Abstract
Ultrasound features related to thyroid lesions structure, shape, volume, and margins are considered to determine cancer risk. Automatic segmentation of the thyroid lesion would allow the sonographic features to be estimated. On the basis of clinical ultrasonography B-mode scans, a multi-output CNN-based semantic segmentation is used to separate thyroid nodules' cystic & solid components. Semantic segmentation is an automatic technique that labels the ultrasound (US) pixels with an appropriate class or pixel category, i.e., belongs to a lesion or background. In the present study, encoder-decoder-based semantic segmentation models i.e. SegNet using VGG16, UNet, and Hybrid-UNet implemented for segmentation of thyroid US images. For this work, 820 thyroid US images are collected from the DDTI and ultrasoundcases.info (USC) datasets. These segmentation models were trained using a transfer learning approach with original and despeckled thyroid US images. The performance of segmentation models is evaluated by analyzing the overlap region between the true contour lesion marked by the radiologist and the lesion retrieved by the segmentation model. The mean intersection of union (mIoU), mean dice coefficient (mDC) metrics, TPR, TNR, FPR, and FNR metrics are used to measure performance. Based on the exhaustive experiments and performance evaluation parameters it is observed that the proposed Hybrid-UNet segmentation model segments thyroid nodules and cystic components effectively.
Keywords: Thyroid ultrasound, Semantic segmentation, SegNet, U-Net, Hybrid-UNet
Introduction
The thyroid is located lower behind the front neck, a small gland in the shape of a butterfly. It regulates human metabolism. It releases a secret hormone that controls human activities, including energy, heat, heart rate, temperature, and oxygen [1, 2]. The human body is harmed when the release of hormones is improper due to the nodule's aberrant growth [1, 2]. Various imaging modalities like mammogram, Ultrasound (US), CT Scan, thermal images, and MRI have been widely used to detect thyroid nodules at an initial stage so that the patient's chance of survival can be increased [3–5]. The limitation of mammogram, MRI, CT, and thermal imaging lie in ionizing, cost, and availability, which might be harmful or easy to use to the patient [3, 4, 6]. US imaging modality is most popular to identify thyroid nodules as an initial screening test over MRI, CT, and thermal imaging [7, 8]. Owing to the disadvantages of the other imaging modalities, the US is considered as a first-line treatment to identify thyroid nodules due to its cost, availability, and no harm to the patient [9]. Low contrast and speckle noise often degrade US images' visual quality, which affect the radiologists' interpretation [10, 11]. The presence of speckle-noise reduces contrast resolution, making it more difficult to identify lesions during diagnosis [12, 13]. Over the past two decades, computer-based research of thyroid tumor US images has been extensively examined. The main objective of this work is to improve detectability during thyroid nodule screening using computer-based algorithms to assist radiologists in abnormality diagnosis [14, 15].
Segmenting thyroid US images may be difficult due to the lack of contrast between different anatomies and the existence of speckle noise [16]. Researchers have proposed different thyroid nodule segmentation techniques to more precisely segment the thyroid nodules according to their size, shape, and location [17]. Most of the segmentation methods are referred to as manual masks to indicate the segmentation algorithms. These algorithms were not suitable for real-time application for diagnosing thyroid nodules. Deep learning (DL)-based algorithms improve the graphics and do not require an initial mask. It is fully automated with an interaction time of just a few seconds and enables real-time implementation. DL-based segmentation algorithms are separated into two categories, i.e., semantic and instance segmentation. Each image pixel belongs to a particular class in the semantic segmentation, while instance segmentation splits distinct objects that belong to the same class [18–21]. Semantic segmentation classifies each pixel with a corresponding class label, i.e., belonging to the background or lesion [19]. Semantic segmentation differs from object detection as it does not predict any bounding boxes around the lesion [22]. A common architecture for semantic segmentation is based on encoder–decoder. It contains of three structure blocks, i.e., convolutional, downsampling, and upsampling. The encoder is a pre-training convolution network, while a decoder consists of a deconvolution layer, interpolation, and upsampling layer [19, 23] 24. The purpose of downsampling is to capture semantic or context information, while upsampling is to recover spatial resolution. The encoder network extracts the feature map, while the decoder network is used to recover channels' resolution. An encoder-decoder structure of segmentation algorithms is widely used to define boundaries between the images. From the executive review of literature, it is observed that semantic segmentation models have been widely used for US images of organs like heart [8, 10], kidney [25–27], breast [28–31], liver [32–34]. In the present work, an ideal despeckling algorithm is used to smoothen the image's homogenous area while preserving the edges of the lesion boundary. Semantic segmentation-based Hybrid-UNet model is proposed to segment the thyroid lesion from the original and pre-processed thyroid US images.
This paper is further arranged as: in Sect. 2 related work and significant findings represented, Sect. 3 elaborates materials and methods, in Sect. 4 result and discussions are presented and in Sect. 5 conclusion of the study is presented.
Related work and significant findings
A brief review of literature for thyroid nodules segmentation using encoder-decoder-based semantic segmentation models is summarized in Table 1.
Table 1.
A brief literature review of thyroid nodules segmentation using semantic segmentation
| Investigator(s) | No. of image(s) | Pre-processing algorithm | Segmentation algorithm | Evaluation metric(s) |
|---|---|---|---|---|
| W. Song et al. (2015) [35] | 4309 | – | VGG16 | mAP—98.2% |
| H. Ravishankar et al. (2016) [36] | 140 | – | Hybrid-CNN | DC—0.9 |
| J. Ma et al. (2017) [37] | 22,123 | – | Self-Design | DR—0.92 |
| Jinlian Ma et al. (2018) [38] | 22,123 | – | Self-Design | DR—0.95 |
| Xuewei Li et al. (2018) [39] | 300 | – | FCN-TN | IoU—91% |
| J. Wang et al. (2018) [40] | 3459 | – | VGG16 | IoU—0.75 |
| S. Zhou et al. (2018) [41] | 893 | – | MG-UNet | DSC—0.94 |
| X. Ying et al. (2018) [42] | 1000 | – | SegNet (VGG19) | IoU—87% |
| P. Poude et al. (2019) [43] | 675 and 1600 | – | U-Net | DSC—0.87 & 0.86 |
| J. Ding et al. (2019) [44] | 1936 | – | ReAgU-Net |
mIoU—0.78 DSC—0.86 |
| V. Kumar et al. (2020) [45] | 914 | – | MPCNN | DSC—0.62 |
| Webb, Jeremy M. (2020) [46] | 120Patients | – | DeepLabv3 + | IoU—0.73 |
| Prabal Poude et al. (2018) [47] | 1416 | HE & MF | U-Net | DC—0.87 |
| Jianguo Sun et al. (2018) [48] | 173 | AMF & HE | FCN-AlexNet | IoU- 0.81 |
| M. Buda et al. (2019) [49] | 1278 |
Contrast stretching |
U-Net | DCS- 0.93 |
| Zihao Guo et al. (2020)[50] | 1400 | HE | DeepLabv3 + | DSC—94.08% |
| Gomes Ataide (2021)[51] | 6066 | Resizing and cropping | ResUNet | DC—0.85 & IoU—0.767 |
Note - HE - histogram equalization, MF - median filter, AMF - adaptive median filter, DCS/DC - dice coefficient, IoU - Intersection of Union, DR - Dice ratio, mAP - mean average precision
From the literature it is observed that different detection methods such as SSD, R-CNN, and YOLO[52–61] are commonly used to detect the bounding box around the lesion (height, width, and position) in thyroid US images. So, it is concluded that thyroid nodules instance detection does not help in the further analysis to design a CAD (computer-based diagnosis system) for characterization of thyroid nodules.
