Abstract
TERT reactivation occurs frequently in human malignancies, especially advanced cancers. However, in vivo functions of TERT reactivation in cancer progression and the underlying mechanism are not fully understood. In this study, we expressed TERT and/or active BRAF (BRAF V600E) specifically in mouse thyroid epithelium. While BRAF V600E alone induced papillary thyroid cancer (PTC), coexpression of BRAF V600E and TERT resulted in poorly differentiated thyroid carcinoma (PDTC). Spatial transcriptome analysis revealed that tumors from mice coexpressing BRAF V600E and TERT were highly heterogeneous, and cell dedifferentiation was positively correlated with ribosomal biogenesis. Mechanistically, TERT boosted ribosomal RNA (rRNA) expression and protein synthesis by interacting with multiple proteins involved in ribosomal biogenesis. Furthermore, we found that CX-5461, an rRNA transcription inhibitor, effectively blocked proliferation and induced redifferentiation of thyroid cancer. Thus, TERT promotes thyroid cancer progression by inducing cancer cell dedifferentiation, and ribosome inhibition represents a potential strategy to treat TERT-reactivated cancers.
Evidence shows TERT induces thyroid cancer progression by promoting ribosomal biogenesis, with promising therapeutic implications.
INTRODUCTION
Telomerase activity is detected in more than 90% of cancers (1). TERT (telomerase reverse transcriptase) and TERC (telomeric RNA component) are two main subunits of telomerase, which respectively provide reverse transcriptase activity and RNA template for telomere elongation (2). TERC is expressed in virtually all somatic cells, while TERT is silenced in most terminally differentiated cells and can only be observed in cells with stemness properties (3). TERT expression can be reactivated through gene amplification or, more frequently, promoter-activating mutation in cancer (4, 5). Reactivation of TERT is associated with poor prognosis in multiple malignancies, including thyroid cancer, melanoma, glioma, and pleural mesothelioma (6). Therapies targeting TERT activity are considered suitable for many cancers since TERT reactivation is a common event in multiple cancer types (7, 8). However, severe adverse effects such as bone marrow failure and cardiovascular damage occur upon TERT inhibition, as stem and progenitor cells are also targeted by telomerase inhibitors (9–11). Therefore, it is important to develop novel approaches targeting TERT reactivation for cancer treatment.
Besides its classical role in telomere length regulation, TERT has nonclassical functions independent of telomerase activity. Recent studies have shown that TERT promotes tumorigenesis via cross-talk with nuclear factor κB (NF-κB), Wnt pathway, FOXO3a, or MYC in multiple cancers (12–14). Moreover, TERT stimulates RNA polymerase I and III (Pol I and III) activity and induces transfer RNA (tRNA) synthesis (15, 16), suggesting functions in ribosomal biogenesis. These mechanisms associated with TERT may be molecularly targeted to treat cancers and are likely to produce fewer side effects compared with direct TERT inhibition.
Thus far, most of the TERT studies have used cancer cell lines that were immortalized by either TERT activation or alternative length of telomere (ALT) (17). Several studies using Tert−/− mice conclude that TERT depletion inhibits cancer progression and tissue renewal (18, 19). Unfortunately, a genetically modified mouse model mimicking TERT promoter C228T or C250T mutation is not available. Ectopic expression of TERT in tumors to some extent may mimic gene reactivation. A TERT transgenic mouse line under the control of the keratin promoter has been successfully established and revealed a role of high TERT expression in the initiation of chemical carcinogen–induced skin cancer (20). Nevertheless, it remains a challenge to investigate the role of TERT reactivation on tumor initiation and progression in vivo in different tissues.
In this study, we established a conditional TERT transgenic mouse model and demonstrated that TERT reactivation accelerated dedifferentiation and progression of thyroid cancer driven by constitutively active BRAF (BRAF V600E). Spatial as well as single-cell transcriptomic analysis and molecular characterization indicated that TERT promoted ribosomal RNA (rRNA) metabolism and translation efficiency. Moreover, targeted inhibition of rRNA transcription effectively repressed tumor progression in cancers with TERT reactivation.
RESULTS
TERT reactivation occurs frequently in multiple cancers and predicted a worse prognosis
TERT reactivation is underestimated in cancer genomics studies because the TERT promoter is not covered by whole-exon sequencing. Recently, with the identification of TERT promoter C228T and C250T mutation, the TERT promoter status has been examined in several large-scale pan-cancer sequencing studies. Three large-scale cohorts, MSK-IMPACT, MSK-MET, and China Origimed2020 cohorts, which include the most abundant cancer types and samples, have analyzed the TERT promoter region. Given that TERT reactivation mostly occurs via promoter mutation or gene amplification (19, 20), we scanned both events in these three cohorts (21), as well as TERT amplification in The Cancer Genome Atlas Program (TCGA) cohort (22, 23). We found frequent occurrence of TERT promoter mutations (C228T or C250T) in thyroid cancer, melanoma, bladder cancer, hepatocellular carcinoma, glioma, as well as head and neck cancer, whereas TERT amplification appeared commonly in lung, ovarian, and bladder cancers (Fig. 1A and fig. S1, A and B). Among different cancer types, melanoma and thyroid cancer exhibited high concordance in different cohorts. Furthermore, in Memorial Sloan Kettering Cancer Center (MSKCC) datasets, TERT promoter mutation predicted a worse prognosis in thyroid cancer (21), glioma (22), and bladder cancer (23) (Fig. 1B and fig. S1, C and D).
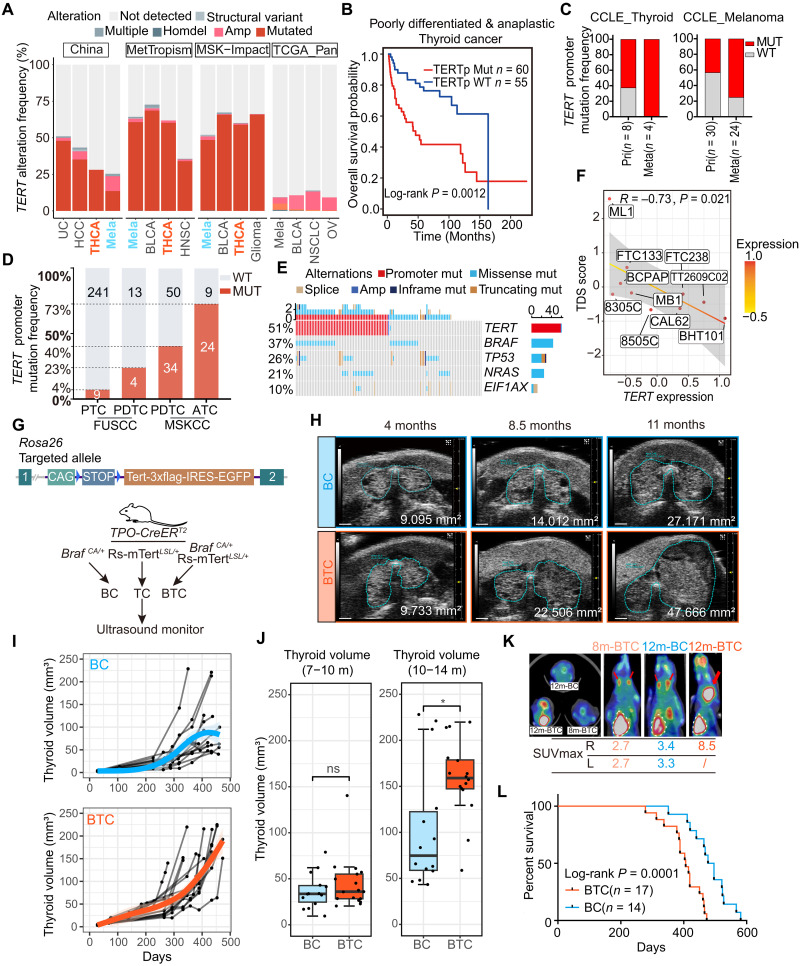
Fig. 1. TERT reactivation predicts a worse prognosis.
(A) Histogram shows TERT alteration frequency in the China Origimed2020, MetTropism, MSK-Impact, and TCGA datasets. UC, urothelial carcinoma; HCC, hepatocellular carcinoma; BLCA, bladder cancer; THCA, thyroid carcinoma; NSCLC, non–small cell lung cancer; OV, ovarian epithelial tumor. (B) Kaplan-Meier curve of PDTC /ATC patients with or without TERT promoter status in MSKCC cohort. (C) TERT promoter mutation frequency of PTC, PDTC, and ATC in Fudan University Shanghai Cancer Center (FUSCC) and MSKCC cohorts. (D) TERT promoter mutation frequency of thyroid cancer and melanoma cell lines derived from primary and metastatic foci in CCLE dataset. (E) Oncoprint shows the co-occurrence of BRAF, TP53, NRAS, and EIF1A with TERT promoter mutation in PDTC and ATC from MSKCC dataset. (F) Correlation between TERT expression and TDS score of thyroid follicular cell lines in CCLE dataset (n = 10). Spearman correlation R = −0.73. (G) Top: Schematic diagram of the constructing strategy of Rs-mTERTLSL/+ mice. Bottom: Experimental flow chart depicting protocol to generate BC, TC, and BTC transgenic mice. Blue triangle, loxP site. (H) Representative serial ultrasound images of BC and BTC thyroid tumors at 4, 8.5, and 9.5 months of age. Scale bar, 1 mm. (I) Growth patterns of BC (n = 14) and BTC (n = 17) thyroid tumors. (J) Thyroid volume (mm3) comparison of BC and BTC thyroid at 7 to 10 months old or 10 to 14 months old. Student’s t test. Boxplot shows the median with interquartile range (IQR) and 1.5 IQR whiskers. *P < 0.05; ns, not significant. (K) 18FDG PET-CT analysis of representative 8m-BTC, 12m-BC, and 12m-BTC mice. Red arrows and white dashed lines indicate thyroid and heart separately. (L) Overall survival of BC (n = 14) and BTC (n = 17) mice.
We also examined 503 cell lines with TERT promoter status defined by whole-genome sequencing and/or targeted sequencing in Cancer Cell Line Encyclopedia (CCLE) dataset (24). Promoter mutation was the dominant alteration for TERT activation in thyroid cancer and melanoma. In both cancer types, cell lines derived from metastasis foci showed a higher frequency of TERT promoter mutation (Fig. 1C), suggesting a potential function of TERT activation in cancer progression and metastasis.
The frequency of TERT promoter mutation (C228 or C250) is associated with the pathological stage and differentiation status of thyroid cancer. In differentiated thyroid cancer, which was associated with favorable survival, TERT promoter mutation occurred at a frequency of 4 to 12% (21, 25). Meanwhile, approximately 50% of poorly differentiated thyroid cancer (PDTC) and anaplastic thyroid cancer (ATC) with poor prognosis harbored TERT promoter mutations (Fig. 1D) (21, 25). Notably, most tumors with TERT promoter mutation also exhibited a mutation in BRAF or NRAS (Fig. 1E) (21). TCGA program has established a thyroid differentiation score (TDS) system to evaluate thyroid cancer differentiation levels using mRNA expression levels of 16 genes (TG, TPO, PAX8, DIO1, DIO2, DUOX1, DUOX2, FOXE1, GLIS3, NKX2-1, SLC26A4, SLC5A5, SLC5A8, THRA, THRB, and TSHR) (26). Expression analysis revealed that TERT expression level was negatively correlated with TDS score in CCLE thyroid cancer cell lines (R = −0.73, Spearman P = 0.021) (Fig. 1F). Together, these results suggest that TERT reactivation, especially promoter mutation, occurs frequently in a variety of cancers and correlates with tumor progression and poor prognosis.
TERT accelerates BRAF V600E–induced thyroid cancer progression
As mentioned above, TERT promoter mutation commonly co-occurs with BRAF or NRAS activating mutations (Fig. 1E). We speculated that instead of initiating carcinogenesis, TERT overexpression may promote cancer progression with existing oncogenic signals. Previous studies showed that TERT induced proliferation, invasion, and many other tumor-like behaviors in vitro. In SV-40–immortalized thyroid cell line NTHY-ori 3-1, which harbors no BRAF or TERT promoter mutation, overexpression of BRAF V600E and TERT collaboratively increased cell migration (fig. S2, A to D). Unexpectedly, in vitro up-regulation of BRAF V600E and TERT did not induce cell proliferation (fig. S2E). On the contrary, the knockdown of TERT in TERT promoter mutant cancer cells inhibited cell proliferation and induced senescence (fig. S2, F to H).
