Abstract
The taxonomy of the genus Saccharomyces has undergone significant changes recently with the use of genotypic rather than phenotypic methods for the identification of strains to the species level. The sequence of rRNA genes has been utilized for the identification of a variety of fungi to the species level. This methodology, applied to species of Saccharomyces, allows unknown Saccharomyces isolates to be assigned to the type strains. It was the aim of the present study to assess whether typing of the intergenic spacer region by using restriction fragment length polymorphisms of PCR products (intergenic transcribed spacer PCR [ITS-PCR] ribotyping) could distinguish among type strains of the 10 accepted species of Saccharomyces and further to assess if this method could distinguish strains that were interspecific hybrids. Cellular DNA, isolated after the lysis of protoplasts, was amplified by PCR using ITS1 and ITS4 primers, purified by liquid chromatography, and digested with restriction endonucleases. Ribotyping patterns using the restriction enzymes MaeI and HaeIII could distinguish all species of Saccharomyces from each other, as well as from Candida glabrata, Candida albicans, and Blastomyces dermatitidis. The only exception to this was the inability to distinguish between Saccharomyces bayanus and S. pastorianus (S. carlsbergensis). Furthermore, interspecific hybrids resulting from the mating of sibling species of Saccharomyces were shown to share the ITS-PCR ribotyping patterns of both parental species. It should now be possible, by this simple PCR-based technique, to accurately identify these strains to the species level, thereby allowing an increase in our understanding of the characteristics required by these interspecific hybrids for their particular ecological niches.
The taxonomy of the genus Saccharomyces has undergone significant changes recently with more importance being placed on genotypic rather than phenotypic methods for the identification of strains to the species level. This has resulted in a change in the number of recognized species of Saccharomyces moving from 41 (15) to 7 (3) or 10 (20).
The DNA that encodes the ribosomal RNA (rDNA) has been utilized by many investigators for the determination of species of a wide variety of yeasts and fungi (1, 7, 11, 14, 21, 27). These methods rely on the conserved nature of rDNA such that isolates from the same species maintain the same sequence, whereas the more phylogenetically diverse the species is, the greater the difference in the sequences of rDNA is. The present methodology has utilized restriction fragment length polymorphisms (RFLPs) of PCR products obtained by using primers that specifically amplify the region spanning from the 3′ end of the 18S rDNA to the 5′ end of the 25S rDNA. This region includes the 5.8S rDNA and the two intergenic transcribed spacer (ITS) regions ITS1 and ITS2 (26).
This methodology has been applied to Saccharomyces species for the authentication of strains in the Saccharomyces cerevisiae complex (10). These authors found that they could separate strains of S. cerevisiae from those of the S. bayanus-S. pastorianus (S. carlsbergensis) cluster; however strains in this cluster could not be separated from each other by this method (10). No other Saccharomyces species were investigated in this previous work.
More recently, Messner and Prillinger (17) have used a similar yet more thorough method for the differentiation of 10 genotypically distinct Saccharomyces species. In their study, the complete chromosomal ribosomal repeat was amplified in two parts, the 18S rDNA, including both ITS regions and the 5.8S rDNA (total length, 2,600 bp), and separately the 25S rDNA (length, 3,300 bp) (17). Restriction fragments generated from these two PCR products by nine restriction endonucleases yielded characteristic patterns by which unknown isolates of Saccharomyces could be assigned to the type strains (17).
Isolates of Saccharomyces are becoming of increasing medical importance due to their recently appreciated pathogenic potential (4, 5, 16). It was the aim of the present study to assess if the simpler PCR-based method (10) could distinguish among type strains of the 10 accepted species of Saccharomyces (20). A further aim was to assess whether this method could distinguish isolates which were interspecific hybrids resulting from the mating of two closely related yet distinct species of Saccharomyces. Isolates that do not sporulate or do not form viable spores cannot be identified to the species level by classical genetic techniques (19), as these strains cannot be separated from nonmating asporogenous strains of either parental species.
