in thousands of deaths worldwide.
A concerted effort to develop effective drugs and vaccines against existing and potential future coronavirus infections and other highly pathogenic virus outbreaks is necessary to reduce overwhelming impacts on human life and worldwide healthcare systems. Given the costly and arduous process involved with clinical drug development, the outbreak of COVID-19 further highlights the value of developing relatively broad-spectrum antiviral drugs and the importance of applying innovative approaches such as artificial intelligence to facilitate drug discovery. Given the lengthy process of new drug development, the current strategy of drug repurposing has become one of the chosen solutions for immediate treatment of SARS-CoV-2 infected individuals. Long-term drug development goals for the pharmaceutical industry include identification of inhibitors aimed at the replication or infection processes associated with SARS-CoV-2 or other related coronaviruses, as well as the symptomatic results of their infections leading to severe disease and/or death. The summarized lists, contained in this report, of small molecule compounds, and additional descriptions of biologics with properties suitable for inhibiting several key coronavirus proteins, could serve as information starting points for drug development. Since vaccines are crucial for prevention of coronavirus-related epidemic diseases in the future, it is reassuring that a number of innovative strategies are already being deployed. Four MERS coronavirus DNA vaccine candidates began phase 1 clinical trials in September of 2019,56 and Moderna Inc. released its first batch of mRNA-1273 in February of 2020, which is an mRNA vaccine against SARS-CoV-2 ready for phase 1 study in the United States.55
Additional collaboration in the areas of antiviral discovery processes and clinical trial performance will enhance patients’ access to drug candidates with improved therapeutic potential and ideally reduce the amount of time required to bring these drugs to market. The abundance of publications and the rapid publication rate associated with the SARS-CoV-2 virus-related disease outbreak, as illustrated in this report, are indicative of the intense effort by research institutes and pharmaceutical industries to address both molecular mechanisms and therapeutic routes useful for treating current and future coronavirus outbreaks.
Acknowledgments
We would like to thank Dr. Gilles George for his encouragement and support for this work. We are grateful to Cristina Tomeo for her advice, and Andrey Sharkov and Cinda Harold for their useful input.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c00272.
Tables of distribution of RNAi patents related to SARS and MERS in the CAS content collection and distribution of vaccine patents related to SARS and MERS in the CAS content collection (PDF)
The authors declare no competing financial interest.
Footnotes
To learn more about CAS proprietary fingerprints: https://www.cas.org/resources/case-studies/data-quality-impacts-machine-learning
Supplementary Material
References
- Gorbalenya A. E.et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 2020, 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K.; Cohen J. Will novel virus go pandemic or be contained?. Science 2020, 367 (6478), 610–611. 10.1126/science.367.6478.610. [DOI] [PubMed] [Google Scholar]
- Coronavirus Disease (COVID-2019) Situation Reports 1–45; World Health Organization, 2020.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- Coronavirus is now expected to curb global economic growth by 0.3% in 2020. https://www.forbes.com/sites/sergeiklebnikov/2020/02/11/coronavirus-is-now-expected-to-curb-global-economic-growth-by-03-in-2020/#5de149ad16da.
- Anthony S. J.; Johnson C. K.; Greig D. J.; Kramer S.; Che X.; Wells H.; Hicks A. L.; Joly D. O.; Wolfe N. D.; Daszak P.; Karesh W.; Lipkin W. I.; Morse S. S.; Mazet J. A. K.; Goldstein T. Global patterns in coronavirus diversity. Virus Evol 2017, 3 (1), vex012. 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Wong G.; Shi W.; Liu J.; Lai A. C.K.; Zhou J.; Liu W.; Bi Y.; Gao G. F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24 (6), 490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382 (8), 727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.; Bragazzi N. L.; Li Q.; Tang S.; Xiao Y.; Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov). Infect Dis Model 2020, 5, 248–255. 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; He Y.; Zhou Y.; Liu S.; Zheng B.-J.; Jiang S. The spike protein of SARS-CoV - A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7 (3), 226–236. 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM structure of the 2019-nCoV Spike in the prefusion conformation. Science 2020, eabb2507. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Kruger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Muller M. A.; Drosten C.; Pohlmann S.. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 10.1016/j.cell.2020.02.052. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E.; Snijder E. J.; Ziebuhr J. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000, 81 (4), 853–879. 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Baez-Santos Y. M.; St. John S. E.; Mesecar A. D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015, 115, 21–38. 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.-W.; Cherney M. M.; Huitema C.; Liu J.; James K. E.; Powers J. C.; Eltis L. D.; James M. N.G. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J. Mol. Biol. 2005, 353 (5), 1137–1151. 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J; Wang D.; Xu W.; Holmes E. C; Gao G. F; Wu G.; Chen W.; Shi W.; Tan W. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J. S.; et al. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem 2020, 21 (5), 730–738. 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W.; Kok K.-H.; Zhu Z.; Chu H.; To K. K.-W.; Yuan S.; Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infect. 2020, 9 (1), 221–236. 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., et al. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China, bioRxiv 2020, 10.1101/2020.01.20.913368. [DOI] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Leist S. R.; Schafer A.; Won J.; Brown A. J.; Montgomery S. A.; Hogg A.; Babusis D.; Clarke M. O.; Spahn J. E.; Bauer L.; Sellers S.; Porter D.; Feng J. Y.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S.. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV Nat. Commun. 2020, Ahead of Print. 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam R. U.; Wilson I. A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (2), 206–214. 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therapeutic options for the 2019 novel coronavirus (2019-nCoV). https://www.nature.com/articles/d41573-020-00016-0. [DOI] [PubMed]
- The Efficacy of Lopinavir Plus Ritonavir and Arbidol Against Novel Coronavirus Infection (ELACOI). https://clinicaltrials.gov/ct2/show/NCT04252885.
