Abstract
Human immunodeficiency virus type 1 (HIV-1) RNA levels in plasma are currently widely used clinically for prognostication and in monitoring antiretroviral therapy. Accurate and reproducible results are critical for patient management. To determine the effects of specimen collection and handling procedures on quantitative measurement of HIV-1 RNA, we compared anticoagulants and sample processing times. Whole blood was collected from 20 HIV-1-infected patients in EDTA, acid citrate dextrose (ACD), and heparin tubes, aliquoted, and stored at room temperature. Plasma was separated from whole-blood aliquots prepared at ≤1, 3, 6, 24, and 48 h postcollection and then stored at −70°C until use. HIV-1 RNA levels were determined by the AMPLICOR HIV-1 MONITOR assay. Heparinized plasma samples, which were pretreated with heparinase prior to analysis, had the lowest baseline HIV-1 RNA levels. In the first 6 h, HIV-1 RNA levels decreased by 10, 20, and 31% in EDTA, ACD, and heparin tubes, respectively. From 6 to 48 h postcollection, HIV-1 RNA levels decreased in all anticoagulants, albeit at a slower, more consistent rate. Our results indicate that EDTA should be the anticoagulant of choice for plasma HIV-1 RNA measurement by reverse transcriptase PCR, but ACD tubes are acceptable if the plasma is separated within 6 h of blood collection. Caution must be applied in the interpretation of absolute HIV-1 RNA copy number values obtained with suboptimal specimen collection and processing procedures.
The level of human immunodeficiency virus type 1 (HIV-1) RNA in plasma of infected individuals serves as an indicator of total viral load and can be used to infer viral replication rates (10, 17). Quantitation of plasma HIV-1 RNA levels has been shown to be an important marker of disease progression, antiretroviral therapy efficacy, and risk of perinatal transmission (6–9, 12). A variety of commercial assays for quantitation of HIV-1 RNA in patient samples have recently become available (4, 16, 20). The availability of standardized laboratory assays for HIV-1 RNA quantitation should help to decrease both intra- and interlaboratory variability in results, which is essential both in clinical trials and for patient management. A better understanding of the factors influencing the outcome of quantitative HIV-1 RNA assays is needed to compare results from studies conducted with the various methodologies in real time or with frozen, batched specimens and to provide clinicians with the most accurate and reproducible results for use in patient management.
The AMPLICOR HIV-1 MONITOR assay uses reverse transcriptase (RT) PCR amplification to achieve high levels of sensitivity (≥400 HIV-1 RNA copies/ml) in the quantitation of HIV-1 RNA levels (16). PCR has limitations as a quantitative assay, however, including decreased efficiency at higher initial copy numbers (5) and inhibition by heparin present in blood samples (1). The accuracy of quantitative HIV-1 RNA measurements in plasma specimens can be affected by both the choice of anticoagulant and specimen-handling procedures (13, 19, 21). In order to determine the effects of specimen collection and handling procedures on quantitation of plasma HIV-1 RNA, we compared anticoagulants and sample processing times with quantitative RT-PCR. We also analyzed possible patient variables influencing HIV-1 RNA loss, including age, gender, pregnancy status, antiretroviral treatment status, initial HIV-1 RNA copy numbers, and CD4 cell counts.
(This work was presented in part at the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy, New Orleans, La., September 1996.)
MATERIALS AND METHODS
Study population.
Twenty HIV-1-seropositive patients monitored at the University of California, Los Angeles, MCIC and CARE clinics were studied, with informed consent obtained under the guidelines of the UCLA Human Subjects Protection Committee. The study population consisted of 12 adults ranging from 22 to 38 years old and 8 children ranging from 2 to 16 years old. The current antiretroviral treatment history, CD4 cell count, and Centers for Disease Control and Prevention (CDC) classification (2, 3) were obtained for each patient at the time of sample collection.
Serological assays.
HIV-1 antibodies were detected by enzyme immunoassay (Abbott Laboratories, North Chicago, Ill.) and confirmed by Western blot assay (Du Pont, Wilmington, Del.).
Sample preparation.
Whole blood was collected into EDTA, acid citrate dextrose (ACD), and heparinized VACUTAINER tubes (Becton Dickinson, Franklin Lakes, N.J.), aliquoted, and stored at room temperature. Plasma was separated from the whole-blood aliquots within 1 h of collection (baseline) and then at 3, 6, 24, and 48 h postcollection. Blood was centrifuged at 400 × g for 10 min in a Sorvall RT 6000B swinging bucket centrifuge (Du Pont) at room temperature. Plasma was transferred to a new tube, and platelets were removed by a second spin at 800 × g for 10 min. The clarified plasma aliquots were stored at −70°C until use.
