Abstract
It is well known that the neuropeptide Y (NPY)/agouti-related peptide (AgRP) neurons increase appetite and decrease thermogenesis. Previous studies demonstrated that optogenetic and/or chemogenetic manipulations of NPY/AgRP neuronal activity alter food intake and/or energy expenditure (EE). However, little is known about intrinsic molecules regulating NPY/AgRP neuronal excitability to affect long-term metabolic function. Here, we found that the G protein-gated inwardly rectifying K+ (GIRK) channels are key to stabilize NPY/AgRP neurons and that NPY/AgRP neuron-selective deletion of the GIRK2 subunit results in a persistently increased excitability of the NPY/AgRP neurons. Interestingly, increased body weight and adiposity observed in the NPY/AgRP neuron-selective GIRK2 knockout mice were due to decreased sympathetic activity and EE, while food intake remained unchanged. The conditional knockout mice also showed compromised adaptation to coldness. In summary, our study identified GIRK2 as a key determinant of NPY/AgRP neuronal excitability and driver of EE in physiological and stress conditions.
It is known that a subset of neurons in the hypothalamus increase appetite and decrease thermogenesis. This study shows that the ion channel GIRK2 functions in hypothalamic NPY/AgRP neurons to increase energy expenditure in mice; activity of this ion channel may help to keep body weight under control.
Introduction
The arcuate nucleus of the hypothalamus (ARH) is home to several distinct types of neurons that control energy homeostasis [1]. In particular, it is well known that neurons co-expressing neuropeptide Y (NPY) and agouti-related peptide (AgRP) (NPY/AgRP neurons) promote food intake [2,3]. NPY/AgRP neurons also decrease energy expenditure (EE), at least in part by suppressing sympathetic tone to the brown adipose tissue (BAT) and inhibiting thermogenesis [4,5]. Consistent with these findings, manipulating the activity of NPY/AgRP neurons using exogenous genetic constructs (e.g., channelrhodopsin and designer receptors) resulted in acute changes in food intake and energy utilization [6,7]. While these studies provided insight into how NPY/AgRP neuronal activity is translated to in vivo metabolic function, we have little information regarding intrinsic molecules that regulate NPY/AgRP neuronal activity per se.
In many excitable cells, the resting membrane potential (RMP) is maintained largely by K+ channels [8]. For example, the “classic” inwardly rectifying K+ (IRK or Kir2) channels maintain RMP of cardiac myocytes [9] and ATP-sensitive K+ (KATP) channels silence pancreatic β-cells [10]. In neurons, KATP channels and G protein-gated inwardly rectifying K+ (GIRK or Kir3) channels have been reported to open at rest to dampen cellular excitability. For example, KATP channel activity hyperpolarizes membrane potential of the pro-opiomelanocortin (POMC) neurons of the ARH [11] and the serotonin 2C receptor-expressing neurons of the lateral parabrachial nucleus [12]. It was also demonstrated that GIRK channels maintain RMP of arcuate POMC neurons [13] and hippocampal CA1 neurons [14]. However, little data is currently available on the identity of K+ channels that regulate RMP of NPY/AgRP neurons.
In this study, we utilized multiple approaches to identify specific K+ channels that regulate NPY/AgRP neuronal activity. Firstly, we found evidence that GIRK2-containing GIRK channels suppress the activity of NPY/AgRP neurons. We subsequently found that GIRK2 ablation in NPY/AgRP neurons results in increased body weight and adiposity when the mice are fed normal chow diet (NCD). Notably, the observed phenotypes were attributed to decreased sympathetic activity and energy expenditure, rather than an increase of food intake. We also found evidence that GIRK2 expressed by NPY/AgRP neurons has a role in cold-induced thermogenesis. Collectively, our results suggest that GIRK2 dampens excitability of the NPY/AgRP neurons to maintain sympathetic tone and thermogenesis in physiological and some stress conditions, which may serve to keep body weight in control independently of appetite.
Results
GIRK channels maintain RMP of NPY neurons
A previous study reported a transcriptome obtained from AgRP neurons [15], which included mRNA of various K+ channels that may contribute to maintenance of RMP. In particular, M-type K+ or M channels, two-pore K+ (K2P) channels, KATP channels, and GIRK channels had significant levels of mRNA expression [15]. Thus, we obtained acute hypothalamic slices from the Npy-hrGFP mice and targeted the fluorescence-labeled NPY neurons within the ARH for whole-cell patch clamp recordings (Fig 1A), where we tested the effects of pharmacological inhibitors of abovementioned K+ channels.
Fig 1. GIRK channels stabilize RMP of NPY neurons.
(A) Brightfield illumination (Brightfield), fluorescent (FITC) illumination (Npy-hrGFP), fluorescent (TRITC) illumination (Alexa Fluor 594), and merged (Merge) images of targeted NPY neuron. Arrows indicate the cell targeted for whole-cell patch clamp recording. (B) Image demonstrates a depolarizing effect of tertiapin-Q. Dotted line indicates RMP. (C) Voltage deflections in response to small hyperpolarizing current steps (from −25 pA to 0 pA by 5 pA increments) before (control, black) and after (tertiapin-Q, red) the perfusion with tertiapin-Q as indicated by arrows in (B). (D) The voltage–current (V-I) relationship demonstrates increased input resistance by tertiapin-Q. Erev = reversal potential. (E) Lines and dots summarize effects of tertiapin-Q on RMP (from −47.7 ± 3.0 mV to −44.9 ± 2.1 mV, n = 11, df = 10, t = 2.787, p = 0.019). Red and black lines indicate changes of membrane potential in depolarized and nonresponsive neurons, respectively. (F) Lines and dots summarize effect of tertiapin-Q on input resistance (from 2.75 ± 0.27 GΩ to 3.03 ± 0.30 GΩ, n = 11, df = 10, t = 4.370, p = 0.001). Red and black lines indicate changes of input resistance in depolarized and nonresponsive neurons, respectively. (G, H) Lines and dots summarize effects of 100 nM tertiapin-Q (G) (from −41.2 ± 0.8 mV to −40.0 ± 1.1 mV, n = 11, df = 10, t = 2.040, p = 0.069) and 500 nM tertiapin-Q (H) (from −42.9 ± 1.2 mV to −40.5 ± 1.1 mV, n = 13, df = 12, t = 3.292, p = 0.006) on RMP. Red and black lines indicate changes of membrane potential in depolarized and nonresponsive neurons, respectively. (I) Histogram summarizes responses (no effects or depolarization) of NPY neurons to different concentrations of tertiapin-Q. (J) Bar graphs and dots summarize effects of K+ channel blockers. Each neuron was tested with only 1 K+ channel blocker. Data are presented as mean ± SEM. Paired t test was used for statistical analyses. *p < 0.05, **p < 0.01. The numerical data for Fig 1D–1J can be found in S1 Data. GIRK, G protein-gated inwardly rectifying K+; NPY, neuropeptide Y; RMP, resting membrane potential.
GIRK channels were demonstrated to maintain RMP of several types of central neurons [13,16,17]. We tested the involvement of GIRK channels and found that bath applications of tertiapin-Q (300 nM), a GIRK channel blocker, depolarized membrane potential in 5 of 11 (approximately 45%) NPY neurons (from −56.3 ± 3.6 mV to −50.6 ± 2.8 mV, n = 5, Fig 1B and 1E, red lines). We applied small hyperpolarizing current steps before and after tertiapin-Q treatments (Fig 1B and 1C) and plotted the amplitudes of voltage responses against the amplitudes of injected currents to obtain a voltage–current (V-I) relationship (Fig 1D). We noted that the depolarizing effects were accompanied by increased input resistance (from 2.49 ± 0.47 GΩ to 2.84 ± 0.50 GΩ, n = 5, Fig 1F, red lines) with a reversal potential (Erev) of −101.3 ± 14.6 mV (n = 5) (Fig 1D). Changes of membrane potential and input resistance by tertiapin-Q were significant when we included all neurons recorded (Fig 1E and 1F). We also tested lower (100 nM) and higher (500 nM) concentrations of tertiapin-Q and found that the depolarizing effects become more significant at higher concentrations (Fig 1G and 1H). In addition, the response rate increased at higher concentrations (Fig 1I). These results suggested the contribution of GIRK channels to the maintenance of RMP in NPY neurons.
Notably, NPY neurons depolarized by tertiapin-Q had significantly lower action potential (AP) firing frequency and hyperpolarized RMP compared to those not responding to tertiapin-Q (S1A and S1B Fig). These data suggest that NPY neurons depolarized by tertiapin-Q have active GIRK channels and therefore are more stable. Consistent with this idea, NPY neurons depolarized by tertiapin-Q had lower input resistance than nonresponsive neurons, although the difference was not significant (S1C Fig). We also noted lower AP threshold in neurons depolarized by tertiapin-Q (S1D Fig), which may result from higher availability of voltage-gated Na+ channels due to more negative RMP.
Subsequently, we tested the effects of M channel blockers (10 μM linopirdine and 10 μM XE991) and observed depolarizing responses (3 mV and 5 mV) in 2 of 12 cells (approximately 17%) tested (Figs 1J, S2A, and S2E). These effects were accompanied by increased input resistance (from 2.98 GΩ to 3.80 GΩ and from 3.56 GΩ to 4.84 GΩ) and Erev of −94.0 mV and −81.0 mV, which suggested the contribution of M channels in a small subpopulation of NPY neurons. We also tested the effects of PK-THPP (1 μM, a TASK-3 channel blocker), spadin (1 μM, a TREK-1 channel blocker), and tolbutamide (100 μM, a KATP channel blocker), but none of these blockers caused significant changes in NPY neuronal membrane potential (Figs 1J, S2B–S2D, and S2F–S2H). Since we included 2 mM of ATP in pipette solutions (see Materials and methods), which may inhibit KATP channels [18], we also tested the effects of tolbutamide using ATP-free pipette solutions but found that RMP still remains unchanged (from −41.1 ± 1.1 mV to −40.9 ± 1.1 mV, p = 0.623, n = 10). Thus, it appears that neither K2P channel nor KATP channel plays a measurable role to maintain RMP of NPY neurons.
Arcuate AgRP neurons preferentially express Girk2 over Girk1
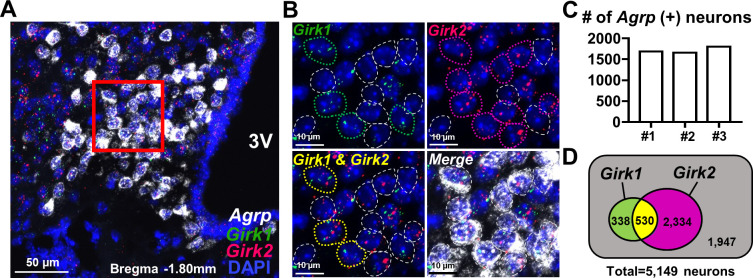
Neuronal GIRK channels contain one or both of GIRK1 and GIRK2 subunits [19], and both Girk1 and Girk2 mRNAs were found in the transcriptome of AgRP neurons [15]. Therefore, we characterized the expression of Girk1 and Girk2 by arcuate AgRP neurons with fluorescence in situ hybridization (FISH) experiments (RNAscope) targeting Agrp, Girk1, and Girk2 mRNA in wild-type mice. As shown in Fig 2A and 2B, Agrp-expressing neurons (white) expressed both Girk1 (green) and Girk2 (magenta) at mRNA levels within the ARH.
Fig 2. Dominant expression of Girk2 over Girk1 by the arcuate AgRP neurons.
(A) Image demonstrates DAPI (blue) and mRNA of Agrp (white), Girk1 (green), and Girk2 (magenta) detected by FISH experiments within the arcuate nucleus. 3V = third ventricle. Scale bar = 50 μm. (B) Magnified images of red rectangular area in (A). Dotted circles indicate Agrp (+) neurons (white) with Girk1 (green), Girk2 (magenta), or both Girk1 and Girk2 (yellow) mRNA. Scale bar = 10 μm. (C) Bar graph demonstrates numbers of Agrp (+) neurons in the arcuate nuclei of 3 wild-type mice. (D) Venn diagram demonstrates the numbers of Girk1- and/or Girk2-expressing Agrp (+) neurons. Data were pooled from neurons of 3 mice shown in (C), and 12 hypothalamic slices from each mouse (from bregma −1.58 mm to −2.02 mm) were included for analyses. The numerical data for Fig 2C can be found in S2 Data. AgRP, agouti-related peptide; FISH, fluorescence in situ hybridization; GIRK, G protein-gated inwardly rectifying K+.
For quantitative analyses, we pooled 5,149 Agrp-positive ARH neurons from 3 mice, each of which had comparable numbers of Agrp-expressing neurons in coronal hypothalamic sections from bregma −1.58 mm to −2.02 mm (Fig 2C). We found that 3,202 neurons (62.2%) express either Girk1 or Girk2 mRNA (Fig 2D), where Girk2 is expressed by a majority (2,864 neurons, 55.6%) but Girk1 by a smaller subpopulation (868 neurons, 16.9%). Moreover, almost two thirds of Girk1-expressing neurons also expressed Girk2 (530 neurons, 10.3%), whereas 338 neurons (6.6%) expressed Girk1 only. The remaining 1,947 Agrp-positive neurons (37.8%) did not express either Girk1 or Girk2. We also noted rather even distribution of both Girk1 and Girk2 along the rostrocaudal axis of the hypothalamus containing Agrp-positive neurons (S3A Fig). These results demonstrate that the arcuate AgRP neurons express both Girk1 and Girk2, but the latter is expressed by a larger subpopulation.
