Abstract
Endemic Burkitt lymphoma (eBL) is a fast-growing germinal center B cell lymphoma, affecting 5–10 per 100,000 children annually, in the equatorial belt of Africa. We hypothesize that co-infections with Plasmodium falciparum (Pf) malaria and Epstein-Barr virus (EBV) impair host natural killer (NK) and T cell responses to tumor cells, and thus increase the risk of eBL pathogenesis. NK cell education is partially controlled by killer immunoglobulin-like receptors and variable expression of KIR3DL1 has been associated with other malignancies. Here, we investigated whether KIR3D-mediated mechanisms contribute to eBL, by testing for an association of KIR3DL1/KIR3DS1 genotypes with the disease in 108 eBL patients and 99 healthy Kenyan children. KIR3DL1 allelic typing and EBV loads were assessed by PCR. We inferred previously observed phenotypes from the genotypes. The frequencies of KIR3DL1/KIR3DL1 and KIR3DL1/KIR3DS1 did not differ significantly between cases and controls. Additionally, none of the study participants was homozygous for KIR3DS1 alleles. EBV loads did not differ by the KIR3DL1 genotypes nor were they different between eBL survivors and non-survivors. Our results suggest that eBL pathogenesis may not simply involve variations in KIR3DL1 and KIR3DS1 genotypes. However, considering the complexity of the KIR3DL1 locus, this study could not exclude a role for copy number variation in eBL pathogenesis.
Introduction
Endemic Burkitt lymphoma (eBL) is a fast-growing germinal center B cell lymphoma, affecting pediatric patients within Papua New Guinea and tropical Africa [1]. It is a multifactorial disease, where risk factors such as genetic, environmental, and childhood infections cooperate to cause pathogenesis [2]. It is well documented that most Burkitt lymphoma (BL) cases over-express the c-myc oncogene, due to chromosomal translocation, t(8:14) [3], which results in uncontrolled cell growth. In Kenya, eBL prevalence is high in the western region where Plasmodium falciparum (Pf) transmission occurs throughout the year [4]. Furthermore, about 90% of eBL cases are infected with Epstein-Barr virus (EBV); [5] a chronic infection usually acquired by the age of 2 years in Africa [6]. Since eBL affects children aged 0–14 years old, these observations have led to the speculation that the early age of co-infections with EBV and repeated malaria increases the risk of eBL tumorigenesis [7, 8]. Malaria induces immune down-regulation that influences immune surveillance over EBV by natural killer (NK) and T cells [9]. This immunomodulation results in the accumulation of a pool of EBV-infected B cells [10], viral reactivation, and higher viral loads contributing to the etiology of eBL [7, 11]. NK cells are lymphocytes involved in antiviral and anti-tumor immunity [12]. Their education and licensing involve the acquisition of inhibitory and activating killer immunoglobulin-like receptors (KIRs) [13], which are also expressed by some T cells [14]. Variations in gene content, alleles and copy numbers of KIR genes influence individuals susceptibility to diseases and treatment outcomes [15, 16]. Interaction of KIRs with human leukocyte antigen class I (HLA-I) ligands on the target cells enables licensed NK cells to recognize and tolerate self or to kill target cells lacking “self” human leukocyte antigen class I [13, 17, 18]. A balance of inhibitory and activating signals is crucial for NK cell education [19]. Therefore interruption of inhibitory signal, through the interaction of mature NK cells with HLA-deficient viral-infected or tumor cells may activate NK cells [20]. Consequently, enhanced NK cell activation is associated with pathogenesis in some virus-associated diseases, probably due to non-specific inflammatory responses [21–24]. Furthermore, blocking KIR3DL1 receptor enhances NK cell cytotoxicity of target cells [25]; explaining the possible role of KIR3DL1 in NK cells self-inhibition. The licensing concept explains how this inhibitory KIR may influence NK cells’ antiviral and antitumor potency, through altered NK cell cytolytic activities [25]. KIR3DL1 is a highly polymorphic KIR whose locus encodes inhibitory (KIR3DL1) and activating (KIR3DS1) allotypes (IPD-KIR sequence database: http://www.ebi.ac.uk/ipd/kir/) [26]. The KIR3DL1/3DS1 gene has over 200 alleles [27]; which can be classified into three genotypes 3DL1/3DL1, 3DL1/3DS1, or 3DS1/3DS1 [28]. Considering the different patterns of expression and homology, the alleles can be categorized into functionally distinct KIR3DL1*High, KIR3DL1*Low, KIR3DL1*Null, and KIR3DS1 (3DS1) genotypes [29, 30]. KIR3DL1 recognizes HLA-Bw4 and HLA-Bw6 ligands and their interactions are categorized into strong, weak, and non-interacting [31]. Their binding avidity may influence NK cell immune responses. KIR3DS1 interacts with HLA-F ligands [32].
Functionally, variations in KIR3DL1 alleles have different influences on diseases. For example, KIR3DL1-High and HLA-Bw4 *057 are associated with better outcomes in HIV infected individuals [33]. Generally, this receptor-ligand interaction would generate strong NK inhibitory signals. KIR3DL1-Low alleles have been implicated in the onset of psoriasis, probably because of enhanced immune responses associated with the disease [34]. In contrast, the same genotype had better outcome in chronic myeloid leukemia, suggesting that decreased inhibitory signal enhanced NK cells cytolytic activities in chronic myeloid leukemia patients [35]. Therefore, given the importance of NK cells in the control of viral infected and tumor cells [36], the functionality of different KIR3DL1 genotypes may provide insights into eBL pathogenesis. Our previous study evaluated the presence and/or absence of 16 KIR genes and reported that individuals with an increased number of activating KIRs had a high risk of eBL pathogenesis [21]. To further our understanding on the role of KIR polymorphisms in eBL pathogenesis, the current study evaluated the association of KIR3DL1 alleles with EBV load, eBL susceptibility and survival in the same Kenyan population.
