Abstract
Understanding variant-specific differences in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral kinetics may explain differences in transmission efficiency and provide insights on pathogenesis and prevention. We evaluated SARS-CoV-2 kinetics from nasal swabs across multiple variants (Alpha, Delta, Epsilon, Gamma) in placebo recipients of the ACTIV-2/A5401 trial. Delta variant infection led to the highest maximum viral load and shortest time from symptom onset to viral load peak. There were no significant differences in time to viral clearance across the variants. Viral decline was biphasic with first- and second-phase decays having half-lives of 11 hours and 2.5 days, respectively, with differences among variants, especially in the second phase. These results suggest that while variant-specific differences in viral kinetics exist, post–peak viral load all variants appeared to be efficiently cleared by the host.
Clinical Trials Registration. NCT04518410.
Keywords: COVID-19, variant, viral kinetics
The coronavirus disease 2019 (COVID-19) pandemic has been fueled by successive waves of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs). Each new variant appears to have distinctive transmission and/or fitness advantages over the previous ones [1]. Understanding the underlying mechanism for this increased transmission efficiency is vital for predicting future variant waves and may provide insights on ways to prevent transmission. Prior studies have reported the impact of key mutations on SARS-CoV-2 viral fitness [2]. There are also reports that certain mutations increase the ability of SARS-CoV-2 variants to bind the human angiotensin-converting enzyme 2 (ACE2) receptor and mediate more rapid cellular entry [3]. However, it remains unclear to what extent differences in the levels or duration of viral shedding may be driving the increasing transmission efficiency. While there have been intriguing reports that VOCs may differ in their viral kinetics during acute COVID-19 [4], other studies have not found a substantial difference in viral shedding among variants [5–8]. Thus, a more comprehensive analysis of viral dynamics across a broad range of SARS-CoV-2 variants is needed.
In this study, we evaluated the SARS-CoV-2 viral load peak and viral decay kinetics across a range of SARS-CoV-2 variants in outpatients with mild to moderate COVID-19 who enrolled in the ACTIV-2/A5401 multicenter phase 2/3 adaptive platform randomized controlled trial.
METHODS
Overview of Study Participants
The study participants included adults with documented acute SARS-CoV-2 infection enrolled in the ACTIV-2/A5401 platform trial of therapeutics for outpatients with mild to moderate COVID-19 (NCT04518410). This analysis was restricted to participants who were enrolled between January and July 2021 in the placebo arms of the phase 2/3 evaluation of amubarvimab plus romlusevimab monoclonal antibodies, and who had information on the infecting SARS-CoV-2 variant [Spike (S) gene sequencing], resulting in 299 participants. In Supplementary Figure 1, we present a flowchart of the number of individuals included in each step of the analyses. The protocol was approved by a central institutional review board, Advarra (Pro00045266), for sites in the United States (US) and by local ethics committees for sites outside the US. All participants provided written informed consent prior to study enrollment.
SARS-CoV-2 Viral Load Testing and Variant Sequencing
Anterior nasal (AN) swabs were self-collected by participants, per protocol, on enrollment (day 0) and daily through study day 14 and at day 28 in phase 2 and on study days 0, 3, 7, 14, and 28 in phase 3. For each participant, we transformed “study days” to “days post–symptom onset” (DPO) using the day of symptom onset for each participant. SARS-CoV-2 viral load from AN swabs were quantified as previously described, with a lower limit of quantification (LLoQ) imputed as 1.7 log10 copies/mL and a lower limit of detection (LLoD) imputed as 0.7 log10 copies/mL, as described before [9]. S gene sequencing was performed for all participants. In brief, viral RNA extraction was performed on 1 mL of swab eluted by TRIzol LSReagent (ThermoFisher). S gene amplification was performed using a nested polymerase chain reaction strategy with an in-house designed primer sets targeting codons 1-814 of S gene [10]. Sequencing was performed on the Illumina MiSeq platform and deep sequencing data analysis was carried out using the Stanford CoV-Resistance Database (RDB) platform [11]. SARS-CoV-2 variant determination was confirmed using 3 different variant-defining platforms, namely, CoV-RDB [11], Scorpio call version 1.2.123 [12], and Nextclade version 1.13.2 [13].
