Significance
Microtubules shape the cell and are involved in many different cellular processes. To do so, microtubules need to be dynamic, and their dynamic behavior is regulated by microtubule-associated proteins, like those interacting with the growing microtubule plus tip. There are hundreds of these proteins at the tiny 500 nm tip; however, how these proteins are organized spatially and temporally is still unclear. We focused on two of such proteins, CLIP-170 and EB3, and show that they can individually, in combination and on microtubules phase separate into biomolecular condensates. Through phase separation, these CLIP-170/EB3-condensates regulate microtubule dynamics. Furthermore, the special organization of the proteins at the growing tip could be explained by a separated liquid phase.
Keywords: microtubule, liquid–liquid phase separation, CLIP-170, EB3, tubulin condensation
Abstract
Regulation of microtubule dynamics is essential for diverse cellular functions, and proteins that bind to dynamic microtubule ends can regulate network dynamics. Here, we show that two conserved microtubule end-binding proteins, CLIP-170 and EB3, undergo phase separation and form dense liquid networks. When CLIP-170 and EB3 act together, the multivalency of the network increases, which synergistically increases the amount of protein in the dense phase. In vitro and in cells, these liquid networks can concentrate tubulin. In vitro, in the presence of microtubules, phase separation of EB3/CLIP-170 can enrich tubulin all along the microtubule. In this condition, microtubule growth speed increases up to twofold and the frequency of depolymerization events are strongly reduced compared to conditions in which there is no phase separation. Our data show that phase separation of EB3/CLIP-170 adds an additional layer of regulation to the control of microtubule growth dynamics.
The microtubule cytoskeleton engages in a plethora of cellular processes, from organelle transport to cell division. To do so, the network dynamically modifies its structure in response to external cues and adapts its architecture to specific cellular functions. Microtubules themselves are highly dynamic polymers that can rapidly cycle between phases of polymerization and depolymerization, a characteristic which is critical for cytoskeletal reorganization (1). In cells, microtubules polymerize at their plus end by addition of GTP–tubulin. After GTP–tubulin addition, GTP is gradually hydrolyzed, resulting in a Guanosine diphosphate (GDP)–tubulin shaft behind the tip. Once the stabilizing GTP–tubulin “cap” disappears from the plus end, the microtubule switches from growing to shrinking, an event termed catastrophe (2–4). Conversely, microtubules can stop shrinking and switch to regrowth, an event termed rescue. Regulation of growth and shrinkage is intimately linked to the local availability of free tubulin at the growing microtubule (5, 6).
In addition to these modes of regulation, microtubule dynamics can be fine-tuned by Tip-Interacting Proteins (+TIPs) (7). These are functionally interdependent and structurally diverse microtubule regulators that concentrate and organize in +TIP networks at growing microtubule ends while exhibiting a weak affinity for the microtubule shaft. It is generally accepted that the unique localization of +TIPs at microtubule ends results from their specific binding to the GTP–tubulin cap or by interacting with proteins that bind to the GTP cap (8).
Key integrators of +TIP networks are the End-Binding proteins (EBs), as they directly bind to GTP–tubulin at growing microtubule ends and recruit a battery of nonautonomously binding +TIPs (9). Within the EB family, higher eukaryotes express EB1, EB2, and EB3. These proteins increase microtubule plus-end dynamics by promoting catastrophes and increasing growth speed in vitro (10–13).
A key accessory protein recruited to plus-ends by the EBs is the Cytoplasmic Linker Protein of 170 kDa (CLIP-170), which increases microtubule rescue frequency and growth speed (10, 14–18). CLIP-170 consists of a microtubule-binding “head” domain in its N terminus, which is composed of two Cap–Gly domains and three serine-rich regions, all of which contribute to its interaction with microtubules (19, 20). Dimerization of CLIP-170 is mediated through a central coiled-coil region, which is followed C-terminally by a zinc–knuckle domain (21, 22). The coiled-coil domain together with the zinc knuckles will hereafter be referred to as the “C-terminal region”. Most studies of CLIP-170 function in vitro have focused on truncated versions, containing only the monomeric head domain (H1) or the head domain with a small extension that allows dimerization (H2). The effect of full-length CLIP-170 on microtubule dynamics has not yet been shown.
Early studies of CLIP-170 expression in cells revealed that in addition to its microtubule plus-end localization, it also formed “cytoplasmic patches” that colocalized with EB1 and the dynein-activating protein dynactin (20, 21). Based on the physical properties of these CLIP-170 cytoplasmic patches, it has been suggested that they form by liquid–liquid phase separation (LLPS) (23, 24). Recently, it has been shown that homologous +TIPs in budding yeast, fission yeast, and higher eukaryotes undergo LLPS (25–27).
LLPS is the process by which molecules spontaneously condense into droplets and demix from their surrounding solution, resulting in the coexistence of two unique liquid phases (28–30). Recently, LLPS has been implicated in driving microtubule-related processes, including spindle assembly (31), nucleation of acentrosomal and branched microtubules (32, 33), and centrosome maturation (34, 35). A key shared feature of these processes is the enrichment of tubulin with LLPS-potent microtubule-associated proteins to catalyze biochemical reactions. Along these lines, condensation of a microtubule +TIP network has been proposed based on transient multivalent interactions between the +TIPs (8, 23, 25–27). Despite this, LLPS of +TIPs and its function are still unclear, particularly their role in regulating microtubules dynamic parameters.
+TIPs concentrate at the growing GTP-microtubule tip. A combination of in vivo, in vitro, and in silico work has indicated that the GTP–tubulin cap size is 1 to 60 tubulins long, indicating a length of <500 nm (5, 6, 36–39). However, in cells, +TIPs bind to a region at the microtubule tip up to fourfold longer than this and form up to 2-µm-long +TIP networks (12, 38, 40). The discrepancies between +TIP profiles in vitro and in cells could be due to different sizes of the GTP cap, but also prompts the question of whether LLPS impacts the shape, organization, and size of +TIP networks, due to the different material properties of a liquid, like viscosity and surface tension.
Interestingly, overexpressed EB3 and CLIP-170 exhibit atypical binding to the growing microtubule tip. Both proteins leave trailing protein foci behind the leading edge of the +TIP network (12, 41–44). One explanation for these “trailing foci” is that they form by binding to tubulin remaining in the GTP state behind the GTP cap (14, 42, 45). As overexpressed CLIP-170 has the potential to undergo LLPS (23–25), another possibility would be that CLIP-170 foci split off from the leading network in a process resembling fission of condensates.
The self-assembly of +TIPs into highly dynamic, multivalent networks allows for the emergence of LLPS. In this study, we provide evidence that LLPS of +TIPs could be a mechanism to contribute to the formation of +TIP networks at the growing microtubule tip. We use tandem in-cell and in vitro approaches to investigate +TIP condensation properties and study the impact of phase-separated +TIPs on microtubule dynamics. We demonstrate that the canonical +TIPs EB3 and CLIP-170 undergo LLPS in vitro at nanomolar concentrations in the absence of crowding agents. We show that cocondensation of EB3 and CLIP-170 depends on specific protein domains at the C-terminal tail of the two proteins. EB3 and CLIP-170 droplets concentrate tubulin, and in the presence of microtubules, tubulin/EB3/CLIP-170 condensates speed up microtubule growth while reducing catastrophe and pausing frequencies. Therefore, +TIPs LLPS adds an additional layer to the regulation of microtubule dynamics.
Results
CLIP-170 Forms Biomolecular Condensates in Cells.
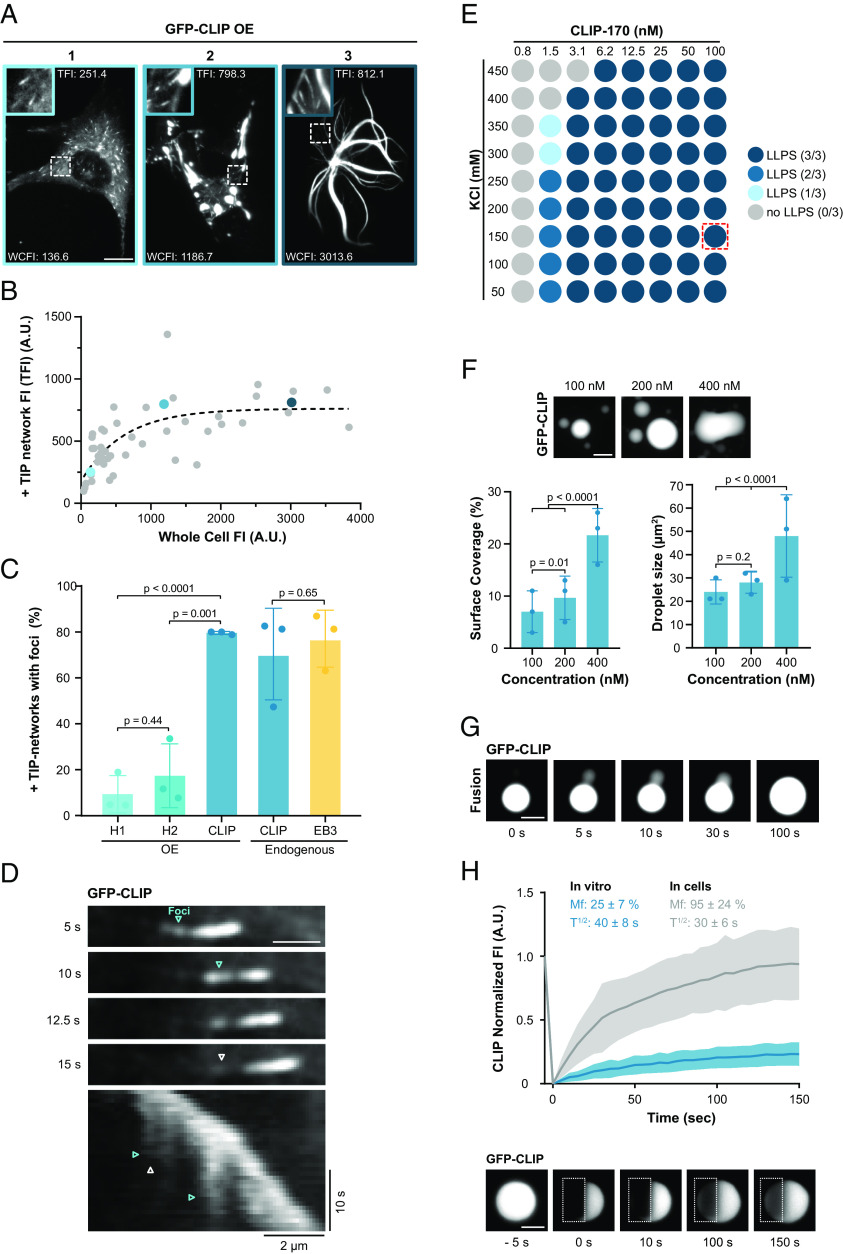
We first studied the condensation properties of the +TIP protein CLIP-170 in RPE-1 cells. By analyzing GFP-CLIP-170 fluorescence intensity at the growing microtubule tip, we observed that with increasing CLIP-170 expression levels, the amount of CLIP-170 at the microtubule tip reached a plateau after an initial phase of accumulation (Fig. 1 A and B). This suggests that at the growing microtubule tip +TIPs are limiting the recruitment of CLIP-170.
