Abstract
Drought stress limits woody species productivity and influences tree distribution. However, dissecting the molecular mechanisms that underpin drought responses in forest trees can be challenging due to trait complexity. Here, using a panel of 300 Chinese white poplar (Populus tomentosa) accessions collected from different geographical climatic regions in China, we performed a genome-wide association study (GWAS) on seven drought-related traits and identified PtoWRKY68 as a candidate gene involved in the response to drought stress. A 12-bp insertion and/or deletion and three nonsynonymous variants in the PtoWRKY68 coding sequence categorized natural populations of P. tomentosa into two haplotype groups, PtoWRKY68hap1 and PtoWRKY68hap2. The allelic variation in these two PtoWRKY68 haplotypes conferred differential transcriptional regulatory activities and binding to the promoters of downstream abscisic acid (ABA) efflux and signaling genes. Overexpression of PtoWRKY68hap1 and PtoWRKY68hap2 in Arabidopsis (Arabidopsis thaliana) ameliorated the drought tolerance of two transgenic lines and increased ABA content by 42.7% and 14.3% compared to wild-type plants, respectively. Notably, PtoWRKY68hap1 (associated with drought tolerance) is ubiquitous in accessions in water-deficient environments, whereas the drought-sensitive allele PtoWRKY68hap2 is widely distributed in well-watered regions, consistent with the trends in local precipitation, suggesting that these alleles correspond to geographical adaptation in Populus. Moreover, quantitative trait loci analysis and an electrophoretic mobility shift assay showed that SHORT VEGETATIVE PHASE (PtoSVP.3) positively regulates the expression of PtoWRKY68 under drought stress. We propose a drought tolerance regulatory module in which PtoWRKY68 modulates ABA signaling and accumulation, providing insight into the genetic basis of drought tolerance in trees. Our findings will facilitate molecular breeding to improve the drought tolerance of forest trees.
Allelic variation in transcription factor PtoWRKY68 affects its transcriptional regulatory activity, resulting in different degrees of drought tolerance through the abscisic acid pathway in Populus.
Introduction
Drought stress is one of the most common abiotic stress factors affecting forest tree growth and productivity (Ragauskas et al. 2006). Plant stem hydraulic conductance and aboveground biomass production decrease substantially under drought stress, resulting in up to 45% reduction in radial growth of many forest trees (Barber et al. 2000). To reduce the adverse effects of drought stress on plant growth and development, plants have evolved multifaceted strategies involving morphological, physiological, and biochemical adaptations (Shinozaki et al. 2003; Bohnert et al. 2006; Mukarram et al. 2021; Zhang et al. 2022). These strategies aim to alleviate or mitigate dehydration stress by increasing water uptake or reducing water loss, to protect plant cells from damage when water becomes scarce and tissue dehydration occurs (Verslues et al. 2006; Yu et al. 2013). Forest trees, in general, are often challenged by mild or moderate drought stress, which requires trees to respond physiologically by reducing the rates of transpiration and photosynthesis to abrogate the acute desiccation that leads to death. Hence, investigation of the mechanisms underpinning the physiological and photosynthetic changes in trees under drought stress could enhance drought tolerance in trees and maintain their growth and productivity (McDowell 2011).
The plant hormone abscisic acid (ABA) is a key factor regulating the plant response to drought or water insufficiency, which mediates stomatal closure and maintains water status (Raghavendra et al. 2010; Wang et al. 2021; Yu et al. 2021). The de novo biosynthesis of ABA is induced by drought stress, and genes involved in this process have been identified. For instance, 9-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (AtNCED3) contributes to drought tolerance by increasing ABA accumulation in Arabidopsis (Arabidopsis thaliana) (Tan et al. 2003). In addition to ABA biosynthesis, catabolism and transport also modulate the accumulation of ABA (Chen et al. 2020). For example, the flowering repressor SVP, a central regulator of ABA catabolism, increases the cellular level of active ABA to respond to drought stress via decreased ABA hydroxylation via repression of CYP707A1/3 and increased ABA-GE hydrolyzation via activation of AtBG1 expression in leaves (Wang et al. 2018). A DTX/MULTIDRUG AND TOXIC COMPOUND EXTRUSION (MATE) family member, AtDTX50, negatively regulates drought tolerance by modulating ABA efflux, and a dtx50 mutant showed increased ABA accumulation and accelerated ABA-induced stomatal closure (Zhang et al. 2014). ABA-dependent transcription networks are important in the response to drought stress. In the ABA signaling pathway, PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) proteins as ABA receptors can interact with PROTEIN PHOSPHATASE 2C (PP2C) family members and repress PP2C activity (Ma et al. 2009). The result is an accumulation of active SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2 (SnRK2), which mediates the direct phosphorylation of the ABA INSENSITIVE5 (ABI5) and ABA-RESPONSE ELEMENT BINDING FACTOR (ABF) transcription factors (TFs) (Wasilewska et al. 2008).
As plant-specific TFs, the WRKY family contains the conserved sequence motif WRKYGQK, which binds to the W-box [(T)TGACC/T] in target gene promoters (Eulgem and Somssich 2007). WRKY TFs play a crucial role in the ABA signaling cascade that underlies the plant response to stress (Rushton et al. 2012; Jiang et al. 2021; Lim et al. 2022). In Arabidopsis, AtWRKY46/54/70 negatively modulates drought tolerance by globally repressing drought-inducible gene expression (Chen et al. 2017; Chen and Yin 2017). Jiang et al. (2021) found that PalWRKY77 acts as a repressor of ABA signaling to negatively regulate the salt stress response in poplar (Populus alba var. pyramidalis) by directly binding to the W-boxes in the promoters of NO APICAL MERISTEM, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR AND CUP-SHAPED COTYLEDON 2 (PalNAC002), and RESPONSIVE TO DESICCATION 26 (PalRD26) to inhibit their expression. However, the ABA-dependent drought-responsive pathway in perennial woody plants is complex and unclear. Understanding the mechanisms that underpin trees to cope with drought stress is needed to enhance ecologic and economic value in the context of climate change.
