Abstract
The basidiomycete Moniliophthora roreri causes frosty pod rot of cacao (Theobroma cacao) in the western hemisphere. Moniliophthora roreri is considered asexual and haploid throughout its hemibiotrophic life cycle. To understand the processes driving genome modification, using long-read sequencing technology, we sequenced and assembled 5 high-quality M. roreri genomes out of a collection of 99 isolates collected throughout the pathogen's range. We obtained chromosome-scale assemblies composed of 11 scaffolds. We used short-read technology to sequence the genomes of 22 similarly chosen isolates. Alignments among the 5 reference assemblies revealed inversions, translocations, and duplications between and within scaffolds. Isolates at the front of the pathogens' expanding range tend to share lineage-specific structural variants, as confirmed by short-read sequencing. We identified, for the first time, 3 new mating type A locus alleles (5 in total) and 1 new potential mating type B locus allele (3 in total). Currently, only 2 mating type combinations, A1B1 and A2B2, are known to exist outside of Colombia. A systematic survey of the M. roreri transcriptome across 2 isolates identified an expanded candidate effector pool and provided evidence that effector candidate genes unique to the Moniliophthoras are preferentially expressed during the biotrophic phase of disease. Notably, M. roreri isolates in Costa Rica carry a chromosome segment duplication that has doubled the associated gene complement and includes secreted proteins and candidate effectors. Clonal reproduction of the haploid M. roreri genome has allowed lineages with unique genome structures and compositions to dominate as it expands its range, displaying a significant founder effect.
Keywords: cacao frosty pod, genome evolution, mating type loci, pathogenomics, Theobroma cacao
Introduction
The basidiomycete Moniliophthora roreri (Mr; causal agent of frosty pod rot) is a major pathogen of cacao (Theobroma cacao) originating in South America. Moniliophthora roreri is a hemibiotroph with an extended biotrophic phase of 45 days or longer (Evans 2016a; Bailey, Evans et al. 2018) and is considered an asexual haploid organism (Evans 2016a). Pod malformation may be the only external symptom of infection during the biotrophic phase. After the biotrophic phase, rapid necrosis occurs, leading to the formation of masses of rhexolytic thallic conidia on the surface of the infected pods (Díaz-Valderrama and Aime 2016b). A single infected cacao pod is capable of producing as many as 7 billion spores (Campuzano Londoño 1976). Spores are spread by wind and rain and, if unrestricted, infect and severely reduce yields. The life cycle of M. roreri is contrasted with that of its sister species, Moniliophthora perniciosa. Moniliophthora perniciosa biotype C is a sexual (homothallic) hemibiotroph causing the disease witches' broom of cacao, also originating in South America. Moniliophthora perniciosa produces basidiocarps, the resulting spores of which infect diverse meristematic tissues including pods, flower cushions, and branches (Evans 2016b).
Genetic recombination among M. roreri isolates is limited due to the apparent lack of a sexual cycle (Ali et al. 2015; Díaz-Valderrama and Aime 2016a; Bailey, Ali et al. 2018). As a result, genetic change in M. perniciosa is expected to be driven by mutations and chromosome rearrangements that are clonally replicated in subsequent generations. Due to its clonal reproduction, M. roreri displays limited diversity at the front of its expanding range in the western hemisphere and is strongly influenced by founder effects as it continues to spread. For example, the nations in Central America have been colonized by genetically similar M. roreri isolates following primarily a single line of descent (Phillips-Mora 2003). These isolates share similar SNP (Ali et al. 2015) and SSR (Díaz-Valderrama and Aime 2016a) profiles in addition to carrying A1B1 mating type alleles (Díaz-Valderrama et al. 2022). Though asexual, M. roreri carries the A and B loci required for tetrapolar mating, but only 2 allelic combinations (A1B1 and A2B2) were identified (Díaz-Valderrama et al. 2022).
We have a reasonable understanding of M. roreri genetic diversity, distribution, and the timing of its spread into new geographic regions (Evans 2016a); however, further investigation is needed to deepen our understanding of the species evolution as it expands to new regions in the absence of sexual reproduction. Our objectives include the assembly of chromosome-scale genomes for multiple genetically and geographically diverse isolates of M. roreri, the structural comparisons of these genomes within the species, and the characterization of processes driving genome modifications and adaptations associated with geographic dispersion. Better genome assemblies will be a fundamental resource of information to expand our understanding of M. roreri, in particular the impact of clonal propagation on genome evolution. Its clonal or near clonal reproduction habits, when combined with recent geographic expansion, allow for potential independent genome evolution due to unique genome structural modifications and mutations. This analysis will also help us gauge M. roreri's potential for change and adaptation to parallel changes in their cacao host as a result of the selection and release of new breeding materials carrying tolerance to infection by M. roreri.
Materials and methods
M. roreri (Mr) and M. perniciosa (Mp) isolates
Most of the M. roreri isolates used in this study were from a collection held by the United States Department of Agriculture (USDA), Beltsville, USA. These isolates were collected from T. cacao fruit in affected areas of South and Central America between 1999 and 2013 (Supplementary Table 1). A SNP study (Ali et al. 2015) separated the diversity found in these isolates into 37 distinct genotypes including 14 “synonymous groups” with multiple members showing identical SNP profiles. Three isolates were collected more recently (2019–2020) from Ecuador (Supplementary Table 1). Isolates were selected for sequencing based on the original geographic location (i.e. represent all regions) and the genetic distance among isolates (i.e. maximize genetic diversity) (Ali et al. 2015). This resulted in isolates from 5 countries (2 from Costa Rica, 14 from Colombia, 4 from Ecuador, 1 from Peru, and 1 from Bolivia) and 8 of 14 SNP synonymous groups being included (Ali et al. 2015). The M. roreri cultures were isolated and maintained as previously described (Ali et al. 2015). Moniliophthora perniciosa isolate MCCS1 was collected in Bahia, Brazil, and maintained there in a similar manner.
Isolation of genomic DNA for short-read and single-molecule real-time (SMRT) sequencing
For DNA extraction, M. roreri and M. perniciosa isolates were grown for 7 days on 20% clarified V8 agar (CV8). From the different cultures, 2–3 agar plugs (0.25 cm2) were transferred to 50-mL falcon tubes containing 20 mL liquid CV8 and grown at 25°C while shaking at 100 rpm. Cultures were incubated for 5 days and homogenized using a Janke & Kunkel Ultra Turrax T25 (IKA, Staufen, Germany) for 30–60 s. The homogenized mycelia (5 ml) were used as starter culture and added to 50 mL liquid CV8 in a 250-mL flask. Mycelia of each isolate were produced by growth at 25°C while shaking at 100 rpm for 7–10 days. The mycelia were washed with sterile water and collected by centrifuging at 20,000 g for 10 min, followed by flash freezing in liquid nitrogen and freeze-dried. Genomic DNA (gDNA) for the short-read and high molecular weight gDNA for SMRT sequencing were extracted from freeze-dried mycelia as previously described in Morales-Cruz et al. (2020). The gDNA concentrations were quantified with a Qubit 3.0 fluorometer using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA). The quality of the extracted gDNA was assessed using a NanoDrop spectrophotometer and 1% (w/v) agarose gel. The high molecular weight gDNA was run on a 0.75% pippin pulse (Sage Science, USA) gel to examine the integrity and molecular weight.
Whole genome library preparations
A total of 22 M. roreri isolates from South and Central America (Supplementary Table 1) were subjected to whole genome Illumina sequencing. A total of 1 μg DNA per sample was used as the input material. DNA-seq libraries were prepared using the Kapa LTP library prep kit (Kapa Biosystems, MA, USA). Libraries were evaluated for quantity and quality with the High Sensitivity chip on a Bioanalyzer 2100 (Agilent Technologies, CA, USA) and sequenced in paired-end 150 bp reads on an Illumina HiSeq4000 (Novogene Corporation Inc, Sacramento, CA, USA).
The genome of M. perniciosa MCCS1 and 5 of the M. roreri isolates were subjected to SMRT sequencing (Table 1 and Supplementary Table 2). High molecular weight gDNA was cleaned with 0.45 × AMPure PB beads (Pacific Biosciences, Menlo Park, CA, USA) before library preparation. SMRTbell template was prepared with 15 µg of sheared DNA using SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer's instructions. SMRTbell template was size-selected using the BluePippin instrument (Sage Science, Beverly, MA, USA) using a cutoff size of 17–50 Kbp. The size-selected library was cleaned with 1 × AMPure PB beads followed by a DNA damage repair treatment and cleaned again with 1 × AMPure PB beads. One SMRTbell library was produced for each of the 5 genotypes, which were sequenced on separate SMRT cells on the PacBio Sequel II platform (DNA Technology Core Facility, University of California, Davis).
Table 1.
Genome sequencing, assembly, and annotation statistics for the 5 M. roreri isolates (MrB3, MrCo8, MrCo84, MrE7, and MrC26) and the M. perniciosa isolate (Mp-MCCS1) sequenced using long-read technology sequencing.
| Metrics | MrB3 | MrCo8 | MrCo84 | MrE7 | MrC26 | Mp-MCCS1 |
|---|---|---|---|---|---|---|
| Sequencing statistics | ||||||
| Sequencing data (Gbp) | 129 | 15 | 140 | 79 | 9 | 128 |
| Coverage (X-fold) | 2,144× | 242× | 2,341× | 1,317× | 156× | 2,600× |
| Assembly statistics | ||||||
| Cumulative length (Mbp) | 57.48 | 58.18 | 56.81 | 57.39 | 59.57 | 48.97 |
| Number of contigs | 11 | 11 | 13 | 21 | 11 | 12 |
| Average sequence length (Mbp) | 5.22 | 5.29 | 4.37 | 2.73 | 5.42 | 4.08 |
| Maximum sequence length (Mbp) | 10.74 | 10.4 | 10.41 | 10.25 | 10.45 | 9.61 |
| N50 length (Mbp) | 5.18 | 5.19 | 4.88 | 4.88 | 5.17 | 5.46 |
| N90 length (Mbp) | 3.46 | 3.58 | 3.61 | 3.49 | 3.58 | 3.55 |
| Annotation statistics | ||||||
| Number of genes | 20,704 | 21,007 | 20,409 | 20,539 | 21,460 | 17,726 |
| Repetitive content | 17.84% | 17.59% | 17.51% | 17.90% | 18.89% | 10.45% |
Isolate MrC26 was also sequenced with additional long-read sequencing using Oxford Nanopore technology. High molecular weight gDNA was cleaned, and libraries were produced (Oxford Nanopore, Oxford, UK) and sequenced using Nanopore MinION technologies. The run produced a total of 2.05 Gbp (∼35× X-fold coverage) in 787,832 reads with an N50 of 6.1 Kbp.
Genome assembly
Genome size was estimated by k-mer content analysis of short-read libraries. K-mer count and frequency were calculated with Jellyfish (ver. 2.2.7, Marçais and Kingsford 2011) using a k-mer length of 21 bp. Genome haploid length and heterozygosity rates were estimated with GenomeScope (ver. 2.0, Ranallo-Benavidez et al. 2020) by fitting the k-mer distribution to a mixture model of 4 evenly spaced negative binomial distributions to the k-mer profile.