Fig. 1.
CLIP-170 condenses into droplets in cells and in vitro. (A) Representative confocal images of RPE-1 cells transfected with GFP-CLIP-170 at three different overexpression levels (1: low, 2: high, and 3: very high overexpression). Zoom-in shows +TIP networks with adjusted contrast for visualization; for images with the same contrast, see (SI Appendix, Fig. S1A). For each cell, the whole cell fluorescence intensity (WCFI) and peak of +TIP network fluorescence intensity (TFI) are indicated; the blue outlining of the image corresponds to colored data points in (B). (Scale bar: 10 µm.) (B) Analysis of A showing the correlation between peak +TIP network fluorescence intensity and WCFI in RPE-1 cells expressing GFP-CLIP-170. The dashed line shows exponential plateau curve fit. Each dot represents 5 analyzed +TIP networks from one cell, data from two independent experiments with a total of 42 cells. (C) Percentage of +TIP networks with trailing foci in fixed cells expressing the indicated CLIP constructs or stained with antibodies to analysis endogenous CLIP-170 or EB3. Mean with SD from three independent experiments. Statistics: one-way ANOVA test. (D) Representative time-lapse TIRF images (Top) and kymograph (Bottom) of +TIP network from GFP-CLIP-170 expressing RPE-1 cell. Cyan and white arrowheads denote trailing foci formation and dissolving, respectively, in both time-lapse images and kymograph. (Scale bar: 2 µm.) (E) Phase diagram of GFP-FL-CLIP in vitro at increasing KCl and protein concentration. The blue shaded dot denotes where phase separation occurred, results of three independent experiments. The red dotted square represents the physiological cell concentration of CLIP-170 where we observe phase separation. (F, Top) representative confocal images of purified GFP-FL-CLIP at indicated concentrations. (Scale bar: 20 µm.) Bottom: quantification of the coverslip surface coverage (Left) and droplet size (Right) for GFP-FL-CLIP condensates at 100, 200, or 400 nM in the absence of PEG. Statistics: two-tailed Student’s t test. Mean with SD from three independent experiments with a total of 27 fields of view per condition. (G) Time-lapse TIRF images of purified GFP-FL-CLIP (1 µM) undergoing fusion. Representative of three experimental replicates. (Scale bar: 10 µm.) (H) Representative TIRF images and recovery curve of purified GFP-FL-CLIP (2 µM) droplets after photobleaching (dashed box). The blue curve shows mean with SD of three individual experiments with a total of 47 condensates. The gray curve shows the FRAP curve for GFP-CLIP droplets in cells. (Scale bar: 5 µm.)
As CLIP-170 becomes saturated at microtubule tips, irregularly shaped CLIP-170 patches started appearing in the cytoplasm, which were also associated with microtubules (Fig. 1A panel 2, Fig. 1B and SI Appendix, Fig. S1 A and B). Consistent with recent results, these cytosolic CLIP-170 patches displayed liquid properties and are hereafter called droplets (23, 24). CLIP-170 droplets underwent fission and fusion within 5 s, implying fast relaxation of the two droplets—a diagnostic feature for a fluid droplet (SI Appendix, Fig. S1 C and D and Movies S1 and S2). Fluorescence recovery after photobleaching of half of the droplets (half-FRAP) showed that CLIP-170 recovered with a half-time of 30 ± 6 s and a mobile fraction of 95 ± 24% (Fig. 1H, SI Appendix, Fig. S1 E and F, and Movie S3), slightly higher values in comparison to previous results (23). Furthermore, at high overexpression levels, CLIP-170 coated the entire microtubule and promoted microtubule bundling (Fig. 1A panel 3), like another microtubule-associated protein undergoing LLPS, tau (32). These properties lead us to conclude that i) CLIP-170 diffuses rapidly within these droplets, and ii) these droplets have a high protein exchange rate with the cytoplasmic pool.
+TIP Networks at the Microtubule Tip Show Liquid-Like Properties.
In WT interphase cells, CLIP-170 is strongly enriched at growing microtubule tips. We hypothesize that due to this local enrichment of CLIP-170, +TIP networks might be droplets. To explore this hypothesis, we studied in RPE-1 cells overexpressing GFP-CLIP-170 whether the profile of +TIP networks with trailing protein foci on the microtubule could result from LLPS (SI Appendix, Fig. S2A). Fluorescence intensity analysis of the GFP-CLIP-170 profiles revealed that 80% of the profiles have trailing foci (Fig. 1C and SI Appendix, Fig. S2A). The appearance of trailing foci could correspond to the fission of a droplet from the +TIP network, where trailing CLIP-170 foci bound to the microtubule shaft dissolved later over time (Fig. 1D and Movie S4).
Using CLIP-170-specific antibodies, we found that ~70% of the CLIP-170 profiles had trailing foci at endogenous protein levels (Fig. 1C and SI Appendix, Fig. S2A). EB3, another prominent +TIP, had ~75% of the endogenous EB3 profiles with trailing foci (Fig. 1C and SI Appendix, Fig. S2A), consistent with a previous overexpression-based study (41). Costaining of endogenous EB3 and CLIP-170 revealed that most +TIP networks displayed fluorescence profiles with distinct EB3 and CLIP-170 trailing foci where both proteins colocalize (SI Appendix, Fig. S2B). Therefore, foci left behind on the microtubule shaft appeared not only upon overexpression but also at endogenous protein levels. In addition, different +TIPs colocalize in these transient trailing foci.
Confirming previous reports, overexpressed C-terminal truncated mutant versions of CLIP-170, the dimeric H2- and monomeric H1, tracked the growing microtubule tip, but did not form any cytosolic droplets, even at high expression levels (SI Appendix, Fig. S2 C and D and Movies S5 and S6) (20, 21). We used these two phase separation–deficient mutants to study whether the appearance of trailing foci on the shaft at the microtubule tip depends on LLPS. Consistent with the idea that foci form by fission from +TIP droplets, the presence of the C-terminal truncated H2 and H1 dramatically reduced the number of trailing foci at the microtubule tip compared to full-length CLIP-170 (Fig. 1C and SI Appendix, Fig. S2E). This suggests that the C-terminal region of CLIP-170 mediates the formation of droplets at microtubule ends. In particular, since H1 and H2 bind to the growing GTP tip, the presence of GTP islands in the shaft alone could not explain the formation of trailing foci. Instead, fission from a liquid droplet at the tip of a growing microtubule may also contribute to the existence of trailing foci. We hypothesize that when the concentration of +TIPs like EB3 and CLIP-170 increases beyond a threshold within the leading droplet, it causes fission of trailing foci from the droplet.
To probe for liquid properties of +TIP networks, we treated cells with the aliphatic alcohol 1,6-hexanediol to reduce hydrophobic interactions, which has been previously reported to interfere with LLPS (46). While untreated cells exhibited 3-µm-long +TIP networks, upon hexanediol treatment, the profile of the +TIP network shortened to ~1.5 µm, and no trailing foci were observed (SI Appendix, Fig. S2 F and G and Movie S7). This indicates that the hydrophobic interactions, characteristic of liquid condensates, influence +TIP network organization. In conclusion, +TIPs interacting with the growing microtubule tips show liquid properties, including sensitivity toward aliphatic alcohol. Also, the excision of trailing foci from the tip resembles fission, a well-established feature of liquid droplets (47).
CLIP-170 Forms Biomolecular Condensates In Vitro.
To study the phase-separation properties of CLIP-170, we purified recombinant full-length human GFP-CLIP-170 (FL-CLIP) from insect cells (SI Appendix, Fig. S3A) and reconstituted droplet formation in vitro. Even in the absence of crowding agents and at physiological salt concentrations (150 mM KCl), FL-CLIP robustly condensed into spheres at concentrations as low as 3.1 nM (Fig. 1E and SI Appendix, Fig. S3B), which is even below the cellular concentration of ~110 nM (48). To quantify the amount of FL-CLIP in the dense phase, we used a high throughput confocal microscopy-based approach. Briefly, i) protein mixtures were incubated in 384-well plates, ii) proteins in the dense phase were centrifuged onto the bottom of the wells, iii) wells were imaged using an automated confocal microscope, and iv) surface coverage of protein in the dense phase and size of the droplets on well-bottoms were measured (SI Appendix, Fig. S3C; see Materials and Methods for details).
Consistent with LLPS properties, the size of the sphere and amount of FL-CLIP in the dense phase increased in a concentration-dependent manner, even in the absence of crowding agents (Fig. 1F). Further hallmarks of the LLPS process include sensitivity toward ionic strength of the buffer and toward the presence of crowding agents (49). Indeed, increasing the ionic strength of the buffer reduced the sphere size (SI Appendix, Fig. S3B), and addition of a crowding agent, 2% polyethylene glycol (PEG), increased the amount of FL-CLIP in the dense phase by 1.5-fold, but reduced the sphere size (SI Appendix, Fig. S3 D and E). To estimate the concentration of FL-CLIP in the spheres with confocal microscopy, we calibrated the fluorescent intensity of GFP. From an initial 200 nM FL-CLIP solution, the protein concentration reached 1.7 µM within the spheres (Table 1). Collectively, these results demonstrate that FL-CLIP undergoes LLPS, and forms droplets at nanomolar concentrations even in the absence of additional proteins or crowding agents.
Table 1.
CLIP-170, EB3, and tubulin concentration inside droplets
| Analyzed protein | Protein concentration | Cdilute (nM) | Cdroplet (nM) | Ratio (Cdroplet/Cdilute) |
|---|---|---|---|---|
| CLIP-170 | CLIP-170 (200 nM) | 68 ± 15 | 1,752 ± 255 | 24 |
| EB3 | mcherry-EB3 (2 µM) + EB3 (8 µM) | 8,678 ± 462 | 31,187 ± 1,954 | 3.5 |
| CLIP-170 | CLIP-170 (200 nM) + mcherry-EB3 (1 µM) | 45 ± 18 | 1,880 ± 301 | 24 |
| EB3 | mcherry-EB3 (1 µM) + CLIP-170 (200 nM) | 396 ± 167 | 2,074 ± 292 | 5 |
| CLIP-170 | CLIP-170 (200 nM) + Tubulin (400 nM) | 44 ± 7 | 1,742 ± 279 | 34 |
| Tubulin | Tubulin (400 nM) + CLIP-170 (200 nM) | 312 ± 38 | Droplet 671 ± 127/Shell 1,180 ± 247 | Droplet 2/Shell 4 |
Protein concentration outside droplets (Cdilute) and inside droplets (Cdroplet), obtained with confocal microscopy by calibrating fluorescent intensities of GFP, mCherry, and Atto565, see calibration in SI Appendix, Fig. S9. Atto565–tubulin and GFP-CLIP were 100% labeled. The ratio of unlabeled EB3 and labeled mcherry-EB3 protein in the assay was considered, when calculating the total protein concentration Cdilute and Cdroplet, see Materials and Methods.
