Abstract
To investigate the diversity of IS6110 fingerprints of Mycobacterium tuberculosis isolates in the United States and to determine if matching IS6110 fingerprints represent recent interstate tuberculosis transmission, we performed restriction fragment length polymorphism analysis of M. tuberculosis isolates from 1,326 patients in three geographically separated states. Seven hundred ninety-five different IS6110 fingerprint patterns were generated, and pattern diversity was similar in each state. Ninety-six percent of the fingerprint patterns were observed in only one state, demonstrating that most IS6110 fingerprint patterns are confined to a single geographic location. Of the IS6110 fingerprint patterns that were shared by isolates from more than one state, most isolates with 1 to 5 IS6110 copies were separable by pTBN12 fingerprinting whereas those with >15 copies were not. One high-copy-number M. tuberculosis strain had identical IS6110 and pTBN12 fingerprints and included 57 isolates from three states. Epidemiological data demonstrated significant recent transmission of tuberculosis within each city but not among the states. This suggests that identical fingerprints of isolates from geographically separate locations most likely reflect interstate tuberculosis transmission in the past, with subsequent intrastate spread of disease. Further evaluation of M. tuberculosis strains that cause outbreaks in different geographic locations will provide insight into the epidemiological and bacteriological factors that facilitate the spread of tuberculosis.
Our understanding of the dynamics of tuberculosis transmission has been greatly enhanced by the development of genotyping methods that allow identification of specific strains of Mycobacterium tuberculosis. The most widely used genotyping method is DNA fingerprinting based on restriction fragment length polymorphism (RFLP) analysis with the insertion element IS6110, which produces extensive fingerprint diversity among M. tuberculosis isolates (6, 9, 12, 13, 16). In some locations, epidemiological and RFLP analyses suggest that identical IS6110 fingerprints among M. tuberculosis isolates are a marker of recent tuberculosis transmission (1, 3, 11, 15), whereas in other areas, identical IS6110 fingerprints do not indicate recent tuberculosis transmission (4, 5). Because most IS6110 fingerprint analyses of M. tuberculosis isolates have been confined to specific geographic locales (1–5, 11, 15), the diversity of IS6110 fingerprints in different parts of the United States has not been systematically evaluated. In addition, it is uncertain if matching fingerprints obtained from patients in different states indicate that the patients are epidemiologically linked by recent tuberculosis transmission. To examine these issues, we performed RFLP analysis of M. tuberculosis isolates obtained from 1,326 patients from three geographically separated states.
MATERIALS AND METHODS
M. tuberculosis strains.
We studied one M. tuberculosis isolate from each of 1,326 patients in California, Texas, and Colorado. One thousand one hundred seventy-six isolates (89%) included all available isolates from public health departments at five geographically separate locations during specific time periods. These locations were in California (Alameda County and Contra Costa County [308 isolates, 1992 to 1994] and central Los Angeles [214 isolates, 1993 to 1995]), Texas (Fort Worth [219 isolates, 1993 to 1995] and El Paso [177 isolates, 1994 to 1995]), and Colorado (Denver, 256 isolates, 1989 to 1994). In addition, we included 150 drug-resistant isolates obtained throughout the state of Texas (1991 to 1995).
All isolates were identified as M. tuberculosis by the submitting laboratories. The isolates were received on Lowenstein-Jensen slants, and each was subcultured in 5 ml of Dubos-Tween 80 medium supplemented with albumin (Difco, Detroit, Mich.) and then incubated at 37°C for 2 to 3 weeks prior to DNA extraction.
DNA fingerprinting.
Chromosomal DNA was prepared by chloroform-isoamyl alcohol extraction (14). One microgram of DNA from each isolate was either restricted with PvuII and then fingerprinted with the IS6110 probe or restricted with AluI and then fingerprinted with the pTBN12 probe. Fingerprinting was performed by standard methods (8, 17). The DNA molecular size standard used for IS6110 fingerprinting was PvuII-restricted chromosomal DNA of M. tuberculosis H37Rv and two additional DNA fragments which hybridize to IS6110 (18). A 1-kb DNA ladder was the DNA molecular size standard for pTBN12 fingerprinting. The DNA probes were prepared as reported previously (18) and labeled with [α-32P]dCTP by the random-primer method (10).