Table 1 shows that several segmentation methods based on self-design, FCN, SegNet and U-Net are used for original thyroid US images. It is observed that Zhou et al. [41] achieved the highest dice coefficient (0.94) using MG-UNet-based algorithm. In literature, very few pre-processing algorithms (only Contrast stretching, adaptive median, median filters, and Histogram equalization) used before the segmentation algorithms for pre-processing of thyroid US images. Most researchers used U-Net (DAG) architecture using simple convolution to segment thyroid nodules using pre-processed thyroid US images. Further it is also observed that limited work is available in the literature on pre-processing thyroid US images for segmentation of thyroid nodule. So, in this work efficient despeckling filters used for enhancing the performance of the segmentation model [62, 63].
Materials and methods
Here, extensive experiments conducted by combining the two publicly accessible benchmark datasets, i.e., DDTI [64] and ultrasoundcases.info (USC) [65]. Out of these two datasets 820 thyroid tumor images selected for experiment purpose. Best despeckle filtering algorithm is selected from a wide range of 64 despeckling algorithms based on diagnostically important features like structure, edge, and margin preservation. The best performing filter is selected objectively from these despeckling filters. The semantic segmentation networks are considered from a void variety of segmentation models, including SegNet (VGG16), U-Net, and proposed Hybrid-UNet. The mean intersection of union (mIoU), mean dice coefficient (mDC), TPR, TNR, FPR, and FNR metrics [66–68] used extensively for the objective assessment of segmentation models.
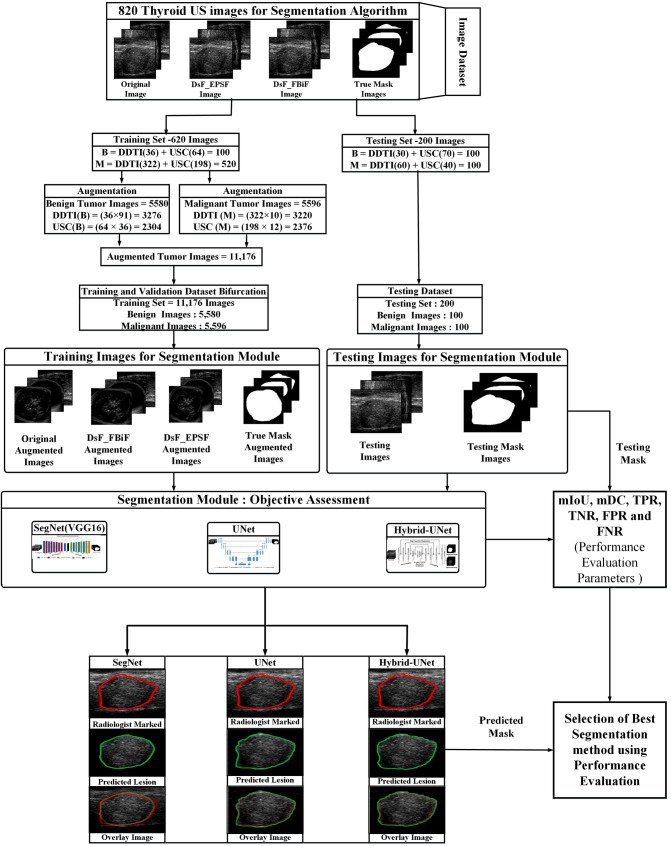
Workflow adopted for the segmentation of thyroid US is presented in Fig. 1. The description of phases used in this work is given below:
Fig. 1.
The workflow adopted for the segmentation of thyroid tumor US images
Phase I: dataset preparation module
Following steps involved in dataset preparation module—(a) Benchmark dataset for thyroid US images (b) Image resizing module for resizing of thyroid US image (c) True mask generation using createMask function and (d) Data/image augmentation.
Benchmark dataset for thyroid US images
820 thyroid US images selected from benchmark datasets of DDTI and ultrasoundcases.info (USC). out of these 820 images 620 images used for training. In 620 training images 36 benign thyroid tumor US images (TTUS) and 322 malignant TTUS images taken from DDTI dataset and 64 benign TTUS and 198 malignant TTUS images taken from USC dataset. For testing purpose 200 images selected from both the datasets. In 200 testing images, 30 benign TTUS and 60 malignant TTUS images taken from DDTI dataset and 70 benign TTUS and 40 malignant TTUS images taken from USC dataset.
Image resizing module
In this module, the unwanted information is removed from the thyroid US images and the resulting US images are resized. The size of the images selected is of 256 × 256 pixels. It is clinically significant to resize the images while maintaining the aspect ratio. Xiaofeng Qi et al. [69] suggested that direct resizing of the images without considering the tumor's shape or lesions and aspect ratio changes is not desirable. In order to preserve the tumor shape by maintaining the aspect ratio, adequate attention is used during resizing the thyroid US images in this work.
True mask generation
In this module, A binary mask defines image pixels belonging to the tumor or background region. The mask’s pixel values outside the lesion is set to 0, and pixel values inside the lesion is set to 1. The binary mask size is the same as that of the input image. The createMask (h) MATLAB function generates a binary mask where h defines the tumor region (ROI object) in this study. There are four different ROI objects available to create the mask h function, i.e. ellipse, point, poly, and rectangle. In the present work, the Poly object is used for generating a binary mask by expanding the interactive polygon to match the shape and size of the tumor region.
Data/image augmentation
Data/image augmentation is a technique that enlarges the dataset by applying certain transformations. 820 thyroid ultrasound images are inadequate to train a DL-based semantic segmentation network. In the present work, rotation (90° and 180°), translation, horizontal and vertical flip, and rotation of flipped images done to enlarge the 620 TTUS images which are selected for training purpose. By applying augmentation methods, 620 TTUS images converted into 11,176 images for training purpose as shown in Fig. 1.
Phase II: Despeckle filtering module
Low-contrast and speckle noise differential diagnosis between these characteristics are complex even for an experienced radiologist [9, 12]. Therefore, controlled despeckling is preferred to preserve the diagnosis information in the images, and controlled despeckling is allowed by eliminating speckle noise from homogenous areas of the region and preserving the edges of the lesion boundary. As proven in the authors' previous study DsF_FBiF and DsF_EPSF filters outperformed to reduce the speckle noise from thyroid US images by preserving edges, structure, and margin[9, 70]. So DsF_FBIF and DsF_EPSF filters used here for despeckling purpose.
Phase III: segmentation module
The main goal of segmentation is to divide an image into several distinct, non-overlapping sections that fully characterize the test object in the image [71–74]. A key task in the care of the subject being examined is the blueprint or dividing of a thyroid nodule from the background [73, 75, 76]. Generally, low SNR in ultrasound images in the form of speckle-noise degrades the margin detection or region obtained from the appropriate method [71]. The segmentation model's performance might be enhanced by despeckling US images. In the case of thyroid ultrasound, several studies have reported using conventional-based segmentation methods [44, 54, 64]. It is observed from the literature that the conventional approach becomes a semi-automatic approach for initializing the mask. Thus, an automatic approach is required. Here, in this study Hybrid-UNet semantic segmentation technique is proposed for automatic mask initialization. Hybrid-UNet is created using SegNet and UNet based semantic segmentation architecture.
Semantic segmentation
The objective of semantic image segmentation is to assign a class to each pixel in an image that corresponds to the object being represented [84–86]. The architecture of semantic segmentation based on encoder-decoder where trainable engine is called an encoder network, and pixel-wise classification is called a decoder network [41, 47, 87].
The SegNet architecture is a deep encoder-decoder model developed by a computer vision research community at Cambridge University for pixel-wise segmentation [19, 88]. It uses VGG16 pre-trained model with a convolution filter of the size of 3 × 3, batch normalization, activation-ReLU (non-linear), 2 × 2 max-pooling, and subsampling on the encoder side. The SegNet architecture is the pre-trained end-to-end network that preserves high-frequency components and decreases the trainable parameters on the decoder side [23, 24, 89].