Consistent with the situation in cells, FL-CLIP spherical droplets displayed liquid properties. This includes droplet fusion, although fusion 5-times slower in vitro than what we observed in cells (Fig. 1G and Movie S8). The slower fusion time together with the roundness of the in vitro FL-CLIP droplets suggests that the droplets are more viscous in vitro than in cells. We further interrogated the liquid properties of these spheres by using half-FRAP, which confirmed that FL-CLIP diffuses within the spheres with a half-time of 40 ± 8 s, which is slower compared to the recovery speed in cells (Fig. 1H and SI Appendix, Fig. S1F). Furthermore, FL-CLIP exchange dynamics were reduced threefold compared to CLIP-170 droplets in cells, with a mobile fraction of 25 ± 7% (Fig. 1H and Movie S9). We hypothesize that the discrepancies between the material properties of the droplets in cells and in vitro are due to the presence of additional proteins and/or CLIP-170 posttranslational modifications in the cellular droplets.
The CLIP-170 C-Terminal Region Drives CLIP-170 into the Dense Phase.
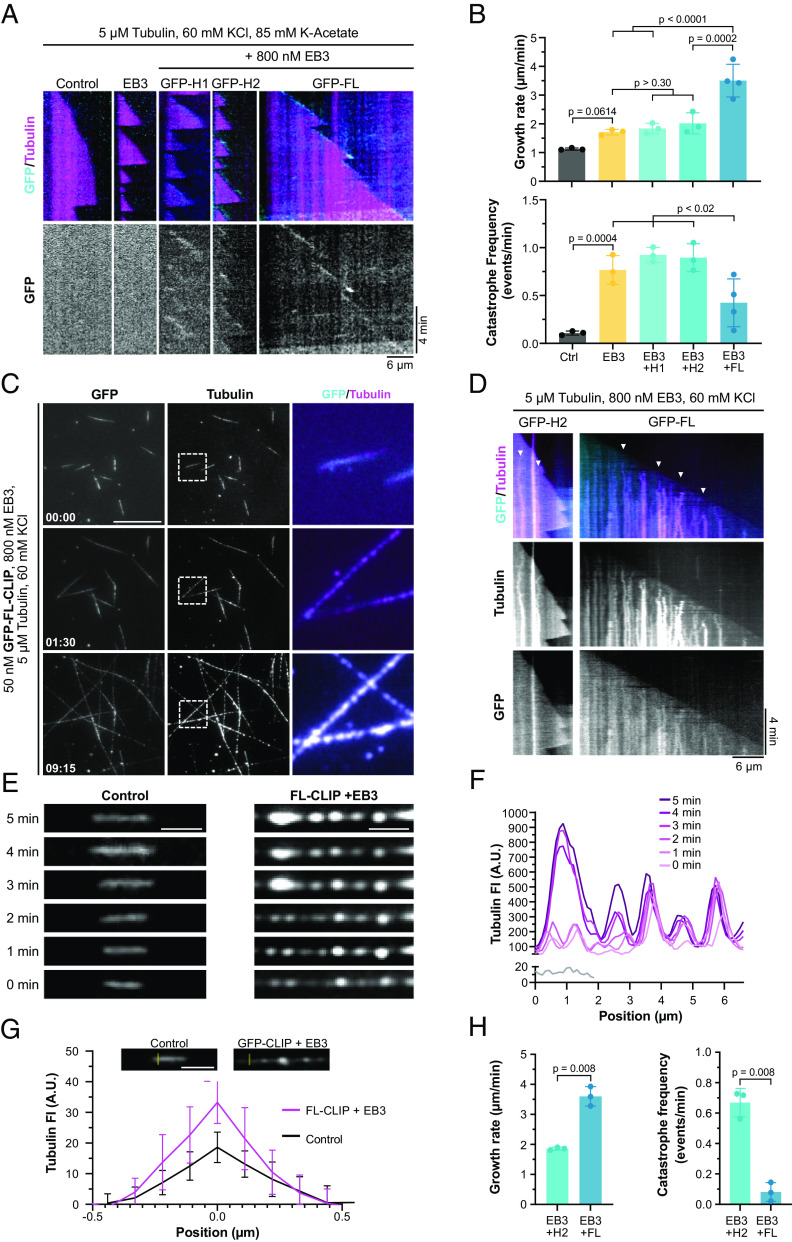
Building on the observation in cells that the C-terminal region impacts droplet formation (SI Appendix, Fig. S2D and Movies S5 and S6) (23), we studied to what extent the C-terminal region is necessary for CLIP-170 to undergo LLPS in vitro. We purified recombinant human truncation mutants of CLIP-170 from bacteria, namely, the dimeric H2 and the monomeric H1 mutant, where the C-terminal region is truncated or fully removed and measured their ability to condense in vitro at nanomolar concentrations (SI Appendix, Fig. S4A). While FL-CLIP phase separated in the absence of other factors, the monomeric H1 showed faint irregularly shaped aggregates but no droplet formation, even in the presence of 10% PEG (Fig. 2B and SI Appendix, Fig. S4 B and C). Contrary to the situation in cells, at nanomolar concentration, the dimeric H2 mutant underwent condensation in vitro, although with a 300-fold reduction of protein surface coverage and a 25-fold reduction in droplet size compared to FL-CLIP (Fig. 2 B–E). Addition of 2% PEG increased H2 protein surface coverage by threefold but did not affect condensate size (SI Appendix, Fig. S4C). Collectively, these results show that while the monomeric H1 is sufficient to track the growing microtubule tip, the dimeric form of CLIP-170 is necessary to undergo LLPS and that the C-terminal region robustly drives CLIP-170 condensation.
Fig. 2.
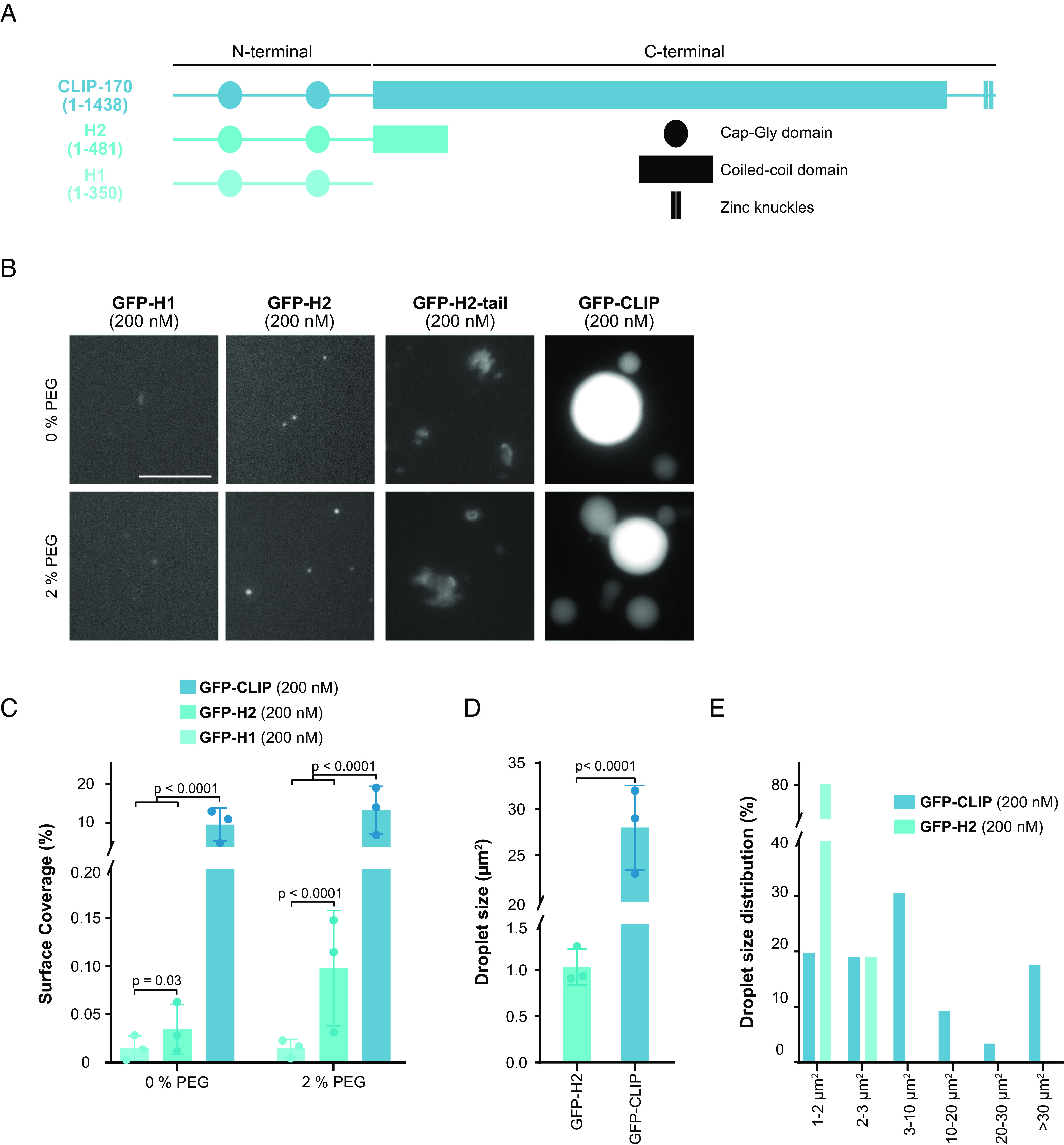
The C-terminal region strongly enhances LLPS of CLIP-170. (A) Secondary structure of CLIP-170 (1 to 1,438), H2 (1 to 481), and H1 (1 to 350) drawn to scale, based on (20, 21, 50). (B) Representative confocal images of purified GFP-H1, GFP-H2, GFP-H2-tail, and GFP-FL-CLIP each at 200 nM in the absence (Top) or presence (Bottom) of 2% PEG. (Scale bar: 20 µm.) (C) Condensate surface coverage of the three constructs at indicated PEG concentrations. Mean with SD from three independent experiments with a total of 27 fields of view per condition. Statistics: two-tailed Student’s t test. (D) Droplet size (area) of GFP-H2 (200 nM) and GFP-FL-CLIP (200 nM). Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test. (E) Size distribution of GFP-FL-CLIP (200 nM) and GFP-H2 (200 nM) droplets in the absence of PEG. The graph shows average size distribution from three independent experiments with a total of 27 fields of view.
Intrinsically disordered domains are common features of proteins that undergo LLPS (28). To parse out the contributions of disordered domains to FL-CLIP’s phase-separation potency, we engineered a mutant containing the H2 domain fused to the far C-terminal region, termed H2-tail (SI Appendix, Fig. S4D). This mutant lacks most of the coiled-coil region but retains many predicted disordered regions within CLIP-170 (SI Appendix, Fig. S4D). Attempts to purify this mutant resulted in insoluble, large protein aggregates in our experimental conditions (Fig. 2B), indicating that the coiled-coil region impacts CLIP-170’s solubility.