- Glowacka I.; Bertram S.; Herzog P.; Pfefferle S.; Steffen I.; Muench M. O.; Simmons G.; Hofmann H.; Kuri T.; Weber F.; Eichler J.; Drosten C.; Pohlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. Journal of Virology 2010, 84 (2), 1198. 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.Compensation of ACE2 function for possible clinical management. Virol. Sin. 2020, 10.1007/s12250-020-00205-6. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M.; Hsieh F.; Baronas E.; Godbout K.; Gosselin M.; Stagliano N.; Donovan M.; Woolf B.; Robison K.; Jeyaseelan R.; Breitbart R. E.; Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000, 87 (5), 426. 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Imai Y.; Kuba K.; Rao S.; Huan Y.; Guo F.; Guan B.; Yang P.; Sarao R.; Wada T.; Leong-Poi H.; Crackower M. A.; Fukamizu A.; Hui C.-C.; Hein L.; Uhlig S.; Slutsky A. S.; Jiang C.; Penninger J. M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436 (7047), 112–116. 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis S. R.; Hooper N. M.; Hyde R.; Karran E.; Christie G.; Turner A. J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275 (43), 33238–33243. 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- De Witt B. J; Garrison E. A; Champion H. C; Kadowitz P. J. L-163,491 is a partial angiotensin AT(1) receptor agonist in the hindquarters vascular bed of the cat. Eur. J. Pharmacol. 2000, 404 (1-2), 213–219. 10.1016/S0014-2999(00)00612-9. [DOI] [PubMed] [Google Scholar]
- Guo D.Old weapon for new enemy: drug repurposing for treatment of newly emerging viral diseases. Virol. Sin. 2020, 10.1007/s12250-020-00204-7. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxmen A. More than 80 clinical trials launch to test coronavirus treatments. Nature 2020, 578 (7795), 347–348. 10.1038/d41586-020-00444-3. [DOI] [PubMed] [Google Scholar]
- Arabi Y. M; Shalhoub S.; Mandourah Y.; Al-Hameed F.; Al-Omari A.; Al Qasim E.; Jose J.; Alraddadi B.; Almotairi A.; Al Khatib K.; Abdulmomen A.; Qushmaq I.; Sindi A. A; Mady A.; Solaiman O.; Al-Raddadi R.; Maghrabi K.; Ragab A.; Al Mekhlafi G. A; Balkhy H. H; Al Harthy A.; Kharaba A.; Gramish J. A; Al-Aithan A. M; Al-Dawood A.; Merson L.; Hayden F. G; Fowler R.. Ribavirin and Interferon Therapy for Critically Ill Patients With Middle East Respiratory Syndrome: A Multicenter Observational Study. Clin. Infect. Dis. 2019, DOI: 10.1093/cid/ciz544. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangji Xinguangzhuang Bingdu Feiyan Zhuanli Xinxi Yanbao. https://tech.sina.cn/2020-02-17/detail-iimxxstf2046715.d.html.