Genetic variation in traits of interest under natural selection, which exist in wild relatives or progenitor species of crop plants, provides vital resources in plant breeding for desirable and complex traits (Han et al. 2018). A genome-wide association study (GWAS) identified several quantitative trait loci (QTLs) and genomic regions harboring single nucleotide polymorphisms/insertion-deletions (SNPs/indels) associated with complex traits. For example, a natural variant of ZmNAC111 was identified in significant association with drought tolerance in maize (Zea mays) seedlings by using GWAS, which governs water-use efficiency under drought stress (Mao et al. 2015). The regulatory hierarchy of complex traits often involves clusters of a few related genes. Although GWAS can provide statistical links from genotypes to phenotypes, it typically cannot uncover functional pathways that encompass multiple related genes (Wang et al. 2010). Omics analyses (e.g. transcriptomics) can bridge the gap between gene and function through genetic regulatory networks which enhances the effectiveness of the GWAS approach (Tang et al. 2021). For example, co-expression networks can be used to infer the functions of genes based on their associations with their network neighbors and integrate transcriptome data to analyze genes that regulate complex traits. Transcriptome analysis, coupled with co-expression network and expression quantitative trait loci (eQTL) mapping, facilitates the integration of potential regulatory networks of complex traits (Langfelder et al. 2011; Majewski and Pastinen 2011; Cubillos et al. 2012; Krouk et al. 2013). The application of these methods singly or in combination provides insight into the underlying genetic regulatory networks for desirable traits in plants (Civelek and Lusis 2014). For instance, the genetic architecture of leaf shape variation and candidate genes associated with leaf development was characterized by GWAS, then co-expression network analysis has been used to prioritize the centrality of GWAS-identified genes, and integrated eQTL enabled the functional characterization of a set of candidate genes in poplar (P. tremula) (Mahler et al. 2020). Tong et al (2022) revealed the transcriptional regulation network of salt-inducible gene PtoRD26 in response to salt stress using co-expression analysis in Chinese white poplar (P. tomentosa Carr. clone 741). Therefore, systematic integration of GWAS, co-expression, and eQTL approaches may facilitate the dissection and rewiring of the gene regulatory hierarchy of complex traits. In this regard, exploiting the causative alleles of complex traits by systems genetics in natural populations provide a rich source of a target for breeding beneficial traits.
Here, we compared drought-related traits of a natural population of P. tomentosa under well-watered and drought conditions and performed a GWAS of the changes in these drought-related traits during drought to mining the candidate genes that were associated with drought tolerance. The most significant locus contains a gene annotated for PtoWRKY68, which featured a 12 bp indel and three nonsynonymous variants in the coding region. These affected its transcriptional regulatory activity and binding to the promoters of the downstream genes PtoABF2.1, PtoRD26.1, and PtoDTX49.1, thereby affecting drought tolerance. Moreover, eQTL mapping identified the MADS-box transcription factor PtoSVP.3 as an upstream regulator of PtoWRKY68 in response to drought stress. Herein we report the identification of a PtoSVP.3-PtoWRKY68-PtoABF2.1/PtoRD26.1/PtoDTX49.1 module that regulates ABA signaling and accumulation to cope with drought stress in Populus.
Results
GWAS for drought tolerance in Populus tomentosa
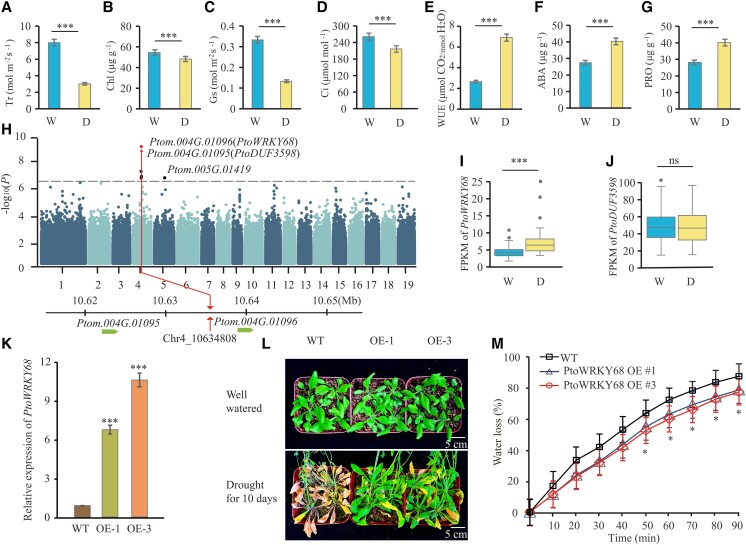
To dissect the genetic architecture of phenotypic variation in drought tolerance, we phenotyped four photosynthetic and three physiological traits in a natural population of P. tomentosa comprising 300 accessions of 1-yr-old trees. Compared to those under well-watered conditions, plants under drought conditions exhibited significantly increased (P < 0.001, t-test) water-use efficiency (WUE; 32.90%), ABA levels (28.41%), and proline level (PRO; 28.46%), and decreased stomatal conductance (Gs; 36.41%), intercellular CO2 concentration (Ci; 27.39%), transpiration rate (Tr; 62.21%), and chlorophyll content (Chl; 13.49%) (Fig. 1). Significant phenotypic variation was identified in these drought-related traits among three climate regions (Supplemental Fig. S1). Accessions from the northwestern (NW) or northeastern (NE) climate regions of China had higher rates of change in these drought-related traits relative to those from the southern (S) region following drought stress (Supplemental Figs. S1 and S2). Based on their high repeatability (h2 > 0.75) and variability among Populus individuals, the percentage reduction or increase in these seven traits during drought was used as drought tolerance indexes.
Figure 1.