EB3 Undergoes LLPS.
To localize to growing microtubule ends, CLIP-170 requires the presence of EBs, of which EB3 has a cellular concentration of ~55 nM (10, 47, 51). This prompted us to study whether purified EB3 could form condensate in vitro (Fig. 3 and SI Appendix, Fig. S5A). Indeed, EB3 can undergo LLPS, albeit at micromolar concentrations: Compared to 48 ± 17 µm2 large droplets observed for 400 nM FL-CLIP, 1 µM EB3 phase separated into numerous small 1.9 ± 0.6 µm2 droplets. This resulted in a 3.5-fold reduced surface coverage compared to FL-CLIP (Fig. 3 A and B). Addition of 2% PEG further increased the surface coverage of EB3 in the dense phase (SI Appendix, Fig. S5B). FRAP revealed that EB3 moves within these droplets faster than FL-CLIP droplets (half-time: 29 ± 4 s compared to 40 ± 8 s), and EB3 showed a 2.5-fold higher mobile fraction compared to FL-CLIP (Fig. 3C and Movie S10). In summary, i) EB3 undergoes LLPS, ii) compared to CLIP-droplets, EB3-droplets are small, which suggests a higher nucleation rate for EB3-droplets, and iii) EB3 moves faster in the droplets, and EB3 has a higher protein exchange rate with the pool outside the droplet compared to FL-CLIP. These properties of the droplet suggest that the composition of +TIPs at the growing microtubule tip can regulate the size and viscosity of a +TIP droplet.
Fig. 3.
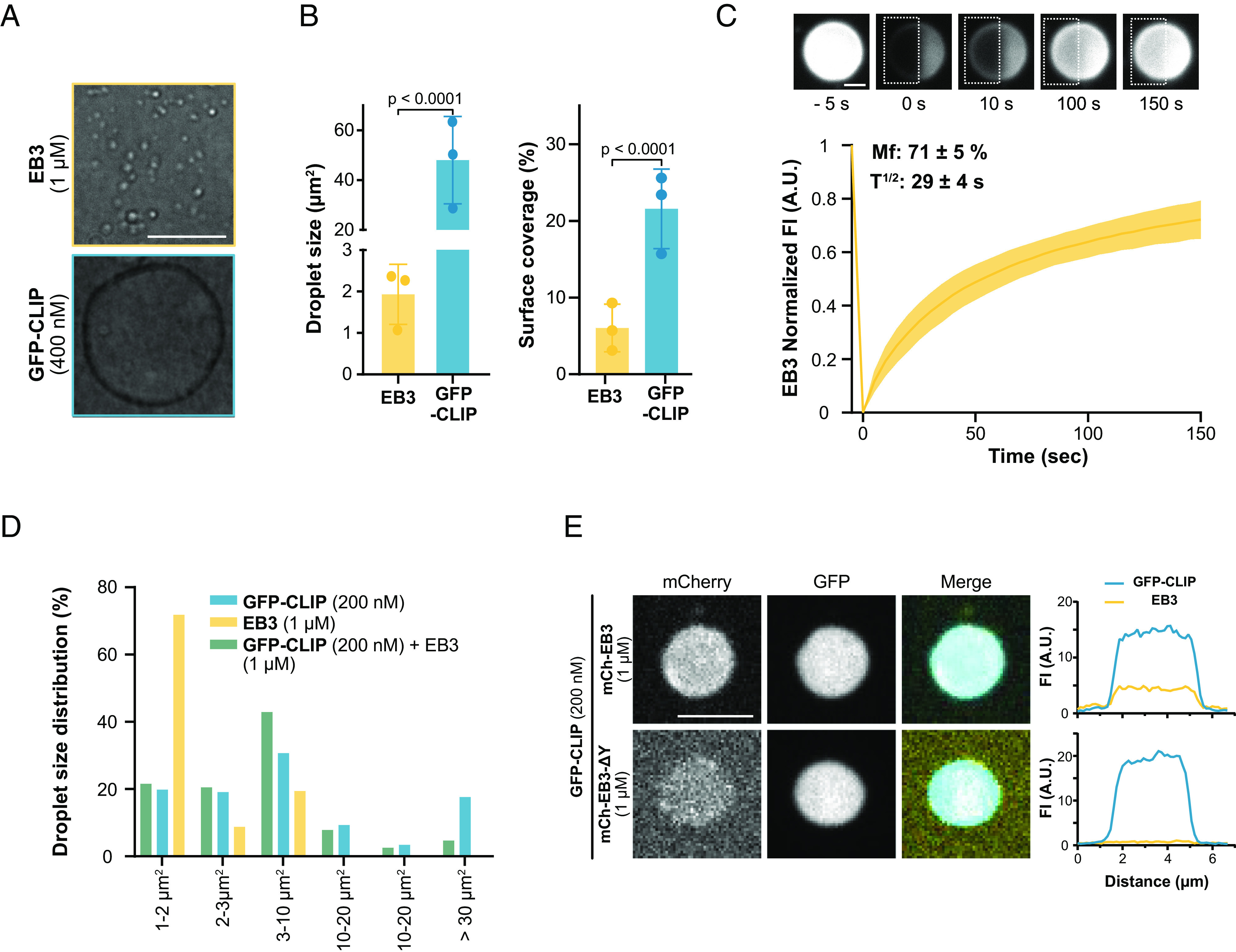
EB3 undergoes LLPS and cocondenses with CLIP-170 in vitro. (A) Representative DIC images of purified EB3 (1 µM) and GFP-FL-CLIP (400 nM) (Scale bar: 20 µm.) (B, Right) droplet size, Left: condensate surface coverage of EB3 (1 µM) and GFP-FL-CLIP (400 nM). Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test. (C) Representative TIRF images and recovery curve of purified EB3 (10 µM) + mCherry-EB3 (100 nM) droplets after photobleaching (dashed box). The curve shows mean with SD of three independent experiments with a total of 32 condensates. (Scale bar: 5 µm.) (D) Size distribution of GFP-FL-CLIP (200 nM), EB3 (1 µM), and the EB3/FL-CLIP droplets in the absence of PEG. Mean size distribution from three independent experiments with a total of 27 fields of view. (E) Representative confocal images of EB3/GFP-FL-CLIP droplets (Top) or EB3-ΔY/GFP-FL-CLIP droplets (Bottom) at denoted concentrations with the corresponding line scan. (Scale bar: 4 µm.)
EB3 Cocondenses with CLIP-170.
Next, we studied the ability of CLIP-170 to cocondense with EB3. Indeed, FL-CLIP and EB3 robustly cocondensed into droplets, in which both proteins showed a homogenous distribution (SI Appendix, Fig. S5C). Cocondensation of EB3 with FL-CLIP increased the number of small droplets such that droplets larger than 40 µm2 were rarely observed (Fig. 3D). This reduced the average droplet size by 20-fold compared to FL-CLIP, despite that the total protein concentration used in the cocondensation assay increased by sixfold compared to the FL-CLIP assay (2 µM FL-CLIP to 12 µM EB3/FL-CLIP) (SI Appendix, Fig. S5D). The method of droplet formation, either by mixing EB3 and FL-CLIP together or by adding one protein to preformed droplets of the other, did not affect the droplet size or protein distribution within the droplets (SI Appendix, Fig. S5E). These results show that EB3 and FL-CLIP robustly cocondense into droplets with a homogenous protein distribution, while addition of EB3 to FL-CLIP changed the material property of the droplets, as indicated by a change in droplet size and number.
To address whether the diffusion coefficient within the droplets is impacted by a multivalent EB3/FL-CLIP network, we performed our half-FRAP experiment with EB3/FL-CLIP cocondensates. Surprisingly, the recovery time of FL-CLIP following photobleaching in EB3/FL-CLIP droplets mirrored that of EB3 in EB3-droplets (Movies S9–S11, Figs. 1H and 3C, and SI Appendix, Fig. S5F). The mobile fraction of FL-CLIP and EB3/FL-CLIP was comparable, while EB3 droplets had a twofold larger mobile fraction. Consistently, the three different droplets exhibited distinct surface interaction properties: EB3 did wet the coverslip resulting in flat droplets, while FL-CLIP and EB3/FL-CLIP droplets remained a similar sphere-like shape when bound to the coverslip (Movies S12–S14). Taken together, these results show that EB3 and FL-CLIP can cocondense and that the multivalency of the network does impact FL-CLIP diffusion within the droplet and the droplet size. We hypothesize that EB3 could nucleate a droplet at the tip through wetting, while addition of CLIP-170 could impact the size and viscosity of the droplet.
Interactions between EB1 and CLIP-170 are driven by the last C-terminal tyrosine of EB1 (10). To probe whether specific protein–protein interactions underlie EB3/FL-CLIP cocondensation, we purified an EB3 tyrosine-deletion mutant (EB3-ΔY) and repeated our colocalization assay. The removal of the last tyrosine drastically reduced EB3’s ability to cocondense with FL-CLIP (Fig. 3E), despite leaving EB3-ΔY’s intrinsic phase-separation properties intact (SI Appendix, Fig. S5G). Therefore, specific binding interactions between EB3 and FL-CLIP are key features in their ability to cocondense.
EB3/FL-CLIP Droplets Cooperatively Phase Separate.
Given that EB3/FL-CLIP droplets were reduced in size compared to FL-CLIP droplets (SI Appendix, Fig. S5D), we quantified whether the total amount of protein in the dense phase is also reduced compared to FL-CLIP. Measuring by confocal microscopy the concentration of FL-CLIP in droplets compared to the dilute phase showed a 24-fold enrichment of FL-CLIP in the droplet (Table 1). Cocondensation of FL-CLIP and EB3 only slightly increased the concentration of FL-CLIP in the droplet, while condensation of EB3 increased 1.4-fold (Table 1). This implies that FL-CLIP enhances the partitioning of EB3 into droplets. To further test this enhancement effect, and to analyze the total amount of protein in the diluted and the dense phase, we performed a droplet-pelleting assay followed by SDS-PAGE analysis (for details, see Materials and Methods). Consistent to what we observed by microscopy, in EB3/FL-CLIP droplets, the amount of condensed EB3 increased by 1.7-fold in the pelleting assay compared to amounts measured for EB3 droplets alone (Fig. 4 A and B). We further studied this enhanced EB3 condensation with our high-throughput microscopy assay. Although the size of EB3/FL-CLIP droplets decreased, the number of droplets increased leading to a surface coverage 2.5-fold higher than FL-CLIP droplets alone (Fig. 4 C and D and SI Appendix, Fig. S5D). This resulted in a 40% increase of surface coverage by EB3/FL-CLIP droplets when compared to the sum of the surface coverages of EB3 droplets alone plus FL-CLIP droplets alone (Fig. 4E). In comparison to the presence of FL-CLIP, repeating the EB3 experiment with H2 or H1 proteins reduced surface coverage by 1.5-fold or 2.5-fold, respectively (Fig. 4 F and G and SI Appendix, Fig. S5H). This shows that CLIP’s C-terminal region is needed for a highly multivalent EB3/FL-CLIP network formation. Taken together, EB3 and CLIP-170 can undergo LLPS both independently, and when acting as an ensemble, the amount of proteins in the dense phase synergistically increases.