PacBio reads sequenced for MrC26, MrB3, MrCO84, MrCO8, MrE7, and M. perniciosa MCCS1 genotypes were assembled separately using Falcon (ver. 2017.06.28–18.01, Chin et al. 2016) assembler testing multiple parameter combinations to achieve the lowest fragmentation. The procedure took advantage of repetitive content identification in both raw and error corrected reads as described in Minio et al. (2019). Assembled contigs were then polished using Arrow (from ver. GCpp 1.0.0-cd36561, “https://github.com/PacificBiosciences/gcpp”). MrC26 and Mp-MCCS1 underwent a scaffolding procedure with SSPACE-LongRead (ver. 1.0, Boetzer and Pirovano 2014). MrC26 scaffold structure was then validated with independent 10 × using Tigmint (ver. 1.2.4, Jackman et al. 2018) and NanoPore sequencing data assembled using CANU (ver. 1.8–14, Koren et al. 2017), Miniasm (ver. 0.3-r179, Li 2016), Shasta (ver. 0.3.0, Shafin et al. 2020), Flye (ver. 2.6-g0d65569, Lin et al. 2016; Kolmogorov et al. 2019), RA (ver. 0.2.1, “https://github.com/lbcb-sci/ra”), and wtdbg2 (ver. 2.3, Ruan and Li 2019). The structure of the scaffolds was then compared with the contigs of the other genotypes to identify syntenic groups of sequences. Pairwise comparisons of all assemblies were performed by aligning the scaffolds of contigs sequences with nucmer (MUMMER v4, ver. 4.0.0beta5, Marçais et al. 2018), local hits were then inserted in an adjacency network, and the highest coverage tiling paths of nonoverlapping hits were identified between all pairs of sequences. Each sequence of each draft assembly was then assigned to a single homolog sequence in each other assembly in a 1-to-1 relationship selecting the overall best of the tiling paths. Pairwise relationships were used to identify syntenic groups and, when possible, draft sequences belonging to the same pseudomolecule as assembled in a single contig in a different genotype.
Illumina reads of each genotype underwent clipping of adapters and quality trimming with Trimmomatic (ver. 0.36, parameters: “ILLUMINACLIP:2:30:10 LEADING:7 TRAILING:7 SLIDINGWINDOW:10:20 MINLEN:36,” Bolger et al. 2014) assembled independently using SPAdes (ver. 3.13.0 edited to allow k-mers up to 251, Bankevich et al. 2012). Different k-mer combinations were tested, and a multiple k-mer set of 105, 115, 125, 135, and 145 was then used to produce the assemblies for each genotype. Assembly completeness was evaluated using BUSCO (v.5, Agaricales odb10 database, Supplementary Table 3, Simão et al. 2015).
Error rates and heterozygosity estimation were performed by calling variants between each genome assembled with SMRT reads and Illumina reads produced for the very same genotype. Short reads were aligned using BWA MEM (ver. 0.7.17, Li 2013) and randomly downsampled to 200 × of coverage with Samtools (ver. 1.9, Li et al. 2009). Alignments were then postprocessed with Picard tools (ver. 2.12.1-SNAPSHOT, Broad institute) to assign reads groups and mark duplicates and variants called. Variant calling was then performed using GATK HaplotypeCaller (ver. 4.0.12.0, parameters: “–ERC GVCF –base-quality-score-threshold 20 –sample-ploidy 2,” Poplin et al. 2017) and genotyped with GenotypeGVCFs (parameters: “–include-non-variant-sites true –sample-ploidy 2”).
To establish phylogenetic relationships among the M. roreri strains, variant sites were collected from nonrepetitive regions of the genome where all samples were genotyped with a depth of coverage 5 ≤ dp ≤ 1000. Phylogeny was calculated with RAxML-NG (ver. 0.9.0, parameters: “–model GTR + G + FO –tree pars{5} –bs-trees 100 –seed 12345,” Kozlov et al. 2019), and tree graph was then produced with FigTree (ver. 1.4.4, “http://tree.bio.ed.ac.uk/software/figtree/”).
Generation of new and acquisition of existing RNA-Seq data
The M. roreri RNA-Seq data used were derived from 3 sources: mycelia grown in vitro (this study), artificially inoculated cacao pods (Meinhardt et al. 2014), and naturally infected cacao pods (Bailey et al. 2014).
For RNA extraction from mycelia, M. roreri isolate C26 was grown as mentioned above. RNA from freeze-dried mycelia were extracted following a described earlier procedure (Bailey et al. 2013). RNA purity was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Hanover Park, IL). The RNA quantity was determined using the RNA broad range kit of the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA). RNA integrity was determined by electrophoresis using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA). Total RNA (300 ng, RNA integrity number > 8.0) was used for cDNA synthesis and library construction. Three RNA-Seq libraries were prepared using the Illumina TruSeq RNA sample preparation kit v.2 (Illumina, CA, USA) following Illumina's low-throughput protocol. This library was evaluated for quantity and quality with the High Sensitivity chip in an Agilent 2100 Bioanalyzer (Agilent Technologies, CA) and was sequenced in 150 bp paired-end reads, using an Illumina HiSeq4000 sequencer (Novogene Corporation Inc, Sacramento, CA, USA).
The RNA-Seq data for artificially inoculated cacao pods at 30 days post infection (DPI) and 60 DPI were obtained from a previously published study (Meinhardt et al. 2014). Data for 3 independent biological replicates for each of the time points were included in the analysis (NCBI BioProject PRJNA229176). Information related to the M. roreri infection assay, RNA isolation, and RNA-Seq analysis can be obtained from the previous study (Meinhardt et al. 2014). Based on the typical progression of M. roreri infection, malformed green pods were selected for RNA-Seq at 30 DPI, corresponding to the biotrophic phase. Necrotic sporulating pods were selected for RNA-Seq at 60 DPI to represent the necrotrophic phase.
The third source of RNA-Seq data was previously discussed in Bailey et al. (2014). The dataset comprises 12 independent cacao pods from 4 different cacao clones (3 per clone) naturally infected and representing a range of symptoms.
Moniliophthora perniciosa mycelia were produced using 15-mm disc plugs from a 5-day-old culture grown on potato dextrose agar (PDA) [20% potato (w/v), 0.2% dextrose (w/v), and 1.5% agar (w/v)] which were transferred to 250 mL potato dextrose broth and incubated in the dark for 4 days at 25°C, shaking at 110–125 rpm orbital shaking. Subsequently, 20 mL mycelia culture was homogenized by Polytron probe for 60 and 10 mL transferred to 250 mL potato dextrose broth and incubated as before. Finally, the entire fungus culture was filtered, and the biomass was washed with sterile distilled water, frozen with liquid nitrogen, and lyophilized.
Biomass of basidiocarps was produced as described by Griffith and Hedger (1993) with some modifications proposed by Macagnan et al. (2006). Briefly, preincubation of the fungus was made at 25°C for 15 days on PDA. Afterwards, agar plugs were transferred to autoclaved compacted bran-based solid medium containing 68 g of triturated cacao dry broom, 18.2 g of oat flakes, and 2.75 g of CaSO4 and brought to 100 mL with distilled water. Once produced, basidiocarps were collected and washed with sterile distilled water, frozen with liquid nitrogen, and lyophilized. Basidiospores were collected from the basidiocarps according to the method of Frias et al. (1995) with some modifications. Before fixing the pile on the Petri dish, the basidiocarps were gently disinfected with a 0.1% streptomycin solution, washed with sterile distilled water, and dried on a paper towel. Basidiospore collection was carried out overnight at 25°C, stirring at 110 rpm.
Biomass of green brooms and necrotic brooms was produced after inoculation as described by Sena et al. (2014). Briefly, apical shoot apexes of 4-week-old seedlings from cacao genotype were inoculated with a 30 μl drop of a basidiospore suspension (2 × 105 basidiospores/mL with >80% germination) in 0.3% agar (Surujdeo-Maharaj et al. 2003). After inoculation, 30 days for green broom and 60 days for dry broom, infected branches were washed with distilled water, frozen in liquid nitrogen and lyophilized, and then preserved in vacuum plastic bags.
Gene prediction
Custom repeat libraries were created for M. roreri and M. perniciosa genomes separately with RepeatMasker (ver. open-4.0.6, Smit et al. 2013) and RepeatModeler (ver. open-1.0.11, Smit and Hubley 2015) starting from homologs to known fungal transposable elements present in RepBase (ver. 20160829).
RNA-Seq reads underwent clipping of adapters and quality trimming with Trimmomatic (ver. 0.36, parameters: “ILLUMINACLIP:2:30:10 LEADING:7 TRAILING:7 SLIDINGWINDOW:10:20 MINLEN:36,” Bolger et al. 2014). Samples were assembled de novo separately using Trinity (ver. v2.2.0, Grabherr et al. 2013; Haas et al. 2013) and used as input to PASA (ver. v2.3.3, Haas et al. 2003, 2008, 2011) to produce a nonredundant training set of protein-coding gene models over MrC26 PacBio genome assembly for M. roreri isolates and Mp-MCCS1 for M. perniciosa. The resulting datasets were used in BRAKER (ver. 1.9, Hoff et al. 2016) to obtain ad hoc models for Augustus (ver. 3.0.3, Stanke et al. 2006). Ab initio prediction of protein-coding gene loci was produced for all PacBio and Illumina-based draft assemblies.
For M. roreri isolates, the collected coding sequences (CDS) of all identified models were then searched for transposable elements related proteins by BLASTp (ver. 2.7.1+, Altschul et al. 1990) search against RefSeq database (retrieved 2017_01_17) and clustered in a nonredundant set of transcripts using EvidentialGene (ver. 2014.05.15, Gilbert 2016). The nonredundant set of transcripts was then mapped with blat (ver. 36 × 2, Kent 2002) on the genomic sequences, and hit regions with identity > 80% and coverage > 50% to the original transcript not overlapping annotated loci were then analyzed with TransDecoder (ver. 3.0.1, “https://github.com/TransDecoder/TransDecoder”) to recover missing protein-coding gene annotations and identifying potential pseudogenes.
The proteins annotated on all genotypes were mapped against RefSeq database (retrieved 2017 January 17) using BLASTp (ver. 2.7.1+, Altschul et al. 1990) with coverage > 50% and identity > 50% to assign a functional description based on sequence homology. CAZyme annotation was performed using HMMer (ver. 3.1b2, Eddy 1998, hmmer.org) with dbCAN database (ver. 4, Yin et al. 2012). Secondary metabolites clusters in all genomes were searched using antiSMASH (ver. 6.0.0, Medema et al. 2011; Blin et al. 2019). For PacBio annotated genes, InterProScan was also performed to further annotate the proteins with functional domains and associate Gene Ontology terms in Blast2GO (ver. 5, Conesa et al. 2005).
Structural variants (SVs), SNP, and short InDel identification among M. roreri isolates
To identify SVs across the different isolates, SPAdes de novo assemblies underwent pairwise comparison to MrC26, MrB3, MrCO84, MrCO8, and MrE7 Falcon assembly. Assembled sequences were mapped using nucmer (MUMMER v4, ver. 4.0.0beta5, Marçais et al. 2018) with default parameters, SNPs and short InDels were identified extracted with show-snps (parameters “-Clr -x 1 –T,” MUMMER v4, ver. 4.0.0beta5, Marçais et al. 2018), and SVs were identified with show-diff (MUMMER v4, ver. 4.0.0beta5, Marçais et al. 2018). For each reference genome, all variants identified across all query genotypes were collected to generate a catalog of breakpoints with a tolerance of 50 bp. Shared breakpoints across all genotypes were collected, and the binary distance between them was calculated as per Rogers and Tanimoto (1960) [S6 coefficient of Gower and Legendre, s4 = (a + d)/(a + 2(b + c) +d)] to generated dendrograms.