Fingerprint analyses and comparison of patterns.
For computer-assisted analyses of IS6110 fingerprints, hybridized blots were exposed to a phosphor screen, the screen was scanned with Image Quant software (Molecular Dynamics, Sunnyvale, Calif.), and the patterns were analyzed with Whole Band Analyzer software (version 3.3; Bioimage, Ann Arbor, Mich.). A band size deviation of up to 2.5% was allowed when matching the patterns. Matching was determined by the average linkage clustering method. Patterns that matched at the 100% level were considered identical, and isolates with identical patterns were considered a cluster.
The geographic source of each IS6110 fingerprint pattern was recorded, and a fingerprint pattern file was developed for each state; this file contained all patterns generated by the isolates from that state. IS6110 fingerprints from different states were compared with each other, as well as with patterns found in a previous study of 210 M. tuberculosis isolates obtained from inmates of the prison system of Madrid, Spain (7). pTBN12 fingerprints in adjacent lanes were visually compared.
RESULTS
IS6110 fingerprints in the study population.
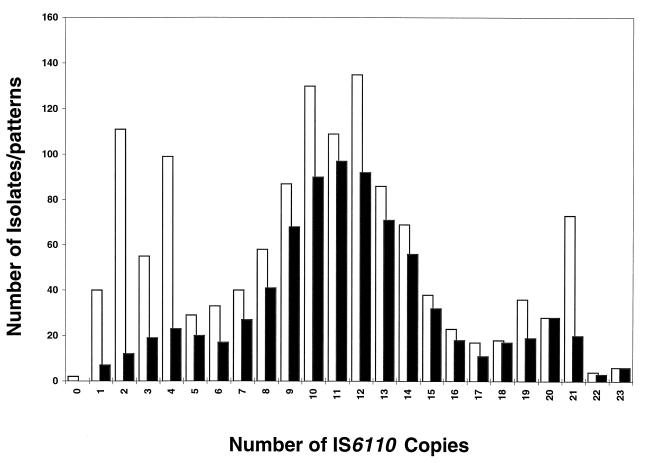
The IS6110 fingerprints for isolates from 1,326 patients were grouped according to the number of copies of IS6110 (Fig. 1). Two isolates with no copies of IS6110 were confirmed to be M. tuberculosis by a DNA probe method (Accuprobe; Gen-Probe, San Diego, Calif.) and conventional biochemical tests. Three hundred thirty-two (25%) isolates contained 1 to 5 copies of IS6110 (low-copy-number strains), 782 isolates (59%) contained 6 to 15 copies, and 212 isolates (16%) had 16 or more copies (high-copy-number strains).
FIG. 1.
Comparison of diversities of IS6110 fingerprints. The light bars show the frequency distribution of IS6110 in M. tuberculosis isolates collected from 1,326 patients in three geographically separated states; the dark bars represent the frequency distribution of 795 IS6110 fingerprint patterns observed in M. tuberculosis isolates from the same patients, excluding two whose isolates were found to lack IS6110.
The two isolates with no copies of IS6110 were excluded, and the remaining 1,324 isolates showed 795 different fingerprint patterns, confirming the extensive polymorphism of IS6110 fingerprints in the United States. Of the 795 patterns, 80 (10%) were low-copy-number strains, 588 (74%) contained 6 to 15 copies of IS6110, and 127 (16%) were high-copy-number strains. As shown in Fig. 1, the diversity of DNA fingerprint patterns was highest in strains with 6 to 15 copies of IS6110, intermediate in high-copy-number strains, and lowest in low-copy-number strains.
Diversity of IS6110 patterns within three states.
In Table 1, the diversities of IS6110 fingerprint patterns in the three states are compared. All three states showed similar percentages of clustered isolates and similar degrees of pattern diversity, as estimated by the mean number of isolates per pattern.