We repeated the same series of experiments for all possible combinations of functional GIRK channels and confirmed that Girk2 expression is higher than any other subunit (S3B–S3D Fig). The Girk2 mRNA was expressed by 55.8 ± 1.7% (n = 3) and 50.9 ± 1.5% (n = 3) of Agrp-positive neurons in 2 independent sets of experiments (S3A and S3D Fig). Expression levels of Girk3 and Girk4 mRNA were comparable to each other: Girk3 mRNA was expressed by 34.5 ± 1.7% (n = 3) and 29.2 ± 1.9% (n = 3) of Agrp-positive neurons in 2 independent sets of experiments (S3B and S3D Fig), while Girk4 mRNA was expressed by 30.1 ± 1.0% (n = 3) of Agrp-positive neurons in 1 set of experiments (S3C Fig). Girk1 mRNA showed the least abundant expression and was expressed by 16.8 ± 0.9% (n = 3), 17.7 ± 1.2% (n = 3) and 16.1 ± 0.5% (n = 3) of Agrp-positive neurons in 3 independent sets of experiments (S3A–S3C Fig). Notably, co-expression of any 2 subunits (i.e., Girk1/Girk2, Girk1/Girk3, Girk1/Girk4, Girk2/Girk3, gray lines in S3A–S3D Fig) was found in a minority of Agrp-positive neuron. The highest level of co-expression was observed for Girk2 and Girk3: These subunits are co-expressed by 18.3 ± 1.4% (n = 3) of Agrp-positive neurons (S3D Fig), while the lowest level of co-expression was observed for Girk1 and Girk3 (co-expression by 7.7 ± 0.8% (n = 3) of Agrp-positive neurons (S3B Fig)). Girk1/Girk2 co-expression and Girk1/Girk4 co-expression was observed in 10.2 ± 0.7% (n = 3) and 8.1 ± 0.6% (n = 3) of Agrp-positive neurons, respectively (S3A and S3C Fig). These results suggest that GIRK2 homomers, followed by GIRK2/GIRK3 heteromers, constitute a majority of GIRK channels in NPY/AgRP neurons.
GIRK2-containing GIRK channels are dispensable for GABAB-activated K+ currents in NPY neurons
GIRK channels are known to mediate slow synaptic inhibition by the stimulation of GABAB receptors [20]. Thus, we performed voltage clamp experiments to determine whether GIRK channels contribute to GABAB-activated currents in NPY neurons. We applied baclofen, a GABAB receptor agonist, to NPY neurons from the Npy-hrGFP transgenic mice using a local perfusion system (see Materials and methods) to record GABAB-activated GIRK currents. At a holding potential of −40 mV, application of 100 μM baclofen caused instantaneous outward currents (S4A Fig). We applied voltage ramp pulses (from −120 mV to −10 mV, 100 mV/s) before and during baclofen applications (arrows “a” and “b” of S4A Fig) to obtain a current–voltage (I-V) relationship of baclofen-activated currents (IBac), where IBac was Ib-Ia (S4B Fig). The I-V relationship of IBac showed inward rectification with Erev close to EK (−88.5 ± 0.7 mV, n = 12), consistent with GIRK channel activation. We also calculated the rectification index (I-120 mV/I-60 mV), the ratio of absolute values of currents at −120 mV (I-120 mV) and −60 mV (I-60 mV) of I-V curve. The average rectification index was 2.5 ± 0.2 (n = 12, S4C Fig).
We next examined currents evoked by baclofen (10 μM and 100 μM) in NPY neurons on WT (NPYG2WT neuron) and GIRK2 KO (NPYG2KO neuron) backgrounds (see Materials and methods). Unexpectedly, we found that GIRK2 ablation did not affect the amplitudes of IBac at 10 μM (1.4 ± 0.1 pA/pF, n = 32, for NPYG2WT neuron and 1.4 ± 0.1 pA/pF, n = 23, for NPYG2KO neuron, p = 0.783) and at 100 μM (1.8 ± 0.1 pA/pF, n = 53, for NPYG2WT neuron and 1.8 ± 0.2 pA/pF, n = 26, for NPYG2KO neuron, p = 0.984), respectively (S4D–S4G Fig). These results demonstrate that GIRK2-containing GIRK channels are not responsible for GABAB-activated K+ currents in NPY neurons.
GIRK2-containing GIRK channels contribute to RMP of NPY neurons
We also performed current clamp experiments to assess the role of GIRK2-containing GIRK channels in the maintenance of NPY neuron RMP. We found that NPYG2KO neurons had significantly depolarized RMP (−44.5 ± 0.7 mV, n = 41, p = 0.012) compared to NPYG2WT neurons (−47.9 ± 0.9 mV, n = 64) (Fig 3A and 3B). NPYG2KO neurons also had significantly higher input resistance (2.78 ± 0.11 GΩ, n = 41, p = 0.011) compared to NPYG2WT neurons (2.35 ± 0.11 GΩ, n = 64) (Fig 3C). In addition, tertiapin-Q (300 nM) depolarized only 1 of 13 (7.7%) NPYG2KO neurons, which was significantly different from the effects of tertiapin-Q on NPYG2WT neurons (S5 Fig). Together with results from S1 and S3 Figs, it seems that GIRK2-containing GIRK channels are open at rest to maintain RMP and decrease input resistance in NPY neurons.
Fig 3. Contribution of GIRK2-containing channels to RMP and GABAB-induced inhibition of NPY neurons.
(A) Traces demonstrate spontaneous firing and RMP of NPYG2WT (black) and NPYG2KO (red) neurons. Dotted line indicates RMP. (B, C) Bar graphs and dots summarize RMP (−47.9 ± 0.9 mV, n = 64, for NPYG2WT and −44.5 ± 0.7 mV, n = 41, for NPYG2KO, df = 103, t = 2.556, p = 0.012) (B) and input resistance (2.35 ± 0.11 GΩ, n = 64, for NPYG2WT and 2.78 ± 0.11 GΩ, n = 41, for NPYG2KO, df = 103, t = 2.590, p = 0.011) (C) of NPYG2WT (n = 64, black) and NPYG2KO (n = 41, red) neurons. (D) Image demonstrates a hyperpolarization of NPYG2WT neuron membrane potential by baclofen (10 μM). Arrows indicate interruptions to apply current step pulses. (E) Small hyperpolarizing current steps (from −50 pA to 0 pA by 10 pA increments) were applied before (control) and after (baclofen) applications of baclofen. (F) Voltage–current relationship demonstrates decreased input resistance and Erev close to EK. (G) Image demonstrates a hyperpolarization of NPYG2KO neuron membrane potential by baclofen (10 μM). (H) Summary of GABAB-induced hyperpolarization of NPYG2WT (black) and NPYG2KO (red) neurons. Changes of membrane potential by 10 μM baclofen was −11.9 ± 2.2 mV for NPYG2WT (n = 14) and −20.9 ± 2.4 mV for NPYG2KO (n = 8) (df = 20, t = 2.655, p = 0.015). Solid lines indicate fitting of dose-response curve (Hill slope = 1.0, Y = Bottom + (Top-Bottom)/(1+10^(logEC50-X)). Both hyperpolarizing and no responses were included for analyses. See Table 1 for hyperpolarizing responses only. Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. *p < 0.05. The numerical data for Fig 3B, 3C, 3F, and 3H can be found in S3 Data. GIRK, G protein-gated inwardly rectifying K+; NPY, neuropeptide Y; RMP, resting membrane potential.
We also examined whether GIRK2-containing GIRK channels have a role in GABAB-induced hyperpolarization of NPY neuronal membrane potential. We noted that treatments of NPYG2WT neurons with CGP54626 (2 μM), a GABAB receptor antagonist, do not affect RMP and input resistance of NPYG2WT neurons (S6 Fig), which suggested that GABAB receptors are not active at rest to affect the membrane potential of NPYG2WT neurons. Subsequently, we found that application of 10 μM baclofen hyperpolarized NPYG2WT neurons by −14.1 ± 1.9 mV (n = 12 of 14 cells) (Fig 3D and Table 1). The hyperpolarizing effects were accompanied by decreased input resistance and Erev of −104.3 ± 5.3 mV (n = 12) based on the V-I relationship calculated from voltage responses to current steps pulses before and after baclofen perfusion (Fig 3E and 3F). We also tried lower (1 μM) and higher (30 μM and 100 μM) concentrations of baclofen and noted dose-dependent effects (Fig 3H and Table 1), where Erev was comparable across all concentrations tested. We conducted the same series of experiments with NPYG2KO neurons and found that NPYG2KO neurons showed significantly augmented hyperpolarization (−20.9 ± 2.4 mV, n = 8, p = 0.035) by 10 μM baclofen (Fig 3G and 3H and Table 1). The observed augmentation of baclofen-induced hyperpolarization is likely due to increased input resistance together with unchanged GABAB-activated GIRK currents in NPYG2KO neurons (S4D–S4G Fig).
Table 1. Summary of GABAB-induced hyperpolarization of arcuate NPY neurons.
| Baclofen Conc. | NPYG2WT neuron | NPYG2KO neuron |
|---|---|---|
| 1 μM | −16.3 ± 3.0 mV (n = 7, 88%) Erev = −94.9 ± 6.1 mV |
−13.1 ± 2.4 mV (n = 6, 67%, p = 0.431) Erev = −99.8 ± 11.1 mV |
| 10 μM | −14.1 ± 1.9 mV (n = 12, 86%) Erev = −104.3 ± 5.3 mV |
−20.9 ± 2.4 mV (n = 8, 100%, p = 0.035) Erev = −94.3 ± 2.0 mV |
| 30 μM | −19.1 ± 2.0 mV (n = 12, 100%) Erev = −99.6 ± 4.8 mV |
−20.8 ± 3.3 mV (n = 9, 100%, p = 0.641) Erev = −83.1 ± 3.8 mV |
| 100 μM | −20.0 ± 2.0 mV (n = 14, 100%) Erev = −92.7 ± 3.1 mV |
−26.7 ± 3.2 mV (n = 9, 100%, p = 0.075) Erev = −89.8 ± 3.7 mV |
Changes of membrane potential are presented as mean ± SEM. Numbers in parentheses indicate the number of responsive cells, response rate, and p values (unpaired t test, NPYG2WT neurons vs. NPYG2KO neurons). Erev = reversal potential. The individual numerical data for changes of membrane potential and reversal potential can be found in S8 Data.
NPY, neuropeptide Y.
GIRK2 ablation, but not GIRK1 ablation, results in a persistent increase of AgRP neuronal activity
Given the higher expression of Girk2 mRNA than Girk1 mRNA (Fig 2) as well as the contribution of GIRK2 subunits to the RMP (Fig 3), we assumed that the GIRK2-containing GIRK channels may play a more important role than GIRK1-containing GIRK channels to maintain AgRP neuronal activity. To test this idea, we labeled AgRP neurons with tdTomato reporter using Agrp-ires-Cre::tdTomato (AgrptdTomato) mice and performed immunohistochemistry (IHC) experiments to measure Fos expression level in arcuate AgRP neurons. We found that 56.0 ± 3.2% (n = 6) of AgRP neurons express Fos when the mice were fasted overnight for 18 h (Fig 4A and 4B).
Fig 4. Deletion of GIRK2, but not GIRK1, leads to increased Fos expression by the arcuate AgRP neurons.
(A) Images demonstrate Fos IHC results from AgrptdTomato, AgrptdTomato/Girk1KO, and AgrptdTomato/Girk2KO mice, as indicated. 3V = third ventricle. Scale bar = 50 μm. (B) Bar graphs and dots summarize proportion of Fos-expressing AgRP neurons in AgrptdTomato (56.0 ± 3.2%, n = 6, black), AgrptdTomato/Girk1KO (64.6 ± 2.6%, n = 4, gray), and AgrptdTomato/Girk2KO (71.7 ± 4.8%, n = 4, red). Twelve hypothalamic slices from each mouse (from bregma −1.46 mm to −2.06 mm) were included for analyses. Data are presented as mean ± SEM. Ordinary one-way ANOVA with Bonferroni correction was used for statistical analyses (df = 2, F2, 11 = 4.961, p = 0.029). *p < 0.05, ns = not significant. The numerical data for Fig 4B can be found in S4 Data. AgRP, agouti-related peptide; GIRK, G protein-gated inwardly rectifying K+; IHC, immunohistochemistry.
To address the contribution of GIRK1 and GIRK2 subunits, we generated conditional knockout mice by breeding Agrp-ires-Cre mice [21] and Girk1flox/flox or Girk2flox/flox mice [22,23]. We performed in situ hybridization (ISH) experiments (BaseScope) to confirm successful deletion of Girk1 in the arcuate AgRP neurons of the Agrp-ires-Cre::Girk1flox/flox (GIRK1AgRP-KO) mice compared to Girk1flox/flox (GIRK1WT) mice (S7A–S7C Fig). We noted that Girk2 expression by the arcuate AgRP neurons was not significantly different between GIRK1WT and GIRK1AgRP-KO mice (S7D Fig). We also examined Girk1 expression in the hippocampus and confirmed that its expression is not affected by AgRP neuron-specific deletions (S8A and S8B Fig), which was further confirmed and quantified by qRT-PCR experiments (S8C Fig). Likewise, we confirmed a significant reduction of Girk2, but comparable expression of Girk1, in the arcuate AgRP neurons of the Agrp-ires-Cre::Girk2flox/flox (GIRK2AgRP-KO) mice compared to Girk2flox/flox (GIRK2WT) mice (S7E–S7H Fig) and observed comparable expression of Girk2 in the hippocampus (S8D–S8F Fig). Together, these results indicate successful generation of conditional GIRK channel knockout models.
Subsequently, we measured Fos expression levels of tdTomato-expressing AgRP neurons in the ARH obtained from Agrp-ires-Cre::tdTomato::Girk1flox/flox (AgrptdTomato/Girk1KO) and Agrp-ires-Cre::tdTomato::Girk2flox/flox (AgrptdTomato/Girk2KO) mice that were fasted overnight for 18 h. We observed significantly increased levels of Fos expression by AgRP neurons from the AgrptdTomato/Girk2KO mice, where 71.7 ± 4.8% (n = 4) of AgRP neurons showed Fos immunoreactivity (p = 0.020, Fig 4A and 4B) compared to results obtained from AgrptdTomato mice. On the other hand, 64.6 ± 2.6% (n = 4) of AgRP neurons from the AgrptdTomato/Girk1KO mice expressed Fos, which was not significantly different from what was observed in the AgrptdTomato mice (p = 0.231, Fig 4A and 4B). These results are consistent with the depolarized RMP of NPYG2KO neurons (Fig 3). Therefore, we suggest that at a population level GIRK2-containing GIRK channels, rather than GIRK1-containing GIRK channels, contribute to maintain AgRP neuronal activity.