Gene expression analysis
Filtered and quality trimmed RNA-Seq reads were mapped using Bowtie2 (ver. 2.3.4.1, parameters: “–non-deterministic -k 1 –very-sensitive –t,” Langmead and Salzberg 2012) on a reference transcriptome consisting of MrC26 and Theobroma cacao Matina1–6 genome v1.1 annotated transcripts. Read counts and differential expression analysis were performed in R using GenomicAlignments (v1.18.1, Lawrence et al. 2013) package and differential expression with DESeq2 package (Love et al. 2014). R implementation of Fisher's exact test (Fisher 1935) was used to calculate odd ratios and significance of upregulated genes in the subsets of all secretome genes, predicted effectors, secreted genes unique to M. roreri, secreted genes common and exclusive to M. roreri and M. perniciosa, or secreted genes in Marasmiaceae.
Orthologous groups and protein-based phylogeny
Ortholog gene groups were identified with OrthoFinder (ver. 2.2.7, Emms and Kelly 2015) using as input BLASTp (ver. 2.7.1+, Altschul et al. 1990) protein-to-protein alignments among M. roreri genotypes only and with an outgroup composed of M. perniciosa isolate MCCS1, Fusarium oxysporum forma specialis Lycopersici 4286, Marasmius fiardii PR-910, Rhizoctonia solani AG-1 IA, and Tetrapyrgos nigripes CBS291.85. The species composing the outgroups were selected for increasing distance in the order of M. perniciosa (Marasmiaceae), M. fiardii PR-910 (Marasmiaceae), T. nigripes (Marasmiaceae), R. solani AG-1 IA (Basidiomycota), and F. oxysporum forma specialis Lycopersici 4286 (Ascomycota).
Single-copy ortholog genes were used to build the phylogenetic tree of the selected species. Proteins of each group underwent multiple alignment with MAFFT (ver 7.475, parameters: “–maxitrate 1000 –localpair,” Katoh and Standley 2013). The alignments were concatenated and trimmed with ClipKIT (ver. 1.4.1, Steenwyk et al. 2020). RAxML-NG (ver 0.9.0, cit., parameters: “–model LG + G8 + F –seed 12345 –tree pars{10} –bs-trees 200,” Kozlov et al. 2019) was used to build a phylogenetic tree. FigTree (ver. 1.4.4, “http://tree.bio.ed.ac.uk/software/figtree/”) was used to produce the graphical representation rooted on M. perniciosa MCCS1 sample as an outgroup.
Secretome and effector prediction
Protein-coding sequences annotated in all M. roreri and M. perniciosa isolates were scanned for possible signal peptides using SignalP, version 5.0 (Almagro Armenteros et al. 2019). The amino acid sequences containing predicted signal peptides were scanned for transmembrane proteins using the TMHMM program (Sonnhammer et al. 1998). Proteins with no more than 1 transmembrane domain were considered potential components of the secretome. All candidate proteins were first clustered with CD-HIT (ver. 4.6.8-2017-0621, parameters: “-g 1 -c 0.99 -s 0.5 -aS 0.5 -p 1,” Li and Godzik 2006) to reduce the dataset redundancy, and cluster representatives were then self-mapped with BLASTp (ver. 2.7.1+, Altschul et al. 1990) and underwent Markov clustering with MCL (ver. 14–137, Enright et al. 2002) following methods described by Cantu et al. (2013).
Fungal effectors among the M. roreri secretome were predicted using the machine learning program EffectorP 2.0 and EffectorP 3.0 (Sperschneider et al. 2018; Sperschneider and Dodds 2022). Each secreted protein was searched for the effector motifs [L/I]xAR, [R/K]CxxCx12H, RxLR, [Y/F/W]xC, YxSL[R/K], and G[I/F/Y][A/L/S/T]R following methods described previously (Cantu et al. 2013). Secreted proteins with <150 amino acids long and a cysteine content higher than 3% were cataloged as small and cysteine rich. Each secreted protein was scored based on the EffectorP 2.0 and 3.0 prediction, similar to previously characterized effector proteins, presence of any effector motifs, absence of any annotation, and small cysteine-rich criteria. The scoring was done following methods previously described by Cantu et al. (2013). A combined score was then used to rank the tribes based on their likelihood of containing potential effector proteins.
Mating type gene identification
Nucleotide CDS sequences were aligned to the genomes using GMAP (ver. 2019-09-12, Wu et al. 2016) and blat (ver. 36 × 2, Kent 2002), and protein sequences were compared with the annotated proteins by BLASTp (ver. 2.7.1+, Altschul et al. 1990) and to the genome with exonerate (ver. 2.2.0, Slater and Birney 2005). Concomitant matches were used to identify the annotated loci ascribed to the HD1, HD2, STE3_Mr4, and Mr_Ph4 mating type genes. CDS for Mr_Ph4 could not be found annotated in isolates MrCo58 and MrCo67. The genomic regions surrounding the B locus were manually inspected, and sequences encoding open reading frames corresponding to the Mr_Ph4 sequence position and length were used for MrCo58 and MrCo67.
The CDS of type genes were aligned with the CDS of mating type genes from Díaz-Valderrama and Aime (Díaz-Valderrama and Aime 2016a). Alignments were conducted with MUSCLE (ver. v3.8.31, Edgar 2004) using the nucleotide-coding sequences. Trees were constructed RAxML (ver. v.8.2.12, Stamatakis 2014) using the GTRGAMMA model with 1000 bootstrap replications.
Results
Assembly of long reads reveals M. roreri genome architecture
SMRT sequencing was used to produce genome assemblies for 5 M. roreri isolates from South and Central America (Table 1 and Supplementary Table 2). With an estimated sequencing depth ranging from 156× to 2,341×, the size of the assemblies produced was 57.89 ± 1.06 Mbp (Table 1). This closely matches the estimated genome size of 57.91 ± 1.95 Mbp based on k-mer distribution analysis. The assemblies contained an average of 20,824 ± 420 predicted protein-coding genes, and 18% of the sequence was identified as repetitive. The accuracy of the genome assemblies was confirmed through comparison with short-read sequences. On average, the genomic sequences had an error rate of 0.0197 ± 0.0076%. Heterozygous variants were rarely found in the genomes (<0.00069% of the bases), confirming isolates are haploid. The assemblies were 96.4 ± 0.3% complete according to BUSCO (Supplementary Table 3) and highly contiguous with an average N50 of 5.05 ± 0.17 Mbp. Sequences of each genome were grouped into 11 syntenic groups. A few contigs covering 0.2 and 1.1% of the MrCO84 and MrE7 genomes, respectively, could not be placed in any of the syntenic scaffolds (Table 1). For 6 of the syntenic groups, it was possible to identify telomeric repeats at both extremities of the sequence for at least 1 genotype, suggesting they may correspond to complete chromosomes. For the remaining 5 groups, telomeric repeats were found only at 1 end of the sequence. Twenty-two genetically diverse M. roreri isolates, collected over the pathogen's geographic range and including the 5 isolates sequenced by SMRT technologies (Supplementary Table 1), were also sequenced using short-read Illumina sequencing technologies (Supplementary Table 2). The average assembly size of Illumina reads was 58.03 ± 1.47 Mbp in 4,223 ± 1,058 contigs with an average contig length of 14,389 ± 2,774 bases (Supplementary Table 2). Analysis using BUSCO models indicated that 94.8 ± 0.1% of the genes are complete, indicating the overall comprehensiveness of these assemblies (Supplementary Table 3).
Structural variability across M. roreri isolates
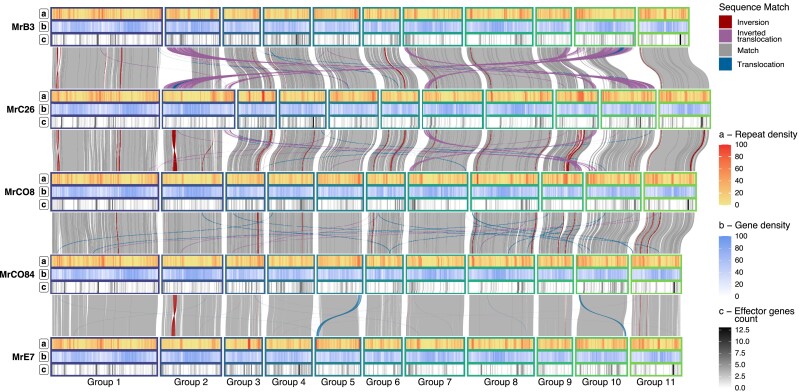
Whole genome alignments of the 5 SMRT assemblies revealed structural rearrangements, primarily involving insertions and duplications (Fig. 1 and Supplementary Fig. 1). On average, variants were found to make up 5.78 ± 1.32 Mbp when comparing each pair of isolates, corresponding to 9.98 ± 2.29% of the total genome assembly (Supplementary Fig. 1a). The MrC26 genome had the highest overall number of variants when compared with the other genotypes, as well as the highest number of genotype-specific variants (Supplementary Fig. 1b). The MrCO84 and MrE7 isolates were the most closely related, with the lowest number of variant bases between them and the highest number of shared SVs when compared with the other isolates (Supplementary Fig. 1a and b).
Fig. 1.
Colinearity between M. roreri isolates syntenic groups. Colinear regions between the 5 isolates have been depicted across the 5 different M. roreri isolates evidencing distant translocation events (blue) and inversions (red). For each of the genotypes, the 3 tracks reported indicate a) the local repeat density in terms of percentage of bases associated to a repetitive element, b) the local gene density in terms of bases ascribed to gene loci, and c) the effector gene density in terms of number of genes in a 100 Kb window.
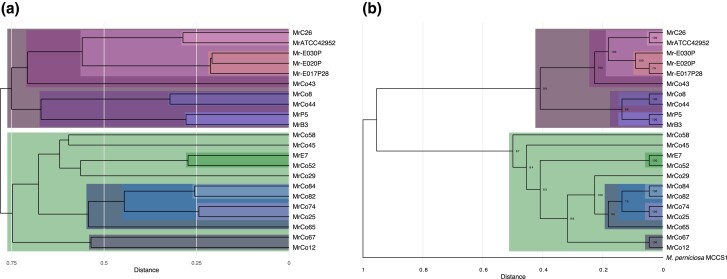
Comparisons of breakpoint/rearrangement patterns between Illumina sequences for 22 isolates and individual PacBio assemblies show that similar SVs are present in independently derived isolates of M. roreri. A distance matrix, based on the presence or absence of SVs, was constructed for all isolates and used to generate dendrograms using each individual SMRT assembly as a reference (Supplementary Fig. 2). This was followed by a combined analysis using breakpoints across all SMRT assemblies (Fig. 2a). These dendrograms show a strong correspondence with the phylogenetic relationships between isolates determined through the multiple alignment of 1,445 single-copy genes (Fig. 2b) and with the synonymous groups identified through SNP analysis of the 22 isolates (Ali et al. 2015). In fact, the Illumina sequences of 6 isolates that were considered unique in the earlier SNP study (Ali et al. 2015) were also identified as unique based on scaffold breakpoints and single gene similarities.
Fig. 2.
Phylogenetic trees relating the M. roreri isolates. Phylogenetic trees describing the relationships present between the M. roreri isolates (A and A) and M. perniciosa (B). Relationships identified with both methods have been highlighted with the same color. a) Tree built on the binary distance between isolates according to the presence of shared breakpoints of the SVs identified between genomes. b) Phylogenetic tree based on amino acid sequence conservation among the single-copy orthologous proteins.