To elucidate the underlying mechanism between TERT reactivation and tumor progression in vivo, we generated a conditional TERT transgenic mouse model by inserting the Loxp-stop-loxp-Tert-3xFlag-IRES-EGFP sequence into the Rosa26 allele (Rs-mTERTLSL/+ mice; Fig. 1G). Mice were then crossed with TPO-CreERT2 mice to generate a thyroid-specific Tert transgenic mouse model (TC mice), which mimicked high TERT expression in tumors. TERT overexpression alone could not induce tumor formation (fig. S3, A to D) (20). We then adopted a previously used thyroid cancer mice model, which was induced by the expression of BRAF V600E in thyroid follicular cells (BC mice), and crossed with Rs-mTERTLSL/+ mice to generate BTC mice (Fig. 1G).
Follow-up ultrasound indicated no notable difference in the growth patterns of BC and BTC thyroid tumors during the first 7 to 10 months after induction (Fig. 1, H to J, and fig. S3, E and F). However, after 10 to 14 months, thyroid tumors from BTC mice grew much faster than those from BC mice (Fig. 1, H to J, and fig. S3, E and F). Tumor progression in BTC and BC mice was also analyzed by 18F-FDG positron emission tomography–computed tomography (PET-CT). The maximum standardized uptake value (SUVmax) of a 12-month BTC thyroid tumor was much higher than those of 12-month BC and 8-month BTC mice, while 12-month BC and 8-month BTC mice had similar FDG SUVmax (Fig. 1K and fig. S3G). In line with the above results, the BTC group showed a worse prognosis with a median survival of 406 days versus 486.5 days of the BC group (Fig. 1L). Together, these results indicate a more rapid tumor progression when both BRAF and TERT are activated.
TERT promotes dedifferentiation of BRAF V600E–induced thyroid cancer
Compared with BC mice, BTC mice displayed accelerated tumor growth and worse prognosis (Fig. 1, J and L). We then characterized the morphological and molecular changes of tumors at different stages. The ultrasound examination, along with the gross appearance following dissection, and hematoxylin and eosin (H&E) pathology, showed that most of the BTC tumors progressed into PDTC with larger sizes in 10 to 14 months (15 of 17, 88.2%). In stark contrast, most BC thyroid tumors remained papillary thyroid cancers (PTCs), despite the progression of tumors from some old BC mice (16 to 18 months) into PDTC (5 of 14, 35.7%) (Fig. 2A and fig. S4, A and B). Immunohistochemistry (IHC) revealed that tumors from BC mice had high expression of thyroid cancer markers, including thyroglobulin (TG), thyroid transcription factor-1 (TTF-1), paired box 8 (PAX8), sodium iodide symporter (NIS; Slc5a5), and CK19. These markers have been widely used in clinic for thyroid cancer identification (27, 28). However, the expression of TG and TTF-1 in BC tumors was lower than in neighboring normal follicular cells (Fig. 2A). On the other hand, tumors from BTC mice expressed low levels of TG, TTF-1, NIS, and PAX8, and high levels of Ki67, indicative of hyperproliferation and dedifferentiation (Fig. 2A and fig. S4B). Tumors from BTC mice originated from thyroid follicular cells, as noted by the expression of ectopic Flag-TERT (Fig. 2A).
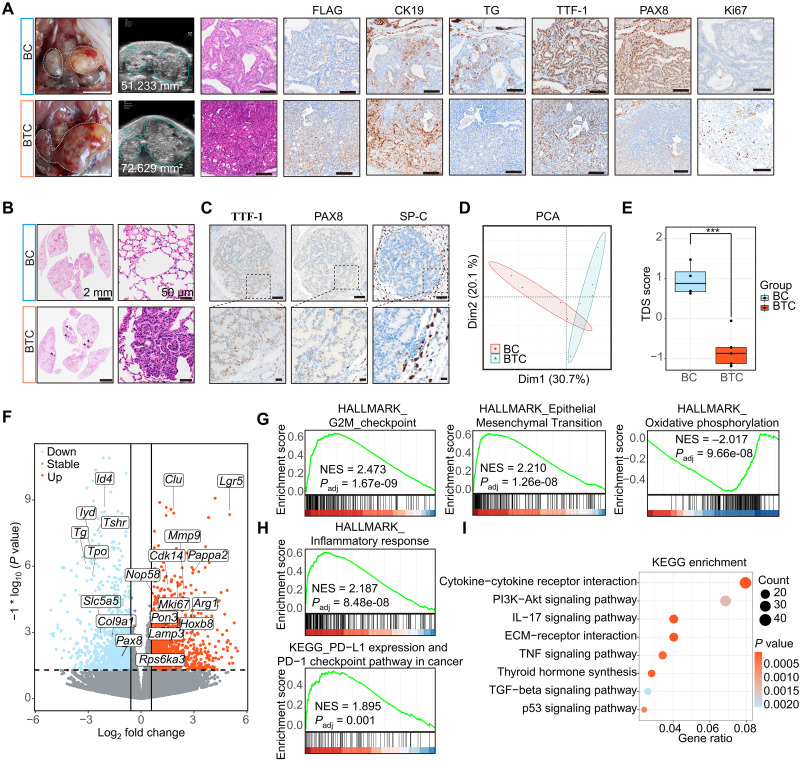
Fig. 2. TERT promotes dedifferentiation of BRAF mutant–induced thyroid cancer.
(A) Photographs of the whole thyroid (white dashed line, thyroid gross appearance; scale bar, 5 mm) and ultrasound imaging (scale bar, 2 mm). H&E and Flag, CK19, TG, TTF1, PAX8, and Ki67 IHC results (scale bar, 100 μm) of BC and BTC mice thyroid. (B) H&E staining of BC and BTC lung sections [scale bars, 2 mm (left) and 50 μm (right)]. (C) TTF-1, PAX8, and SP-C IHC results of BTC mice lung metastasis foci [scale bars, 100 μm (top) and 20 μm (bottom)]. (D) PCA result of BC (n = 4) and BTC (n = 5) transcriptome data. (E) Boxplot shows TDS scores of BC and BTC. Median with IQR and 1.5 IQR whiskers. Student’s t test, ***P < 0.001. (F) Volcano plot indicates up-regulated and down-regulated genes in BTC and BC mice. (G and H) GSEA results show that BTC mice positively enriched in G2-M checkpoint, epithelial-mesenchymal transition, the inflammatory response (HALLMARK), and PD-L1 and PD-1 checkpoint pathway in cancer (KEGG), and negatively enriched in oxidative phosphorylation of hallmark. NES, normalized enrichment score. (I) Dot plot shows KEGG pathway enrichment of differentially expressed genes between BTC and BC mice.
Advanced thyroid cancer frequently metastasizes to distant organs, including the lung (29). We observed widespread metastasis foci inside the BTC lung (Fig. 2B). Cells in metastasis foci were negative for SP-C (a marker of AT2 lung cells) and TG, positive for TTF-1, and weakly positive for PAX8 and NIS, indicating a thyroid origin of these metastasized cells (Fig. 2C and fig. S4C).
The transcriptomes of BC and BTC tumors were analyzed to identify potential mechanisms underlying cell dedifferentiation and tumor metastasis. Tumors from BC and BTC mice (n = 4 and 5, respectively) were collected and subjected to RNA sequencing (RNA-seq). Principal components analysis (PCA) indicated that BTC and BC thyroid tumors showed notably different gene expression profiles (Fig. 2D). Global assessment displayed a much lower TDS score for BTC tumors (Fig. 2E and fig. S4D). In BTC tumors, thyroid-specific genes such as Tg, Pax8, Tshr, and Tpo were down-regulated, whereas multiple tumor genes such as Lgr5, Clu, Mmp9, Pappa2, Mki67, and Arg1 were up-regulated (Fig. 2F). We were able to isolate and immortalize primary tumor cells from BTC tumor (312BTC). On the other hand, cells from BC thyroid tumors developed senescence rapidly after several passages (fig. S4E). These results are in agreement with IHC analysis, indicating a hyperproliferative and dedifferentiation phenotype associated with tumors from BTC mice.
To identify key cancer pathways associated with the progression from PTC to PDTC in BTC mice, gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) for hallmark gene sets were performed (30). Several cancer hallmarks, such as E2F targets, G2-M checkpoints, epithelial-mesenchymal transition, inflammatory response, programmed cell death 1 ligand 1 (PD-L1) expression, and programmed cell death protein 1 (PD-1) checkpoint pathway, were activated, whereas oxidative phosphorylation and fatty acid metabolism were down-regulated in BTC tumors (Fig. 2, G and H, and fig. S4F). In addition, Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of differentially expressed genes indicated that cytokine-cytokine receptor interaction, phosphatidylinositol 3-kinase (PI3K)–AKT signaling pathway, interleukin-17 (IL-17) signaling pathway, extracellular matrix (ECM)–receptor interaction, and tumor necrosis factor (TNF) signaling pathway were enriched in BTC tumors (Fig. 2I).
Spatial transcriptomic analysis of BTC tumor reveals rRNA metabolism–associated dedifferentiation
To further explore the function of TERT in dedifferentiation of BRAF V600E–induced thyroid cancer, a tumor from BTC mice with profound heterogeneity was subjected to Visium 10X spatial transcriptome (ST) analysis. The tumor had regions with features of normal follicular cells, PTC, and PDTC, which potentially represented the whole development trajectory of thyroid cancers (Fig. 3A and fig. S5, A and B). After removing low-quality spots, a total of 2552 spots covering the thyroid, esophagus, and trachea regions were subjected to further analysis (fig. S5, C and D). Using the t-distributed stochastic neighbor embedding (t-SNE) method, all these spots are divided into 17 clusters. Combined with the spatial pathological features, the 17 clusters were further categorized into the following 8 cell types: thyroid epithelial cells (Epcam, Tg, Tpo), salivary gland cells (Pigr, Msln, Agr2), adipocyte cells (Adipoq, Cidec, Cfd), immune cells (Ptprcap, Cd79b, Ccl5, Cxcl9), muscle cells (Actn2, Trdn, Mypz1), erythroid cells (Hbb-bt, Hba-a1, Hba-a2), and esophagus epithelial cells (Mt4, Lgals7, Krt78, Fam25c) (fig. S5, E to L).
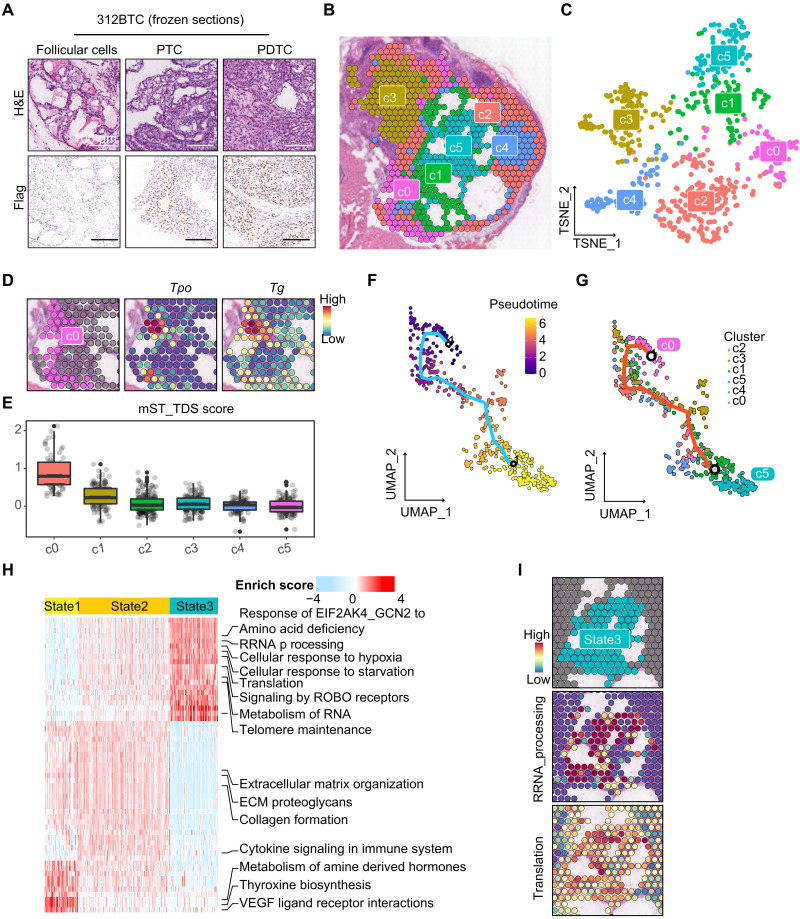
Fig. 3. Spatial transcriptomic analysis reveals heterogeneity of BTC mice thyroid.