Deletion of GIRK2 subunits in AgRP neurons increases adiposity and body weight independently of food intake
In order to delineate the metabolic function of GIRK2 subunits expressed by AgRP neurons, we measured body weight and food intake of GIRK2AgRP-KO and GIRK2WT mice once a week and found that GIRK2AgRP-KO mice gained more body weight than GIRK2WT mice on NCD (Fig 5A). The difference of body weight became more pronounced week by week to be statistically significant when the mice were 16 weeks old (Fig 5A). The nuclear magnetic resonance (NMR) analyses of body compositions, which was performed when the mice were 20 weeks old, demonstrated that the weight gain was due to increased fat mass (Fig 5B), while lean mass or body fluids were similar between genotypes (Fig 5C and 5D). Consistent with these findings, hematoxylin and eosin (HE) staining revealed infiltration of fat into the liver as well as increased size of adipocytes within the inguinal white (IGW) and perigonadal white (PGW) fat tissues of GIRK2AgRP-KO mice (Fig 5E). We noted that differences in food consumption do not explain the increased adiposity, since cumulative food intake was not different between GIRK2WT mice and GIRK2AgRP-KO mice (Fig 5F). We also found that food intake was not influenced by GIRK2 deletion when the mice (21- to 22-week-old) were refed after overnight fasting (Fig 5G).
Fig 5. GIRK2AgRP-KO mice show increased body weight and adiposity independently of food intake.
(A) Body weights of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice on NCD. Two-way repeated measures ANOVA with Bonferroni correction, gene (df = 1, F1, 28 = 5.251, and p = 0.030), time (df = 13, F13, 364 = 416.6, and p < 0.0001), and interaction (df = 13, F13, 364 = 3.372, and p < 0.0001). (B–D) Bar graphs and dots summarize fat mass (4.1 ± 0.3 g, n = 14, for GIRK2WT and 5.4 ± 0.3 g, n = 16, for GIRK2AgRP-KO, df = 28, t = 2.627, p = 0.014) (B), lean mass (21.5 ± 0.3 g, n = 14, for GIRK2WT and 22.0 ± 0.4 g, n = 16, for GIRK2AgRP-KO, df = 28, t = 1.113, p = 0.275) (C), and body fluids (2.1 ± 0.1 g, n = 14, for GIRK2WT and 2.3 ± 0.1 g, n = 16, for GIRK2AgRP-KO, df = 28, t = 1.648, p = 0.111) (D) of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice by NMR spectrometer analyses. (E) Images demonstrate HE staining results of liver, IGW, and PGW obtained from GIRK2WT and GIRK2AgRP-KO mice. Scale bar = 100 μm. (F) Cumulative food intake of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice. Two-way repeated measures ANOVA with Bonferroni correction, gene (df = 1, F1, 28 = 0.007, and p = 0.934), time (df = 13, F13, 364 = 2196, and p < 0.0001), and interaction (df = 13, F13, 364 = 0.0389, and p > 0.9999). (G) Food intake of GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice in fast-refeeding experiments. Two-way repeated measures ANOVA with Bonferroni correction, gene (df = 1, F1, 15 = 0.269, and p = 0.612), time (df = 4, F4, 60 = 960.6, and p < 0.0001), and interaction (df = 4, F4, 60 = 0.713, and p = 0.587). Data are presented as mean ± SEM. Two-way repeated measures ANOVA with Bonferroni correction (A, F, G) and unpaired t test (B–D) were used for statistical analyses. *p < 0.05, **p < 0.01, ns = not significant. The numerical data for Fig 5A–5D, 5F, and 5G can be found in S5 Data. GIRK, G protein-gated inwardly rectifying K+; HE, hematoxylin and eosin; IGW, inguinal white; NCD, normal chow diet; NMR, nuclear magnetic resonance; PGW, perigonadal white.
GIRK2-containing GIRK channels expressed by AgRP neurons are required for normal sympathetic activity and BAT function
Given no changes in food intake, we hypothesized that the body weight gain observed in GIRK2AgRP-KO would be caused by decreased energy expenditure. To test this idea, we measured oxygen consumption (VO2) and carbon dioxide production (VCO2) with an indirect calorimetry from 20-week-old GIRK2WT and GIRK2AgRP-KO mice. We observed significantly decreased VO2 and VCO2 in GIRK2AgRP-KO mice compared to GIRK2WT mice (Fig 6A, left and middle). The calculated EE was also significantly decreased in the GIRK2AgRP-KO mice (Fig 6A, right). During the indirect calorimetry measurements, we also measured ambulatory movements and rearing activities, but there was no difference between genotypes (S9A and S9B Fig). Both GIRK2WT mice and GIRK2AgRP-KO mice (21- to 22-weeks-old) moved similar distance when they were allowed to move freely in chambers designed for an open field test (OFT) (S9C and S9D Fig). AgRP neurons were shown to regulate anxiety level [24], but our OFT results demonstrated similar levels of anxiety regardless of genotypes, based on their comparable preference to the center zone and the outer zone in the chamber (S9E–S9G Fig). Thus, the decreases in EE observed in GIRK2AgRP-KO mice are likely due to reduced basal metabolic rate.
Fig 6. GIRK2AgRP-KO mice show decreased EE associated with BAT dysfunction and decreased sympathetic activity.
(A) Bar graphs and dots summarize oxygen consumption (VO2) (left, dark cycle: 3.23 ± 0.07 L/h/kg, n = 14, for GIRK2WT and 2.98 ± 0.09 L/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 2.088, p = 0.046; light cycle: 2.57 ± 0.06 L/h/kg, n = 14, for GIRK2WT and 2.38 ± 0.07 L/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 1.842, p = 0.076), carbon dioxide production (VCO2) (middle, dark cycle: 2.99 ± 0.06 L/h/kg, n = 14, for GIRK2WT and 2.66 ± 0.08 L/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 3.198, p = 0.003; light cycle: 2.33 ± 0.04 L/h/kg, n = 14, for GIRK2WT and 2.09 ± 0.07 L/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 2.847, p = 0.008), and EE (right, dark cycle: 15.9 ± 0.3 kcal/h/kg, n = 14, for GIRK2WT and 14.7 ± 0.4 kcal/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 2.140, p = 0.041; light cycle: 12.6 ± 0.3 kcal/h/kg, n = 14, for GIRK2WT and 11.7 ± 0.4 kcal/h/kg, n = 16, for GIRK2AgRP-KO, df = 28, t = 1.972, p = 0.059) of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice measured by indirect calorimetry. (B) Images demonstrate HE (upper), oil red O (middle) staining, and UCP1 (lower) immunostaining results of BAT obtained from GIRK2WT (left) and GIRK2AgRP-KO (right) mice. Scale bar = 20 μm. (C, D) Images on the left demonstrate IHC of ChAT (red), Fos (green), and DAPI (blue) in upper (T1-T6) thoracic spinal cords of GIRK2WT (C) and GIRK2AgRP-KO (D) mice at a lower magnification. Scale bar = 100 μm. Areas of IML in the rectangles are shown on the right at a higher magnification. In merged images, gray arrowheads indicate Fos (−) and ChAT (+) neurons, and yellow arrowheads indicate Fos (+) and ChAT (+) neurons. Scale bar = 10 μm. (E) Bar graphs and dots summarize proportion of Fos-expressing ChAT neurons in IML of GIRK2WT (52.4 ± 3.9%, n = 6) and GIRK2AgRP-KO (32.4 ± 2.6%, n = 4) mice (df = 8, t = 3.79, p = 0.005). A total of 48 spinal cord slices from each mouse (levels T1-T6) were included for analyses. Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. *p < 0.05, **p < 0.01. The numerical data for Fig 6A and 6E can be found in S6 Data. BAT, brown adipose tissue; ChAT, choline acetyltransferase; EE, energy expenditure; GIRK, G protein-gated inwardly rectifying K+; HE, hematoxylin and eosin; IHC, immunohistochemistry; IML, intermediolateral column.
Decreased BAT thermogenesis is often a major cause of reduced basal metabolic rate and energy expenditure [25]. Indeed, we noted increased adiposity and triacylglycerol level in the BAT from the GIRK2AgRP-KO mice by HE and oil red O staining (Fig 6B, top and middle). In addition, uncoupling protein-1 (UCP-1) immunoreactivity was markedly decreased in the BAT of GIRK2AgRP-KO mice (Fig 6B, bottom). Since BAT thermogenesis is regulated by sympathetic tone [26] and NPY/AgRP neurons are known to decrease sympathetic activity [5,27,28], we predicted that increased activity of NPY/AgRP neurons would result in decreased sympathetic activity of GIRK2AgRP-KO mice. To test this idea, we performed IHC experiments and measured Fos levels in the cholinergic sympathetic preganglionic neurons of the intermediolateral column (IML) of T1 to T6 spinal cords. We found in GIRK2AgRP-KO mice a significantly lower percentage (32.4 ± 2.6%, n = 4, p = 0.005) of choline acetyltransferase (ChAT)-positive IML neurons expressing Fos compared to observations in the GIRK2WT mice (52.4 ± 3.9%, n = 6) (Fig 6C–6E) at 8 to 12 weeks of age. Together, these results suggest that decreased sympathetic activity and BAT thermogenesis lead to decreased energy expenditure and body weight gain in GIRK2AgRP-KO mice.
GIRK2 expressed by AgRP neurons is necessary for prompt adaptation to a cold temperature
Our results suggested that GIRK2 subunits expressed by AgRP neurons contribute to maintain body weight by promoting EE in non-stress conditions. To explore if GIRK2 subunits also have a role in stress conditions, we intraperitoneally (i.p.) injected 10-week-old GIRK2WT and GIRK2AgRP-KO mice with ghrelin (0.4 mg/kg) and measured food intake for 4 h after injections. We expected that ghrelin produces hunger-induced stress, but found that ghrelin-induced increase of food intake was similar between GIRK2WT and GIRK2AgRP-KO mice (S10 Fig). GIRK2WT and GIRK2AgRP-KO mice used for this experiment weighed 27.7 ± 1.0 g (n = 4) and 28.1 ± 1.2 g (n = 4), respectively (p > 0.5 by unpaired t test).
In a different set of experiments, we exposed 10-week-old GIRK2WT and GIRK2AgRP-KO mice to a cold environment (5°C) to challenge the mice with cold stress. There was no significant difference in body weight or body composition between the genotypes (Fig 7A–7D). When the temperature dropped from 25°C to 5°C, GIRK2WT mice showed a prompt increase of VO2 and VCO2, which reached a new steady state after approximately 4 h (Fig 7E and 7F). GIRK2AgRP-KO mice also showed increase of VO2 and VCO2 in response to the cold exposure, but there was a significant delay in the rising phase of VO2 and VCO2 (Fig 7E and 7F). The calculated EE were also significantly different in the rising phase between the genotypes (Fig 7G). We noted no significant differences in ambulatory movement or rearing activity of GIRK2WT and GIRK2AgRP-KO mice (Fig 7H and 7I), suggesting that the increases of VO2 and VCO2 are likely from increased BAT thermogenesis. Taken together, we propose that GIRK2 expressed by AgRP neurons is dispensable for ghrelin-induced feeding, but is necessary for prompt adaptation to a cold environment.
Fig 7. GIRK2AgRP-KO mice show compromised adaptation to cold exposure.
(A–D) Bar graphs and dots summarize body weight (27.1 ± 0.6 g, n = 10, for GIRK2WT and 26.8 ± 0.6 g, n = 8, for GIRK2AgRP-KO, df = 16, t = 0.380, p = 0.706) (A), fat mass (3.1 ± 0.2 g, n = 10, for GIRK2WT and 3.5 ± 0.4 g, n = 8, for GIRK2AgRP-KO, df = 16, t = 0.967, p = 0.348) (B), lean mass (19.7 ± 0.6 g, n = 10, for GIRK2WT and 19.0 ± 0.3 g, n = 8, for GIRK2AgRP-KO, df = 16, t = 0.936, p = 0.364) (C), and body fluids (1.6 ± 0.1 g, n = 10, for GIRK2WT and 1.7 ± 0.0 g, n = 8, for GIRK2AgRP-KO, df = 16, t = 0.587, p = 0.566) (D) of 10-week-old male GIRK2WT mice (n = 10) and GIRK2AgRP-KO mice (n = 8) on NCD before cold exposure. (E–I) Graphs summarize oxygen consumption (VO2) (E), carbon dioxide production (VCO2) (F), EE (G), ambulatory movement (H), and rearing activity (I) of 10-week-old male GIRK2WT mice (n = 10) and GIRK2AgRP-KO (n = 8) in response to cold exposure (from 25°C to 5°C). Mice were acclimated for 2 days before experiments. Unpaired t test was used for statistical analyses (A–D), and two-way repeated measures ANOVA with Bonferroni correction was used for statistical analysis (E–I). VO2 (E): gene (df = 1, F1,16 = 1.670, p = 0.215), time (df = 11, F11, 176 = 267.1, p < 0.0001), and interaction (df = 11, F11, 176 = 3.237, p = 0.0005). VCO2 (F): gene (df = 1, F1,16 = 1.565, p = 0.229), time (df = 11, F11, 176 = 284.1, p < 0.0001), and interaction (df = 11, F11, 176 = 3.226, p = 0.0005). EE (G): gene (df = 1, F1,16 = 0.110, p = 0.744), time (df = 11, F11, 176 = 257.1, p < 0.0001), and interaction (df = 11, F11, 176 = 3.075, p = 0.0008). Ambulatory movement (H): gene (df = 1, F1,16 = 1.38, p = 0.257), time (df = 11, F11, 176 = 12.87, p < 0.0001), and interaction (df = 11, F11, 176 = 0.787, p = 0.653). Rearing activity (I): gene (df = 1, F1,16 = 2.391, p = 0.142), time (df = 11, F11, 176 = 11.51, p < 0.0001), and interaction (df = 11, F11, 176 = 0.569, p = 0.852). ***p < 0.001, ****p < 0.0001, ns = not significant. The numerical data for Fig 7A–7I can be found in S7 Data. EE, energy expenditure; GIRK, G protein-gated inwardly rectifying K+; NCD, normal chow diet.