Materials and methods
Study participants
This retrospective study analyzed available DNA from 108 eBL patients and 99 healthy children (HC) from Kenya. Sample size was determined logically, based on available DNA. Patients with eBL were enrolled at Jaramogi Oginga Odinga Teaching and Referral Hospital, a regional referral hospital in western Kenya, between 2007 and 2012, and were aged 0–13.5 years old. The eBL diagnosis was performed from fine-needle aspirates as previously described [37]. The eBL patients were treated with a combination of cyclophosphamide, vincristine, methotrexate, prednisone, and Adriamycin (CHOP), which is the standard therapy for eBL [38]. The healthy controls were conveniently sampled from children aged 0–12 years old, with a healthy medical history and no known history of cancer. They were living in the same malaria-endemic areas of western Kenya, as the eBL patients between 2005 and 2012.
Ethical approval
This study was approved by the Scientific and Ethics Review Unit at the Kenya Medical Research Institute and the Institutional Review Board at the University of Massachusetts Chan Medical School, Worcester, USA. Written informed consent was obtained from the parents and/or guardians before enrollment. Assent was sought from children aged 13 years and above as per the local institutional review board guidelines.
DNA extraction and KIR3DL1 genotyping
Genomic DNA was extracted from blood samples using QIAGEN QIAamp® (Valencia CA, USA). KIR3DL1 typing was performed by customized nested real-time PCR applying Taqman genotyping assay, using the probes and PCR conditions adapted from a published protocol [39]. We distinguished the alleles which were functionally expressed on the surface of NK cells (including *001, *002, *008, *009, *015, *020, *029, *035, *005, *007, and *053 and 3DS1) from the non-functional KIR3DL1 alleles (*004 and *019, which were retained intracellularly) [30]. Only KIR3DL1 alleles with a frequency greater than 1% in the African population were investigated, those with a frequency less than 1% were ignored [40]. Taqman probes (Applied Biosystems, Streetsville, ON, Canada) were used to differentiate between common KIR3DL1 genotypes. The allele expression levels were inferred from the genotypes as previously described [39]. KIR3DS1 and KIR3DL1 were considered alleles of the same locus. KIR3DL1 locus with high sequence similarities was analyzed by a nested real-time PCR strategy. Amplification was performed on an Eppendorf flexlid nexus gradient Mastercycler as instructed by the manufacturer. The PCR amplification conditions were: 90s at 94°C, 30s at 94°C (30 cycles), 30s at 56°C—plate read, 30s at 72°C. The PCR reactions were optimized as previously described [39]. The product of amplification was verified by gel electrophoresis and then diluted one in 106, to provide a template for custom Taqman genotyping assays. All assays were performed according to the manufacturer’s instructions for Taqman Genotyping Master Mix (Applied Biosystems).
Definition for KIR3DL1 allelic polymorphisms
KIR3DL1*High alleles are densely expressed on the cell surface and strongly inhibit NK cell-mediated lysis of tumor cells. KIR3DL1*Low are lowly expressed on the surface of NK cells and generate weak inhibition signals to NK-cell mediated cytolysis. KIR3DL1*Null alleles are not expressed on the cell surface, but have intracellular retention, with minimal inhibitory signaling. KIR3DS1 generates activating signals. The KIR3DL1 phenotypes were inferred from the observed genotypes as previously reported [39]. Individuals carrying at least one high allele but no low allele were considered as KIR3DL1*High carriers, while individuals carrying at least one low allele were considered KIR3DL1*Low carriers. If an individual had one null allele and no copy of either a low or high allele, they were considered a *Null phenotype [29].
Determination of EBV load
EBV load was determined by qPCR as previously described [41]. Briefly, DNA was extracted from 200μl whole blood using the Qiagen DNA easy kit (Qiagen) following the manufacturer’s instructions and stored at -20°C until use. Standard curve dilutions were made by adding DNA to each tube sequentially. A volume of 2μl of sample and 13μl of master mix (BioRad Laboratories, Hercules, CA Cat. No.170-8860) were added to the bottom of the center of the well to bring the total well volume to 15μl. Amplification was done in a BioRad CFX96 Real-Time System with a C1000 Thermocycler base for the primers and probes. The PCR amplification conditions were: 180s at 95°C, 10s at 95°C, 30s at 63.5°C, 10s at 95°C (39 cycles).
Statistical methods
This genetic association study applied the STREGA assessment (STrengthening the REporting of Genetic Association Studies) [42]. KIR3DL1/KIR3DS1 frequencies were calculated by direct counting and expressed as the percentage of the study population having the trait in eBL patients and healthy controls. Fisher’s exact tests were used to test the association between KIR3DL1 genotypes and the risk of eBL. Comparisons were done using the HC as the reference group. Survival was defined as the interval between the hospital admission and the date of last follow-up or death and was computed using the Kaplan-Meier method. Differences between subgroups were tested by log-rank tests. Kruskal–Wallis test and Mann-Whitney test were used to compare log-transformed EBV load between the genotypes and between eBL survivors and non-survivors respectively. The statistical significance of associations was assessed using odds ratios (OR) with 95% confidence intervals (CI). Statistical analyses were performed in R version 3.6.1 (The R Foundation for Statistical Computing) and Graphpad Prism version 8.0.2 (GraphPad Software, La Jolla, CA). The results are reported using the median and p-value. A p-value less than or equal to 0.05 was considered statistically significant, while a p-value greater than 0.05 was indicated as non-significant (ns).
