Abstract
Learning high-quality, self-supervised, visual representations is essential to advance the role of computer vision in biomedical microscopy and clinical medicine. Previous work has focused on self-supervised representation learning (SSL) methods developed for instance discrimination and applied them directly to image patches, or fields-of-view, sampled from gigapixel whole-slide images (WSIs) used for cancer diagnosis. However, this strategy is limited because it (1) assumes patches from the same patient are independent, (2) neglects the patient-slide-patch hierarchy of clinical biomedical microscopy, and (3) requires strong data augmentations that can degrade downstream performance. Importantly, sampled patches from WSIs of a patient’s tumor are a diverse set of image examples that capture the same underlying cancer diagnosis. This motivated HiDisc, a data-driven method that leverages the inherent patient-slide-patch hierarchy of clinical biomedical microscopy to define a hierarchical discriminative learning task that implicitly learns features of the underlying diagnosis. HiDisc uses a self-supervised contrastive learning framework in which positive patch pairs are defined based on a common ancestry in the data hierarchy, and a unified patch, slide, and patient discriminative learning objective is used for visual SSL. We benchmark HiDisc visual representations on two vision tasks using two biomedical microscopy datasets, and demonstrate that (1) HiDisc pretraining outperforms current state-of-the-art self-supervised pretraining methods for cancer diagnosis and genetic mutation prediction, and (2) HiDisc learns high-quality visual representations using natural patch diversity without strong data augmentations.
1. Introduction
Biomedical microscopy is an essential imaging method and diagnostic modality in biomedical research and clinical medicine. The rise of digital pathology and whole-slide images (WSIs) has increased the role of computer vision and machine learning-based approaches for analyzing microscopy data [51]. Improving the quality of visual representation learning of biomedical microscopy is critical to introducing decision support systems and automated diagnostic tools into clinical and laboratory medicine.
Biomedical microscopy and WSIs present several unique computer vision challenges, including that image resolutions can be large (10K×10K pixels) and annotations are often limited to weak slide-level or patient-level labels. Moreover, even weak annotations are challenging to obtain in order to protect patient health information and ensure patient privacy [54]. Additionally, data that predates newly developed or future clinical testing methods, such as genomic or methylation assays, also lack associated weak annotations. Because of large WSI sizes and weak annotations, the majority of computer vision research in biomedical microscopy has focused on WSI classification using a weakly supervised, patch-based, multiple instance learning (MIL) framework [2, 7, 20, 37, 38, 48]. Patches are arbitrarily defined fields-of-view (e.g., 256×256 pixels) that can be used for model input. The classification tasks include identifying the presence of cancerous tissue, such as breast cancer metastases in lymph node biopsies [13], differentiating specific cancer types [7, 11, 18], predicting genetic mutations [11, 26, 32], and patient prognostication [8, 29]. A limitation of end-to-end MIL frameworks for WSI classification is the reliance on weak annotations to train a patch feature extractor and achieve high-quality patch-level representation learning. This limitation, combined with the challenge of obtaining fully annotated, high-quality WSIs, necessitates better methods for self-supervised representation learning (SSL) of biomedical microscopy.
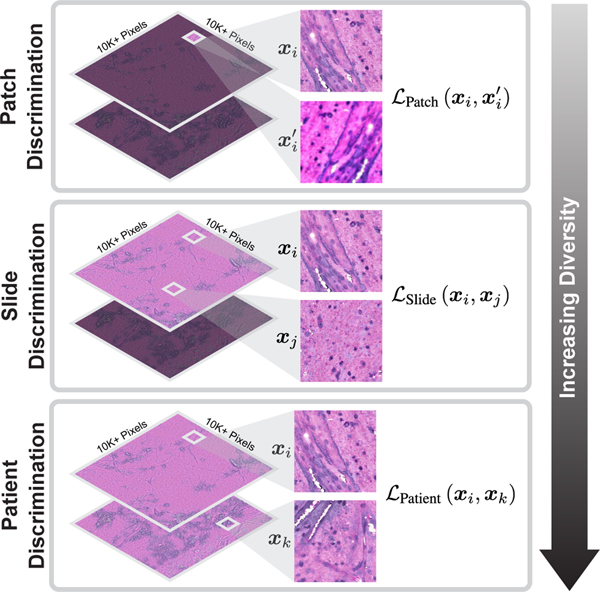
To date, research into improving the quality and efficiency of patch-level representation learning without annotations has been limited. Previous studies have focused on using known SSL methods, such as contrastive learning [35, 47, 50], and applying them directly to WSI patches for visual pretraining. These SSL methods are not optimal because the majority use instance (i.e., patch) discrimination as the pretext learning task [5, 9, 10, 15, 55]. Patches belonging to the same slide or patient are correlated, which can decrease the learning efficiency. Instance discrimination alone does not account for patches from a common slide or patient being different and diverse views of the same underlying pathology. Moreover, previous SSL methods neglect the inherent patient-slide-patch data hierarchy of clinical biomedical microscopy as shown in Figure 1. This hierarchical data structure is not used to improve representation learning when training via a standard SSL objective. Lastly, most SSL methods require strong data augmentations for instance discrimination tasks [9]. However, strong and domain-agnostic augmentations can worsen representation learning in microscopy images by corrupting semantically important and discriminative features [21, 50].
Figure 1. Hierarchical self-supervised discriminative learning for visual representations.
Clinical biomedical microscopy has a hierarchical patch-slide-patient data structure. HiDisc combines patch, slide, and patient discrimination into a unified self-supervised learning task.
Here, we introduce a method that leverages the inherent patient-slide-patch hierarchy of clinical biomedical microscopy to define a self-supervised hierarchical discriminative learning task, called HiDisc. HiDisc uses a self-supervised contrastive learning framework such that positive patch pairs are defined based on a common ancestry in the data hierarchy, and a combined patch, slide, and patient discriminative learning objective is used for visual SSL. By sampling patches across the data hierarchy, we introduce increased diversity between the positive examples, allowing for better visual representation learning and bypassing the need for strong, out-of-domain data augmentations. While we examine the HiDisc learning objective in the context of contrastive learning, it can be generalized to any siamese representation learning method [10].
We benchmark HiDisc self-supervised pretraining on two computer vision tasks using two diverse biomedical microscopy datasets: (1) multiclass histopathologic cancer diagnosis using stimulated Raman scattering microscopy [41] and (2) molecular genetic mutation prediction using light microscopy of hematoxylin and eosin (H&E)-stained cancer specimens [30]. These tasks are selected because of their clinical importance and they represent examples of how deep learning-based computer vision methods can push the limits of what is achievable through biomedical microscopy [18, 24, 26, 31]. We benchmark HiDisc in comparison to several state-of-the-art SSL methods, including SimCLR [9], BYOL [15], and VICReg [1]. We demonstrate that HiDisc has superior performance compared to other SSL methods across both datasets and computer vision tasks. Our results demonstrate how hierarchical discriminative learning can improve self-supervised visual representations of biomedical microscopy.