GWAS for drought tolerance in a Populus natural population. A to G) Seven drought-related traits under well-watered (W) and drought D) conditions. Transpiration rate (A, Tr), chlorophyll (B, Chl), stomatal conductance (C, Gs), intercellular CO2 concentration (D, Ci), water-use efficiency (E, WUE), F (ABA), and proline (G, PRO) under the two conditions. Each trait was measured in 300 P. tomentosa accessions. Error bars are ± SD; significant differences were determined using a t-test, ***P < 0.001. H) Manhattan plot for GWAS with the percentage increase in stomatal conductance (Gs) under well-watered and drought conditions. Red dashed horizontal line, Bonferroni-adjusted significance threshold (P < 3.3 × 10–7). SNPs in candidate genes, as identified by GWAS, are shown as red dots. I to J) FPKM values of two candidate genes in the P. tomentosa population under well-watered and drought-stress conditions. In box plots, center line represents the median, box limits denote the upper and lower quartiles, whickers indicate the interquartile range, and dots are outliers. Significant differences were determined using a t-test, ***P < 0.001. ns, no significant difference. K) RT-qPCR analysis of PtoWRKY68 transcript level in two independent transgenic lines were relative to wild-type (WT) plants, 35S:PtoWRKY68–1 (OE1) and 35S:PtoWRKY68–3 (OE3). Error bars are ± SD from three biological replicates (n = 3 plants for each replicate); significant differences were determined using a t-test, ***P < 0.001. L) Images of the drought-stress phenotypes of WT, OE-1, and OE-3 plants. Twenty-eight-day-old plants were subjected to drought stress for 10 d. Well-watered represents a 95% relative gravimetric soil water content (rSWC), and drought represents a 17% rSWC. Scale bar, 5 cm. M) Time course of water loss from detached leaves of 28-d-old WT, OE-1, and OE-3 transgenic plants. Water loss is shown as a percentage of the initial fresh weight. Three independent experiments were carried out, each involving three plants. Error bars are ± SD; significant differences were determined using a t-test, *P < 0.05.
Using 3,002,432 SNPs from genome resequencing of a natural population of P. tomentosa, we performed GWAS to identify the genetic loci underlying drought tolerance. At the whole-genome scale, based on the population structure (Q) and kinship (K) of the population, a total number of 22 SNPs were significantly associated with the seven drought-related traits (P < 3.3 × 10−7; 1/n, Bonferroni test; Fig. 1H; Supplemental Fig. S3). These SNPs were annotated into 16 candidate genes distributed on chromosomes 3, 4, and 15, and they were independent of each other based on linkage disequilibrium (LD) analysis (Supplemental Table S1). The most significant SNP, Chr4_10634808 (C/T) (P = 5.7 × 10−10), located 8.0 kb downstream of the termination codon of Ptom.004G.01095 and 1.9 kb upstream of the start codon of Ptom.004G.01096, was significantly associated with Gs (Fig. 1H). Ptom.004G.01095, which harbors the Pfam domain DUF3598 (PtoDUF3598) is a homolog of an Arabidopsis gene annotated as “Biogenesis Factor Required for ATP Synthase 1″ based on phylogenetic analysis (Supplemental Fig. S4A) (Zhang et al. 2018). Ptom.004G.01096 encodes a Group I WRKY protein that has two WRKY domains and a CCHC (represent amino acid sequence, where C is cysteine, H is histidine) zinc finger motif, which homolog in P. trichocarpa had been designated as PtrWRKY68, thus we named it PtoWRKY68 (Jiang et al. 2014). Phylogenetic analysis of Group I WRKY members indicated that Ptom.004G.01096 is a homolog of AtWRKY3, which is involved in the regulation of defense response to pathogen and salt stress (Supplemental Fig. S4B) (Lai et al. 2008; Li et al. 2021). PtoWRKY68 expression was significantly increased in leaves, in contrast to the unaffected PtoDUF3598, upon drought stress (Fig. 1, I and J). Therefore, PtoWRKY68 might involve in drought responses in P. tomentosa.
PtoWRKY68 is a positive regulator of drought tolerance
To assess the involvement of PtoWRKY68 in drought tolerance, we ectopically overexpressed PtoWRKY68 in Arabidopsis, and two independently generated homozygous transgenic lines, OE-1 and OE-3, were selected and subjected to drought stress (Fig. 1K). The transgenic lines were significantly more tolerant to drought stress than Columbia-0 wild-type (WT) plants. Compared to WT, phenotyping showed that the rosette leaves of the two OE transgenic plants had alleviated wilting or the chlorosis phenotype upon exposure to drought stress for 10 d (Fig. 1L). In addition, photosynthetic parameters Gs (43.87%) and Tr (15.16%) were significantly decreased in OE lines compared to WT plants, the Ci (5.41%) and Chl content (10.90%) were also slightly reduced but not obviously in transgenic lines (Supplemental Fig. S5, A to D). However, WUE (26.62%), ABA (42.7%), and PRO (18.33%) in the OE plant leaves were elevated relative to the WT following drought treatment for 10 d (Supplemental Fig. S5, E to G). Therefore, PtoWRKY68 may enhance the drought tolerance of Arabidopsis transgenic plants. Further, the detached leaves of OE lines showed a significantly lower rate of water loss than the WT at multiple time points (Fig. 1M), consistent with the reduction in Gs and the improvements in the phenotypic performance of the OE lines under drought treatment. Taken together, these results implicate PtoWRKY68 in the positive regulation of drought tolerance.