Results
Patients’ characteristics
To assess the association of KIR3DL1 polymorphisms with eBL, we typed 108 Kenyan patients with eBL and 99 healthy controls. The median age of eBL patients at diagnosis in our study population was 8.2 years, (interquartile range (IQR):6.2–10.3), and 66.7% of eBL patients were males. Controls were healthy volunteers derived from the same community, without a history of childhood cancers. Their median age at the time of enrollment into the study was 6.1 years, (IQR:3.4–8.3), and were 56.1% males. The peak onset of eBL ranges from 5–9 years of age (8, 9). Malaria positivity was 20.0% (18/90) and 39.0% (30/77) in eBL patients and HC, respectively. Among eBL patients, 35.0% (36/103) individuals died, while the rest were alive at the last follow-up. The patients were followed for 2 years post-diagnosis.
Distribution of KIR3DL1 alleles in eBL patients and healthy controls
Typing of KIR3DL1 allowed us to distinguish 3 distinct functional genotypes, with different allele combinations. We observed that none of the genotypes were associated with eBL. Additionally, no specific combination of KIR3DL1/S1 alleles conferred protection or risk to eBL (Table 1).
Table 1. Frequency of KIR3DL1 alleles with different predicted cell expression levels in eBL patients and healthy Kenyan children.
| KIR3DL1/S1 Alleles | |||||
|---|---|---|---|---|---|
| Allele 1 | Allele 2 | eBL | HC | Odds Ratio | p- value |
| n = 108 (%) | n = 104 (%) | (95% Confidence Interval) | |||
| High genotype | |||||
| 3DL1*High | 3DL1*High | 55 (50.93) | 53 (53.54) | 0.901 (0.521–1.550) | 0.781 |
| 3DL1*High | 3DL1*Null | 10 (9.26) | 6 (6.06) | 1.582 (0.599–3.930) | 0.444 |
| 3DL1*High | 3DS1 | 10 (9.26) | 6 (6.06) | 1.582 (0.599–4.520) | 0.444 |
| Low genotype | |||||
| 3DL1*High | 3DL1 * Low | 26 (24.07) | 20 (20.20) | 1.252 (0.638–2.399) | 0.616 |
| 3DL1 * Low | 3DL1 * Low | 2 (1.85) | 1 (1.01) | 1.849 (0.212–27.040) | 1.000 |
| 3DL1 * Low | 3DL1*Null | 1 (0.93) | 2 (2.02) | 0.453 (0.031–3.960) | 0.607 |
| 3DL1 * Low | 3DS1 | 2 (1.85) | 5 (5.05) | 0.355 (0.070–1.380) | 0.262 |
| Null genotype | |||||
| 3DL1*Null | 3DL1*Null | 2 (1.85) | 2 (2.02) | 0.915 (0.141–5.930) | 1.000 |
| 3DL1*Null | 3DS1 | 0 | 4 (4.04) | 0.000 (0.000–0.680) | 0.060 |
| 3DS1 genotype | |||||
| 3DS1 | 3DS1 | 0 | 0 | N/A | 0 |
Abbreviation: n, number of subjects
Distribution of KIR3DL1 genotypes in eBL patients and healthy controls
All the study participants carried at least one KIR3DL1 allele (n = 207), hence we did not observe any homozygotes for KIR3DS1 (KIR3DS1/KIR3DS1) genotype (i.e. individuals who do not carry any copies of KIR3DL1). In addition, there were no individuals negative for both alleles. The frequency of homozygous KIR3DL1 (KIR3DL1/KIR3DL1) was 88.89% and 84.85%, while heterozygous KIR3DS1 (KIR3DL1/KIR3DS1) had a frequency of 11.11% and 15.15% in eBL patients and HC respectively. The KIR3DL1*High, KIR3DL1*Low, and KIR3DL1*Null genotypes represented 69.44% vs 65.65%, 28.70% vs 28.28%, and 1.85% vs 6.06% in eBL patients and HC respectively (Table 2). No KIR3DL1 genotypes were associated with eBL.
Table 2. Analysis of diploid KIR3DL1 genotypes in eBL patients and healthy Kenyan children.
| Diploid KIR3DL1 | eBL | HC | Odds Ratio | p- value | |
|---|---|---|---|---|---|
| n = 108 (%) | n = 99 (%) | (95% Confidence Interval) | |||
| KIR3DL1/KIR3DL1 | 96 (88.89) | 84 (84.85) | 1.430 (0.657–3.320) | 0.415 | |
| KIR3DL1/KIR3DS1 | 12 (11.11) | 15 (15.15) | |||
| KIR3DS1/KIR3DS1 | 0 | 0 | |||
| KIR3DL1 genotype | |||||
| KIR3DL1*High | 75 (69.44) | 65 (65.65) | 1.19 (0.668–2.120) | 0.656 | |
| KIR3DL1*Low | 31 (28.70) | 28 (28.28) | 1.02 (0.549–1.830) | 1.000 | |
| KIR3DL1*Null | 2 (1.85) | 6 (6.06) | 0.292 (0.059–1.220) | 0.156 | |
Abbreviation: n, number of subjects
KIR3DL1 genotypes and survival of eBL patients
To evaluate whether KIR3DL1 genotypes were associated with a patient’s survival, we analyzed the KIR3DL1 genotypes between eBL survivors and the non-survivors (Table 3).