Fig. 4.
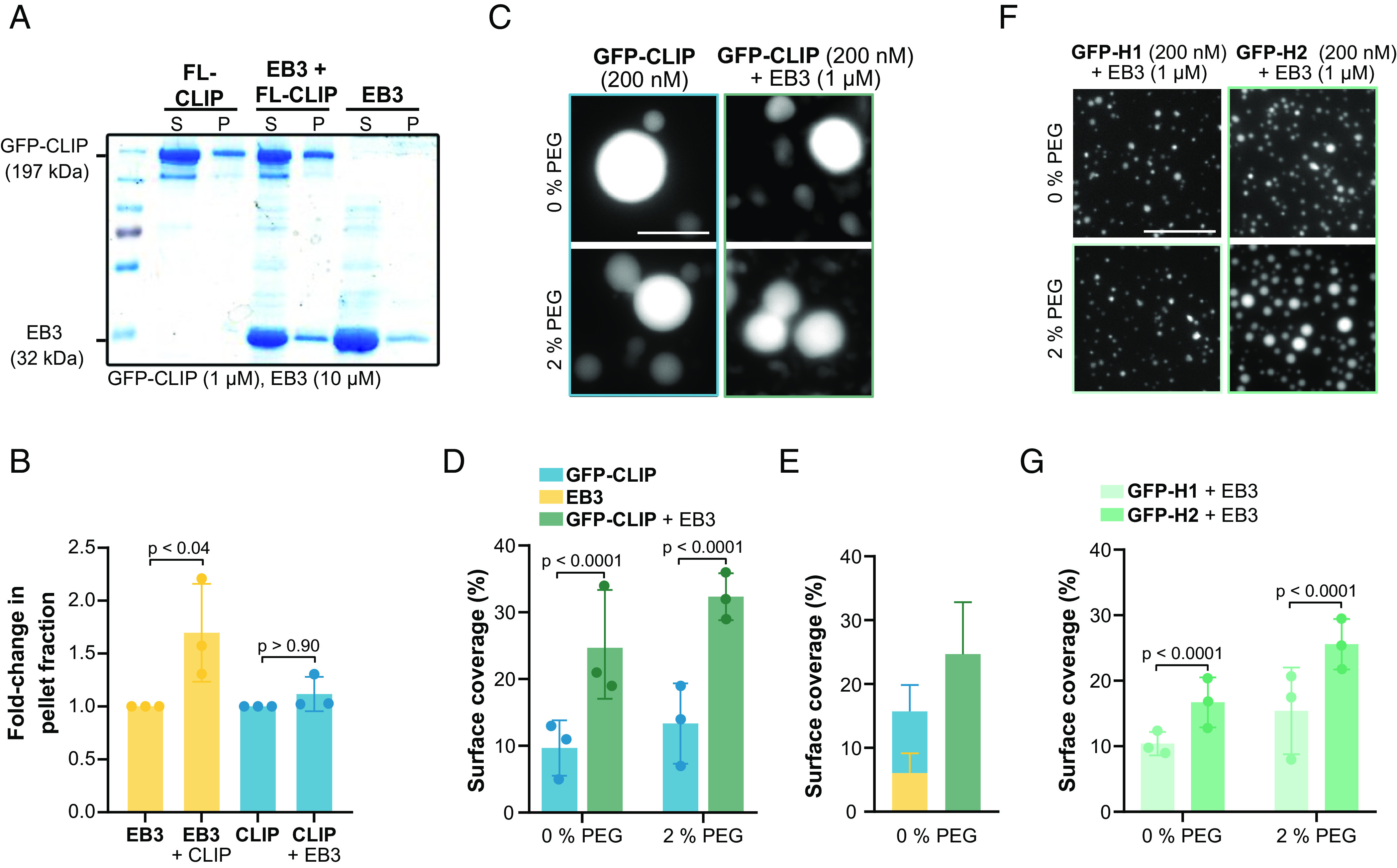
Synergistic condensation of CLIP-170 and EB3. (A) Representative SDS-PAGE analysis from the droplet-pelleting assay showing protein fractions in supernatant dilute phase (S) or pellet dense phase (P) under each condition: GFP-FL-CLIP (1 µM); EB3 (10 µM); EB3/GFP-FL-CLIP (10 µM + 1 µM). (B) Quantification of SDS-PAGE analysis showing the fold-change of protein in the pellet fraction at the three conditions. Mean with SD from three independent experiments. Statistics: one-way ANOVA. (C) Representative fluorescence confocal images of purified GFP-FL-CLIP in the absence (Left) or presence (Right) of EB3 and in the absence (Top) or presence (Bottom) of 2% PEG. (Scale bar: 20 µm.) (D) Condensate surface coverage of purified GFP-FL-CLIP (200 nM) in the absence (Left) or presence (Right) of EB3 (1 µM) at indicated PEG concentrations. Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test. (E) Quantification of droplet surface coverage of EB3 (1 µM) and GFP-FL-CLIP (200 nM) alone compared to surface coverage of EB3/GFP-FL-CLIP (1 µM/200 nM) droplet formation when undergoing synergistic LLPS in the absence of PEG. (F) Representative fluorescence confocal images of purified GFP-H1 (Left) or GFP-H2 (Right) in the presence of EB3 and in the absence (Top) or presence (Bottom) of 2% PEG. (Scale bar: 20 µm.) (G) Quantification of condensate surface coverage of indicated GFP-H1 (200 nM) or GFP-H2 (200 nM) in the presence of EB3 (1 µM) and in the presence of the indicated PEG concentrations. Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test.
Tubulin Is a Client Protein of EB3/CLIP-170 Droplets In Vitro.
A CLIP-170 dimer has up to 8 tubulin binding sites, can bind tubulin in vitro, and colocalizes with tubulin in cells (Fig. 5A) (14, 20, 23, 52). Therefore, we studied whether CLIP-170 droplets can enrich tubulin in vitro. Tubulin alone did not form droplets at micromolar concentrations; even in the presence of 5% PEG, only aggregation was observed (SI Appendix, Fig. S6A). However, when 200 nM FL-CLIP was mixed with 400 nM Atto565–tubulin, the two proteins phase separated into 26.8 ± 12 µm2 droplets (SI Appendix, Fig. S6 B and C). We repeated these experiments with H1 and found irregular-shaped aggregates with tubulin but no droplet formation (SI Appendix, Fig. S6 B and C), while EB3 enriched tubulin in droplets measuring 1 ± 0.07 µm2 droplets (Fig. 5 B and C). Consistently, EB3/FL-CLIP networks concentrated tubulin in intermediate-sized droplets (Fig. 5 B and C). These results demonstrate that FL-CLIP, EB3, and EB3/FL-CLIP droplets effectively enriched tubulin.
Fig. 5.
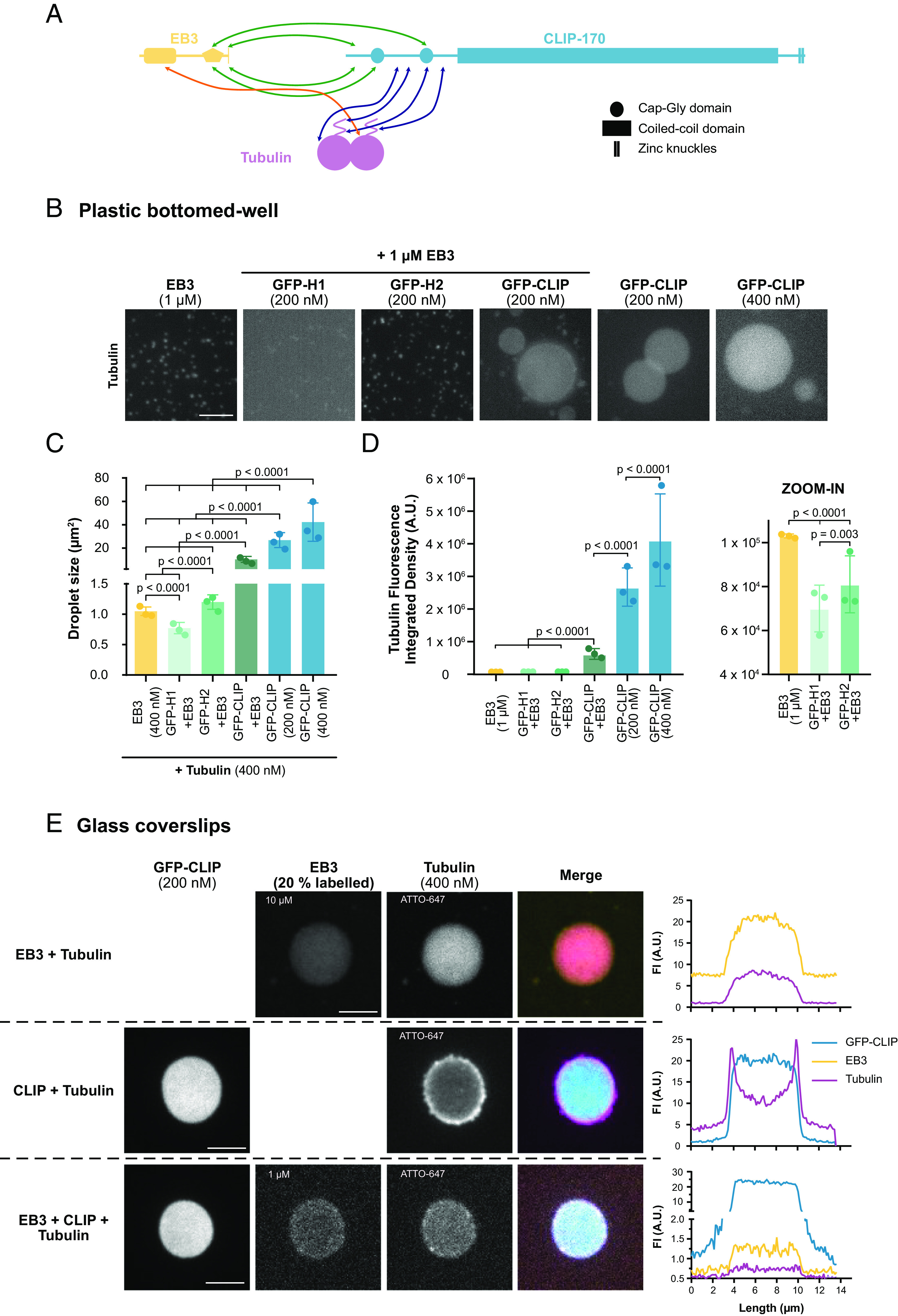
CLIP-170 and EB3 droplets concentrate tubulin. (A) Cartoon schematic of domain interactions between EB3, CLIP-170, and tubulin based primarily on (52–54). Interaction sites between EB3 and CLIP-170, EB3 and tubulin, and CLIP-170 and tubulin are shown with green, orange, and blue arrows, respectively. For simplification, monomers of EB3 and CLIP-170 are shown. (B) Representative confocal images with the same contrast settings, of Atto565–tubulin (400 nM) in the presence of purified EB3 (1 µM), GFP-H1 (200 nM) + EB3 (1 µM), GFP-H2 (200 nM) + EB3 (1 µM), GFP-FL-CLIP (200 nM) + EB3 (1 µM), and GFP-FL-CLIP (200 nM and 400 nM) alone. (Scale bar: 20 µm.) Note that the treated plastic surface used for this experiment did not allow for a tubulin shell around the FL-CLIP droplet as shown in (E) for details see Materials and Methods. (C) Quantification of the droplet size from (B) under denoted conditions. Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test. (D) Quantification of the integrated density of tubulin fluorescence under denoted conditions with zoom in for the first three conditions. Mean with SD from three independent experiments with a total of 27 fields of view. Statistics: two-tailed Student’s t test. (E) Representative images of EB3/mCherry-EB3/tubulin-647 droplets (8 µM/2 µM/4 µM), GFP-FL-CLIP/tubulin-647 droplets (200 nM/400 nM), and EB3/mCherry-EB3/GFP-FL-CLIP/tubulin-647 droplets (900 nM/100 nM/200 nM/400 nM), with the corresponding line scans. (Scale bar: 5 µm.)