The U-Net architecture was designed by Ronneberger, Fischer, and Bronx [90]. The U-Net architecture has two paths (a) contraction/encoder and (b) expansion/decoder path [87]. The contraction path followed by the expansion path delivers a U-shaped network. The convolution and the max-pooling layers reduce the spatial information while preserving the feature map [87, 90].
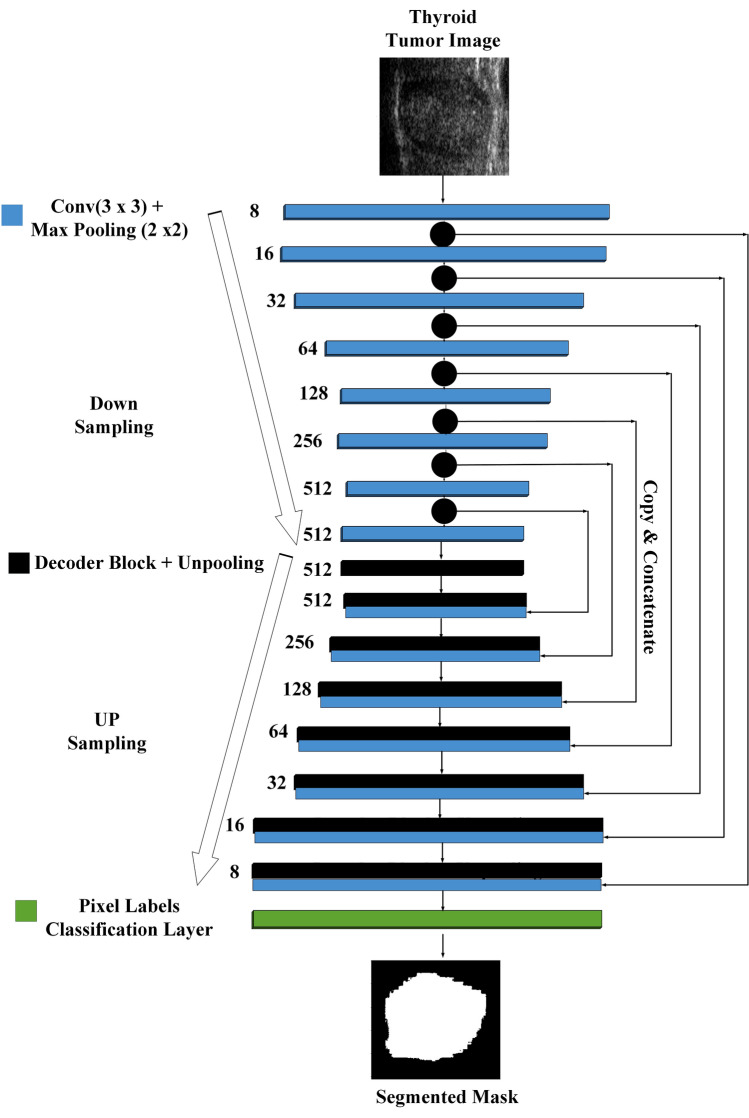
Semantic segmentation is based on FCN or classical architecture like SegNet and U-Net is trendy segmentation method for medical images. A general U-Net architecture consisting of a complete path follows a convolution network with the max-pooling operation and continued down sample feature map. As encoder in U-net, we used a relatively similar CNN architecture of SegNet (VGG-16) that consists 16 sequential layers. The entire pooling layer feature map is transferred between the encoder and decoder sides but with two convolution layers at each stage and then concatenated to perform the convolution operation. The first convolution has 8 channels, then doubles after each max pooling operation until it reaches 512. Here, Hybrid-UNet design is proposed for segmentation by using a combination of U-Net and SegNet(VGG16) [20]. The architecture of Hybrid-UNet applied in this study is presented in Fig. 2.
Fig. 2.
Hybrid-UNet architecture
The SegNet, U-Net and Hybrid-UNet models are trained via Adam optimizer with 32 mini-batch sizes and learning rates ϵ {10–3, 10–4, 10–5, 10–6}. The mini-batch size and learning rates are carefully selected so that number of images split in training so that the complete training dataset is passed in the models during training epochs and data cannot be discarded. Segmentation models trained for 30 epochs, and overfitting is avoided using early stopping criteria. These segmentation models (SegNet, U-Net, and Hybrid-UNet) are implemented on NVIDIA 1070Ti GPU with 2,432 CUDA cores.
Phase IV: Performance evaluation of proposed segmentation module
For the performance evaluation of the semantic segmentation methods researchers used different evaluation metrics like accuracy [37, 91], Precision [92, 93], Dice coefficient [73, 94], Jaccard index [95, 96], TPR [97], TNR [97], FPR [98], FNR [97], F-measure [99], Hausdorff distance [73], average distance [100], Mahalanobis distance [101] mutual information and variation of information [102]. Here the performance of SegNet, UNet and Hybrid-UNet segmentation models is calculated using the overlay region between the lesion marked by experienced participating radiologists and the lesion obtained from the segmentation model [68].
In this work, the area of interaction between ground truth (SR) and predicted mask (SA) calculated using the mIoU, mDC, TPR, FPR, TNR, and FNR [69–70]. The performance evaluation parameters used for the assessment of proposed segmentation algorithms are given as:
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
The sample confusion matrix obtained from the proposed Hybrid-UNet segmentation method is shown in Fig. 3.
Fig. 3.
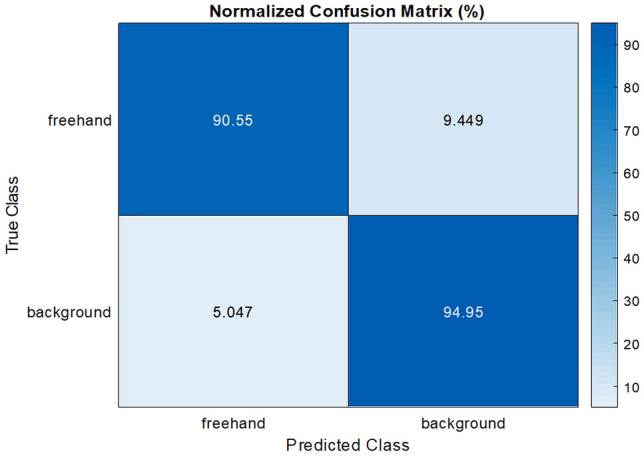
Sample confusion matrix
Result and discussions
The experiments utilized to evaluate the segmentation models are listed below in Table 2.
Table 2.
Experiments conducted for the assessment of segmentation models
| Experiment 1 | Objective assessment of segmentation models using original thyroid US images |
| Experiment 2 | Objective assessment of segmentation models using pre-processed thyroid US images using DsF_FBiF |
| Experiment 3 | Objective assessment of segmentation models using pre-processed thyroid US images using DsF_EPSF |
The performance of SegNet, UNet and proposed Hybrid-UNet segmentation models using original and despeckled thyroid US Images by DsF_FBiF and DsF_EPSF filters is tabulated in Table 3, and the best outcome is marked in grey.
Table 3.
