Abstract
Tissue macrophages, including microglia, are notoriously resistant to genetic manipulation. Here, we report the creation of Adeno-associated viruses (AAV) variants that efficiently and widely transduce microglia and tissue macrophages in vivo following intravenous delivery, with transgene expression of up to 80%. We use this technology to demonstrate manipulation of microglia gene expression and microglial ablation, thereby providing invaluable research tools for the study of these important cells.
Keywords: microglia, AAV, gene therapy, brain
Microglia are tissue-resident macrophages (TRMs) of the Central nervous system (CNS) (1), central to development, homeostasis, and the pathogenesis of many CNS diseases including Alzheimer’s disease, multiple sclerosis, cancer, trauma, and infection (2–6), making them attractive therapeutic targets (7–8). However, viral vector-based gene therapy for the innate immune system is challenging since viral particles need to evade multiple defense strategies and, in the case of microglia, are required to penetrate the blood–brain barrier. It has recently been demonstrated that AAVs targeting microglia can be delivered directly into the CNS with low pathogenicity, but their distribution is limited (9–11).
To overcome these challenges, we conducted target viral evolution against microglia to develop AAV-vectors that were capable of targeting microglia and other TRMs (SI Appendix, Materials and Methods). We identified capsid sequences that can deliver transgenes to microglia and other TRMs (lung, liver, intestine, heart, and spleen) with up to 80% efficiency after intravenous administration.
Results
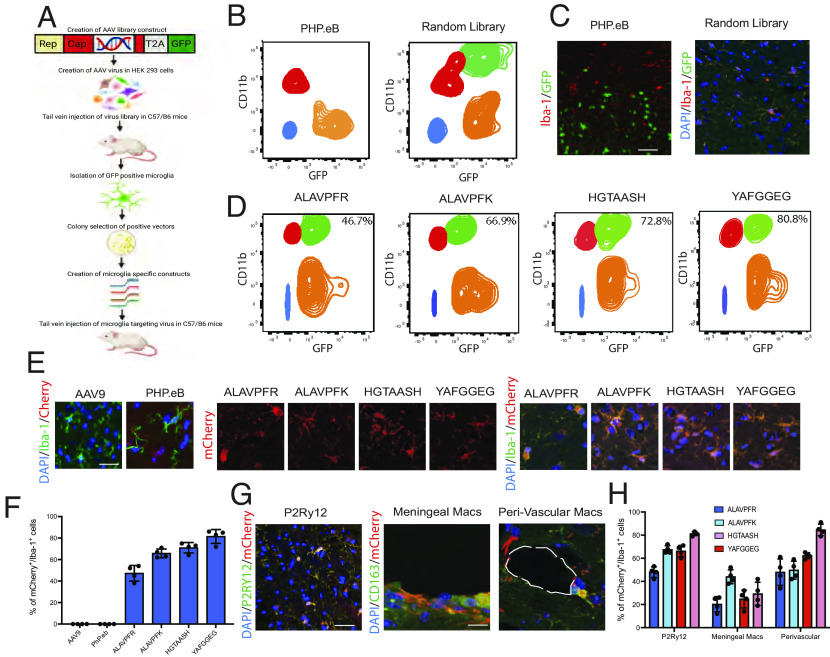
We created a random library of AAV capsids by inserting a random 21-mer library between the nucleotides encoding for amino acids 588 and 589 of the AAV9 capsid (Fig. 1A). This approach has been recently used to create capsids for neuronal and muscle-specific AAV after systemic injection (12, 13). The AAV library was injected intravenously into 8-wk-old C57/Bl6 mice at a concentration of 1 × 1012 viral genomes (vg) per mouse. After 3 wk, Iba1+/GFP+ microglia were found throughout the CNS. In contrast, no GFP+ cells were found with PHP.eB control vector (Fig. 1 B and C).
Fig. 1.
(A) Schematic for the process of microglia AAV derivation. (B) Recovery of GFP-positive microglia from PHP.eB and Random Library infected microglia. The green population indicated GFP+/CD11b+ microglia, the orange population indicates GFP+/CD11b− cells, the red population indicates GFP−/CD11b+ microglia, and the blue population indicates double-negative cells. (C) Immunohistochemistry of PHP.eB and random library virus infection using Iba-1 antibodies for microglia identification (red) and GFP for fluorescent infection. (D) Four AAV-innate sequences demonstrated positive infection between 46.7% and 80.8%. The quantification presented is indicative of the green population of GFP+/CD11b+ cells. (E) Immunohistochemistry was used to confirm microglia infection using an additional mCherry construct in all four AAV-innate vectors using AAV9 and PHP.eB as controls. (F) Quantification of the percentage of mCherry+/Iba-1+ cells. (G) Immunohistochemical confirmation of microglia infection levels was performed using an antibody against P2RY12. Meningeal and perivascular macrophages are characterized using an antibody against CD163. (H) Quantification of the percentage of mCherry+/P2RY12+ cells and mCherry+/CD163+ cells. Data show mean ± SEM. Fluorescence-activated cell sorting (FACS) data are gated on singlets. Example images are taken from the cerebral cortex. (Scale bars: C, 50 µm, E, 10 µm, and G, 50 µm and 10 µm.)