(A) H&E and anti-Flag staining results of 312BTC thyroid presented normal follicular cells, PTC, and PDTC contents. Scale bar, 100 μm. (B and C) Spatial and t-SNE projection show different clusters of 312BTC thyroid spots. (D) Spot plots show high expression of Tg and Tpo in c0. (E) Boxplot and scatter plots show the TDS score of each thyrocyte cluster. (F and G) Monocle3 trajectory visualization of thyrocytes, colored by pseudotime and clusters. (H) Heatmap presents Reactome pathway enrichment scores of state1/2/3 spots calculated by AUCell. (I) Spatial feature plots show high enrichment of rRNA processing and translation in state3.
To explore the transcriptional heterogeneity of thyrocytes, we reclustered the 637 thyroid epithelial spots (Fig. 3, B and C). Cells in cluster 0 (c0) express high levels of Tg and Tpo, marking well-differentiated follicular cells (Fig. 3D). Several TDS signature genes, such as Slc5a5, Thrb, Duox1, Duox2, and Tshr, were not detected in ST analysis; hence, we defined an mST_TDS (murine ST TDS) with Tg, Tpo, Dio1, Fhl1, and Sorbs2 as signature genes (31). The mST_TDS score showed that c0 was the best-differentiated cluster, followed by c1 and c2/c3/c4/c5 with poor differentiation (Fig. 3E and fig. S6A).
To trace the evolutionary dynamics of the thyroid clusters, we performed trajectory analysis using Monocle3 and set c0, the well-differentiated follicular cells, as the trajectory starting root (Fig. 3F). In the uniform manifold approximation and projection (UMAP) projection, thyrocytes exhibited an evolution pattern of “c0-c1/c2/c3/c4-c5,” with c0 and c5 separately collected at the start and end point of the trajectory route, and the other four clusters distributed in between (Fig. 3G). Therefore, we could postulate that thyroid cells evolved from c0 (represented as state1), through the middle clusters c2/c3/c4 (state2), and finally dedifferentiated into c5 (state3). The mST_TDS score presented a decreasing trend from state1 to state3 (fig. S6B). GSVA enrichment of hallmark gene sets indicated that state3 was enriched in MYC targets V1, mammalian (or mechanistic) target of rapamycin complex 1 (MTORC1) signaling, and unfolded protein response pathways (fig. S6C). To further identify molecular pathway alterations, Reactome enrichment scores of the three states were calculated using AUCell (Fig. 3H). Consistent with the c0 normal thyroid manifestations, state1 enriched in the thyroxine biosynthesis pathway. State2 presented an intermediate condition between state1 and state3, mainly enriched ECM and immune-associated pathways. Intriguingly, state3 was more active than state1/2 in telomere maintenance, and enriched in rRNA processing and translation pathways, which reflected the ribosome biogenesis and activities (Fig. 3, H and I). Moreover, the physical stress such as cellular response to hypoxia and starvation were also enriched in state3 cells (Fig. 3H and fig. S6D). Pseudo-temporal gene expression dynamics during dedifferentiation suggested that multiple ribosomal proteins, such as Rpl10 and Rps18, were gradually increased with pseudotime, whereas Actb remained stable (fig. S6, E and F). The observation indicates an important role of ribosome biogenesis in c05/state3 cells.
TERT induces rRNA synthesis and ribosomal activity
Extracellular signal–regulated kinase (ERK) activation was considered the upstream signaling of rRNA transcription, likely via phosphorylation of upstream binding transcription factor (UBF) (32). Moreover, TERT has been shown to induce rRNA and tRNA transcription (15, 16). We evaluated the pre-45S rRNA levels in NTHY-CTL, NTHY-B (BRAF V600E), and NTHY-BT (BRAF V600E and TERT) cells. We found that pre-45S rRNA transcription was up-regulated by BRAF V600E and further induced by TERT (fig. S7A). Hence, TERT and BRAF V600E may work together to enhance ribosomal biogenesis.
In cancer cells, ribosomes are frequently deregulated, resulting in uncontrolled proliferation and metastasis (33). To explore whether TERT reactivation was associated with ribosomal activities in cancer, we analyzed genes coexpressed with TERT in the datasets of CCLE. The genes that positively correlated with TERT expression (Spearman R > 0.25, P < 0.01) were involved in cell cycle progression and rRNA metabolism (Fig. 4A). The ribosome consists of rRNA and ribosomal proteins, and ribosomal biogenesis needs the participation of all RNA polymerases (Pol I/II/III), as well as small nucleolar RNA (snoRNA) and multiple other enzymes involved in the rRNA process and maturation (34, 35). We found that TERT positively correlated with the rRNA transcription regulators UBTF, POLR3E, and FBL, and the rRNA exonuclease EXOSC2 in the CCLE database (Fig. 4B and fig. S7B). GSEA also indicated that the TERT high-expression group had higher rRNA metabolism and ribosome biogenesis (fig. S7C). We then analyzed our recent thyroid cancer single-cell RNA-seq (scRNA-seq) dataset (31). Since TERT was not effectively detected in scRNA-seq, we used the Reactome_telomere_maintenance enrichment score as a surrogate for TERT activity. In thyroid cells (n = 42,708 cells), Reactome_telomere_maintenance enrichment scores were positively associated with rRNA processing (R = 0.53, P < 2.2 × 10−16) and translation (R = 0.56, P < 2.2 × 10−16) (Fig. 4C). These results demonstrate a tight association between TERT and ribosomal metabolism.
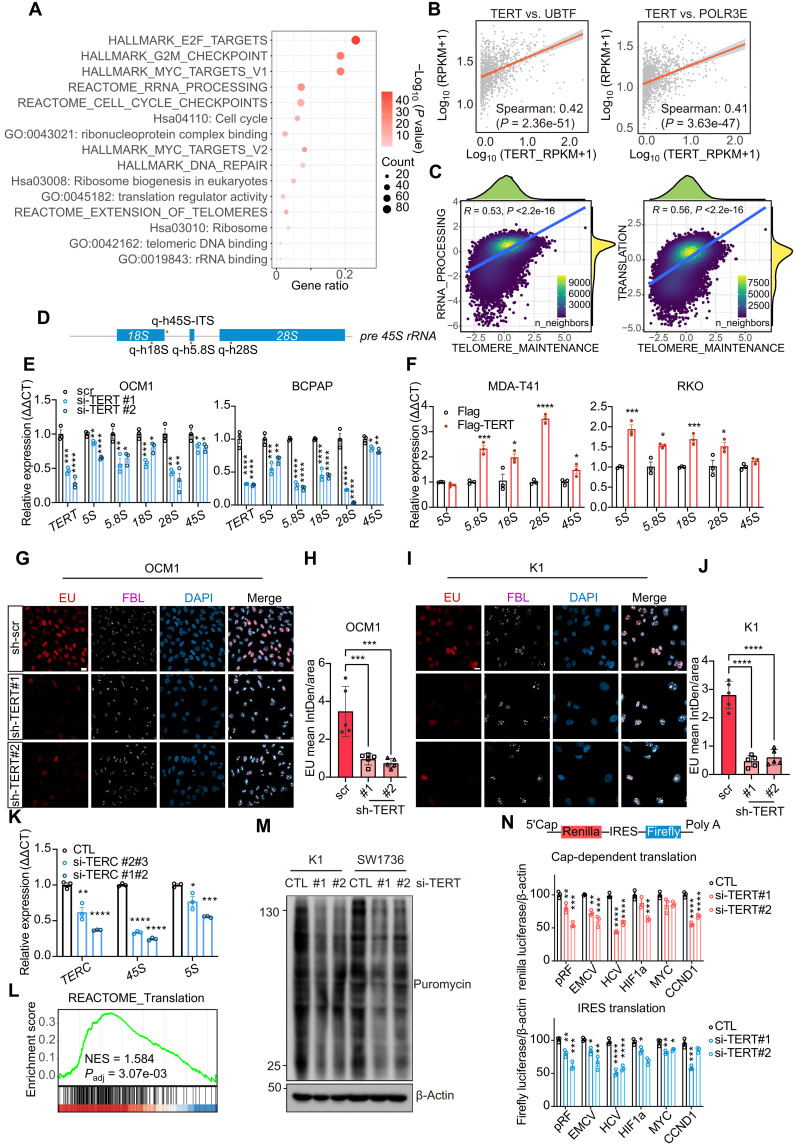
Fig. 4. TERT induces rRNA synthesis and ribosomal activity.
(A) Enrichment results of TERT-positive correlation genes (R > 0.25) in CCLE database (n = 1156). (B) Correlation between TERT and UBTF, POLR3E (n = 1156). (C) Correlation of Reactome: telomere maintenance and rRNA processing/translation enrichment scores in THCA single-cell data of thyroid cells (n = 42,708 cells). (D) Schematic diagram of pre-45S rRNA and the positions of qPCR primers. (E and F) qPCR results of rRNA 5S, 5.8S, 18S, 28S, and 45S change after TERT knockdown [(D); in OCM1 and BCPAP cells] and TERT overexpression [(E); in MDA-T41 and RKO cells]. Data represent means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test in (D) and Student’s t test in (E). (G and I) EU (red) staining and FBL (white) immunofluorescence results after TERT knockdown. Scale bar, 20 μm. (H and J) Quantification results of (G) and (I). EU staining results were quantified using ImageJ software (n = 5). Data represent means ± SD. ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test. (K) qPCR results of TERC, 45S, and 5S after TERC knockdown in BCPAP cells. Data represent means ± SEM (n = 3). (L) GSEA result of between TERT high-expression (n = 538) and low-expression (n = 538) cells in CCLE datasets. (M) SUnSET assay results after TERT siRNA treatment of K1 and SW1736. (N) pRF, EMCV, HCV, HIF-1a, MYC, and CCND1 bicistronic reporter assay results of HEK293T after TERT siRNA treatment. The ratios represented the relative luciferase reads normalized by separative β-actin protein levels. Data represent means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test.
We then examined the effect of TERT on rRNA in different cancer cells. A dramatic decrease in rRNA levels was observed after TERT knockdown in K1 (thyroid cancer), OCM1 (uveal melanoma), and BCPAP (thyroid cancer) cells (all with TERT promoter C228T or C250T mutation) (Fig. 4, D to F, and fig. S7, D and E). The down-regulation of rRNA occurred not only in the Pol I–transcribed 18S, 5.8S, and 28S but also in the Pol III–transcribed 5S rRNA (Fig. 4E and fig. S7E). On the contrary, rRNA expression was induced notably by ectopic TERT in thyroid cell line MDA-T41 and colon cancer cell lines RKO and HT-29 [all with wild-type (WT) TERT promoter] (Fig. 4F and fig. S7F). As more than 80% of cellular RNA was rRNA, ethynyl uridine (EU) assay was performed to label nascent RNA, especially rRNA. In K1 and OCM1 cells, TERT knockdown notably repressed nascent rRNA synthesis in the nucleus, whereas fibrillarin (FBL; a Cajal body component) was not changed (Fig. 4, G to J, and fig. S7G). Together, these results indicate that TERT is required for optimum rRNA synthesis.
As TERT is one of the main subunits of telomerase, we wondered whether telomerase activity was involved in rRNA synthesis. We found that the knockdown of TERC, another main component of telomerase, in BCPAP cells led to a reduction of rRNA expression (Fig. 4K), suggesting that the regulation of rRNA synthesis by TERT was at least partially dependent on telomerase activity.
The major function of ribosomes is translation, in which protein is synthesized using mRNA as a template. As TERT could enhance rRNA transcription, we wondered whether TERT could regulate the translation process. In our ST data and CCLE dataset, high TERT expression was associated with translation (Figs. 3H and 4L). To validate the role of TERT in translation, we adopted two assays to examine translational efficiency. Intriguingly, the surface sensing of translation (SUnSET) assay demonstrated that TERT knockdown resulted in decreased nascent protein production in BRAF and TERT promoter mutated K1 and SW1736 cells (Fig. 4M). In the bicistronic reporter assay, knockdown of TERT inhibited both the 5′ cap– and internal ribosomal entry site (IRES)–dependent translation efficiencies (Fig. 4N and fig. S7H). Hence, the protein synthesis appears to be dependent on TERT in TERT-reactivated cancer cells.