Table 3. The percentage of KIR3DL1 genotypes in eBL survivors compared to non-survivors.
| KIR3DL1 genotype | eBL survivors | eBL non-survivors | Odds Ratio | p- value |
|---|---|---|---|---|
| n = 67 (%) | n = 36 (%) | (95% Confidence Interval) | ||
| KIR3DL1*High | 47 (70.15) | 25 (69.44) | 1.030 (0.432–2.490) | 1.000 |
| KIR3DL1*Low | 18 (26.87) | 11 (30.56) | 0.835 (0.337–2.030) | 0.819 |
| KIR3DL1*Null | 2 (2.99) | 0 | 0.541 |
Abbreviation: n, number of subjects
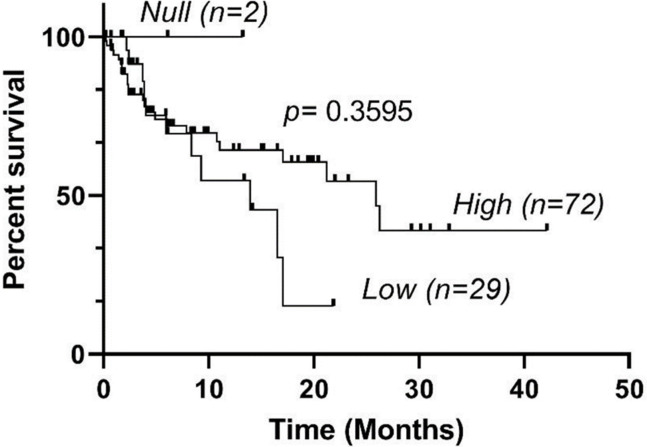
In the study population, 65% (67/103) of eBL patients survived and 35% died (36/103). We did not have 2-year survival data for 5 eBL patients. We observed no significant differences in survival among eBL patients with the High, Low and Null KIR3DL1 genotypes (Fig 1).
Fig 1. KIR3DL1 genotypes and overall survival (OS) in patients with eBL.
OS of eBL patients encoding KIR3DL1*High, KIR3DL1*Low, and KIR3DL1*Null genotypes were analyzed using the Kaplan-Meier method.
KIR3DL1 genotypes and EBV loads in eBL patients and healthy controls
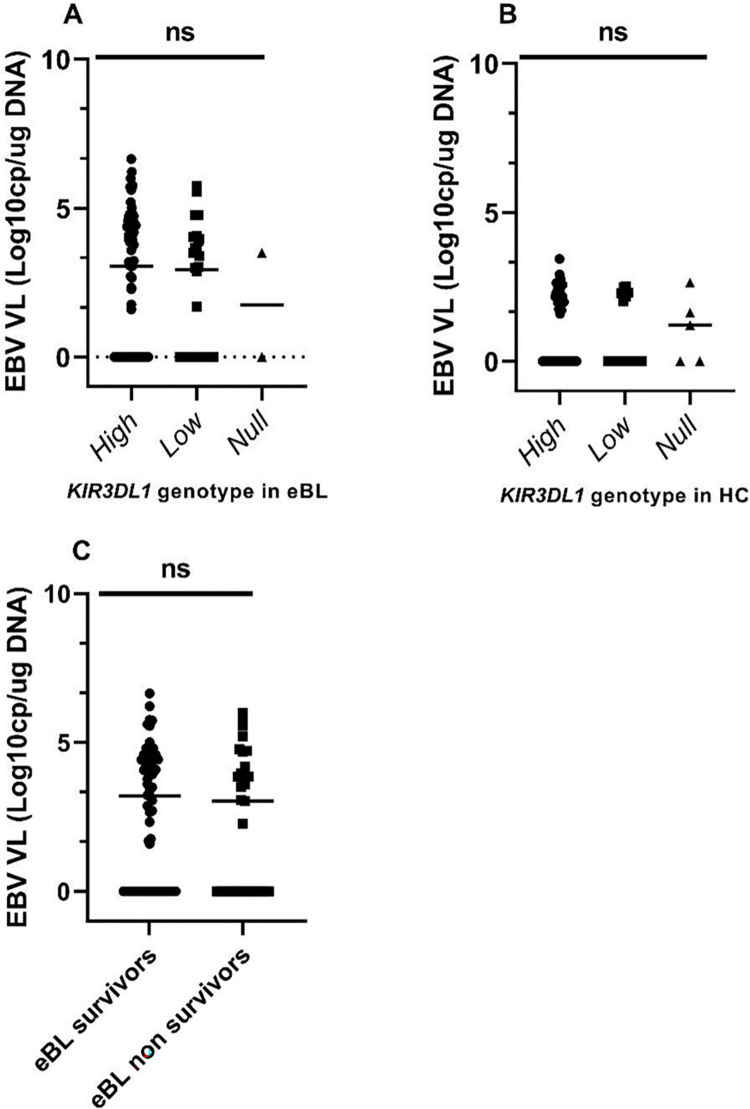
To determine whether EBV loads differed by the KIR3DL1 genotypes, we analyzed the available viral loads for 102 Kenyan patients with eBL and 78 HC. As expected, children with eBL had significantly higher viremia (median 1099.54 EBV copies/ug of DNA) compared to healthy children (median 0 EBV copies/ug of DNA) (p-value <0.0001) (9). However, the median EBV load was not different between KIR3DL1 genotypes (Fig 2A and 2B). As previously reported, high EBV load was associated with an increased risk of eBL (OR = 3.072; 95% CI 1.108–5.6814; p = 0.006), [43] but not independently associated with the patient’s survival (Fig 2C).