Strikingly, FL-CLIP droplets enriched tubulin 40-fold more than EB3 droplets, even at fivefold lower FL-CLIP concentrations compared to EB3 (Fig. 5D). This difference in concentrated tubulin cannot be solely explained by the 3.5-fold difference in surface coverage between EB3 and FL-CLIP but indicates a difference in the potency of the two proteins in concentrating tubulin (Fig. 4E). We further studied whether addition of EB3 to FL-CLIP droplets alters the multivalency of the network such that it affects its ability to concentrate tubulin. Indeed, we observed that EB3/FL-CLIP droplets concentrated tubulin fourfold less efficiently than FL-CLIP alone (Fig. 5D). While FL-CLIP droplets effectively enriched tubulin, EB3 can directly bind and recruit other proteins to the growing microtubule tip. Therefore, in a scenario where tubulin is enriched at the growing microtubule tip, the two proteins FL-CLIP and EB3 would need to cooperate to efficiently concentrate tubulin at the growing microtubule tip.
Consistent with the different potency of the droplets to enrich tubulin, the distribution of tubulin within the droplet changed. Tubulin was enriched at the interface of the FL-CLIP droplets, where the concentration of tubulin was twice as high as within the droplet (Fig. 5E and Table 1). This phenomenon was independent of the fluorophore used to label tubulin, as antibody staining of unlabeled tubulin revealed the same distribution (SI Appendix, Fig. S6D). These results suggest that tubulin, as a client protein, is energetically more stable at the droplet interface due to incomplete wetting by the dense phase of FL-CLIP (55). In contrast to FL-CLIP droplets, tubulin was homogenously distributed in EB3 and EB3/FL-CLIP droplets (Fig. 5E). Therefore, the addition of EB3 to FL-CLIP droplets allows for complete wetting of tubulin, consistent with the wetting properties of EB3 (Movie S12).
Tubulin Is a Client Protein of CLIP-170 Condensates in Cells.
We next addressed whether tubulin can be a client protein of CLIP-170 droplets in cells. To do so, we depolymerized the microtubule network with 5 µM nocodazole in GFP-CLIP-170 overexpressing RPE-1 cells and observed robust tubulin colocalization with CLIP-170 droplets as previously reported (SI Appendix, Fig. S6 E, Top) (14, 23). Under these conditions, tubulin was concentrated 2.4-fold more in the CLIP-170 droplets compared to the rest of the cytosol (SI Appendix, Fig. S6F). To further study whether CLIP-170 could concentrate tubulin at its endogenous concentration, we used antibody staining after depolymerizing the microtubule network. In WT cells, small clusters of endogenous CLIP-170 were frequently observed after microtubule network depolymerization, and these clusters showed local enrichment of tubulin (SI Appendix, Fig. S6 E, Bottom). In cells depleted of CLIP-170 and in CLIP-170 knockdown cells rescued with H2, we did not observe tubulin/FL-CLIP clusters (SI Appendix, Fig. S6G). Contrary to our in vitro experiments, such tubulin clusters were not observed in nocodazole-treated cells overexpressing H2 or EB3 (SI Appendix, Fig. S6H). We conclude that CLIP-170, but not EB3 by itself, can concentrate tubulin in cells.
Phase Separation–Potent +TIP Networks Increase Microtubule Growth.
The absence of either EB3 or CLIP-170 at the microtubule tip does not decrease growth speeds in cells (12, 17, 56). However, combined siRNA knockdown of EB3 and CLIP-170 reduced microtubule growth speeds by 20% (SI Appendix, Fig. S7 A–D). To understand whether LLPS of +TIPs could affect microtubule tip dynamics, we turned to the in vitro reconstitution assay. We first reconstituted microtubule growth in the presence of +TIP networks with either reduced LLPS activity (50 nM H2 + 800 nM EB3) or minimal LLPS activity (50 nM H1 + 800 nM EB3) (Fig. 4F), implying reduced or minimal enrichment of tubulin at the growing tip (Fig. 5D). To our knowledge, these CLIP mutants preserve binding domains to interact with EB3 and tubulin, implying that the difference between these networks lies primarily in their phase-separation potency. Consistent with previous observations, EB3 alone increased microtubule growth speeds by 1.5-fold and catastrophe frequency by eightfold (Fig. 6 A and B) (12, 13). Addition of H1 or H2 to the assay with EB3 resulted in tip-tracking behavior by EB3/H1 and EB3/H2 networks but did not further alter any of the dynamic parameters compared to EB3 alone (Fig. 6 A and B and SI Appendix, Fig. S8A). These results show that CLIP/EB3 networks with reduced or minimal capacity to concentrate tubulin have the same impact on microtubule tip dynamics as EB3 alone.
Fig. 6.
LLPS of +TIPs regulates microtubule dynamics through local tubulin enrichment. (A) Representative microtubule kymographs of denoted +TIP networks in higher salt buffer (60 mM KCl and 85 mM K-acetate, see Materials and Methods). Note that tip-tracking efficiency (GFP channel) is weaker at 5 µM tubulin than at higher tubulin concentrations (SI Appendix, Fig. S8B). (B) Microtubule growth rate (Top) and catastrophe frequency (Bottom) in high salt buffer. Tubulin (5 µM), EB3 (800 nM), H1 (50 nM), H2 (50 nM), and FL-CLIP (50 nM). Mean with SD of minimum of three independent experiments with the following number of analyzed microtubules: control—48; EB3—31; EB3/H1—28; EB3/H2—28; EB3/FL-CLIP—60. Statistics: One-way ANOVA Fisher’s LSD test. (C) Representative time-lapse TIRF images of GFP-FL-CLIP (50 nM) with tubulin (5 µM) in the presence of unlabeled EB3 (800 nM) and 60 mM KCl. Time denoted in minutes: seconds. (Scale bar: 20 µm.) The zoom-ins (white dashed box) show representative droplet formation along microtubules. (D) Representative microtubule kymographs in the presence of GFP-H2 or GFP-FL-CLIP (50 nM), EB3 (800 nM), tubulin (5 µM), and 60 mM KCl. Arrowheads denote areas of robust tubulin/FL-CLIP condensation on growing microtubule shaft. (E) Representative TIRF images of Atto565–tubulin (5 µM) microtubules growing in the absence (Left, control) and in the presence of EB3/FL-CLIP (800 nM/50 nM) (Right). Right images show tubulin enrichment along microtubule over time. (Scale bar: 2 µm.) (F) Corresponding line scan of E with the gray line scan representing the 5 µM tubulin control condition and the magenta line scans of 5 µM tubulin in the presence of EB3/FL-CLIP (800 nM/50 nM). (G) Tubulin fluorescence intensity in tip-proximal regions in 5 µM tubulin control condition and in the presence of EB3/FL-CLIP. Quantification of perpendicular line scans in tip-proximal regions. Mean with SD from two independent experiments for each condition and a total of 20 microtubules; (scale bar: 2 µm.) (H) Quantification of microtubule growth rate (Left) and catastrophe frequency (Right) in the presence of EB3/H2-networks (800 nM/50 nM) and EB3/FL-CLIP droplets (800 nM/50 nM) in experiments from D (5 µM tubulin and 60 mM KCl). Mean with SD of three independent experiments with the following number of analyzed microtubules: EB3/H2—29; EB3/FL-CLIP—59. Statistics: paired t test.
When we repeated the reconstitution experiments with EB3/FL-CLIP, microtubules grew at a speed of 3.5 µm/min, a 1.5-fold increase compared to analogous experiments with EB3/H2 and EB3/H1, and nearly fourfold increase compared to controls with tubulin alone (Fig. 6 A and B and Movie S15). Furthermore, catastrophe events were reduced under these conditions (Fig. 6 A and B), and, when they occurred, these were rapidly followed by rescue events (SI Appendix, Fig. S8A). To reconstitute this fast microtubule growth in the presence of a phase separation–deficient EB3/H2 network, we increased the tubulin concentration while keeping the EB3/H2 concentration constant. We measured microtubule growth speeds and found that a growth speed of 3.5 µm/min (the speed achieved by EB3/FL-CLIP in the presence of 5 µM tubulin) corresponds to 12.8 µM tubulin in the presence of EB3/H2 (SI Appendix, Fig. S8 B and C). These results show that phase separation–potent EB3/FL-CLIP networks increase the growth rate and reduce the catastrophe events compared to EB3/H2 networks. A phase-separated +TIP network might impact the growing microtubule tip due to changes in viscosity, or by concentrating tubulin at the tip, based on our observation that EB3/FL-CLIP can concentrate tubulin (Fig. 5B).
+TIPs Undergo LLPS and Concentrate Tubulin on Microtubules, Promoting Microtubule Growth.
To understand whether LLPS of EB3/FL-CLIP networks could enrich tubulin on microtubules, we used microtubules as a platform to induce LLPS. A recent study showed that addition of cell lysate containing overexpressed CLIP-170 to microtubules formed CLIP-170-containing droplets along microtubules (24). We studied whether this droplet formation is due to the crowding environment of the cell lysate or a general property of FL-CLIP. Under assay conditions with lower ionic strength (Materials and Methods), we repeated the in vitro reconstitution assay described above to study whether purified EB3 together with FL-CLIP could phase separate along microtubules. When we incubated purified CLIP-170 and EB3 with dynamic microtubules in vitro, we found that purified EB3/FL-CLIP formed droplets along the shaft (Fig. 6C and Movie S16).