Quantitative RNA PCR.
Quantitative HIV-1 RNA PCR was performed on batched plasma samples with the AMPLICOR HIV-1 MONITOR assay according to the manufacturer’s instructions (Roche Molecular Systems, Somerville, N.J.). RNA was prepared from EDTA- and ACD-treated plasma according to the MONITOR assay protocol. Briefly, plasma samples (200 μl) were added to lysis buffer (600 μl) containing guanidinium thiocyanate and an internal quantitation standard. After a 10-min incubation, RNA was precipitated by the addition of 800 μl of isopropanol. RNA was pelleted by centrifugation at 16,000 × g for 15 min in a microcentrifuge (Brinkman Instruments, Westbury, N.Y.), washed once with 70% ethanol, and pelleted again at 16,000 × g for 5 min. RNA was resuspended in a low-ionic-strength diluent (400 μl). Heparinized samples were pretreated for 1 h at room temperature with 7.5 units of heparinase I (Sigma Chemical Company, St. Louis, Mo.) in 1× heparinase buffer (15 mM Tris base, 3 mM CaCl2, 45 mM NaCl, 0.02% bovine serum albumin [pH 7.5]). The heparinase I reaction was stopped by the addition of EDTA to a final concentration of 10 mM prior to extraction by the MONITOR assay protocol.
Fifty microliters of each prepared RNA sample was used for PCR. Following amplification and detection of the PCR product, the starting HIV-1 RNA copy number in each sample was calculated by comparison with the internal quantitation standard, and results were expressed as the numbers of HIV-1 RNA copies per milliliter of plasma. Baseline (≤1 h postcollection) HIV-1 RNA levels were determined in duplicate and the results were averaged.
Statistics.
Descriptive statistics are provided as means ± standard deviations (SD). Correlations between patient variables were determined with the Spearman rank correlation coefficient. Virologic variables between groups were compared by the Mann-Whitney and Kruskal-Wallis tests. Statistical significance was defined as a P value of less than 0.05.
RESULTS
Study population.
The average CD4 cell count for the 12 adults enrolled in this study was 275 ± 185/mm3 (range, 22 to 522/mm3). Among the 12 adults in the study there were 4 men, 4 nonpregnant women, and 4 pregnant women. Nine of these 12 adults were receiving zidovudine (ZDV) monotherapy at the time of sample collection. All 12 adults had symptomatic HIV-1 disease (CDC classes B2 through C3). Among the eight children in our study, the average CD4 count was 527 ± 478/mm3 (range, 14 to 1,429/mm3). Six of the eight children had symptomatic disease (CDC classes A2 through C3), and five were being treated with ZDV monotherapy at the time of sample collection. A total of 360 quantitative HIV-1 RNA PCRs were run on the samples from the 20 patients enrolled in this study.
Effects of anticoagulants on baseline plasma HIV-1 RNA levels.
The HIV-1 RNA copy numbers measured at baseline varied over a wide range for all three anticoagulants, as would be predicted from the range in CD4 cell counts among study participants (Fig. 1A). Baseline (within 1 h postcollection) HIV-1 RNA levels were highest in plasma collected in VACUTAINER tubes with EDTA as the anticoagulant. HIV-1 RNA levels in plasma collected in EDTA VACUTAINER tubes were, on average, 14% higher than those measured in ACD tubes and 31% higher than those measured in heparinized tubes (P < 0.0001) (Fig. 1B) at baseline.
FIG. 1.
(A) Baseline (≤1 h) HIV-1 RNA copy numbers in plasma collected from infected individuals with three anticoagulants. HIV-1 RNA levels are expressed as the numbers of HIV-1 RNA copies per milliliter of plasma. Symbols: ○, patients who were being treated with ZDV at the time of specimen collection; •, patients who were not receiving ZDV at the time of specimen collection. Horizontal bars indicate the means of the measured variables. (B) Comparison of HIV-1 RNA copy numbers, as determined by RT-PCR, of EDTA-, ACD-, and heparin-treated plasma at baseline. Results are expressed as percentages of the mean EDTA baseline HIV-1 RNA copy number. Horizontal bars indicate SD. P values were determined by the Kruskal-Wallis test.
Stability of HIV-1 RNA in whole blood.