2. Related Work
Biomedical microscopy and computational pathology
Biomedical microscopy refers to a diverse set of microscopy methods used in both clinical medicine and biomedical research. The most common clinical use of biomedical microscopy is light microscopy combined with H&E staining of clinical tissue specimens, such as tissue biopsies for cancer diagnosis [34]. The introduction of fast and efficient whole-slide digitization resulted in a rapid increase in the availability of WSIs and accelerated the field of computational pathology [42,51]. Computational pathology aims to discover and characterize histopathologic features within biomedical microscopy data for cancer diagnosis, prognostication, and to estimate response to treatment.
The introduction of deep learning to WSI has resulted in clinical-grade computational pathology with diagnostic performance on par with board-certified pathologists [2, 18, 20]. See [43] for a comprehensive survey of deep learning-based methods in computational pathology. Ilse et al. presented MIL framework using an attention-based global pooling operation for slide-level inference [22]. Campanella et al. extended the strategy of a trainable aggregation operation using a recurrent neural network for gigapixel WSI classification tasks [2]. Lu et al. updated the attention-based MIL method to allow for better interpretability and efficient weakly supervised training using transformers [38]. HiDisc is complementary to any MIL framework and can be used as an effective self-supervised pretraining strategy.
Other biomedical microscopy methods have an increasing role in clinical medicine. Electron [39], fluorescence [44, 46, 53], and stimulated Raman scattering microscopy [14, 41] are a few examples of imaging methods that generate microscopy images used for patient diagnosis. Several studies have applied deep learning-based methods to these modalities for image analysis [18, 19, 23, 24, 44, 53].
Self-supervised pretraining in biomedical microscopy
Self-supervised pretraining has been used in computational pathology to improve patch-level representation learning [7, 28, 43, 45, 57]. Generally, a two-stage approach is used where first an SSL method is applied for patch-level feature learning using instance discrimination, and then the patch-level features are aggregated for slide- or patient-level diagnosis. SSL patch pretraining can reduce the amount of data needed compared to end-to-end MIL training [43]. Contrastive predictive coding [36], SimCLR [28], MoCo [45], VQ-VAE [6], and VAE-GAN [57] are examples of deep self-supervised visual representation learning methods applied to biomedical microscopy images [43].
Chen et al. presented a study using vision transformers and self-supervised pretraining at different image scales within individual WSIs [7]. They aim to represent the hierarchical structure of visual histopathologic tokens (e.g., cellular features, supracellular structures, etc.) at varying image resolutions using a transformer pyramid, resulting in a single slide-level representation. Knowledge distillation was used for SSL at each image resolution [5]. HiDisc is complementary to their method and can be used for SSL at any image resolution or, more generally, field-of-view pretraining for any MIL method for slide-level representations.
3. Methods
3.1. The Patient-Slide-Patch Hierarchy
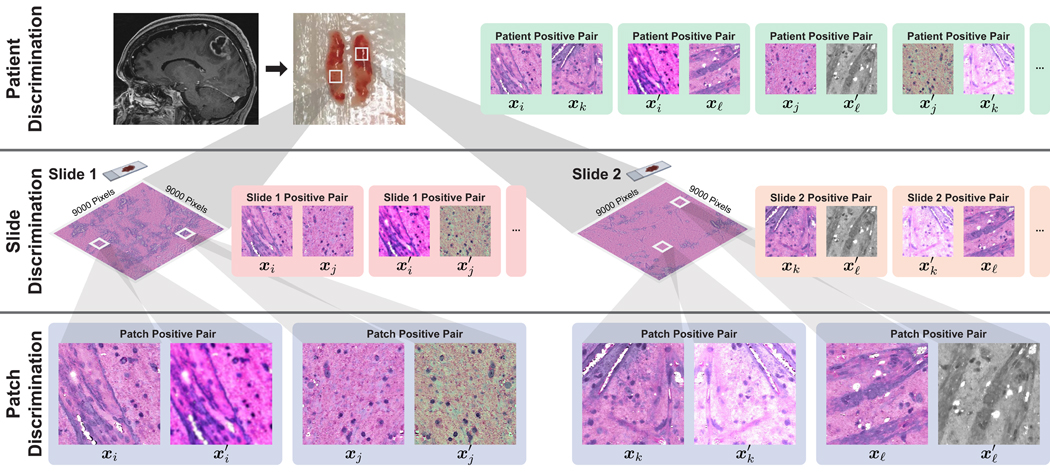
The motivation for HiDisc is that fields-of-view from clinical WSIs, sampled from within a patient’s tumor, are a diverse set of image examples that capture the same underlying cancer diagnosis. Our approach focuses on how to use these diverse fields-of-view in the context of the known clinical patient-slide-patch hierarchical structure to improve visual representation learning. Most patients included in public cancer histopathology datasets, including The Cancer Genome Atlas (TCGA) [4] and OpenSRH [24], contain multiple WSIs as part of their clinical cancer diagnosis. These WSIs may be sampled from different locations in the patient’s tumor, or different regions within the same tumor specimen. Both histopathologic and molecular heterogeneity has been well described within human cancers, encouraging clinicians to obtain multiple specimens/samples/views of the patient’s tumor [49]. To leverage the hierarchy shown in Figure 1, we create positive pairs at the patch-, slide-, and patient-level to define different discriminative learning tasks with a corresponding increase in visual feature diversity:
Patch discrimination: Positive pairs are created from different random augmentations of the same patch. This strategy is similar to existing work on SSL via instance discrimination [1, 9, 10].
Slide discrimination: Positive pairs are created from different augmented patches sampled from the same WSI. These pairs capture local feature diversity within the same specimen. Regional differences in cytologic and histoarchitectural features can be captured at this hierarchical level.
Patient discrimination: Positive pairs are created from different WSIs from the same patient. Patches from different WSIs have the same underlying cancer diagnosis, but can have the greatest degree of feature diversity due to spatially separated tumor specimens. Additionally, diversity in specimen quality, processing, and staining, etc., is captured at this level.
An overview of the hierarchical discrimination tasks is shown in Figure 2.
Figure 2. HiDisc Overview.
Motivated by the patient-slide-patch data hierarchy of clinical biomedical microscopy, HiDisc defines a patient, slide, and patch discriminative learning objective to improve visual representations. Because WSI and microscopy data are inherently hierarchical, defining a unified hierarchical loss function does not require additional annotations or supervision. Positive patch pairs are defined based on a common ancestry in the data hierarchy. A major advantage of HiDisc is the ability to define positive pairs without the need to sample from or learn a set of strong image augmentations, such as random erasing, shears, color inversion, etc. Because each field-of-view in a WSI is a different view of a patient’s underlying cancer diagnosis, HiDisc implicitly learns image features that predict that diagnosis.
3.2. Hierarchical Discrimination (HiDisc)
The formulation of the HiDisc loss function is based on NT-Xent [9] and inspired by [27, 56] for the purpose of multiple positive pairs. The fundamental difference is that no class labels are used during training with a HiDisc loss.
Algorithm 1.
HiDisc Pseudocode in PyTorch style
 |
HiDisc utilizes the natural hierarchy inherent to biomedical microscopy images to improve visual SSL.