Impact of structural variation on the gene space
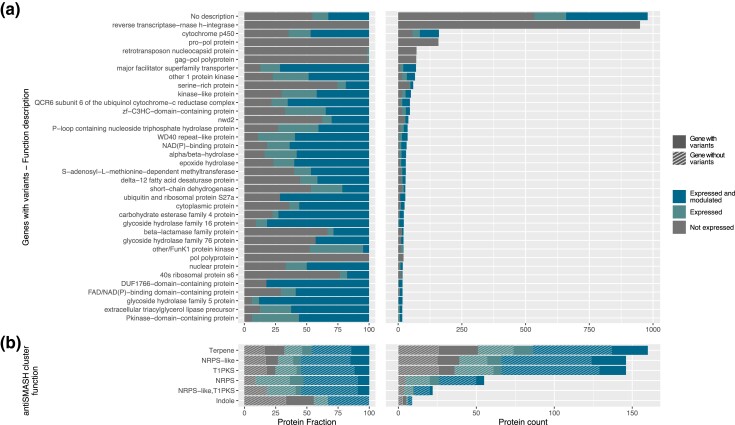
Clustering of the M. roreri proteins annotated in all 22 isolates identified 23,437 orthogroups. Of these clusters, 62.2% (14,576) were conserved across all isolates and consisted of single-copy genes, while 1,102 were found to have a variable number of copies (Supplementary Fig. 4). The remaining 7,759 groups were not shared between all isolates (Supplementary Fig. 3), with 408 groups being specific to a single isolate. The variability in gene content confirms the significant impact of structural variability on the gene space (Fig. 3a). When comparing the SVs to the gene annotation, the highest number of genes affected by variants were associated with transposable elements (e.g. reverse transcriptase-rnase h-integrase, gag−pol polyprotein, retrotransposon nucleocapsid protein, and pol polyprotein, Supplementary Fig. 5), further emphasizing the major role of TE mobility in the variation of the genomic structure of M. roreri isolates. Other major categories affected by structural variability were associated with secondary metabolism (e.g. cytochrome p450), cellular transport (e.g. MFS transporters), and cell wall modifications (Carbohydrate-Active Enzymes, CAZymes, Supplementary Fig. 6), which may play a role in pathogenicity. RNA-Seq analysis of MrC26 showed that most of these genes were also expressed and modulated across different sampling conditions (Fig. 3a and Supplementary Fig. 7). The results of our secondary metabolites cluster analysis provided a similar observation. We found that the long-read assembled genomes exhibited an exceptional level of consistency in terms of functional categories, cluster number, and cluster size (Supplementary Fig. 8). However, the impact of SVs was evident in several clusters, affecting all functional categories that we identified. In particular, we found that a significant number of expressed and modulated genes were affected by these SVs (Fig. 3b).
Fig. 3.
Function of genes affected by SNPs and SVs. a) Distribution of the gene functions associated with the genes affected by SVs in long-read assemblies when comparing MrC26 with the other 4 isolates. Most of the genes are associated with transposable elements, no homolog in RefSeq database, or no biological function, often related to fungal pathogenicity (Pusztahelyi et al. 2015; Macheleidt et al. 2016). Expression and modulation in the RNA-Seq samples have been also reported, showing that many of the genes related to the secondary metabolism are active, with DE meaning significantly differentially expressed in at least 1 condition, EX meaning expressed in at least 1 condition even if not significantly modulated, and NO meaning not expressed. b) Secondary metabolite clusters found in MrC26 with antiSMASH. Expression and modulation in the RNA-Seq samples have been also reported as long as the presence of SVs in the locus.
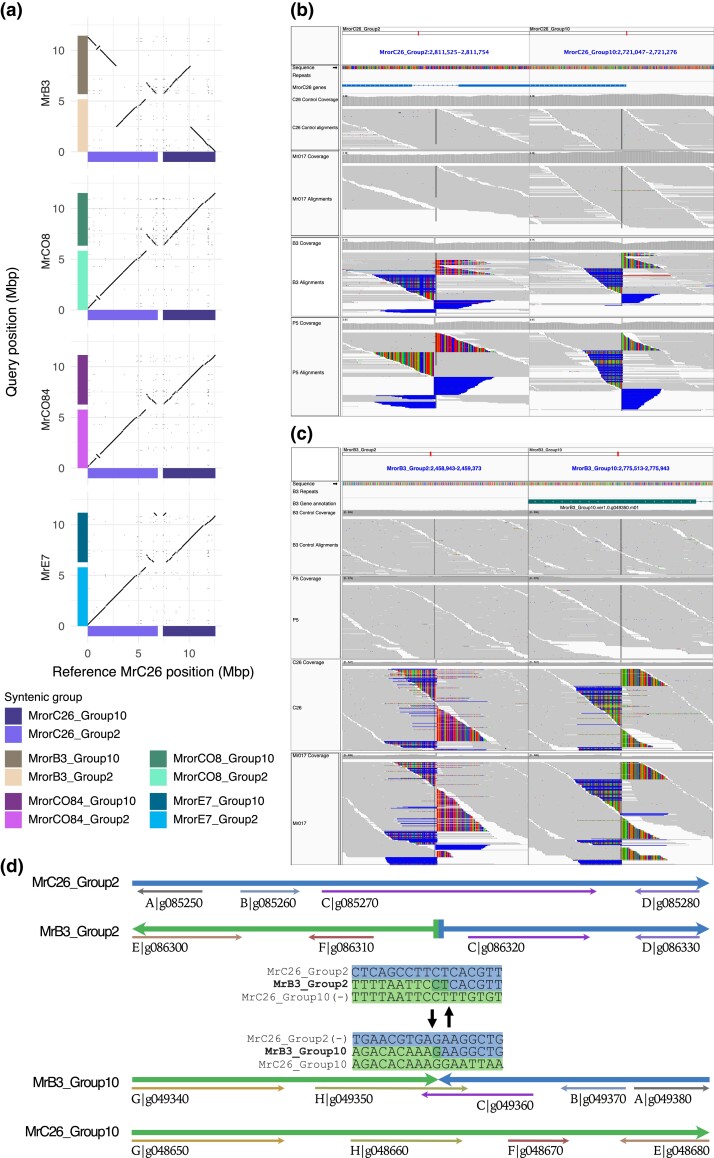
Syntenic groups 2 and 10 exhibit some of the most notable SVs across the different isolates. A large translocation is present in the MrB3 isolate between syntenic groups 2 and 10, as compared with the other 4 M. roreri isolates (Fig. 4a). This was verified through the detection of identical breakpoints/sequence in contigs from independently obtained short sequence assemblies and in the sequenced reads (Fig. 4b).
Fig. 4.
SVs identified between syntenic groups 2 and group 10 in the different M. roreri isolates. Syntenic group 2 and syntenic group 10 show the largest SV identified among the M. roreri isolates assembled with long reads. a) The dotplots evidence the presence of a translocation event specific of MrB3 and a duplication event specific of MrC26 among the 5 long-read assemblies. b) and c) Raw read alignments confirm the translocation event in MrB3 and evidence that is shared only with the closely related isolate MrP5 using both MrC26 (b) and MrB3 (c) genome assemblies as reference. d) Detailed view of the translocation breakpoint and the effect on the gene content of MrC26 and MrB3.
Specifically, MrP5 and MrB3 share the same breakpoint, while MrC26, MrAT42952, and all other isolates do not show evidence of this translocation event. The SV appears to have originated in the ancestors of MrB3 and causes a modification of the gene content of the locus by breaking in a gene-rich area (Fig. 4c). RNA-Seq conducted on MrC26 showed that the genes around the breakpoint are expressed (Supplementary Table 4). Additionally, the MrC26 genome shows a duplication between syntenic groups 2 and 10 (Fig. 4a). This duplication involves 2 32,849 bp tandem repeats at the terminal end, composed of retroelement sequences as well as at least 290 non–retroelement-related genes, at least 140 of which are expressed. This results in the duplication of 15 secreted proteins, 10 of which are candidate effectors. Two of these are unique to the Marasmiaceae, and 2 are unique to M. roreri and M. perniciosa (Mr/Mp) among the comparisons made. One Mr/Mp-unique candidate effector, an adhesin protein called Mad1-related protein, is preferentially expressed during the biotrophic phase.
Candidate effector profiles
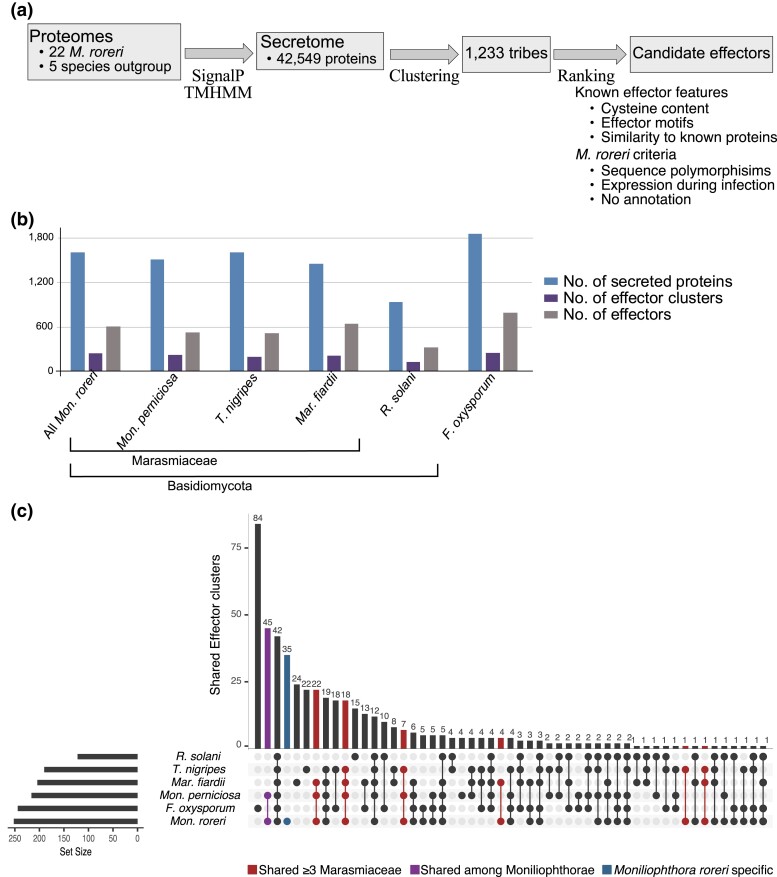
A search for effectors identified an average of 1,601.1 ± 16.9 secreted proteins for each of 22 M. roreri genomes (Fig. 5a and b and Supplementary Tables 5 and 6). Among the secreted proteins, an average 739.5 ± 9.5 for each M. roreri isolate was classified as candidate effectors. Analysis of the effector sequences in the M. roreri isolates and 6 other species identified 263 effector tribes in M. roreri. Of these, 53 tribes were unique to the 4 Marasmiaceae (including M. roreri and 1 other Marasmiaceae species other than M. perniciosa), 45 tribes were unique to M. roreri and M. perniciosa (Mr/Mp), and 35 tribes were unique to M. roreri only (Fig. 5c). Genes with upregulated expression at 60 days compared with 30 DPI were enriched in secreted genes, effector genes, and Marasmiaceae-specific effector genes (Fisher's exact test, P ≤ 0.005; Table 2). None of the Marasmiaceae-specific effector genes were upregulated at 30 DPI compared with 60 DPI. Genes upregulated at 30 DPI compared with 60 DPI were only enriched in Mr/Mp-specific effector genes (Table 2). The small pool of Mr-specific effector genes did not show a preference for up- or downregulation at either phase of infection (Table 2).