Discussion
In this study, we found evidence that GIRK2 subunits are key to regulating the long-term baseline activity of arcuate NPY/AgRP neurons. In agreement, GIRK2 ablation in arcuate NPY/AgRP neurons resulted in increased adiposity and body weight. This phenotype was associated with decreased sympathetic activity and reduced energy expenditure, but not changes in food intake. We also demonstrated that GIRK2 ablation in arcuate NPY/AgRP neurons delays the initial phase of cold-induced thermogenesis. Together, our findings identified GIRK2 as a regulator of arcuate NPY/AgRP neuron activity that maintains sympathetic activity and burns fat, which should help to maintain homeostasis in physiological (normal caloric or non-stressed) and some stressed conditions.
In vivo metabolic effects of AgRP neuron activity
Previous studies demonstrated that the optogenetic or chemogenetic activation of AgRP neurons results in increased food intake and/or decreased energy expenditure [6,7,29], which occurred within hours. The activation of AgRP neurons using the “capsaicin-Trpv1” system also resulted in rapid decreases of energy expenditure and thermogenesis [30]. On the other hand, chemogenetic inhibition of AgRP neurons for 14 days led to decreased body weight, decreased food intake, and fat burning [31]. It was also shown that chemogenetic inhibition of AgRP neurons reverses diabetes-induced hyperphagia and hyperglycemia within a few hours [32]. Therefore, available data suggested that modulation of AgRP neuronal activity can significantly affect food intake and energy expenditure in time frames of hours to days.
Since acute activation of AgRP neurons resulted in rapid metabolic effects regardless of activating methods, we may expect that long-term activation of AgRP neurons would also produce similar phenotypes. A recent study overexpressed bacterial sodium channel (NachBac) or Kir2.1 channel selectively in AgRP neurons to achieve long-term activation and inhibition of neuronal activity, respectively [33]. The authors reported that NachBac overexpression resulted in massive obesity accompanied by increased food intake but no changes of energy expenditure, but that Kir2.1 overexpression did not produce any phenotype. In this study, we genetically deleted GIRK2 subunits selectively in the AgRP neurons, which presumably increased AgRP neuronal activity for longer periods (approximately 5 months). While we noted significantly increased body weight in GIRK2AgRP-KO mice, cumulative food intake was not different between genotypes. We also noted that fasting- and ghrelin-induced feeding were not different between genotypes. This finding was quite surprising given the prominent role of AgRP neurons in the regulation of food intake. Instead, O2 consumption and CO2 production were significantly decreased in GIRK2AgRP-KO mice, which were associated with decreased activity of sympathetic preganglionic neurons and decreased UCP-1 expression by BAT. GIRK2AgRP-KO mice also showed a significant delay in cold-induced increase of energy expenditure compared to GIRK2WT mice (Fig 7), which further suggested that thermogenic response is compromised in GIRK2AgRP-KO mice.
It is not clear why persistently increased activity of AgRP neurons decreased energy expenditure but did not regulate food intake in our study. One hypothesis is that GIRK2-expressing AgRP neurons preferentially regulate energy expenditure, like leptin receptor-expressing POMC neurons [34]. In this scenario, food intake and energy expenditure are regulated by distinct subpopulations of AgRP neurons, as previously suggested for POMC neurons [35]. An alternative possibility is that GIRK2-expressing AgRP neurons also regulate food intake, but this effect is discernable only in short-term time frames. In other words, AgRP neurons can regulate both food intake and energy expenditure, but a compensatory anorexia (due to decreased energy expenditure) may develop over time to mask increased food intake. In either case, the inhibition of energy expenditure by GIRK2-ablated AgRP neurons looks large enough. It is also important to note that we deleted GIRK2 subunits before birth. As GIRK channels can be assembled in 5 different compositions [36,37], other GIRK channel subunits may take the role of GIRK2 subunits in AgRP neurons in GIRK2AgRP-KO mice. Therefore, we may need a strategy to delete GIRK2 subunits postnatally to delineate the actual function of GIRK2 subunits expressed by AgRP neurons. Indeed, a previous study suggested that a compensatory mechanism may develop before birth to overcome the severe anorexia observed in AgRP-ablated mice [38]. We suggest that future studies are directed to delineate the metabolic functions designated to individual AgRP neurons, which will help to understand variable phenotypes obtained from AgRP neuron-specific conditional knockout mouse models.
Metabolic function of K+ channels expressed by NPY/AgRP neurons
A few previous studies reported the role of K+ channels expressed by the AgRP neurons in the regulation of energy balance. For example, a study demonstrated that down-regulation of the small conductance Ca2+-activated K+ (SK) channels contribute to fasting-induced activation of AgRP neurons [39]. It was noted that SK3 channel deletion resulted in increased firing rate of AgRP neurons without changes in RMP, which makes sense given the role of SK channel in the regulation of afterhyperpolarization. AgRP neuron-specific deletions of SK3 channels resulted in a transient obesity in NCD-fed mice, but profoundly exacerbated HFD-induced obesity. The increased susceptibility to HFD was associated with increased food intake and decreased EE, while the locomotive activity remained unchanged. More recently, it was shown that CRISPR knockdown of Kcnq3, an M channel subunit, in NPY/AgRP neurons did not affect RMP but increased input resistance and decreased rheobase current, which suggested increased response to external stimuli [40]. However, it was noted that in NCD conditions KCNQ3 deficiency resulted in no changes of food intake and body weight, while locomotive activity was decreased. Similar results were obtained in HFD-fed mice except that there was an increase of abdominal fat mass. Together, it is suggested that M channels expressed by NPY/AgRP neurons are largely dispensable for the control of energy balance whether the mice are given NCD or HFD.
In this study, we found a predominant role for GIRK channels to maintain RMP of NPY/AgRP neurons (Fig 1). Notably, knockout of GIRK2 resulted in a depolarized RMP (approximately 3.5 mV), amplitude of which was comparable to the depolarizing effects of the GIRK channel blocker (approximately 4 mV). We also noted a few cells depolarized by M channel blockers, but a majority of NPY/AgRP neurons did not respond (S2 Fig), which is consistent with the limited in vivo function of KCNQ3 expressed by NPY/AgRP neurons [40]. While SK3 and KCNQ3 expressed by the AgRP neurons were largely dispensable in NCD-fed mice [39,40], GIRK2 ablation in AgRP neurons led to significantly increased body weight in NCD-fed mice (Fig 5). Together, it appears that GIRK2-containing GIRK channels have a unique role in the physiological NCD-fed conditions to regulate NPY/AgRP neuronal excitability and energy balance.
Functional GIRK channel subunit composition in NPY/AgRP neurons
GIRK1/GIRK2 heterotetramers are the prototypes of neuronal GIRK channels, and loss of either GIRK1 or GIRK2 is usually sufficient to eliminate most or all of GIRK channel activity in central neurons [36,37]. A notable exception is the GIRK channel of midbrain dopaminergic neurons. They lack GIRK1 subunits, and GIRK2 homotetramers and/or GIRK2/GIRK3 heterotetramers are believed to be the subunit composition of GIRK channels in these neurons [36,37]. In this study, we found that GIRK2 is the major GIRK channel subunit expressed by arcuate AgRP neurons (Figs 2 and S3). Analyses of our FISH data suggest that GIRK2 homotetramer (and GIRK2/GIRK3 heterotetramer) may constitute a majority of functional GIRK channels in AgRP neurons (S3 Fig). Therefore, GIRK channel subunit composition of AgRP neurons appear to be similar to that of midbrain dopaminergic neurons.
We unexpectedly found that ablation of GIRK2 did not affect GABAB-activated K+ currents in NPY/AgRP neurons (S4 Fig). On the other hand, GIRK2-containing GIRK channels contributed to long-term control of NPY/AgRP neuronal excitability and in vivo energy balance (Figs 4–6). Together, these data may suggest that GIRK2-containing GIRK channels are open at rest to regulate in vivo metabolic function but are not functionally coupled to GABAB receptor stimulation. In this case, GIRK1/GIRK3 and/or GIRK1/GIRK4 heterotetramers may be responsible for GABAB-activated K+ currents. However, we also need to consider the possibility that the loss of GIRK2 subunits and the contribution to GABAB-activated K+ currents was replaced by other GIRK subunits through a compensatory mechanism in our model. This is especially so given that almost all NPY/AgRP neurons generate outward currents by baclofen (S4 Fig) but Girk1/Girk3 (7.7 ± 0.7%, n = 3) and Girk1/Girk4 (8.1 ± 0.2%, n = 3) co-expression levels in NPY/AgRP neurons are quite low (S3B and S3C Fig). It remains to be tested whether GIRK2-containing GIRK channels are coupled to other Gi/o protein-coupled receptors. Overall, more investigations are necessary to delineate the subunit composition of functional GIRK channels of NPY/AgRP neurons.
Materials and methods
Ethics statement
All experiments were performed in accordance with the guidelines established by the Korea Advanced Institute of Science and Technology (KAIST) Institutional Animal Care and Use Committee (IACUC) (Protocol No. KA2021-126). KAIST IACUC follows the standard operating guidelines for IACUC established by the Animal and Plant Quarantine Agency and the Ministry of Food and Drug Safety of South Korea.
Mice
All mice used for breeding and experiments in this study were housed in a light-dark (12 h on/off; lights on at 7:00 AM) and temperature-controlled environment with food and water available ad libitum in the KAIST facilities. Npy-hrGFP mice were obtained from the Jackson laboratory (#006417). For some patch clamp experiments GIRK2 KO mice [41], used with the permission from Dr. Markus Stoffel (ETH Zurich), were crossed with Npy-hrGFP mice. Agrp-ires-Cre mice (Jackson laboratory, #012899) were crossed with tdTomato reporter mice (Jackson laboratory, #007914), GIRK1flox/flox mice [22] or GIRK2flox/flox mice [23] for FISH, ISH, IHC, and in vivo metabolic experiments. Mice were fed standard NCD (Teklad global 18% protein 2018S, ENVIGO).
Electrophysiology
Five- to 13-week-old male Npy-hrGFP mice were used for all patch clamp experiments in order to identify NPY-expressing neurons in the ARH. Npy-hrGFP mice were fasted for 18 h before being killed for experiments. Whole-cell patch clamp recordings from hrGFP-expressing neurons were maintained in acute hypothalamic slice preparations as previously described [42]. In brief, mice were deeply anesthetized with isoflurane inhalation and transcardially perfused with a modified ice-cold artificial CSF (ACSF) (described below), in which an equiosmolar amount of sucrose was substituted for NaCl. The mice were then decapitated, and the entire brain was removed from the skull and immediately submerged in ice-cold, carbogen-saturated (95% O2 and 5% CO2) ACSF (123 mM NaCl, 26 mM NaHCO3, 2.8 mM KCl, 1.25 mM NaH2PO4, 1.2 mM MgSO4, 2.5 mM CaCl2, and 10 mM glucose). A brain block containing the hypothalamus was made. Coronal sections (250 μm) were cut with a Leica VT1200S vibrating microtome and then incubated in oxygenated ACSF at 34°C for at least 1 h before recording. Brain slices were transferred to the recording chamber and allowed to equilibrate for 10 to 20 min before recording. The slices were bathed in oxygenated ACSF (32°C to 34°C) at a flow rate of approximately 2 ml/min. The pipette solution was modified to include an intracellular dye (Alexa Fluor 594) for whole-cell patch clamp recording: 120 mM K-gluconate, 10 mM KCl, 10 mM HEPES, 1 mM CaCl2, 1 mM MgCl2, 5 mM EGTA, 2 mM Mg-ATP, and 0.03 mM Alexa Fluor 594 hydrazide dye (pH 7.3). Epifluorescence was briefly used to target fluorescent cells, at which time the light source was switched to infrared differential interference contrast imaging to obtain the whole-cell recording (Nikon Eclipse FN1 equipped with a fixed stage and an optiMOS scientific CMOS camera). Recording electrodes had resistances of 3 to 5 MΩ when filled with the K-gluconate internal solution.
In current clamp experiments, input resistance was assessed by measuring amplitudes of voltage deflections in response to hyperpolarizing rectangular current step pulses (500 ms, −25 pA to 0 pA by 5 pA increments or −50 pA to 0 pA by 10 pA increments) which was applied at a stable membrane potential before and after drug application. AP threshold was determined from averaged AP traces of firing neurons. The voltage at the last minimum of dV/dt preceding the spike (within 2 ms preceding 10 V/s) was estimated to be AP threshold, as described previously [43]. A drug effect was required to be associated temporally with drug application, and the responses had to be stable within a few minutes. We determined membrane potential before (control) and during drug applications (drug) by averaging membrane potential for 10 s in each condition. A neuron was considered to be depolarized or hyperpolarized if a change in membrane potential was larger than 2 mV in amplitude. Membrane potentials were not compensated for liquid junction potentials (−8 mV).
For voltage clamp experiments, we used the same K-gluconate pipette solutions described above and added 0.5 μM tetrodotoxin (TTX) and synaptic blockers (50 μM picrotoxin and 1 mM kynurenic acid) to bath solutions. We held the membrane potential at −40 mV and locally applied baclofen using micropipettes attached to the Picospritzer III microinjection dispense system (Parker Hannifin). We placed micropipettes 10 to 20 μm away from soma and ejected small volume (15 to 20 pL) of ACSF containing baclofen and the blocker cocktail with a pressure of 16 to 18 psi for 15 s. Baclofen-activated currents (IBac) were normalized by cell capacitance. Voltage ramp pulses (from −120 mV to −10 mV, 100 mV/s) were applied before and after baclofen applications from a holding potential of −40 mV to obtain I-V relationships of IBac.