TABLE 1.
IS6110 fingerprint patterns of M. tuberculosis isolates from three statesa
| State | No. of isolates | No. (%) of isolates in clusters | No. of patterns | No. of clusters | Mean no. of isolates per pattern |
|---|---|---|---|---|---|
| Texas | 546 | 216 (40) | 381 | 50 | 1.4 |
| California | 522 | 242 (46) | 348 | 48 | 1.5 |
| Colorado | 256 | 120 (47) | 189 | 30 | 1.4 |
When DNA fingerprint patterns of the three states were compared, it was found that 762 (96%) of the 795 patterns were restricted to one state and only 33 (4%) were not, demonstrating that most IS6110 patterns in this set are confined to a specific geographic location. Although the number of IS6110 patterns found in two or more states was small, the number of isolates with these patterns was large, including 397 (30%) of the 1,326 isolates. Of these 397 isolates, 245 were low-copy-number strains comprising 12 fingerprint patterns and 152 had 6 or more copies of IS6110 comprising 21 fingerprint patterns. The distribution of these 21 patterns in the three states is shown in Table 2.
TABLE 2.
Isolates of M. tuberculosis obtained from more than one state that shared fingerprint patterns having six or more IS6110 copiesa
| IS6110 copy no. | Fingerprint pattern | No. of isolates by state of origin
|
||
|---|---|---|---|---|
| California | Colorado | Texas | ||
| 6 | 6-1 | 1 | 0 | 1 |
| 7 | 7-1 | 0 | 1 | 4 |
| 8 | 8-1 | 0 | 2 | 1 |
| 8 | 8-2 | 0 | 1 | 1 |
| 9 | 9-1 | 2 | 1 | 4 |
| 10 | 10-1 | 0 | 1 | 1 |
| 10 | 10-2 | 0 | 1 | 8 |
| 10 | 10-3 | 1 | 3 | 1 |
| 10 | 10-4 | 4 | 0 | 3 |
| 12 | 12-1 | 1 | 0 | 1 |
| 12 | 12-2 | 12 | 3 | 4 |
| 12 | 12-3 | 0 | 1 | 4 |
| 12 | 12-4 | 1 | 2 | 2 |
| 12 | 12-5 | 0 | 1 | 2 |
| 12 | 12-6 | 1 | 1 | 0 |
| 12 | 12-7 | 1 | 0 | 5 |
| 13 | 13-1 | 0 | 2 | 2 |
| 13 | 13-2 | 0 | 1 | 1 |
| 14 | 14-1 | 0 | 1 | 3 |
| 17 | 17-1 | 1 | 2 | 0 |
| 21 | 21-1 | 39 | 2 | 16 |
Evaluation of IS6110 patterns shared by isolates from more than one state.
To determine if the M. tuberculosis isolates from different states with identical IS6110 fingerprints represented the same genotype, we studied isolates from 32 of the 33 interstate clusters. One isolate from each state in the cluster was randomly selected, including 30 low-copy-number strains comprising 12 patterns and 46 isolates with 6 or more IS6110 copies comprising 20 patterns. The IS6110 fingerprints of these 76 isolates were reexamined after electrophoresis of restriction digests in adjacent lanes on the same gel. This analysis confirmed that the IS6110 fingerprints were identical in all cases (data not shown).
The 76 isolates were then subjected to secondary fingerprinting with the pTBN12 probe. This probe was used because it distinguishes M. tuberculosis strains with identical low-copy-number IS6110 fingerprints (5, 8, 18) and because pTBN12-based fingerprinting results have been validated by epidemiological studies (3, 5, 18). The percentage of isolates that were differentiated by pTBN12 fingerprinting varied inversely with the number of IS6110 copies. Twenty-four (80%) of 30 low-copy-number strains were no longer clustered after pTBN12 typing, compared to none of 6 high-copy-number strains (Table 3). This finding is consistent with the intuitive concept that the random probability of two isolates having identical IS6110 fingerprints increases as the number of IS6110 copies falls.