Analyses of Viral Load Data
We analyzed differences in baseline (study entry) participant characteristics by variant using general linear models for continuous variables, namely, age, days post–symptom onset (DPO) to entry into the study, and baseline viral load, and χ2 tests or Fisher exact tests (if any expected count was <1 or >20% of expected counts <5) for categorical variables (sex, race, study phase).
Maximum viral load post–symptom onset was defined as the highest viral load recorded in each participant. The time to viral load maximum is then the time from symptom onset to the time of the maximum observed viral load. The duration of viral shedding since symptom onset was defined as the time from symptom onset until the first time of viral load below the limit of quantification, and no subsequent larger viral load. We excluded 26 participants because they had all viral load measurements above the LLoQ (Supplementary Figure 1). The maximum viral load, time to viral load maximum, and duration of viral shedding were compared across variants using general linear models, adjusting for potential baseline confounders, namely, age, sex, race, study phase, and time since symptom onset at study entry.
We defined viral rebound after >10 DPO as ≥1 log10 increase from the preceding viral load measurement, and a viral load reaching at least 3 log10. As an alternative definition we also analyzed similar cases but with an increase of ≥0.5 log10.
Analyses of Viral Decay Rate Postpeak
We quantified the rate of viral decline after the maximum observed viral load, assuming an exponential decay in the viral load. This assumption is consistent with visual inspection of the data. The model we fit is a biexponential (ie, biphasic) decay given by:
where V is the viral load, V0 is its maximum value, A is the fraction of V that decays in the first phase at rate λ1, and (1 – A) is the fraction that decays in the second phase at rate λ2. We tested if a biphasic or a single-phase decay is better by setting A = 1 in the expression above, which then causes V to decay as a single exponential, and λ2 is not estimated. The selection of the best model was based on the corrected Bayesian information criterion (cBIC), where a smaller value signifies a statistically preferred fit.
We fitted this model using nonlinear mixed effects, implemented in Monolix 2021R1 (lixoft.com/products/monolix/), to participants with at least 2 viral load measurements above the LLoQ during the decay phase (ie, decrease from maximum viral load) (n = 204). Each estimated parameter was assumed to follow a given distribution in the population (V0 is lognormal, A is logit normal, and λ1 and λ2 are lognormal), and the parameter value for an individual i can be expressed (if lognormal) as where θ is the median value of the population distribution and ηi is the individual random effect, assumed to be normally distributed as N(0, ω2), accounting for variability between individuals. Data below the LLoQ and below the LLoD were handled as censored data.
We then tested whether the estimated model parameters differed by variant using general linear models to adjust for baseline confounders, as above, and analyzed correlation between parameters using Pearson correlation.
RESULTS
Of 299 participants included in this study, 83 (28%) were infected with the SARS-CoV-2 Delta variant, 53 (18%) with Gamma, 42 (14%) with Alpha, 28 (9%) with Epsilon, and 93 (31%) with “other” variants (these included Wuhan, non-VOCs, and <12 each of Beta, Iota, Lambda, and Mu variants). Demographics and baseline characteristics are described in Supplementary Table 1.
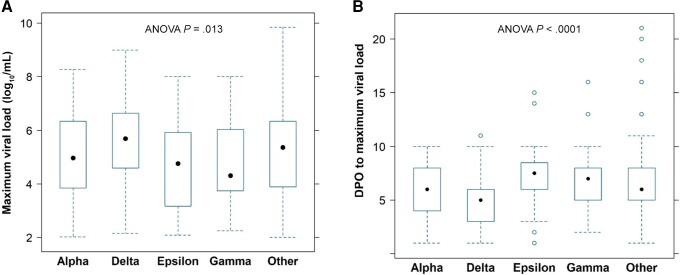
We found evidence that the maximum observed viral load differed among the variants (P = .03, Figure 1A, Supplementary Figure 2A). The maximal viral load for Delta infection (median, 5.69 log10 SARS-CoV-2 RNA copies/mL) was the highest compared to the other variants, including Alpha (4.96 log10 copies/mL), Epsilon (4.76 log10 copies/mL), Gamma (4.31 log10 copies/mL), and other (5.36 log10 copies/mL). This difference among variants remained significant (P = .013) even in a model adjusting for baseline confounders, including age, sex, race, study phase, and time since symptom onset at study entry. With this model, we also found that maximum viral load increased by 0.016 log10 copies/mL per year of age (P = .01). We then analyzed the time from symptom onset to maximum viral load, which in 19% of individuals occurred at a timepoint after baseline. We found that the time to maximum viral load was significantly different between variants (P < .0001, adjusted for baseline confounders) with Delta demonstrating the shortest period (Figure 1B, Supplementary Figure 2B). Of note, 1 of the potential confounders that we adjusted for in this multivariate model was the time post-onset of symptoms at study entry, which was positively correlated with time to maximum viral load. Even after taking into consideration the timing of symptoms, there was still a significant effect of variants on time to maximum viral load (P < .0001).