Fig 2. EBV load stratified by KIR3DL1 genotypes and by survival in eBL patients.
Cellular EBV levels were compared for (A) eBL patients (n = 102) and (B) HC (n = 78) after stratification by KIR3DL1 genotypes. In addition, EBV levels were compared between eBL survivors and non-survivors (C). Analyses were performed by Kruskal–Wallis and Mann-Whitney tests. p≤0.05 was considered statistically significant. VL is viral load. Data are median, ns, not significant.
Discussion
NK cells destroy tumor cells lacking HLA ligands with varying efficiency [44], depending on receptor-ligand binding affinity, avidity, and cell surface density [45]. KIR3DL1 alleles have variable inhibitory strength upon ligation to HLA-Bw4 and HLA-Bw6 ligands [31]. We previously demonstrated that NK cells from eBL patients have increased density of inhibitory KIR3DL1, and have limited ability to kill target K562 tumor cells in vitro relative to healthy controls [9]. The current study investigated the possible association of KIR3DL1 genotypes with pathogenesis and survival in eBL patients. Overall, the Kenyan study population had the KIR3DL1 genotype, and no individual had the KIR3DS1 genotype. Unlike non-African populations where diversifying selection has maintained the KIR3DL1-High, KIR3DL1-Low, and KIR3DS1 genotypes at equivalence, directional selection favors the KIR3DL1-High genotype among the African population [40] Consequently, KIR3DL1-Low is uncommon while KIR3DS1 is rare among Africans [40, 46]. Consistent with these findings, KIR3DL1-High alleles had a higher frequency than KIR3DL1-Low alleles in the Kenyan study population. Previous studies have demonstrated that the density of KIR3DL1 impacts NK cell functions where KIR3DL1-High interacts with its HLA-ligand to generate a strong inhibitory signal [31]. Since NK cells are sensitive to cells with decreased expression of HLA-I, the strength of interaction of KIR3DL1 and HLA-B subtypes determines the degree of NK inhibition and hence their anti-tumor activities [40]. Consequently, the stronger the inhibition that prevents an NK cell from attacking healthy cells, the stronger its response toward unhealthy cells [18, 47]. Natural selection would therefore select for NK cells with stronger responses to infections [40]. Exposure of Africans to acute and chronic infections [48], might favor the selection of individuals with potent NK cells that are sensitive to decreased expression of HLA-I in the African population [40]. Considering this hypothesis, we expected KIR3DL1*High to provide a selective advantage through enhanced clearance of tumor cells, thus protecting individuals from eBL pathogenesis. We would therefore expect eBL patients to be less associated with the KIR3DL1*High genotype. On the contrary, the frequencies of KIR3DL1 genotypes were not different between eBL patients and healthy controls in our study population.
KIR3DL1 alleles have been reported to differentially influence outcomes within the context of other cancers. While poor overall survival was observed in weakly interacting and non-interacting alleles in metastatic colorectal cancer patients on chemotherapy [49], the same phenotypes were associated with favorable outcomes in patients with neuroblastoma, treated with anti-GD2 monoclonal antibody [31]. Furthermore, strongly interacting KIR3DL1 genotypes were associated with poor overall survival [31, 45]. Individuals with KIR3DL1*High genotypes have an increased percentage of NK cells abundantly expressing KIR3DL1 phenotype [50]. The strong interaction of this genotype with its HLA-Bw4 ligands would strongly inhibit NK cells under normal circumstances [29, 39], hence weakening their immune responses. In some instances, the lack of NK inhibition may be beneficial for successful NK responses [51], as evidenced by enhanced survival in patients with lymphoma receiving rituzimab treatment [52]. Furthermore, blockade of KIR3DL1-high and Bw4 interaction has been shown to restore the effector functions in vitro [31]. In contrast, our results did not demonstrate significant differences in overall survival between eBL patients with different KIR3DL1 genotypes. However, since KIR3DL1 is a complex locus with sequence and copy number variations [53], future micro-array experiments will be needed to evaluate duplications, deletions and rearrangements which results in varying numbers of copies of KIR3DL1 and KIR3DS1 in each chromosome and their impact on eBL etiology.
Highly educated NK cells are important effectors in antiviral, anti-tumor, and antimalarial immune responses. Infections with Pf alter the NK cells subsets and the KIR/HLA repertoire, subsequently affecting NK cell responses to malaria [48]. In the African population, chronic malaria infections down-regulate NK and T cell responses, resulting in elevated EBV loads [9, 11]. Consequently, evolutionary pressure from malaria pathogens might have selected KIR/HLA combinations that protect against severe malaria but which increase the risk of other diseases [48]. In the context of other infectious diseases, the role of KIR3DL1 and their HLA ligands is controversial, with some studies associating them with protection from HIV and AIDs [20, 33] and others with disease severity in COVID-2019 [54]. Interestingly, malaria-exposed children and eBL patients express the KIR3DL1*High phenotype [9], a proposed mechanism by which malaria subverts NK-cell mediated immune responses [9]. In contrast, our analysis, while requiring replication, did not associate the predicted KIR3DL1*High phenotype with increased risk of eBL in the same study population. Considering EBV infections, the association between EBV and outcome in lymphomas is controversial, with some studies reporting an association [55] and others no association [56]. In our study, EBV loads were not different between the KIR3DL1 genotypes in the study population.