The performance of semantic segmentation models using original and despeckled thyroid US Images by DsF_FBiF and DsF_EPSF Filters
| Experiment (Image Type) |
P.E.P | mIoU (%) | mIoU (B) (%) | mIoU (M) (%) | mDC (%) | DC (B) (%) | DC (M) (%) | TPR (%) | FNR (%) | TNR (%) | FPR (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Seg. Algo | |||||||||||
| Experiment 1 (OTTUS) |
Seg Net |
78.4 ± 16.1 | 80.3 ± 15.0 | 77.4 ± 14.1 | 87.2 ± 06.4 | 87.3 ± 07.4 | 86.4 ± 06.1 | 85.2 ± 05.8 | 14.8 ± 03.8 | 87.6 ± 4.9 | 10.4 ± 3.4 |
| UNet | 80.9 ± 11.2 | 81.4 ± 09.2 | 80.6 ± 11.1 | 89.1 ± 05.4 | 88.1 ± 04.2 | 87.4 ± 05.3 | 86.9 ± 06.9 | 13.1 ± 03.7 | 89.9 ± 3.1 | 10.1 ± 2.9 | |
| Hybrid-UNet | 83.1 ± 07.8 | 83.4 ± 08.7 | 82.2 ± 06.9 | 90.1 ± 02.9 | 91.4 ± 03.2 | 89.4 ± 0.02 | 88.2 ± 04.9 | 11.8 ± 02.8 | 90.8 ± 3.2 | 09.2 ± 2.3 | |
|
Experiment 2 (DTTUS by DsF_FBiF Filter) |
SegNet | 79.4 ± 13.2 | 80.3 ± 11.1 | 79.1 ± 12.4 | 88.7 ± 05.8 | 89.4 ± 06.2 | 88.1 ± 04.8 | 87.6 ± 04.9 | 12.4 ± 03.1 | 88.2 ± 4.1 | 08.8 ± 2.9 |
| UNet | 82.1 ± 09.7 | 84.0 ± 07.1 | 82.8 ± 09.2 | 89.3 ± 05.1 | 89.9 ± 05.7 | 88.8 ± 04.3 | 87.9 ± 05.9 | 12.1 ± 03.8 | 89.1 ± 3.1 | 10.9 ± 2.5 | |
| Hybrid-UNet | 85.2 ± 06.2 | 86.2 ± 05.8 | 84.4 ± 05.7 | 90.6 ± 02.8 | 90.7 ± 0.02 | 89.4 ± 0.03 | 89.7 ± 04.1 | 10.3 ± 02.7 | 90.8 ± 3.2 | 09.2 ± 2.1 | |
|
Experiment 3 (DTTUS by DsF_EPSF Filter) |
SegNet | 80.4 ± 12.8 | 81.4 ± 11.7 | 79.3 ± 15.4 | 90.0 ± 05.1 | 90.4 ± 04.9 | 89.9 ± 05.2 | 89.1 ± 04.9 | 10.9 ± 03.0 | 90.4 ± 4.1 | 09.6 ± 3.1 |
| UNet | 83.2 ± 09.7 | 85.1 ± 08.5 | 83.6 ± 11.1 | 92.9 ± 03.9 | 93.1 ± 03.8 | 91.9 ± 04.1 | 92.3 ± 04.9 | 07.7 ± 02.8 | 93.2 ± 3.3 | 06.8 ± 2.9 | |
| Hybrid-UNet | 86.6 ± 09.8 | 88.8 ± 8.7 | 85.6 ± 09.5 | 93.2 ± 03.1 | 93.9 ± 02.8 | 92.8 ± 03.2 | 90.5 ± 03.1 | 09.5 ± 02.6 | 94.9 ± 2.7 | 05.1 ± 2.0 |
Note - P.E.P. - performance evaluation parameters, OTTUS - Original Thyroid Tumor US Images, DTTUS - Despeckled Thyroid Tumor US Images, B - benign, M - malignant
In Table 3 the performance of semantic segmentation models (i.e. SegNet, U-Net and Hybrid-UNet using original and despeckled thyroid US images is presented in terms of mIoU, mDC, TPR, TNR, FPR, and FNR. It is noticed that proposed Hybrid-UNet segmentation model perform better segmentation, with the highest values of mIoU (86.6%) & mDC (93.2%) for thyroid US images filtered by DsF_EPSF while separately in case of benign tumor images 88.8% mIoU 93.9% mDC and for malignant tumor images 85.6% mIoU & 92.8% mDC noticed. As shown in Table 4, the impact of the despeckling algorithm on the performance of the proposed Hybrid-UNet segmentation model is also examined.
Table 4.
Effect of despeckling algorithm on the performance of proposed Hybrid-UNet segmentation model
| Type of Images | Assessment Metric for Despeckle filter (SEPI) | Overall mIoU (%) | Overall mDC (%) | TPR (%) | FNR (%) | TNR (%) | FPR (%) |
|---|---|---|---|---|---|---|---|
| Original | – | 83.1 ± 7.8 | 90.1 ± 2.9 | 88.2 ± 4.9 | 11.8 ± 2.8 | 90.8 ± 3.2 | 09.2 ± 2.3 |
| DsF_FBiF | 0.98 ± 0.02 | 85.2 ± 6.2 | 90.6 ± 2.8 | 89.7 ± 4.1 | 10.3 ± 2.7 | 90.8 ± 3.2 | 09.2 ± 2.1 |
| DsF_EPSF | 0.99 ± 0.01 | 86.6 ± 9.8 | 93.2 ± 3.1 | 90.5 ± 3.1 | 9.5 ± 2.6 | 94.9 ± 2.7 | 05.1 ± 2.0 |
In Fig. 4, the sample thyroid tumor US original image(a1), despeckled image by DsF_FBiF(b1) and DsF_EPSF(c1), and corresponding tumor marked by radiologist, true mask, predicted mask, predicted lesion and overlay diagram between true TTUS lesion and predicted TTUS lesion using SegNet, UNet and Hybrid-UNet models are presented.
Fig. 4.