TERT interacts with ribosomal scaffolds and enhances MTORC1 activity
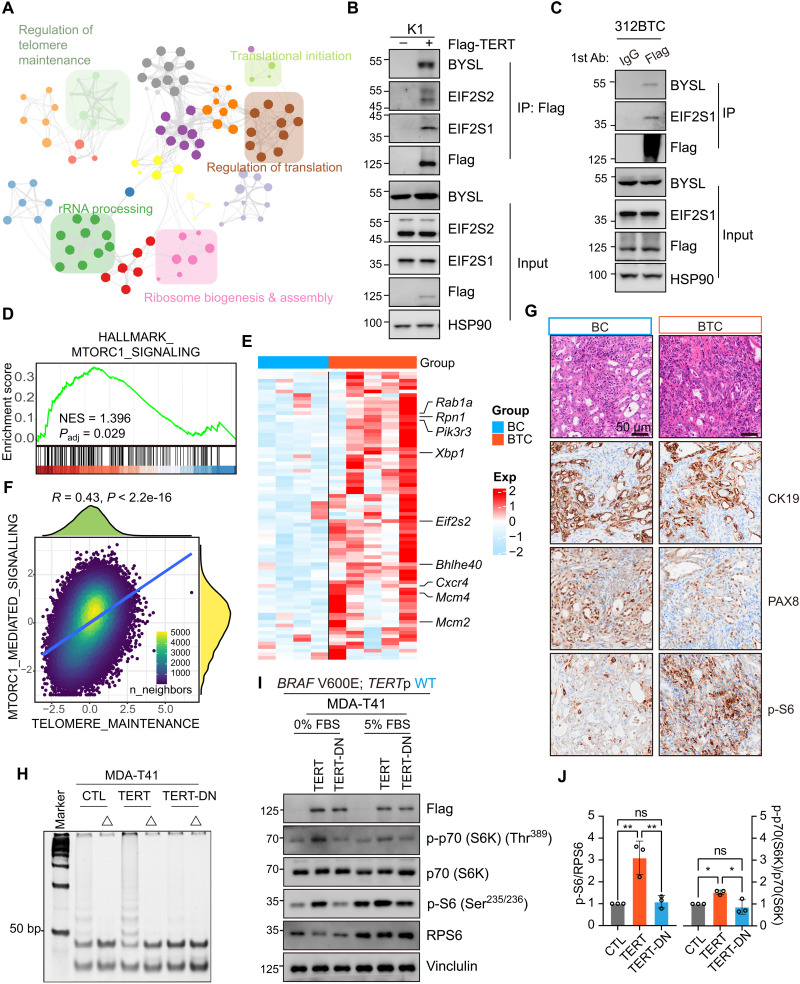
The functional telomerase complex includes multiple components besides TERT and TERC, such as dyskerin pseudouridine synthase 1 (DKC1) and pescadillo ribosomal biogenesis factor 1 (PES1). DKC1 and PES1 have been linked to ribosome function (36–39). Accordingly, we asked whether TERT participates in the ribosome pathway by interacting with rRNA metabolism–associated proteins. We performed immunoprecipitation and mass spectrometry to identify potential binding partners of TERT. Several proteins involved in RNA catabolic process, ribosome biogenesis, translational control, rRNA processing, and telomere maintenance have thus been identified (Fig. 5A and fig. S8, A and B). We further confirmed in coimmunoprecipitation assays that TERT interacted with EXOSC2/7/8, EIF2S1/2, and POLR1C (fig. S8, C and D). In addition, the interaction between TERT and Bystin Like (BYSL), which participates in pre-rRNA processing in yeast and human, was identified in both human embryonic kidney (HEK) 293T and thyroid cancer cells K1 (Fig. 5B and fig. S8D) (40). Furthermore, we validated the existence of protein interaction between TERT and BYSL as well as EIF2 subunits in K1, HEK293T, and a murine primary thyroid cancer cell line 312BTC (Fig. 5C and fig. S8D). Collectively, these results suggest that TERT may regulate ribosome biogenesis by interacting with known ribosome regulators.
Fig. 5. TERT interacts with ribosomal scaffolds and enhances MTORC1 activity.
(A) TERT binding protein networks identified by immunoprecipitation (IP)/mass spectrometry and visualized in Metascape (http://metascape.org). (B and C) Coimmunoprecipitation analysis of protein extracted from K1 and 312BTC cells. (D) GSEA shows positive enrichment of MTORC1 signaling with BTC mice thyroid (n = 5) compared to BC mice thyroid (n = 4). (E) Heatmap shows that multiple genes involved in the MTORC1 pathway were highly expressed in BTC thyroid (n = 5) compared to BC thyroid (n = 4). (F) Correlation of telomere maintenance and MTORC1 mediated signaling enrichment scores in THCA single-cell data of thyroid cells (n = 42,708 cells). (G) H&E morphology and anti-CK19, PAX8, and p-S6 IHC staining in BC and BTC thyroid. (H) Telomere repeat amplification protocol (TRAP) results of MDA-T41 (CTL/TERT/TERT-DN) protein lysate. Δ, heat shock at 85°C, 5 min for telomerase inactivation. (I) Western blot shows in 0 and 5% FBS media, P-p70 (S6K), p70 (S6K), P-S6, and RPS6 change in MDA-T41 (CTL/ TERT/ TERT-DN) cells. (J) Quantification results of (I). Data presented as means ± SD (n = 3). *P < 0.05; **P < 0.01. One-way ANOVA and Tukey’s multiple comparisons test.
PI3K-AKT and MTORC1 signaling pathways play a major role in regulating protein synthesis at the ribosome (41, 42). In our bulk and spatial transcriptomic analysis, PI3K-AKT and MTORC1 signaling pathways were enriched in BTC tumors (Fig. 5D and figs. S4F and S6C). Consistently, multiple genes involved in the MTORC1 pathway, such as Rpn1, Xbp1, Eif2s2, were up-regulated in the BTC group (Fig. 5E). In addition, the scRNA-seq data of thyroid cancer showed a notable correlation between telomere maintenance and MOTRC1-mediated signaling in thyroid cells (R = 0.43, P < 2.2 × 10−16) (Fig. 5F). Moreover, as a readout of MTORC1 kinase activity, p-S6 level was notably enhanced in tumors from BTC mice (Fig. 5G). To further investigate the role of TERT in MTORC1 signaling in thyroid cancer, we expressed WT-TERT or DN-TERT (mutant without telomerase activity) (15) in MDA-T41 cells and analyzed the phosphorylation of known downstream effectors of MTORC1 (Fig. 5, H and I). In agreement with in vivo results, the expression of WT-TERT effectively induced phosphorylation of S6 and S6K, especially at serum-starved conditions (Fig. 5, I and J). Hence, MTORC1 may also participate in the activation of ribosomes by TERT.
Targeted inhibition of rRNA transcription blocks tumor progression
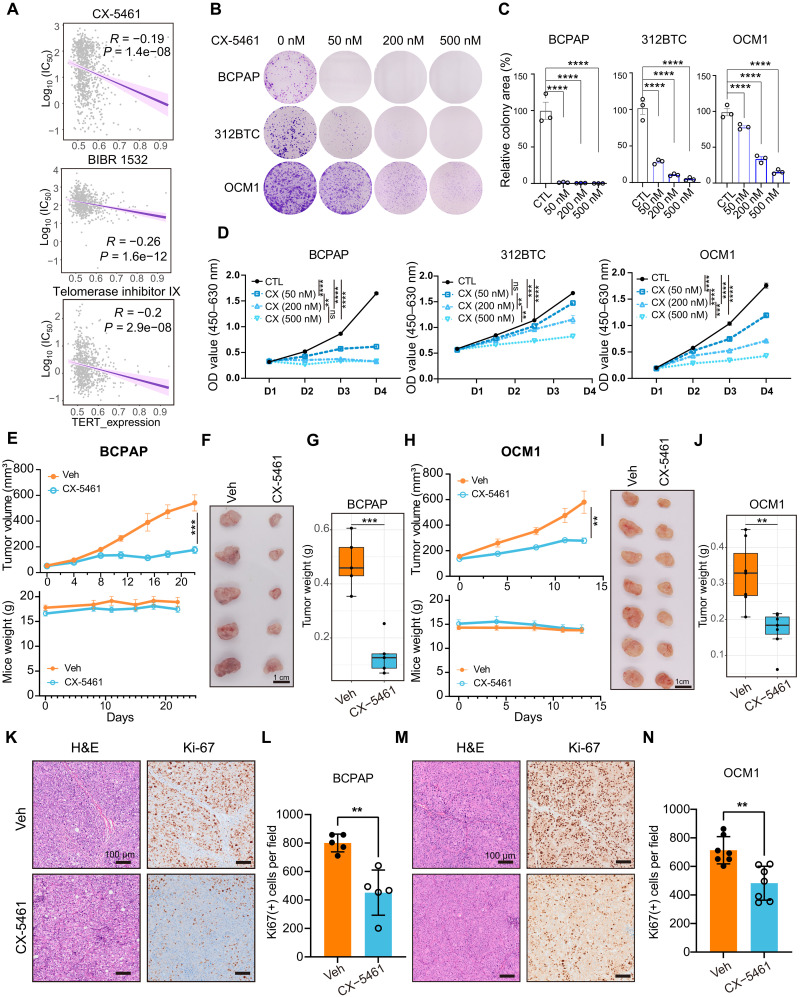
Targeting mechanisms downstream of TERT, such as ribosome biogenesis and MTORC1 signaling, may serve as alternative antitumor strategies with reduced side effects compared with TERT inhibitors. We analyzed Genomics of Drug Sensitivity in Cancer (GDSC) datasets (43) and observed that cells with higher TERT expression levels were more sensitive to CX-5461, an inhibitor of Pol I–mediated rRNA transcription, which was tolerated in normal somatic cells. CX-5461 presented a pattern like previous telomerase-targeted drugs, such as BIBR 1532 and telomerase inhibitor IX (Fig. 6A) (44–48). BRAF V600E and TERT promoter co-mutant BCPAP and OCM1 cells, and mouse 312BTC cells, were used to test the effect of CX-5461. CX-5461 inhibited cancer cell proliferation in a dose-dependent manner (Fig. 6, B to D), comparable to the effects of TERT knockdown (fig. S2G). BCPAP, OCM1, and murine 611BTPC (312BTC cells with Trp53 inactivation) subcutaneous tumor models were then established, and CX-5461 was orally administrated to test the antitumor effect of CX-5461 in vivo. We found that CX-5461 repressed tumor growth effectively in all three models and showed no or mild body weight loss in the mice tested (Fig. 6, E to J, and fig. S9, A to C). Moreover, Ki67 expression in tumors was also reduced upon CX-5461 treatment (Fig. 6, K to N). Together, these results indicate that CX-5461 is effective in inhibiting the proliferation of cells with BRAF mutation and TERT reactivation.
Fig. 6. CX-5461 inhibits multiple tumor progression.
(A) Correlation between TERT expression and log10 (IC50, median inhibitory concentration) of CX-5461, BIBR 1532, and telomerase inhibitor IX in GDSC datasets. Spearman correlation test. (B) Effects of CX-5461 on colony formation of BCPAP, 312BTC, and OCM1 cells. (C) Quantifications results of (B). (D) Effects of CX-5461 on survival of BCPAP, 312BTC, and OCM1 cells in CCK8 assay. Data represent means ± SEM (n = 3). **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA and Dunnett’s multiple comparisons test. (E to G) Growth curve, mice weight, tumor gross appearance, and tumor weight of BCPAP xenografts treated by vehicle or CX-5461. n = 5 for each group. Oral administration for five times. (H to J) Growth curve, mice weight, tumor gross appearance, and tumor weight of OCM1 xenografts treated by vehicle or CX-5461. n = 7 for each group. Oral administration for four times. (K and M) H&E and Ki67 staining of BCPAP and OCM1 xenografts with vehicle or CX-5461. Scale bar, 100 μm. (L and N) Quantification results of Ki67 [(K) and (M)] per field (10× objective). Data assessed by Student’s t test. **P < 0.01. [(E) and (H)] Data represent means ± SEM. [(G) and (J)] Data represent the median with IQR and 1.5 IQR whiskers. [(L) and (N)] Data represent means ± SD.
CX-5461 induces thyroid cancer redifferentiation
Radioactive iodine (RAI) therapy is regarded as the first-line treatment for metastatic thyroid cancers (49). However, nearly all PDTC and ATC, as well as some of the well-differentiated thyroid cancer, do not efficiently uptake iodine because of the reduced expression of functional thyroid follicular markers especially iodine transporter NIS. These patients are classified as RAI refractory, and RAI refraction is one of the leading causes of thyroid cancer–related deaths (50). Given that the major role of TERT in tumor progression is to induce tumor dedifferentiation, blocking TERT or its downstream effectors may lead to cancer differentiation.