MATERIALS AND METHODS
The yeast strains examined in this study are listed in Table 1. All strains were obtained from the American Type Culture Collection, except for the interspecific hybrid strains and the Candida glabrata and Candida albicans isolates, which were wild-type (clinical) isolates collected by the California Institute for Medical Research laboratory and identified to the species level by routine phenotypic methods.
TABLE 1.
Lanes, species, and reference numbers of the strains utilized for the assessment of ITS-PCR ribotyping for the identification of Saccharomyces isolates to the species level
| Lane no. | Species or hybrid | Strain reference no.a |
|---|---|---|
| 1 | S. cerevisiae | 52530 |
| 2 | S. cerevisiae | 26108 |
| 3 | S. pastorianus (S. carlsbergensis) | 12752 |
| 4 | S. pastorianus (S. carlsbergensis) | 76670 |
| 5 | S. paradoxus | 76856 |
| 6 | S. paradoxus | 76858 |
| 7 | S. bayanus | 76513 |
| 8 | S. bayanus | 76515 |
| 9 | S. paradoxus/S. cerevisiae | YJM 508 |
| 10 | S. bayanus/S. cerevisiae | YJM 334 |
| 11 | S. kluyveri | 22512 |
| 12 | S. servazzii | 58439 |
| 13 | S. unisporus | 58440 |
| 14 | S. castellii | 76901 |
| 15 | S. dairensis | 10597 |
| 16 | S. exiguus | 60477 |
| 17 | C. glabrata | 96-111 |
| 18 | C. albicans | 96-139 |
| 19 | B. dermatitidis | 56920 |
American Type Culture Collection reference numbers, apart from lanes 9 and 10 (interspecific hybrids) and lanes 17 and 18 (clinical isolates).
Cellular DNA was isolated by previously described methods (22, 23).
Primers for the amplification of the 5.8S rDNA were ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′). DNA was amplified in the buffer supplied by the Taq polymerase manufacturer (Gibco BRL, Grand Island, N.Y.) in a 100-μl volume containing 1 μM primers, 1.5 mM MgCl2, 2.5 U of Taq polymerase, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, and 200 μM dTTP. The reactions were performed in an automated thermal cycler (GeneMate; Lab-Line Instruments, Melrose Park, Ill.). DNA samples were denatured by incubation for 3 min at 94°C before 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 2.5 min. After PCR, amplified DNA was purified by liquid chromatography (Wizard PCR Preps; Promega, Madison, Wis.).
For RFLP analysis, the purified PCR products were digested with one of the following restriction enzymes: MaeI, HaeIII, CfoI, DdeI, BglII, BamHI, HindIII, EcoRI, SmaI, or PstI (Boehringer Mannheim, Indianapolis, Ind.). Digestions were done by overnight incubation with 10 U of enzyme at 37°C. Aliquots (10 μl) of the PCR products, with and without endonuclease digestion, were then analyzed by electrophoresis through a 3% (wt/vol) agarose gel (2% Nusieve GTG, 1% SeaKem Gold [FMC BioProducts, Philadelphia, Pa.]) in TAE buffer (40 mM Tris-acetate, 0.2 mM EDTA) for 2 h at 10 V/cm and visualized by UV transillumination at 302 nm after ethidium bromide staining.
RESULTS
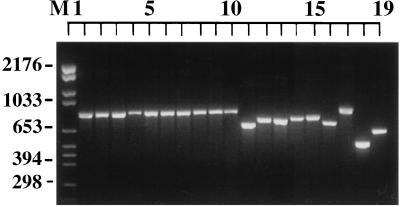
The ITS region amplified by PCR showed a product of approximately 850 bp for S. cerevisiae, S. pastorianus (S. carlsbergensis), S. paradoxus, and S. bayanus and for the interspecific hybrids S. paradoxus/S. cerevisiae and S. bayanus/S. cerevisiae (Fig. 1, lanes 1 to 10). The ITS-PCR products for the remainder of the Saccharomyces species varied between approximately 660 and 760 bp (Fig. 1, lanes 11 to 16). The size of the ITS-PCR products for C. glabrata, C. albicans, and Blastomyces dermatitidis were approximately 850, 520, and 640 bp, respectively (Fig. 1, lanes 17 to 19).