Figure 1.
A, Maximum viral load during study follow-up by variant. B, Days post–symptom onset to maximum viral load by variant. The boxplots represent the 25th and 75th percentiles (bottom and top edge of the box), the circle in the box represents the median, and whiskers extending from the edges of the box represent the smallest (bottom) or largest (top) value no further than 1.5 times the interquartile range. Outliers are represented as open circles. The P values shown correspond to analyses after adjusting for baseline covariates. Abbreviations: ANOVA, analysis of variance; DPO, days post–symptom onset.
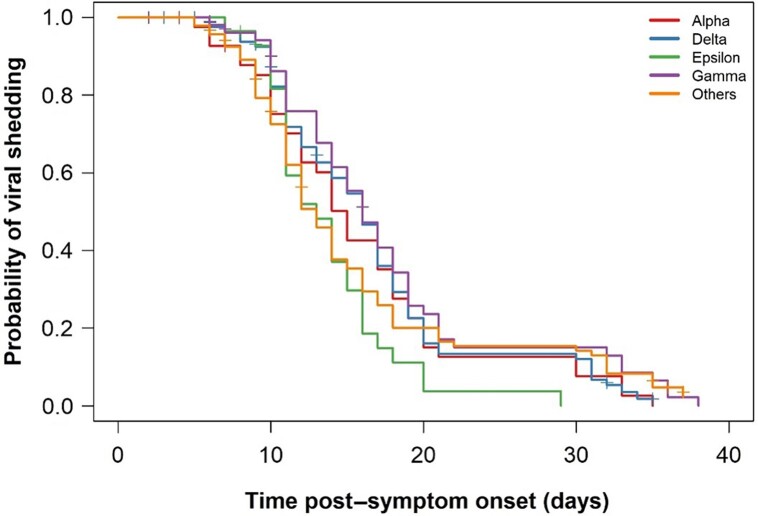
Next, we analyzed the duration of viral shedding from symptom onset to below LLoQ and found that this was not significantly different among the variants (P = .1) (median duration: Alpha = 15 days, Delta = 16 days, Epsilon = 13 days, Gamma = 16 days, and other = 13 days; Figure 2 and Supplementary Figure 3). In a multivariable model (n = 273, see Supplementary Figure 1), adjusting for baseline confounders, the duration of shedding was still not significantly different by variant (P = .12), but it was 2.3 days longer in males (P = .014).
Figure 2.
Kaplan-Meier plot of duration of viral shedding by variant (N = 299). The y-axis denotes the probability of continuing to shed virus at different times and the x-axis denotes time post–symptom onset. The vertical tick marks correspond to 26 individuals lost to follow-up.
We analyzed cases of viral rebound using a stringent definition (see “Analyses of Viral Load Data” in Methods) and found that 18 (of 299) individuals showed a rebound of at least 1 log10 in viral load after >10 DPO (Supplementary Figure 4A). If we define rebound as at least a 0.5-log10 increase, we find 21 individuals with a rebound (Supplementary Figure 4B) [14].