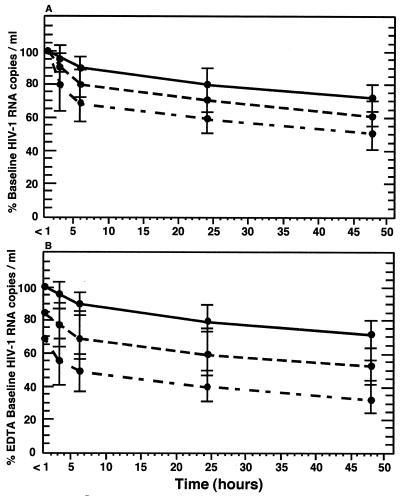
Whole-blood aliquots were made within 1 h of blood collection, and the aliquots were held at room temperature for either 3, 6, 24, or 48 h before plasma processing. Plasma HIV-1 RNA levels decreased over time in all anticoagulant tubes tested but were most stable when collected in EDTA tubes (Fig. 2A). At 24 h postcollection, HIV-1 RNA levels had decreased by 20, 29, and 40% in EDTA, ACD, and heparinized plasma, respectively (P ≤ 0.0001) (Table 1). In comparison to baseline EDTA plasma, ACD and heparinized plasma samples had 39 and 60% less HIV-1 RNA, respectively, when measured at 24 h postcollection (P ≤ 0.0001) (Fig. 2B; Table 1).
FIG. 2.
(A) Stability of HIV-1 RNA in whole blood stored at room temperature. Results are expressed as mean percentages of baseline copy numbers for EDTA (top curve), ACD (middle curve), and heparinized (bottom curve) plasma. Horizontal bars represent SD. (B) Stability of HIV-1 RNA in whole blood stored at room temperature in comparison to mean baseline (≤1 h) EDTA-treated specimens. Results are expressed as mean percentages of EDTA baseline copy numbers for EDTA (top curve), ACD (middle curve), and heparinized (bottom curve) plasma. Horizontal bars represent SD.
TABLE 1.
Stability of HIV-1 RNA in whole blood stored at room temperaturea
| Time post- collection (h) | % of baseline HIV-1 RNA in:
|
% of EDTA baseline HIV-1 RNA in:
|
|||
|---|---|---|---|---|---|
| EDTA | ACD | Heparin | ACD | Heparin | |
| <1 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 86 ± 12 | 69 ± 13 |
| 3 | 95 ± 8 | 90 ± 10 | 80 ± 14 | 77 ± 13 | 54 ± 13 |
| 6 | 90 ± 7 | 80 ± 10 | 69 ± 12 | 69 ± 14 | 48 ± 12 |
| 24 | 80 ± 8 | 71 ± 9 | 60 ± 12 | 61 ± 12 | 41 ± 11 |
| 48 | 72 ± 8 | 61 ± 8 | 51 ± 11 | 53 ± 10 | 34 ± 12 |
Baseline, <1 h. Values are means ± SD based on quantitation of copies of HIV-1 RNA per milliliter.
The average rate of HIV-1 RNA loss in whole blood was 0.8%/h for EDTA, 1.2%/h for ACD, and 1.7%/h for heparinized blood samples held at room temperature for 24 h (P < 0.0001). However, as shown in Fig. 2, the rate of HIV-1 RNA loss was greater during the first 6 h postcollection for all three anticoagulants tested. The decay rate during the first 6 h of room temperature storage was 1.8%/h for EDTA samples, 3.3%/h for ACD samples, and 5.0%/h for heparinized samples (P ≤ 0.0001) (Fig. 2). Between 6 and 48 h postcollection, the HIV-1 RNA decay rate decreased dramatically and remained consistent for all three anticoagulants. This suggests the presence of a population of highly labile HIV-1 virions in the blood of infected patients which is degraded rapidly in the first few hours postcollection.
Correlation between patient variables and HIV-1 RNA loss.
We also correlated a variety of patient parameters with the average rate of HIV-1 RNA loss over time. The stability of HIV-1 RNA in plasma was not influenced by the status of the patient as adult versus child (RNA loss, 0.849%/h versus 0.761%/h [P = 0.643]) or male versus female (0.761%/h versus 0.617%/h [P = 0.671]). Among the eight adult women in our study, four were pregnant and four were not pregnant at the time of sample collection; however, this did not influence the stability of HIV-1 RNA levels (RNA loss, 0.795%/h versus 0.880%/h [P = 0.797]). No correlation was seen between the use or nonuse of antiretroviral therapy and the rate of HIV-1 RNA loss over time (0.772%/h versus 0.912%/h [P = 0.322]). Neither patient CD4 count (r = 0.085; P = 0.716) nor initial HIV-1 RNA copy number (r = 0.196; P = 0.395) correlated with the stability of HIV-1 RNA in plasma stored at room temperature.