We randomly sample a minibatch of patients, slides from each patient, patches from each slide, and we augment each patch times. For patients with less than slides, the slides are repeated and sampled. We assume numbers of patches available for WSI sampling. Note that if a patient has only one WSI, then patient discrimination degenerates to slide discrimination.
The HiDisc loss consists of the sum of three losses, each of which corresponds to a discrimination task at a different level of the patch-slide-patient hierarchy. Similar to the supervised contrastive learning loss [27], the component HiDisc losses are designed to fit more than one pair of positives within each level of the hierarchy. We define the HiDisc loss at the level to be:
| (1) |
where is the level of discrimination, and is the set of all images in the minibatch. is the set of all images in except for the anchor image ,
| (2) |
and is a set of images that are positive pairs of at the -level,
| (3) |
where is the -level ancestry for an augmented patch in the batch. For example, patches and from the same patient would have the same patient ancestry, i.e., .
Patch-, slide-, and patient-level HiDisc losses share the same overall contrastive objective, but capture positive pairs at different levels in the hierarchy. Each loss has a different number of positive pairs within a minibatch. Details about this relationship are shown in Table 1.
Table 1. Batch composition at all discrimination levels.
The number of samples in the batch treated as independent and the number of positive pairs at each discrimination level., number of patients in the batch, , number of slides sampled per patient, , number of patches sampled per slide, , number of augmentations performed on each patch.
| Discrimination level | Number of samples treated as independent | Number of Positive pairs |
|---|---|---|
| Patch | ||
| Slide | ||
| Patient |
Finally, the complete HiDisc loss is the sum of the patch-, slide-, and patient-level discrimination losses defined above:
| (4) |
where is a weighting hyperparameter for level in the total loss. The pseudocode in PyTorch-style detailing the training process is shown in Algorithm 1.
4. Experiments
We evaluate HiDisc using two different computational histopathology tasks: multiclass histopathologic cancer diagnosis and molecular genetic mutation prediction. We present quantitative classification performance metrics, as well as qualitative evaluation with visualizations [52] of the learned patch-level representations.
4.1. Datasets
Stimulated Raman histology (SRH)
We validate HiDisc on a multiclass histopathological cancer diagnosis task using an SRH dataset. Stimulated Raman histology is an optical microscopy method that provides rapid, label-free, sub-micron resolution images of unprocessed biological tissues [14, 41]. The SRH dataset includes specimens from patients who underwent brain tumor biopsy or tumor resection. Patients were consecutively and prospectively enrolled at the University of Michigan for intraoperative SRH imaging, and this study was approved by the Institutional Review Board (HUM00083059). Informed consent was obtained for each patient prior to SRH imaging and approved the use of tumor specimens for research and development. The SRH dataset consists of 852K patches from 3560 slide images from 896 patients with classes consisting of normal brain tissue and 6 different brain tumor diagnoses: high-grade glioma (HGG), low-grade glioma (LGG), meningioma, pituitary adenoma, schwannoma, and metastatic tumor. All slides are divided into 300×300 patches, and they are preprocessed to exclude the empty or non-diagnostic regions using a segmentation model [18].
TCGA diffuse gliomas
We additionally validate HiDisc using WSIs from The Cancer Genome Atlas (TCGA) dataset. We focus on WSIs from brain tumor patients diagnosed with diffuse gliomas, the most common and deadly primary brain tumor [34]. Molecular genetic mutation classification is used as the evaluation task. The most important genetic mutation that defines lower grade versus high grade diffuse gliomas is isocitrate dehydrogenase-1/2 (IDH) mutational status [3]. IDH-mutant tumors are known to have a better prognosis and overall survival (median survival ~10 years) compared to IDH-wildtype tumors (~1.5 years). The classification task is to predict IDH mutational status using formalin-fixed, paraffin-embedded H&E-stained WSI images at 20× magnification from the TCGA dataset. Predicting IDH mutational status from WSIs is currently not feasible for board-certified neuropathologists [3, 12]; genetic mutation prediction from WSIs could avoid time-consuming and expensive laboratory techniques like genetic sequencing. WSIs are divided into 300×300 pixel fields-of-view, and blank patches are excluded. The rest of the patches are stain normalized using the Macenko algorithm [40]. The TCGA data we included consists of a total of 879 patients and 1703 slides, and 11.3M patches.
4.2. Implementation details
We train HiDisc using ResNet-50 [17] as the backbone feature extractor and a one-layer MLP projection head to project the embedding to 128-dimensional latent space. We use an AdamW [33] optimizer with a learning rate of 0.001 and a cosine decay scheduler after warmup in the first 10% of the iterations. For a fair comparison, we control the total number of images in one minibatch as 512 and 448 for SRH and TCGA data, respectively. We train HiDisc with patient discrimination by setting , slide discrimination by setting , , and patch discrimination by setting , . The number of patients sampled from each batch is adjusted accordingly. for each level of discriminating loss is set to 1, and temperature is set as 0.7 for all experiments. We define weak augmentation as random horizontal and vertical flipping. The strong augmentations are similar as [9], including random erasing, color jittering, and affine transformation (for details, see Appendix A). We train HiDisc till convergence for both datasets (100K and 60K iterations for SRH and TCGA, respectively) with three random seeds. Training details for all baselines, including SimCLR [9], SimSiam [10], BYOL [15], VICReg [1], and SupCon [27], are similar to HiDisc.
4.3. Evaluation protocols
evaluation
Standard protocols to evaluate self-supervised representation learning include linear and fine-tuning evaluation. However, both methods are sensitive to hyperparameters, such as learning rate [5]. Therefore, we use the nearest neighbor () classifier for quantitative evaluation. We freeze the pretrained backbone to compute the representation vectors for both training and testing data, and the nearest neighbor classifier is used to match each patch in the testing dataset to the most similar patches in the training set based on cosine similarity. It also outputs a prediction score measured by the cosine similarity between each test image and its nearest neighbors. Due to the size of the TCGA dataset, we randomly sample 400 patches from each slide for evaluation across three different random seeds. Using the classifier, we can compute accuracy (ACC), mean class accuracy (MCA), and area under receiver operating characteristic curve (AUROC) for patch metrics. We use MCA for SRH dataset because it is a multiclass classification problem and the classes are imbalanced. The AUROC is used for the TCGA dataset since it is a balanced binary classification task.
Slide and patient metrics
In contrast to patch-level evaluation, slide and patient predictions are more practical for cancer diagnosis and other clinical uses [2]. After getting patch prediction scores by evaluation, we use average pooling over the scores within each WSI or patient to obtain an aggregated prediction score. Compared to most MIL methods, this non-parametric method directly evaluates representation learning without additional training.
