Abstract
The feasibility of the major peripheral blood leukocyte (PBL) subsets for use in qualitative and quantitative PCR to monitor secondary cytomegalovirus (CMV) infection and ganciclovir therapy was assessed with 188 blood samples derived from 40 CMV immunoglobulin G-positive renal-allograft recipients. In pp65 antigen-positive patients all leukocyte fractions, but only 79.5% of plasma preparations, were PCR positive. In pp65 antigen-negative samples from patients after antiviral treatment only 7.3% of polymorphonuclear cell (PMNL) samples, but 81.8% of peripheral blood mononuclear cells (PBMC), and 10.9% of plasma samples remained PCR positive. Similarly, in patients with latent infections only 5.0% of PMNL, but 51.7% of PBMC preparations, and 8.0% of plasma samples were PCR positive. Regarding patients with active CMV infection, CMV DNA copy numbers in PMNL correlated significantly with pp65 antigen-positive cell counts before and after onset of ganciclovir therapy. Significant differences in CMV DNA copy numbers in PMNL and plasma were observed (i) between patients with symptomatic infection and those with asymptomatic infection and (ii) between patients with active infection and those with latent infection. In contrast, PBMC harbored equally low CMV DNA levels both in patients with active infection and those with latent infections, and no decline of CMV DNA load in PBMC was observed during antiviral treatment. We conclude that detection of CMV DNA in PMNL, not in PBMC, is associated with active infections and is more sensitive than detection of CMV DNA in plasma. Negative PCR results for PMNL after antiviral therapy indicate recovery, and fewer unwanted positive results occur compared to PBMC and plasma. Therefore, purified PMNL should be preferred for analysis by qualitative CMV PCR to avoid unwanted positive results. The CMV DNA load in PBMC compared with that in PMNL is negligible during active infection, so mixed PBL are sufficient for use in quantitative PCR.
The level of viremia plays an important role in the course of cytomegalovirus (CMV) infection. The pp65 antigenemia assay and PCR methods for detection of CMV DNA in peripheral blood leukocytes (PBL) and plasma have been increasingly utilized in recent years for the rapid diagnosis of active CMV infection in immunocompromised hosts and for monitoring antiviral treatment (for reviews, see references 29 and 36). However, the identification of patients at risk for clinically relevant disease is problematic for various reasons. Firstly, low-level antigenemia is not necessarily indicative of symptomatic infections (36). On the other hand, positive PCR results from leukocytes are often obtained in pp65 antigen-negative samples of patients after successful antiviral therapy and even in CMV immunoglobulin G (IgG)-positive immunosuppressed individuals with latent infection (1, 2, 6, 20, 40, 42), although CMV DNA is only occasionally detectable in healthy seropositive blood donors (33–35). Plasma has been shown to yield fewer unwanted positive PCR results than PBL (10), but the lower sensitivity might be problematic with respect to early detection of CMV relapse or therapy monitoring (9, 10).
To overcome these problems, protocols for quantitative assessment of antigenemia and DNAemia have been devised, which are now successfully used for predicting symptomatic infection and for monitoring antiviral therapy (29). However, the antigenemia assay is error-prone due to rapid decline of pp65 antigens in blood samples processed with delay, whereas CMV DNA copy numbers in PBL remain at constant levels for at least 72 h (30). On the other hand, quantitative PCR requires elaborate techniques which limit its routine use. Thus, from the diagnostic standpoint it remains desirable to improve the clinical utility of qualitative CMV PCR. As PCR with PBL is the most sensitive, the selective detection of CMV DNA associated with active infection in PBL would be highly interesting.
Reports on the mechanisms of interaction between CMV and circulating leukocytes are still controversial (5, 12, 14, 17, 19, 22, 25, 26, 37, 39). Polymorphonuclear leukocytes (PMNL) are the predominant carriers of pp65 antigen and also harbor CMV DNA during active infections (5, 12, 15–17, 27). A positive correlation has been demonstrated between pp65 antigen-positive cell counts and the overall amount of CMV DNA present in mixed PBL during active infection (23, 38). However, only little information exists regarding the quantitative distribution of CMV DNA among leukocyte subpopulations (3, 27). The exact tropism and state of CMV genomes found in pp65-negative patients after successful antiviral therapy and in latently infected immunosuppressed patients are still poorly defined. CMV DNA occasionally detected in seropositive healthy blood donors is probably due to persistent viral genomes in peripheral blood mononuclear cells (PBMC) (33–35).
If the detection of CMV genomes in different leukocyte subsets reflects different states of the virus, the fraction chosen for PCR analysis should strongly influence the value of PCR in discriminating between active and latent CMV infection or in monitoring antiviral therapy. Therefore, we have compared qualitative and quantitative CMV PCRs for the major leukocyte subpopulations with plasma PCR for 40 CMV IgG-positive renal-allograft recipients (i) with active CMV infections before, during, and after ganciclovir treatment and (ii) with latent CMV infections.
MATERIALS AND METHODS
Patients.
One hundred eighty-eight blood samples from 40 CMV IgG-positive renal-allograft recipients were analyzed. Only patients without current antiviral treatment at the time of the first investigation were enrolled in the study. Patients without evidence of active CMV infection who were prophylactically treated with ganciclovir during steroid-resistant graft rejection therapy (anti-lymphocyte globulin [ATG] and/or OKT3) were excluded as well. Samples were collected weekly during the primary hospitalization phase (between 21 and 50 days after transplantation). Patients treated with ganciclovir during primary hospitalization were monitored every other day for 2 weeks and at weekly intervals for up to 5 further weeks. From five patients samples were drawn every 2 days for 2 weeks upon readmission to the ward due to active CMV infections between days 90 and 160 posttransplantation. All samples were analyzed prospectively.