DISCUSSION
Determination of HIV-1 RNA levels in plasma is now used widely in laboratory evaluations of HIV disease status and prognosis (15, 18). Caution must be applied in the interpretation of absolute HIV-1 RNA values determined in real time for measurement of antiretroviral effects and patient management, as a variety of factors can influence the outcome of quantitative HIV-1 RNA measurements, including the method used to make the determination, the genotype of the virus, and specimen-handling procedures (13). We compared different anticoagulants and various specimen processing times in order to determine optimal handling procedures for plasma HIV-1 RNA quantitation by RT-PCR.
The differences between baseline HIV-1 RNA levels measured in EDTA and ACD plasma can be accounted for by the increased volume of anticoagulant present in ACD VACUTAINER tubes, resulting in dilutions of plasma HIV-1 RNA levels in these tubes. However, it is not clear why heparinized plasma showed the lowest baseline HIV-1 RNA levels. Because heparin has been shown to inhibit PCR (11), we attempted to inactivate heparin by pretreating the plasma samples with heparinase. The significantly lower levels of HIV-1 RNA measured in heparinized plasma at baseline may, therefore, reflect incomplete inactivation by heparinase.
The fact that plasma HIV-1 RNA levels decreased over time in blood stored in all three anticoagulants but were most stable in EDTA-treated whole blood is of major importance both for in-house assays and in clinical trials which may involve shipment of whole-blood specimens overnight at ambient temperatures to control laboratories. HIV-1 RNA levels showed a significant decline from baseline in heparin-treated samples after as little as 24 h of storage, indicating that heparinized blood is not suitable for studies involving shipment of blood at ambient temperatures. Interestingly, plasma HIV-1 RNA levels decreased more rapidly in the first 6 h postcollection than in hours 6 to 48 with all three anticoagulants tested. This may reflect the presence of defective virions in which RNA may be degraded by RNase present in plasma soon after blood draw.
In this study, we found that the stability of HIV-1 RNA in whole blood was not influenced by a variety of individual patient parameters, including gender, age, and pregnancy status. We also saw no correlation between antiretroviral treatment status, initial CD4 count, or initial HIV-1 RNA copy number and the rate of HIV-1 RNA loss over time. These data strongly suggest that anticoagulants and specimen processing times can influence RNA stability, while host factors seem to be less important.
Proper specimen-handling procedures are of major importance in the design of new clinical trials and the interpretation of previous trials involving quantitative HIV-1 RNA analysis. Samples collected during such studies may not have been processed within optimal time limits or with EDTA used as an anticoagulant. Direct comparison of previous reports shows that HIV-1 RNA levels measured in patients with similar profiles by different methods can give rise to severalfold-different absolute copy numbers (14, 17, 20). Our results indicate strongly that valid comparisons of study results will occur only if all studies involved use the same specimen-handling procedures and quantitate HIV-1 RNA levels with consistent methodologies.
In summary, samples collected in EDTA VACUTAINER tubes provided the highest baseline HIV-1 RNA levels and showed the best stability of HIV-1 RNA levels in whole blood stored up to 48 h at room temperature. These results indicate that EDTA should be the anticoagulant of choice for quantitative HIV-1 RNA PCR performed either in-house or following overnight shipment to a testing facility. Samples collected in ACD tubes are acceptable for quantitation of HIV-1 RNA levels if plasma is separated within 6 h of blood draw. In contrast, heparinized plasma is poorly suited to HIV-1 RNA quantitation by RT-PCR due both to difficulties in specimen processing and to the poor quality of results generated regardless of the length of storage. Our results indicate clearly that tube type and sample processing time should be consistent in order to ensure optimal and reproducible results with quantitative HIV-1 RNA PCR.
ACKNOWLEDGMENTS
This research was supported in part by grants HD30629 and HD26621 from the National Institute of Child Health and Human Development, by grants ACTG AI27550, AI30629, and AI32440 from the National Institute of Allergy and Infectious Diseases, by Universitywide AIDS Research Program grant R91-LA-152, and by Clinical Research Center grant RR-00-865.