5. Results
5.1. Quantitative metrics
In this section, we evaluate the representations learned by self-supervised HiDisc pretraining using the training and evaluation protocols described in Section 4. We compare HiDisc and SSL baselines on the testing set of both datasets in Table 2. Since HiDisc-Patch is most similar to SimCLR, we observe similar performances. After we add slide and patient discrimination in HiDisc-Slide and HiDisc-Patient, we observe a significant boost in patch accuracy (+6.1 and +6.6 on SRH, +5.5 and +5.9 on TCGA), and a similar increase in other performance metrics as well. Among all methods, HiDisc-Patient has the best performance and outperforms the best baseline, BYOL, with an improvement of 3.9% and 3.1% in classification accuracy for SRH and TCGA, respectively. The GPU wall time needed to train BYOL is roughly 1.5x longer than HiDisc because it requires updating the weights of the target network using exponential moving average. Appendix B shows additional model evaluation metrics.
Table 2. Main results.
We compare HiDisc to state-of-the-art SSL. Supervised refers to models trained with supervision from class labels. Standard deviations are reported in parentheses. HiDisc-Slide and HiDisc-Patient outperform all baseline methods in all metrics for both tasks. Our HiDisc benchmark outperforms previously reported fully supervised baselines from the existing literature on the same genetic mutation classification task on TCGA [30]. MCA, mean class accuracy, AUROC, area under the receiver operating characteristic curve.
| SRH - Patch | SRH - Patient | TCGA - Patch | TCGA - Patient | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Accuracy | MCA | Accuracy | MCA | Accuracy | AUROC | Accuracy | AUROC | |
| SimCLR [9] | 81.0 (0.1) | 73.9 (0.2) | 83.1 (0.7) | 78.4 (0.6) | 77.8 (0.0) | 85.2 (0.1) | 80.7 (0.6) | 88.9 (0.3) |
| SimSiam [10] | 80.3 (1.9) | 73.6 (2.7) | 82.3 (1.7) | 77.0 (4.0) | 68.4 (0.3) | 74.1 (0.3) | 76.6 (0.6) | 82.4 (0.3) |
| BYOL [15] | 83.5 (0.1) | 78.2 (0.2) | 84.8 (1.0) | 82.7 (1.0) | 80.0 (0.1) | 87.5 (0.1) | 83.1 (0.6) | 89.8 (0.2) |
| VICReg [1] | 82.1 (0.3) | 76.0 (0.4) | 82.1 (0.7) | 78.7 (1.9) | 75.5 (0.1) | 82.9 (0.1) | 77.0 (0.5) | 86.0 (0.3) |
|
| ||||||||
| HiDisc-Patch | 80.8 (0.0) | 73.5 (0.1) | 82.6 (0.3) | 77.9 (0.3) | 77.2 (0.1) | 84.7 (0.1) | 81.0 (0.5) | 88.1 (0.2) |
| HiDisc-Slide | 86.9 (0.2) | 83.2 (0.2) | 87.6 (0.5) | 87.0 (1.4) | 82.7 (0.2) | 89.3 (0.2) | 84.3 (0.3) | 92.3 (0.3) |
| HiDisc-Patient | 87.4 (0.1) | 83.5 (0.2) | 87.9 (0.5) | 86.4 (0.6) | 83.1 (0.1) | 90.1 (0.1) | 83.6 (0.3) | 91.8 (0.2) |
|
| ||||||||
| Supervised | 88.9 (0.3) | 86.3 (0.3) | 88.5 (0.5) | 89.1 (0.5) | 85.1 (0.3) | 91.7 (0.2) | 88.3 (0.4) | 95.2 (0.2) |
5.2. Qualitative evaluation
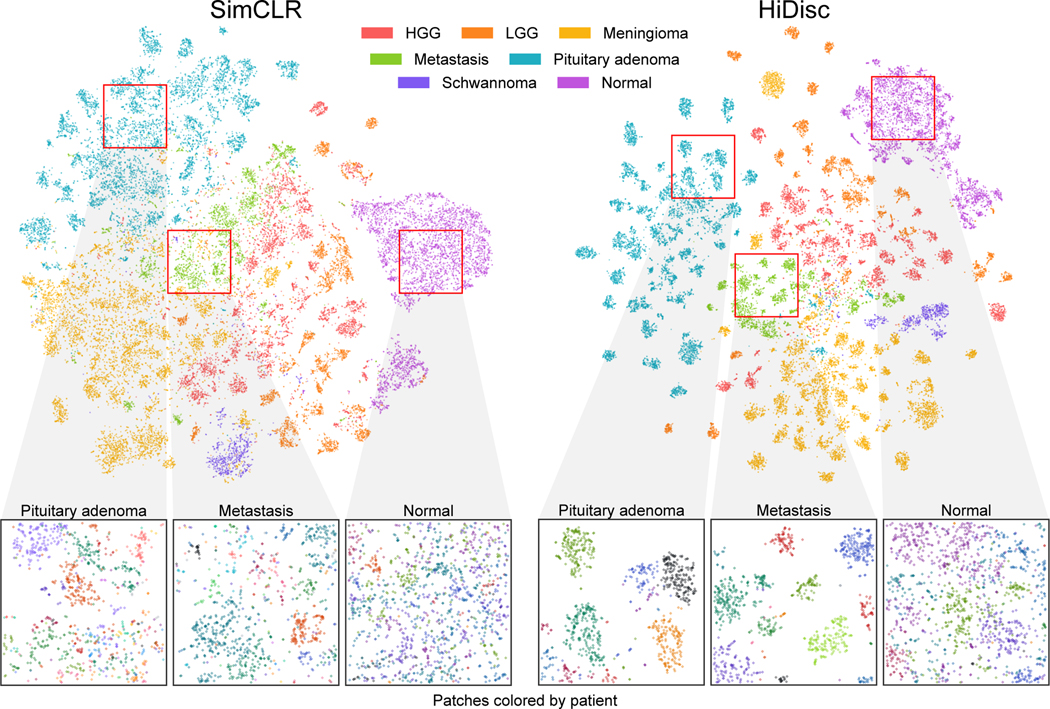
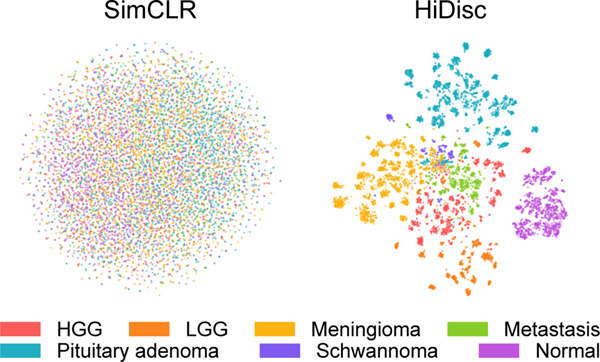
We also qualitatively evaluate the learned patch representations with [52] colored by class and patient label for both SimCLR and HiDisc. Figures 3 and 4 show the learned representations for the SRH and TCGA datasets, respectively. Here, we randomly sample patches from the validation set and plot them by class membership (tumor or molecular subtype). We observe that HiDisc learns better representations for both classification tasks, with more discernible clusters for each class. We also observe better patient clusters within each tumor class in the representations learned by HiDisc. Furthermore, patient clusters are not observed in normal brain tissues for both SimCLR and HiDisc-Patch, as there are minimal differentiating microscopic features between patients.
Figure 3. Visualization of learned SRH representations using SimCLR and HiDisc.