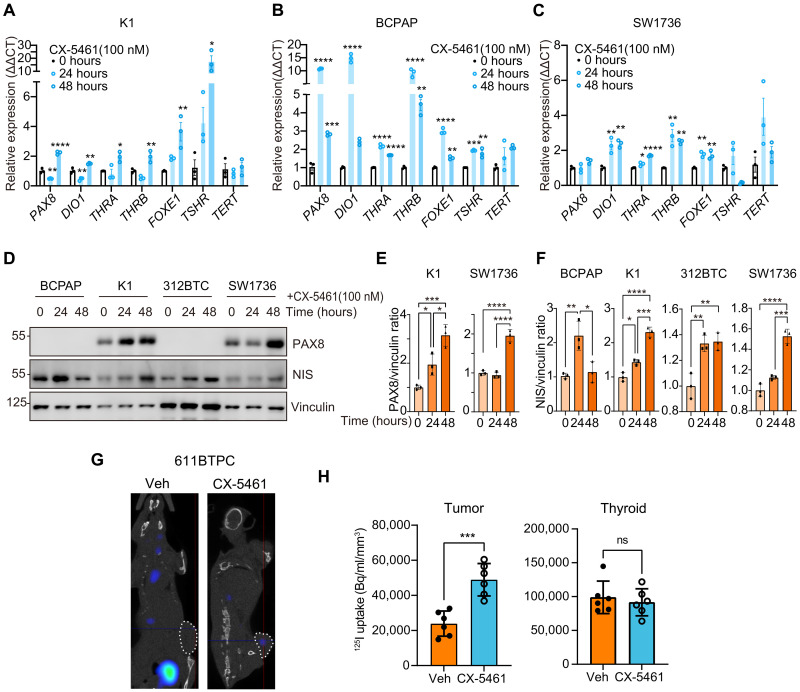
Thus, the effect of CX-5461 on differentiation markers in thyroid cancer cells was evaluated. The expression levels of PAX8, DIO1, THRA, THRB, FOXE1, and TSHR in K1, BCPAP, and SW1736 cells were notably up-regulated after 24 or 48 hours of CX-5461 treatment (Fig. 7, A to C). NIS and PAX8 protein expression levels were also elevated in K1 and SW1736 cells after treatment with CX-5461 (Fig. 7, D to F). Consistently, high NIS expression was observed in the BCPAP tumors with CX-5461 treatment (fig. S9D). We then tested whether CX-5461 treatment could increase iodine uptake in thyroid tumors. The 611BTPC syngeneic tumor model was used to detect differences in tumor iodine uptake before and after CX-5461 administration. The results showed that CX-5461 treatment induced iodine uptake in subcutaneous 611BTPC tumors, while iodine uptake in the native thyroid remained unchanged (Fig. 7, G and H). The results of gamma counting also indicated that CX-5461 induced iodine uptake in tumors, without notable effect on thyroid, stomach, and salivary glands (fig. S9E). Furthermore, we obtained tumor tissues with both TERT promoter and BRAF V600E mutations from two patients and constructed miniPDX in nude mice (fig. S9G). Mice with miniPDX were treated with or without CX-5461 and then subjected to [125I] uptake assay (fig. S9H). The results showed that after treatment with CX-5461, the gamma counting was robustly increased (fig. S9I). These results together suggest that rRNA transcription blockade is an effective way to induce thyroid cancer redifferentiation.
Fig. 7. CX-5461 induces thyroid cancer redifferentiation.
(A to C) Representative thyroid differentiation–associated genes PAX8, DIO1, THRA, THRB, FOXE1, and TSHR qPCR results of K1, BCPAP, and SW1736 after CX-5461 (100 nM) treatment for 0, 24, and 48 hours. Data represent means ± SEM (n = 3). (D) Western blots show PAX8 and NIS change of K1, BCPAP, 312BTC, and SW1736 treated by CX-5461 (100 nM) for 0, 24, and 48 hours. (E and F) Quantification results of PAX8 and NIS change in (D). Data represent means ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus 0 hours by one-way ANOVA and Tukey’s multiple comparisons test. (G) Typical results of [125I] SPECT/CT. White dashed lines indicate the subcutaneous tumors. (H) Biodistribution of [125I] in subcutaneous tumor and thyroid, measured by VivoQuant software. Data are assessed by Student’s t test and represent means ± SD (n = 6).
DISCUSSION
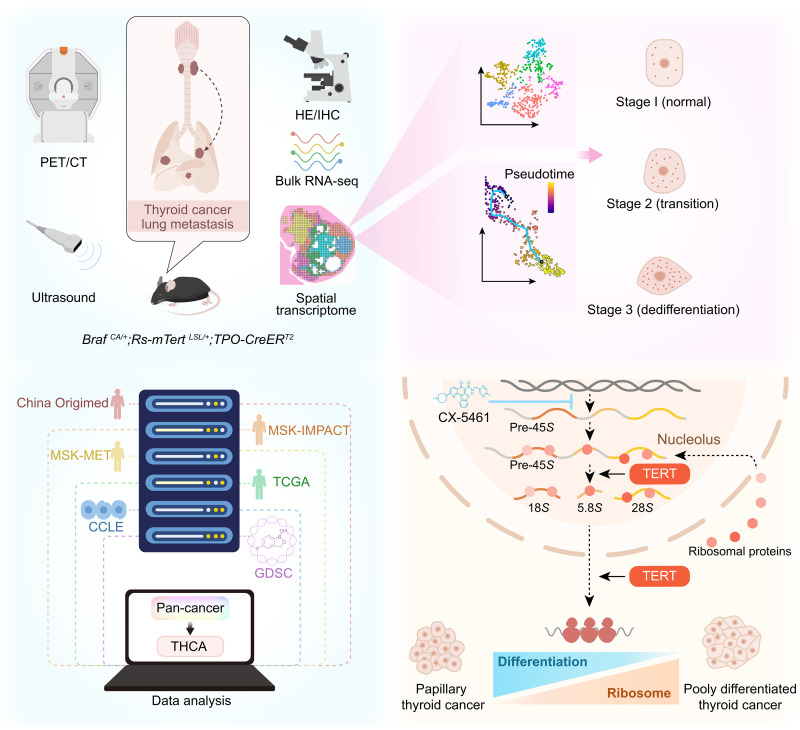
We demonstrated in this study that conditional transgenic expression of TERT promoted dedifferentiation and cancer metastasis driven by BRAF V600E mutation in mouse thyroid. The expression of TERT in both mouse and human BRAF V600E tumor cells induced rRNA transcription and ribosomal biogenesis. The rRNA transcription inhibitor CX-5461 induced tumor differentiation and slowed down cancer progression (Fig. 8).
Fig. 8. Schematic diagram of the present study.
Comprehensive integration of animal models, ST sequencing, databases analysis, and experiments reveals the role of TERT in ribosomal pathways.
A large portion of human PTC remains stable or progresses rather slowly during active surveillance, while PDTC and ATCs usually exhibit a sudden increase in tumor size and abrupt deterioration (51). Previous studies have modeled thyroid cancer in mice, mainly by transgenic or conditional expression of BRAF V600E. The Tg-Braf mice (transgenic expression of BRAF V600E under Thyroglobulin promoter) exhibited elevated thyroid stimulating hormone (TSH) levels and impaired thyroid function after birth, and developed PDTC at 3 months old. In this model, a high dose of BRAF V600E may contribute to early tumor formation and dedifferentiation (52, 53). In the BRAF V600E; TPO-CreERT2 (BC) model, BRAF V600E expression was induced by one dose of tamoxifen in 2-month-old mice to mimic BRAF mutation in human thyroid cancer, whereas no lung metastasis or profound tumor dedifferentiation was observed (54). In our study, tumor progression in BC and BTC mouse models was monitored by ultrasound imaging. Tumors in BTC mice exhibited a sudden increase in tumor size and metastasis (Figs. 1 and 2). Spatial transcriptomic analysis of a tumor from a BTC mouse revealed a continuous trajectory of dedifferentiation of thyroid cancer cells (Fig. 3). The BRAF-activating mutation presents in both PTC and PDTC, while TERT reactivation occurs more frequently in PDTC (55). Both molecular and phenotypic transitions from PTC to PDTC in human thyroid cancers were mimicked in our BTC mouse model. The histological and molecular features of tumors from BC and BTC mice were similar to those from PTC and PDTC, respectively. Studies of BC and BTC mouse models indicated an essential role of TERT in the progression of thyroid cancer into the advanced stage. A Tert promoter mutation and BRAF V600E expression murine model has recently been disclosed, and the transition from PTC to PDTC is also captured in Tert promoter mutant mice (56).
TERT reactivation is observed frequently in various cancer types. Cross-talk between TERT and important signaling pathways, such as NF-κB, Wnt pathway, FOXO3a, and MYCs, has been previously reported (12–14). In addition, TERT stimulates Pol I activity and tRNA transcription (15, 16). Our study highlights the important role of rRNA transcription and ribosomal biogenesis in cancer progression upon TERT reactivation (Figs. 4 and 5). TERT activates MTORC1 signaling, which may induce ribosomal biogenesis at both rRNA transcription and translation levels (Fig. 5) (42). Moreover, several ribosome scaffold proteins interact with TERT, which may also boost ribosome activity (Fig. 5, B and C). Further investigations are required to understand the precise mechanism linking TERT and ribosome biogenesis.
Ribosomal biogenesis is crucial for various cancer cell activities including survival, proliferation, differentiation, and metastasis. Ribosomes are therefore attractive molecular targets for cancer drugs (34, 57, 58). CX-5461 is a small molecule that targets rRNA transcription, which has been reported to repress the proliferation of melanoma, breast cancer, and ovarian cancer (44–47). There is an ongoing clinical trial (NCT04890613) exploring the role of CX-5461 in BRCA1/2, PALB2, or homologous recombination deficiency mutant solid tumors. We have shown that CX-5461 effectively inhibited the proliferation of different cancer cells with BRAF V600E and TERT promoter mutations (Fig. 6). Moreover, CX-5461 successfully induced the expression of thyroid differentiation markers including PAX8, THRA, THRB, and NIS in human and murine thyroid tumors and restored the iodine uptake of thyroid tumors. These results provide evidence supporting the effectiveness of CX-5461 in treating advanced thyroid tumors, especially those with high TERT expression or dedifferentiated thyroid cancers insensitive to radioiodine therapies. Moreover, it would also be critical to test the effect of CX-5461 on patient-derived xenograft models or BTC mice with spontaneous thyroid tumors in the future.
METHODS
Mouse models and administration strategy
In the C57BL6/J background, C57BL/6-Gt(ROSA)26Sorem1(CAG-LSL-Tert-3xFlag-IRES-EGFP-WPRE-polyA)Smoc conditional TERT overexpression model was generated by loxp-stop-loxp-Tert-3xflag-IRES-EGFP-polyA sequence insertion in mouse Rosa26 foci. Tg (TPO-cre/ERT2)1139Tyj (strain no.: 026512) and B6;129-Braf tm1Mmcm/+ (strain no.: 017837) mice were obtained from The Jackson Laboratory. Genotyping of Braf and TPO-CreERT2 mice was performed separately according to Jackson protocol. Primers for Rs26-mTert mice genotyping were described in table S1. Expression of the transgene was induced using tamoxifen (20 mg/ml; MCE, HY-13757A) by intraperitoneal administration four times at 1 month of age. All animals were housed in specific pathogen–free environment. Animal care and experimental procedures were performed with approval by Fudan University Animal Ethics Community.
Syngeneic and xenograft transplantations and drug administration
Human thyroid cancer cell line BCPAP and human uveal melanoma cell line OCM1 were used to construct a subcutaneous tumor model. In total, 5 × 106 cells (100-μl volume per injection) per mouse were subcutaneously injected into BALB/c nude mice (4 to 5 weeks old). Murine 611BTPC cells (1 × 106) were subcutaneously injected into C57BL/6 mice (4 to 5 weeks old). Mice with tumors were randomized in the vehicle and CX-5461 treatment groups when the average tumor size reached about 60 to 150 mm3. CX-5461 (50 mg/kg; MCE, HY-13323A) was administrated by oral gavage twice a week. Mice were sacrificed when the average tumor size of the vehicle group reached 600 mm3.
Cell culture
HEK293T (CVCL_0063), OCM1 (CVCL_6934), and BCPAP (CVCL_0153) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; BasalMedia, L110KJ), and NTHY-ori 3-1 (CVCL_2659), MDA-T41 (CVCL_W914), SW1736 (CVCL3883), and RKO (CVCL_0504) were maintained in RPMI 1640 (BasalMedia, L210KJ). Media were supplemented with 10% fetal bovine serum (FBS; ExCell Bio, FSP500) and penicillin-streptomycin (50 mg/ml; Meilunbio, MA0110) unless otherwise stated in the text. All the cells were incubated at 37°C with 5% CO2.
Establishment of primary mouse thyroid cell line
Primary thyroid cells were separated and cultured as previously described (59). After dissection using scissors, thyroid tissues were digested in F-12 medium (Gibco, 11765-054) with the following regents: collagenase A (100 U/ml; Gibco, 10103578001), dispase II (1 mg/ml; Solarbio, D6430), 10% FBS, and penicillin-streptomycin (50 mg/ml) at 37°C for 1 hour and filtered with 70-μm cell strainer.