To retrieve the capsid sequences capable of microglial infection, we FACS-isolated Cd11b+/GFP+ microglia. Cells were gated against the nontransfected brain to identify GFP− fractions in the context of autofluorescence in the context of CD11b+/− fractions. The PHP.eB serotype variant encoding for Cytomegalovirus - Enhanced green fluorescent protein (CMV-eGFP) provided a control to distinguish GFP+ and GFP− cells in the sample containing the capsid library. Viral DNA recovered from GFP+ microglia was sequenced, and four AAV sequences were selected for characterization: ALAVPFR, ALAVPFK, HGTAASH, and YAFGGEG, which we collectively refer to as AAV-innate. To confirm the ability of capsid variants containing these insertions to infect microglia and to determine their individual efficiency, we created four separate AAVs. The transfection efficiency of each variants was assessed by FACS 21 d after intravenous injection, was ALAVPFR 46.7 ± 1.8%, ALAVPFK 66.9 ± 2.32%, HGTAASH 72.8 ± 2.1%, and YAFGGEG 80.8 ± 1.9% (Fig. 1 D–F). Microglia infection was further characterized using an antibody against P2RY12. Infection of meningeal and perivascular macrophages was characterized using an antibody against CD163 (Fig. 1 G and H).
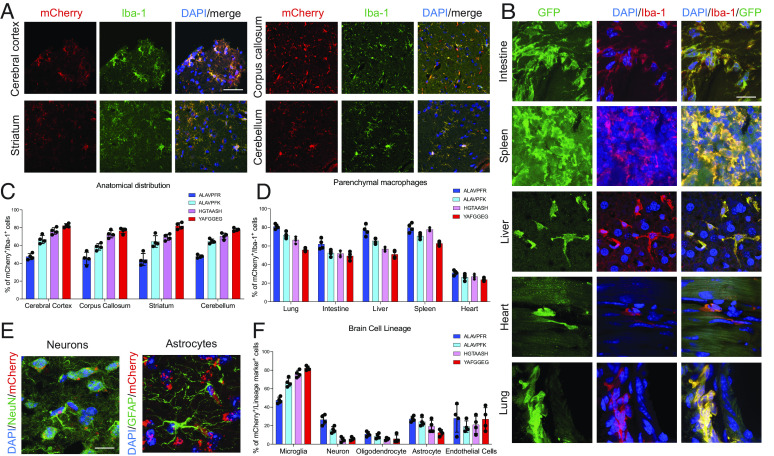
Following intravenous injection of all four AAV-innate vectors, Iba-1+/mCherry+ microglia were found in the cerebral cortex, corpus callosum, striatum, and cerebellum (Fig. 2 A and C). We then asked whether AAV-innate was capable of transfecting TRMs in other organs and found high levels of infection of TRM in the lungs, heart, liver, spleen, and gut (Fig. 2 B and D). The AAVs which had lower efficacy in microglia (ALAVPFR and ALAVPFK) had higher levels of infection in other parenchymal macrophages. ALAVPFR had the greatest ability to infect lung macrophages (81.0 ± 1.3), splenic macrophages (79.8 ± 2.4%), liver macrophages (76.8 ± 2.9%), intestinal macrophages (62.0 ± 2.8%), and cardiac macrophages (31.3 ± 1.3%). Declaration of positive fluorescence was based on full cytoplasmic fluorescence in microglia/macrophages.
Fig. 2.
(A) Immunohistochemistry of the mouse brain labeled with Iba-1 antibody (green) and mCherry expression representing infected microglia, delivered using the HGTAASH capsid in the cerebral cortex, corpus callosum, striatum, and cerebellum. (B) Immunohistochemistry of GFP delivery using the HGTAASH capsid in the lung, intestine, liver, heart, and spleen in the context of Iba-1 labeling (red) of TRMs. (C) Quantification of mCherry+/Iba-1+ cells after systemic delivery in the cerebral cortex, corpus callosum, striatum, and cerebellum using the HGTAASH capsid. (D) Quantification of GFP+ fluorescence in Iba-1+ TRM (red) in the lung, intestine, liver, heart, and spleen. (E) Representative image of mCherry infection in neurons and astrocytes. (F) Quantification of infection of different cell lineages in the brain. Data show mean average ± SEM. (Scale bars: A, 50 µm, B, 10 µm, and E, 10 µm.)