- Warren T. K.; Wells J.; Panchal R. G.; Stuthman K. S.; Garza N. L.; Van Tongeren S. A.; Dong L.; Retterer C. J.; Eaton B. P.; Pegoraro G.; Honnold S.; Bantia S.; Kotian P.; Chen X.; Taubenheim B. R.; Welch L. S.; Minning D. M.; Babu Y. S.; Sheridan W. P.; Bavari S. Protection Against Filovirus Diseases by a Novel Broad-Spectrum Nucleoside Analogue BCX4430. Nature 2014, 508 (7496), 402–405. 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.; Griffin I.; Tucker C.; Smith D.; Oechsle O.; Phelan A.; Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395 (10223), e30–e31. 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud E. J.; Hayden F. G.; Hurt A. C. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res. 2019, 169, 104545. 10.1016/j.antiviral.2019.104545. [DOI] [PubMed] [Google Scholar]
- Zeldin R. K. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Antimicrob. Chemother. 2003, 53 (1), 4–9. 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- Ortiz-Alcantara J.; et al. Small molecule inhibitors of the SARS-CoV NSP15 endoribonuclease. Virus Adapt. Treat. 2010, 2, 125–133. 10.2147/VAAT.S12733. [DOI] [Google Scholar]
- Zhou Y.; Vedantham P.; Lu K.; Agudelo J.; Carrion R.; Nunneley J. W.; Barnard D.; Pohlmann S.; McKerrow J. H.; Renslo A. R.; Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015, 116, 76–84. 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.; Shin J. S.; Shie J.-J.; Ku K. B.; Kim C.; Go Y. Y.; Huang K.-F.; Kim M.; Liang P.-H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 2017, 141, 101–106. 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K. Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 2003, 300 (5626), 1763–1767. 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Li F. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309 (5742), 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Bisht H.; Roberts A.; Vogel L.; Subbarao K.; Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology 2005, 334 (2), 160–165. 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. Lancet 2020, 395, 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y.; Huang Y.; Ganesh L.; Leung K.; Kong W.-P.; Schwartz O.; Subbarao K.; Nabel G. J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein andenhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004, 78, 5642–5650. 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A.; Gramberg T.; Simmons G.; Moller P.; Rennekamp A. J.; Krumbiegel M.; Geier M.; Eisemann J.; Turza N.; Saunier B.; Steinkasserer A.; Becker S.; Bates P.; Hofmann H.; Pohlmann S. DC-SIGN and DCSIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol 2004, 78, 12090–12095. 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The overview of Interferon. https://www.cusabio.com/c-20629.html.
- Zeng J.Cross Kingdom small RNAs among Animals, Plants and Microbes Cells 2019, 8( (4), ) 371, 10.3390/cells8040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B. microRNAs in Cardiovascular Disease: Small Molecules but Big Roles. Current Topics in Medicinal Chemistry 2019, 19 (21), 1918–1947. 10.2174/1568026619666190808160241. [DOI] [PubMed] [Google Scholar]
- Liu J.; Guo B. RNA-based therapeutics for colorectal cancer: Updates and future Directions. Pharmacol. Res. 2020, 152, 104550. 10.1016/j.phrs.2019.104550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U. J.; Bukreyev A.; Yang L.; Lamirande E. W.; Murphy B. R.; Subbarao K.; Collins P. L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (26), 9804–9809. 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GlaxoSmithKline press release on 2/24/20. https://www.gsk.com/en-gb/media/press-releases/clover-and-gsk-announce-research-collaboration-to-evaluate-coronavirus-covid-19-vaccine-candidate-with-pandemic-adjuvant-system.
- Generex press release on 2/27/20. https://storage.googleapis.com/wzukusers/user-26831283/documents/5e57ed391b286sVf68Kq/PR_Generex_Coronavirus_Update_2_27_2020.pdf.
- Novavax press release on 2/26/20. http://ir.novavax.com/news-releases/news-release-details/novavax-advances-development-novel-covid-19-vaccine.
- Moderna press release on 2/24/2020. https://investors.modernatx.com/news-releases/news-release-details/moderna-ships-mrna-vaccine-against-novel-coronavirus-mrna-1273.
- Modjarrad K.; Roberts C. C; Mills K. T; Castellano A. R; Paolino K.; Muthumani K.; Reuschel E. L; Robb M. L; Racine T.; Oh M.-d.; Lamarre C.; Zaidi F. I; Boyer J.; Kudchodkar S. B; Jeong M.; Darden J. M; Park Y. K; Scott P. T; Remigio C.; Parikh A. P; Wise M. C; Patel A.; Duperret E. K; Kim K. Y; Choi H.; White S.; Bagarazzi M.; May J. M; Kane D.; Lee H.; Kobinger G.; Michael N. L; Weiner D. B; Thomas S. J; Maslow J. N Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019, 19 (9), 1013–1022. 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inovio Accelerates Timeline for COVID-19 DNA Vaccine INO-4800. http://ir.inovio.com/news-and-media/news/press-release-details/2020/Inovio-Accelerates-Timeline-for-COVID-19-DNA-Vaccine-INO-4800/default.aspx.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.