Active CMV infection was confirmed by detection of pp65 antigens in PBL. Latent infection was assumed when leukocyte samples remained pp65 antigen negative and no organ involvement (see below) was detectable throughout the observation period. Positive CMV DNA PCR alone was not rated as active infection.
Patients suffering from symptomatic CMV infections or experiencing active CMV infection combined with steroid-resistant graft rejection were treated with ganciclovir for a minimum of 14 days at 10 mg/kg of body weight daily with adjustment for renal function. Symptomatic infection was defined by two CMV-associated symptoms (unexplained fever >38°C for >3 days, arthralgia, hyperhidrosis, graft dysfunction without histological evidence of rejection, leukopenia, thrombocytopenia, or liver enzyme elevation) and/or confirmed organ involvement (pneumonitis or gastrointestinal ulceration with virus isolation from biopsies) (21).
Isolation of leukocytes.
Ten milliliters of whole blood was collected in EDTA tubes and processed within 4 h postdrawing. Mixed PBL were isolated by centrifugation of 2 ml of whole blood at 700 × g for 3 min and subsequent lysis of erythrocytes with 0.8% ammonium chloride. Pelleted PBL were washed with phosphate-buffered saline, pH 7.4, and quantitated with a hematological cell counter. The proportions of PMNL and PBMC were determined, and aliquots containing 105 cells were subjected to antigenemia assay and PCR, respectively.
PBMC (lymphocytes and monocytes) and PMNL were semipurified by Ficoll-metrizoate density centrifugation using Lymphoprep and Polymorphprep (Nycomed, Oslo, Norway) according to the manufacturer’s instructions and quantitated. PBMC fractions contained about 5% PMNL, and PMNL fractions were contaminated by 5% lymphocytes and monocytes on average as determined by Giemsa staining and morphological analysis. Subfractions were adjusted with phosphate-buffered saline to the number of PMNL and PBMC per milliliter determined in 106 mixed PBL.
Antigenemia assay.
A total of 105 mixed PBL (100 μl) were centrifuged onto glass slides, fixed, and permeabilized with 5% paraformaldehyde–0.5% Nonidet P-40, and pp65 antigen was detected in PMNL by indirect immunofluorescence as described previously (11) using Clonab CMV (Biotest, Dreieich, Germany) by following the manufacturer’s instructions. Results were expressed as the average of two pp65-positive cell counts with deviations ≤20% for cell counts ≥100 and deviations ≤30% for counts ≤50 (data not shown).
DNA extraction from leukocytes and plasma.
DNA was extracted from 500 μl of mixed PBL, PMNL, and PBMC preparations (equivalent to the number of cells present in 5 × 105 mixed PBL) by digestion with 100 μg of proteinase K (8, 28) per ml in a 50-μl reaction volume. After being boiled for 10 min, a 10-μl supernatant sample was subjected to PCR.
Plasma was obtained by centrifugation of 1 ml of whole blood at 700 × g for 10 min to remove cells. Supernatants (approximately 500 μl) were centrifuged for a second time to remove cell debris and then extracted once with 1 volume of phenol-chloroform and subsequently with 1 volume of chloroform-isoamyl alcohol (24:1). DNA from 100 μl of the aqueous phase was precipitated with 0.1 volume of sodium acetate, pH 5.2, and 2.5 volumes of absolute ethanol. After centrifugation at 15,000 × g for 30 min, pellets were washed with 70% ethanol and resuspended in 100 μl of H2O. A 10-μl sample was subjected to PCR.
CMV PCR.
CMV and β-globin target sequences were quantitated by competitive PCR using cloned standard (ST) sequences as described elsewhere (20, 28) with slight modifications. The CMV ST sequence was generated in PCR by site-directed mutagenesis and subcloned into the vector pSPT19 (Boehringer Mannheim, Mannheim, Germany) (20). It contained three successive point mutations compared to the amplified coding region of glycoprotein B derived from the CMV laboratory strain AD169 (4). The cloning of the β-globin ST was previously described (18).
Ten-microliter samples of DNA extracts from leukocyte and plasma preparations were added to PCR mixtures (8, 28) to a total volume of 100 μl containing 2 × 105 copies of the β-globin ST and either 103 copies (high ST) or 50 copies (low ST) of the CMV ST. CMV target sequences were amplified in 20 cycles with the external CMV-specific primers E1 (5′-TCCAACACCCACTAGACCGGT-3′) and E2 (5′-CGGAAACGATGGTGTAGTTGG-3′) (8). Ten microliters each of the external reaction mixture was reamplified in a second round of PCR for 30 cycles with the internal CMV-specific primers TGGE1B (5′-CCGGATCCCGCCGCCCGCCCCGCGCCCGCCGCGGCAGCACCTGGCT-3′) and TGGE2E (5′-GCGAATTCGTAAACCACATCACCGTGGA-3′) and the β-globin-specific primers 1aB (5′-CCGGATCCCGCCGCCCGCCCCGCGCCCCTGCCGTTACTGCCCTGT-3′) and 1bE (5′-GCGAATCCTATTGGTCTCCTTAAACCTG-3′), respectively. ST and wild-type CMV and β-globin PCR amplimers were quantitated by hybridization to a strand-specifically labeled ST sequence, separation by temperature gradient gel electrophoresis, and densitometric analysis of autoradiographs (28). For products with ≥500 CMV DNA copies in 10 μl the values from the high-ST reaction were used, and for products with <500 CMV copies those from the low-ST reaction were used. Results were expressed as the average value of two measurements, which differed by ≤15% (data not shown). Exact quantification was possible within the ranges of 5 to 104 CMV wild-type genome and 2 × 103 to 2 × 105 β-globin copies per PCR.