TABLE 3.
Relationship of IS6110 copy number to results of pTBN12 fingerprinting of M. tuberculosis isolatesa
| IS6110 copy no. | No. of isolates in IS6110 clusters (no. of clusters) | Isolates differentiated by pTBN12 fingerprinting (%) |
|---|---|---|
| 1–5 | 30 (12) | 24 (80) |
| 6–10 | 20 (9) | 7 (35) |
| 12–14 | 20 (9) | 3 (14) |
| 17–21 | 6 (2) | 0 (0) |
The identical IS6110 and pTBN12 patterns observed for some M. tuberculosis isolates obtained from different states could have arisen by chance alone, or they could represent interstate transmission of tuberculosis. To evaluate these possibilities, we compared the 795 fingerprint patterns of the 1,326 isolates from the United States with 79 fingerprint patterns previously found for 210 isolates from prisoners in Madrid, Spain (7). This population was selected because these two groups of patients were extremely unlikely to be epidemiologically linked. Only one pattern with three IS6110 copies was shared by one Spanish isolate and four Texas isolates. pTBN12 patterns were identical for the Texas isolates and different from the Spanish isolate. These findings suggest that identical IS6110 and pTBN12 fingerprints in different geographic locations are unlikely to arise by chance alone. Therefore, the isolates with identical fingerprint patterns common to two or more states may well have resulted from interstate transmission of tuberculosis.
Evaluation of the largest interstate cluster.
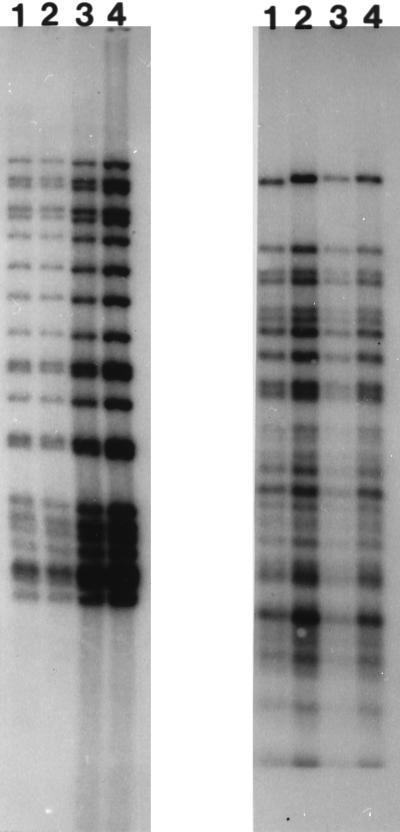
To determine if interstate transmission of tuberculosis in the recent past accounted for clustering of isolates from different states, we performed a more detailed fingerprinting and epidemiological analysis of the largest interstate cluster, which included 57 isolates with 21 copies of IS6110 (cluster 21-1 in Table 2). Thirty-nine patients were from California (38 from Los Angeles and 1 from Alameda-Contra Costa County), 16 patients were from Fort Worth, Tex., and two patients were from Denver, Colo. Twenty-six of these isolates were randomly selected for pTBN12 secondary fingerprinting (one isolate from Alameda-Contra Costa County, Calif.; 19 isolates from Los Angeles, Calif.; one isolate from Denver, Colo.; and 5 isolates from Fort Worth, Tex.), and all fingerprints were identical, suggesting that these isolates represent the same genotype. Examples of these IS6110 and pTBN12 fingerprints are shown in Fig. 2.
FIG. 2.
IS6110 (left panel) and pTBN12 (right panel) fingerprint patterns of four M. tuberculosis isolates from the largest cluster found in more than one state. The isolates were obtained from Denver, Colo. (lane 1), Alameda-Contra Costa County, Calif. (lane 2), Los Angeles, Calif. (lane 3), and Fort Worth, Tex. (lane 4).