Identification of drought-tolerant/sensitive alleles of PtoWRKY68
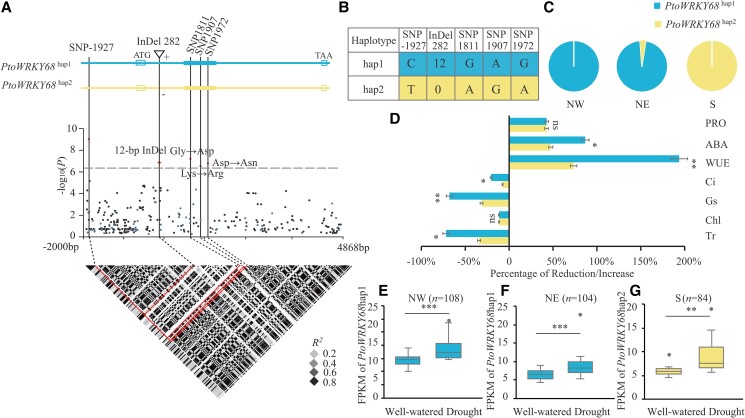
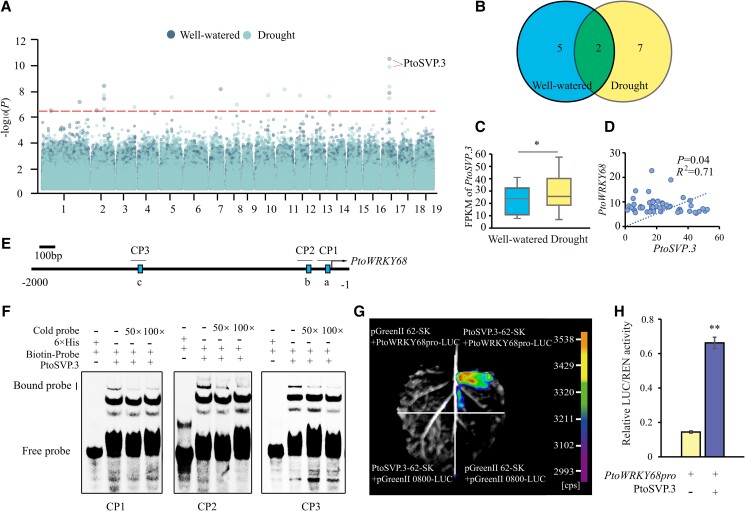
To determine the molecular basis of the natural variation in PtoWRKY68 in P. tomentosa, a 6.8 kb genomic DNA fragment encompassing the entire coding sequence (CDS) of PtoWRKY68 and its 2 kb upstream regulatory region was analyzed in the re-sequenced natural population of 300 P. tomentosa accessions. Totals of 225 SNPs and 75 indels were identified in the population and subjected to sequence variation association analysis (Supplemental Data Set 1). A 12 bp indel upstream of the WRKY domain and three nonsynonymous variants (MAF ≥ 0.05) in the WRKY domain were significantly associated with Gs (P < 3.3 × 10−7; Fig. 2A). These variants were in strong LD (r2 > 0.8) with the peak signal (Chr4_10634808) in the PtoWRKY68 promoter region (Fig. 2A). The 12 bp indel occurs in a stretch of Asn-Thr repeats; PtoWRKY68hap1 and PtoWRKY68hap2 have six and four Asn-Thr repeats, respectively (Supplemental Fig. S6).
Figure 2.
Natural variation in PtoWRKY68 was significantly associated with drought tolerance in Populus tomentosa. A) Association analysis of genetic variation in PtoWRKY68 with the percentage increase in stomatal conductance (Gs) and the pattern of pairwise LD of DNA polymorphisms. Black dots denote SNPs and blue dots represent insertions and/or deletions (indels). A significant SNP, three nonsynonymous variants, and a 12 bp indel are indicated by dots and a triangle above the horizontal dashed line, respectively, and are connected to the pairwise LD diagram by a dashed line (middle). Red lines (bottom) highlight the strong LD of these variations. B) Haplotypes of PtoWRKY68 among Populus natural variations. The PtoWRKY68 haplotype groups were categorized by these five variants. C) The distribution of PtoWRKY68 haplotypes in Populus climate regions. NW, NE, and S represent northwest, northeast, and southern climate regions, respectively. D) Seven drought-related traits of the PtoWRKY68hap1 and PtoWRKY68hap2 alleles. Transpiration rate (Tr), chlorophyll (Chl), stomatal conductance (Gs), intercellular CO2 concentration (Ci), water-use efficiency (WUE), ABA, and proline (PRO) were measured among 300 Populus genotypes under well-watered and drought-stress conditions. The percentage reduction/increase represents the change in measured traits between the two conditions. Error bars are ± SD; significant differences were determined using the t-test, *P < 0.05, **P < 0.01. ns, no significant difference. E to G) FPKM values of PtoWRKY68 in NW E), NE F), and S G) under well-watered and drought-stress conditions. In box plots, center line represents the median, box limits denote the upper and lower quartiles, whickers indicate the interquartile range, and dots are outliers. n denotes the number of genotypes belonging to each haplotype group. Statistical significance was determined using the t-test, **P < 0.01, ***P < 0.001.
The 300 accessions of P. tomentosa were classified into two haplotype groups, PtoWRKY68hap1 (overexpressed in Arabidopsis above) and PtoWRKY68hap2, based on these five significant variants (Fig. 2B). PtoWRKY68hap1 was detected mainly in the accessions from the NE and NW regions whereas PtoWRKY68hap2 occurred mostly in the S region, as revealed by allele frequency investigation (Fig. 2C). We subsequently evaluated whether these polymorphisms of PtoWRKY68 alter the drought tolerance of natural P. tomentosa varieties. The accessions with PtoWRKY68hap1 were significantly more responsive to drought than those with PtoWRKY68hap2, in terms of changes in WUE, ABA, PRO, and photosynthetic parameters (P < 0.05) (Fig. 2D). PtoWRKY68hap1 and PtoWRKY68hap2 were significantly upregulated by drought stress in the accessions from the three climate regions, and the increase in the transcript level of PtoWRKY68hap1 in the NW (n = 108, P = 8.69 × 10−4) and NE (n = 104, P = 8.65 × 10−4) groups was comparable with PtoWRKY68hap2 in the S group (n = 84, P = 9.49 × 10−3) (Fig. 2, E to G). These results suggested that the difference in drought responses between the two haplogroups is not due to the difference expression level of PtoWRKY68 alleles.
To explore whether allelic variations in PtoWRKY68 confer drought tolerance, we overexpressed PtoWRKY68hap2 in Arabidopsis (Supplemental Fig. S7A). In a drought stress assay, transgenic plants carrying PtoWRKY68hap2 (PtoWRKY68hap2OE) displayed more drought tolerance than WT plants but an enhanced wilting phenotype compared to transgenic plants carrying PtoWRKY68hap1 (PtoWRKY68hap1OE) after 10-d drought stress (Supplemental Fig. S7B). Regarding photosynthetic and physiological traits, PtoWRKY68hap1OE plants had higher WUE, ABA, and PRO values but lower Gs and Tr values compared to WT and PtoWRKY68hap2OE plants under drought stress (Supplemental Fig. S7, C to I). In addition, PtoWRKY68hap2OE plants exhibited faster water loss than PtoWRKY68hap1OE plants under drought stress (Supplemental Fig. S7J). These results suggest that PtoWRKY68hap1 improves drought tolerance and that this improvement is associated with the allelic variation. Moreover, we identified Asn-Thr repeats and nonsynonymous variants in the PtoWRKY68 homolog PtrWRKY68 (P. trichocarpa) and PagWRKY68 (P. alba × P. glandulosa, 84 K), with similar haplotypes to PtoWRKY68hap1 or PtoWRKY68hap2 (Supplemental Fig. S8). Therefore, the different responses to drought stress in Populus might be attributable to allelic variation in PtoWRKY68, with PtoWRKY68hap1 and PtoWRKY68hap2 designated as the drought-tolerant and drought-sensitive alleles, respectively.