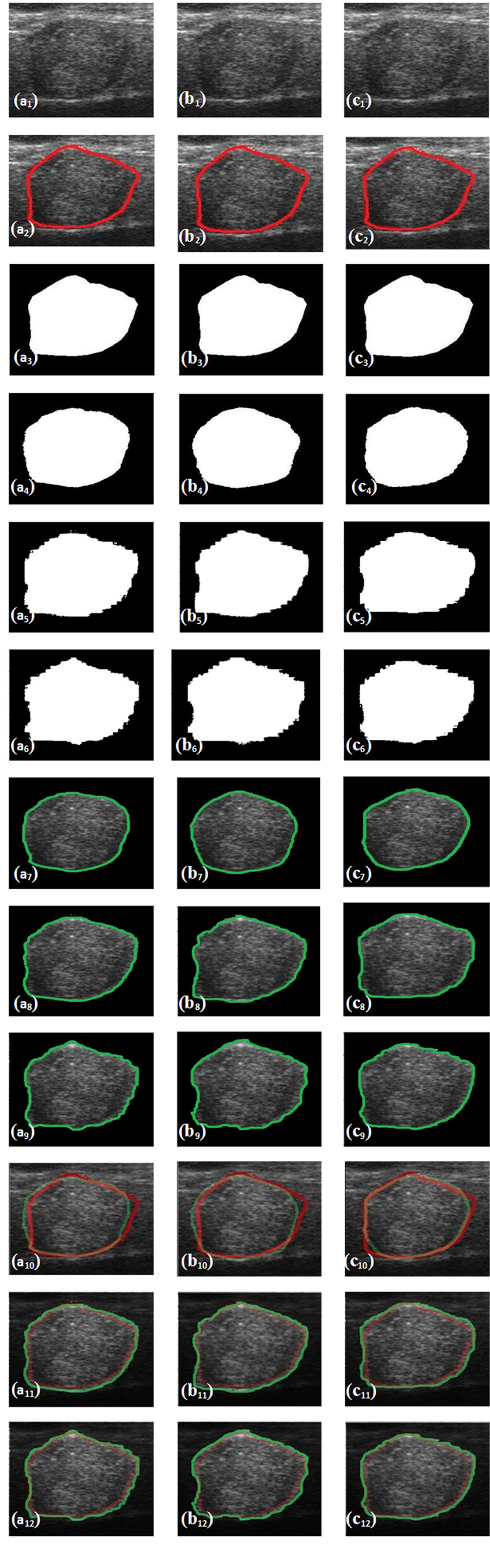
a1 Original Thyroid Tumor Ultrasound (TTUS) image, b1 despeckled TTUS image by DsF_FBiF, c1 Despeckled TTUS image by DsF_EPSF filter, a2–c2 original TTUS images with tumor marked by radiologist, a3–c3 True TTUS masked images, a4–a6 predicted masks (applied on Original TTUS) images using SegNet, UNet and Hybrid-UNet, b4–b6 predicted masked (applied on despeckled TTUS by DsF_FBiF filter) images using SegNet, UNet and Hybrid-UNet, c4–c6 predicted masked (applied on despeckled TTUS by DsF_EPSF filter) images using SegNet, UNet and Hybrid-UNet, a7–a9 predicted lesions (applied on Original TTUS) images using SegNet, UNet and Hybrid-UNet, b7–b9 predicted lesions (applied on despeckled TTUS by DsF_FBiF filter) images using SegNet, UNet and Hybrid-UNet, c7–c9 predicted lesions (applied on despeckled TTUS by DsF_EPSF filter) images using SegNet, UNet and Hybrid-UNet, a10–a12 overlay diagram between true TTUS lesion and predicted TTUS lesion (applied on Original TTUS) images using SegNet, UNet and Hybrid-UNet, b10–b12 overlay diagram between true TTUS lesion and predicted TTUS lesion (applied on despeckled TTUS by DsF_FBiF filter) images using SegNet, UNet and Hybrid-UNet, c10–c12 overlay diagram between true TTUS lesion and predicted TTUS lesion (applied on despeckled TTUS by DsF_EPSF filter) images using SegNet, UNet and Hybrid-UNet
From Table 4, it is observed that the values of performance evaluation parameters are improved using despeckled thyroid US images. The result obtained from the objective assessment of segmentation models using original and despeckled thyroid US images indicates that the DsF_EPSF filter yield better segmentation results in terms of mIoU, mDC, TPR, TNR, FPR, and FNR by using proposed Hybrid-UNet segmentation model.
A comparison of the proposed methodology Hybrid-UNet segmentation model with other existing techniques for thyroid tumor segmentation is tabulated in Table 5.
Table 5.
Comparison of the proposed methodology (Hybrid-UNet segmentation model with other existing techniques for thyroid tumor segmentation
| Investigator (s) | Number of images | Technique used for pre-processing | Segmentation model(s) | Evaluation metrics |
|---|---|---|---|---|
| Case 1: Using Original TTUS Images | ||||
| S. Zhou et al. [41] | 893 | – | MG-U-Net | Dice Coefficient -0.9 |
| J. Ma et al. [37] | 10,357 | – | Self-design | Dice ratio- 0.9 |
| Proposed method (Hybrid_UNet) | 820 | – | Hybrid-UNet | mIoU—83.1 ± 7.8 and mDC = 90.1 ± 2.9 |
| Case 2: Using pre-processed TTUS Images | ||||
| Prabal Poude et al. [47] | 416 | Median Filter and Histogram Equalization | UNet | Dice Coefficient = 0.876 |
| J. Sun et al. [48] | 173 | Adaptive Median Filter and Histogram Equalization | FCN-AlexNet | mIoU = 0.81 |
| M. Buda et al. (2019) [49] | 1278 | Contrast stretching | U-Net | Dice Coefficient = 0.93 |
| Zihao Guo et al. (2020)[50] | 1400 | Histogram Equalization | DeepLabv3 + | Dice Coefficient = 0.94 |
| Gomes Ataide et al. (2021)[51] | 6066 | Resizing and cropping | ResUNet | mDC—0.857 & mIoU—0.767 |
| Proposed method (Hybrid_UNet) | 820 | DsF_EPSF | Hybrid-UNet | mIoU—86.6 ± 9.8 and mDC = 93.2 ± 3.1 |
From Table 5 it is concluded that, Hybrid-UNet performs better in comparison to other techniques when pre-processed TTUS images using DsF_EPSF are used as input to the proposed Hybrid-UNet segmentation model.
Conclusion
In this study, extensive experiments performed to segment the thyroid tumor US images using SegNet, UNet and Hybrid-UNet using original and despeckled thyroid tumor US images using DsF_EPSF and DsF_FBiF filters. The performance of Hybrid-UNet segmentation model is compared with existing segmentation models (i.e. SegNet and UNet) in terms of mIoU, mDC, TPR, TNR, FPR, and FNR metrics. From the Tables 3 and 4 it is noticed that the Hybrid-UNet segmentation method yields better segmentation in terms of shape, margin, composition, and echogenic characteristics exhibited by lesions. In case of segmentation of original thyroid US images using proposed segmentation model 83.1% mIoU and 90.1% mDC achieved while 86.6% mIoU and 93.2% mDC achieved when despeckled TTUS images by DsF_EPSF filter are given as input to the proposed method.
The proposed model can be used to make things simpler i.e. extracting the thyroid organ with region of interest and transform the thyroid US images into meaningful subject. An efficient CAD tool for analysis and classification of thyroid US Images can be designed to take as a second opinion during the clinical treatment. It is concluded that Hybrid-UNet segmentation using DsF_EPSF filtered images, yields more clinically acceptable enhanced segmented diagnostic information concerning the proper shape, size, and margins of thyroid tumor.
Acknowledgements
The authors would like to thanks Dr. Jyotsna Sen, Sr. Professor, department of radiodiagnosis, Pt. B. D. Sharma Postgraduate Institute of Medical Sciences, Rohtak for stimulating discussions regarding different sonographic characteristics exhibited by various types of benign and malignant thyroid tumors. The first author acknowledge “National Project Implementation Unit (NPIU), a unit of Ministry of Human Resource Development, Government of India” for the financial assistantship through TEQIP-III project as Deenbandhu Chhotu Ram University of Science and Technology, Murthal, Haryana, India.
Funding
This study and authors not received any funding from other sources.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Niranjan Yadav, Email: niranjanyadav97@gmail.com.
Rajeshwar Dass, Email: rajeshwardas10@gmail.com.
Jitendra Virmani, Email: jitendra.virmani@gmail.com.