Fig. 5.
Secreted protein in M. roreri. a) Secretome identification pipeline used for M. roreri. b) Number of secreted proteins, effectors, and effector clusters identified in the 22 M. roreri isolates, in the M. perniciosa Mp-MCCS1 isolate, and in the species used as outgroup (F. oxysporum forma specialis Lycopersici 4286, M. fiardii PR-910, R. solani AG-1 IA, and T. nigripes CBS291.85), c) Effector clusters shared across the different species; the effectors specific of M. roreri are evidenced in blue, the effector clusters shared between the Moniliophthorae in violet, and the effector clusters shared between M. roreri and at least 1 other Marasmiaceae species with or without M. perniciosa and not F. oxysporum or R. solani in red.
Table 2.
Expression modulation of secreted genes during biotrophic or necrotrophic phases.
| Subseta | Phaseb | In subset | Not in subset | Odds ratio estimatec | |||
|---|---|---|---|---|---|---|---|
| Upregulated | Not modulated | Upregulated | Not modulated | P-valued | |||
| Secreted proteins | |||||||
| (1622 genes) | Biotrophic | 62 | 1,339 | 824 | 17,770 | 1.00 | 1 |
| Necrotrophic | 221 | 1,339 | 1,244 | 17,770 | 2.36 | <2.2 × 10−16 | |
| Effector proteins | |||||||
| (599 genes) | Biotrophic | 25 | 493 | 861 | 18,616 | 1.10 | 0.664 |
| Necrotrophic | 81 | 493 | 1,384 | 18,616 | 2.21 | 2.874 × 10−9 | |
| M. roreri secreted proteins | |||||||
| (33 genes) | Biotrophic | 2 | 30 | 884 | 19,079 | 1.44 | 0.652 |
| Necrotrophic | 1 | 30 | 1,464 | 19,079 | 0.43 | 0.723 | |
| M. roreri and M. perniciosa secreted proteins | |||||||
| (74 genes) | Biotrophic | 10 | 55 | 876 | 19,054 | 3.95 | 5.428 × 10−4 |
| Necrotrophic | 9 | 55 | 1,456 | 19,054 | 2.14 | 0.0455 | |
| Marasmiaceae secreted proteins | |||||||
| (97 genes) | Biotrophic | 0 | 77 | 886 | 19,032 | 0.00 | 0.051 |
| Necrotrophic | 20 | 77 | 1,445 | 19,032 | 5.58 | 1.058 × 10−8 | |
The modulation of 5 different subsets of secreted proteins is compared with the rest of the annotated genes in MrC26 isolate (21,460 annotated gene loci) during the transition between biotrophic phase (30 DPI) and the necrotrophic phase (60 DPI) in cocoa-infected pods. aSubsets include all secretome genes, predicted effectors, secreted genes unique to M. roreri (Mr), secreted genes in both M. roreri and M. perniciosa (Mr/Mp), or secreted genes in Marasmiaceae (Mar)—the latter 3 categories are mutually exclusive. bPhase indicates upregulated at 30 DPI vs 60 DPI (Biotrophic) or upregulated at 60 DPI vs 30 DPI (Necrotrophic). cValues >1 and <1 indicate the proportions of genes in the subset upregulated are greater or less than the proportions of genes not in the subset upregulated, respectively. dP-values < 0.05 are in bold.
Among the genes in the Marasmiaceae-specific effector pool, 20 were upregulated at 60 DPI (Table 2). These necrotrophy-specific genes encode components of the fungal cell surface or enzymes expected to modify surfaces. Of the 74 genes in the MrC26/Mp-specific gene pool, 10 were upregulated at 30 DPI compared with 60 DPI, while 9 genes were upregulated at 60 DPI (Table 2). The biotrophic phase–specific genes encode putative adhesion-related proteins, a Pr-1-like protein, an immunomodulatory protein, several unknown proteins, and others. The necrotrophic-specific genes encode putative mucin-2-like isoform X2-related proteins and a necrosis-inducing-like protein (npp1), among others. Only 2 of the 33 MrC26-specific candidate effector genes (MrorC26_Group4.ver1.0.g127640.m01 and MrorC26_Group9.ver1.0.g204140.m01, genes predicted to encode a fungal hydrophobin and an integral membrane protein, respectively) were upregulated at 30 days vs 60 days, and 1 of the 33 MrC26-specific candidate effector genes (MrorC26_Group10.ver1.0.g055790.m01, a gene predicted to encode a cytochrome P450) was upregulated at 60 days vs 30 days (Table 2).
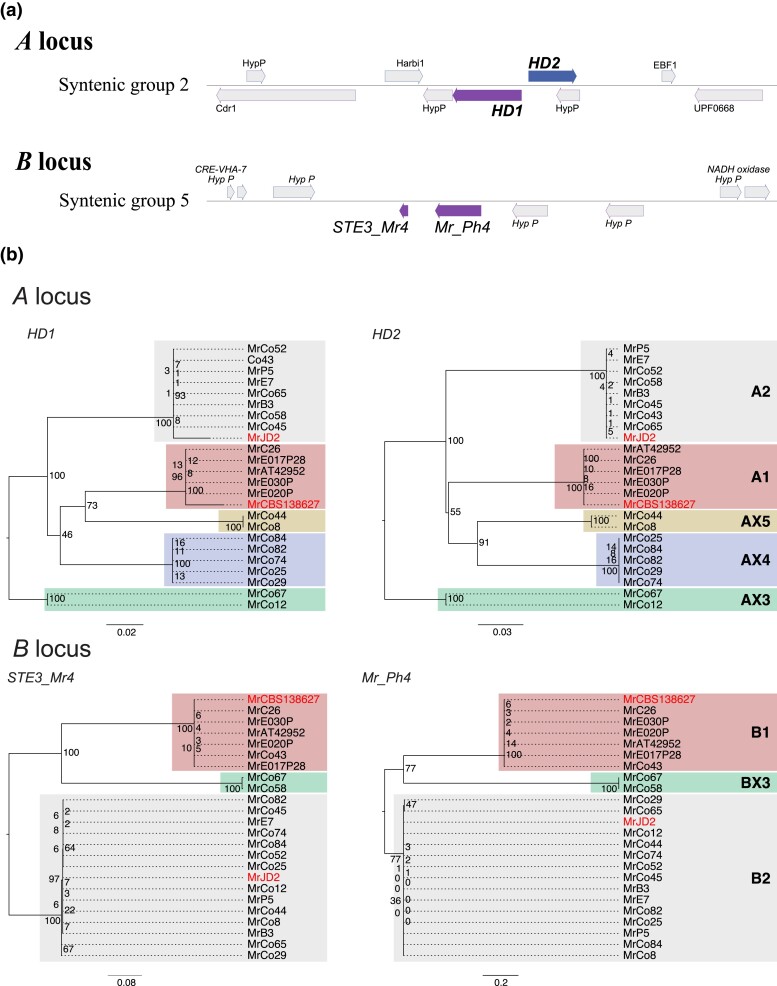
Identification of new mating type loci variants
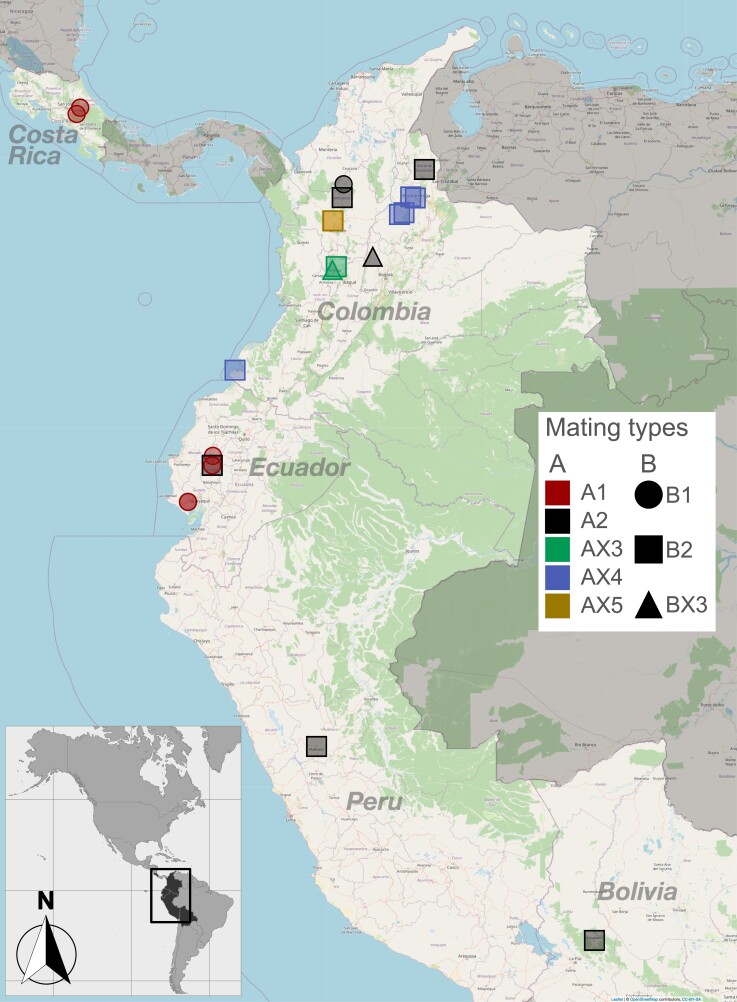
Despite being asexual and haploid, M. roreri carries the genes required for tetrapolar mating at the A and B loci, located on syntenic group 2 and group 5, respectively (Fig. 6a). The A locus consists of 2 consecutive genes, HD1 and HD2, arranged in opposite orientation with neighboring gene starts in the middle of the locus (Fig. 6a). Variation between alleles is primarily located along the 2 start sequences (Fig. 7a). The B locus also consists of 2 consecutive gene loci, Ph4 and STE3, both oriented on the negative strand of the syntenic group 5 (Fig. 6a), with sequence variation extending across both gene sequences (Fig. 7b). The regions bordering the 2 mating type loci showed close similarity among isolates carrying the same allele, but varied between isolates carrying different alleles, with the degree of dissimilarity depending on the alleles being compared (Fig. 7b). Two allelic configurations were previously reported for M. roreri, namely A1B1 and A2B2 (Díaz-Valderrama and Aime 2016a). In our population, 5 isolates found between Costa Rica and Ecuador could be assigned to the A1B1 allelic form (Fig. 8); 6 isolates spreading across Colombia, Ecuador, Peru, and Bolivia could be assigned to the A2B2 allelic form (Fig. 8). A single A2B1 isolate was identified in Colombia (Fig. 8). By assembling the genomes of all isolates in our population, we were able to identify 3 previously unreported alleles for the A locus (called AX3, AX4, and AX5) and a new allele for the B locus (called BX3). Notably, the BX3 locus was not identified by gene prediction tools during annotation procedure, the locus manually identified through a focused search of an open reading frame in the expected genomic region. The isolates carrying the newly identified alleles for the A locus (2 AX3 isolates, 5 AX4 isolates, and 2 AX5 isolates) and the 2 isolates carrying the BX3 allele were collected in Colombia. The similarity of the alleles of each gene loci is shown in Fig. 7b. After analyzing the sequence similarity among single-copy genes across their genomes (Fig. 7b), the 5 A1B1 isolates from the far ends of the pathogen's range (Costa Rica and Ecuador) showed some diversity but still clustered together. Isolates carrying the A2B2 allelic combination showed varied groupings, intersecting with other A locus allelic variants but excluding the B1 allele (Fig. 6b).