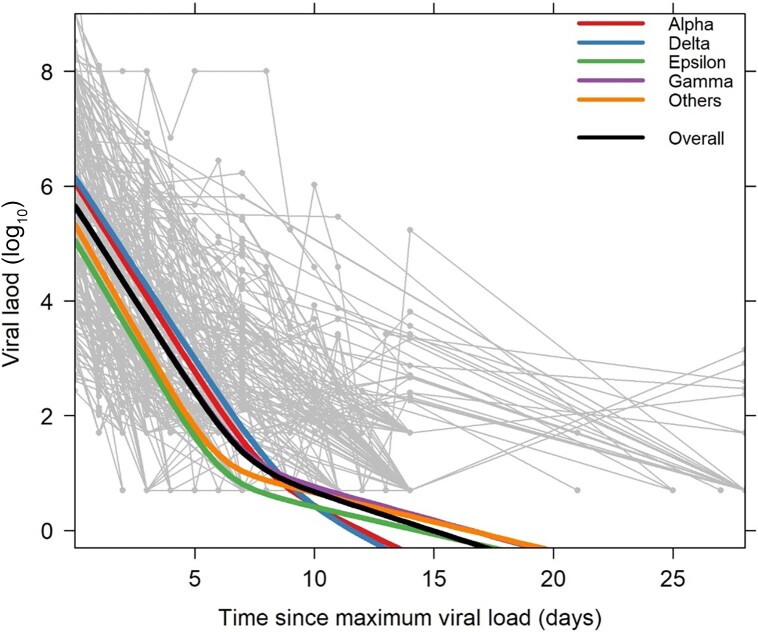
Finally, we modeled the viral decline after the observed maximum viral load. We could only fit the model to participants with sufficient data (n = 204). We verified that the distribution of variants across these 204 participants was similar to distribution in the full dataset (P = .99). A biphasic decay fitted the data better than a single exponential decay model (cBIC: 2999 vs 3098, respectively). There was no significant difference in the fraction of virus that decays in the first phase among the variants (P = .22). The overall estimates of the first- and second-phase viral decay half-lives (t1/2) were 11.0 hours (95% confidence interval [CI], 10.2–11.9 hours) and 2.5 days (95% CI, 1.5–3.4 days), respectively (Figure 3). There was a significant difference in the first phase of decay among the variants with Delta and Gamma variants showing the longest half-lives (P = .016; median t1/2: Alpha = 10.9 hours, Delta = 11.4 hours, Epsilon = 10.3 hours, Gamma = 11.4 hours, and other = 9.9 hours), although these small differences are of unclear clinical significance. In the second phase there was also a significant difference (P = .002), with Alpha (median t1/2 = 36 hours) and Delta (median t1/2 = 34 hours) variants having shorter second-phase half-lives than the other variants (median t1/2: Epsilon = 76 hours, Gamma = 63 hours, and other = 72 hours). This difference is clearly visible in Figure 3. Interestingly, we found a strong correlation (r = 0.90, P < .001) between the second-phase decay rate and the initial viral load at the start of decay (V0 in the model).
Figure 3.
Spaghetti plot of the decay in viral load (light gray lines and symbols) for each participant (n = 204), with the best population fit represented as a thick black line. The decay dynamics estimated from the model for each variant is shown as thick lines in the following order from top to bottom at time 0: Delta, Alpha, Overall, Others, Epsilon (note that the line for Gamma is barely visible under the Overall/Others lines). The y-axis denotes log10 severe acute respiratory syndrome coronavirus 2 RNA copies/mL and the x-axis denotes time in days since maximum viral load.
DISCUSSION
In this study, we performed a modeling analysis of longitudinal SARS-CoV-2 viral kinetics across a range of variants in untreated outpatients with mild to moderate COVID-19. The results demonstrate significant variant-specific differences in both the maximal observed viral loads and timing of the viral load maximum from AN swabs, with the Delta variant reaching a higher viral load and at an earlier time compared to prior variants. On the other hand, the duration of viral shedding (days to below LLoQ from symptom onset) was not different across variants. Our findings suggest that viral decay was better fit by a biphasic model than a simple exponential decay, as reported before in the context of a different protocol [9]. Our results estimated a first-phase decay t1/2 of 11 hours and a second-phase decay t1/2 of 60 hours. Understanding viral kinetics using mathematical modeling of SARS-CoV-2 infection is important for understanding SARS-CoV-2 transmission and to better inform public health responses to these outbreaks [15, 16].