We characterized the expression of AAV-innate in vivo in other brain cell lineages and found low levels of expression in NeuN+ neurons (5.75 ± 1.85%), Olig2+ oligodendrocyte lineage cells (5.75 ± 2.1%), and GFAP+ astrocytes (12.5 ± 1.71%) (Fig. 2 E and F). To minimize the risk of false-negative characterization in nonmacrophage lineage cells, any puncta of fluorescence observed in the cell was included in the analysis.
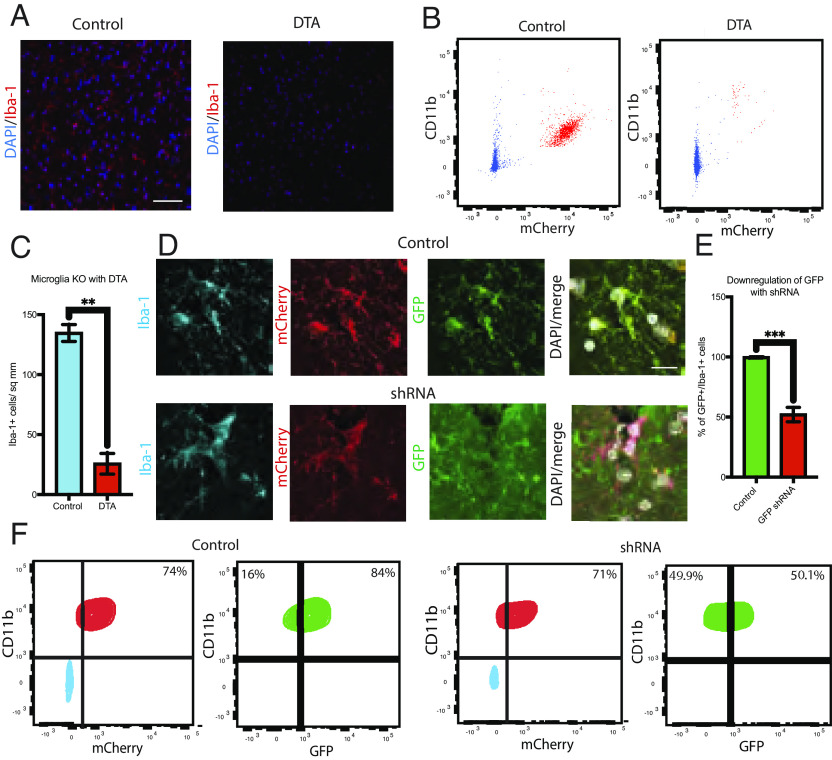
To determine whether the capsid sequences allowed genetic manipulation of microglia in vivo, we injected viruses with the HGTAASH capsid encoding for diphtheria toxin A (DTA) under the control of a phagocyte-specific Cd11b promoter into wild-type mice. The brains were prepared into a single-cell suspension and Cd11b+ microglia selected by flow cytometry. The proportion of microglia decreased 10-fold by 28 d following intravenous injection in DTA-infected brains compared to a mCherry expressing control virus (Fig. 3 A–C). Microglia depletion by DTA was also confirmed using immunohistochemistry (Fig. 3A). To test whether the HGTAASH capsid AAV could be used to suppress microglial gene expression, we used a viral construct encoding for an shRNA targeting GFP in transgenic mice that express GFP in all cells. After 28 d, 75% of Iba-1+ microglia were infected, judged by the expression Cd11b driven mCherry, of which 50% demonstrated decreased expression of GFP (Fig. 3 D–F).
Fig. 3.
(A) Immunohistochemistry for Iba-1+ (red) microglia after injection with control AAV (CD11b promoter-mCherry) and DTA (CD11b promoter-DTA) using the HGTAASH capsid. (B) Quantification of Iba-1+ cells control AAV and DTA AAV delivery. (C) Flow cytometry of DTA-mediated microglial depletion in vivo. FACS data are gated on singlets. (D) shRNA targeting GFP was delivered to the tail vein of mice using the HGTAASH capsid. Immunohistochemistry of the mouse brain labeled with Iba-1 antibody (blue) to detect microglia mCherry (red) as a positive control for infection and constitutive expression of GFP (green) in CMV-eGFP mice. (E) Quantification of GFP+/Iba-1+ cells with GFP expression after control or GFP shRNA vector. (F) GFP silencing characterized using flow cytometry. Data show mean average ± SEM with statistical significance determined by unpaired t tests. FACS data are gated on singlets and then CD11b+/mCherry+ to analyze the GFP+ cohort. Example images are taken from the cerebral cortex. (Scale bars: A, 100 µm, and D, 10 µm.)