Top. Randomly sampled patch representations are visualized after SimCLR versus HiDisc pretraining using tSNE [52]. Representations are colored based on brain tumor diagnosis. HiDisc qualitatively achieves higher quality feature learning and class separation compared to SimCLR. Expectedly, HiDisc shows within-diagnosis clustering that corresponds to patient discrimination. Bottom. Magnified cropped regions of the above visualizations show subclusters that correspond to individual patients. Patch representations in magnified crops are colored according to patient membership. We see patient discrimination within the different tumor diagnoses. Importantly, we do not see patient discrimination within normal brain tissue because there are minimal-to-no differentiating microscopic features between patients. This demonstrates that in the absence of discriminative features at the slide- or patient-level, HiDisc can achieve good feature learning using patch discrimination without overfitting the other discrimination tasks. HGG, high grade glioma; LGG, low grade glioma; Normal, normal brain tissue.
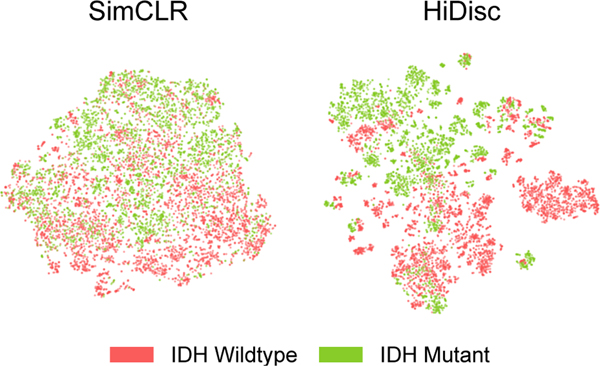
Figure 4. Visualization of learned TCGA representations using SimCLR and HiDisc.
We randomly sample patches from the validation set, and visualize these representations using [52]. Representations on the plots are colored by IDH mutational status. Qualitatively, we can observe that HiDisc forms better representations compared to SimCLR, with clusters within each mutation that corresponds to patient membership. This observation is consistent with the visualizations for the SRH dataset in Figure 3.
5.3. Ablation Studies
Weak Augmentation
We demonstrate that HiDisc is capable of preserving excellent performance without the use of strong, domain-agnostic data augmentations as shown in Table 3. SimCLR suffers from dimensional collapse with weak augmentations [25]. Similar to SimCLR, HiDisc-Patch collapses because it is limited to the diversity from augmentations alone. HiDisc-Slide and HiDisc-Patient performance remain high across both datasets and tasks. HiDisc-Patient outperforms HiDisc-Slide, especially when evaluating at the patch level. We hypothesize that this is a result of additional diversity between positive pairs contributed by patient-level discrimination. We also provide supervised contrastive learning (SupCon) baselines [27] as an upper performance bound. Figure 5 shows SimCLR fails to learn semantically meaningful features while HiDisc achieves high-quality representations. Overall, we observe that HiDisc, especially HiDisc-Patient, performs well regardless of whether strong augmentations are used.
Table 3. Results for weak augmentation.
Comparison of HiDisc with the SimCLR and SupCon on SRH and TCGA datasets with weak augmentation (random flipping). Both SimCLR and HiDisc-Patch patch collapse since weak augmentation will make the pretext task trivial. SupCon, on the other hand, is not sensitive to data augmentations since its positive pairs are defined by class labels. HiDisc-Slide and HiDisc-Patient are only slightly affected by the augmentation and achieve performance close to the supervised method.
| SRH - Patch | SRH - Patient | TCGA - Patch | TCGA - Patient | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Accuracy | MCA | Accuracy | MCA | Accuracy | AUROC | Accuracy | AUROC | |
| SimCLR [9] | 31.5 (2.3) | 23.1 (1.9) | 40.2 (6.9) | 28.9 (4.5) | 57.1 (1.1) | 58.4 (2.2) | 58.1 (1.1) | 72.8 (3.2) |
|
| ||||||||
| HiDisc-Patch | 31.3 (0.6) | 22.2 (0.5) | 47.4 (2.1) | 33.1 (1.6) | 59.0 (0.8) | 61.5 (1.3) | 61.2 (4.2) | 75.8 (2.5) |
| HiDisc-Slide | 82.8 (0.2) | 77.4 (0.3) | 84.2 (0.5) | 82.3 (0.4) | 79.6 (0.1) | 86.3 (0.2) | 77.7 (0.4) | 85.3 (0.3) |
| HiDisc-Patient | 84.9 (0.2) | 78.9 (0.1) | 84.7 (0.5) | 80.9 (1.4) | 82.9 (0.2) | 89.6 (0.2) | 82.3 (0.3) | 90.3 (0.3) |
|
| ||||||||
| SupCon [27] | 90.0 (0.2) | 87.4 (0.3) | 90.0 (0.5) | 90.3 (0.4) | 85.4 (0.4) | 92.0 (0.2) | 88.4 (0.8) | 95.2 (0.4) |
Figure 5. Hierarchical self-supervised discriminative learning without strong data augmentations.
Randomly sampled patch representations are shown after SimCLR and HiDisc-Patient pretraining without the use of strong, domain-agnostic data augmentations on SRH dataset. HiDisc achieves high-quality representation learning, while SimCLR is unable to learn semantically meaningful features via instance discrimination alone. HGG, high grade glioma; LGG, low grade glioma; Normal, normal brain tissue
Other Ablation studies
Additional experiments are included in Appendix C. We perform ablation studies on the weighting factor for each level of discrimination. We also train HiDisc with different number of iterations, learning rate and batch size. Some settings may marginally improve model performance but cost a significant amount of computation resources.
6. Conclusion
We present HiDisc, a unified, hierarchical, self-supervised representation learning method for biomedical microscopy. HiDisc is able to outperform other state-of-the-art SSL methods for visual representation learning. The performance increase is driven by leveraging the inherent patient-slide-patch hierarchy of clinical WSIs. The inherent data hierarchy provides image diversity by defining positive patch pairs based on a common ancestry in the hierarchy and does not require strong, domain-agnostic augmentations. By combining patch, slide, and patient discrimination into a single learning objective, HiDisc implicitly learns image features of the patient’s underlying diagnosis without the need for patient-level annotations or supervision.
Limitations
Like other WSI classification methods, HiDisc representation learning is currently limited to single-resolution fields-of-view that are arbitrarily defined. Expanding HiDisc to include multiple image resolutions could improve its ability to capture multiscale image features of the patient’s underlying diagnosis. Also, we have limited the evaluation to a contrastive learning framework and HiDisc can also be evaluated using other siamese learning frameworks [1, 5, 10].
Broader Impacts
We have limited this investigation to biomedical microscopy and WSIs. However, many imaging medical modalities, such as magnetic resonance imaging and fundoscopy [16], have a clinical hierarchical data structure that could benefit from a similar hierarchical representation learning framework. We hope that hierarchical discriminative learning will extend beyond microscopy to other medical and non-medical imaging domains.
Supplementary Material
Acknowledgements and Competing Interests
We would like to thank Karen Eddy, Lin Wang, Andrea Marshall, Eric Landgraf, and Alexander Gedeon for their support in data collection and programming.