While there have been conflicting results of how viral kinetics differ among variants [17–19], our results suggest that differences in viral kinetics could contribute to enhanced transmission. Specifically, faster increases in viral loads and higher maximal peak viral loads are likely to have contributed to Delta's rapid spread and replacement of prior variants. Starting with the introduction of the D614G mutation, it became clear that changes in spike protein could lead to increased viral fitness [20]. Mutations within the spike protein of the Delta variant, including L452R, T478K, and P681R, have been associated with increased viral infectivity, pathogenesis, and transmission [21, 22]. The combination of L452R and T478K appears to have synergistic interactions, resulting in enhanced ACE2 binding [23]. In vitro studies have shown that the Delta variant demonstrated a fitness advantage and increased infectiousness compared with the prior Alpha variant across physiologically relevant systems, including human alveolar epithelial and 3-dimensional airway organoid systems. This replication advantage was linked to differences in the Delta spike protein conformation, with a higher proportion of Spike found in a cleaved state compared to Alpha spike. This led to highly efficient cellular entry that was more resistant to neutralizing antibody inhibition compared to wild-type spike. The lack of significant differences in the time from symptom onset to viral clearance among the variants, despite differences in the maximum viral load, suggests that the effectiveness of the immune responses is not different across variants. We did find that the second-phase clearance rate was faster in participants with higher maximum viral load. This would explain why despite differences in maximum viral loads across variants, there was no difference in the time to reach viral loads below the limit of quantification. However, it is also possible that participants with larger maximal viral loads allowed better quantification of the second-phase decay rate.
Another interesting observation is that the duration of shedding was significantly longer in males than females. This finding was not linked to higher maximum viral loads in men but is consistent with our previous report in an overlapping population [24]. The underlying etiology for sex-based differences in viral shedding kinetics remains unclear but could be mediated in part by differential anti-coronavirus immune responses, related to both SARS-CoV-2 [25] and seasonal coronaviruses [26].
One limitation of our study is that time of symptom onset is self-reported, and enrollment occurred a median of 5 days after symptom onset (Supplementary Table 1), which means that we may have missed the true viral load peak in many participants. Thus, our analyses refer to the maximum observed viral load, as defined in the Methods. However, fitting a dynamic model to compare the kinetics of viral load across variants makes efficient use of all of the data and reduces biases that may arise from missing the peak viral load, which is not needed to estimate the decay. Another issue is that we are studying a largely unvaccinated population infected with pre-Omicron variants. Nevertheless, Delta and Omicron variants have been reported to have similar shedding kinetics [27], and vaccination also does not seem to substantially alter viral decay kinetics in breakthrough Delta infection after vaccination [6, 27, 28], although time since vaccination can be an important modulator of viral load levels [29]. When evaluating viral infectivity, there are also factors outside of viral load kinetics that may affect transmission potential, including escape from host immune pressure [28]. One of the strengths of this study is the uniform and frequent sampling of AN swabs that was performed within a rigorous randomized controlled trial setting, as well as the use of a validated quantitative SARS-CoV-2 viral load assay to assess levels of viral shedding [30]. However, even more frequent viral sampling and larger group sizes for individuals infected with the different variants would provide more power to detect dynamic differences among variants and allow more precise estimates of viral shedding.
In summary, we demonstrate that Delta variant infection led to, on average, the highest maximum viral load and shortest time from symptom onset to maximum viral load. We also found no significant differences for time to viral clearance among variants, with the first- and second-phase viral decays having overall half-lives of 11 hours and 2.5 days, respectively. These results suggest that while variant-specific differences in viral kinetics exist, all variants appeared to be cleared efficiently by the host.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Ruy M Ribeiro, Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, New Mexico.
Manish C Choudhary, Division of Infectious Diseases, Brigham & Women's Hospital, Harvard Medical School, Cambridge, Massachusetts.
Rinki Deo, Division of Infectious Diseases, Brigham & Women's Hospital, Harvard Medical School, Cambridge, Massachusetts.
Mark J Giganti, Center for Biostatistics in AIDS Research, Harvard T. H. Chan School of Public Health, Boston, Massachusetts.