In mixed PBL, CMV/cellular DNA ratios were expressed as CMV copies/2 × 105 copies of β-globin (the theoretical maximum DNA yield from 105 cells). To allow precise comparison of CMV DNA levels in PMNL and PBMC, CMV DNA copies were related to the expected β-globin DNA copy numbers present in the mixed PBL population. For example, if 6 × 104 PMNL were counted in 105 mixed PBL, the theoretical maximum cellular DNA yield from PMNL was 1.2 × 105 copies of β-globin. Thus, in this case the CMV/β-globin ratio in the PMNL fraction was expressed as CMV copies/1.2 × 105 copies of β-globin.
Plasma CMV copy numbers were expressed as CMV genome equivalents present in 10 μl of plasma (approximately equivalent to the volume of whole blood containing 105 PBL).
Statistical analysis.
Frequencies of PCR-positive results among different leukocyte subsets were compared by McNemar’s test. Frequencies of PCR-positive results for plasma were compared by the χ2 test. Correlation of pp65 antigen-positive cells and CMV DNA levels in leukocyte subsets was determined by Spearman regression analysis. Differences in pp65 antigen-positive cell counts and in CMV copy numbers between different groups of patients were tested for significance by the Mann-Whitney U test. In all test procedures P values <0.05 were considered to be statistically significant.
RESULTS
Active CMV infections.
Twenty-three of the 40 CMV IgG-positive patients developed CMV infections as confirmed by pp65 antigenemia. Eighteen patients, of whom 10 were considered to be symptomatic, were treated with ganciclovir. Four patients (of whom three showed other CMV-associated symptoms) experienced steroid-resistant graft rejection concomitant with pp65 antigenemia and were therefore additionally treated with ATG and OKT3.
A total of 43 pp65 antigen-positive specimens were obtained prior to antiviral treatment. Thirty additional samples were antigen positive after onset of ganciclovir therapy.
Sixty samples were obtained from the remaining 17 patients with no evidence of active CMV infection.
Qualitative CMV PCR.
All 73 antigen-positive samples (before or during therapy) tested positive by qualitative PCR for all leukocyte fractions (Table 1). All patients became pp65 antigen negative within the first week of ganciclovir therapy, which paralleled clinical recovery within 10 days. Of 55 pp65-negative samples analyzed up to 7 weeks after onset of treatment, 47 mixed-PBL samples (85.5%) (Table 1) and 45 PBMC samples (81.8%) were still CMV PCR positive. In contrast, consistent with clinical recovery, only four PMNL specimens (7.3%) remained positive (P < 0.001).
TABLE 1.
Frequencies of positive CMV PCR results in leukocyte subsets and plasma
| Group | No. of patients | Total no. of samples | No. of samples positivea (%)
|
|||
|---|---|---|---|---|---|---|
| Mixed PBL | PBMC | PMNL | Plasmab | |||
| Active infection | ||||||
| Before therapy, pp65 positive | 23 | 43 | 43 (100) | 43 (100) | 43 (100) | 39 (90.7) |
| Therapy | ||||||
| pp65 positive | 18 | 30 | 30 (100) | 30 (100) | 30 (100) | 19 (63.3) |
| pp65 negativec | 18 | 55 | 47 (85.5) | 45 (81.8) | 4 (7.3) | 6 (10.9) |
| Latent infectiond | 17 | 60 | 35 (58.3) | 31 (51.7) | 3 (5.0) | 5 (8.0) |
For leukocytes, ≥5 copies/105 total PBL; for plasma, ≥5 CMV DNA copies/10 μl.
pp65-positive samples before versus during therapy, P < 0.05 (χ2 test); pp65-positive samples before therapy versus pp65-negative samples after therapy, P < 0.001; pp65-positive samples before therapy versus latent infection, P < 0.001.
Mixed PBL versus PBMC, no significant difference (McNemar’s test); mixed PBL versus PMNL, P < 0.001; PBMC versus PMNL, P < 0.001.
Mixed PBL versus PBMC, no significant difference (McNemar’s test); mixed PBL versus PMNL, P < 0.001; PBMC versus PMNL, P < 0.001.
Of the 60 samples drawn from latently CMV-infected patients, 35 mixed-PBL samples (58.3%) and 31 PBMC samples (51.7%) were PCR positive, whereas only 3 (5.0%) of the PMNL fractions contained detectable CMV DNA (P < 0.001). All samples PMNL positive by PCR were also PBMC positive (data not shown).
For plasma, 39 (90.7%) of the 43 pp65-positive samples obtained before onset of therapy were positive by CMV PCR (Table 1). In contrast, only 19 (63.3%) (P < 0.05) of the 30 samples pp65 positive after onset of therapy, 6 (10.9%) (P < 0.001) of the 55 samples pp65 negative after therapy, and 5 (8.3%) (P < 0.001) of the 60 samples from latently infected patients were positive by plasma PCR.