FIG. 1.
Photograph of ethidium bromide-stained, UV-transilluminated PCR products after electrophoresis within a 3% agarose gel. The order of the isolates is given in Table 1.
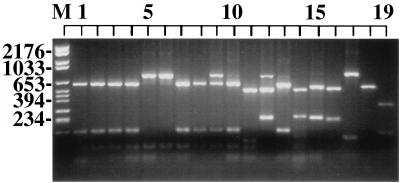
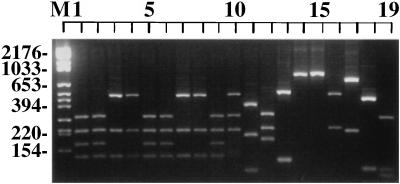
Of the 10 restriction endonucleases used, only two were able to discriminate among all species tested. Utilization of the RFLPs generated by these enzymes, MaeI and HaeIII, made it possible to distinguish all species of Saccharomyces from each other as well as from C. glabrata, C. albicans, and B. dermatitidis, with the exception of the nonseparation of S. pastorianus (S. carlsbergensis) from S. bayanus (Fig. 2 and 3, lanes 7 and 8 and lanes 4 and 5, respectively).
FIG. 2.
Photograph of ethidium bromide-stained, UV-transilluminated MaeI-digested PCR products after electrophoresis within a 3% agarose gel. The DNA from the PCR (Fig. 1) was purified by the Wizard PCR Preps purification system prior to overnight digestion with 10 U of the restriction endonuclease MaeI. The lane order is the same as that in Fig. 1.
FIG. 3.
Photograph of ethidium bromide-stained, UV-transilluminated, HaeIII-digested PCR products after electrophoresis within a 3% agarose gel. The DNA from the PCR (Fig. 1) was purified by the Wizard PCR Preps purification system prior to overnight digestion with 10 U of the restriction endonuclease HaeIII. The lane order is the same as that in Fig. 1.
The ITS-PCR ribotyping pattern obtained with the restriction enzyme MaeI was found to separate all species of Saccharomyces with the exception of three pairs, S. cerevisiae-S. bayanus, S. cerevisiae-S. pastorianus (S. carlsbergensis), and S. pastorianus (S. carlsbergensis)-S. bayanus (Fig. 2).
The ITS-PCR ribotyping pattern obtained with HaeIII could separate all species of Saccharomyces with the exception of three pairs of species, S. cerevisiae-S. paradoxus, S. pastorianus (S. carlsbergensis)-S. bayanus, and S. castellii-S. dairensis (Fig. 3).
Therefore, the combined information from both of these restriction enzymes, MaeI and HaeIII, made it possible to separate all species of Saccharomyces except S. pastorianus (S. carlsbergensis) and S. bayanus (Fig. 2 and 3) from each other. The inability to separate these two species was found with all 10 restriction endonucleases assessed.
The ITS-PCR ribotyping of interspecific hybrids was then determined. The RFLPs generated by MaeI showed that the interspecific hybrid S. paradoxus/S. cerevisiae shared banding patterns with both of the parental species (Fig. 2) and was therefore distinguishable from both these parental species as well as the other species of Saccharomyces. The RFLPs generated by HaeIII showed that the interspecific hybrid S. bayanus/S. cerevisiae shared banding patterns with both of these species (Fig. 3) and was therefore distinguishable from both its parental species as well as from other species.
DISCUSSION
The results of the present study show that ITS-PCR ribotyping is a simple method that can distinguish among all Saccharomyces species other than S. bayanus-S. pastorianus (see below). These findings extend the usefulness of this technique from previous investigations (10) and confirm the findings of Messner and Prillinger (17) while simplifying the methodology by markedly decreasing the size of the PCR amplicon required to successfully differentiate these species of Saccharomyces.