Carlee Moser, Center for Biostatistics in AIDS Research, Harvard T. H. Chan School of Public Health, Boston, Massachusetts.
Justin Ritz, Center for Biostatistics in AIDS Research, Harvard T. H. Chan School of Public Health, Boston, Massachusetts.
Alexander L Greninger, Department of Laboratory Medicine and Pathology, University of Washington, Seattle.
James Regan, Division of Infectious Diseases, Brigham & Women's Hospital, Harvard Medical School, Cambridge, Massachusetts.
James P Flynn, Division of Infectious Diseases, Brigham & Women's Hospital, Harvard Medical School, Cambridge, Massachusetts.
David A Wohl, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill.
Judith S Currier, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles.
Joseph J Eron, Department of Medicine, School of Medicine, University of North Carolina at Chapel Hill.
Michael D Hughes, Center for Biostatistics in AIDS Research, Harvard T. H. Chan School of Public Health, Boston, Massachusetts.
Davey M Smith, Division of Infectious Diseases and Global Public Health, University of California, San Diego, La Jolla, California.
Kara W Chew, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles.
Eric S Daar, Lundquist Institute, Harbor-UCLA Medical Center, Torrance, California.
Alan S Perelson, Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, New Mexico.
Jonathan Z Li, Division of Infectious Diseases, Brigham & Women's Hospital, Harvard Medical School, Cambridge, Massachusetts.
for the ACTIV-2/A5401 Study Team:
Lara Hosey, Jhoanna Roa, Nilam Patel, Grace Aldrovandi, William Murtaugh, Frontier Science, Marlene Cooper, Howard Gutzman, Kevin Knowles, Rachel Bowman, Bill Erhardt, and Stacey Adams
Notes
ACTIV-2/A5401 Study Team
Kara Chew, MD, MS, Co-Chair, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA
David (Davey) Smith, MD, MAS, Co-Chair, University of California, San Diego, La Jolla, CA, USA
Eric Daar, MD, Vice Chair, Lundquist Institute at Harbor-UCLA Medical Center, Torrance, CA, USA
David Wohl, MD, Vice Chair, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill NC, USA
Judith Currier, MD, MSc, Protocol Investigator and ACTG Chair, David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA, USA
Joseph Eron, MD, Protocol Investigator and ACTG Vice Chair, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill NC, USA
Arzhang Cyrus Javan, MD, MPH, DTM&H, NIH Division of AIDS (DAIDS) Clinical Representative, National Institutes of Health, Rockville, MD, USA
Michael Hughes, PhD, Lead Statistician, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Carlee Moser, PhD, Statistician, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Mark Giganti, PhD, Statistician, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Justin Ritz, MS, Statistician, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Lara Hosey, MA, Clinical Trials Specialist, AIDS Clinical Trials Group (ACTG) Network Coordinating Center, Social & Scientific Systems, a DLH Company, Silver Spring, MD, USA
Jhoanna Roa, MD, Clinical Trials Specialist, AIDS Clinical Trials Group (ACTG) Network Coordinating Center, Social & Scientific Systems, a DLH Company, Silver Spring, MD, USA
Nilam Patel, Clinical Trials Specialist, AIDS Clinical Trials Group (ACTG) Network Coordinating Center, Social & Scientific Systems, a DLH Company, Silver Spring, MD, USA
Kelly Colsh, PharmD, DAIDS Pharmacist, NIH/DAIDS Pharmaceutical Affairs Branch, Rockville, MD, USA
Irene Rwakazina, PharmD, DAIDS Pharmacist, NIH/DAIDS Pharmaceutical Affairs Branch, Rockville, MD, USA
Justine Beck, PharmD, DAIDS Pharmacist, NIH/DAIDS Pharmaceutical Affairs Branch, Rockville, MD, USA
Scott Sieg, PhD, Protocol Immunologist, Case Western Reserve University, Cleveland, OH, USA
Jonathan Li, MD, MMSc, Protocol Virologist, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
Courtney Fletcher, PharmD, Protocol Pharmacologist, University of Nebraska Medical Center, Omaha, NE, USA