In situ hybridization
In situ hybridization experiments were performed using RNAscope or BaseScope assays available from the Advanced Cell Diagnostics Inc. (Hayward, California, United States of America). Briefly, 8- to 12-week-old male mice were deeply anesthetized with isoflurane and transcardially perfused with a DEPC-treated PBS and subsequently with a 4% paraformaldehyde (163–20145, FUJIFILM Wako). Brains were removed from the skull and submerged in cold 4% paraformaldehyde solutions for 24 h (post-fixation). Brains were then transferred to a series of sucrose solutions of gradients (4 h in 10% sucrose, 12 h in 20% sucrose, and 24 h in 30% sucrose) at 4°C. Brain slices with 10 μm thickness were obtained using a cryostat (Leica) and were stored in a cryo-protectant. We collected coronal sections that contain arcuate nucleus based on the shape of third ventricle, median eminence, and hippocampus referring to the Allen Brain Atlas. Brain slices were transferred to a well plate (20 wells, 4 rows × 5 columns) immediately after sectioning one by one in a rostrocaudal order. We used rows from left to right and columns from up to down. After collecting the first 20 slices, we repeated the same procedure 5 to 7 times so that one well contains 5 to 7 brain slices each of which is spaced by 200 μm. We used 1 column for 1 set of experiments. Brain slices were mounted on glass slides (Superfrost Plus Microscope Slides, Thermo Fisher) for the RNAscope or the BaseScope assays. All reagents for these assays were purchased from Advanced Cell Diagnostics.
AgRP probe (target region 11–764, accession # NM_001271806.1), Girk1 probe (target region 658–1679, accession # NM_008426.2), Girk2 probe (target region 282–1456, accession # NM_001025584.2), Girk3 probe (target region 84–1276, accession # NM_008429.2), and Girk4 probe (target region 523–1781, accession # NM_010605.4) were used for the triple RNAscope Multiplex Fluorescent Assay. AgRP probe (target region 61–214, 2 pairs, accession # NM_007427.3), Girk1 probe (target region 1351–1479, accession # NM_001355118.1), and Girk2 probe (target region 560–678, accession # NM_001025585.2) were used for the BaseScope Duplex Assay. Girk1 and Girk2 probes were custom-designed based on the targeted sequences of the Girk1flox/flox and the Girk2flox/flox mice [22,23]. Images of the RNAscope and BaseScope assays were obtained with a confocal microscope (LSM 780, Carl Zeiss) and a slide scanner (Axio Scan. Z1, Carl Zeiss), respectively, and were analyzed with ZEN lite (ZEN Microscopy software) and the Image J software. We lined up all stained images from individual mice, based on the reference brain atlas (Allen Brain Atlas), and selected 12 or 16 brain slices (from each mouse) that are available from all mice to be included for analyses.
RNA extraction and qRT-PCR
Brain blocks were prepared and submerged in ice-cold ACSF. Coronal brain slices (500 μm-1 mm thickness) were obtained using Leica VT1200S vibrating microtome. The brain slices were transferred to DEPC-based PBS on ice, and hippocampal regions containing dentate gyrus and CA1 were punched out using a blunt-end 16 gauge needle (#28110, STEMCELL Technologies) under a stereomicroscope. Hippocampal tissues were transferred to E-tubes on dry ice and preserved at −80°C. RNA was extracted from the hippocampal tissues by using RNeasy Lipid Tissue Mini Kit (74804, Qiagen).
qRT-PCR operation and analysis were performed as reported previously [44]. Briefly, 2 μg of RNA was added to master mix (SuperiorScript III RT Master Mix, RT300S, Enzynomics) and autoclaved de-ionized water to be 20 μl volume of mixture in total. We used a polymerase chain reaction (PCR) protocol, 25°C (10 min)– 42°C (1 h)– 85°C (5 min), for the synthesis of first-strand complimentary DNA (cDNA) synthesis. Product of PCR was diluted with autoclaved de-ionized water to be 100 μl. Approximately 1 μl of cDNA was mixed with SYBR Green Realtime PCR Master Mix (QPK-201, TOYOBO), target primers, and autoclaved de-ionized water to be 20 μl volume. Cycle threshold (Ct) method was used for quantitative analysis of target gene mRNA. Relative expressional levels of target genes were determined by comparing the level to that of 2 housekeeping genes: GAPDH and 18S genes. Primers targeting GIRK channel subunits and protocols for amplification were prepared as described previously [45]. The primers used are as follows: Girk1(forward) 5′-GAGGGACGGAAAACTCACTCT-3′; Girk1(reverse) 5′-TCAGGTGTCTGCCGAGATT-3′; Girk2(forward) 5′-CGTGGAGTGAATTATTGAATCT-3′; Girk2(reverse) 5′-GTCATTTCTTCTTTGTGCTTTT-3′. We used an amplification protocol of 95°C (5 min, 1 cycle)– 45 cycles of 95°C (10 s)– 60°C (30 s)– 72°C (10 s).
Immunohistochemistry
Fos activity in the hypothalamus and the spinal cord was detected using IHC experiments. Briefly, 8- to 12-week-old male mice were deeply anesthetized with isoflurane and transcardially perfused with a PBS and subsequently with a 4% paraformaldehyde (163–20145, FUJIFILM Wako).
Brains were obtained from the AgrptdTomato, AgrptdTomato/Girk1KO, and AgrptdTomato/Girk2KO mice and submerged in cold 4% paraformaldehyde for 24 h (post-fixation). Brains were then transferred to a series of sucrose solutions (4 h in 10% sucrose, 12 h in 20% sucrose, and 24 h in 30% sucrose) at 4°C. Brain slices with 15 μm thickness were obtained using a cryostat (Leica) and were prepared for incubation with primary antibodies. Collection of brain slices were performed as described in the methods for in situ hybridization experiments, except that brain slices in 1 well are spaced by 300 μm. Brain slices were mounted on glass slides (Superfrost Plus Microscope Slides, Thermo Fisher) and were treated with PBS containing 10% BSA and 0.3% Triton X-100 (blocking solution). The slices were subsequently incubated with anti-Fos (1:2,000, ab190289, Abcam) antibodies overnight at 4°C. Brain slices were then washed 3 times with PBS (10 min each) and were incubated with Alexa Fluor 647 donkey anti-rabbit secondary antibodies (Thermo Fisher Scientific) for 1 h at room temperature (RT) for immunofluorescence detection. The slices were incubated with DAPI for 10 min, washed 3 times with PBS (10 min each), and cover-slipped using fluorescence mounting medium (DAKO). Images were obtained with a confocal microscope (LSM 780, Carl Zeiss) and were analyzed with ZEN lite (ZEN Microscopy software) and Image J software.
Thoracic spinal cords obtained from GIRK2WT and GIRK2AgRP-KO mice were submerged in cold 4% paraformaldehyde for 12 h (post-fixation). Spinal cords were then transferred to 30% sucrose solutions for 24 h at 4°C. Spinal cord slices with 40 μm thickness were obtained using a cryostat (Leica). We collected coronal sections beginning from the cervical enlargements to preserve T1 level. Spinal cord slices were transferred to 24-well plates, placing 5 consecutive slices in 1 well. Since thoracic spinal cord (T1-L1) is about 18.2 mm in length [46], we needed 4 plates for 1 mouse spinal cord. We randomly selected 1 slice from 1 well, which allows average spacing of 200 μm between slices. Selected slices were washed 3 times with PBS (10 min each) to be prepared for incubation with primary antibodies. The slices were mounted on adhesive microscope slides (TruBond 380, Electron Microscopy Science) and were washed 3 times with PBS (10 min each). The slices then underwent heat-induced epitope retrieval at 60°C for 30 min and were treated with PBS containing 5% normal donkey serum (NDS) and 0.3% Triton X-100 (blocking solution). Subsequently, the slices were treated with anti-ChAT (1:100, AB144P, Sigma) antibody diluted with the blocking solution overnight at 4°C, which was followed by 1 h treatment with Alexa Fluor 488 donkey anti-goat secondary antibodies (Thermo Fisher Scientific) at RT. After that, the slices were treated with the blocking solution for 1 h, and then with anti-Fos (1:500, ab190289, Abcam) antibodies overnight at 4°C. The slices were then treated with Alexa Fluor 647 donkey anti-rabbit secondary antibodies (Thermo Fisher Scientific) for 1 h at RT. The slices were incubated with DAPI for 10 min and were washed 3 times with PBS (10 min each). Then, the slices were cover-slipped using fluorescence mounting medium (DAKO). Images were obtained with a confocal microscope LSM 780 (Carl Zeiss) and were analyzed with ZEN lite (ZEN Microscopy software) and Image J software. We examined the images to look for red fluorescence-expressing neurons in the IML since the first slice with positive fluorescence is T1. We proceeded starting from that slice to determine the level of each spinal cord section. We selected 48 spinal cord slices (approximately 9.6 mm in length, T1-T6, from each mouse) to be included for analyses.
UCP1 in the BAT was detected using IHC experiments. Briefly, 22- to 23-week-old male mice were deeply anesthetized with isoflurane. BAT was obtained from GIRK2WT and GIRK2AgRP-KO mice and was immediately submerged into formalin (HT501128, Sigma) for at least 24 h. Sections of BAT were treated with recombinant anti-UCP1 antibody (1:1,000, ab234430, Abcam), which was followed by incubation with horseradish peroxidase (HRP) secondary antibodies (Envision kit HRP, DAKO). Images were obtained with a slide scanner (Axio Scan. Z1, Carl Zeiss).
Body weight and food intake
Body weight and food intake were measured from male mice as indicated in results. Each conditional knockout mouse (GIRK2AgRP-KO) had its littermate control mouse (GIRK2WT), and 21- to 22-week-old male mice were fasted for 18 h for fast-refeeding experiments. Refeeding started at 10 AM. For some experiments, 10-week-old male mice were i.p. injected with saline or ghrelin (0.4 mg/kg) in fed state (10 AM).
Energy expenditure, physical activity, and body composition
Energy expenditure and physical activity were measured from 20-week-old male mice by an indirect calorimetric chamber (CLAMS 12; Columbus Instruments, Columbus, Ohio, USA). After acclimation for 2 days, O2 consumption (VO2) and CO2 production (VCO2) were measured for 2 days to determine the energy expenditure. Simultaneously, physical activity was determined using a multidimensional infrared light beam system with beams installed on bottom and top levels of cage. Ambulatory movement was defined as breaks of any 2 different light beams at bottom level of cage, while rearing was recorded once the mouse broke any light beam at the top level. Body composition was measured by Time Domain (TD) NMR spectrometer (Minispec LF50, Bruker biospin, Rheinstetten, Germany). For some experiments, we exposed mice (10 weeks old) to cold environment by changing the temperature to 5°C from 25°C, which occurred at 10:30 AM. The mice were allowed to acclimate for 2 to 3 days at 25°C before the transition.
Open field test
Single-housed male mice (21 to 22 weeks old) with access to water and food were adapted for 30 min to a custom-made chamber (40 cm × 40 cm × 40 cm) in a ventilated soundproof booth. A camera was installed on the ceiling of the soundproof booths, and mice were allowed to move freely within the chamber for another 30 min. Light intensity was 120 to 140 lux. Light cycle experiment (3:00 to 4:00 PM) and dark cycle experiment (8:30 to 9:30 PM) were performed 4 days apart. A square-shaped area (20 cm × 20 cm) in the center was defined as the center zone, and the remaining area was defined as the outer zone. Locomotion was analyzed by EthoVision XT 15 (Noldus Wageningen, the Netherlands).
Tissue staining
After the end of metabolic and behavioral experiments, all mice (22 to 23 weeks old) were deeply anesthetized with isoflurane and killed to harvest tissues. Liver, brown fat, epididymal fat, and inguinal fat tissues were isolated and fixed in neutralized formaldehyde solution (HT501128, Sigma). Paraffin-embedded tissue sections were stained with HE, or oil red O. Images were obtained with a slide scanner (Axio Scan. Z1, Carl Zeiss).
Drugs
Tertiapin-Q (STT-170, Alomone Labs), CGP54626 (1088, Tocris), baclofen (0417, Tocris), linopirdine (1999, Tocris), XE991 (2000, Tocris), PK-THPP (5338, Tocris), spadin (5594, Tocris), and tolbutamide (T0891, dissolution with alcohol, Sigma) were used in whole-cell patch clamp mode. Kynurenic acid (K3375, Sigma), picrotoxin (1128, Tocris), and tetrodotoxin (T-550, Alomone Labs) were used to block synaptic currents. All solutions used in this study were made according to manufacturer’s specifications, and stock solutions of all drugs were dissolved in autoclaved de-ionized water unless specifically stated.
Data analysis
Statistical analysis was done using GraphPad Prism 7 (GraphPad Software). Statistical data are expressed as mean ± SEM, where n represents the number of cells or mice studied. The significance of differences between groups was evaluated using two-tailed paired or unpaired Student’s t test, with a confidence level of p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). For some analyses, we used ordinary one-way ANOVA (to compare values between 3 experimental groups) or two-way repeated measures ANOVA (to compare time-dependent changes of values between groups) with a confidence level of p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), or p < 0.0001 (****). We used the Bonferroni correction for all post hoc tests of ANOVA.
Supporting information
Related to Fig 1. Bar graphs and dots summarize action potential (AP) frequency (A), RMP (B), input resistance (C), and AP threshold (D) of cells depolarized by 100 nM, 300 nM, or 500 nM tertiapin-Q (Depol) vs. cells that did not respond (No Response). (A) AP frequency was 1.9 ± 0.5 Hz (n = 15) and 3.5 ± 0.3 Hz (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 2.798, p = 0.009). (B) RMP was −47.4 ± 2.2 mV (n = 15) and −41.2 ± 0.7 mV (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 3.045, p = 0.005). (C) Input resistance was 2.39 ± 0.22 GΩ (n = 15) and 2.89 ± 0.19 GΩ (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 1.739, p = 0.091). (D) AP threshold was −30.7 ± 0.7 mV (n = 12) and −28.3 ± 0.4 mV (n = 20) in “Depol” and “No Response” cells, respectively (df = 30, t = 3.128, p = 0.004). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. **p < 0.01. The numerical data for S1A–S1D Fig can be found in S1 Data.