Epidemiological information regarding the patients in Los Angeles and Fort Worth, who comprised 95% (54 of 57) of the largest cluster, was obtained. Of the 38 patients from central Los Angeles, 37 were homeless or marginally housed, often eating and spending time at homeless shelters. Of the 34 patients who were interviewed, 28 spent long periods at one of three homeless shelters, suggesting that these were sites of disease transmission. None of the patients had lived in or near Fort Worth. Three patients were born in Texas and had moved to Los Angeles 5, 36, and 50 years prior to development of tuberculosis.
Of the 16 patients from Fort Worth, five were homeless or marginally housed persons who lived in the same area. Four patients lived in a small African-American community within Fort Worth. These two groups were epidemiologically linked by drug users who were members of both communities. The other seven patients were from different economic and ethnic groups and were not epidemiologically linked. Two of the 16 patients had lived in Los Angeles. One patient lived 10 miles from central Los Angeles for 4 months in 1990, 3 years prior to being diagnosed with tuberculosis. Her spouse was a drug user who often brought others to visit the residence. The second patient was homeless in central Los Angeles at various periods from 1983 to 1990 and developed tuberculosis in 1993. None of the patients from Fort Worth or Los Angeles was listed in the tuberculosis registry of the other city or had traveled to the other city within 3 years of diagnosis.
DISCUSSION
Our findings confirm and extend previous observations regarding the extensive diversity of IS6110 fingerprints among M. tuberculosis isolates in the United States. For isolates from 1,324 patients from three states, 795 IS6110 fingerprint patterns were generated, and similar degrees of pattern diversity were observed in each state. Ninety-six percent of the fingerprint patterns were found in only one state, demonstrating that most IS6110 patterns are confined to a specific geographic location. Of the IS6110 fingerprint patterns that were shared by more than one state, most low-copy-number patterns were separable by pTBN12 fingerprinting whereas high-copy-number patterns were not. In one large cluster, epidemiological data demonstrated extensive intrastate transmission of tuberculosis and suggested the potential for interstate disease transmission. Since none of the patients in Fort Worth or Los Angeles had traveled to the other city within 3 years of the diagnosis of tuberculosis, a more plausible hypothesis is that transmission of this strain between these areas occurred a few years ago, with subsequent local transmission of the disease. No evidence of recent interstate transmission was found, demonstrating that matching IS6110 and pTBN12 fingerprints of isolates from geographically separate locations are not indicative of a recent tuberculosis outbreak.
pTBN12 fingerprinting distinguishes M. tuberculosis isolates with identical IS6110 fingerprints comprised of five or fewer fragments, and pTBN12 fingerprinting results correlate well with epidemiological data suggestive of tuberculosis transmission (18). In previous studies, M. tuberculosis isolates from the same locale with identical fingerprints containing six or more copies of IS6110 were not further discriminated by pTBN12 fingerprinting (5, 8). In contrast, we found that the percentage of isolates that were differentiated by pTBN12 fingerprinting varied inversely with the number of IS6110 copies (Table 3). More than one-third of our isolates with identical fingerprints containing 6 to 10 copies of IS6110 were separated by pTBN12 fingerprinting. Therefore, in evaluating patients from different geographic regions, M. tuberculosis isolates should be considered part of a cluster only when the isolates have matching IS6110 and pTBN12 fingerprints. pTBN12 fingerprinting is less essential for confirmation of clustering among isolates with six or more IS6110 copies in the same geographic location, because the probability of disease transmission is higher.
Our findings provide strong evidence that a single M. tuberculosis strain caused disease in a substantial number of patients in more than one state. The isolates from different states were high-copy-number strains with identical IS6110 and pTBN12 fingerprints, and epidemiological data confirmed significant transmission of this strain within the cities of Fort Worth, Tex., and Los Angeles, Calif. Epidemiological investigations did not reveal recent contact between tuberculosis patients in these cities, indicating that identical fingerprint patterns of M. tuberculosis isolates from different locations do not necessarily indicate recent disease transmission between these locations. It is more likely that our findings reflect interstate transmission of tuberculosis a few years ago with subsequent intrastate spread of disease, and our epidemiological data suggested the potential for such linkages. Population groups at increased risk for tuberculosis, such as homeless persons and drug users, are highly mobile, favoring continued spread of M. tuberculosis across state borders.