No significant differences were observed between symptomatic and asymptomatic patients in the frequencies of positive PCR results for leukocytes or plasma before, during, or after therapy (data not shown).
CMV DNA levels in patients with active CMV infection.
For the 73 pp65 antigen-positive samples CMV DNA copy numbers in the leukocyte subsets and plasma were determined (Table 2). Antigen-positive counts varied between 4 and 563 (median, 152) cells/105 PBL in the 43 samples drawn from patients before onset of ganciclovir treatment and were significantly lower in the 30 samples obtained during therapy (range, 5 to 122; median, 26) (P < 0.001).
TABLE 2.
CMV DNA load in leukocyte subsets and plasma
| Group | No. of patients | No. of samples | Median no. of pp65-positive cellsf (range) | Median no. of CMV DNA copiesa (range)
|
|||
|---|---|---|---|---|---|---|---|
| Mixed PBLb | PBMCc | PMNLd | Plasmae | ||||
| Active infection | |||||||
| Before therapy, pp65 positive | 23 | 43 | 152 (4–563) | 950 (10–2,580) | 35 (10–60) | 920 (5–2,520) | 45 (10–105) |
| Therapy | |||||||
| pp65 positive | 18 | 30 | 26 (5–122) | 190 (10–870) | 40 (10–55) | 170 (5–805) | 25 (5–45) |
| pp65 negative | 18 | 55 | NAg | 35 (5–60) | 30 (10–55) | 7.5 (5–15) | 10 (5–20) |
| Latent infection | 17 | 60 | NA | 30 (10–65) | 35 (5–60) | 10 (5–20) | 10 (5–15) |
Per 105 total PBL (leukocytes) or per 10 μl (plasma). Differences in CMV DNA levels between groups were analyzed by the Mann-Whitney U test.
pp65-positive samples before versus after onset of therapy, P < 0.001; pp65-positive samples versus pp65-negative samples after onset of therapy, P < 0.001; pp65-negative samples after onset of therapy versus latent infection, no significant difference.
No significant differences.
Before versus during therapy, P < 0.001. Only four samples after antiviral treatment and three samples in patients without active infection were PCR positive (Table 1).
Before versus during therapy, P < 0.001. Only six samples after antiviral treatment and five samples in patients without active infection were PCR positive (Table 1).
pp 65 antigen-positive cell counts: before versus after therapy, P < 0.001. All samples were pp65 antigen negative when ganciclovir therapy was discontinued.
NA, not applicable.
CMV DNA copies in mixed PBL ranged from 10 to 2,580 (median, 950) copies/105 PBL before onset of therapy and from 10 to 870 (median, 190) copies/105 PBL during treatment (P < 0.001). Similarly, in PMNL the differences in CMV DNA levels before (5 to 2,520 copies/105 total PBL [median, 920]) and during (5 to 805/105 total PBL [median, 170]) therapy were significant (P < 0.001). In contrast, no significant differences in CMV DNA copy numbers in PBMC were seen in samples before (10 to 60 copies/105 total PBL [median, 35]) or during (10 to 55 copies/105 total PBL [median, 30]) therapy.
Similarly to CMV in mixed PBL and PMNL, plasma CMV copy numbers significantly declined during ganciclovir therapy (10 to 105 copies/10 μl [median, 45] in pp65-positive samples prior to treatment versus 5 to 45 copies/10 μl [median, 25] after onset of therapy) (P < 0.001).
In the 55 samples which were pp65 antigen negative in patients following treatment with ganciclovir, CMV DNA load was significantly lower in mixed PBL and PMNL compared to samples of pp65 antigen-positive patients (P < 0.001). Again, no difference in the numbers of CMV DNA copies in pp65-positive and pp65-negative samples was observed in PBMC. Analogously to mixed PBL and PMNL, CMV DNA load in plasma obtained from pp65-negative samples was significantly lower (P < 0.001) than that in pp65-positive samples.
Although patients with symptomatic CMV infection had significantly higher peaks of pp65 antigen-positive cell counts (P < 0.001) and of CMV DNA copies in mixed PBL (P < 0.001), PMNL (P < 0.001), and plasma (P < 0.001) than patients without symptoms, there were no differences between the two groups with respect to the reduction kinetics of antigenemia and DNAemia during antiviral treatment. In PBMC, no differences in CMV DNA levels were notable at all between symptomatic and asymptomatic patients (data not shown).
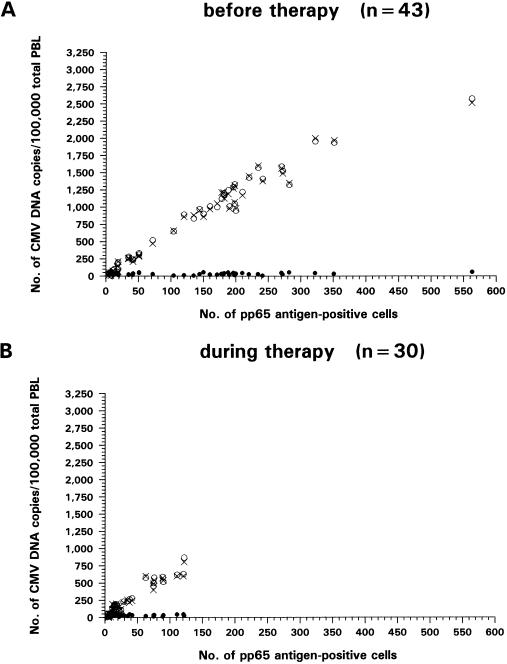
Spearman regression analysis revealed a high correlation of pp65 antigen-positive cells before onset of therapy with CMV DNA levels in mixed PBL (rS = 0.979; P < 0.001) and in PMNL (rS = 0.973; P < 0.001) but no correlation with CMV DNA copies in PBMC (rS = −0.150; P = 0.20) (Fig. 1A). The same relationship was observed during antiviral treatment between pp65 antigen-positive cell counts and CMV DNA copies in mixed PBL (rS = 0.964; P < 0.001) and PMNL (rS = 0.958; P < 0.001) but not in PBMC (rS = 0.068; P = 0.25) (Fig. 1B). The higher the CMV DNA load in PMNL, the more negligible was the difference in CMV copies in mixed PBL, due to the constant low levels of CMV genome copies in PBMC.