The first three passages were cultured in F-12 medium with transferrin (5 μg/ml), bovine insulin (10 μg/ml; Biosharp, BS001), hydrocortisone (3.5 ng/ml; Solarbio, G8450), glycyl-l-histidyl-l-lysine (2 ng/ml; MCE, HY-P0046), and somatostatin (10 ng/ml; Meilunbio, MB1225). After three passages, cells were cultured in DMEM with 10% FBS.
Plasmids and lentivirus package
For overexpression, the target mRNA coding sequence was cloned using PrimeSTAR Max (TAKARA, R045) or Q5 High-Fidelity DNA Polymerase (NEB, M0491). After separation by agarose gel electrophoresis and retraction, the fragments were subcloned into the pLVX-puro vector (Takara, no. 632164) with Ha- or Flag-tag. Short hairpin RNAs (shRNAs) targeting TERT were cloned into pLKO.1. Transfection was performed using PolyJet DNA In Vitro Transfection Reagent (Signagen, no. SL100688) or PEI (Polysciences Inc., 23966-2) following the manufacturer’s instructions. For lentivirus production, pLVX-based plasmids were cotransfected into HEK293T cells together with psPAX2 and pMD.2g.
Transfection of plasmids and siRNA
For small interfering RNA (siRNA) transfection, control siRNA, TERT siRNA, and TERC siRNA were separately transfected into cultured cells using the riboFECT CP Transfection Kit (RIBOBIO, C10511-05). The sequences of siRNAs were listed in table S1.
Animal ultrasound imaging and PET-CT
Fujifilm Vevo 2100 was adopted to monthly monitor the thyroid morphology of mice. Briefly, after anesthesia by isoflurane and dehairing, an MS400 detector was used to scan the neck region. All images were collected using the same preset.
Micro-PET/CT (Inveon, Siemens) scanning was performed at the Department of Nuclear Medicine, Fudan University Shanghai Cancer Center. After fasting overnight, 18F-FDG was intraperitoneally injected for 45 min before scanning. The PET and CT images were fused using Inveon Research Workplace software (Siemens Medical Solutions). Data were expressed in the form of standardized uptake value (SUV).
In vivo [125I] uptake experiments
Mice were fed with water containing 0.1% potassium iodide (Sangon, A100512) for 7 days before [125I] uptake experiments. Imaging was performed with a small animal single photon emission computed tomography (SPECT/CT) system (X-Cube and γ-Cube, Molecubes, Gent, Belgium). Each mouse was given a tail vein injection of 1 mCi [125I] and underwent SPECT γ-Cube scanning 10 min after the injection. This was followed by CT scanning using X-Cube. Images were reconstructed and exported with Invicro VivoQuant 2021. All images of SPECT/CT in an assay were adjusted to a fixed threshold. Subcutaneous tumors, thyroid, salivary glands, and stomach of mice were measured for counts per minute using a gamma counter.
IHC staining
Formalin-fixed, paraffin-embedded sections were baked at 65°C for 1 hour. After deparaffinization and hydration, sodium citrate buffer (Beyotime, P0083) was adopted for heat-treated antigen and boiled for 15 min at 100°C. Subsequently, endogenous peroxidase activities were blocked using 3% H2O2 for 20 min. After blocking with 3% bovine serum albumin (BSA) in Tris buffered saline with Tween 20 (TBST) for 1 hour at room temperature, the primary antibodies were incubated at the optimized concentration at 4°C overnight. DAKO (K4003) and GenTech (GK500705) secondary horseradish peroxidase–conjugated antibodies were separately incubated for 1 hour and reacted with 3, 3'-diaminobenzidine (DAB). Hematoxylin was counterstained for nuclear staining. Sections were scanned using Olympus VS200.
RNA extraction, cDNA synthesis, and RT-qPCR
Total RNA of adherent cells was isolated with the EZB Kit (EZB, B0004DP), and the total RNA of mouse thyroid tissues was isolated with the Tiangen Kit (Tiangen, DP419) and then reverse-transcribed to cDNA using One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, AU311). The amplification and fluorescence detection of reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed in Q6 (Invitrogen) using TAKARA TB Green Premix (RR420). The relative expression level of target genes was calculated with the ΔΔCT method, with β-actin as the internal control. The primer sequences were listed in table S1.
Bulk RNA-seq
The total RNA samples (1 μg) were used for the following library preparation. Double-stranded cDNA was purified and treated to repair both ends and add adaptors to them. Each sample was then amplified by PCR using P5 and P7 primers, and the PCR products were validated. Libraries with different indexes were multiplexed and loaded on Illumina Novaseq 6000 instrument for sequencing. Clean data of BTC and BC mice thyroid were aligned to reference genome via software Hisat2 (v2.0.1). GRCm39.104 was used as a reference genome in alignment. Gene expression levels were estimated using HTseq (v0.6.1), and differential expression analysis used the DEseq2 (60) Bioconductor package. PCA analysis and visualization were performed with R packages FactoMineR (v2.4) and factoextra (v1.0.7). Single-sample GSVA and GSEA were performed to determine the Hallmarks of Cancer phenotypes of BTC and BC thyroid cancers. We used BC as the control group for GSEA. Visualizations were generated using ggplot2 (v3.3.6) and pheatmap (v1.0.12) R packages. The counts data were uploaded as table S4.
ST sample preparation, library construction, and sequencing
Fresh thyroid tissues of BTC mice were fast-frozen in the precooled isopentane and embedded with the optimal cutting compound (OCT) (Sakura, 4583). Tissues were stored at −80°C. The optimal permeabilization time was found to be 12 min using 10X Genomics Visium Tissue Optimization Kit. Spatially tagged cDNA libraries were constructed with the 10X Genomics Visium Spatial Gene Expression 3′ Library Construction V1 Kit and then sequenced on Novaseq (Illumina).
ST data processing and visualization
Spaceranger-1.1.0 pipeline was used to process raw fastq files with mm10 reference genome, and the output files were integrated using Seurat (v4.1.0) R package (61). Seurat SCTransform function was used for normalization. FindNeighbors and FindClusters functions of Seurat were used for downstream graph-based clustering, and t-SNE dimensionality reduction was then used for projection visualization (62). To identify the marker genes of the clusters, FindAllMarkers function was performed (logFC.threshold = 0.25, test.use = “wilcox”). Seurat’s AddModuleScore function was applied to quantify the differentiation scores of 312BTC thyroid spots.
Trajectory analysis
Trajectory inference and pseudotime analysis were performed using Monocle3 (v1.2.9). All spots of the 312BTC sample were integrated using new_cell_data_set function, and thyrocytes were chosen using the choose_cells function. Cluster c0 was indicated as the root.
Thyroid cancer single-cell dataset analysis
The 10X scRNA datasets were generated by our previous work (31). The quality control, dimension reduction, and cell annotation were all the same as previously reported.
Single-cell or ST enrichment analysis
We adopted AUCell (v1.16.0) to calculate the enrichment score of each spot or cell with the Reactome database of Molecular Signatures Database (MSigDB) (63, 64). Visualization was performed with ComplexHeatmap (v2.10.0).
EU assay
To label nascent RNA, Cell-Light EU RNA Imaging Kit (RIBOBIO, C10316) was adopted. For OCM1 and K1 cells, EU treatment lasted for 3 hours, and then the EU-labeled RNA was detected following the protocol. To visualize the nucleolar region, the anti-FBL immunofluorescence was costained.
Immunoblotting
Whole-cell lysates were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were transferred onto nitrocellulose membranes. Following blocking in 5% nonfat milk, membranes were incubated with primary antibodies in 3% BSA overnight at 4°C and then with secondary antibodies in 5% milk for 1 hour at room temperature. High-sig ECL Western Blotting Substrate (Tanon, no. 180-501) was adopted for chemiluminescence detection with Tanon 5200S imaging system or Bio-Rad XRS+ imaging system.
Immunoprecipitation
Cells were lysed with mild lysis buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, and 1.5 mM Na3VO4] containing EDTA-free protease inhibitor cocktail (Bimake, B14012). After sonication and centrifugation, the supernatants were incubated with primary antibodies for 1 hour at 4°C. Protein A beads were then added and incubated for additional 2 hours, followed by four times centrifugation at 2000 rpm and washing with mild lysis buffer at 4°C. Proteins precipitated were dissolved in 1× SDS-PAGE sample buffer and followed by immunoblotting.
Immunoprecipitation and liquid chromatography–mass spectrometry
For the identification of the FLAG-TERT interaction proteomics, about 107 HEK293T cells were transfected with 5 μg of pLVXyu-FLAG-TERT plasmid. The immunoprecipitated proteins were separated by SDS-PAGE using the 4 to 15% tris/glycine gel. After in-gel digestion, the peptides were analyzed by online nanoflow liquid chromatography tandem mass spectrometry performed on an EASY-nanoLC 1200 system (Thermo Fisher Scientific, MA, USA) connected to a Q Exactive Plusmass spectrometer (Thermo Fisher Scientific, MA, USA). Tandem mass spectra were processed by PEAKS Studio version X+ (Bioinformatics Solutions Inc., Waterloo, Canada). Interacting proteins identified are shown in table S3.
Telomere repeat amplification protocol assay
The telomerase activity was measured using the TRAPeze Telomerase Detection Kit (Millipore, S7700). Briefly, 106 cells were lysed in 200 μl of 1× CHAPS buffer with ribonuclease inhibitor. After centrifugation and bicinchoninic acid protein quantification, the telomerase activity to length telomere template was detected using the PCR method with the following procedures: 30°C, 30 min, 1 cycle; 95°C, 2 min, 1 cycle; 94°C, 15 s, 59°C, 30 s, 72°C, 1 min, 32 cycles; 4°C keep. The PCR products were separated using the 10% acis/bis gel, followed by staining with GelRed (Tsingke, TSJ003), using 50–base pair (bp) DNA ladder (Generay, DL0501) as the reference.
Surface sensing of translation
The SUnSET assay was performed just as previously reported (65). Briefly, cells without puromycin resistance were labeled with puromycin (10 μg/ml; InvivoGen, ant-pr) for 10 min, followed by a 1-hour chase. Then, the anti-puromycin primary antibody (Sigma-Aldrich, MABE343) was used to detect the puromycin incorporation with the immunoblot assay.
Bicistronic dual luciferase reporter
The plasmids used in the bicistronic reporter assay were gifts from M. T. Bedford laboratory. After 24 hours of transfection of reporter plasmids, 105 HEK293T cells were seeded in a 24-well plate. Then, 48 hours after siRNA transfection, cell lysates were treated with the dual luciferase measurement kit (Transgene, FR201), and the firefly and renilla luciferase activities were measured using BioTek Synergy LX, with β-actin protein level (by Western blot) as the internal control.
Public dataset analysis
MSK-IMPACT, MSK-MET, China Origimed2020 cohorts, TCGA, and CCLE datasets were accessed through cbioportal (https://cbioportal.org/datasets). CCLE counts data were downloaded from Xena. Visualization was performed with ggplot2 (v3.3.6) R package.
Colony formation
Cells (1000 to 3000) were seeded in 12-well plates. After culture for 1 to 2 weeks, the cells were fixed by 4% paraformaldehyde for 10 min, followed by phosphate-buffered saline washing and 0.1% crystal violet (Servicebio, G1014) staining for 15 min. Colony areas were measured using ImageJ.
Cell counting kit-8
About 5000 to 10,000 cells were seeded in each well of 96-well plates. For cell counting kit-8 (CCK8) assay, the cells were added with CCK8 solution (meilunbio, MA0218) and incubated for 1 hour at 37°C. The absorbance was measured at 450 nm and at a reference wavelength of 630 nm with BioTek Synergy LX.
OncoVee MiniPDX of thyroid cancer patients
OncoVee MiniPDX involves implanting thyroid tumors into specially designed hollow fiber capsules with micropores, which are then transplanted under the skin of nude mice. The experimental details were as described in previous studies (66–68). This experiment has been approved by the ethics committee of Shanghai Cancer Center, Fudan University.
Statistics
Comparisons between two groups were performed using Student’s two-sided t test. One-way analysis of variance (ANOVA) and Dunnett’s or Tukey’s multiple comparisons test were adopted for multiple comparisons. All statistical analyses were performed using GraphPad Prism v9.0.0 or R package ggsignif (v0.6.3). P value less than 0.05 was considered as significant. The number of replicates was noted in the figures and legends.