References
- 1.Gesing A. The thyroid gland and the process of aging; what is new? Thyroid Res. 2015;8:A8. doi: 10.1186/1756-6614-8-S1-A8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kongburan W, Padungweang P, Krathu W, Chan JH. Semi-automatic construction of thyroid cancer intervention corpus from biomedical abstracts. Eighth Int Conf Adv Comput Intell. 2016 doi: 10.1109/ICACI.2016.7449819. [DOI] [Google Scholar]
- 3.Chung R, Kim D. Imaging of thyroid nodules. Appl Radiol. 2019;48:16–26. doi: 10.37549/AR2555. [DOI] [Google Scholar]
- 4.Hoang JK, Sosa JA, Nguyen XV, et al. Imaging thyroid disease. updates, imaging approach, and management pearls. Radiol Clin N Am. 2014;53:145–161. doi: 10.1016/j.rcl.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Botta F, Raimondi S, Rinaldi L, et al. Association of a CT-based clinical and radiomics score of non-small cell lung cancer (NSCLC) with lymph node status and overall survival. Cancers (Basel) 2020 doi: 10.3390/cancers12061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaglan P, Dass R, Duhan M. Breast cancer detection techniques: issues and challenges. J Inst Eng Ser B. 2019;100:379–386. doi: 10.1007/s40031-019-00391-2. [DOI] [Google Scholar]
- 7.Chaudhary V, Bano S. Thyroid ultrasound. Indian J Endocrinol Metab. 2013;17:219–227. doi: 10.4103/2230-8210.109667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brillantino C, Rossi E, Baldari D, et al. Duodenal hematoma in pediatric age: a rare case report. J Ultrasound. 2022;25:349–354. doi: 10.1007/s40477-020-00545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadav N, Dass R, Virmani J. Despeckling filters applied to thyroid ultrasound images: a comparative analysis. Multimed Tools Appl. 2022 doi: 10.1007/s11042-022-11965-6. [DOI] [Google Scholar]
- 10.Biradar N, Dewal ML, Rohit MK, et al. Blind source parameters for performance evaluation of Despeckling filters. Hindawi Publ Corp J Biomed Imaging. 2016;2016:1–12. doi: 10.1155/2016/3636017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitale V, Rossi E, Di M, et al. Pediatric encephalic ultrasonography: the essentials. J Ultrasound. 2020;23:127–137. doi: 10.1007/s40477-018-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriti VJ, Agarwal R. Assessment of despeckle filtering algorithms for segmentation of breast tumours from ultrasound images. Biocybern Biomed Eng. 2019;39:100–121. doi: 10.1016/j.bbe.2018.10.002. [DOI] [Google Scholar]
- 13.Biradar N, Dewal ML, Rohit MK. Speckle Noise Reduction in B-Mode Echocardiographic Images: a comparison. IETE Tech Rev (Institution Electron Telecommun Eng India) 2015;32:435–453. doi: 10.1080/02564602.2015.1031714. [DOI] [Google Scholar]
- 14.Koundal D, Gupta S, Singh S. Speckle reduction method for thyroid ultrasound images in neutrosophic domain. IET Image Process. 2016;10:167–175. doi: 10.1049/iet-ipr.2015.0231. [DOI] [Google Scholar]
- 15.Brillantino C, Rossi E, Pirisi P, et al. Pseudopapillary solid tumour of the pancreas in paediatric age: description of a case report and review of the literature. J Ultrasound. 2022;25:251–257. doi: 10.1007/s40477-021-00587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brillantino C, Rossi E, Bifano D, et al. An unusual onset of pediatric acute lymphoblastic leukemia. J Ultrasound. 2021;24:555–560. doi: 10.1007/s40477-020-00461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav N, Dass R, Virmani J. Machine learning-based CAD system for thyroid tumour characterisation using ultrasound images. Int J Med Eng Inform. 2022;1:1–13. doi: 10.1504/IJMEI.2022.10049164. [DOI] [Google Scholar]
- 18.Dai J, He K, Sun J. Instance-aware Semantic Segmentation via Multi-task Network Cascades. CVPR. 2015 doi: 10.1109/CVPR.2016.343. [DOI] [Google Scholar]
- 19.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 20.Iglovikov V (2018) TernausNet: U-Net with VGG11 Encoder pre-trained on imagenet for image segmentation. Comput Vis Pattern Recognition (arXiv180105746v1) pp 1–5
- 21.Jaglan P, Dass R, Duhan M. An automatic and efficient technique for tumor location identification and classification through breast MR images. Expert Syst Appl. 2021;185:115580. doi: 10.1016/j.eswa.2021.115580. [DOI] [Google Scholar]
- 22.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift fur Medizinische Phys. 2018;29:102–127. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Siam M, Gamal M, Abdel-Razek M, et al. A comparative study of real-time semantic segmentation for autonomous driving. CVPR Work. 2018 doi: 10.1109/CVPRW.2018.00101. [DOI] [Google Scholar]
- 24.Lian S, Luo Z, Zhong Z, et al. Attention guided U-Net for accurate iris segmentation. J Vis Commun Image Represent. 2018;56:296–304. doi: 10.1016/j.jvcir.2018.10.001. [DOI] [Google Scholar]
- 25.Yin S, Zhang Z, Li H, et al (2019) Fully-automatic segmentation of kidneys in clinical ultrasound images using a boundary distance regression network. In: IEEE 16th Int Symp Biomed Imaging, pp 1741–1744. 10.1109/isbi.2019.8759170 [DOI] [PMC free article] [PubMed]
- 26.Tabrizi PR, Mansoor A, Cerrolaza JJ, et al (2018) Automatic kidney segmentation in 3D pediatric ultrasound images using deep neural networks and weighted fuzzy active shape model. In: IEEE 15th Int Symp Biomed Imaging (ISBI 2018) 2018-April, pp1170–1173. 10.1109/ISBI.2018.8363779
- 27.Yin S, Peng Q, Li H, et al (2018) Automatic kidney segmentation in ultrasound images using subsequent boundary distance regression and pixelwise classification networks. Comput Vis Pattern Recognit 1–22 [DOI] [PMC free article] [PubMed]
- 28.Almajalid R, Shan J, Du Y, Zhang M (2019) Development of a deep-learning-based method for breast ultrasound image segmentation. In: Proceedings of the 17th IEEE Int Conf Mach Learn Appl ICMLA 2018, pp 1103–1108. 10.1109/ICMLA.2018.00179
- 29.Yap M, Goyal M, Osman F, et al. Breast ultrasound lesions recognition: end-to-end deep learning approaches. J Med Imaging. 2018;6:1. doi: 10.1117/1.jmi.6.1.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Guo Y, Wang Y, et al. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med Phys. 2019;46:215–228. doi: 10.1002/mp.13268. [DOI] [PubMed] [Google Scholar]
- 31.Kumar V, Webb JM, Gregory A, et al. Automated and real-time segmentation of suspicious breast masses using convolutional neural network. PLoS ONE. 2018 doi: 10.1371/journal.pone.0195816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu P, Wu F, Peng J, et al. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676–8698. doi: 10.1088/1361-6560/61/24/8676. [DOI] [PubMed] [Google Scholar]
- 33.Reddy DS, Bharath R, Rajalakshmi P (2018) A novel computer-aided diagnosis framework using deep learning for classification of fatty liver disease in ultrasound imaging. In: 2018 IEEE 20th Int Conf e-Health Networking, Appl Serv Heal 2018, pp 1–5. 10.1109/HealthCom.2018.8531118
- 34.Yadav N, Dass R, Virmani J. Texture analysis of liver ultrasound images. Emergent Converging Technol Biomed Syst Lect Notes Electr Eng. 2022;841:575–585. doi: 10.1007/978-981-16-8774-7_48. [DOI] [Google Scholar]
- 35.Song W, Li S, Liu J, et al. Multi-task cascade convolution neural networks for automatic thyroid nodule detection and recognition. IEEE J Biomed Heal Inform. 2015;14:1–11. doi: 10.1109/JBHI.2018.2852718. [DOI] [PubMed] [Google Scholar]