This work is supported in part by grants NIH/NIGMS T32GM141746, NIH K12 NS080223, NIH R01CA226527, Cook Family Brain Tumor Research Fund, Mark Trauner Brain Research Fund: Zenkel Family Foundation, Ian’s Friends Foundation, and the Investigators Awards grant program of Precision Health at the University of Michigan.
C.W.F. is an employee and shareholder of Invenio Imaging, Inc., a company developing SRH microscopy systems. D.A.O. is an advisor and shareholder of Invenio Imaging, Inc, and T.C.H. is a shareholder of Invenio Imaging, Inc.
References
- [1].Bardes Adrien, Ponce Jean, and LeCun Yann. VI-CReg: Variance-Invariance-Covariance regularization for Self-Supervised learning. Feb. 2022.
- [2].Campanella Gabriele, Matthew G Hanna Luke Geneslaw, Miraflor Allen, Silva Vitor Werneck Krauss, Busam Klaus J, Brogi Edi, Reuter Victor E, Klimstra David S, and Fuchs Thomas J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med, 25(8):1301–1309, Aug. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse Lower-Grade gliomas. N. Engl. J. Med, 372(26):2481–2498, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cancer Genome Atlas Research Network, Weinstein John N, Collisson Eric A, Mills Gordon B, Shaw Kenna R Mills, Ozenberger Brad A, Ellrott Kyle, Shmulevich Ilya, Sander Chris, and Stuart Joshua M. The cancer genome atlas Pan-Cancer analysis project. Nat. Genet, 45(10):1113–1120, Oct. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Caron Mathilde, Touvron Hugo, Misra Ishan, Hervé Jégou Julien Mairal, Bojanowski Piotr, and Joulin Armand. Emerging properties in self-supervised vision transformers. pages 9650–9660, Apr. 2021.
- [6].Chen Chengkuan, Lu Ming Y, Williamson Drew F K, Chen Tiffany Y, Schaumberg Andrew J, and Mahmood Faisal. Fast and scalable search of whole-slide images via self-supervised deep learning. Nat Biomed Eng, Oct. 2022. [DOI] [PMC free article] [PubMed]
- [7].Richard J Chen Chengkuan Chen, Li Yicong, Chen Tiffany Y, Trister Andrew D, Krishnan Rahul G, and Mahmood Faisal Scaling vision transformers to gigapixel images via hierarchical Self-Supervised learning. June 2022.
- [8].Chen Richard J, Lu Ming Y, Williamson Drew F K, Chen Tiffany Y, Lipkova Jana, o Zahra, Shaban Muhammad, Shady Maha, Williams Mane, Joo Bumjin, and Mahmood Faisal. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell, 40(8):865–878.e6, Aug. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Ting, Kornblith Simon, Norouzi Mohammad, and Hinton Geoffrey. A simple framework for contrastive learning of visual representations. In Iii Hal Dauméand Singh Aarti, editors, Proceedings of the 37th International Conference on Machine Learning, volume 119 of Proceedings of Machine Learning Research, pages 1597–1607. PMLR, 2020. [Google Scholar]
- [10].Chen Xinlei and He Kaiming. Exploring simple siamese representation learning. Nov. 2020.
- [11].Coudray Nicolas, Paolo Santiago Ocampo Theodore Sakellaropoulos, Narula Navneet, Snuderl Matija, Fenyö David, Moreira Andre L, Razavian Narges, and Tsirigos Aristotelis. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med, 24(10):1559–1567, Oct. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Eckel-Passow Jeanette E, Lachance Daniel H, Molinaro Annette M, Walsh Kyle M Decker Paul A, Sicotte Hugues, Pekmezci Melike, Rice Terri, Kosel Matt L, Smirnov Ivan V, Sarkar Gobinda, Caron Alissa A, Kollmeyer Thomas M, Praska Corinne E, Chada Anisha R, Halder Chandralekha, Hansen Helen M, McCoy Lucie S, Bracci Paige M, Marshall Roxanne, Zheng Shichun, Reis Gerald F, Pico Alexander R, O’Neill Brian P, Buckner Jan C, Giannini Caterina, Huse Jason T, Perry Arie, Tihan Tarik, Berger Mitchell S, Chang Susan M, Prados Michael D, Wiemels Joseph, Wiencke John K, Wrensch Margaret R, and Jenkins Robert B. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med, 372(26):2499–2508, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Babak Ehteshami Bejnordi Mitko Veta, Paul Johannes van Diest, Bram van Ginneken, Nico Karssemeijer, Geert Litjens, Jeroen A W M van der Laak, the CAMELYON16 Consortium, Meyke Hermsen, Quirine F Manson, Maschenka Balkenhol, Oscar Geessink, Nikolaos Stathonikos, Marcory Crf van Dijk, Peter Bult, Francisco Beca, Andrew H Beck, Dayong Wang, Aditya Khosla Rishab Gargeya, Humayun Irshad, Aoxiao Zhong, Qi Dou, Quanzheng Li, Hao Chen, Huang-Jing Lin, Pheng-Ann Heng, Christian Haß, Elia Bruni, Quincy Wong, Ugur Halici, Mustafa Ümit Öner, Rengul Cetin-Atalay, Matt Berseth, Vitali Khvatkov, Alexei Vylegzhanin, Oren Kraus, Muhammad Shaban, Nasir Rajpoot, Ruqayya Awan, Korsuk Sirinukunwattana, Qaiser Talha, Yee-Wah Tsang, David Tellez, Jonas Annuscheit, Peter Hufnagl, Mira Valkonen, Kimmo Kartasalo, Leena Latonen, Pekka Ruusuvuori, Kaisa Liimatainen, Shadi Albarqouni, Bharti Mungal, George Ami, Stefanie Demirci, Navab Nassir, Seiryo Watanabe, Shigeto Seno, Yoichi Takenaka, Hideo Matsuda, Hady Ahmady Phoulady, Vassili Kovalev, Alexander Kalinovsky, Vitali Liauchuk, Gloria Bueno, M Milagro Fernandez-Carrobles, Ismael Serrano, Oscar Deniz, Daniel Racoceanu, and Rui Venâncio. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA, 318(22):2199–2210, Dec. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christian W Freudiger Wei Min, Brian G Saar Sijia Lu, Gary R Holtom Chengwei He, Jason C Tsai, Jing X Kang, and X Sunney Xie. Label-free biomedical imaging with high sensitivity by stimulated raman scattering microscopy. Science, 322(5909):1857–1861, Dec. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Grill Jean-Bastien, Strub Florian, Florent Altché Corentin Tallec, Pierre H Richemond Elena Buchatskaya, Doersch Carl, Bernardo Avila Pires Zhaohan Daniel Guo, Mohammad Gheshlaghi Azar Bilal Piot, Kavukcuoglu Koray, Munos Rémi, and Valko Michal. Bootstrap your own latent: A new approach to self-supervised learning. June 2020.