Acknowledgments
We thank R. Liu (Sun Yat-sen University) for providing the MDA-T41 cell line and H. Ji (University of Chinese Academy of Sciences) for providing the Trp53 fl/fl mice and advice. We also appreciate the help from S. Song, J. Zhang, and D. Wang (Nuclear Medicine, Fudan University Shanghai Cancer Center) on the iodine uptake assay.
Funding: This work was supported by grants from the Ministry of Science and Technology of China (National Key R&D program, 2018YFA0800304 and 2020YFA0803202), the Science and Technology Commission of Shanghai Municipality (21S11905000), and the Shanghai Municipal Health Commission (2022XD049) to F.-X.Y., and grants from the National Natural Science Foundation of China (81972501) and Shanghai Shenkang Hospital Development Center (SHDC22021201) to Y.-L.W. This work is also supported by the Medical Science Data Center in Shanghai Medical College of Fudan Univeristy.
Author contributions: Conceptualization: Y.-L.W., F.-X.Y., and N.Q. Experiments: P.Y., R.Z., J.H., L.T., C.F., Y.L., J.L., and Chenxi He. Bioinformatics: P.Y., P.H., J.W., C.F., and X.S. Pathological analysis: H.G., Y.-L.W., W.W., and X.S. Validation: J.H., P.Y., Cong He, L.T., and J.H. Visualization: P.Y. and P.H. Writing—original draft: P.Y., F.-X.Y., J.H., N.Q., and Cong He. Writing—review and editing: Y.-L.W., F.-X.Y., Y.W., and W.W. Supervision: Y.-L.W., F.-X.Y., F.L., and Q.J.
Competing interests: A patent “A construction method and its application of a conditional mTERT overexpression mouse model” (ZL 2020 1 1359691.6) was authorized by China National Intellectual Property Administration. The authors declare no other competing interests.
Data and materials availability: The Rs26-mTert model can be provided by Y.-L.W. and F.-X.Y. pending scientific review and a completed material transfer agreement. Requests for the Rs26-mTert model should be submitted to: yulongwang@fudan.edu.cn and fxyu@fudan.edu.cn. The raw sequence data reported here have been deposited in the Genome Sequence Archive (69) in National Genomics Data Center (70), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA010824) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, J. W. Shay, Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Y. He, Y. Wang, B. Liu, C. Helmling, L. Sušac, R. Cheng, Z. H. Zhou, J. Feigon, Structures of telomerase at several steps of telomere repeat synthesis. Nature 593, 454–459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. Hohaus, M. T. Voso, E. Orta-La Barbera, S. Cavallo, A. Bellacosa, S. Rutella, C. Rumi, M. Genuardi, G. Neri, G. Leone, Telomerase activity in human hematopoietic progenitor cells. Haematologica 82, 262–268 (1997). [PubMed] [Google Scholar]
- 4.X. Li, X. Qian, B. Wang, Y. Xia, Y. Zheng, L. Du, D. Xu, D. Xing, R. A. DePinho, Z. Lu, Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nat. Cell Biol. 22, 282–288 (2020). [DOI] [PubMed] [Google Scholar]
- 5.R. J. A. Bell, H. T. Rube, A. Kreig, A. Mancini, S. D. Fouse, R. P. Nagarajan, S. Choi, C. Hong, D. He, M. Pekmezci, J. K. Wiencke, M. R. Wrensch, S. M. Chang, K. M. Walsh, S. Myong, J. S. Song, J. F. Costello, Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. Pirker, A. Bilecz, M. Grusch, T. Mohr, B. Heidenreich, V. Laszlo, P. Stockhammer, D. Lötsch-Gojo, J. Gojo, L. Gabler, S. Spiegl-Kreinecker, B. Dome, A. Steindl, T. Klikovits, M. A. Hoda, M. Jakopovic, M. Samarzija, K. Mohorcic, I. Kern, B. Kiesel, L. Brcic, F. Oberndorfer, L. Müllauer, W. Klepetko, W. M. Schmidt, R. Kumar, B. Hegedus, W. Berger, Telomerase reverse transcriptase promoter mutations identify a genomically defined and highly aggressive human pleural mesothelioma subgroup. Clin. Cancer Res. 26, 3819–3830 (2020). [DOI] [PubMed] [Google Scholar]
- 7.A. M. Amen, C. Fellmann, K. M. Soczek, S. M. Ren, R. J. Lew, G. J. Knott, J. E. Park, A. M. McKinney, A. Mancini, J. A. Doudna, J. F. Costello, Cancer-specific loss of TERT activation sensitizes glioblastoma to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 118, e2008772118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. N. Guterres, J. Villanueva, Targeting telomerase for cancer therapy. Oncogene 39, 5811–5824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Ruden, N. Puri, Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 39, 444–456 (2013). [DOI] [PubMed] [Google Scholar]
- 10.K. Ait-Aissa, J. D. Ebben, A. O. Kadlec, A. M. Beyer, Friend or foe? Telomerase as a pharmacological target in cancer and cardiovascular disease. Pharmacol. Res. 111, 422–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Y. Rha, E. Izbicka, R. Lawrence, K. Davidson, D. Sun, M. P. Moyer, G. D. Roodman, L. Hurley, D. Von Hoff, Effect of telomere and telomerase interactive agents on human tumor and normal cell lines. Clin. Cancer Res. 6, 987–993 (2000). [PubMed] [Google Scholar]
- 12.C. M. Koh, E. Khattar, S. C. Leow, C. Y. Liu, J. Muller, W. X. Ang, Y. Li, G. Franzoso, S. Li, E. Guccione, V. Tergaonkar, Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Invest. 125, 2109–2122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Ghosh, G. Saginc, S. C. Leow, E. Khattar, E. M. Shin, T. D. Yan, M. Wong, Z. Zhang, G. Li, W.-K. Sung, J. Zhou, W. J. Chng, S. Li, E. Liu, V. Tergaonkar, Telomerase directly regulates NF-κB-dependent transcription. Nat. Cell Biol. 14, 1270–1281 (2012). [DOI] [PubMed] [Google Scholar]
- 14.C. Hu, Z. Ni, B. Li, X. Yong, X. Yang, J. Zhang, D. Zhang, Y. Qin, M. Jie, H. Dong, S. Li, F. He, S. Yang, hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut 66, 31–42 (2017). [DOI] [PubMed] [Google Scholar]
- 15.E. Khattar, P. Kumar, C. Y. Liu, S. C. Akıncılar, A. Raju, M. Lakshmanan, J. J. P. Maury, Y. Qiang, S. Li, E. Y. Tan, K. M. Hui, M. Shi, Y. H. Loh, V. Tergaonkar, Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 126, 4045–4060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O. G. Gonzalez, R. Assfalg, S. Koch, A. Schelling, J. K. Meena, J. Kraus, A. Lechel, S.-F. Katz, V. Benes, K. Scharffetter-Kochanek, H. A. Kestler, C. Günes, S. Iben, Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat. Commun. 5, 4599 (2014). [DOI] [PubMed] [Google Scholar]
- 17.J. D. Henson, A. A. Neumann, T. R. Yeager, R. R. Reddel, Alternative lengthening of telomeres in mammalian cells. Oncogene 21, 598–610 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Z. Ding, C.-J. Wu, M. Jaskelioff, E. Ivanova, M. Kost-Alimova, A. Protopopov, G. C. Chu, G. Wang, X. Lu, E. S. Labrot, J. Hu, W. Wang, Y. Xiao, H. Zhang, J. Zhang, J. Zhang, B. Gan, S. R. Perry, S. Jiang, L. Li, J. W. Horner, Y. A. Wang, L. Chin, R. A. DePinho, Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148, 896–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. A. Strong, S. L. Vidal-Cardenas, B. Karim, H. Yu, N. Guo, C. W. Greider, Phenotypes in mTERT+/− and mTERT−/− mice are due to short telomeres, not telomere-independent functions of telomerase reverse transcriptase. Mol. Cell. Biol. 31, 2369–2379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.E. González-Suárez, E. Samper, A. Ramírez, J. M. Flores, J. Martín-Caballero, J. L. Jorcano, M. A. Blasco, Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 20, 2619–2630 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.I. Landa, T. Ibrahimpasic, L. Boucai, R. Sinha, J. A. Knauf, R. H. Shah, S. Dogan, J. C. Ricarte-Filho, G. P. Krishnamoorthy, B. Xu, N. Schultz, M. F. Berger, C. Sander, B. S. Taylor, R. Ghossein, I. Ganly, J. A. Fagin, Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J. Clin. Invest. 126, 1052–1066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. Jonsson, A. L. Lin, R. J. Young, N. M. DiStefano, D. M. Hyman, B. T. Li, M. F. Berger, A. Zehir, M. Ladanyi, D. B. Solit, A. G. Arnold, Z. K. Stadler, D. Mandelker, M. E. Goldberg, J. Chmielecki, M. Pourmaleki, S. Q. Ogilvie, S. S. Chavan, A. T. McKeown, M. Manne, A. Hyde, K. Beal, T. J. Yang, C. P. Nolan, E. Pentsova, A. Omuro, I. T. Gavrilovic, T. J. Kaley, E. L. Diamond, J. B. Stone, C. Grommes, A. Boire, M. Daras, A. F. Piotrowski, A. M. Miller, P. H. Gutin, T. A. Chan, V. S. Tabar, C. W. Brennan, M. Rosenblum, L. M. DeAngelis, I. K. Mellinghoff, B. S. Taylor, Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin. Cancer Res. 25, 5537–5547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. Zehir, R. Benayed, R. H. Shah, A. Syed, S. Middha, H. R. Kim, P. Srinivasan, J. Gao, D. Chakravarty, S. M. Devlin, M. D. Hellmann, D. A. Barron, A. M. Schram, M. Hameed, S. Dogan, D. S. Ross, J. F. Hechtman, D. F. DeLair, J. Yao, D. L. Mandelker, D. T. Cheng, R. Chandramohan, A. S. Mohanty, R. N. Ptashkin, G. Jayakumaran, M. Prasad, M. H. Syed, A. B. Rema, Z. Y. Liu, K. Nafa, L. Borsu, J. Sadowska, J. Casanova, R. Bacares, I. J. Kiecka, A. Razumova, J. B. Son, L. Stewart, T. Baldi, K. A. Mullaney, H. Al-Ahmadie, E. Vakiani, A. A. Abeshouse, A. V. Penson, P. Jonsson, N. Camacho, M. T. Chang, H. H. Won, B. E. Gross, R. Kundra, Z. J. Heins, H.-W. Chen, S. Phillips, H. Zhang, J. Wang, A. Ochoa, J. Wills, M. Eubank, S. B. Thomas, S. M. Gardos, D. N. Reales, J. Galle, R. Durany, R. Cambria, W. Abida, A. Cercek, D. R. Feldman, M. M. Gounder, A. A. Hakimi, J. J. Harding, G. Iyer, Y. Y. Janjigian, E. J. Jordan, C. M. Kelly, M. A. Lowery, L. G. T. Morris, A. M. Omuro, N. Raj, P. Razavi, A. N. Shoushtari, N. Shukla, T. E. Soumerai, A. M. Varghese, R. Yaeger, J. Coleman, B. Bochner, G. J. Riely, L. B. Saltz, H. I. Scher, P. J. Sabbatini, M. E. Robson, D. S. Klimstra, B. S. Taylor, J. Baselga, N. Schultz, D. M. Hyman, M. E. Arcila, D. B. Solit, M. Ladanyi, M. F. Berger, Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Ghandi, F. W. Huang, J. Jané-Valbuena, G. V. Kryukov, C. C. Lo, E. R. McDonald, J. Barretina, E. T. Gelfand, C. M. Bielski, H. Li, K. Hu, A. Y. Andreev-Drakhlin, J. Kim, J. M. Hess, B. J. Haas, F. Aguet, B. A. Weir, M. V. Rothberg, B. R. Paolella, M. S. Lawrence, R. Akbani, Y. Lu, H. L. Tiv, P. C. Gokhale, A. de Weck, A. A. Mansour, C. Oh, J. Shih, K. Hadi, Y. Rosen, J. Bistline, K. Venkatesan, A. Reddy, D. Sonkin, M. Liu, J. Lehar, J. M. Korn, D. A. Porter, M. D. Jones, J. Golji, G. Caponigro, J. E. Taylor, C. M. Dunning, A. L. Creech, A. C. Warren, J. M. McFarland, M. Zamanighomi, A. Kauffmann, N. Stransky, M. Imielinski, Y. E. Maruvka, A. D. Cherniack, A. Tsherniak, F. Vazquez, J. D. Jaffe, A. A. Lane, D. M. Weinstock, C. M. Johannessen, M. P. Morrissey, F. Stegmeier, R. Schlegel, W. C. Hahn, G. Getz, G. B. Mills, J. S. Boehm, T. R. Golub, L. A. Garraway, W. R. Sellers, Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.P.-C. Yu, L.-C. Tan, X.-L. Zhu, X. Shi, R. Chernikov, A. Semenov, L. Zhang, B. Ma, Y. Wang, X.-Y. Zhou, Q.-H. Ji, W.-J. Wei, Y.-L. Wang, Arms-qPCR improves detection sensitivity of earlier diagnosis of papillary thyroid cancers with worse prognosis determined by coexisting BRAF V600E and tert promoter mutations. Endocr. Pract. 27, 698–705 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network , Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. C. Cheung, S. Ezzat, J. L. Freeman, I. B. Rosen, S. L. Asa, Immunohistochemical diagnosis of papillary thyroid carcinoma. Mod. Pathol. 14, 338–342 (2001). [DOI] [PubMed] [Google Scholar]