REFERENCES
- 1.Beutler E, Gelbart T, Kuhl W. Interference of heparin with polymerase chain reaction. BioTechniques. 1990;90:166–167. [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1994 revised classification system for human immunodeficiency virus infection in children less than 13 years of age. Morbid Mortal Weekly Rep. 1994;43:1–9. [Google Scholar]
- 3.Centers for Disease Control. Guidelines for prophylaxis against Pneumocystis carinii pneumonia for persons infected with human immunodeficiency virus. Morbid Mortal Weekly Rep. 1989;5:1–9. [PubMed] [Google Scholar]
- 4.Dewar R L, Highbarger H C, Sarmiento M D, Todd J A, Vasudevachari M B, Davey R T, Jr, Kovacs J A, Salzman N P, Lane H C, Urdea M S. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis. 1994;170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 5.Dickover R E, Donovan R M, Goldstein E, Dandekar S, Bush C E, Carlson J R. Quantitation of human immunodeficiency virus DNA by using the polymerase chain reaction. J Clin Microbiol. 1990;28:2130–2133. doi: 10.1128/jcm.28.9.2130-2133.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickover R E, Garratty E M, Herman S A, Sim M S, Plaeger S, Boyer P J, Keller M, Deveikis A, Stiehm E R, Bryson Y J. Identification of levels of maternal HIV-1 RNA associated with risk of perinatal transmission. Effect of maternal zidovudine treatment on viral load. JAMA. 1996;275:599–605. [PubMed] [Google Scholar]
- 7.Fang G, Burger H, Grimson R, Tropper P, Nachman S, Mayers D, Weislow O, Moore R, Reyelt C, Hutcheon N, Baker D, Weiser B. Maternal plasma human immunodeficiency virus type 1 RNA level: a determinant and projected threshold for mother-to-child transmission. Proc Natl Acad Sci USA. 1995;92:12100–12104. doi: 10.1073/pnas.92.26.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galetto-Lacour A, Yerly S, Perneger T V, Baumberger C, Hirschel B, Perrin L the Swiss HIV Cohort Study Group. Prognostic value of viremia in patients with long standing human immunodeficiency virus infection. J Infect Dis. 1996;173:1388–1393. doi: 10.1093/infdis/173.6.1388. [DOI] [PubMed] [Google Scholar]
- 9.Henrard D R, Phillips J F, Muenz L R, Blattner W A, Wiesner D, Eyster M E, Goedert J J. Natural history of HIV-1 cell-free viremia. JAMA. 1995;274:554–558. [PubMed] [Google Scholar]
- 10.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 11.Holodniy M, Kim S, Katzenstein D A, Konrad M, Groves E, Merigan T C. Inhibition of human immunodeficiency virus gene amplification by heparin. J Clin Microbiol. 1991;29:676–679. doi: 10.1128/jcm.29.4.676-679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holodniy M, Mole L, Margolis D, Moss J, Dong H, Boyer E, Urdea M, Kolberg J, Eastman S. Determination of human immunodeficiency virus RNA in plasma and cellular viral DNA genotypic zidovudine resistance and viral load during zidovudine-didanosine combination therapy. J Virol. 1995;69:3510–3516. doi: 10.1128/jvi.69.6.3510-3516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holodniy M, Mole L, Yen-Lieberman B, Margolis D, Starkey C, Carroll R, Spahlinger T, Todd J, Jackson J B. Comparative stabilities of quantitative human immunodeficiency virus RNA in plasma from samples collected in VACUTAINER CPT, VACUTAINER PPT, and standard VACUTAINER tubes. J Clin Microbiol. 1995;33:1562–1566. doi: 10.1128/jcm.33.6.1562-1566.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in the plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 16.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 18.Shearer W T, Quinn T C, LaRussa P, Lew J F, Almy M L S, Rich K, Handelsman E, Diaz C, Pagano M, Smeriglio V, Kalish L A. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N Engl J Med. 1997;336:1337–1342. doi: 10.1056/NEJM199705083361901. [DOI] [PubMed] [Google Scholar]
- 19.van Bueren J, Simpson R A, Jacobs P, Cookson B D. Survival of human immunodeficiency virus in suspension and dried onto surfaces. J Clin Microbiol. 1994;32:571–574. doi: 10.1128/jcm.32.2.571-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 21.Winters M A, Tan L B, Katzenstein D A, Merigan T C. Biological variation and quality control of plasma human immunodeficiency virus type 1 RNA quantitation by reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1993;31:2960–2966. doi: 10.1128/jcm.31.11.2960-2966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]