William Fischer MD, Protocol Critical Care Specialist, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill NC, USA
Teresa Evering, MD, MS, Protocol Investigator, Weill Cornell Medicine, New York, NY, USA
Rachel Bender Ignacio, MD, MPH, Protocol Investigator, University of Washington, Seattle, WA, USA
Sandra Cardoso, MD, PhD, Protocol Investigator, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil
Katya Corado, MD, Lundquist Institute at Harbor-UCLA Medical Center, Torrance, CA, USA
Prasanna Jagannathan, MD, Protocol Investigator, Stanford University School of Medicine, Palo Alto, CA, USA
Nikolaus Jilg, MD, PhD, Protocol Investigator, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
Alan Perelson, PhD, Protocol Investigator, Los Alamos National Laboratory, Los Alamos, NM, USA
Sandy Pillay, MB, CHB, Protocol Investigator, Enhancing Care Foundation, Durban, KwaZulu-Natal, South Africa
Cynthia Riviere, MD, Protocol Investigator, GHESKIO Center, Port-au-Prince, Haiti
Upinder Singh, MD, Protocol Investigator, Stanford University School of Medicine, Palo Alto, CA, USA
Babafemi Taiwo, MBBS, MD, Protocol Investigator, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Joan Gottesman, BSN, RN, CCRP, Field Representative, Vanderbilt University Medical Center, Nashville, TN, USA
Matthew Newell, BSN, RN, CCRN, Field Representative, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill NC, USA
Susan Pedersen, BSN, RN, Field Representative, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill NC, USA
Joan Dragavon, MLM, Laboratory Technologist, University of Washington, Seattle, WA, USA
Cheryl Jennings, BS, Laboratory Technologist, Northwestern University, Chicago, IL, USA
Brian Greenfelder, BA, Laboratory Technologist, Ohio State University, Columbus, OH, USA
William Murtaugh, MPH, Laboratory Specialist, ACTG Laboratory Center, University of California, Los Angeles, Los Angeles, CA, USA
Jan Kosmyna, MIS, RN, CCPR, ACTG Community Scientific Subcommittee Representative, Case Western University Clinical Research Site, North Royalton, OH, USA
Morgan Gapara, MPH, International Site Specialist, ACTG Network Coordinating Center, Social & Scientific Systems, a DLH Company, Durham, NC, USA
Akbar Shahkolahi, PhD, International Site Specialist, ACTG Network Coordinating Center, Social & Scientific Systems, a DLH Company, Silver Spring, MD, USA
Paul Klekotka, MD, PhD, Industry Representative, Lilly Research Laboratories, San Diego, CA, USA
Acknowledgments. We thank the study participants, site staff, site investigators, and the entire ACTIV-2/A5401 study team; the AIDS Clinical Trials Group (ACTG), including Lara Hosey, Jhoanna Roa, and Nilam Patel; the ACTG Laboratory Center, including Grace Aldrovandi and William Murtaugh; Frontier Science, including Marlene Cooper, Howard Gutzman, Kevin Knowles, and Rachel Bowman; the Harvard Center for Biostatistics in AIDS Research and ACTG Statistical and Data Analysis Center; the National Institute of Allergy and Infectious Diseases (NIAID), Division of AIDS; Bill Erhardt; the Foundation for the National Institutes of Health (NIH) and the Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) partnership, including Stacey Adams; and the PPD clinical research business of ThermoFisher Scientific.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Financial support. This work was supported by the NIAID/NIH (award numbers UM1AI068634 to M. D. H., UM1AI068636 to J. S. C., and UM1AI106701 to J. Z. L.); the NIH (grant number R01-HL143541); and Los Alamos National Laboratory (Laboratory Directed Research and Development grant numbers 20200743ER and 20200695ER).
Supplement sponsorship. This article appears as part of the supplement “Findings From the ACTIV-2/AIDS Clinical Trials Group A5401 Adaptive Platform Trial of Investigational Therapies for Mild-to-Moderate COVID-19,” sponsored by the National Institutes of Health through a grant to the University of California, Los Angeles.