- [16].Gulshan Varun, Peng Lily, Coram Marc, Martin C Stumpe Derek Wu, Narayanaswamy Arunachalam, Venugopalan Subhashini, Widner Kasumi, Madams Tom, Cuadros Jorge, Kim Ramasamy, Raman Rajiv, Nelson Philip C, Mega Jessica L, and Webster Dale R. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA, 316(22):2402–2410, Dec. 2016. [DOI] [PubMed] [Google Scholar]
- [17].He K, Zhang X, Ren S, and Sun J. Deep residual learning for image recognition. Proc. IAPR Int. Conf. Pattern Recogn, 2016.
- [18].Hollon Todd C, Pandian Balaji, Adapa Arjun R, Urias Esteban, Save Akshay V, Khalsa Siri Sahib S, Eichberg Daniel G, D’Amico Randy S, Farooq Zia U, Lewis Spencer, Petridis Petros D, Marie Tamara, Shah Ashish H, Garton Hugh J L, Maher Cormac O, Heth Jason A, McKean Erin L, Sullivan Stephen E, Hervey-Jumper Shawn L, Patil Parag G, Thompson B Gregory, Sagher Oren, McKhann Guy M 2nd, Komotar Ricardo J, Ivan Michael E, Snuderl Matija, Otten Marc L, Johnson Timothy D, Sisti Michael B, Bruce Jeffrey N, Muraszko Karin M, Trautman Jay, Freudiger Christian W, Canoll Peter, Lee Honglak, Camelo-Piragu Sandra, and Orringer Daniel A. Near real-time intraoperative brain tumor diagnosis using stimulated raman histology and deep neural networks. Nat. Med, Jan. 2020. [DOI] [PMC free article] [PubMed]
- [19].Hollon Todd C, Pandian Balaji, Urias Esteban, Save Akshay V, Adapa Arjun R, Srinivasan Sudharsan, Jairath Neil K, Farooq Zia, Marie Tamara, Al-Holou Wajd N, Eddy Karen, Heth Jason A, Khalsa Siri Sahib S, Conway Kyle, Sagher Oren, Bruce Jeffrey N, Canoll Peter, Freudiger Christian W, Camelo-Piragua Sandra, Lee Honglak, and Orringer Daniel A. Rapid, label-free detection of diffuse glioma recurrence using intraoperative stimulated raman histology and deep neural networks. Neuro. Oncol, July 2020. [DOI] [PMC free article] [PubMed]
- [20].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, and Saltz JH. Patch-Based convolutional neural network for whole slide tissue image classification. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 2424–2433, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hussain Zeshan, Gimenez Francisco, Yi Darvin, and Rubin Daniel. Differential data augmentation techniques for medical imaging classification tasks. AMIA Annu. Symp. Proc, 2017:979–984, 2017. [PMC free article] [PubMed] [Google Scholar]
- [22].Ilse Maximilian, Tomczak Jakub, and Welling Max. Attention-based deep multiple instance learning. In Jennifer Dy and Andreas Krause, editors, Proceedings of the 35th International Conference on Machine Learning, volume 80 of Proceedings of Machine Learning Research, pages 2127–2136. PMLR, 2018. [Google Scholar]
- [23].Jiang Cheng, Bhattacharya Abhishek, Linzey Joseph R, Joshi Rushikesh S, Cha Sung Jik, Srinivasan Sudharsan, Alber Daniel, Kondepudi Akhil, Urias Esteban, Pandian Balaji, Al-Holou Wajd N, Sullivan Stephen E, Thompson B Gregory, Heth Jason A, Freudiger Christian W, Khalsa Siri Sahib S, Pacione Donato R, Golfinos John G, Camelo-Piragua Sandra, Orringer Daniel A, Lee Honglak, and Hollon Todd C. Rapid automated analysis of skull base tumor specimens using intraoperative optical imaging and artificial intelligence. Neurosurgery, Mar. 2022. [DOI] [PMC free article] [PubMed]
- [24].Jiang Cheng, Chowdury Asadur, Hou Xinhai, Kondepudi Akhil, Freudiger Christian, Conway Kyle, Camelo-Piragua Daniel Orringer Sandra, Lee Honglak, and Hollon Todd. OpenSRH: optimizing brain tumor surgery using intraoperative stimulated raman histology. In Advances in Neural Information Processing Systems, volume 35, pages 28502–28516, 2022. [PMC free article] [PubMed] [Google Scholar]
- [25].Jing Li, Vincent Pascal, Yann LeCun, and Yuandong Tian. Understanding dimensional collapse in contrastive self-supervised learning. Oct. 2021.
- [26].Kather Jakob Nikolas, Heij Lara R, Grabsch Heike I, Loeffler Chiara, Echle Amelie, Muti Hannah Sophie, Krause Jeremias, Niehues Jan M, Sommer Kai A J, Bankhead Peter, Kooreman Loes F S, Schulte Jefree J, Cipriani Nicole A, Buelow Roman D, Boor Peter, Ortiz-Brüchle Nadi-Na, Hanby Andrew M, Speirs Valerie, Kochanny Sara, Patnaik Akash, Srisuwananukorn Andrew, Brenner Hermann, Hoffmeister Michael, van den Brandt Piet A, Jäger Dirk, Trautwein Christian, Pearson Alexander T, and Luedde Tom. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer, 1(8):789–799, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khosla Prannay, Teterwak Piotr, Wang Chen, Sarna Aaron, Tian Yonglong, Isola Phillip, Maschinot Aaron, Liu Ce, and Krishnan Dilip. Supervised contrastive learning. Apr. 2020.
- [28].Li Bin, Li Yin, and Eliceiri Kevin W. Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. Conf. Comput. Vis. Pattern Recognit. Workshops, 2021:14318–14328, June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lipkova Jana, Chen Tiffany Y, Lu Ming Y, Chen Richard J, Shady Maha, Williams Mane, Wang Jingwen, Noor Zahra, Mitchell Richard N, Turan Mehmet, Coskun Gulfize, Yilmaz Funda, Demir Derya, Nart Deniz, Basak Kayhan, Turhan Nesrin, Ozkara Selvinaz, Banz Yara, Odening Katja E, and Mahmood Faisal. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat. Med, 28(3):575–582, Mar. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu Sidong, Shah Zubair, Sav Aydin, Russo Carlo, Berkovsky Shlomo, Qian Yi, Coiera Enrico, and Di Ieva Antonio. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep, 10(1):1–11, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu Xiaoxuan, Samantha Cruz Rivera David Moher, Melanie J Calvert, and Alastair K Denniston. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med, 26(9):1364–1374, Sept. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu Zhijie, Su Wei, Ao Jianpeng, Wang Min, Jiang Qiuli, He Jie, Gao Hua, Lei Shu, Nie Jinshan, Yan Xuefeng, Guo Xiaojing, Zhou Pinghong, Hu Hao, and Ji Minbiao. Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated raman histology. Nat. Commun, 13(1):4050, July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Loshchilov Ilya and Hutter Frank. Decoupled weight decay regularization. Nov. 2017.