FIG. 1.
Correlation of pp65 antigen-positive cells with CMV DNA levels in mixed PBL (○), PMNL (x) and PBMC (•) of patients with active CMV infection determined by Spearman regression analysis. (A) Forty-three samples from 23 patients were analyzed before onset of ganciclovir therapy. Significant correlation was observed for mixed PBL (rS = 0.979; P < 0.001) and PMNL (rS = 0.973; P < 0.001) but not for PBMC (rS = −0.150; P = 0.20). (B) Thirty samples were obtained from 18 patients during antiviral treatment. Again, CMV DNA load in mixed PBL (rS = 0.964; P < 0.001) and PMNL (rS = 0.958; P < 0.001), but not in PBMC (rS = 0.068; P = 0.25), correlated significantly with pp65 antigen-positive cell counts.
CMV DNA levels in patients with latent CMV infection.
In PBMC, no differences in CMV DNA load were detectable between patients with active CMV infection and those with latent CMV infection (Table 2). In contrast, the CMV DNA copy numbers were significantly lower in mixed PBL (P < 0.001) and PMNL (P < 0.001) of latently infected patients than in those of patients with active CMV infection and positive pp65 antigenemia. When pp65 antigenemia was negative after ganciclovir therapy of patients with active CMV infection, no significant differences in CMV DNA copy numbers were found in mixed PBL, PMNL, and PBMC compared to patients with latent infections.
Similarly to CMV DNA levels in mixed PBL and PMNL, in plasma samples from patients with latent infection CMV DNA levels were significantly lower (P < 0.001) than in pp65 antigen-positive samples (Table 2). No significant differences were noted in comparison to pp65 antigen-negative samples after ganciclovir treatment.
DISCUSSION
In our setting PBMC (monocytes and/or lymphocytes) must be regarded as the main source of unwanted positive results by qualitative PCR in pp65 antigen-negative samples. In pp65 antigen-negative patients who had clinically recovered after antiviral therapy as well as in latently infected patients the rate of detection CMV DNA in PMNL was significantly lower than the rate of detection in PBMC, which is compatible with reports that in latently infected patients PBMC harbor CMV DNA in the nuclei more frequently than PMNL (5, 22, 37). Even if PMNL were not infected at all, small-scale contamination of the Ficoll-purified PMNL fractions with monocytes and/or lymphocytes could cause positive PCR results.
CMV DNA copy numbers in PBMC determined by quantitative PCR were equally low in pp65 antigen-negative patients and in actively infected individuals, and no reduction kinetics were seen during antiviral therapy. This suggests that the detected CMV genomes reflected abortive infection. However, deviant observations have been reported (3, 27): some immunocompromised patients with active CMV infection showed high CMV DNA copy numbers in PBMC. Although not statistically significant, ganciclovir induction treatment has been shown to reduce the number of CMV genome copies in PBMC (3). The reasons for these discrepant findings are unclear. One possibility is that patients with primary CMV infections were enrolled in the above studies, whereas in our setting only patients with secondary CMV infections were monitored. During primary virus dissemination PBMC might play a more important role than in secondary infection. On the other hand, differences in cell tropism among viral strains should also be considered.
A high correlation has been described between pp65 antigen-positive cell counts and overall CMV DNA copies in mixed PBL detected by a hybrid capture system (23, 38). However, the quantitative distribution of CMV DNA among leukocyte subpopulations was not considered in these previous investigations. Utilizing quantitative PCR, we observed that solely CMV DNA copy numbers in the PMNL were responsible for the positive correlation with pp65 antigenemia. Firstly, pp65-positive samples showed significantly higher CMV DNA load in mixed PBL and in PMNL than pp65-negative samples. The mean differences in CMV DNA load in mixed PBL and PMNL were negligible due to the constantly low CMV DNA copy numbers in PBMC. Furthermore, the sharp decline of CMV DNA genomes in the mixed PBL fraction during antiviral therapy, which has also been noted by others (3, 9, 10), accompanied by rapid clinical recovery of patients with symptomatic infections, could be attributed to PMNL alone as well. Thus, CMV DNA copies in PMNL of pp65 antigen-negative patients following therapy showed no difference from the levels in PMNL of latently infected patients. Since the same high correlation between CMV DNA levels in mixed PBL or PMNL and pp65 antigen-positive cells could be observed before and during therapy, this would suggest that elimination kinetics are similar for both parameters. Considering the much higher stability of CMV DNA compared to pp65 antigens in blood samples processed with delay (30), CMV DNA quantitation in mixed PBL must be regarded as a promising alternative to the pp65 antigenemia assay.