Identification of the target genes of PtoWRKY68 for regulation of drought tolerance
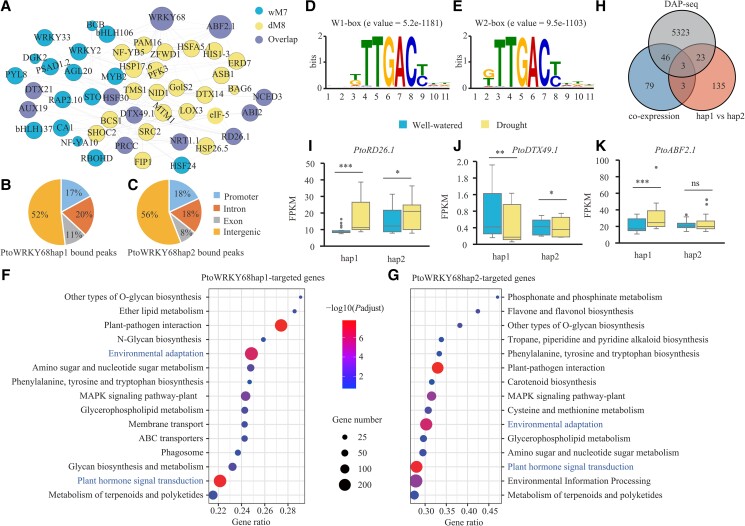
To investigate the potential gene interaction networks of PtoWRKY68 in response to drought stress, we constructed co-expression networks for the differentially expressed genes (DEGs) between well-watered and drought-stress conditions. The well-watered and drought stress networks encompassed 17 and 23 modules (Supplemental Fig. S9, A and B), respectively, suggesting rearrangement of the co-expression network in response to drought. We analyzed the correlation between the module eigengenes with the phenotypic values of each accession. The well-watered network module 7 (wM7) and the drought-stressed module 8 (dM8), containing PtoWRKY68, were negatively correlated with Gs, Ci, Tr, and Chl but positively correlated with WUE, ABA, and PRO (Supplemental Fig. S9, A and B). To prioritize causal genes, we focused on genes in these two modules with high gene significance (absolute value >0.80) measured using drought-related traits and high module membership (absolute value >0.20) with the module eigengenes (Supplemental Fig. S9C; Supplemental Data Sets S2 and S3) (Liu et al. 2019). Gene annotation analysis of well-watered and drought-stress networks led to the identification of 131 conserved hub genes with high connectivity (Supplemental Table S2). Among these conserved hub genes, 11 overlap genes were found in wM7 and dM8 (Fig. 3A), which are involved in environmental information processing and signal transduction, such as ABC transporter, respiratory burst oxidase homolog protein, and ABA-responsive element binding factor.
Figure 3.
Identification of genome-wide direct targets of PtoWRKY68hap1 and PtoWRKY68hap2. A) Genes identified in the well-watered network module 7 (wM7), the drought-stressed module 8 (dM8) (containing PtoWRKY68) and overlapping genes under two conditions (Overlap) with putative Arabidopsis orthologs are shown in network view. Node size indicates module membership. The larger nodes are highly connected within a module. B and C) Whole-genome distribution of bound genes by PtoWRKY68hap1B) and PtoWRKY68hap2C) obtained by DAP-seq with P < 0.05. Promoter regions were defined as the binding peaks within 2 kb upstream of ATG. D and E) Binding motifs of PtoWRKY68hap1 (D, W1-box) and PtoWRKY68hap2 (E, W2-box) protein by MEME-ChIP. F and G) The top KEGG-enriched terms of PtoWRKY68hap1F) and PtoWRKY68hap2G) bound genes by DAP-seq. P-value by Fisher's test. H) Venn diagram showing that three overlapping genes were considered PtoWRKY68 alleles direct genes, were co-expressed with PtoWRKY68, were differentially expressed in PtoWRKY68hap1 vs. PtoWRKY68hap2, and were directly bound by PtoWRKY68 alleles. I to K) FPKM values of PtoRD26.1I), PtoDTX49.1J), and PtoABF2.1K) in hap1 and hap2 haplogroup under well-watered and drought-stress conditions. In box plots, center line represents the median, box limits denote the upper and lower quartiles, whickers indicate the interquartile range, and dots are outliers. Significant differences were determined using the t-test, *P < 0.05, **P < 0.01, ***P < 0.001. ns, no significant difference.
To define the potential transcriptional targets of PtoWRKY68hap1 or PtoWRKY68hap2 in response to drought stress, we analyzed DEGs between the PtoWRKY68hap1 and PtoWRKY68hap2 groups under drought stress. In all, 164 genes were differentially expressed in two haplogroups based on a significant difference (P < 0.05) and an at least twofold change (absolute log2 fold change >1, Supplemental Data Set 4), suggesting that their expression is affected by allelic variation in PtoWRKY68. To identify direct targets of PtoWRKY68hap1 and PtoWRKY68hap2, DNA affinity purification sequencing (DAP-seq) was employed (Bartlett et al. 2017). A total of 58,960 and 36,899 notable peaks were identified in genomic regions of PtoWRKY68hap1 or PtoWRKY68hap2, respectively. Among these peaks, 17% and 18% were located within the promoters (2 kb regions upstream of ATG), respectively (Supplemental Fig. S3, B and C). Using the MEME suite, the core sequence and the W-box motif of (T/G)TTGAC(C/T) (W1-box, e-value = 5.2e−1181) and (T/G)TTGAC(C/T) (W2-box, e-value = 9.5e−1103) were substantially enriched among the PtoWRKY68hap1 and PtoWRKY68hap2 binding regions, respectively (Supplemental Fig. S3, D and E). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that PtoWRKY68 target genes were primarily enriched in defense response and phytohormone signaling pathway (Fig. 3, F and G).