- 36.Ravishankar H, Prabhu S, Vaidya V, Singhal N (2016) Hybrid approach for automatic segmentation of fetal abdomen from ultrasound images using deep learning. In: IEEE 13th Int Symp Biomed Imaging, pp 779–782. 10.1109/ISBI.2016.7493382
- 37.Ma J, Wu F, Jiang T, et al. Ultrasound image-based thyroid nodule automatic segmentation using convolutional neural networks. Int J Comput Assist Radiol Surg. 2017;12:1895–1910. doi: 10.1007/s11548-017-1649-7. [DOI] [PubMed] [Google Scholar]
- 38.Jinlian M, Dexing K. Deep learning models for segmentation of lesion based on ultrasound images. Adv Ultrasound Diagnosis Ther. 2018;2:82. doi: 10.37015/audt.2018.180804. [DOI] [Google Scholar]
- 39.Li X, Wang S, Wei X, et al (2018) Fully Convolutional Networks for Ultrasound Image Segmentation of Thyroid Nodules. In: 2018 IEEE 20th Int Conf High Perform Comput Commun IEEE 16th Int Conf Smart City; IEEE 4th Int Conf Data Sci Syst 886–890. 10.1109/HPCC/SmartCity/DSS.2018.00147
- 40.Wang J, Li S, Song W, et al. Learning from weakly-labeled clinical data for automatic thyroid nodule classification in ultrasound images. IEEE Int Conf Image Process. 2018 doi: 10.1109/ICIP.2018.8451085. [DOI] [Google Scholar]
- 41.Zhou S, Wu H, Gong J, et al (2018) Mark-guided segmentation of ultrasonic thyroid nodules using deep learning. In: Proc 2nd Int Symp Image Comput Digit Med, pp 21–26. 10.1145/3285996.3286001
- 42.Ying X, Yu Z, Ry B, et al. Thyroid nodule segmentation in ultrasound images based on cascaded convolutional neural network. Int Conf Neural Inf Process. 2018;2:373–384. doi: 10.1007/978-3-030-04224-0. [DOI] [Google Scholar]
- 43.Poudel P, Illanes A (2019) Performance evaluation of U-Net convolutional neural network on different percentages of training data for thyroid ultrasound image segmentation. In: 41st Annu Int Conf IEEE Eng Med Biol Soc, pp 2–5 [DOI] [PubMed]
- 44.Ding J, Huang Z, Shi M, Ning C (2019) Automatic thyroid ultrasound image segmentation based on U-shaped network. In: 12th Int Congr Image Signal Process Biomed Eng Informatics, CISP-BMEI 1–5. 10.1109/CISP-BMEI48845.2019.8966062
- 45.Kumar V, Webb J, Gregory A, et al. Automated segmentation of thyroid nodule, gland, and cystic components from ultrasound images using deep learning. IEEE Access. 2020;8:63482–63496. doi: 10.1109/ACCESS.2020.2982390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb JM, Meixner DD, Adusei SA, et al. Automatic deep learning semantic segmentation of ultrasound thyroid Cineclips using recurrent fully convolutional networks. IEEE Access. 2021;9:5119–5127. doi: 10.1109/ACCESS.2020.3045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poudel P, Illanes A, Sheet D, Friebe M. Evaluation of commonly used algorithms for thyroid ultrasound images segmentation and improvement using machine learning approaches. Hindawi J Healthc Eng. 2018;2018:1–13. doi: 10.1155/2018/8087624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Sun T, Yuan Y, et al (2018) Automatic diagnosis of thyroid ultrasound image based on FCN-AlexNet and transfer learning. In: IEEE 23rd Int Conf Digit Signal Process, pp 1–5. 10.1109/ICDSP.2018.8631796
- 49.Wildman-Tobriner BCK. Deep learning-based segmentation of nodules in thyroid ultrasound: improving performance by utilizing markers present in the images. Ultrasound Med Biol. 2019 doi: 10.1016/j.ultrasmedbio.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Guo Z, Zhou J, Zhao D (2020) Thyroid nodule ultrasonic imaging segmentation based on a deep learning model and data augmentation. In: IEEE 4th Inf Technol Autom Control Conf (ITNEC 2020), pp 549–554. 10.1109/ITNEC48623.2020.9085093
- 51.Gomes Ataide EJ, Agrawal S, Jauhari A, et al. Comparison of deep learning algorithms for semantic segmentation of ultrasound thyroid nodules. Curr Dir Biomed Eng. 2021;7:879–882. doi: 10.1515/cdbme-2021-2224. [DOI] [Google Scholar]
- 52.Ke W, Wang Y, Wan P, Liu W. An ultrasonic image recognition method for papillary thyroid carcinoma based on depth convolution neural network. Neural Inf ICONIP 2017 Process Lect Notes Comput Sci. 2017;10635:82–91. doi: 10.1007/978-3-319-70096-0_9. [DOI] [Google Scholar]
- 53.Wang Y, Ke W, Wan P. A method of ultrasonic image recognition for thyroid papillary carcinoma based on deep convolution neural network. NeuroQuantology. 2018;16:757–768. doi: 10.14704/nq.2018.16.5.1306. [DOI] [Google Scholar]
- 54.Li H, Weng J, Shi Y, et al. An improved deep learning approach for detection of thyroid papillary cancer in ultrasound images. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-25005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T, Guo Q, Lian C, et al. Automated detection and classification of thyroid nodules in ultrasound images using clinical-knowledge-guided convolutional neural networks. Med Image Anal. 2019;58:101555. doi: 10.1016/j.media.2019.101555. [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Yang S, Yang S, et al. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J Surg Oncol. 2019;17:1–9. doi: 10.1186/s12957-019-1558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie S, Yu J, Liu T, et al (2019) Thyroid nodule detection in ultrasound images with convolutional neural networks. In: Proc 14th IEEE Conf Ind Electron Appl ICIEA 2019, pp 1442–1446. 10.1109/ICIEA.2019.8834375
- 58.Yu X, Wang H, Ma L. Detection of thyroid nodules with ultrasound images based on deep learning. Curr Med Imaging. 2020;16:174–180. doi: 10.2174/1573405615666191023104751. [DOI] [PubMed] [Google Scholar]
- 59.Abdolali F, Kapur J, Jaremko JL, et al. Automated thyroid nodule detection from ultrasound imaging using deep convolutional neural networks. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103871. [DOI] [PubMed] [Google Scholar]
- 60.Wang L, Zhang L, Zhu M, et al. Automatic diagnosis for thyroid nodules in ultrasound images by deep neural networks. Med Image Anal. 2020;61:101665. doi: 10.1016/j.media.2020.101665. [DOI] [PubMed] [Google Scholar]
- 61.Yao S, Yan J, Wu M, et al. Texture synthesis based thyroid nodule detection from medical ultrasound images: interpreting and suppressing the adversarial effect of in-place manual annotation. Front Bioeng Biotechnol. 2020;8:1–11. doi: 10.3389/fbioe.2020.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dass R. Speckle noise reduction of ultrasound images using BFO cascaded with wiener filter and discrete wavelet transform in homomorphic region. Procedia Comput Sci. 2018;132:1543–1551. doi: 10.1016/j.procs.2018.05.118. [DOI] [Google Scholar]
- 63.Dass R, Vikash R. Comparative analysis of threshold based, K-means and level set segmentation algorithms. IJCST. 2013;4:93–95. [Google Scholar]
- 64.Pedraza L, Vargas C, Narváez F, et al (2015) An open access thyroid ultrasound image database. 10th Int Symp Med Inf Process Anal 9287:92870W1–6. 10.1117/12.2073532
- 65.(2018) https://www.ultrasoundcases.info/cases/head-and-neck/thyroid-gland/. access In: September, 2018
- 66.Rezatofighi H, Tsoi N, Gwak J, et al. Generalized intersection over union: a metric and a loss for bounding box regression. CVPR. 2019;2019:1–9. [Google Scholar]