Our findings add to the evidence that viral DNA detected in PMNL is associated with active CMV infection. This is supported by the fact that in patients who became symptomatic significantly higher peak CMV DNA levels were observed in the PMNL. However, de novo virus production seems to occur mainly at surrounding sites of replication (12, 14, 17, 22) and not, or probably to a much lesser extent (13, 39), in the PMNL themselves. Although only 1 to 4% of pp65 antigen-positive PMNL have been reported to harbor CMV DNA (17), we found CMV DNA by PCR in all PMNL preparations from actively infected patients. The fact that CMV genomes were detected in all PMNL fractions of low-level antigenemic samples may have been due in part to the presence of abortively infected PMNL or to small-scale contamination with infected PBMC. On the other hand, endothelium-derived cells carrying CMV DNA, which have been found in Ficoll-purified PMNL and PBMC fractions (15) during active CMV infection, also might have contributed to the positive PCR results. However, this was not likely to be a major reason for the high correlation of pp65 antigen with CMV DNA we observed in PMNL, since we found no increased percentage of atypical cells in Giemsa stains of PMNL fractions from high-level pp65-antigenemic patients.
In recent years CMV PCR for plasma has been proposed by various investigators (7, 24, 32). Advantages over analysis of leukocytes are seen in the fact that plasma is comparatively easy to process and that higher specificity of positive results with respect to active CMV infection has been reported. However, the exact character of DNA detectable in plasma is still controversial. Although infectious virus has occasionally been recovered from plasma (10, 32), shedding of CMV DNA from lysed leukocytes into the plasma is also a possible explanation (41). Although the CMV DNA remains detectable after ultrafiltration, cellular DNA is also routinely detected in the same samples (10). It is probable that the outcome of plasma PCR strongly varies with the DNA preparation method used. Therefore, more investigations are needed to precisely characterize the CMV DNA detectable in plasma samples.
In our setting positive CMV PCR from plasma was highly associated with active CMV infection. Although less sensitive than mixed-PBL PCR for identifying active infection, quantitative analysis revealed significantly higher peak levels in symptomatic than in asymptomatic patients, similar to leukocyte DNAemia. This has been also described by others for AIDS patients (10, 31), although for unknown reasons Gerna et al. (10) observed only low-level plasma DNAemia in solid-organ transplant recipients with CMV disease.
During antiviral therapy significant decline of plasma DNAemia was detectable. Furthermore, unwanted positive results in pp65-negative samples occurred in plasma with lower frequency than in mixed PBL. However, in comparing PCR for plasma with PCR for the purified leukocyte fractions the situation was different. PCR for PMNL was more sensitive for active infection than PCR for plasma and produced positive results in all samples still positive for pp65 antigens during antiviral treatment, whereas negative plasma PCR results were obtained in 38% of these samples.
Interestingly, in pp65-negative samples (after therapy or in latently CMV-infected patients) the frequency of positive PCR results was lower in PMNL than in plasma, although the difference was not statistically significant. The reasons for this are not clear, but the most likely explanation is that cell debris of latently CMV-infected PBMC contaminated plasma fractions, since the CMV copy numbers detected in plasma of pp65-negative samples were low and we did not subject plasma samples to ultrafiltration. Low copy numbers of cellular DNA in the plasma were also regularly detectable.
In summary, we demonstrated that analysis of leukocytes by qualitative CMV PCR to monitor secondary CMV infection and antiviral therapy in renal-allograft recipients is feasible only if purified PMNL are used. In our setting we observed higher sensitivity of PMNL PCR in pp65-positive samples but fewer unwanted positive PCR results in pp65 antigen-negative samples compared to corresponding plasma preparations.
On the other hand, mixed PBL are sufficient for use in quantitative PCR, since during active infections the differences between mixed PBL and PMNL in CMV DNA load become negligible due to the low copy numbers of nonpredictive CMV DNA in PBMC. Furthermore, due to the higher sensitivity, PBL might be superior to plasma for use in quantitative CMV PCR to monitor CMV therapy and to screen for CMV relapse.
ACKNOWLEDGMENT
We thank Dietrich Mack for critically reviewing the manuscript.