- 28.R. D. Chernock, Immunohistochemistry of thyroid gland carcinomas: Clinical utility and diagnostic pitfalls. Diagn. Histopathol. 22, 184–190 (2016). [Google Scholar]
- 29.N. Besic, B. Gazic, Sites of metastases of anaplastic thyroid carcinoma: Autopsy findings in 45 cases from a single institution. Thyroid 23, 709–713 (2013). [DOI] [PubMed] [Google Scholar]
- 30.A. Liberzon, C. Birger, H. Thorvaldsdóttir, M. Ghandi, J. P. Mesirov, P. Tamayo, The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.W. Pu, X. Shi, P. Yu, M. Zhang, Z. Liu, L. Tan, P. Han, Y. Wang, D. Ji, H. Gan, W. Wei, Z. Lu, N. Qu, J. Hu, X. Hu, Z. Luo, H. Li, Q. Ji, J. Wang, X. Zhang, Y.-L. Wang, Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat. Commun. 12, 6058 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.V. Y. Stefanovsky, G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, T. Moss, An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8, 1063–1073 (2001). [DOI] [PubMed] [Google Scholar]
- 33.R. Y. Ebright, S. Lee, B. S. Wittner, K. L. Niederhoffer, B. T. Nicholson, A. Bardia, S. Truesdell, D. F. Wiley, B. Wesley, S. Li, A. Mai, N. Aceto, N. Vincent-Jordan, A. Szabolcs, B. Chirn, J. Kreuzer, V. Comaills, M. Kalinich, W. Haas, D. T. Ting, M. Toner, S. Vasudevan, D. A. Haber, S. Maheswaran, D. S. Micalizzi, Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Pelletier, G. Thomas, S. Volarević, Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63 (2018). [DOI] [PubMed] [Google Scholar]
- 35.R. Martin, P. Hackert, M. Ruprecht, S. Simm, L. Brüning, O. Mirus, K. E. Sloan, G. Kudla, E. Schleiff, M. T. Bohnsack, A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 20, 1173–1182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. N. Guerrieri, F. Zacchini, C. Onofrillo, S. Di Viggiano, M. Penzo, A. Ansuini, I. Gandin, Y. Nobe, M. Taoka, T. Isobe, D. Treré, L. Montanaro, DKC1 overexpression induces a more aggressive cellular behavior and increases intrinsic ribosomal activity in immortalized mammary gland cells. Cancer 12, 3512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Cheng, B. Yuan, S. Ying, C. Niu, H. Mai, X. Guan, X. Yang, Y. Teng, J. Lin, J. Huang, R. Jin, J. Wu, B. Liu, S. Chang, E. Wang, C. Zhang, N. Hou, X. Cheng, D. Xu, X. Yang, S. Gao, Q. Ye, PES1 is a critical component of telomerase assembly and regulates cellular senescence. Sci. Adv. 5, eaav1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Y. R. Lapik, C. J. Fernandes, L. F. Lau, D. G. Pestov, Physical and functional interaction between pes1 and bop1 in mammalian ribosome biogenesis. Mol. Cell 15, 17–29 (2004). [DOI] [PubMed] [Google Scholar]
- 39.D. Ruggero, S. Grisendi, F. Piazza, E. Rego, F. Mari, P. H. Rao, C. Cordon-Cardo, P. P. Pandolfi, Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299, 259–262 (2003). [DOI] [PubMed] [Google Scholar]
- 40.W. Chen, J. Bucaria, D. A. Band, A. Sutton, R. Sternglanz, Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31, 690–699 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. X. T. Nguyen, B. S. Mitchell, Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc. Natl. Acad. Sci. U.S.A. 110, 20681–20686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V. Iadevaia, R. Liu, C. G. Proud, mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 36, 113–120 (2014). [DOI] [PubMed] [Google Scholar]
- 43.W. Yang, J. Soares, P. Greninger, E. J. Edelman, H. Lightfoot, S. Forbes, N. Bindal, D. Beare, J. A. Smith, I. R. Thompson, S. Ramaswamy, P. A. Futreal, D. A. Haber, M. R. Stratton, C. Benes, U. McDermott, M. J. Garnett, Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. Sanij, K. M. Hannan, J. Xuan, S. Yan, J. E. Ahern, A. S. Trigos, N. Brajanovski, J. Son, K. T. Chan, O. Kondrashova, E. Lieschke, M. J. Wakefield, D. Frank, S. Ellis, C. Cullinane, J. Kang, G. Poortinga, P. Nag, A. J. Deans, K. K. Khanna, L. Mileshkin, G. A. McArthur, J. Soong, E. M. J. J. Berns, R. D. Hannan, C. L. Scott, K. E. Sheppard, R. B. Pearson, CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 11, 2641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. Xu, M. Di Antonio, S. McKinney, V. Mathew, B. Ho, N. J. O’Neil, N. D. Santos, J. Silvester, V. Wei, J. Garcia, F. Kabeer, D. Lai, P. Soriano, J. Banáth, D. S. Chiu, D. Yap, D. D. Le, F. B. Ye, A. Zhang, K. Thu, J. Soong, S.-C. Lin, A. H. C. Tsai, T. Osako, T. Algara, D. N. Saunders, J. Wong, J. Xian, M. B. Bally, J. D. Brenton, G. W. Brown, S. P. Shah, D. Cescon, T. W. Mak, C. Caldas, P. C. Stirling, P. Hieter, S. Balasubramanian, S. Aparicio, CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 8, 14432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R. Cornelison, K. Biswas, D. C. Llaneza, A. R. Harris, N. G. Sosale, M. J. Lazzara, C. N. Landen, CX-5461 treatment leads to cytosolic DNA-mediated STING activation in ovarian cancer. Cancer 13, 5056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D. Drygin, A. Lin, J. Bliesath, C. B. Ho, S. E. O’Brien, C. Proffitt, M. Omori, M. Haddach, M. K. Schwaebe, A. Siddiqui-Jain, N. Streiner, J. E. Quin, E. Sanij, M. J. Bywater, R. D. Hannan, D. Ryckman, K. Anderes, W. G. Rice, Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71, 1418–1430 (2011). [DOI] [PubMed] [Google Scholar]
- 48.M. Haddach, M. K. Schwaebe, J. Michaux, J. Nagasawa, S. E. O’Brien, J. P. Whitten, F. Pierre, P. Kerdoncuff, L. Darjania, R. Stansfield, D. Drygin, K. Anderes, C. Proffitt, J. Bliesath, A. Siddiqui-Jain, M. Omori, N. Huser, W. G. Rice, D. M. Ryckman, Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 3, 602–606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.B. R. Haugen, E. K. Alexander, K. C. Bible, G. M. Doherty, S. J. Mandel, Y. E. Nikiforov, F. Pacini, G. W. Randolph, A. M. Sawka, M. Schlumberger, K. G. Schuff, S. I. Sherman, J. A. Sosa, D. L. Steward, R. M. Tuttle, L. Wartofsky, 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid 26, 1–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L. Fugazzola, R. Elisei, D. Fuhrer, B. Jarzab, S. Leboulleux, K. Newbold, J. Smit, 2019 European thyroid association guidelines for the treatment and follow-up of advanced radioiodine-refractory thyroid cancer. Eur. Thyroid J. 8, 227–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. Sugitani, Y. Ito, D. Takeuchi, H. Nakayama, C. Masaki, H. Shindo, M. Teshima, K. Horiguchi, Y. Yoshida, T. Kanai, M. Hirokawa, K. Y. Hames, I. Tabei, A. Miyauchi, Indications and strategy for active surveillance of adult low-risk papillary thyroid microcarcinoma: Consensus statements from the japan association of endocrine surgery task force on management for papillary thyroid microcarcinoma. Thyroid 31, 183–192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.L. P. Fernández, A. López-Márquez, P. Santisteban, Thyroid transcription factors in development, differentiation and disease. Nat. Rev. Endocrinol. 11, 29–42 (2015). [DOI] [PubMed] [Google Scholar]
- 53.J. A. Knauf, M. A. Sartor, M. Medvedovic, E. Lundsmith, M. Ryder, M. Salzano, Y. E. Nikiforov, T. J. Giordano, R. A. Ghossein, J. A. Fagin, Progression of BRAF-induced thyroid cancer is associated with epithelial-mesenchymal transition requiring concomitant MAP kinase and TGFβ signaling. Oncogene 30, 3153–3162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. G. McFadden, A. Vernon, P. M. Santiago, R. Martinez-McFaline, A. Bhutkar, D. M. Crowley, M. McMahon, P. M. Sadow, T. Jacks, p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc. Natl. Acad. Sci. U.S.A. 111, E1600–E1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.I. Landa, I. Ganly, T. A. Chan, N. Mitsutake, M. Matsuse, T. Ibrahimpasic, R. A. Ghossein, J. A. Fagin, Frequent somatic TERT promoter mutations in thyroid cancer: Higher prevalence in advanced forms of the disease. J. Clin. Endocrinol. Metab. 98, E1562–E1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.I. Landa, C. E. Thornton, B. Xu, J. Haase, G. P. Krishnamoorthy, J. Hao, J. A. Knauf, Z. T. Herbert, M. A. Blasco, R. Ghossein, J. A. Fagin, Telomerase reactivation induces progression of mouse BrafV600E-driven thyroid cancers without telomere lengthening. bioRxiv 2023.01.24.525280 [Preprint]. 24 January 2023. 10.1101/2023.01.24.525280. [DOI]
- 57.Q. Zhang, N. A. Shalaby, M. Buszczak, Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343, 298–301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Panda, A. Yadav, H. Yeerna, A. Singh, M. Biehl, M. Lux, A. Schulz, T. Klecha, S. Doniach, H. Khiabanian, S. Ganesan, P. Tamayo, G. Bhanot, Tissue- and development-stage-specific mRNA and heterogeneous CNV signatures of human ribosomal proteins in normal and cancer samples. Nucleic Acids Res. 48, 7079–7098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. T. Jeker, M. Hejazi, C. L. Burek, N. R. Rose, P. Caturegli, Mouse thyroid primary culture. Biochem. Biophys. Res. Commun. 257, 511–515 (1999). [DOI] [PubMed] [Google Scholar]
- 60.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.L. van der Maaten, G. Hinton, Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 63.A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.A. Liberzon, A. Subramanian, R. Pinchback, H. Thorvaldsdóttir, P. Tamayo, J. P. Mesirov, Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.E. K. Schmidt, G. Clavarino, M. Ceppi, P. Pierre, SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Y. Ge, X. Zhang, W. Liang, C. Tang, D. Gu, J. Shi, X. Wei, OncoVee-MiniPDX-guided anticancer treatment for gastric cancer patients with synchronous liver metastases: A retrospective cohort analysis. Front. Oncol. 11, 757383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.J. Wang, J. Huang, H. Wang, W. Yang, Q. Bai, Z. Yao, Q. Li, H. Lv, B. Chen, C. Nie, W. Xu, S. Tu, H. Li, X. Chen, Personalized treatment of advanced gastric cancer guided by the MiniPDX model. J. Oncol. 2022, 1987705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.F. Zhang, W. Wang, Y. Long, H. Liu, J. Cheng, L. Guo, R. Li, C. Meng, S. Yu, Q. Zhao, S. Lu, L. Wang, H. Wang, D. Wen, Characterization of drug responses of mini patient-derived xenografts in mice for predicting cancer patient clinical therapeutic response. Cancer Commun. Lond. Engl. 38, 60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.T. Chen, X. Chen, S. Zhang, J. Zhu, B. Tang, A. Wang, L. Dong, Z. Zhang, C. Yu, Y. Sun, L. Chi, H. Chen, S. Zhai, Y. Sun, L. Lan, X. Zhang, J. Xiao, Y. Bao, Y. Wang, Z. Zhang, W. Zhao, The genome sequence archive family: Toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 19, 578–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.CNCB-NGDC Members and Partners , Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 50, D27–D38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for tables S1 to S4
Tables S1 to S4