- 67.Rahman MA, Wang Y (2016) Optimizing intersection-over-union in deep neural networks for image segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 10072 LNCS, pp 234–244. 10.1007/978-3-319-50835-1_22
- 68.Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging. 2015 doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qi X, Zhang L, Chen Y, et al. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal. 2019;52:185–198. doi: 10.1016/j.media.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 70.Dass R, Yadav N. Image quality assessment parameters for despeckling filters. Procedia Comput Sci. 2020;167:2382–2392. doi: 10.1016/j.procs.2020.03.291. [DOI] [Google Scholar]
- 71.Zaitoun Nida M, Aqel MJ. Survey on Image Segmentation Techniques. Procedia Comput Sci. 2015;65:797–806. doi: 10.1016/j.procs.2015.09.027. [DOI] [Google Scholar]
- 72.Litjens G, Bejnordi BE, Arindra A, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017 doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 73.Koundal D, Gupta S, Singh S. Computer aided thyroid nodule detection system using medical ultrasound images. Biomed Signal Process Control. 2018;40:117–130. doi: 10.1016/j.bspc.2017.08.025. [DOI] [Google Scholar]
- 74.Snekhalatha U, Gomathy V. Ultrasound thyroid image segmentation, feature extraction, and classification of disease using feed forward back propagation network. Prog Adv Comput Intell Eng. 2018;563:89–98. doi: 10.1007/978-981-10-6872-0_9. [DOI] [Google Scholar]
- 75.Ying X, Yu Z, B RY, et al (2018) Thyroid Nodule Segmentation in Ultrasound Images Based on Cascaded Convolutional Neural Network. In: Neural Inf Process ICONIP 2018 Lect Notes Comput Sci 11306:373–384. 10.1007/978-3-030-04224-0_32
- 76.Gireesha HSN. Thyroid nodule segmentation and classification in ultrasound images. Int J Eng Res Technol. 2014;3:2252–2256. [Google Scholar]
- 77.Dass R, Priyanka DS. Image segmentation techniques. IJECT. 2012;3:66–70. [Google Scholar]
- 78.Chang C, Huang H, Chen S. Automatic thyroid nodule segmentation and component analysis in ultrasound images. Biomed Eng Appl Basis Commun. 2010;22:81–89. doi: 10.4015/S1016237210001803. [DOI] [Google Scholar]
- 79.Gireesha HM, Nanda S. Thyroid nodule segmentation and classification in ultrasound images. Int J Eng Res Technol. 2014;3:2252–2256. [Google Scholar]
- 80.Singh N, Jindal A. A segmentation method and comparison of classification methods for thyroid ultrasound images. Int J Comput Appl. 2012;50:43–49. doi: 10.5120/7818-1115. [DOI] [Google Scholar]
- 81.Abbasian Ardakani A, Bitarafan-Rajabi A, Mohammadzadeh A, et al. A hybrid multilayer filtering approach for thyroid nodule segmentation on ultrasound Images. J Ultrasound Med. 2019;38:629–640. doi: 10.1002/jum.14731. [DOI] [PubMed] [Google Scholar]
- 82.Nugroho HA, Nugroho A, Choridah L. Thyroid nodule segmentation using active contour bilateral filtering on ultrasound images. Int Conf Qual Res. 2015;2015:43–46. doi: 10.1109/QiR.2015.7374892. [DOI] [Google Scholar]
- 83.Selvathi D, Sharnitha VVSS. Thyroid classification and segmentation in ultrasound images using machine learning algorithms. Int Conf Signal Process Commun Comput Netw Technol. 2011;2011:836–841. doi: 10.1109/ICSCCN.2011.6024666. [DOI] [Google Scholar]
- 84.Huang K, Cheng HD, Zhang Y, et al (2018) Medical Knowledge Constrained Semantic Breast Ultrasound Image Segmentation. In: 2018 24th Int Conf Pattern Recognit, pp 1193–1198
- 85.Wang J, Li S, Song W, et al (2018) Learning from weakly-labeled clinical data for automatic thyroid nodule classification in ultrasound images. In: 2018 25th IEEE Int Conf Image Process, pp 3114–3118. 10.1109/ICIP.2018.8451085
- 86.Ravishankar H, Sudhakar P, Venkataramani R, et al (2016) Understanding the mechanisms of deep transfer learning for medical images. In: Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 10008 LNCS, pp 188–196. 10.1007/978-3-319-46976-8_20
- 87.Yao W, Zeng Z, Lian C, Tang H. Pixel-wise Regression using U-net and its application on pansharpening. Neurocomputing. 2018 doi: 10.1016/j.neucom.2018.05.103. [DOI] [Google Scholar]
- 88.Rezaee M, Zhang Y, Mishra R, et al (2018) Using a VGG-16 Network for Individual Tree Species Detection with an Object-Based Approach. In: 2018 10th IAPR Work Pattern Recognit Remote Sens, pp 1–7. 10.1109/PRRS.2018.8486395
- 89.Liu X, Deng Z, Yang Y. Recent progress in semantic image segmentation. Artif Intell Rev. 2018 doi: 10.1007/s10462-018-9641-3. [DOI] [Google Scholar]
- 90.Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. MICCAI 2015(arXiv:150504597) 1–8
- 91.Brito JP, Gionfriddo MR, Al NA, et al. The accuracy of thyroid nodule ultrasound to predict thyroid cancer: systematic review and meta-analysis. J Clin Endocrinol Metab. 2014;99:1253–1263. doi: 10.1210/jc.2013-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choi H (2017) Deep Learning in Nuclear Medicine and Molecular Imaging: Current Perspectives and Future Directions. Nucl Med Mol Imaging 2016. DOI: 10.1007/s13139-017-0504-7 [DOI] [PMC free article] [PubMed]
- 93.Girshick R, Donahue J, Darrell T, et al. Rich feature hierarchies for accurate object detection and semantic segmentation. IEEE Conf Comput Vis Pattern Recognit. 2012;2014:2–9. doi: 10.1109/CVPR.2014.81. [DOI] [Google Scholar]
- 94.De Brébisson A, Montana G (2015) Deep neural networks for anatomical brain segmentation. IEEE Comput Soc Conf Comput Vis Pattern Recognit Work 2015-Octob:20–28. 10.1109/CVPRW.2015.7301312
- 95.Xu Y, Wang Y, Yuan J, et al. Medical breast ultrasound image segmentation by machine learning. Ultrasonics. 2019;91:1–9. doi: 10.1016/j.ultras.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 96.China D, Illanes A, Poudel P, et al. Anatomical structure segmentation in ultrasound volumes using cross frame belief propagating iterative random walks. IEEE J Biomed Heal Inform. 2019;23:1110–1118. doi: 10.1109/JBHI.2018.2864896. [DOI] [PubMed] [Google Scholar]
- 97.Lestari DP, Madenda S, Ernastuti WEP. Comparison of three segmentation methods for breast ultrasound images based on level set and morphological operations. Int J Electr Comput Eng. 2017;7:383–391. doi: 10.11591/ijece.v7i1.pp383-391. [DOI] [Google Scholar]
- 98.Prabha DS, Kumar JS. Performance evaluation of image segmentation using objective methods. Indian J Sci Technol. 2016;9:1–8. doi: 10.17485/ijst/2016/v9i8/87907. [DOI] [Google Scholar]
- 99.Yap MH, Pons G, Martí J, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Heal Informatics. 2018;22:1218–1226. doi: 10.1109/JBHI.2017.2731873. [DOI] [PubMed] [Google Scholar]
- 100.Lu F, Wu F, Hu P, et al. Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg. 2017;12:171–182. doi: 10.1007/s11548-016-1467-3. [DOI] [PubMed] [Google Scholar]
- 101.Da Nóbrega RVM, Peixoto SA, Da Silva SPP, Filho PPR. Lung nodule classification via deep transfer learning in CT lung images. Proc IEEE Symp Comput Med Syst. 2018;2018:244–249. doi: 10.1109/CBMS.2018.00050. [DOI] [Google Scholar]
- 102.Hermessi H, Mourali O, Zagrouba E. Deep feature learning for soft tissue sarcoma classification in MR images via transfer learning. Expert Syst Appl. 2019;120:116–127. doi: 10.1016/j.eswa.2018.11.025. [DOI] [Google Scholar]