Fig. 6.
Phylogeny of mating type loci. a) Location and structure of mating type loci. Moniliophthora roreri mating type locus A genes (HD1 and HD2) are located in syntenic group 2, and locus B genes (STE3_Mr4 and Mr_Ph4) are located in syntenic group 5. b) Phylogenetic trees describing the relationships present between the mating type genes identified in the 22 M. roreri isolates. The HD1 and HD2 (A locus) and STE3_Mr4 and Mr_Ph4 (B locus) proteins previously described in Díaz-Valderrama and Aime (2016a) for A1B1 and A2B2 mating types are evidenced in red. A1, A2, and alleles for A1 locus were assigned using HD1 and HD2 gene sequence homology, and X3, X4, and X4 IDs were assigned to the newly identified alleles. B locus alleles B1 and B2 were assigned based on homology of STE3_Mr4 and Mr_Ph4 genes alleles, and the newly identified locus was denominated BX3.
Fig. 7.
Moniliophthora roreri mating type loci variability across 22 isolates. Comparison of mating type loci structure across different isolates in relation to the phylogeny calculated on the conservation of single-copy ortholog proteins. By aligning the raw Illumina reads of each isolate on the different A locus structures (a) and B locus structures (b) identified for the long-read assemblies, it is possible to identify the closest allele structure. The location of both genes of each mating type locus is highlighted in gray in the plot. B locus shows a lower amount of variation among the isolates and the presence of 3 different alleles, and A locus counts 5 alleles that do closely follow the relationships described by the single-copy orthologs.
Fig. 8.
Moniliophthora roreri mating type loci allele geographical distribution. Geographical distribution of the different mating type alleles. Boxes sharing the same color carry the same mating type alleles. Color coding has been used to distinguish the mating type A locus alleles (A1, A2, and the newly identified AX3, AX4, and AX5); icon shape was used to distinguish the B locus alleles (B1, B2, and the newly identified BX3).
Discussion
Moniliophthora roreri is thought to have evolved in the mountain valleys of Colombia and Ecuador, possibly on Theobroma species other than cacao (Evans 2007, 2016a). An alternative hypothesis suggests it originated in the upper Amazon and was introduced to Colombia and Ecuador (Díaz-Valderrama et al. 2022). Moniliophthora roreri continues to spread to new areas, including Jamaica (Johnson et al. 2017) and western Brazil (da Silva et al. 2022). The timing of M. roreri's transition from sexual to asexual reproduction remains unclear. History, dating back as far as 1817 in Colombia, has always described M. roreri based on the asexual spores produced on pods (Phillips-Mora 2003; Evans 2016a). These spores, rhexolytic thallic conidia, are the primary means of infection and dispersal of M. roreri (Díaz-Valderrama and Aime 2016b). While some conidia contain only 1 haploid nucleus, most have 2 or more identical homokaryotic haploid nuclei formed from multinucleated mycelia, which are derived from mycelia that are typically mononucleated (Bailey, Ali et al. 2018). Multiple genome sequence assemblies consistently show the absence of heterozygosity in the M. roreri genome (Meinhardt et al. 2014; Díaz-Valderrama and Aime 2016a).
Structural changes in the M. roreri genome in association with geographic dispersion
The ability of pathogens to adapt to changes in their hosts through genetic modification is critical to maintaining pathogen/host relationships (Karasov et al. 2014). Sexual reproduction generates new genotypes through recombination while maintaining species identity through maintenance of chromosome structure (Heng 2007). On the other hand, homothallism and asexual reproduction contribute to genome evolution and speciation through accumulation and stabilization of chromosome modifications and repetitive sequence (Whittle et al. 2011). The genome of M. roreri was estimated to be 52 Mbp with 13.5% of repetitive content (Meinhardt et al. 2014). Our multiple single-molecule real-time long-read sequences assemblies of the M. roreri genome revealed an average genome size of nearly 58 Mbp with 11 scaffolds (Table 1). The number of M. roreri chromosomes may be smaller than the number of scaffolds identified. Karyotyping estimated the chromosome number of M. perniciosa to be between 8 and 10 for cacao isolates (Rincones et al. 2006), but karyotyping has not been performed on M. roreri. The genome of M. roreri shows an accumulation of repeats across multiple isolates (>17.5%), and multiple isolate-specific scaffold rearrangements (Table 1 and Fig. 1). The repeat sequences are organized into relatively dense blocks with little associated functional gene structure. This is quite different than in some pathogens, for example, the Phytophthoras, where repeat sequence can be extensive and associated with increased numbers of virulence-associated genes (Poplin et al. 2017; Morales-Cruz et al. 2020).
The rearrangements, found and validated among 5 SMRT assemblies, include translocations, duplications, and inversions (Supplementary Fig. 1). The scaffold rearrangements were verified by comparison of breakpoints in multiple Illumina sequence assemblies of related isolates (Supplementary Fig. 2). The large reciprocal translocation between syntenic groups 2 and 10 found in isolates B3 as compared with other isolates is a unique finding (Figs. 2 and 4). Isolate MrB3 was previously placed into synonymous group 2 (Ali et al. 2015), the only synonymous SNP group found in Ecuador without a representative found in Colombia. Synonymous group 2 was composed of isolates from a single isolate from Manabi, Ecuador, and multiple isolates from Peru and Bolivia at the southern edge of M. roreri range. Breakpoints associated with the reciprocal translocation were shared with Peruvian isolate MrP5 (also synonymous group 2).
MrC26 is representative of the narrow genetic diversity found throughout Central America (Ali et al. 2015; Díaz-Valderrama et al. 2022) at the northern edge of M. roreri range and falls within synonymous group 1 which includes members from Colombia and Ecuador. The MrC26 genome assembly is unusual in that it carries a duplicated segment on the end of syntenic group 2 originating from syntenic group 10 (Fig. 4). It is through duplications and deletions that gene composition may be most directly altered. This is the case in MrC26, as the duplicated segment includes more than 270 potentially functional genes along with a large tandem repeat of retroelement-associated sequence.
New mating type variants were identified in Colombia
Forced asexual reproduction can result in genome degradation in sexual fungi (Horgen et al. 1996; Saleh et al. 2012) and loss of sexual reproduction in nature. For example, Saleh et al. (2012) suggested the movement of Magnaporthe oryzae into new areas resulted in increased asexual reproduction due to mating type isolation and permanent loss of sexual reproduction potential outside its Asian center of origin. Due to the discontinuous distribution of Theobroma species in their natural habitats, both M. roreri and M. perniciosa were likely under pressure to eliminate or simplify their tetrapolar mating system (Kües and Navarro-González 2010; Díaz-Valderrama and Aime 2016a). In M. roreri, the structures of the unlinked mating type locus A (syntenic group 2) and B (syntenic group 5) have been described in detail (Díaz-Valderrama and Aime 2016a). Díaz-Valderrama and Aime (2016a) found only 2 mating type combinations (A1B1 and A2B2) in their initial characterization of the M. roreri mating gene loci, but sampling inside Colombia was limited to 5 isolates from a single region, Antioquia. Forty-one isolates from outside Colombia were included in the study with 10 isolates from a total of 6 locations in Ecuador (Díaz-Valderrama and Aime 2016a). The isolates sequenced in the current study were selected from observed M. roreri SNP diversity (Ali et al. 2015) originally including more than 60 isolates collected throughout Colombia as well as 11 isolates from Ecuador and additional isolates from Peru, Bolivia, and Venezuela. We identified 3 new alleles (5 alleles total) for the A locus and 1 potentially new allele (3 alleles total) for the B locus (Figs. 7 and 8) occurring in 8 AB locus allelic combinations. All the A and B locus allele combinations differing from the A1B1 and A2B2 combinations were found within Colombia (Fig. 6). This supports Colombia as a center of diversity and possible center of origin for M. roreri and establishes the possibility of a more complex historical mating system in M. roreri than originally proposed (Díaz-Valderrama and Aime 2016a).
Although we have 5 A locus alleles and 3 B locus alleles, most of the non-A1B1 mating type allele combinations we identified show potential for combining with only the A1B1 variant in a traditional tetrapolar mating system, A2B1 and AX3BX3 isolates being exceptions. The BX3 allele of the B locus was not called by gene annotation software and may represent the nonfunctional evolution of the locus. This is interesting as Díaz-Valderrama and Aime (2016a) highlighted observations that in the Agaricomycetes bipolarity has always emerged because of “loss of mating type specificity and polymorphism in the B mating locus.” The identification of a single A2B1 isolate, something not observed by Díaz-Valderrama and Aime (2016a), at least alludes to possible sexual recombination in the past or at least some form of recombination. The 2 mating loci are on different scaffolds and would be expected to sort independently sexual reproduction processes. The observation that MrC26 (A1B1) and MrE7 (A2B2) share an inversion located on syntenic group 2 (Fig. 4) suggests they share lineages despite the different mating types providing additional evidence of recombination in the past having taken place after the inversion.
Currently, all M. roreri isolates collected outside Colombia are considered A1B1 or A2B2 [here and Díaz-Valderrama and Aime (2016a)]. In many places, we can clearly identify the time frame of the movement into new areas (Evans 2016a): for example, Costa Rica (all A1B1) in the late 1970s and in Peru in the late 1980s (both A1B1 and A2B2, Díaz-Valderrama and Aime 2016a) and in Bolivia (A2B2) by at least 2015 (Phillips-Mora et al. 2015). Note the 3 A1B1 isolates from Ecuador (Supplementary Table 1) all have SNP profiles matching synonymous group 4 (Ali et al. 2015), a group with isolates from both Ecuador and Colombia, while the 2 A1B1 isolates from Costa Rica fall within synonymous group 1, a group having isolates from Colombia, Costa Rica, and Ecuador (Ali et al. 2015). It is also worth noting isolate MrAT42952 was collected from Costa Rica in 1978 and shares genome structure (Fig. 2), locus sequence (Fig. 7), and SNP profile (Supplementary Table 1) with isolate MrC26 collected in Costa Rica 21 years later. Mating type variants other than A1B1 and A2B2 have been found close to the Colombia border with Ecuador in the Tumaco, Narino region (Fig. 6c), and it is likely additional mating type variants will be found outside and, possibly, within Colombia in the future (Espinoza-Lozano et al. 2022).