(TIF)
Related to Fig 1. (A) Trace demonstrates depolarizing effects of linopirdine and XE991, M channels blockers. (B) Trace demonstrates no effects of PK-THPP, a TASK-3 channel blocker. (C) Trace demonstrates no effects of spadin, a TREK-1 channel blocker. (D) Trace demonstrates no effects of tolbutamide, a KATP channel blocker. (E–H) Bar graphs and dots summarize effects on RMP change of linopirdine and XE991 (from −40.4 ± 0.7 mV to −39.5 ± 0.7 mV, n = 12, df = 11, t = 1.650, p = 0.127) (E), PK-THPP (from −42.5 ± 1.0 mV to −42.1 ± 0.8 mV, n = 12, df = 11, t = 0.890, p = 0.393) (F), spadin (from −41.9 ± 1.1 mV to −42.3 ± 1.0 mV, n = 13, df = 12, t = 1.866, p = 0.087) (G), and tolbutamide (from −42.2 ± 0.7 mV to −41.7 ± 0.8 mV, n = 13,df = 12, t = 1.879, and p = 0.085) (H). Red and black lines indicate changes of membrane potential in depolarized and nonresponsive neurons, respectively. Data are presented as mean ± SEM. Paired t test was used for statistical analyses. ns = not significant. The numerical data for S2E–S2H Fig can be found in S1 Data.
(TIF)
Related to Fig 2. (A) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk2. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk2 (magenta): Girk2-containing Agrp (+) neurons; Girk1 and Girk2 (gray): Agrp (+) neurons containing both Girk1 and Girk2. n = 3. (B) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk3. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk3 (cyan): Girk3-containing Agrp (+) neurons; Girk1 and Girk3 (gray): Agrp (+) neurons containing both Girk1 and Girk3. n = 3. (C) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk4. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk4 (orange): Girk4-containing Agrp (+) neurons; Girk1 and Girk4 (gray): Agrp (+) neurons containing both Girk1 and Girk4. n = 3. (D) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk2 and/or Girk3. Girk2 (magenta): Girk2-containing Agrp (+) neurons; Girk3 (cyan): Girk3-containing Agrp (+) neurons; and Girk2 and Girk3 (gray): Agrp (+) neurons containing both Girk2 and Girk3. n = 3. Data are presented as mean ± SEM. Twelve hypothalamic slices from each mouse (from bregma −1.58 mm to −2.02 mm) were included for analyses. See text for specific values. The numerical data for S3A–S3D Fig can be found in S2 Data.
(TIF)
Related to Fig 3. (A) Image demonstrates outward currents by local application of 100 μM baclofen. Voltage ramp pulses (from −120 mV to −10 mV, 100 mV/s) were applied as indicated by arrows, a and b, to obtain current responses, Ia and Ib. (B) Image demonstrates current–voltage (I-V) relationship of baclofen-activated currents (IBac); IBac was calculated by subtracting current responses (Ib- Ia) obtained in (A). (C) Rectification index was calculated by obtaining the ratio of amplitudes at −120 mV (I-120 mV) and −60 mV (I-60 mV) in 12 NPY neurons. (D, E) Images demonstrate IBac recorded from NPYG2WT (black) and NPYG2KO (red) neurons using 10 μM (D) or 100 μM (E) baclofen. (F, G) Image summarizes normalized amplitudes of IBac recorded from NPYG2WT (black) and NPYG2KO (red) neurons using 10 μM baclofen (1.4 ± 0.1 pA/pF, n = 32, for NPYG2WT and 1.4 ± 0.1 pA/pF, n = 23, for NPYG2KO, df = 53, t = 0.276, p = 0.783) (F) and 100 μM baclofen (1.8 ± 0.1 pA/pF, n = 53, for NPYG2WT and 1.8 ± 0.2 pA/pF, n = 26, for NPYG2KO, df = 77, t = 0.021, and p = 0.984) (G). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S4C, S4F, and S4G Fig can be found in S3 Data.
(TIF)
Related to Fig 3. (A, B) Lines and dots summarize effects of tertiapin-Q (300 nM) on RMP (from −44.8 ± 1.8 mV to −44.4 ± 1.7 mV, n = 13, df = 12, t = 0.856, p = 0.409) (A) and input resistance (from 2.85 ± 0.24 GΩ to 2.80 ± 0.30 GΩ, n = 13, df = 12, t = 0.299, p = 0.770) (B) of NPYG2KO neurons. Red and black lines indicate changes of membrane potential or input resistance in depolarized and nonresponsive neurons, respectively. (C) Bar graphs and dots summarize changes of membrane potentials by tertiapin-Q (300 nM) in NPYG2WT neurons and NPYG2KO neurons (2.8 ± 1.0 mV, n = 11, for NPYG2WT and 0.4 ± 0.4 mV, n = 13, for NPYG2KO, df = 22, t = 2.354, p = 0.028). Data are presented as mean ± SEM. Paired t test (A and B) and unpaired t test (C) were used for statistical analyses. *p < 0.05, ns = not significant. The numerical data for S5A–S5C Fig can be found in S3 Data.
(TIF)
Related to Fig 3. (A) Image demonstrates no effects of CGP54626 on NPYG2WT neurons. Dotted line indicates RMP. (B) Lines and dots summarize effects of CGP54626 on RMP (from −42.9 ± 0.8 mV to −43.2 ± 0.8 mV, n = 12, df = 11, t = 2.191, p = 0.051). (C) Lines and dots summarize effect of CGP54626 on input resistance (from 2.68 ± 0.20 GΩ to 2.71 ± 0.21 GΩ, n = 12, df = 11, t = 0.519, p = 0.614). Paired t test was used for statistical analyses. ns = not significant. The numerical data for S6B and S6C Fig can be found in S3 Data.
(TIF)
Related to Fig 4. (A, B) Left panels demonstrate mRNA of Agrp (magenta) and Girk1 (cyan) detected by in situ hybridization (ISH) experiments within the arcuate nucleus of GIRK1WT (A) and GIRK1AgRP-KO mice (B). 3V = third ventricle. Scale bar = 50 μm. Right panels of (A) and (B) demonstrate magnified images of black rectangular area in the left panels of (A) and (B). Girk1 (+) Agrp-expressing neurons are marked by red arrowheads, and Girk1 (-) Agrp-expressing neurons are marked by black arrowheads. Scale bar = 10 μm. (C, D) Bar graphs and dots summarize the proportion of Girk1-expressing AgRP neurons (25.5 ± 2.8%, n = 3, for GIRK1WT and 6.7 ± 2.0%, n = 3, for GIRK1AgRP-KO, df = 4, t = 5.489, p = 0.005) (C) and Girk2-expressing AgRP neurons (46.2 ± 4.3%, n = 3, for GIRK1WT and 47.6 ± 3.0%, n = 3, for GIRK1AgRP-KO, df = 4, t = 0.248, p = 0.817) (D) in GIRK1WT (n = 3) and GIRK1AgRP-KO (n = 3) mice. (E, F) Left panels demonstrate mRNA of Agrp (magenta) and Girk2 (cyan) detected by ISH experiments within the arcuate nucleus of GIRK2WT (E) and GIRK2AgRP-KO mice (F). 3V = third ventricle. Scale bar = 50 μm. Right panels of (E) and (F) demonstrate magnified images of black rectangular area in the left panels of (E) and (F). Girk2 (+) Agrp-expressing neurons are marked by red arrowheads, and Girk2 (-) Agrp-expressing neurons are marked by black arrowheads. Scale bar = 10 μm. (G, H) Bar graphs and dots summarize the proportion of Girk2-expressing AgRP neurons (57.1 ± 3.8%, n = 3, for GIRK2WT and 13.2 ± 2.1%, n = 3, for GIRK2AgRP-KO, df = 4, t = 10.08, p = 0.0005) (G) and Girk1-expressing AgRP neurons (32.0 ± 5.1%, n = 3, for GIRK2WT and 25.7 ± 4.1%, n = 3, for GIRK2AgRP-KO, df = 4, t = 0.972, p = 0.386) (H) in GIRK2WT (n = 3) and GIRK2AgRP-KO (n = 3) mice. A total of 16 hypothalamic slices from each mouse (from bregma −1.46 mm to −2.06 mm) were included for analyses. Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. **p < 0.01, ***p < 0.001, ns = not significant. The numerical data for S7C, S7D, S7G, and S7H Fig can be found in S4 Data.
(TIF)
Related to Fig 4. (A, B) Images demonstrate Girk1 mRNA (cyan) detected by ISH experiments in the hippocampus of GIRK1WT (A) and GIRK1AgRP-KO (B) mice. Scale bar = 200 μm. (C) Bar graphs and dots summarize normalized mRNA levels of Girk1 by qRT-PCR of hippocampus in GIRK1WT mice (WT, n = 5) and GIRK1AgRP-KO mice (KO, n = 6) (1.01 ± 0.06, n = 5, for GIRK1WT and 1.04 ± 0.08, n = 6, for GIRK1AgRP-KO, df = 9, t = 0.279, p = 0.787 in left graph; 1.00 ± 0.03, n = 5, for GIRK1WT and 0.89 ± 0.15, n = 6, for GIRK1AgRP-KO, df = 9, t = 0.682, p = 0.513 in right graph). (D, E) Images demonstrate Girk2 mRNA (cyan) detected by ISH experiments in the hippocampus of GIRK2WT (D) and GIRK2AgRP-KO (E) mice. Scale bar = 200 μm. (F) Bar graphs and dots summarize normalized mRNA levels of Girk2 by qRT-PCR of hippocampus in GIRK2WT mice (WT, n = 5) and GIRK2AgRP-KO mice (KO, n = 6) (1.02 ± 0.11, n = 5, for GIRK2WT and 1.05 ± 0.09, n = 6, for GIRK2AgRP-KO, df = 9, t = 0.190, p = 0.854 in left graph; 1.00 ± 0.04, n = 5, for GIRK2WT and 0.87 ± 0.15, n = 6, for GIRK2AgRP-KO, df = 9, t = 0.819, and p = 0.434 in right graph). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S8C and S8F Fig can be found in S4 Data.
(TIF)
Related to Fig 6. (A) Bar graphs and dots summarize ambulatory movement of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice (260.8 ± 48.5 counts, n = 14, for GIRK2WT and 256.9 ± 48.8 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.057, p = 0.955 in dark cycle; 58.0 ± 10.3 counts, n = 14, for GIRK2WT and 51.6 ± 8.1 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.4921, p = 0.627 in light cycle). (B) Bar graphs and dots summarize rearing activity of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice (148.3 ± 27.1 counts, n = 14, for GIRK2WT and 146.3 ± 32.0 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.049, p = 0.962 in dark cycle; 26.7 ± 9.6 counts, n = 14, for GIRK2WT and 18.7 ± 3.9 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.804, p = 0.428 in light cycle). (C) Trajectory of freely moving GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice in the OFT chamber in dark and light cycles. (D) Bar graphs and dots summarize total moving distance of GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice (95.1 ± 9.0 m, n = 8, for GIRK2WT and 108.1 ± 4.1 m, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.370, p = 0.191 in dark cycle; 113.8 ± 6.6 m, n = 8, for GIRK2WT and 123.9 ± 7.8 m, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.980, p = 0.343 in light cycle). (E) Image demonstrates a view of chamber by a camera that is installed on the ceiling of sound-proof booths. (F) Heat-maps demonstrate zone preference of GIRK2WT and GIRK2AgRP-KO mice in the chamber. (G) Bar graphs and dots summarize proportions of duration in center and outer zones of GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice (10.6 ± 1.8%, n = 8, for GIRK2WT and 8.4 ± 0.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.224, p = 0.240 in dark cycle and center; 13.4 ± 1.4%, n = 8, for GIRK2WT and 12.3 ± 1.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.523, p = 0.609 in light cycle and center; 89.4 ± 1.8%, n = 8, for GIRK2WT and 91.6 ± 0.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.224, p = 0.240 in dark cycle and outer; 86.6 ± 1.4%, n = 8, for GIRK2WT and 87.8 ± 1.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.523, p = 0.609 in light cycle and outer). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S9A, S9B, S9D, and S9G Fig can be found in S6 Data.
(TIF)
Related to Fig 7. Graph demonstrates food intake of GIRK2WT mice (black, n = 4) and GIRK2AgRP-KO mice (red, n = 4) after i.p. injections of saline (filled circles) or ghrelin (0.4 mg/kg, empty circles). Mice were injected at 10 AM and food intake was measured for the next 4 h. Data are presented as mean ± SEM. Two-way repeated measures ANOVA with Bonferroni correction was used for statistical analyses. Group (df = 3, F3, 12 = 12.03, p = 0.0006), time (df = 4, F4, 48 = 23.99, p < 0.0001), interaction (df = 12, F12, 48 = 4.83, p < 0.0001). *p < 0.05; **, ##p < 0.01; ****, ####p < 0.0001. ns = not significant. Saline, GIRK2WT vs. Ghrelin, GIRK2WT (*). Saline, GIRK2AgRP-KO vs. Ghrelin, GIRK2AgRP-KO (#). Saline, GIRK2WT vs. Saline, GIRK2AgRP-KO (ns). Ghrelin, GIRK2WT vs. Ghrelin, GIRK2AgRP-KO (ns). The numerical data for S10 Fig can be found in S7 Data.
(TIF)
Each tab includes data for individual panels of Figs 1 and S1 and S2.
(XLSX)
Each tab includes data for individual panels of Figs 2 and S3.
(XLSX)
Each tab includes data for individual panels of Figs 3 and S4–S6.
(XLSX)
Each tab includes data for individual panels of Figs 4 and S7 and S8.
(XLSX)
Each tab includes data for individual panels of Fig 5.
(XLSX)
Each tab includes data for individual panels of Figs 6 and S9.
(XLSX)
Each tab includes data for individual panels of Figs 7 and S10.
(XLSX)
Numbers represent individual numerical data for changes of membrane potential and reversal potential in Table 1.
(XLSX)
Acknowledgments
We thank staff members of core facilities at KAIST for their help with our experiments. We also thank all members of the Sohn lab for the constructive discussion and comments on this work.