Based on the results of co-expression, DEG, and DAP-seq analyses, we prioritized three genes co-expressed with PtoWRKY68, differentially expressed in PtoWRKY68hap1 vs. PtoWRKY68hap2, and directly bound by PtoWRKY68 alleles (Fig. 3H). Ptom.014G.00172 encoding a basic leucine zipper [bZIP] transcription factor, which homolog in Arabidopsis was AtABF2 and thus named PtoABF2.1 (Supplemental Fig. S10). Ptom.011G.00876 encoded an NAC domain-containing protein (PtoRD26.1), which homolog in Arabidopsis, AtRD26 (or else named NAC072), regulates ABA-dependent stress-response signaling (Supplemental Fig. S11) (Fujita et al. 2004). Ptom.003G.00934 encoded the MATE family protein DETOXIFICATION, which was named PtoDTX49.1 based on phylogenetic analysis (Supplemental Fig. S12). A homolog in Arabidopsis, AtDTX50, functions as an ABA efflux transporter and negatively regulates the drought response (Zhang et al. 2014). Under drought stress in Populus, the FPKM values of PtoABF2.1 and PtoRD26.1 showed significantly greater upregulation in the PtoWRKY68hap1 group than in the PtoWRKY68hap2 group under drought stress (Supplemental Fig. S3, I and J), whereas PtoDTX49.1 expression was lower in the PtoWRKY68hap1 group than in the PtoWRKY68hap2 group (P < 0.05) (Fig. 3K). These data suggest PtoWRKY68-mediated transcriptional regulation of PtoDTX49.1, PtoABF2.1, and PtoRD26.1 in response to drought stress.
Allelic variation in PtoWRKY68 affects its binding affinity to downstream target genes
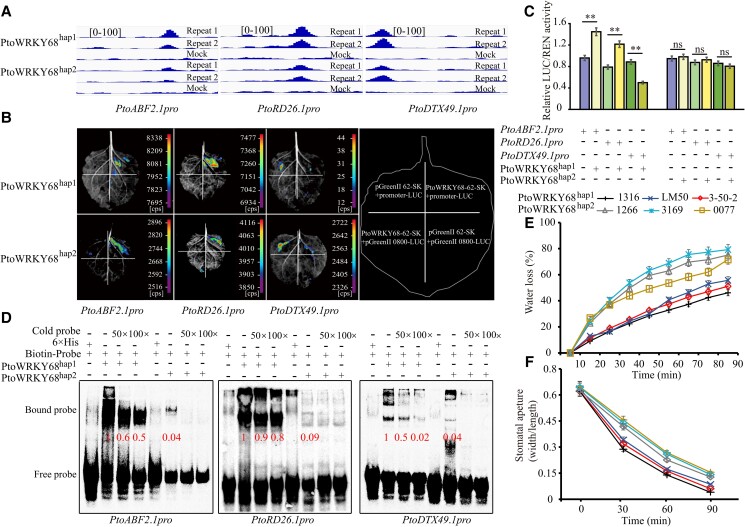
To investigate whether the variation in PtoWRKY68 alleles affects its binding affinity to downstream targets, we analyzed the DAP-seq data. The binding peaks of PtoWRKY68hap1 were larger than those of PtoWRKY68hap2 (located in the same region of downstream promoters) (Fig. 4A). Next, we evaluated the interaction between PtoWRKY68 and downstream genes using a dual-luciferase reporter assay (DLRA). Luciferase (LUC) luminescence was observed when PtoABF2.1 and PtoRD26.1 pro-LUC were each transformed into Nicotiana benthamiana leaves, and it was significantly enhanced by co-transforming PtoWRKY68hap1 compared to co-transforming PtoWRKY68hap2 (Fig. 4, B and C). By contrast, the LUC activity from the PtoDTX49.1 promoters decreased substantially when the reporter was co-transfected with PtoWRKY68hap1 compared to co-transforming PtoWRKY68hap2 (Fig. 4, B and C). Electrophoretic mobility shift assays (EMSAs) were performed to investigate the effects of allelic variation on PtoWRKY68 binding activity. Biotin labeling showed that both PtoWRKY68hap1 and PtoWRKY68hap2 bound to W-box probes derived from the PtoABF2.1, PtoDTX49.1, and PtoRD26.1 promoters, and the former generated a stronger signal (Fig. 4D), suggestive of higher affinity for target gene promoters. Taken together, these results indicate that PtoWRKY68hap1 and PtoWRKY68hap2 activate or repress downstream genes by binding to their promoters and that natural variants in PtoWRK58 are responsible for the differential binding ability of PtoWRKY68 to the promoters of three downstream genes. Reverse transcription quantitative PCR (RT-qPCR) analysis showed that compared to WT and PtoWRKY68hap2OE plants, the expression of AtABF2 and AtRD26 were significantly elevated in PtoWRKY68hap1OE plants after drought stress (Supplemental Fig. S13, A and B), whereas the transcript level of AtDTX50 was markedly reduced in PtoWRKY68hap1OE plants (Supplemental Fig. S13C). The expression levels of several ABA- or drought-inducible genes—including AtNCED3 (Iuchi et al. 2001; Tan et al. 2003), AtRD20 (Aubert et al. 2010), AtRD29B (Nakashima et al. 2006), LINKER HISTONE H1–3 (AtHIS1–3) (Ascenzi and Gantt 1997), AtABI2 (Ma et al. 2009), and AtSnRK2.6 (Zhu et al. 2020)—were enhanced in PtoWRKY68hap1OE plants compared to PtoWRKY68hap2OE plants and WT plants under drought conditions (Supplemental Fig. S13D). Most of these genes were co-expressed with PtoWRKY68 (Fig. 3A; Supplemental Table S2), suggesting them to be downstream targets in the PtoWRKY68-dependent drought-responsive pathway. These up-/downregulations of ABA- or drought-inducible genes were consistent with the result that the drought tolerance of PtoWRKY68hap1OE plants was significantly elevated compared to WT and PtoWRKY68hap2OE plants (Supplemental Fig. S13D). ABA-sensitivity assays showed that PtoWRKY68 is involved in the ABA signaling pathway. On 1/2 MS medium without ABA, the two transgenic lines germinated without discernible morphological changes compared to WT plants in a time-course analysis (Supplemental Fig. S13, E and F). On 1/2 MS medium containing ABA (0.5 and 1 μM), the PtoWRKY68hap1OE and PtoWRKY68hap2OE lines showed a lower germination rate compared to WT plants at both ABA concentrations (Supplemental Fig. S13, G and H). When 5-d-old seedlings were transferred onto 1/2 MS medium supplemented with 1 or 3 μM ABA, PtoWRKY68hap1OE plants displayed shorter primary roots than WT and PtoWRKY68hap2OE plants. Moreover, stomatal aperture assays showed that stomata closed more rapidly in PtoWRKY68hap1OE than in WT and PtoWRKY68hap2OE under ABA treatment (Supplemental Fig. S13I), implicating PtoWRKY68 alleles were functioned in ABA-dependent stomatal closure. These observations suggest that overexpression of PtoWRKY68hap1 substantially enhanced drought tolerance, which is mediated by amplified drought-stress signaling and an increased cellular ABA level as a result of increased ABA signaling and decreased ABA efflux in leaves.