Unique effector candidates show increased regulation in response to the biotrophic/necrotrophic shift
The manuscript describing the first M. roreri genome focused on the secretome (Meinhardt et al. 2014) identifying 1,535 candidate genes in a 52.3 Mb genome. Some genes differentially expressed between the biotrophic/necrotrophic phases were discussed as potential effectors: Pr1-like, cerato-platanins, and “biotrophy-associated” secreted proteins. Subsequent work by Barbosa et al. (2018) identified 243 candidate effectors in a 45.2 Mb M. roreri genome composed of 2,994 contigs, but lacked expression data. Here, multiple genome assemblies of M. roreri have identified a large number of candidate effectors, with estimated in the range from 1,535 to 2,994. Our highly contiguous genome assembly identified 1,622 candidate secreted proteins and 599 candidate effectors, using a combination of different prediction tools and focusing on small, secreted proteins with high cysteine content. The genome analysis identified numerous potential effector proteins that are unique to M. roreri, M. perniciosa, or Marasmiaceae species. Some of these proteins show preferential expression during the biotrophic or necrotrophic phases. Many of the proteins that show differential expression between the biotrophic and necrotrophic phases are associated with the cell surface or surface modification. For example, Pr-1-like proteins and necrosis-inducing protein-like Npp1s may be important in plant–pathogen interactions and have been found to be under positive selection in the Moniliophthoras (Vasconcelos et al. 2021) or to have been acquired through gene transfer (Tiburcio et al. 2010), respectively. It appears many genes encoding secreted proteins/candidate effectors are downregulated during the biotrophic phase, potentially limiting pathogen recognition, but are upregulated during the necrotrophic phase to contribute to tissue necrosis and pathogen growth. On the other hand, a unique set of genes encoding candidate effectors in M. roreri and M. perniciosa are upregulated during the biotrophic phase, potentially contributing to the avoidance or overcoming of plant defense responses, allowing for the establishment of infections and early tissue colonization. It is notable the large segment duplication found in the genome of MrC26 carries 10 candidate effectors including 2 effectors unique to Mr/Mp (Fig. 5). These unique genes, an undescribed gene and a Mad1-like adhesin protein, were upregulated in the biotrophic phase. Adhesins function in the adhesion of spores to plants and insects (Epstein and Nicholson 2006) and may respond to exudates from their host in the context of the prolonged M. roreri biotrophic phase. A GH18 protein which was previously identified as an active effector in M. roreri (Fiorin et al. 2018) was not identified as a candidate effector by our structure-based screening approach. This finding indicates that the identification of fungal effectors remains challenging due to their structural diversity.
In conclusion, the current research presents 4 focused and unique findings. We assembled 5 near-complete M. roreri genome assemblies allowing new discussion of the pathogen's genome evolution. A maximum of 11 scaffolds are indicated, although the chromosome number may be smaller. Moniliophthora roreri isolates are accumulating chromosomal rearrangements, changes in repeat sequence composition, and changes in independent line of descent clonal lineages. Scaffold rearrangements identified include large and small segmental translocations and duplications between and within scaffolds, along with inversions. We also identify multiple additional mating type alleles expanding what was considered 2 mating type allele pattern combinations (A1B1 and A2B2). A total of 5 A locus alleles and 3 B locus alleles were identified. The new mating types identified were found in isolates from Colombia, a proposed center of diversity for the pathogen. The use of multiple genome assemblies enabled us to broaden the pool of candidate effectors beyond what was previously estimated. Moreover, they provided evidence that genes encoding unique effector candidates for the Moniliophthora species have been preferentially expressed during the biotrophic phase of the disease. At the same time, genes encoding many secreted proteins and candidate effectors are repressed during the biotrophic phase, possibly to avoid pathogen recognition. Taken together, the clonal propagation of M. roreri, combined with its haploid genome, has allowed lineages with unique genome structures and gene compositions to dominate in areas as it expands its range displaying a significant founder effect.
Supplementary Material
Contributor Information
Andrea Minio, Department of Viticulture and Enology, University of California Davis, Davis 95616, CA, USA; Genome Center, University of California Davis, 95616 Davis, CA, USA.
Rosa Figueroa-Balderas, Department of Viticulture and Enology, University of California Davis, Davis 95616, CA, USA.
Stephen P Cohen, Sustainable Perennial Crops Laboratory, USDA/ARS, Beltsville 20705, MD, USA.
Shahin S Ali, Sustainable Perennial Crops Laboratory, USDA/ARS, Beltsville 20705, MD, USA.
Denny Carriel, Mars La Chola (MLCH), Mars Inc., Guayaquil 090103, Ecuador.
Dahyana Britto, Mars Center for Cocoa Science, Mars Inc., Fazenda Almirante, Caixa Postal 55, Itajuípe, BA, CEP 45630-000, Brazil.
Conrad Stack, Mars Digital Technologies, Mars Inc., Chicago 60642, IL, USA.
Indrani K Baruah, Sustainable Perennial Crops Laboratory, USDA/ARS, Beltsville 20705, MD, USA.
Jean-Philippe Marelli, Mars Wrigley Plant Sciences Laboratory, Mars Inc., 95616 Davis, CA, USA.
Dario Cantu, Department of Viticulture and Enology, University of California Davis, Davis 95616, CA, USA.
Bryan A Bailey, Sustainable Perennial Crops Laboratory, USDA/ARS, Beltsville 20705, MD, USA.
Data availability
Supplemental files are available at figshare: https://doi.org/10.25387/g3.22713796. Supplementary Fig. 1 contains the variant distribution across the 5 M. roreri SMRT-read assemblies, Supplementary Fig. 2 contains the phylogeny between M. roreri isolates based on shared SVs, Supplementary Fig. 3 contains the breakdown of non–single-copy genes shared between the different isolates, Supplementary Fig. 4 contains the shared orthogroups between the 22 M. roreri isolates, Supplementary Fig. 5 contains the counts of genes assigned to different CAZymes families and subfamilies, Supplementary Fig. 6 contains the counts of genes affected by variants categorized by gene function, Supplementary Fig. 7 contains the function distribution of the genes affected by SNPs and structural variants in MrC26, Supplementary Fig. 8 contains the comparison of secondary metabolites clusters identified in the different isolates, Supplementary Table 1 contains the geographic origin of the M. roreri isolates used in this study, Supplementary Table 2 contains the complete statistics for the assembly results, Supplementary Table 3 reports the genome assembly completeness assessment, Supplementary Table 4 reports the results of the RNA-Seq and differential expression analysis, Supplementary Table 5 reports the count and expression analysis results for the secretome genes, and Supplementary Table 6 contains the secretome gene cluster analysis results. Raw sequencing data were deposited at NCBI under BioProject number PRJNA925193, and RNA-Seq expression analysis results were deposited to GEOarchive under the accession numbers GSE226814 and GSE226813 for M. roreri and M. perniciosa respectively. Genome assemblies (FASTA), variant identified between accessions (VCF), gene annotation (GFF3), and repeat annotation (GFF3) files are deposited at Zenodo under the accession https://zenodo.org/record/7872498.
.
Funding
This work was supported by the Mars-USDA TRUST FUND COOPERATIVE AGREEMENT No. 58-6038-6-004 and the Mars-UC Davis agreement A18-1698. References to a company and/or product by the USDA are only for the purposes of information and do not imply approval or recommendation of the product to the exclusion of others that may also be suitable. USDA is an equal opportunity lender, provider, and employer. The funder had the following involvement with the study: conceptualization and project administration.
Literature cited
- Ali SS, Shao J, Strem MD, Phillips-Mora W, Zhang D, Meinhardt LW, Bailey BA. Combination of RNAseq and SNP nanofluidic array reveals the center of genetic diversity of cacao pathogen Moniliophthora roreri in the upper Magdalena Valley of Colombia and its clonality. Front Microbiol. 2015;6:850. doi: 10.3389/fmicb.2015.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37(4):420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey BA, Ali SS, Strem MD, Meinhardt LW. Morphological variants of Moniliophthora roreri on artificial media and the biotroph/necrotroph shift. Fungal Biol. 2018;122(7):701–716. doi: 10.1016/j.funbio.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Bailey BA, Crozier J, Sicher RC, Strem MD, Melnick R, Carazzolle MF, Costa GG, Pereira GA, Zhang D, Maximova S, et al. . Dynamic changes in pod and fungal physiology associated with the shift from biotrophy to necrotrophy during the infection of Theobroma cacao by Moniliophthora roreri. Physiol Mol Plant Pathol. 2013;81:84–96. doi: 10.1016/j.pmpp.2012.11.005. [DOI] [Google Scholar]
- Bailey BA, Evans HC, Phillips-Mora W, Ali SS, Meinhardt LW. Moniliophthora roreri, causal agent of cacao frosty pod rot. Mol Plant Pathol. 2018;19(7):1580–1594. doi: 10.1111/mpp.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey BA, Melnick RL, Strem MD, Crozier J, Shao J, Sicher R, Phillips-Mora W, Ali SS, Zhang D, Meinhardt L. Differential gene expression by Moniliophthora roreri while overcoming cacao tolerance in the field. Mol Plant Pathol. 2014;15(7):711–729. doi: 10.1111/mpp.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa CS, Fonseca RR, Batista TM, Barreto MA, Argolo CS, Carvalho MR, Amaral DO, Silva EM, Arévalo-Gardini E, Hidalgo KS, et al. . Genome sequence and effectorome of Moniliophthora perniciosa and Moniliophthora roreri subpopulations. BMC Genomics. 2018;19(1):509. doi: 10.1186/s12864-018-4875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47(W1):W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15(1):1–9. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano Londoño H. Fluctuación de poblaciones de esporas de Monilia roreri cif. & par. y viabilidad durante un ciclo completo de afección. Noticias Fitopatológicas (Colombia). 1976;5(2):107. [Google Scholar]
- Cantu D, Segovia V, MacLean D, Bayles R, Chen X, Kamoun S, Dubcovsky J, Saunders DG, Uauy C. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics. 2013;14(1):270. doi: 10.1186/1471-2164-14-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. . Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13(12):1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- da Silva NJ, Reis SP, Diorato VS, Rocha JS, Barbosa CS, Ciampi-Guillardi M, Patrocínio NG, Niella GR, Solis K, Peñaherrera S, et al. . A molecular diagnostic for Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao, that differentiates it from its sister taxon Moniliophthora roreri. Crop Protection. 2022;158:106003. doi: 10.1016/j.cropro.2022.106003. [DOI] [Google Scholar]
- Díaz-Valderrama JR, Aime MC. The cacao pathogen Moniliophthora roreri (Marasmiaceae) possesses biallelic A and B mating loci but reproduces clonally. Heredity (Edinb). 2016a;116(6):491–501. doi: 10.1038/hdy.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Valderrama JR, Aime MC. The cacao pathogen Moniliophthora roreri (Marasmiaceae) produces rhexolytic thallic conidia and their size is influenced by nuclear condition. Mycoscience. 2016b;57(3):208–216. doi: 10.1016/j.myc.2016.01.004. [DOI] [Google Scholar]
- Díaz-Valderrama JR, Zambrano R, Cedeño-Amador S, Córdova-Bermejo U, Casas GG, García-Zurita N, Sánchez-Arévalo JA, Arévalo-Gardini E, Dávila D, Ruiz J, et al. . Diversity in the invasive cacao pathogen Moniliophthora roreri is shaped by agriculture. Plant Pathol. 2022;71(8):1721–1734. doi: 10.1111/ppa.13603. [DOI] [Google Scholar]
- Eddy SR.. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. Orthofinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16(1):1–14. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L, Nicholson RL. Adhesion and Adhesives of Fungi and Oomycetes. In: Smith AM, Callow JA, editors. Biological Adhesives. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. p. 41–62. [Google Scholar]
- Espinoza-Lozano F, Amaya-Márquez D, Pinto CM, Villavicencio-Vásquez M, Sosa del Castillo D, Pérez-Martínez S. Multiple introductions of Moniliophthora roreri from the Amazon to the Pacific region in Ecuador and shared high azoxystrobin sensitivity. Agronomy. 2022;12(5):1119. doi: 10.3390/agronomy12051119. [DOI] [Google Scholar]
- Evans HC. Cacao diseases—the trilogy revisited. Phytopathology®. 2007;97(12):1640–1643. doi: 10.1094/PHYTO-97-12-1640. [DOI] [PubMed] [Google Scholar]
- Evans HC. Frosty Pod Rot (Moniliophthora roreri). In: Bailey BA, Meinhardt LW, editors. Cacao Diseases: A History of Old Enemies and New Encounters. Cham: Springer International Publishing; 2016a. p. 63–96. [Google Scholar]
- Evans HC. Witches’ Broom Disease (Moniliophthora perniciosa): History and Biology. In: Bailey BA, Meinhardt LW, editors. Cacao Diseases: A History of Old Enemies and New Encounters. Cham: Springer International Publishing; 2016b. p. 137–177. [Google Scholar]
- Fiorin GL, Sanchéz-Vallet A, de Toledo Thomazella DP, do Prado PF, do Nascimento LC, de Oliveira Figueira AV, Thomma BP, Pereira GA, Teixeira PJ. Suppression of plant immunity by fungal chitinase-like effectors. Curr Biol. 2018;28(18):3023–3030.e5. doi: 10.1016/j.cub.2018.07.055. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The logic of inductive inference. Journal of the Royal Statistical Society. 1935;98(1):39–82. doi: 10.2307/2342435. [DOI] [Google Scholar]
- Frias G, Purdy L, Schmidt R. An inoculation method for evaluating resistance of cacao to Crinipellis perniciosa. Plant Disease. 1995;79(8):787–791. doi: 10.1094/PD-79-0787. [DOI] [Google Scholar]
- Gilbert D. Accurate & complete gene construction with EvidentialGene. Galaxy Community Conference F1000Research. 2016;5:1567. [Google Scholar]
- Grabherr MG, Brian N, Haas J, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, et al. . Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2013;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith GM, Hedger JN. A novel method for producing basidiocarps of the cocoa pathogen Crinipellis perniciosa using a bran-vermiculite medium. Netherlands Journal of Plant Pathology. 1993;99(4):227–230. doi: 10.1007/BF01974667. [DOI] [Google Scholar]
- Haas BJ. https://github.com/TransDecoder/TransDecoder.
- Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):1–22. doi: 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. . De novo transcript sequence reconstruction from RNA-Seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR. Approaches to fungal genome annotation. Mycology. 2011;2(3):118–141. doi: 10.1080/21501203.2011.606851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HHQ. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome. 2007;50(5):517–524. doi: 10.1139/G07-039. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32(5):767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgen PA, Carvalho D, Sonnenberg A, Li A, Van Griensven LJLD. Chromosomal abnormalities associated with strain degeneration in the cultivated mushroom, Agaricus bisporus. Fungal Genet Biol. 1996;20(3):229–241. doi: 10.1006/fgbi.1996.0038. [DOI] [Google Scholar]
- Jackman SD, Coombe L, Chu J, Warren RL, Vandervalk BP, Yeo S, Xue Z, Mohamadi H, Bohlmann J, Jones SJ, et al. . Tigmint: correcting assembly errors using linked reads from large molecules. BMC Bioinformatics. 2018;19(1):393. doi: 10.1186/s12859-018-2425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Rutherford MA, Edgington S, Flood J, Crozier J, Cafá G, Buddie AG, Offord L, Elliott SM, Christie KV. First report of Moniliophthora roreri causing frosty pod rot on Theobroma cacao in Jamaica. New Disease Reports. 2017;36(1):2–2. doi: 10.5197/j.2044-0588.2017.036.002. [DOI] [Google Scholar]
- Karasov TL, Horton MW, Bergelson J. Genomic variability as a driver of plant-pathogen coevolution? Curr Opin Plant Biol. 2014;18:24–30. doi: 10.1016/j.pbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U, Navarro-González M. Mating-type orthologous genes in the primarily homothallic Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao. J Basic Microbiol. 2010;50(5):442–451. doi: 10.1002/jobm.201000013. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997. 10.48550/arXiv.1303.3997, 16 March 2013, preprint: not peer reviewed. [DOI]
- Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32(14):2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yuan J, Kolmogorov M, Shen MW, Chaisson M, Pevzner PA. Assembly of long error-prone reads using de bruijn graphs. Proc Natl Acad Sci USA. 2016;113(52):E8396–E8405. doi: 10.1073/pnas.1604560113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagnan D, da Romeiro RS, de Souza JT, Pomella AWV. Isolation of actinomycetes and endospore-forming bacteria from the cacao pod surface and their antagonistic activity against the witches’ broom and black pod pathogens. Phytoparasitica. 2006;34(2):122–132. doi: 10.1007/BF02981312. [DOI] [Google Scholar]
- Macheleidt J, Mattern DJ, Fischer J, Netzker T, Weber J, Schroeckh V, Valiante V, Brakhage AA. Regulation and role of fungal secondary metabolites. Annu Rev Genet. 2016;50(1):371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14(1):e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, De Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(suppl_2):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt LW, Costa GG, Thomazella D, Teixeira PJ, Carazzolle MF, Schuster SC, Carlson JE, Guiltinan MJ, Mieczkowski P, Farmer A, et al. . Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. BMC Genomics. 2014;15(1):1–25. doi: 10.1186/1471-2164-15-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minio A, Massonnet M, Figueroa-Balderas R, Castro A, Cantu D. Diploid genome assembly of the wine grape carménère. G3 (Bethesda). 2019;9(5):1331–1337. doi: 10.1534/g3.119.400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Cruz A, Ali SS, Minio A, Figueroa-Balderas R, García JF, Kasuga T, Puig AS, Marelli JP, Bailey BA, Cantu D. Independent whole-genome duplications define the architecture of the genomes of the devastating West African Cacao black pod pathogen Phytophthora megakarya and its close relative Phytophthora palmivora. G3 (Bethesda). 2020;10(7):2241–2255. doi: 10.1534/g3.120.401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Mora W. Origin, Biogeography, Genetic Diversity and Taxonomic Affinities of Cacao (Theobroma cacao L.) Fungus Moniliophthora roreri (cif.) Evans et al. as Determined Using Molecular, Phytopathological and Morpho-Physiological Evidence. Reading: University of Reading, School of Plant Sciences; 2003. [Google Scholar]
- Phillips-Mora W, Baqueros F, Melnick Rl, Bailey Ba. First report of frosty pod rot caused by Moniliophthora roreri on cacao in Bolivia. New Disease Reports. 2015;31(1):29–29. doi: 10.5197/j.2044-0588.2015.031.029. [DOI] [Google Scholar]
- Picard Toolkit . Broad Institute, GitHub Repository. 2018. http://broadinstitute.github.io/picard/.
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. . 2017. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178. 10.1101/201178, 24 July 2018, preprint: not peer reviewed. [DOI]
- Pusztahelyi T, Holb I, Pócsi I. Secondary metabolites in fungus-plant interactions. Front Plant Sci. 2015;6:1–23. doi: 10.3389/fpls.2015.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. http://tree.bio.ed.ac.uk/software/figtree/.
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. Genomescope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-14998-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincones J, Mazotti GD, Griffith GW, Pomela A, Figueira A, Leal Jr GA, Queiroz MV, Pereira JF, Azevedo RA, Pereira GA, et al. . Genetic variability and chromosome-length polymorphisms of the witches’ broom pathogen Crinipellis perniciosa from various plant hosts in South America. Mycol Res. 2006;110(7):821–832. doi: 10.1016/j.mycres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Rogers DJ, Tanimoto TT. A computer program for classifying plants. Science. 1960;132(3434):1115–1118. doi: 10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H. 2019Fast and accurate long-read assembly with wtdbg2. bioRxiv 530972. 10.1101/530972, 26 January 2019, preprint: not peer reviewed. [DOI]
- Saleh D, Milazzo J, Adreit H, Tharreau D, Fournier E. Asexual reproduction induces a rapid and permanent loss of sexual reproduction capacity in the rice fungal pathogen Magnaporthe oryzae: results of in vitro experimental evolution assays. BMC Evol Biol. 2012;12(1):42. doi: 10.1186/1471-2148-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena K, Alemanno L, Gramacho KP. 2014. The infection process of Moniliophthora perniciosa in cacao. Plant Pathol 63:1272–1281. [Google Scholar]
- Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, Bosworth C, Armstrong J, Tigyi K, Maurer N, Koren S, et al. . Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol. 2020;38(9):1044–1053. doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(1):31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R. RepeatModeler Open-1.0. 2015.
- Smit AFA, Hubley R, Green P. 2013. RepeatMasker Open-4.0. http://www.repeatmasker.org 2013–2015.
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN. Effectorp 3.0: prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. MPMI. 2022;35(2):146–156. doi: 10.1094/MPMI-08-21-0201-R. [DOI] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 2018;19(9):2094–2110. doi: 10.1111/mpp.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34(Web Server):W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ, Li Y, Shen X-X, Rokas A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 2020;18(12):e3001007. doi: 10.1371/journal.pbio.3001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surujdeo-Maharaj S, Umaharan P, Butler DR, Sreenivasan TN. An optimized screening method for identifying levels of resistance to Crinipellis perniciosa in cocoa (Theobroma cacao). Plant Pathol. 2003;52(4):464–475. doi: 10.1046/j.1365-3059.2003.00865.x. [DOI] [Google Scholar]
- Tiburcio RA, Costa GG, Carazzolle MF, Mondego JM, Schuster SC, Carlson JE, Guiltinan MJ, Bailey BA, Mieczkowski P, Meinhardt LW, et al. . Genes acquired by horizontal transfer are potentially involved in the evolution of phytopathogenicity in Moniliophthora perniciosa and Moniliophthora roreri, two of the major pathogens of cacao. J Mol Evol. 2010;70(1):85–97. doi: 10.1007/s00239-009-9311-9. [DOI] [PubMed] [Google Scholar]
- Vasconcelos AA, José J, Tokimatu PM, Camargo AP, Teixeira PJ, Thomazella D, do Prado PF, Fiorin GL, Costa JL, Figueira A, et al. . Adaptive evolution of Moniliophthora PR-1 proteins towards its pathogenic lifestyle. BMC Ecol Evo. 2021;21(1):84. doi: 10.1186/s12862-021-01818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Nygren K, Johannesson H. Consequences of reproductive mode on genome evolution in fungi. Fungal Genet Biol. 2011;48(7):661–667. doi: 10.1016/j.fgb.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol Biol. 2016;1418:283–334. doi: 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40(W1):W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplemental files are available at figshare: https://doi.org/10.25387/g3.22713796. Supplementary Fig. 1 contains the variant distribution across the 5 M. roreri SMRT-read assemblies, Supplementary Fig. 2 contains the phylogeny between M. roreri isolates based on shared SVs, Supplementary Fig. 3 contains the breakdown of non–single-copy genes shared between the different isolates, Supplementary Fig. 4 contains the shared orthogroups between the 22 M. roreri isolates, Supplementary Fig. 5 contains the counts of genes assigned to different CAZymes families and subfamilies, Supplementary Fig. 6 contains the counts of genes affected by variants categorized by gene function, Supplementary Fig. 7 contains the function distribution of the genes affected by SNPs and structural variants in MrC26, Supplementary Fig. 8 contains the comparison of secondary metabolites clusters identified in the different isolates, Supplementary Table 1 contains the geographic origin of the M. roreri isolates used in this study, Supplementary Table 2 contains the complete statistics for the assembly results, Supplementary Table 3 reports the genome assembly completeness assessment, Supplementary Table 4 reports the results of the RNA-Seq and differential expression analysis, Supplementary Table 5 reports the count and expression analysis results for the secretome genes, and Supplementary Table 6 contains the secretome gene cluster analysis results. Raw sequencing data were deposited at NCBI under BioProject number PRJNA925193, and RNA-Seq expression analysis results were deposited to GEOarchive under the accession numbers GSE226814 and GSE226813 for M. roreri and M. perniciosa respectively. Genome assemblies (FASTA), variant identified between accessions (VCF), gene annotation (GFF3), and repeat annotation (GFF3) files are deposited at Zenodo under the accession https://zenodo.org/record/7872498.
.