- [34].David N Louis Arie Perry, Wesseling Pieter, Brat Daniel J, Cree Ian A, Figarella-Branger Dominique, Hawkins Cynthia, Ng HK, Pfister Stefan M, Reifenberger Guido, Soffietti Riccardo, von Deimling Andreas, and Ellison David W. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro. Oncol, June 2021. [DOI] [PMC free article] [PubMed]
- [35].Lu Ming Y, Chen Richard J, Wang Jingwen, Dillon Debora, and Mahmood Faisal. Semi-Supervised histology classification using deep multiple instance learning and contrastive predictive coding. Oct. 2019.
- [36].Lu Ming Y, Chen Richard J, Wang Jingwen, Dillon Debora, and Mahmood Faisal. Semi-Supervised histology classification using deep multiple instance learning and contrastive predictive coding. ArXiv, 2019.
- [37].Lu Ming Y, Chen Tiffany Y, Williamson Drew F K, Zhao Melissa, Shady Maha, Lipkova Jana, and Mahmood Faisal. AI-based pathology predicts origins for cancers of unknown primary. Nature, 594(7861):106–110, June 2021. [DOI] [PubMed] [Google Scholar]
- [38].Lu Ming Y, Williamson Drew F K, Chen Tiffany Y, Chen Richard J, Barbieri Matteo, and Mahmood Faisal. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng, 5(6):555–570, June 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Luse SA. Electron microscopic studies of brain tumors. Neurology, 10:881–905, Oct. 1960. [DOI] [PubMed] [Google Scholar]
- [40].Macenko Marc, Niethammer Marc, J S Marron David Borland, John T Woosley Xiaojun Guan, Schmitt Charles, and Thomas Nancy E. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pages 1107–1110, June 2009. [Google Scholar]
- [41].Orringer Daniel A, Pandian Balaji, Niknafs Yashar S, Hollon Todd C, Boyle Julianne, Lewis Spencer, Garrard Mia, Hervey-Jumper Shawn L, Garton Hugh J L, Maher Cormac O, Heth Jason A, Sagher Oren, Wilkinson D Andrew, Snuderl Matija, Venneti Sriram, Ramkissoon Shakti H, McFadden Kathryn A, Fisher-Hubbard Amanda, Lieberman Andrew P, s Timothy D, Xie X Sunney, Trautman Jay K, Freudiger Christian W, and Camelo-Piragua Sandra. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated raman scattering microscopy. Nat Biomed Eng, 1, Feb. 2017. [DOI] [PMC free article] [PubMed]
- [42].Parwani Anil V. Whole Slide Imaging. Springer International Publishing. [Google Scholar]
- [43].Qu Linhao, Liu Siyu, Liu Xiaoyu, Wang Manning, and Song Zhijian. Towards label-efficient automatic diagnosis and analysis: a comprehensive survey of advanced deep learning-based weakly-supervised, semi-supervised and self-supervised techniques in histopathological image analysis. Phys. Med. Biol, 67(20), Oct. 2022. [DOI] [PubMed] [Google Scholar]
- [44].Rivenson Yair, Wang Hongda, Wei Zhensong, Kevin de Haan Yibo Zhang, Wu Yichen, Günaydın Harun, Zuckerman Jonathan E, Chong Thomas, Sisk Anthony E, Westbrook Lindsey M, Wallace W Dean, and Aydogan Ozcan. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng, 3(6):466–477, June 2019. [DOI] [PubMed] [Google Scholar]
- [45].Saillard Charlie, Dehaene Olivier, Marchand Tanguy, Moindrot Olivier, Kamoun Aurélie, Schmauch Benoit, and Jegou Simon. Self supervised learning improves dMMR/MSI detection from histology slides across multiple cancers. Sept. 2021.
- [46].Sanai Nader, Snyder Laura A, Honea Norissa J, Coons Stephen W, Eschbacher Jennifer M, Smith Kris A, and Spetzler Robert F. Intraoperative confocal microscopy in the visualization of 5-aminolevulinic acid fluorescence in low-grade gliomas. J. Neurosurg, 115(4):740–748, Oct. 2011. [DOI] [PubMed] [Google Scholar]
- [47].Schirris Yoni, Gavves Efstratios, Nederlof Iris, Hugo Mark Horlings, and Jonas Teuwen. DeepSMILE: Contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med. Image Anal, 79:102464, July 2022. [DOI] [PubMed] [Google Scholar]
- [48].Shao Zhuchen, Bian Hao, Chen Yang, Wang Yifeng, Zhang Jian, Ji Xiangyang, and Zhang Yongbing. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification. May 2021.
- [49].Sottoriva Andrea, Spiteri Inmaculada, Sara GM Piccirillo, Anestis Touloumis, V Peter Collins, Marioni John C, Christina Curtis, Colin Watts, and Simon Tavaré. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A, 110(10):4009–4014, Mar. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stacke Karin, Unger Jonas, Claes Lundström, and Gabriel Eilertsen. Learning representations with contrastive Self-Supervised learning for histopathology applications. Machine Learning for Biomedical Imaging, 1(August 2022):1–10, Aug. 2022. [Google Scholar]
- [51].Hamid Reza Tizhooshand Liron Pantanowitz. Artificial intelligence and digital pathology: Challenges and opportunities. J. Pathol. Inform, 9:38, Nov. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].van der Laurens Maatenand Geoffrey Hinton Visualizing data using t-SNE. J. Mach. Learn. Res., 9(86):2579–2605, 2008. [Google Scholar]
- [53].Weigert Martin, Schmidt Uwe, Boothe Tobias, Andreas Müller Alexandr Dibrov, Jain Akanksha, Wilhelm Benjamin, Schmidt Deborah, Broaddus Coleman, Culley Siân, Mauricio Rocha-Martins Fabián Segovia-Miranda, Norden Caren, Henriques Ricardo, Zerial Marino, Solimena Michele, Rink Jochen, Tomancak Pavel, Royer Loic, Jug Florian, and Eugene W Myers. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods, 15(12):1090–1097, Dec. 2018. [DOI] [PubMed] [Google Scholar]
- [54].Wiens Jenna, Saria Suchi, Sendak Mark, Ghassemi Marzyeh, Vincent X Liu Finale Doshi-Velez, Jung Kenneth, Heller Katherine, Kale David, Saeed Mohammed, Pilar N Ossorio Sonoo Thadaney-Israni, and Goldenberg Anna. Do no harm: a roadmap for responsible machine learning for health care. Nat. Med, 25(9):1337–1340, Sept. 2019. [DOI] [PubMed] [Google Scholar]
- [55].Wu Xiong, Yu, and Lin. Unsupervised feature learning via non-parametric instance discrimination. In 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), volume 0, pages 3733–3742, June 2018. [Google Scholar]
- [56].Zhang Shu, Xu Ran, Xiong Caiming, and Ramaiah Chetan. Use all the labels: A hierarchical Multi-Label contrastive learning framework. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 16660–16669, 2022. [Google Scholar]
- [57].Zhao Yu, Yang Fan, Fang Yuqi, Liu Hailing, Zhou Niyun, Zhang Jun, Sun Jiarui, Yang Sen, Menze Bjoern, Fan Xinjuan, and Yao Jianhua. Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pages 4836–4845, June 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.