Consistent with our observations that EB3/FL-CLIP droplets concentrated tubulin (Fig. 5B), these networks enriched tubulin all along the microtubule and concentrated tubulin over time into droplets on the shaft, even more efficiently than free-floating droplets (Fig. 6 C–G). In this condition where tubulin is concentrated along the microtubule shaft, we observed rapid microtubule growth speeds of 3.6 µm/min and very few catastrophe events or pauses in the growth phase (Fig. 6 D and H and SI Appendix, Fig. S8D). This increase in growth speed and reduction in catastrophe events is consistent with our results from above where EB3/FL-CLIP was located at the microtubule tip. When we repeated these experiments with less potent phase-separating EB3/H2-networks, we only occasionally observed condensate formation along the microtubule (Fig. 6 D, Left). In the presence of EB3/H2-networks, growth speeds were twofold slower than for EB3/FL-CLIP, catastrophe frequencies were increased sevenfold, and pauses increased 30-fold (Fig. 6H and SI Appendix, Fig. S8D). In the absence of EB3, we did not observe microtubule binding or increased growth speeds for any CLIP constructs (SI Appendix, Fig. S8 E and F). Collectively, these experiments demonstrate that EB3/FL-CLIP networks undergo LLPS on microtubules, can concentrate tubulin, and drive microtubule growth.
Discussion
Rearrangements of the microtubule network architecture require spatiotemporal regulation of microtubule growth. A large body of work has highlighted the role of +TIPs that act as microtubule polymerases (such as XMAP215) or increase microtubule tip dynamics (such as the EBs) in promoting these highly regulated changes (10, 11, 56–60). While these studies showed the impact of +TIPs on microtubule dynamics, it has remained unclear how these proteins self-assemble to form highly dynamic and multivalent networks. LLPS allows for network formation of proteins based on highly multivalent interactions. We now provide evidence that LLPS of +TIPs could be a mechanism to explain the formation of +TIP networks at the growing microtubule tip. This mechanism could be a basis to locally recruit and enrich diverse +TIPs.
Here, we propose a mechanism by which LLPS of +TIPs regulate microtubule growth. Our model of LLPS-driven microtubule growth is based on the following observations: i) the +TIPs EB3 and CLIP-170 have the capacity to undergo LLPS in vitro and in cells (Figs. 1–4), ii) phase-separated EB3/FL-CLIP droplets can concentrate tubulin (Fig. 5), iii) microtubule growth speed is increased and catastrophe frequency reduced when EB3/FL-CLIP undergo phase separation on microtubules (Fig. 6H), and iv) tubulin enriched along the microtubule in the presence of phase-separated EB3/FL-CLIP (Fig. 6 C–G). We hypothesize that this tubulin enrichment may be a mechanism to increase local tubulin availability at microtubule ends, uncoupled from cytoplasmic tubulin concentrations. Microtubule dynamics might be further modulated by the viscosity of the droplets, such as sterically constraining protofilaments in a manner that accelerates growth or prevents catastrophes.
Droplet fission is not a spontaneous process, leading us to question how trailing foci separate from the leading +TIP droplet. What drives the process is unknown, but several mechanisms could cause droplet fission at the growing microtubule tip. One possibility is that a fraction of the droplet simply becomes “stuck” at the lattice. Alternatively, these trailing foci may appear by binding to GTP–tubulin, that could remain behind the tip in the GDP shaft. It is also possible that a combination of parameters like microtubule growth speed, a specific droplet size at the tip, and droplet viscosity might generate drag force causing the droplet to split from the tip droplet.
Microtubules could serve as a general platform to concentrate proteins locally to an LLPS-sufficient concentration. We propose that LLPS initiation at the growing tip depends on conformational properties of the growing microtubule tip. This could lead to differential binding of EB3 to the tip over the shaft (61), followed by CLIP-170 recruitment to form a multivalent network. As LLPS is a concentration-dependent process, the localized recruitment of EB3 and CLIP-170 could increase their concentration compared to the solution, prompting them to undergo LLPS. We hypothesize that once a critical CLIP-170 concentration is reached, the +TIP network undergoes LLPS. This +TIP droplet could then potentially concentrate tubulin and drive microtubule growth. Upon GTP-cap hydrolysis, the +TIP droplet dissolves. This implies that the shape of the +TIP droplets would correspond to the decaying GTP–tubulin profile at the growing microtubule tip. Whether the stoichiometry between EB3 and CLIP-170 has an impact on the LLPS process, as well as the distinct fluid properties and dynamics of an EB3/CLIP-170 droplet, merits investigation in future work.
An EB3/FL-CLIP droplet has distinct properties that differ from EB3- or FL-CLIP droplets in terms of mobile fraction and tubulin interaction. These characteristics may be important for the interaction between microtubule and EB3/FL-CLIP droplets. Interestingly, we observed that EB3/FL-CLIP droplets on microtubules strongly enriched tubulin, whereas tubulin was less concentrated in free-floating EB3/FL-CLIP droplets. This finding suggests that the microtubule surface can change the propensity of the EB3/FL-CLIP droplets to enrich tubulin.
We hypothesize that LLPS of +TIPs serves an organizational purpose, allowing for the formation of a highly concentrated protein network at microtubule ends. Our observation that +TIPs concentrate soluble tubulin into droplets and along microtubules, leads us to postulate that this process contributes to the regulation of microtubule dynamics in cells. Interestingly, previous reports showed that depletion of CLIP-170 or EB3 from cells only moderately affects microtubule dynamics in cells (17), while depletion of EB3 and CLIP-170 reduced the growth speed by 20% (SI Appendix, Fig. S7). This suggests that there is some redundancy in the functions of EBs in regulating microtubule dynamics in cells (12, 62). Additionally, considering the observation that diverse +TIPs partition into CLIP-170 condensates (23), we favor the hypothesis that the condensation of the +TIP network can be driven by multiple different proteins. In this scenario, the depletion of CLIP-170 or EB3 alone would not significantly impede microtubule growth, as other +TIPs could compensate for driving +TIP network phase separation. Further studies that compare the phase-separation potencies of other +TIPs, as well as combinatorial depletions of LLPS-potent +TIPs in cells, will be of interest in addressing this question.
Is phase separation a common feature of +TIPs? Studies performed in parallel to our work demonstrate that this phenomenon is conserved across evolution: +TIPs in budding yeast, fission yeast, and higher eukaryotes have recently been demonstrated to undergo phase separation (23–27). Intriguingly, while the LLPS potency of EB homologs varied between organisms, the yeast studies confirmed our results that the CLIP-170 homolog played a key role in the phase-separation process. The role of different mammalian EB family members in regulating LLPS will be an interesting direction for future studies. The ability of +TIP networks to phase separate depends on intrinsically disordered regions (25, 27) and multivalent interaction modules (26), consistent with the observation that these features are highly evolutionarily conserved across +TIPs (23). Further studies will be necessary to investigate whether additional +TIPs contribute to the formation and regulation of +TIP droplets.
Our work here and other recent studies demonstrate that +TIP networks can behave like liquid condensates (23–27). This work adds to the growing list of microtubule-related processes that are driven by LLPS and provides an exciting paradigm for how cells can spatiotemporally control microtubule dynamics through local tubulin concentration (25–27, 31–35). Exploring the mechanical properties and composition of +TIP droplets, as well as studying their regulation throughout the cell cycle, will be exciting avenues for future research.
Materials and Methods
For extended Materials and Methods section see also SI Appendix, Methods.
Cell Culture and Treatments.
Parental RPE-1 and CRISPR/Cas9 knock-in RPE-1–GFP–tubulin cells were cultured, transiently transfected with FL-CLIP, H1, H2, and EB3, knocked down for CLIP-170 or EB3 (Santa Cruz, 43281; Thermo Fisher, s22683), and treated with 5 µM nocodazole as described in SI Appendix.
Immunofluorescence.
Cells were fixed with 100% methanol for 5 min at −20 °C and then for 15 min with 3% paraformaldehyde at room temperature. Cells were incubated with antibodies targeting tubulin (Sigma T6199, DM1α, 1:1,000, mouse), EB1 (Millipore AB 6057, rabbit, 1:1,000), EB3 (Santa Cruz Biotechnology sc-101475, KT36, rat, 1:200), or CLIP-170 (Santa Cruz Biotechnology sc-28325, F3, mouse, 1:500).
Protein Purification.
Tubulin was purified and labeled from the fresh bovine brain by two subsequent polymerization/depolymerization cycles as described previously (63). EB3, mCherry-EB3, EB3-ΔY, mCherry-EB3-ΔY, GFP-H1, GFP-H2 and GFP-H2-tail was purified from Escherichia coli BL21 (DE3) (for details, see SI Appendix, Methods). FL-CLIP-170-GFP was purified from insect cells as described previously (64).
Phase-Separation Assay.
For in vitro phase-separation assays, proteins were diluted to the appropriate concentration in BRB80 pH 6.9 supplemented with 50 mM potassium chloride and PEG 4000 (0, 2, 5, or 10% by weight) in Eppendorf tubes. To establish a phase diagram we used protein concentrations from 0.8 to 100 nM and salt concentrations from 50 to 450 mM. After thorough mixing, reactions were transferred to 384-well plates (plastic well-bottom treated for cell culture, Falcon) and incubated for 7 min. The plate was then centrifuged to sediment proteins in the dense phase on the well bottoms prior to acquisition.
Microtubule Dynamics Assays In Vitro.
Microtubule seeds and flow chambers were prepared as described previously (63). Assays involving FL-CLIP-170 were carried out as noted above, but within the first 5 h postpurification as FL-CLIP-170 activity is poorly preserved after freezing (64). For “low salt” assays (Fig. 6 E and F), the reaction buffer was supplemented with 60 mM potassium chloride. For “high salt” assays (Fig. 6 A and B), the reaction buffer was supplemented with 60 mM potassium chloride and 85 mM potassium acetate to facilitate tip tracking as described previously (64).
Supplementary Material
Appendix 01 (PDF)
Representative RPE-1 expressing mCherry-CLIP. Inset highlights fission of mCherry-CLIP droplets.
Representative RPE-1 expressing mCherry-CLIP. Inset highlights fusion of mCherry-CLIP droplets.
Fluorescence recovery after photobleaching of a mCherry-CLIP droplet in a representative RPE-1 cell.
Representative foci formation resembling fission in GFP-CLIP overexpressing RPE-1 cells. Scale bar: 2 μm.
Representative RPE-1 (magenta) expressing mCherry-H1 (cyan).
Representative RPE-1 (magenta) expressing mCherry-H2 (cyan).
Representative RPE-1 cell expressing mCherry-EB3 and treated with 5% 1,6-hexanediol (added at 00:30).
Fusion of two GFP-CLIP droplets (1μM) in vitro.
Fluorescence recovery after photobleaching of a purified GFP-CLIP droplet (2 μM) in vitro.
Fluorescence recovery after photobleaching of a purified EB3 (10 μM) labeled with GFP-EB3 (100 nM) droplet in vitro.
Fluorescence recovery after photobleaching of a purified EB3 (10 μM) labeled with GFP-EB3 (100 nM) mixed with GFP-CLIP (2 μM) droplet in vitro.