By this method it was not possible to distinguish between the type strains of S. pastorianus (S. carlsbergensis) and S. bayanus. This finding is consistent with earlier studies showing that these two species are very closely related (2, 10, 24). Our results do not support the previously proposed hypothesis that S. pastorianus (S. carlsbergensis) is a separate species or an interspecific hybrid of S. cerevisiae with another sibling species (8, 25). Previous research has shown that S. pastorianus (S. carlsbergensis) carries two copies of certain genes, one that is identical to that of S. cerevisiae and a second gene, which is distinct. The genes that have been most extensively studied are MET2, which encodes homoserine acetyltransferase (9), the gene encoding acetyl-coenzyme A-binding protein (6), and ATF1, which encodes alcohol acetyltransferase (13). The results from this previous research are consistent with the proposal that S. pastorianus (S. carlsbergensis) originated as a hybrid between S. cerevisiae and a sibling species, such as S. monacensis or S. bayanus (6, 9, 13). However, the results of the present study show a clear difference between the ITS-PCR ribotyping pattern of the interspecific hybrid S. cerevisiae/S. bayanus and the ITS-PCR ribotyping patterns of S. pastorianus, S. bayanus, and S. cerevisiae. Other studies support such differentiation (18). It may well be that deletion of one set of the rDNA has occurred in the type strains of S. pastorianus (S. carlsbergensis) used in the present study, resulting in an ITS-PCR ribotyping pattern identical to that of S. bayanus. This process has been previously postulated to occur in species of Saccharomyces (12). Part of the difficulty in interpreting these data arises from the terminology used by various authors. For example, the species S. monacensis has been abolished and replaced by S. pastorianus (S. carlsbergensis) (3). Future studies, utilizing sequence data from a number of regions of the rDNA, are necessary to definitively establish the taxonomic relationship between the species of Saccharomyces.
The finding of the present study that interspecific hybrids resulting from the mating of two closely related yet distinct species of Saccharomyces share the ITS-PCR ribotyping patterns of both parental species allows the previously not possible differentiation of isolates (19). It may well be that some strains of Saccharomyces occurring in nature, used commercially in industry, promoted for human consumption for health reasons, or acting as human pathogens are the result of interspecific mating. These hybrid strains may be phenotypically better adapted for certain ecological niches and have hitherto been designated as S. cerevisiae due to the difficulty in the conventional methodologies required for correct identification to the species level. It should now be possible, by this simple PCR-based species identification technique, to accurately delineate these strains, thereby allowing an increase in our understanding of the characteristics required for each of these particular ecological niches.
ACKNOWLEDGMENTS
This research was funded in part by a Fellowship from the Commonwealth AIDS Research Grants Committee of the National Health and Medical Research Council of the Australian Federal Government.
REFERENCES
- 1.Baleiras Couto M M, Vogels J T, Hofstra H, Huis in’t Veld J H, Vossen J M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food-borne yeasts. J Appl Bacteriol. 1995;79:525–535. doi: 10.1111/j.1365-2672.1995.tb03173.x. [DOI] [PubMed] [Google Scholar]
- 2.Banno I, Kaneko Y. A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast. 1989;5:S373–S377. [PubMed] [Google Scholar]
- 3.Barnett J A. The taxonomy of the genus Saccharomyces Meyen ex Rees: a short review for the non-taxonomists. Yeast. 1992;8:1–23. [Google Scholar]
- 4.Clemons K V, McCusker J H, Davis R W, Stevens D A. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis. 1994;169:859–867. doi: 10.1093/infdis/169.4.859. [DOI] [PubMed] [Google Scholar]