Abbreviations
- AgRP
agouti-related peptide
- AP
action potential
- ARH
arcuate nucleus of the hypothalamus
- BAT
brown adipose tissue
- cDNA
complimentary DNA
- ChAT
choline acetyltransferase
- EE
energy expenditure
- FISH
fluorescence in situ hybridization
- GIRK
G protein-gated inwardly rectifying K+
- HE
hematoxylin and eosin
- HRP
horseradish peroxidase
- IACUC
Institutional Animal Care and Use Committee
- IGW
inguinal white
- IHC
immunohistochemistry
- IML
intermediolateral column; i.p., intraperitoneally
- ISH
in situ hybridization
- KAIST
Korea Advanced Institute of Science and Technology
- NCD
normal chow diet
- NDS
normal donkey serum
- NMR
nuclear magnetic resonance
- NPY
neuropeptide Y
- OFT
open field test
- PCR
polymerase chain reaction
- PGW
perigonadal white
- POMC
pro-opiomelanocortin
- RMP
resting membrane potential
- RT
room temperature
- TTX
tetrodotoxin
- UCP-1
uncoupling protein-1
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the national research foundation of Korea (NRF-2019R1A2C2005161 and NRF-2022R1A2C3005613 to J.-W.S.) and from the US National Institutes of Health (DA034696 and AA027544 to K.W.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Waterson MJ, Horvath TL. Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metab. 2015;22(6):962–970. doi: 10.1016/j.cmet.2015.09.026 . [DOI] [PubMed] [Google Scholar]
- 2.Erickson JC, Clegg KE, Palmiter RD. Sensitivity to leptin and susceptibility to seizures of mice lacking neuropeptide Y. Nature. 1996;381(6581):415–421. doi: 10.1038/381415a0 . [DOI] [PubMed] [Google Scholar]
- 3.Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, et al. Agouti-related peptide–expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8(10):1289–1291. doi: 10.1038/nn1548 . [DOI] [PubMed] [Google Scholar]
- 4.Billington CJ, Briggs JE, Grace M, Levine AS. Effects of intracerebroventricular injection of neuropeptide Y on energy metabolism. Am J Physiol. 1991;260(2 Pt 2):R321–R327. doi: 10.1152/ajpregu.1991.260.2.R321 . [DOI] [PubMed] [Google Scholar]
- 5.Egawa M, Yoshimatsu H, Bray GA. Neuropeptide Y suppresses sympathetic activity to interscapular brown adipose tissue in rats. Am J Physiol. 1991;260(2 Pt 2):R328–R334. doi: 10.1152/ajpregu.1991.260.2.R328 . [DOI] [PubMed] [Google Scholar]
- 6.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14(3):351–355. doi: 10.1038/nn.2739 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121(4):1424–1428. doi: 10.1172/JCI46229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly Rectifying Potassium Channels: Their Structure, Function, and Physiological Roles. Physiol Rev. 2010;90(1):291–366. doi: 10.1152/physrev.00021.2009 . [DOI] [PubMed] [Google Scholar]
- 9.Zaritsky JJ, Redell JB, Tempel BL, Schwarz TL. The consequences of disrupting cardiac inwardly rectifying K+ current (IK1) as revealed by the targeted deletion of the murine Kir2.1 and Kir2.2 genes. J Physiol. 2001;533(3):697–710. doi: 10.1111/j.1469-7793.2001.t01-1-00697.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rorsman P, Eliasson L, Kanno T, Zhang Q, Gopel S. Electrophysiology of pancreatic β-cells in intact mouse islets of Langerhans. Prog Biophys Mol Biol. 2011;107(2):224–235. doi: 10.1016/j.pbiomolbio.2011.06.009 . [DOI] [PubMed] [Google Scholar]
- 11.Parton LE, Ye CP, Coppari R, Enriori PJ, Choi B, Zhang C-Y, et al. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature. 2007;449(7159):228–232. doi: 10.1038/nature06098 . [DOI] [PubMed] [Google Scholar]
- 12.Park S, Williams KW, Liu C, Sohn J-W. A neural basis for tonic suppression of sodium appetite. Nat Neurosci. 2020;23(3):423–432. doi: 10.1038/s41593-019-0573-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohn J-W, Xu Y, Jones JE, Wickman K, Williams KW, Elmquist JK. Serotonin 2C receptor activates a distinct population of arcuate pro-opiomelanocortin neurons via TRPC channels. Neuron. 2011;71(3):488–497. doi: 10.1016/j.neuron.2011.06.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Johnston D. Constitutively active G-protein-gated inwardly rectifying K+ channels in dendrites of hippocampal CA1 pyramidal neurons. J Neurosci. 2005;25(15):3787–3792. doi: 10.1523/JNEUROSCI.5312-04.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife. 2015;4:e09800. doi: 10.7554/eLife.09800 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrade R, Malenka RC, Nicoll RA. A G protein couples serotonin and GABAB receptors to the same channels in hippocampus. Science. 1986;234(4781):1261–1265. doi: 10.1126/science.2430334 . [DOI] [PubMed] [Google Scholar]
- 17.Kim CS, Johnston D. A1 adenosine receptor-mediated GIRK channels contribute to the resting conductance of CA1 neurons in the dorsal hippocampus. J Neurophysiol. 2015;113(7):2511–2523. doi: 10.1152/jn.00951.2014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn JW, Ho WK. Cellular and systemic mechanisms for glucose sensing and homeostasis. Pflugers Arch. 2020;472(11):1547–1561. doi: 10.1007/s00424-020-02466-2 . [DOI] [PubMed] [Google Scholar]
- 19.Luján R, Fernandez M, de Velasco E, Aguado C, Wickman K. New insights into the therapeutic potential of Girk channels. Trends Neurosci. 2014;37(1):20–29. doi: 10.1016/j.tins.2013.10.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz HG, Ivanova T, Lunn ML, Stoffel M, Slesinger PA, Luscher C. Bi-directional effects of GABA(B) receptor agonists on the mesolimbic dopamine system. Nat Neurosci. 2004;7(2):153–159. doi: 10.1038/nn1181 . [DOI] [PubMed] [Google Scholar]
- 21.Tong Q, Ye C-P, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11(9):998–1000. doi: 10.1038/nn.2167 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez M, de Velasco E, Carlblom N, Xia Z, Wickman K. Suppression of inhibitory G protein signaling in forebrain pyramidal neurons triggers plasticity of glutamatergic neurotransmission in the nucleus accumbens core. Neuropharmacology. 2017;117:33–40. doi: 10.1016/j.neuropharm.2017.01.021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotecki L, Hearing M, McCall NM, Fernandez M, de Velasco E, Pravetoni M, et al. GIRK Channels Modulate Opioid-Induced Motor Activity in a Cell Type- and Subunit-Dependent Manner. J Neurosci. 2015;35(18):7131–7142. doi: 10.1523/JNEUROSCI.5051-14.2015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietrich MO, Zimmer MR, Bober J, Horvath TL. Hypothalamic Agrp Neurons Drive Stereotypic Behaviors beyond Feeding. Cell. 2015;160(6):1222–1232. doi: 10.1016/j.cell.2015.02.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamsi F, Wang C-H, Tseng Y-H. The evolving view of thermogenic adipocytes—ontogeny, niche and function. Nat Rev Endocrinol. 2021;17(12):726–744. doi: 10.1038/s41574-021-00562-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H, Ding X, Cao Y, Wang H, Zeng W. Dense Intra-adipose Sympathetic Arborizations Are Essential for Cold-Induced Beiging of Mouse White Adipose Tissue. Cell Metab. 2017;26(4):686–692. doi: 10.1016/j.cmet.2017.08.016 . [DOI] [PubMed] [Google Scholar]
- 27.Lundberg JM, Rudehill A, Sollevi A. Pharmacological characterization of neuropeptide Y and noradrenaline mechanisms in sympathetic control of pig spleen. Eur J Pharmacol. 1989;163(1):103–113. doi: 10.1016/0014-2999(89)90401-9 . [DOI] [PubMed] [Google Scholar]
- 28.Shi Z, Madden CJ, Brooks VL. Arcuate neuropeptide Y inhibits sympathetic nerve activity via multiple neuropathways. J Clin Invest. 2017;127(7):2868–2880. doi: 10.1172/JCI92008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke LK, Darwish T, Cavanaugh AR, Virtue S, Roth E, Morro J, et al. mTORC1 in AGRP neurons integrates exteroceptive and interoceptive food-related cues in the modulation of adaptive energy expenditure in mice. eLife. 2017;6:e22848. doi: 10.7554/eLife.22848 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan H-B, Dietrich Marcelo O, Liu Z-W, Zimmer Marcelo R, Li M-D, Singh Jay P, et al. O-GlcNAc Transferase Enables AgRP Neurons to Suppress Browning of White Fat. Cell. 2014;159(2):306–317. doi: 10.1016/j.cell.2014.09.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodd GT, Andrews ZB, Simonds SE, Michael NJ, DeVeer M, Brüning JC, et al. A Hypothalamic Phosphatase Switch Coordinates Energy Expenditure with Feeding. Cell Metab. 2017;26(2):375–393. doi: 10.1016/j.cmet.2017.07.013 . [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Bartolome CL, Low CS, Yi X, Chien CH, Wang P, et al. Genetic identification of leptin neural circuits in energy and glucose homeostases. Nature. 2018;556(7702):505–509. doi: 10.1038/s41586-018-0049-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu C, Jiang Z, Xu Y, Cai ZL, Jiang Q, Xu Y, et al. Profound and redundant functions of arcuate neurons in obesity development. Nat Metab. 2020;2(8):763–774. doi: 10.1038/s42255-020-0229-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berglund ED, Vianna CR, Donato J Jr, Kim MH, Chuang JC, Lee CE, et al. Direct leptin action on POMC neurons regulates glucose homeostasis and hepatic insulin sensitivity in mice. J Clin Invest. 2012;122(3):1000–1009. doi: 10.1172/JCI59816 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sohn JW, Williams KW. Functional heterogeneity of arcuate nucleus pro-opiomelanocortin neurons: implications for diverging melanocortin pathways. Mol Neurobiol. 2012;45(2):225–233. doi: 10.1007/s12035-012-8240-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo H, Velasco EMF, Wickman K. Neuronal G protein-gated K+ channels. Am J Physiol Cell Physiol. 2022;323(2):C439–C460. doi: 10.1152/ajpcell.00102.2022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Gameiro-Ros I, Glaaser IW, Slesinger PA. Advances in Targeting GIRK Channels in Disease. Trends Pharmacol Sci. 2021;42(3):203–215. doi: 10.1016/j.tips.2020.12.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310(5748):683–685. doi: 10.1126/science.1115524 . [DOI] [PubMed] [Google Scholar]
- 39.He Y, Shu G, Yang Y, Xu P, Xia Y, Wang C, et al. A Small Potassium Current in AgRP/NPY Neurons Regulates Feeding Behavior and Energy Metabolism. Cell Rep. 2016;17(7):1807–1818. doi: 10.1016/j.celrep.2016.10.044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stincic TL, Bosch MA, Hunker AC, Juarez B, Connors AM, Zweifel LS, et al. CRISPR knockdown of Kcnq3 attenuates the M-current and increases excitability of NPY/AgRP neurons to alter energy balance. Mol Metab. 2021;49:101218. doi: 10.1016/j.molmet.2021.101218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Signorini S, Liao YJ, Duncan SA, Jan LY, Stoffel M. Normal cerebellar development but susceptibility to seizures in mice lacking G protein-coupled, inwardly rectifying K+ channel GIRK2. Proc Natl Acad Sci U S A. 1997;94(3):923–927. doi: 10.1073/pnas.94.3.923 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sohn JW, Oh Y, Kim KW, Lee S, Williams KW, Elmquist JK. Leptin and insulin engage specific PI3K subunits in hypothalamic SF1 neurons. Mol Metab. 2016;5(8):669–679. doi: 10.1016/j.molmet.2016.06.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heigele S, Sultan S, Toni N, Bischofberger J. Bidrectional GABAergic control of action potential firing in newborn hippocampal granule cells. Nat Neurosci. 2016;19(2):263–270. doi: 10.1038/nn.4218 . [DOI] [PubMed] [Google Scholar]
- 44.Park SE, Lee D, Jeong JW, Lee SH, Park SJ, Ryu J, et al. Gut Epithelial Inositol Polyphosphate Multikinase Alleviates Experimental Colitis via Governing Tuft Cell Homeostasis. Cell Mol Gastroenterol Hepatol. 2022;14(6):1235–1256. doi: 10.1016/j.jcmgh.2022.08.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez M, de Velasco E, Hearing M, Xia Z, Victoria NC, Lujan R, et al. Sex differences in GABABR-GIRK signaling in layer 5/6 pyramidal neurons of the mouse prelimbic cortex. Neuropharmacology. 2015;95:353–360. doi: 10.1016/j.neuropharm.2015.03.029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison M, O’Brien A, Adams L, Cowin G, Ruitenberg MJ, Sengul G, et al. Vertebral landmarks for the identification of spinal cord segments in the mouse. Neuroimage. 2013;68:22–29. doi: 10.1016/j.neuroimage.2012.11.048 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Fig 1. Bar graphs and dots summarize action potential (AP) frequency (A), RMP (B), input resistance (C), and AP threshold (D) of cells depolarized by 100 nM, 300 nM, or 500 nM tertiapin-Q (Depol) vs. cells that did not respond (No Response). (A) AP frequency was 1.9 ± 0.5 Hz (n = 15) and 3.5 ± 0.3 Hz (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 2.798, p = 0.009). (B) RMP was −47.4 ± 2.2 mV (n = 15) and −41.2 ± 0.7 mV (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 3.045, p = 0.005). (C) Input resistance was 2.39 ± 0.22 GΩ (n = 15) and 2.89 ± 0.19 GΩ (n = 20) in “Depol” and “No Response” cells, respectively (df = 33, t = 1.739, p = 0.091). (D) AP threshold was −30.7 ± 0.7 mV (n = 12) and −28.3 ± 0.4 mV (n = 20) in “Depol” and “No Response” cells, respectively (df = 30, t = 3.128, p = 0.004). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. **p < 0.01. The numerical data for S1A–S1D Fig can be found in S1 Data.