REFERENCES
- 1.Bitsch A, Kirchner H, Dennin R, Hoyer J, Fricke L, Steinhoff J, Sack K, Bein G. Long persistence of CMV DNA in the blood of renal transplant patients after recovery from CMV infection. Transplantation. 1993;56:108–113. doi: 10.1097/00007890-199307000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Bitsch A, Kirchner H, Dupke R, Bein G. Cytomegalovirus transcripts in peripheral blood leukocytes of actively infected transplant patients detected by reverse transcription-polymerase chain reaction. J Infect Dis. 1993;167:740–743. doi: 10.1093/infdis/167.3.740. [DOI] [PubMed] [Google Scholar]
- 3.Boivin G, Quirk M R, Kringstad B A, Germain M, Jordan M C. Early effects of ganciclovir therapy on the quantity of cytomegalovirus DNA in leukocytes of immunocompromised patients. Antimicrob Agents Chemother. 1997;41:860–862. doi: 10.1128/aac.41.4.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cranage M P, Kouzarides T, Bankier A T, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell S E, Minson A C, Smith G L. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dankner W M, McCutchan J A, Richman D D, Hirata K, Spector S A. Localization of human cytomegalovirus in peripheral blood leukocytes by in situ hybridization. J Infect Dis. 1990;161:31–36. doi: 10.1093/infdis/161.1.31. [DOI] [PubMed] [Google Scholar]
- 6.Delgado R, Lumbreras C, Alba C, Pedraza M A, Otero J R, Gomez R, Moreno E, Noriega A R, Paya C V. Low predictive value of polymerase chain reaction for diagnosis of cytomegalovirus disease in liver transplant recipients. J Clin Microbiol. 1992;30:1876–1878. doi: 10.1128/jcm.30.7.1876-1878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freymuth F, Gennetay E, Petitjean J, Eugene G, De Ligny B H, Ryckelynck J-P, Legoff C, Hazera P, Bazin C. Comparison of nested PCR for detection of DNA in plasma with pp65 leukocytic antigenemia procedure for diagnosis of human cytomegalovirus infection. J Clin Microbiol. 1994;32:1614–1618. doi: 10.1128/jcm.32.6.1614-1618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gass P, Kiessling M, Schäfer P, Mester C, Schmitt H P, Kühn J E. Detection of human cytomegalovirus DNA in paraffin sections of human brain by polymerase chain reaction and the occurrence of false negative results. J Neurol Neurosurg Psychiatry. 1993;56:211–214. doi: 10.1136/jnnp.56.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D the Italian Foscarnet Study Group. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerna G, Furione M, Baldanti F, Sarasini A. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol. 1994;32:2709–2717. doi: 10.1128/jcm.32.11.2709-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerna G, Zipeto D, Percivalle E, Parca M, Revello M G, Maccario R, Peri G, Milanesi G. Human cytomegalovirus infection of the major leukocyte subpopulations and evidence for initial viral replication in polymorphonuclear leukocytes from viremic patients. J Infect Dis. 1992;166:1236–1244. doi: 10.1093/infdis/166.6.1236. [DOI] [PubMed] [Google Scholar]
- 13.Gozlan J, Salord J-M, Chouaïd C, Duvivier C, Picard O, Meyohas M-C, Petit J-C. Human cytomegalovirus (HCMV) late-mRNA detection in peripheral blood of AIDS patients: diagnostic value for HCMV disease compared with those of viral culture and HCMV DNA detection. J Clin Microbiol. 1993;31:1943–1945. doi: 10.1128/jcm.31.7.1943-1945.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grefte J M M, Harmsen M C, van der Giessen M, Knollema S, van Son W J, The T H. Presence of human cytomegalovirus (HCMV) immediate early mRNA but not ppUL83 (lower matrix protein pp65) mRNA in polymorphonuclear and mononuclear leukocytes during active HCMV infection. J Gen Virol. 1994;75:1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 15.Grefte J M M, van der Giessen M, van Son W J, The T H. Circulating cytomegalovirus-infected endothelial cells in patients with an active cytomegalovirus infection. J Infect Dis. 1993;167:270–277. doi: 10.1093/infdis/167.2.270. [DOI] [PubMed] [Google Scholar]
- 16.Grefte J M M, van der Gun B T F, Schmolke S, van der Giessen M, van Son W J, Plachter B, Jahn G, The T H. The lower matrix protein pp65 is the principal viral antigen present in peripheral blood leukocytes during an active HCMV infection. J Gen Virol. 1992;73:2923–2932. doi: 10.1099/0022-1317-73-11-2923. [DOI] [PubMed] [Google Scholar]
- 17.Hackstein H, Kirchner H, Jahn G, Bein G. The intracellular localization of human cytomegalovirus DNA in peripheral blood leukocytes during active infections by high-resolution fluorescence in situ hybridization. Arch Virol. 1996;141:1293–1305. doi: 10.1007/BF01718831. [DOI] [PubMed] [Google Scholar]
- 18.Henco K, Heibey M. Quantitative PCR: the determination of template copy numbers by temperature gradient gel electrophoresis (TGGE) Nucleic Acids Res. 1990;18:6733–6734. doi: 10.1093/nar/18.22.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell C L, Miller M J, Martin W J. Comparison of rates of virus isolation from leukocyte populations separated from blood by conventional and Ficoll-Paque/Macrodex methods. J Clin Microbiol. 1979;10:533–537. doi: 10.1128/jcm.10.4.533-537.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühn J E, Wendland T, Schäfer P, Möhring K, Wieland U, Elgas M, Eggers H J. Monitoring of renal allograft recipients by quantitation of human cytomegalovirus genomes in peripheral blood leukocytes. J Med Virol. 1994;44:398–405. doi: 10.1002/jmv.1890440416. [DOI] [PubMed] [Google Scholar]
- 21.Ljungman P, Plotkin S A. Workshop on CMV disease; definitions, clinical severity scores, and new syndromes. Scand J Infect Dis Suppl. 1995;99:87–89. [Google Scholar]