Discussion
The benchmark for CNS-penetrating vectors, the PHP family of AAVs, has demonstrated the ability to infect the CNS with high efficiency (7–8, 12). Several rounds of selection resulted in the ability to infect 69% of cortical and 55% of striatal neurons with the AAV-PHP.eB and 82% of dorsal root ganglion neurons but do not infect microglia (12). While AAV vectors that can infect microglia have recently been described, these are only effective when injected directly into the CNS. Here, we describe a family of AAV vectors, AAV-innate, that can transfect resident microglia with up to 80% efficiency following systemic (intravenous) delivery. This latter feature has obvious potential therapeutic advantages and will be an invaluable tool for the experimental study of microglia biology, avoiding problems that come with local injections (9–13).
Complementary to the CNS penetrating AAV-innate variants, we demonstrate that members of the family can be used to target other TRM with high efficiency. Specifically, the low CNS-penetrant vector ALAVPFR can be used to target non-CNS TRM, which currently remains unreported to our knowledge. Moreover, as infection is driven by the capsid binding arm, there is scope to modulate specificity over the TRM target of interest by driving transgene expression under cell-specific promoters.
Our technology improves current technologies in two ways. First, AAV-innate enables microglia to be targeted throughout the parenchyma and not just at the site of injection. Second, the AAV-innate family is freely translatable to human cells, potentially expediting the translation of microglia therapies into humans. In the future, it will be important to explore mutations that improve expression levels in microglia protecting from intracellular degradation (14). The AAVs we report here will open up avenues in the research and possibly therapy development of neurological disorders.
Methods
AAV production and purification followed the previously described protocol (15). A total of 1 × 1012 vg per virus were injected into the tail vein of 8-wk-old mice. Eight-wk-old WT mice were injected with AAVs encoding for DTA. CMV-eGFP mice were injected with AAVs containing shRNA targeting GFP; both were driven by a human Cd11b promoter. Microglia were isolated using antibodies against CD11b. Immunohistochemistry was performed against Iba-1 and P2RY12.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
A.Y. was supported by a Wellcome Trust Clinicians PhD Fellowship (RRZD/029). B.N. was supported by a Kim and Julianna Silverman research fellowship. R.J.M.F was supported by the Dr. Miriam and Shelson G. Adelson Medical Research Foundation, the UK Multiple Sclerosis Society (MS50), and a core grant from the Wellcome Trust and MRC (203151/Z/16/Z).
Author contributions
A.Y., B.N., M.S., and R.J.M.F. designed research; A.Y., B.N., M.S., C.Z.-Y.C., and P.T. performed research; B.N. and M.S. contributed new reagents/analytic tools; A.Y., B.N., M.S., and R.J.M.F. analyzed data; and A.Y., B.N., and R.J.M.F. wrote the paper.
Competing interests
This work has been filed as an international patent WO 2022/023773 A1.
Data, Materials, and Software Availability
All data, code, and materials used in the analysis will be made available to readers in a public repository upon publication. DOIURL: not currently available.
Supporting Information
References
- 1.Li Q., Barres B. A., Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18, 225–242 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Young A. M. H., et al. , A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rock R. B., et al. , Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17, 942–64 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loane D. J., Kumar A., Microglia in the TBI brain: The good, the bad, and the dysregulated. Exp. Neurol. 275, 316–327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangoso E., et al. , Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 184, 2454–2470.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda T., et al. , Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Bedbrook C. N., Deverman B. E., Gradinaru V., Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci. 41, 1–26 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Jüttner J., et al. , Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 22, 1345–1356 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Lin R., et al. , Directed evolution of adeno-associated virus for efficient gene delivery to microglia. Nat. Methods 19, 976–985 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Cucchiarini M., Ren X. L., Perides G., Terwilliger E. F., Selective gene expression in brain microglia mediated via adeno-associated virus type 2 and type 5 vectors. Gene. Ther. 10, 657–667 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Rosario A. M., et al. , Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol. Ther. Methods Clin. Dev. 3, 16026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deverman B. E., et al. , Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabebordbar M., et al. , Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 184, 4919–4938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty D., Monahan P., Samulski R., Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene. Ther. 8, 1248–1254 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Challis R. C., et al. , Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 14, 379–414 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data, code, and materials used in the analysis will be made available to readers in a public repository upon publication. DOIURL: not currently available.