Figure 4.
PtoWRKY68hap1 and PtoWRKY68hap2 directly bind the promoters of PtoABF2.1, PtoRD26.1, and PtoDTX49.1 to activate their transcription. A) Binding peaks (repeats 1 and 2) and negative control (mock) of PtoWRKY68hap1 and PtoWRKY68hap2 in PtoABF2.1, PtoRD26.1, and PtoDTX49.1 by DAP-seq. The [0 to 100] shows the scale bar for binding peak heights. B) DLRA of the interaction between PtoWRKY68 and the promoters of ABA-related genes. A schematic of transient co-expression of effectors and reporters is at the right; an image of luciferase activity is at the left. pGREEN 62-SK + pGREEN 0800-LUC indicates transient co-expression of empty effector (without PtoWRKY68) and empty reporter (without promoter); PtoWRKY68-62-SK + pGREEN 0800-LUC indicates transient co-expression of effector and empty reporter; pGREEN 62-SK + promoter-LUC indicates transient co-expression of empty effector and reporter; and promoter-LUC + PtoWRKY68 62-SK indicates co-expression of effector and reporter. C) Relative luciferase activity according to a DLRA assay of N. benthamiana leaves. Quantification was performed by normalizing firefly luciferase (LUC) activity to that of Renilla luciferase (REN), 35S:REN was used as the internal control. Relative luciferase activities using PtoWRKY68-pGreenII62-SK as the effector were compared to the control effector (pGreenII62-SK empty vector). Error bars are ± SD. Statistical analysis was performed using the t-test (n = 8) and statistically significant differences are indicated by *P < 0.05, **P < 0.01. ns, no significant difference. Three independent transfection experiments were performed. D) EMSA. The probe sequence was isolated from three downstream genes and consisted of a W1-box or W2-box motif. “+” and “−” indicate presence or absence of reagents in the lane during protein electrophoresis. His-labeled probe with PtoWRKY68hap1 and PtoWRKY68hap2 proteins are shown. The bound probe indicates binding affinity between the PtoWRKY68hap1/PtoWRKY68hap2 alleles and the promoters of downstream genes. Protein concentration was 2 µg/µL. Reciprocal competitive EMSA to evaluate the binding of recombinant PtoWRKY68hap1/PtoWRKY68hap2 protein to the W-box motifs using the indicated biotin-labeled probes and unlabeled competitors. For each probe, 50× and 100× excess competitor was added. E) Time course of water loss from detached leaves of three 6-week-old PtoWRKY68hap1P. tomentosa accessions (1316, LM50, and 3–50–2) and three PtoWRKY68hap2P. tomentosa accessions (1266, 3169, and 0077). Water loss is shown as a percentage of the initial fresh weight. Three independent experiments were carried out (n = 3 plants for each experiment). Error bars are ± SD. F) Stomatal aperture of six P. tomentosa accessions (as in E) in the presence of 1 μM ABA. Epidermal strips were peeled and photographs were taken under a microscope. Three independent experiments were carried out (n = 60 guard cells for each experiment). Error bars are ± SD.
To confirm the association between allelic variation in PtoWRKY68 and drought tolerance in an ecological context, we selected six Populus tomentosa accessions: three with PtoWRKY68hap1 and three with PtoWRKY68hap2. We measured the water loss rate and stomatal movement in response to ABA in detached leaves from these two haplogroups. As shown in Fig. 4, E and F, accessions with PtoWRKY68hap1 had a slower water loss and faster stomatal closure. Accessions with PtoWRKY68hap2 had a faster water loss and slower stomatal closure, indicating lower drought stress tolerance. Collectively, PtoWRKY68hap1 plays an important role in the modulation of ABA efflux and ABA signaling in to response drought stress.
PtoSVP.3 regulates PtoWRKY68 response to drought stress and ABA signaling
To explore the regulatory pathway of PtoWRKY68, we conducted eQTL analysis using the expression of PtoWRKY68 as a parameter to analyze the upstream regulatory hierarchy under well-watered and drought-stress conditions. Eight eQTLs were identified under well-watered conditions and 12 were identified under drought-stress conditions, which were annotated to five and seven candidate genes, respectively (Fig. 5, A and B; Table 1). Notably, Chr17_7792185 was significantly associated with PtoWRKY68 expression under both well-watered (P = 1.38 × 10−11) and drought stress (P = 1.41 × 10−6) conditions. This SNP is in the exon region of Ptom.017G.00699, a MADS-box transcription factor, named PtoSVP.3 based on phylogenetic analysis (Supplemental Fig. S14A). A homolog of the Arabidopsis, AtSVP, positively regulates ABA accumulation and ABA levels increase during the response to drought stress (Wang et al. 2018). The transcription of PtoSVP.3 was induced by drought stress, and its FPKM was positively correlated with that of PtoWRKY68 (Fig. 5, C and D), suggesting that PtoSVP.3 as a candidate for regulation of PtoWRKY68.