References
- 1. Carabelli AM, Peacock TP, Thorne LG, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol 2023; 21:162–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Obermeyer F, Jankowiak M, Barkas N, et al. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 2022; 376:1327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramanathan M, Ferguson ID, Miao W, Khavari PA. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis 2021; 21:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat Commun 2022; 13:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahashi K, Ishikane M, Ujiie M, et al. Duration of infectious virus shedding by SARS-CoV-2 Omicron variant-infected vaccinees. Emerg Infect Dis 2022; 28:998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boucau J, Marino C, Regan J, et al. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N Engl J Med 2022; 387:275–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hay JA, Kissler SM, Fauver JR, et al. Quantifying the impact of immune history and variant on SARS-CoV-2 viral kinetics and infection rebound: a retrospective cohort study. Elife 2022; 11:e81849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ke R, Martinez PP, Smith RL, et al. Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Nat Microbiol 2022; 7:640–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chew KW, Moser C, Daar ES, et al. Antiviral and clinical activity of bamlanivimab in a randomized trial of non-hospitalized adults with COVID-19. Nat Commun 2022; 13:4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choudhary MC, Chew KW, Deo R, et al. Emergence of SARS-CoV-2 escape mutations during bamlanivimab therapy in a phase II randomized clinical trial. Nat Microbiol 2022; 7:1906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tzou PL, Tao K, Pond SLK, Shafer RW. Coronavirus resistance database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons. PLoS One 2022; 17:e0261045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rambaut A, Holmes EC, O’Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ivan Aksamentov CR, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
- 14. Deo R, Choudhary MC, Moser C, et al. Symptom and viral rebound in untreated SARS-CoV-2 infection. Ann Intern Med 2023; 176:348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Overton CE, Stage HB, Ahmad S, et al. Using statistics and mathematical modelling to understand infectious disease outbreaks: COVID-19 as an example. Infect Dis Model 2020; 5:409–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ke R, Zitzmann C, Ho DD, Ribeiro RM, Perelson AS. In vivo kinetics of SARS-CoV-2 infection and its relationship with a person’s infectiousness. Proc Natl Acad Sci U S A 2021; 118:e2111477118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kissler SM, Fauver JR, Mack C, et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med 2021; 385:2489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teyssou E, Delagreverie H, Visseaux B, et al. The Delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J Infect 2021; 83:e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Wintersdorff CJH, Dingemans J, van Alphen LB, et al. Infections with the SARS-CoV-2 Delta variant exhibit fourfold increased viral loads in the upper airways compared to Alpha or non-variants of concern. Sci Rep 2022; 12:13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L, Jackson CB, Mou H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 2020; 11:6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishra A, Kumar N, Bhatia S, et al. SARS-CoV-2 Delta variant among Asiatic Lions, India. Emerg Infect Dis 2021; 27:2723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cherian S, Potdar V, Jadhav S, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms 2021; 9:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan KC, Song Y, Xu Z, Shang C, Zhou R. SARS-CoV-2 Delta variant: interplay between individual mutations and their allosteric synergy. Biomolecules 2022; 12:1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moser C, Li JZ, Eron JJ, et al. Predictors of SARS-CoV-2 RNA from nasopharyngeal swabs and concordance with other compartments in nonhospitalized adults with mild to moderate COVID-19. Open Forum Infect Dis 2022; 9:ofac618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi T, Ellingson MK, Wong P, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020; 588:315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood R, Thomson E, Galbraith R, et al. Sharing a household with children and risk of COVID-19: a study of over 300 000 adults living in healthcare worker households in Scotland. Arch Dis Child 2021; 106:1212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kandel CE, Banete A, Taylor M, et al. Similar duration of viral shedding of the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) Delta variant between vaccinated and incompletely vaccinated individuals. Infect Control Hosp Epidemiol 2023; 44:1002–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mlcochova P, Kemp SA, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021; 599:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bramante CT, Proper JL, Boulware DR, et al. Vaccination against SARS-CoV-2 is associated with a lower viral load and likelihood of systemic symptoms. Open Forum Infect Dis 2022; 9:ofac066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berg MG, Zhen W, Lucic D, et al. Development of the RealTime SARS-CoV-2 quantitative laboratory developed test and correlation with viral culture as a measure of infectivity. J Clin Virol 2021; 143:104945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.