Our study has some limitations. First, although KIR/HLA combinations influence individual’s susceptibility to diseases and may exert selective pressure in populations [57], due to limited genomic material, our study focused only on analysis of KIR3L1/3DS1 alleles using the available DNA. We therefore did not perform HLA typing experiments for this study population. Convenience sampling of the HC led them to being younger when compared with eBL patients. However, since KIR3DL1 alleles do not differ by age, we believe that convenient sampling would not bias the findings. In addition, our conclusions were limited by the small number of study participants with the KIR3DL1 High, Low, and Null genotypes.
Conclusion
Our findings, while requiring replication, add to the existing body of literature on KIRs and eBL pathogenesis. While our previous study associated activating KIRs with eBL pathogenesis [21], the current study extends these observations to exclude the association of inhibitory KIR3DL1 alleles in eBL pathogenesis. Since the frequencies of KIR3DL1 and KIR3DS1 alleles are not significantly different in eBL patients compared to healthy Kenyan controls, they do not appear to independently increase the risk of eBL. In addition, the alleles do not influence EBV viral loads, suggesting the possibility of other mechanisms inhibiting NK cell antiviral and anti-tumor activities in eBL patients. Considering that maximum NK cell education is dependent on a high density of receptors and ligands and their binding strength [45] and that the KIR3DL1 alleles impact the strength of interaction with its HLA-Bw4 and HLA-Bw6 ligands, further studies with larger sample sizes are required to explore the influence of varying strengths of receptor-ligand interactions on NK cell anti-tumor and anti-viral activities in eBL patients.
Supporting information
(XLS)
Acknowledgments
We thank the Kenyan children and their families for participating in this study. We also wish to thank the Director of the Kenya Medical Research Institute for approving this manuscript for publication.
Data Availability
All relevant data are within the paper and its supporting information files.
Funding Statement
BMM was supported by the US National Institutes of Health, National Cancer Institute R01 CA189806 (AMM), and DELTAS Africa grant (DEL-15-007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z: Awandare) and the UK government. The views expressed in this publication are those of the author(s) and not those of the funders. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kafuko GW, Burkitt DP. Burkitt’s lymphoma and malaria. International Journal of Cancer. 1970;6(1):1–9. doi: 10.1002/ijc.2910060102 [DOI] [PubMed] [Google Scholar]
- 2.Brady G, MacArthur G, Farrell P. Epstein–Barr virus and Burkitt lymphoma. Postgraduate medical journal. 2008;84(993):372–7. doi: 10.1136/jcp.2007.047977 [DOI] [PubMed] [Google Scholar]
- 3.Magrath IT. African Burkitt’s lymphoma: history, biology, clinical features, and treatment. Journal of Pediatric Hematology/Oncology. 1991;13(2):222–46. [PubMed] [Google Scholar]
- 4.Rainey JJ, Omenah D, Sumba PO, Moormann AM, Rochford R, Wilson ML. Spatial clustering of endemic Burkitt’s lymphoma in high‐risk regions of Kenya. International journal of cancer. 2007;120(1):121–7. doi: 10.1002/ijc.22179 [DOI] [PubMed] [Google Scholar]
- 5.Kaymaz Y, Oduor CI, Yu H, Otieno JA, Ong’echa JM, Moormann AM, et al. Comprehensive transcriptome and mutational profiling of endemic Burkitt lymphoma reveals EBV type–specific differences. Molecular Cancer Research. 2017;15(5):563–76. doi: 10.1158/1541-7786.MCR-16-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piriou E, Asito AS, Sumba PO, Fiore N, Middeldorp JM, Moormann AM, et al. Early age at time of primary Epstein–Barr Virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the Etiology of Endemic Burkitt Lymphoma. Journal of Infectious Diseases. 2012;205(6):906–13. doi: 10.1093/infdis/jir872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkitt D. Etiology of Burkitt’s lymphoma—an alternative hypothesis to a vectored virus. Journal of the National Cancer Institute. 1969;42(1):19–28. [PubMed] [Google Scholar]
- 8.Orem J, Mbidde EK, Lambert B, De Sanjose S, Weiderpass E. Burkitt\’s lymphoma in Africa, a review of the epidemiology and etiology. African health sciences. 2007;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forconi C, Cosgrove CP, Saikumar-Lakshmi P, Nixon CE, Foley J, Ong’echa JM, et al. Poorly cytotoxic terminally differentiated CD56negCD16pos NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood advances. 2018;2(10):1101–14. doi: 10.1182/bloodadvances.2017015404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittle HC, Brown J, Marsh K, Greenwood BM, Seidelin P, Tighe H, et al. T-cell control of Epstein–Barr virus-infected B cells is lost during P. falciparum malaria. Nature. 1984;312(5993):449–50. doi: 10.1038/312449a0 [DOI] [PubMed] [Google Scholar]
- 11.Moormann A, Chelimo K, Sumba OP, Lutzke ML, Ploutz-Snyder R, Newton D, et al. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. The Journal of infectious diseases. 2005;191(8):1233–8. doi: 10.1086/428910 [DOI] [PubMed] [Google Scholar]