3D representation of EB3 (8 μM) labelled with mCherry- EB3 (2 μM) droplets in vitro. Note that floating droplets moved during image acquisition.
3D representation of GFP-CLIP (2 μM) droplets in vitro.
3D representation of GFP-CLIP (2 μM) mixed with EB3 (8 μM) labelled with mCherry- EB3 (2 μM) droplets in vitro. Note that floating droplets moved during image acquisition.
Representative in vitro microtubule dynamics assay of Atto-565 tubulin (magenta) polymerized in the presence of 50 nM GFP-FL-CLIP (cyan) and 800 nM EB3 in high salt buffer.
Representative in vitro microtubule dynamics assay of Atto-565 tubulin (magenta) polymerized in the presence of 50 nM GFP-FL-CLIP (cyan) and 800 nM EB3 in low salt buffer.
Acknowledgments
We thank Thomas Surrey (Centre for Genomic Regulation, Barcelona, Spain) for the FL-CLIP-170 insect cells expression vector and Michel Steinmetz (Paul Scherrer Institute, Villigen PSI, Switzerland) for providing vectors for EB3 purification. We thank Peter Bieling (MPI of Molecular Physiology, Dortmund, Germany) for discussing FL-CLIP purification and Maria Hondele (University of Basel, Switzerland) for discussing phase-separation experiments. We thank ACCESS Geneva for their help with high-content microscopy and data analysis. We thank the Protein Platform (University of Geneva, Switzerland) for their support in FL-CLIP-170 expression. We thank Karsten Kruse and Marcos Gaitan-Gonzales for critically reading the manuscript. J.M. has been supported by the SNSF (Swiss NSF), 31003A_182473; R.T.W. has been supported by the NCCR Chemical Biology program; C.A. has been supported by the DIP of the Canton of Geneva.
Author contributions
J.M. and C.A. designed research; J.M., R.T.W., M.-C.V., and C.A. performed research; J.M., R.T.W., and C.A. analyzed data; and J.M., R.T.W., and C.A. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. H.V.G. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Brouhard G. J., Rice L. M., Microtubule dynamics: An interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 19, 451–463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howard J., Hyman A. A., Growth, fluctuation and switching at microtubule plus ends. Nat. Rev. Mol. Cell Biol. 10, 569–574 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Brouhard G. J., Dynamic instability 30 years later: Complexities in microtubule growth and catastrophe. Mol. Biol. Cell 26, 1207–1210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudimchuk N. B., McIntosh J. R., Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 22, 777–795 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Walker R. A., et al. , Dynamic instability of individual microtubules analyzed by video light microscopy: Rate constants and transition frequencies. J. Cell Biol. 107, 1437–1448 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voter W. A., O’Brien E. T., Erickson H. P., Dilution-induced disassembly of microtubules: Relation to dynamic instability and the GTP cap. Cell Motil. Cytoskeleton 18, 55–62 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova A., Steinmetz M. O., Microtubule +TIPs at a glance J. Cell Sci. 123, 3415–3419 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Akhmanova A., Steinmetz M. O., Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Galjart N., Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528–R537 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Bieling P., et al. , CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183, 1223–1233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitre B., et al. , EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10, 415–421 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Komarova Y., et al. , Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184, 691–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montenegro Gouveia S., et al. , In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20, 1717–1722 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Perez F., Diamantopoulos G. S., Stalder R., Kreis T. E., CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517–527 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Arnal I., Heichette C., Diamantopoulos G. S., Chrétien D., CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 14, 2086–2095 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Bieling P., et al. , Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100–1105 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Komarova Y. A., Akhmanova A. S., Kojima S.-I., Galjart N., Borisy G. G., Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589–599 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komarova Y., et al. , EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell 16, 5334–5345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta K. K., et al. , Minimal plus-end tracking unit of the cytoplasmic linker protein CLIP-170*. J. Biol. Chem. 284, 6735–6742 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierre P., Pepperkok R., Kreis T. E., Molecular characterization of two functional domains of CLIP-170 in vivo. J. Cell Sci. 107, 1909–1920 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Goodson H. V., et al. , CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil. Cytoskeleton 55, 156–173 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Pierre P., Scheel J., Rickard J. E., Kreis T. E., CLIP-170 links endocytic vesicles to microtubules. Cell 70, 887–900 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Wu Y.-F.O., et al. , Overexpression of the microtubule-binding protein CLIP-170 induces a +TIP network superstructure consistent with a biomolecular condensate. PLoS One 16, e0260401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jijumon A. S., et al. , Lysate-based pipeline to characterize microtubule-associated proteins uncovers unique microtubule behaviours. Nat. Cell Biol. 24, 253–267 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Maan R., et al. , Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus-ends. Nat. Cell Biol. 25, 68–78 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier S. M., et al. , Multivalency ensures persistence of a +TIP body at specialized microtubule ends. Nat. Cell Biol. 25, 56–67 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X., et al. , Phase separation of EB1 guides microtubule plus-end dynamics. Nat. Cell Biol. 25, 79–91 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Boeynaems S., et al. , Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Jiang H., et al. , Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernández-Vega A., et al. , Local nucleation of microtubule bundles through tubulin concentration into a condensed Tau phase. Cell Rep. 20, 2304–2312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King M. R., Petry S., Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun. 11, 270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodruff J. B., et al. , The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Jiang X., et al. , Condensation of pericentrin proteins in human cells illuminates phase separation in centrosome assembly. J. Cell Sci. 134, jcs258897 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drechsel D. N., Kirschner M. W., The minimum GTP cap required to stabilize microtubules. Curr. Biol. 4, 1053–1061 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Schek H. T., Gardner M. K., Cheng J., Odde D. J., Hunt A. J., Microtubule assembly dynamics at the nanoscale. Curr. Biol. 17, 1445–1455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seetapun D., Castle B. T., McIntyre A. J., Tran P. T., Odde D. J., Estimating the microtubule GTP cap size in vivo. Curr. Biol. 22, 1681–1687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rickman J., Duellberg C., Cade N. I., Griffin L. D., Surrey T., Steady-state EB cap size fluctuations are determined by stochastic microtubule growth and maturation. Proc. Natl. Acad. Sci. U.S.A. 114, 3427–3432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth D., Fitton B. P., Chmel N. P., Wasiluk N., Straube A., Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties. J. Cell Sci. 132, jcs219550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mustyatsa V. V., et al. , Fine structure and dynamics of EB3 binding zones on microtubules in fibroblast cells. Mol. Biol. Cell 30, 2105–2114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henrie H., et al. , Stress-induced phosphorylation of CLIP-170 by JNK promotes microtubule rescue. J. Cell Biol. 219, e201909093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura S., et al. , Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS One 7, e51442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohan R., et al. , End-binding proteins sensitize microtubules to the action of microtubule-targeting agents. Proc. Natl. Acad. Sci. U.S.A. 110, 8900–8905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Forges H., et al. , Localized mechanical stress promotes microtubule rescue. Curr. Biol. 26, 3399–3406 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Kroschwald S., Alberti S., Gel or die: Phase separation as a survival strategy. Cell 168, 947–948 (2017). [DOI] [PubMed] [Google Scholar]
- 47.McSwiggen D. T., Mir M., Darzacq X., Tjian R., Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 33, 1619–1634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiśniewski J. R., Hein M. Y., Cox J., Mann M., A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics 13, 3497–3506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Diamantopoulos G. S., et al. , Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144, 99–112 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixit R., et al. , Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. U.S.A. 106, 492–497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta K. K., et al. , Probing interactions between CLIP-170, EB1, and microtubules. J. Mol. Biol. 395, 1049–1062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J., et al. , α-tubulin tail modifications regulate microtubule stability through selective effector recruitment, not changes in intrinsic polymer dynamics. Dev. Cell 56, 2016–2028.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjelić S., et al. , Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued). J. Struct. Biol. 177, 160–167 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Böddeker T. J., et al. , Non-specific adhesive forces between filaments and membraneless organelles. Nat. Phys. 18, 571–578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Straube A., Merdes A., EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr. Biol. 17, 1318–1325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gard D. L., Kirschner M. W., A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 105, 2203–2215 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brouhard G. J., et al. , XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanic M., Widlund P. O., Hyman A. A., Howard J., Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15, 688–693 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Srayko M., Kaya A., Stamford J., Hyman A. A., Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell 9, 223–236 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Zhang R., Alushin G. M., Brown A., Nogales E., Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell 162, 849–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang C., et al. , EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J. Cell Biol. 216, 3179–3198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andreu-Carbó M., Fernandes S., Velluz M.-C., Kruse K., Aumeier C., Motor usage imprints microtubule stability along the shaft. Dev. Cell 57, 5–18.e8 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Telley I. A., Bieling P., Surrey T., Reconstitution and quantification of dynamic microtubule end tracking in vitro using TIRF microscopy. Methods Mol. Biol. 777, 127–145 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Representative RPE-1 expressing mCherry-CLIP. Inset highlights fission of mCherry-CLIP droplets.
Representative RPE-1 expressing mCherry-CLIP. Inset highlights fusion of mCherry-CLIP droplets.
Fluorescence recovery after photobleaching of a mCherry-CLIP droplet in a representative RPE-1 cell.
Representative foci formation resembling fission in GFP-CLIP overexpressing RPE-1 cells. Scale bar: 2 μm.
Representative RPE-1 (magenta) expressing mCherry-H1 (cyan).
Representative RPE-1 (magenta) expressing mCherry-H2 (cyan).
Representative RPE-1 cell expressing mCherry-EB3 and treated with 5% 1,6-hexanediol (added at 00:30).
Fusion of two GFP-CLIP droplets (1μM) in vitro.
Fluorescence recovery after photobleaching of a purified GFP-CLIP droplet (2 μM) in vitro.
Fluorescence recovery after photobleaching of a purified EB3 (10 μM) labeled with GFP-EB3 (100 nM) droplet in vitro.
Fluorescence recovery after photobleaching of a purified EB3 (10 μM) labeled with GFP-EB3 (100 nM) mixed with GFP-CLIP (2 μM) droplet in vitro.
3D representation of EB3 (8 μM) labelled with mCherry- EB3 (2 μM) droplets in vitro. Note that floating droplets moved during image acquisition.
3D representation of GFP-CLIP (2 μM) droplets in vitro.
3D representation of GFP-CLIP (2 μM) mixed with EB3 (8 μM) labelled with mCherry- EB3 (2 μM) droplets in vitro. Note that floating droplets moved during image acquisition.
Representative in vitro microtubule dynamics assay of Atto-565 tubulin (magenta) polymerized in the presence of 50 nM GFP-FL-CLIP (cyan) and 800 nM EB3 in high salt buffer.
Representative in vitro microtubule dynamics assay of Atto-565 tubulin (magenta) polymerized in the presence of 50 nM GFP-FL-CLIP (cyan) and 800 nM EB3 in low salt buffer.
Data Availability Statement
All data are included in the manuscript and/or supporting information.