Identification of M. tuberculosis strains that cause outbreaks in different geographic locations is important for at least two reasons. First, it improves our understanding of the epidemiological factors that facilitate the spread of tuberculosis. Second, it may allow identification of biological characteristics of M. tuberculosis strains that favor disease transmission. An understanding of these factors will enhance our understanding of the pathogenesis of tuberculosis and permit development of more-effective tuberculosis control strategies.
ACKNOWLEDGMENTS
This study was supported by a CDC-VA interagency agreement for the Tuberculosis Genotyping and Surveillance Network and by the National Institutes of Health (grants 1R01A135265 and AI35222). Peter F. Barnes holds the Margaret E. Byers Cain Chair for Tuberculosis Research.
We thank Don Cunningham, Stephen Kincaid, and Susan Rowland for excellent technical assistance.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Ducker E, Bloom B R. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Barnes P F, El Hajj H, Preston-Martin S, Cave M D, Jones B E, Otaya M, Pogoda J, Eisenach K D. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. [PubMed] [Google Scholar]
- 3.Barnes P F, Yang Z, Preston-Martin S, Pogoda J M, Jones B E, Otaya M, Eisenach K D, Knowles L, Harvey S, Cave M D. Patterns of tuberculosis transmission in central Los Angeles. JAMA. 1997;278:1159–1163. [PubMed] [Google Scholar]
- 4.Braden C R, Templeton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of M. tuberculosis isolates from a stable, relatively rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 5.Burman W J, Reves R R, Hawkes A P, Rietmeijer C A, Yang Z, El Hajj H, Bates J H, Cave M D. DNA fingerprinting with two probes decreases clustering of M. tuberculosis. Am J Respir Crit Care Med. 1997;155:1140–1146. doi: 10.1164/ajrccm.155.3.9117000. [DOI] [PubMed] [Google Scholar]
- 6.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 7.Chaves F, Dronda F, Cave M D, Alonso-Sanz M, Gonzalez-Lopez A, Eisenach K D, Ortega A, Lopez-Cubero L, Fernandez-Martin I, Catalan S, Bates J H. A longitudinal study of transmission of tuberculosis in a large prison population. Am J Respir Crit Care Med. 1997;155:719–725. doi: 10.1164/ajrccm.155.2.9032218. [DOI] [PubMed] [Google Scholar]
- 8.Chaves F, Yang Z, El Hajj H, Alonso M, Burman W J, Eisenach K D, Dronda F, Bates J H, Cave M D. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenach K D, Crawford J T, Bates J H. Repetitive DNA sequences as probes for Mycobacterium tuberculosis. J Clin Microbiol. 1988;26:2240–2245. doi: 10.1128/jcm.26.11.2240-2245.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg A P, Vogelstein B. A technique for radiolabelling DNA restriction fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 11.Genewein A, Telenti A, Bernasconi C, Mordasini C, Weiss S, Maurer A-M, Rieder H L, Schopfer K, Bodmer T. Molecular approach to identifying route of transmission of tuberculosis in the community. Lancet. 1993;342:841–844. doi: 10.1016/0140-6736(93)92698-s. [DOI] [PubMed] [Google Scholar]
- 12.Hermans P W M, van Soolingen D, Dale J W, Schuitema A R J, McAdam R A, Catty D, van Embden J D A. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J Clin Microbiol. 1990;28:2051–2058. doi: 10.1128/jcm.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAdam R A, Hermans P W M, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D A, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 14.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med. 1994;30:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 16.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendation for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Chaves F, Barnes P F, Burman W J, Koehler J, Eisenach K D, Bates J H, Cave M D. Evaluation of method for secondary DNA typing of Mycobacterium tuberculosis with pTBN12 in epidemiologic study of tuberculosis. J Clin Microbiol. 1996;34:3044–3048. doi: 10.1128/jcm.34.12.3044-3048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]