- 12.Bhat R, Rommelaere J. Emerging role of Natural killer cells in oncolytic virotherapy. ImmunoTargets and therapy. 2015:65–77. doi: 10.2147/ITT.S55549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodoen M, Lanier L. Viral modulation of NK cell immunity. Nature Reviews Microbiology. 2005;3(1):59–69. doi: 10.1038/nrmicro1066 [DOI] [PubMed] [Google Scholar]
- 14.Battistini L, Borsellino G, Sawicki G, Poccia F, Salvetti M, Ristori G, et al. Phenotypic and cytokine analysis of human peripheral blood gamma delta T cells expressing NK cell receptors. Journal of immunology (Baltimore, Md: 1950). 1997;159(8):3723–30. [PubMed] [Google Scholar]
- 15.Hanson A, Sahhar J, Ngian G-S, Roddy J, Walker J, Stevens W, et al. Contribution of HLA and KIR alleles to systemic sclerosis susceptibility and immunological and clinical disease subtypes. Frontiers in Genetics. 2022;13:913196. doi: 10.3389/fgene.2022.913196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khakoo S, Carrington M. KIR and disease: a model system or system of models? Immunological reviews. 2006;214(1):186–201. doi: 10.1111/j.1600-065X.2006.00459.x [DOI] [PubMed] [Google Scholar]
- 17.Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–42. doi: 10.1016/j.immuni.2006.06.013 [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song Y-J, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–13. doi: 10.1038/nature03847 [DOI] [PubMed] [Google Scholar]
- 19.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 20.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, et al. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. The journal of immunology. 2010;184(4):2057–64. doi: 10.4049/jimmunol.0902621 [DOI] [PubMed] [Google Scholar]
- 21.Muriuki B, Forconi CS, Oluoch PO, Bailey JA, Ghansah A, Moormann AM, et al. Association of killer cell immunoglobulin-like receptors with endemic Burkitt lymphoma in Kenyan children. Scientific Reports. 2021;11(1):11343. doi: 10.1038/s41598-021-90596-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiang Q, Zhengde X, Chunyan L, Zhizhuo H, Junmei X, Junhong A, et al. Killer cell immunoglobulin‐like receptor gene polymorphisms predispose susceptibility to Epstein‐Barr virus associated hemophagocytic lymphohistiocytosis in Chinese children. Microbiology and immunology. 2012;56(6):378–84. doi: 10.1111/j.1348-0421.2012.00443.x [DOI] [PubMed] [Google Scholar]
- 23.Merino AM, Dugast A-S, Wilson CM, Goepfert PA, Alter G, Kaslow RA, et al. KIR2DS4 promotes HIV-1 pathogenesis: new evidence from analyses of immunogenetic data and natural killer cell function. PLoS one. 2014;9(6):e99353. doi: 10.1371/journal.pone.0099353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernal E, Gimeno L, Alcaraz MJ, Quadeer AA, Moreno M, Martínez-Sánchez MV, et al. Activating killer-cell immunoglobulin-like receptors are associated with the severity of coronavirus disease 2019. The Journal of infectious diseases. 2021;224(2):229–40. doi: 10.1093/infdis/jiab228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binyamin L, Alpaugh RK, Hughes TL, Lutz CT, Campbell KS, Weiner LM. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. The Journal of Immunology. 2008;180(9):6392–401. doi: 10.4049/jimmunol.180.9.6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, et al. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? The Journal of Immunology. 2008;180(10):6743–50. doi: 10.4049/jimmunol.180.10.6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison GF, Leaton LA, Harrison EA, Kichula KM, Viken MK, Shortt J, et al. Allele imputation for the killer cell immunoglobulin-like receptor KIR3DL1/S1. PLoS Computational Biology. 2022;18(2):e1009059. doi: 10.1371/journal.pcbi.1009059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. The Journal of Immunology. 2001;166(5):2992–3001. doi: 10.4049/jimmunol.166.5.2992 [DOI] [PubMed] [Google Scholar]
- 29.Ahn R, Moslehi H, Martin M, Abad‐Santos M, Bowcock A, Carrington M, et al. Inhibitory KIR3DL1 alleles are associated with psoriasis. British Journal of Dermatology. 2016;174(2):449–51. doi: 10.1111/bjd.14081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zvyagin I, Mamedov IZ, Britanova OV, Staroverov DB, Nasonov EL, Bochkova AG, et al. Contribution of functional KIR3DL1 to ankylosing spondylitis. Cellular & Molecular Immunology. 2010;7(6):471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forlenza C, Boudreau JE, Zheng J, Le Luduec J-B, Chamberlain E, Heller G, et al. KIR3DL1 allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. Journal of Clinical Oncology. 2016;34(21):2443. doi: 10.1200/JCO.2015.64.9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK, et al. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS One. 2016;11(9):e0163297. doi: 10.1371/journal.pone.0163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin M, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nature genetics. 2007;39(6):733–40. doi: 10.1038/ng2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni Martin, Carrington, editors. The Yin and Yang of HLA and KIR in human disease. Seminars in immunology; 2008: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shindo T, Ureshino H, Kojima H, Tanaka H, Kimura S. Allelic polymorphisms of KIRs and antitumor immunity against chronic myeloid leukemia. Immunological Medicine. 2021;44(2):61–8. doi: 10.1080/25785826.2020.1796062 [DOI] [PubMed] [Google Scholar]