- 22.Martin D C, Katzenstein D A, Yu G S M, Jordan M C. Cytomegalovirus viremia detected by molecular hybridization and electron microscopy. Ann Intern Med. 1984;100:222–225. doi: 10.7326/0003-4819-100-2-222. [DOI] [PubMed] [Google Scholar]
- 23.Mazzulli T, Wood S, Chua R, Walmsley S. Evaluation of the Digene Hybrid Capture System for detection and quantitation of human cytomegalovirus viremia in human immunodeficiency virus-infected patients. J Clin Microbiol. 1996;34:2959–2962. doi: 10.1128/jcm.34.12.2959-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel R, Smith T F, Espy M, Wiesner R H, Krom R A F, Portela D, Paya C V. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J Clin Microbiol. 1994;32:1431–1434. doi: 10.1128/jcm.32.6.1431-1434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinaldo C R, Black P H, Hirsch M S. Interaction of cytomegalovirus with leukocytes from patients with mononucleosis due to cytomegalovirus. J Infect Dis. 1977;136:667–676. doi: 10.1093/infdis/136.5.667. [DOI] [PubMed] [Google Scholar]
- 26.Saltzman R L, Quirk M R, Jordan M C. Disseminated cytomegalovirus infection: molecular analysis of virus and leukocyte interactions in viremia. J Clin Invest. 1988;81:75–81. doi: 10.1172/JCI113313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saltzman R L, Quirk M R, Jordan M C. High levels of circulating cytomegalovirus DNA reflect visceral organ disease in viremic immunosuppressed patients other than marrow recipients. J Clin Invest. 1992;90:1832–1838. doi: 10.1172/JCI116059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schäfer P, Braun R W, Möhring K, Henco K, Kang J, Wendland T, Kühn J E. Quantitative determination of human cytomegalovirus target sequences in peripheral blood leukocytes by nested polymerase chain reaction and temperature gradient gel electrophoresis. J Gen Virol. 1993;74:2699–2707. doi: 10.1099/0022-1317-74-12-2699. [DOI] [PubMed] [Google Scholar]
- 29.Schäfer P, Laufs R. Experience with quantitative PCR for the management of HCMV disease. Intervirology. 1996;39:204–212. doi: 10.1159/000150496. [DOI] [PubMed] [Google Scholar]
- 30.Schäfer P, Tenschert W, Gutensohn K, Laufs R. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J Clin Microbiol. 1997;35:741–744. doi: 10.1128/jcm.35.3.741-744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinkai M, Bozzette S A, Powderly W, Frame P, Spector S A. Utility of urine and leukocyte cultures and plasma DNA polymerase chain reaction for identification of AIDS patients at risk for developing human cytomegalovirus disease. J Infect Dis. 1997;175:302–308. doi: 10.1093/infdis/175.2.302. [DOI] [PubMed] [Google Scholar]
- 32.Spector S A, Merrill R, Wolf D, Dankner W M. Detection of human cytomegalovirus in plasma of AIDS patients during acute visceral disease by DNA amplification. J Clin Microbiol. 1992;30:2359–2365. doi: 10.1128/jcm.30.9.2359-2365.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanier P, Taylor D L, Kitchen A D, Wales N, Tryhom Y, Tyms A S. Persistence of cytomegalovirus in mononuclear cells in peripheral blood from blood donors. Br Med J. 1989;299:897–898. doi: 10.1136/bmj.299.6704.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor-Wiedeman J, Hayhurst G P, Sissons J G, Sinclair J H. Polymorphonuclear cells are not sites of persistence of human cytomegalovirus in healthy individuals. J Gen Virol. 1993;74:265–268. doi: 10.1099/0022-1317-74-2-265. [DOI] [PubMed] [Google Scholar]
- 35.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 36.The T H, van den Berg A P, Harmsen M C, van der Bij W, van Son W J. The cytomegalovirus antigenemia assay: a plea for standardization. Scand J Infect Dis Suppl. 1995;99:25–29. [PubMed] [Google Scholar]
- 37.Turtinen L W, Saltzman R, Jordan M C, Haase A T. Interactions of human cytomegalovirus with leukocytes in vivo: analysis by in situ hybridization. Microb Pathog. 1987;3:287–297. doi: 10.1016/0882-4010(87)90062-3. [DOI] [PubMed] [Google Scholar]
- 38.Veal N, Payan C, Fray D, Sarol L, Blanchet O, Kouyoumdjian S, Lunel F. Novel DNA assay for cytomegalovirus detection: comparison with conventional culture and pp65 antigenemia assay. J Clin Microbiol. 1996;34:3097–3100. doi: 10.1128/jcm.34.12.3097-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Laer D, Serr A, Meyer-König U, Kirste G, Hufert F T, Haller O. Human cytomegalovirus immediate early and late transcripts are expressed in all major leukocyte populations in vivo. J Infect Dis. 1995;172:365–370. doi: 10.1093/infdis/172.2.365. [DOI] [PubMed] [Google Scholar]
- 40.Weber B, Nestler U, Ernst W, Rabenau H, Braner J, Birkenbach A, Scheuermann E-H, Schoeppe W, Doerr H W. Low correlation of human cytomegalovirus DNA amplification by polymerase chain reaction with cytomegalovirus disease in organ transplant recipients. J Med Virol. 1994;43:187–193. doi: 10.1002/jmv.1890430217. [DOI] [PubMed] [Google Scholar]
- 41.Zipeto D, Morris S, Hong C, Dowling A, Wolitz R, Merigan T C, Rasmussen L. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J Clin Microbiol. 1995;33:2607–2611. doi: 10.1128/jcm.33.10.2607-2611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zipeto D, Revello M G, Silini E, Parea M, Percivalle E, Zavattoni M, Milanesi G, Gerna G. Development and clinical significance of a diagnostic assay based on the polymerase chain reaction for detection of human cytomegalovirus DNA in blood samples from immunocompromised patients. J Clin Microbiol. 1992;30:527–530. doi: 10.1128/jcm.30.2.527-530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]