- 5.Clemons K V, McCusker J H, Davis R W, Stevens D A. Saccharomyces cerevisiae and the host-fungus interplay. In: Vanden Bossche H, Stevens D A, Odds F C, editors. Host-fungal interplay. Bethesda, Md: National Foundation for Infectious Diseases; 1997. pp. 193–198. [Google Scholar]
- 6.Fujii T, Nagasawa N, Iwamatsu A, Bogaki T, Tamai Y, Hamachi M. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl Environ Microbiol. 1994;60:2786–2792. doi: 10.1128/aem.60.8.2786-2792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond J R M. Brewer’s yeasts. In: Rose A H, Harrison J S, editors. The yeasts. New York, N.Y: Academic Press; 1993. pp. 8–11. [Google Scholar]
- 9.Hansen J, Kielland-Brandt M C. Saccharomyces carlsbergensis contains two MET2 alleles similar to homologues from S. cerevisiae and S. monacensis. Gene. 1994;140:33–40. doi: 10.1016/0378-1119(94)90727-7. [DOI] [PubMed] [Google Scholar]
- 10.Huffman J L, Molina F I, Jong S C. Authentication of ATCC strains in the Saccharomyces cerevisiae complex by PCR fingerprinting. Exp Mycol. 1992;16:316–319. [Google Scholar]
- 11.Johnston C G, Aust S D. Detection of Phanerochaete chrysosporium in soil by PCR and restriction enzyme analysis. Appl Environ Microbiol. 1994;60:2350–2354. doi: 10.1128/aem.60.7.2350-2354.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kielland-Brandt M C, Nilsson-Tillgren T, Gjermansen C, Holmberg S, Pedersen M B. Genetics of brewing yeasts. In: Wheals A E, Rose A H, Harrison J S, editors. The yeasts. New York, N.Y: Academic Press; 1995. pp. 223–254. [Google Scholar]
- 13.Knudsen J, Faergeman N J, Skott H, Hummel R, Borsting C, Rose T M. Yeast acyl-CoA-binding protein:acyl-CoA-binding affinity and effect on intracellular acyl-CoA pool size. J Biochem. 1994;302:479–485. doi: 10.1042/bj3020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumeda Y, Asao T. Single-strand conformation polymorphism analysis of PCR-amplified ribosomal DNA internal transcribed spacers to differentiate species of Aspergillus Section Flavi. Appl Environ Microbiol. 1996;62:2947–2952. doi: 10.1128/aem.62.8.2947-2952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodder J, Kreger-van Rij N J W. The yeasts, a taxonomic study. Amsterdam, The Netherlands: North Holland Publishing Company; 1952. [Google Scholar]
- 16.McCusker J H, Clemons K V, Stevens D A, Davis R W. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messner R, Prillinger H. Saccharomyces species assignment by long range ribotyping. Antonie Leeuwenhoek. 1995;67:363–370. doi: 10.1007/BF00872936. [DOI] [PubMed] [Google Scholar]
- 18.Molnar O, Messner R, Prillinger H, Stahl U, Slavikova E. Genotypic identification of Saccharomyces species using random amplified polymorphic DNA analysis. Syst Appl Microbiol. 1995;18:136–145. [Google Scholar]
- 19.Naumov G I. Genetic basis for classification and identification of the ascomycetous yeast. Studies Mycol. 1987;30:469–475. [Google Scholar]
- 20.Naumov G I, Naumov E S, Gailadrin C. Genetic and karyotypic identification of wine Saccharomyces bayanus yeast isolated in France and Italy. Syst Appl Microbiol. 1993;16:274–279. [Google Scholar]
- 21.O’Donnell K, Gray L E. Phylogenetic relationships of the soybean sudden death syndrome pathogen Fusarium solani f. sp. phaseoli inferred from rDNA sequence data and PCR primers for its identification. Mol Plant-Microbe Interact. 1995;8:709–716. doi: 10.1094/mpmi-8-0709. [DOI] [PubMed] [Google Scholar]
- 22.Philippsen P, Stotz A, Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 23.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan-Martini A, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 25.Vaughan-Martini A, Martini A, Cardinali G. Electrophoretic karyotyping as a taxonomic tool in the genus Saccharomyces. Antonie Leeuwenhoek. 1993;63:145–156. doi: 10.1007/BF00872389. [DOI] [PubMed] [Google Scholar]
- 26.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1996. pp. 315–322. [Google Scholar]
- 27.Williams D W, Wilson M J, Lewis M A, Potts A J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]