- 36.Trinchieri G, Santoli D. Enhancement of human natural killer cell activity by interferon. J Immunol. 1978;120:1845–50. [Google Scholar]
- 37.Movassagh M, Oduor C, Forconi C, Moormann AM, Bailey JA. Sensitive detection of EBV microRNAs across cancer spectrum reveals association with decreased survival in adult acute myelocytic leukemia. Scientific Reports. 2019;9(1):20321. doi: 10.1038/s41598-019-56472-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckle G, Maranda L, Skiles J, Ong’echa JM, Foley J, Epstein M, et al. Factors influencing survival among Kenyan children diagnosed with endemic Burkitt lymphoma between 2003 and 2011: A historical cohort study. International journal of cancer. 2016;139(6):1231–40. doi: 10.1002/ijc.30170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berinstein J, Pollock R, Pellett F, Thavaneswaran A, Chandran V, Gladman DD. Association of variably expressed KIR3dl1 alleles with psoriatic disease. Clinical rheumatology. 2017;36:2261–6. doi: 10.1007/s10067-017-3784-5 [DOI] [PubMed] [Google Scholar]
- 40.Norman P, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nature genetics. 2007;39(9):1092–9. doi: 10.1038/ng2111 [DOI] [PubMed] [Google Scholar]
- 41.Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, et al. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. Journal of Clinical Microbiology. 1999;37(1):132–6. doi: 10.1128/JCM.37.1.132-136.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, Von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement. Genetic Epidemiology: The Official Publication of the International Genetic Epidemiology Society. 2009;33(7):581–98. [DOI] [PubMed] [Google Scholar]
- 43.Mulama DH, Bailey JA, Foley J, Chelimo K, Ouma C, Jura WG, et al. Sickle cell trait is not associated with endemic Burkitt lymphoma: An ethnicity and malaria endemicity‐matched case–control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. International journal of cancer. 2014;134(3):645–53. doi: 10.1002/ijc.28378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. The Journal of Immunology. 2005;175(8):5222–9. doi: 10.4049/jimmunol.175.8.5222 [DOI] [PubMed] [Google Scholar]
- 45.Boudreau J, Mulrooney TJ, Le Luduec J-B, Barker E, Hsu KC. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. The Journal of Immunology. 2016;196(8):3398–410. doi: 10.4049/jimmunol.1502469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson J, Marsh SG. IPD: the immuno polymorphism database. Immunoinformatics: Predicting Immunogenicity In Silico. 2007:61–74. [DOI] [PubMed] [Google Scholar]
- 47.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nature Reviews Immunology. 2006;6(7):520–31. doi: 10.1038/nri1863 [DOI] [PubMed] [Google Scholar]
- 48.Tukwasibwe S, Nakimuli A, Traherne J, Chazara O, Jayaraman J, Trowsdale J, et al. Variations in killer-cell immunoglobulin-like receptor and human leukocyte antigen genes and immunity to malaria. Cellular & Molecular Immunology. 2020;17(8):799–806. doi: 10.1038/s41423-020-0482-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Re V, Caggiari L, De Zorzi M, Talamini R, Racanelli V, Andrea MD, et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS one. 2014;9(1):e84940. doi: 10.1371/journal.pone.0084940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yawata M, Yawata N, Draghi M, Little A-M, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. The Journal of experimental medicine. 2006;203(3):633–45. doi: 10.1084/jem.20051884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–4. doi: 10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
- 52.Du J, Lopez-Verges S, Pitcher BN, Johnson J, Jung S-H, Zhou L, et al. CALGB 150905 (Alliance): rituximab broadens the antilymphoma response by activating unlicensed NK cells. Cancer immunology research. 2014;2(9):878–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erer B, Takeuchi M, Ustek D, Tugal-Tutkun I, Seyahi E, Özyazgan Y, et al. Evaluation of KIR3DL1/KIR3DS1 polymorphism in Behçet’s disease. Genes & Immunity. 2016;17(7):396–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajeer A, Jawdat D, Massadeh S, Aljawini N, Abedalthagafi MS, Arabi YM, et al. Association of KIR gene polymorphisms with COVID-19 disease. Clinical Immunology. 2022;234:108911. doi: 10.1016/j.clim.2021.108911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarrett RF, Stark GL, White J, Angus B, Alexander FE, Krajewski AS, et al. Impact of tumor Epstein-Barr virus status on presenting features and outcome in age-defined subgroups of patients with classic Hodgkin lymphoma: a population-based study. Blood. 2005;106(7):2444–51. doi: 10.1182/blood-2004-09-3759 [DOI] [PubMed] [Google Scholar]
- 56.Satou A, Asano N, Nakazawa A, Osumi T, Tsurusawa M, Ishiguro A, et al. Epstein-Barr virus (EBV)-positive sporadic burkitt lymphoma: an age-related lymphoproliferative disorder? The American Journal of Surgical Pathology. 2015;39(2):227–35. doi: 10.1097/PAS.0000000000000332 [DOI] [PubMed] [Google Scholar]
- 57.Yindom L-M, Mendy M, Bodimeade C, Chambion C, Aka P, Whittle HC, et al. KIR content genotypes associate with carriage of hepatitis B surface antigen, e antigen and HBV viral load in Gambians. PloS one. 2017;12(11):e0188307. doi: 10.1371/journal.pone.0188307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
Data Availability Statement
All relevant data are within the paper and its supporting information files.