(TIF)
Related to Fig 1. (A) Trace demonstrates depolarizing effects of linopirdine and XE991, M channels blockers. (B) Trace demonstrates no effects of PK-THPP, a TASK-3 channel blocker. (C) Trace demonstrates no effects of spadin, a TREK-1 channel blocker. (D) Trace demonstrates no effects of tolbutamide, a KATP channel blocker. (E–H) Bar graphs and dots summarize effects on RMP change of linopirdine and XE991 (from −40.4 ± 0.7 mV to −39.5 ± 0.7 mV, n = 12, df = 11, t = 1.650, p = 0.127) (E), PK-THPP (from −42.5 ± 1.0 mV to −42.1 ± 0.8 mV, n = 12, df = 11, t = 0.890, p = 0.393) (F), spadin (from −41.9 ± 1.1 mV to −42.3 ± 1.0 mV, n = 13, df = 12, t = 1.866, p = 0.087) (G), and tolbutamide (from −42.2 ± 0.7 mV to −41.7 ± 0.8 mV, n = 13,df = 12, t = 1.879, and p = 0.085) (H). Red and black lines indicate changes of membrane potential in depolarized and nonresponsive neurons, respectively. Data are presented as mean ± SEM. Paired t test was used for statistical analyses. ns = not significant. The numerical data for S2E–S2H Fig can be found in S1 Data.
(TIF)
Related to Fig 2. (A) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk2. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk2 (magenta): Girk2-containing Agrp (+) neurons; Girk1 and Girk2 (gray): Agrp (+) neurons containing both Girk1 and Girk2. n = 3. (B) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk3. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk3 (cyan): Girk3-containing Agrp (+) neurons; Girk1 and Girk3 (gray): Agrp (+) neurons containing both Girk1 and Girk3. n = 3. (C) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk1 and/or Girk4. Girk1 (green): Girk1-containing Agrp (+) neurons; Girk4 (orange): Girk4-containing Agrp (+) neurons; Girk1 and Girk4 (gray): Agrp (+) neurons containing both Girk1 and Girk4. n = 3. (D) Graph demonstrates percentage of Agrp (+) neurons that express mRNA of Girk2 and/or Girk3. Girk2 (magenta): Girk2-containing Agrp (+) neurons; Girk3 (cyan): Girk3-containing Agrp (+) neurons; and Girk2 and Girk3 (gray): Agrp (+) neurons containing both Girk2 and Girk3. n = 3. Data are presented as mean ± SEM. Twelve hypothalamic slices from each mouse (from bregma −1.58 mm to −2.02 mm) were included for analyses. See text for specific values. The numerical data for S3A–S3D Fig can be found in S2 Data.
(TIF)
Related to Fig 3. (A) Image demonstrates outward currents by local application of 100 μM baclofen. Voltage ramp pulses (from −120 mV to −10 mV, 100 mV/s) were applied as indicated by arrows, a and b, to obtain current responses, Ia and Ib. (B) Image demonstrates current–voltage (I-V) relationship of baclofen-activated currents (IBac); IBac was calculated by subtracting current responses (Ib- Ia) obtained in (A). (C) Rectification index was calculated by obtaining the ratio of amplitudes at −120 mV (I-120 mV) and −60 mV (I-60 mV) in 12 NPY neurons. (D, E) Images demonstrate IBac recorded from NPYG2WT (black) and NPYG2KO (red) neurons using 10 μM (D) or 100 μM (E) baclofen. (F, G) Image summarizes normalized amplitudes of IBac recorded from NPYG2WT (black) and NPYG2KO (red) neurons using 10 μM baclofen (1.4 ± 0.1 pA/pF, n = 32, for NPYG2WT and 1.4 ± 0.1 pA/pF, n = 23, for NPYG2KO, df = 53, t = 0.276, p = 0.783) (F) and 100 μM baclofen (1.8 ± 0.1 pA/pF, n = 53, for NPYG2WT and 1.8 ± 0.2 pA/pF, n = 26, for NPYG2KO, df = 77, t = 0.021, and p = 0.984) (G). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S4C, S4F, and S4G Fig can be found in S3 Data.
(TIF)
Related to Fig 3. (A, B) Lines and dots summarize effects of tertiapin-Q (300 nM) on RMP (from −44.8 ± 1.8 mV to −44.4 ± 1.7 mV, n = 13, df = 12, t = 0.856, p = 0.409) (A) and input resistance (from 2.85 ± 0.24 GΩ to 2.80 ± 0.30 GΩ, n = 13, df = 12, t = 0.299, p = 0.770) (B) of NPYG2KO neurons. Red and black lines indicate changes of membrane potential or input resistance in depolarized and nonresponsive neurons, respectively. (C) Bar graphs and dots summarize changes of membrane potentials by tertiapin-Q (300 nM) in NPYG2WT neurons and NPYG2KO neurons (2.8 ± 1.0 mV, n = 11, for NPYG2WT and 0.4 ± 0.4 mV, n = 13, for NPYG2KO, df = 22, t = 2.354, p = 0.028). Data are presented as mean ± SEM. Paired t test (A and B) and unpaired t test (C) were used for statistical analyses. *p < 0.05, ns = not significant. The numerical data for S5A–S5C Fig can be found in S3 Data.
(TIF)
Related to Fig 3. (A) Image demonstrates no effects of CGP54626 on NPYG2WT neurons. Dotted line indicates RMP. (B) Lines and dots summarize effects of CGP54626 on RMP (from −42.9 ± 0.8 mV to −43.2 ± 0.8 mV, n = 12, df = 11, t = 2.191, p = 0.051). (C) Lines and dots summarize effect of CGP54626 on input resistance (from 2.68 ± 0.20 GΩ to 2.71 ± 0.21 GΩ, n = 12, df = 11, t = 0.519, p = 0.614). Paired t test was used for statistical analyses. ns = not significant. The numerical data for S6B and S6C Fig can be found in S3 Data.
(TIF)
Related to Fig 4. (A, B) Left panels demonstrate mRNA of Agrp (magenta) and Girk1 (cyan) detected by in situ hybridization (ISH) experiments within the arcuate nucleus of GIRK1WT (A) and GIRK1AgRP-KO mice (B). 3V = third ventricle. Scale bar = 50 μm. Right panels of (A) and (B) demonstrate magnified images of black rectangular area in the left panels of (A) and (B). Girk1 (+) Agrp-expressing neurons are marked by red arrowheads, and Girk1 (-) Agrp-expressing neurons are marked by black arrowheads. Scale bar = 10 μm. (C, D) Bar graphs and dots summarize the proportion of Girk1-expressing AgRP neurons (25.5 ± 2.8%, n = 3, for GIRK1WT and 6.7 ± 2.0%, n = 3, for GIRK1AgRP-KO, df = 4, t = 5.489, p = 0.005) (C) and Girk2-expressing AgRP neurons (46.2 ± 4.3%, n = 3, for GIRK1WT and 47.6 ± 3.0%, n = 3, for GIRK1AgRP-KO, df = 4, t = 0.248, p = 0.817) (D) in GIRK1WT (n = 3) and GIRK1AgRP-KO (n = 3) mice. (E, F) Left panels demonstrate mRNA of Agrp (magenta) and Girk2 (cyan) detected by ISH experiments within the arcuate nucleus of GIRK2WT (E) and GIRK2AgRP-KO mice (F). 3V = third ventricle. Scale bar = 50 μm. Right panels of (E) and (F) demonstrate magnified images of black rectangular area in the left panels of (E) and (F). Girk2 (+) Agrp-expressing neurons are marked by red arrowheads, and Girk2 (-) Agrp-expressing neurons are marked by black arrowheads. Scale bar = 10 μm. (G, H) Bar graphs and dots summarize the proportion of Girk2-expressing AgRP neurons (57.1 ± 3.8%, n = 3, for GIRK2WT and 13.2 ± 2.1%, n = 3, for GIRK2AgRP-KO, df = 4, t = 10.08, p = 0.0005) (G) and Girk1-expressing AgRP neurons (32.0 ± 5.1%, n = 3, for GIRK2WT and 25.7 ± 4.1%, n = 3, for GIRK2AgRP-KO, df = 4, t = 0.972, p = 0.386) (H) in GIRK2WT (n = 3) and GIRK2AgRP-KO (n = 3) mice. A total of 16 hypothalamic slices from each mouse (from bregma −1.46 mm to −2.06 mm) were included for analyses. Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. **p < 0.01, ***p < 0.001, ns = not significant. The numerical data for S7C, S7D, S7G, and S7H Fig can be found in S4 Data.
(TIF)
Related to Fig 4. (A, B) Images demonstrate Girk1 mRNA (cyan) detected by ISH experiments in the hippocampus of GIRK1WT (A) and GIRK1AgRP-KO (B) mice. Scale bar = 200 μm. (C) Bar graphs and dots summarize normalized mRNA levels of Girk1 by qRT-PCR of hippocampus in GIRK1WT mice (WT, n = 5) and GIRK1AgRP-KO mice (KO, n = 6) (1.01 ± 0.06, n = 5, for GIRK1WT and 1.04 ± 0.08, n = 6, for GIRK1AgRP-KO, df = 9, t = 0.279, p = 0.787 in left graph; 1.00 ± 0.03, n = 5, for GIRK1WT and 0.89 ± 0.15, n = 6, for GIRK1AgRP-KO, df = 9, t = 0.682, p = 0.513 in right graph). (D, E) Images demonstrate Girk2 mRNA (cyan) detected by ISH experiments in the hippocampus of GIRK2WT (D) and GIRK2AgRP-KO (E) mice. Scale bar = 200 μm. (F) Bar graphs and dots summarize normalized mRNA levels of Girk2 by qRT-PCR of hippocampus in GIRK2WT mice (WT, n = 5) and GIRK2AgRP-KO mice (KO, n = 6) (1.02 ± 0.11, n = 5, for GIRK2WT and 1.05 ± 0.09, n = 6, for GIRK2AgRP-KO, df = 9, t = 0.190, p = 0.854 in left graph; 1.00 ± 0.04, n = 5, for GIRK2WT and 0.87 ± 0.15, n = 6, for GIRK2AgRP-KO, df = 9, t = 0.819, and p = 0.434 in right graph). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S8C and S8F Fig can be found in S4 Data.
(TIF)
Related to Fig 6. (A) Bar graphs and dots summarize ambulatory movement of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice (260.8 ± 48.5 counts, n = 14, for GIRK2WT and 256.9 ± 48.8 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.057, p = 0.955 in dark cycle; 58.0 ± 10.3 counts, n = 14, for GIRK2WT and 51.6 ± 8.1 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.4921, p = 0.627 in light cycle). (B) Bar graphs and dots summarize rearing activity of GIRK2WT (n = 14) and GIRK2AgRP-KO (n = 16) mice (148.3 ± 27.1 counts, n = 14, for GIRK2WT and 146.3 ± 32.0 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.049, p = 0.962 in dark cycle; 26.7 ± 9.6 counts, n = 14, for GIRK2WT and 18.7 ± 3.9 counts, n = 16, for GIRK2AgRP-KO, df = 28, t = 0.804, p = 0.428 in light cycle). (C) Trajectory of freely moving GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice in the OFT chamber in dark and light cycles. (D) Bar graphs and dots summarize total moving distance of GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice (95.1 ± 9.0 m, n = 8, for GIRK2WT and 108.1 ± 4.1 m, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.370, p = 0.191 in dark cycle; 113.8 ± 6.6 m, n = 8, for GIRK2WT and 123.9 ± 7.8 m, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.980, p = 0.343 in light cycle). (E) Image demonstrates a view of chamber by a camera that is installed on the ceiling of sound-proof booths. (F) Heat-maps demonstrate zone preference of GIRK2WT and GIRK2AgRP-KO mice in the chamber. (G) Bar graphs and dots summarize proportions of duration in center and outer zones of GIRK2WT (n = 8) and GIRK2AgRP-KO (n = 9) mice (10.6 ± 1.8%, n = 8, for GIRK2WT and 8.4 ± 0.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.224, p = 0.240 in dark cycle and center; 13.4 ± 1.4%, n = 8, for GIRK2WT and 12.3 ± 1.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.523, p = 0.609 in light cycle and center; 89.4 ± 1.8%, n = 8, for GIRK2WT and 91.6 ± 0.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 1.224, p = 0.240 in dark cycle and outer; 86.6 ± 1.4%, n = 8, for GIRK2WT and 87.8 ± 1.6%, n = 9, for GIRK2AgRP-KO, df = 15, t = 0.523, p = 0.609 in light cycle and outer). Data are presented as mean ± SEM. Unpaired t test was used for statistical analyses. ns = not significant. The numerical data for S9A, S9B, S9D, and S9G Fig can be found in S6 Data.
(TIF)
Related to Fig 7. Graph demonstrates food intake of GIRK2WT mice (black, n = 4) and GIRK2AgRP-KO mice (red, n = 4) after i.p. injections of saline (filled circles) or ghrelin (0.4 mg/kg, empty circles). Mice were injected at 10 AM and food intake was measured for the next 4 h. Data are presented as mean ± SEM. Two-way repeated measures ANOVA with Bonferroni correction was used for statistical analyses. Group (df = 3, F3, 12 = 12.03, p = 0.0006), time (df = 4, F4, 48 = 23.99, p < 0.0001), interaction (df = 12, F12, 48 = 4.83, p < 0.0001). *p < 0.05; **, ##p < 0.01; ****, ####p < 0.0001. ns = not significant. Saline, GIRK2WT vs. Ghrelin, GIRK2WT (*). Saline, GIRK2AgRP-KO vs. Ghrelin, GIRK2AgRP-KO (#). Saline, GIRK2WT vs. Saline, GIRK2AgRP-KO (ns). Ghrelin, GIRK2WT vs. Ghrelin, GIRK2AgRP-KO (ns). The numerical data for S10 Fig can be found in S7 Data.
(TIF)
Each tab includes data for individual panels of Figs 1 and S1 and S2.
(XLSX)
Each tab includes data for individual panels of Figs 2 and S3.
(XLSX)
Each tab includes data for individual panels of Figs 3 and S4–S6.
(XLSX)
Each tab includes data for individual panels of Figs 4 and S7 and S8.
(XLSX)
Each tab includes data for individual panels of Fig 5.
(XLSX)
Each tab includes data for individual panels of Figs 6 and S9.
(XLSX)
Each tab includes data for individual panels of Figs 7 and S10.
(XLSX)
Numbers represent individual numerical data for changes of membrane potential and reversal potential in Table 1.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.