Figure 5.
eQTL analysis of PtoWRKY68 expression under well-watered and drought-stress conditions. A) Manhattan plots of eQTL analysis with expression of PtoWRKY68 under well-watered and drought-stress conditions. Horizontal dashed line, significance threshold (P < 3.3 × 10–7). Arrowhead, candidate gene. B) Venn diagram showing the overlap of well-watered and drought-stress genes identified by eQTL analysis. C) FPKM values of PtoSVP.3 under well-watered and drought-stress conditions. In box plots, center line represents the median, box limits denote the upper and lower quartiles, whickers indicate the interquartile range, and dots are outliers. Error bars are ± SD. Statistical analysis was performed using the t-test, *P < 0.05. D) Scatter diagram of the correlation between the expression of PtoSVP.3 and PtoWRKY68 under drought stress. Dashed line represents linear regression. E) Schematic of the locations of CArG motifs in the promoter of PtoWRKY68. Solid squares, CArG motifs; short black lines, CArG Probe 1 (CP1); CP2 and CP3 indicate the probes used in F). F) EMSA. Relative binding affinity of SVP.3 to CArG motifs. Reciprocal competitive EMSA of the binding of recombinant SVP.3 protein to CArG motifs using the indicated biotin- labeled probes and unlabeled competitors. For each probe, 50× and 100× excess competitor was added. G) DLRA of the interaction between PtoSVP.3 and the PtoWRKY68 promoter. H) Relative luciferase activity by DLRA in N. benthamiana leaves. Quantification was performed by normalizing firefly luciferase (LUC) activity to that of Renilla luciferase (REN), 35S:REN was used as the internal control. Relative luciferase activities using PtoSVP.3-pGreenII62-SK as the effector compared to the control effector (pGreenII62-SK empty vector). Error bars are ± SD. Statistical analysis was performed using the t-test (n = 8); statistically significant differences are indicated by **P < 0.01. Three independent transfection experiments were performed.
Table 1.
Details of significant SNPs associated with expression of PtoWRKY68 in the association population of Populus tomentosa
| Traits | Associated SNP | P-value | Allele | Position | Gene annotation | Description |
|---|---|---|---|---|---|---|
| Well-watered | Chr1_9411429 | 2.85E−07 | A/T | — | — | — |
| Well-watered | Chr2_13185574 | 2.97E−08 | G/A | Genebody | Ptom.002G.01839 | G-box binding factor 3 (GBF3) |
| Well-watered | Chr2_13186727 | 2.52E−09 | C/T | Genebody | Ptom.002G.01839 | G-box binding factor 3 (GBF3) |
| Well-watered | Chr17_7792185 | 1.38E−11 | T/A | Genebody | Ptom.017G.00699 | MADS-box protein SVP-like (SVP.3) |
| Well-watered | Chr17_7792203 | 9.78E−09 | T/C | Genebody | Ptom.017G.00699 | MADS-box protein SVP-like (SVP.3) |
| Well-watered | Chr7_11563877 | 4.73E−09 | T/C | Genebody | Ptom.007G.01183 | Formin homology 2 domain-containing protein (FH2) |
| Well-watered | Chr2_5270080 | 2.80E−07 | C/T | Genebody | Ptom.002G.00819 | STAY-GREEN-like protein (SGRL) |
| Well-watered | Chr1_40730777 | 5.51E−08 | A/C | Genebody | Ptom.001G.03606 | Solanesyl diphosphate synthase (SPS) |
| Drought | Chr2_13186528 | 1.53E−08 | C/A | Genebody | Ptom.002G.01839 | G-box binding factor 3 (GBF3) |
| Drought | Chr2_13187307 | 2.22E−07 | G/C | Genebody | Ptom.002G.01839 | G-box binding factor 3 (GBF3) |
| Drought | Chr3_18942646 | 1.46E−07 | C/A | Genebody | Ptom.003G.01892 | Large subunit ribosomal protein (RP1) |
| Drought | Chr5_12016609 | 2.23E−08 | C/T | — | — | — |
| Drought | Chr8_11395818 | 9.20E−08 | G/A | — | — | — |
| Drought | Chr10_13813433 | 1.52E−08 | A/G | Genebody | Ptom.010G.01912 | Cyclopropane-fatty-acyl-phospholipid synthase |
| Drought | Chr11_8137577 | 4.93E−09 | A/T | Genebody | Ptom.011G.00545 | Interleukin-1 receptor-associated kinase 4 |
| Drought | Chr12_4257409 | 1.60E−08 | C/T | Genebody | Ptom.012G.00356 | Vesicle-mediated transport |
| Drought | Chr14_1273549 | 1.90E−08 | C/T | Genebody | Ptom.014G.00168 | Basic helix-loop-helix (bHLH) family protein |
| Drought | Chr17_7793142 | 6.56E−11 | T/A | Genebody | Ptom.017G.00699 | MADS-box protein SVP-like (SVP.3) |
| Drought | Chr17_7785531 | 2.76E−08 | A/G | Genebody | Ptom.017G.00699 | MADS-box protein SVP-like (SVP.3) |
| Drought | Chr17_7792192 | 2.88E−09 | C/T | Genebody | Ptom.017G.00699 | MADS-box protein SVP-like (SVP.3) |
Previous study has reported that AtSVP positively regulates the drought response by directly binding to the CArG motif of a downstream promoter in Arabidopsis (Wang et al. 2018). To assess whether PtoWRKY68 is a direct target of PtoSVP.3, we analyzed a 2 kb fragment of the upstream regulatory sequence of PtoWRKY68 and identified three CArG motifs (Fig. 5E). Reciprocal competitive EMSA showed strong and specific binding of PtoSVP.3 to the CArG motifs in the promoter regions of PtoWRKY68 (Fig. 5F). The in vivo physical interaction between PtoSVP.3 and these CArG motifs was analyzed by DLRA. The negative controls were recombined without the PtoSVP.3 or PtoWRKY68 promoter. Luciferase luminescence was observed when PtoWRKY68 pro-LUC and PtoSVP.3 were co-transformed into N. benthamiana leaves, which showed higher luminescence than the negative controls (Fig. 5, G and H). DLRA confirmed that PtoSVP.3 activates the PtoWRKY68 promoter in N. benthamiana leaves. To determine whether SVP regulates the expression of PtoWRKY68 homolog AtWRKY3 in Arabidopsis, we obtained the Atsvp T-DNA insertion mutant from the Arabidopsis Biological Resource Center (ABRC) (Supplemental Fig. S14, B and C), and examined its AtWRKY3 expression. RT-qPCR analysis indicated that the expression level of AtWRKY3 was significantly lower in svp mutant than WT plants (Supplemental Fig. S14D), suggesting the transcript level of AtWRKY3 in svp mutant was markedly inhibited. Taken together, our results show that PtoSVP.3 enhances the expression of PtoWRKY68 under drought stress by binding to CArG motifs in its promoter, thereby positively regulating drought tolerance in a manner involving the ABA signaling pathway rather than ABA catabolism.
Discussion
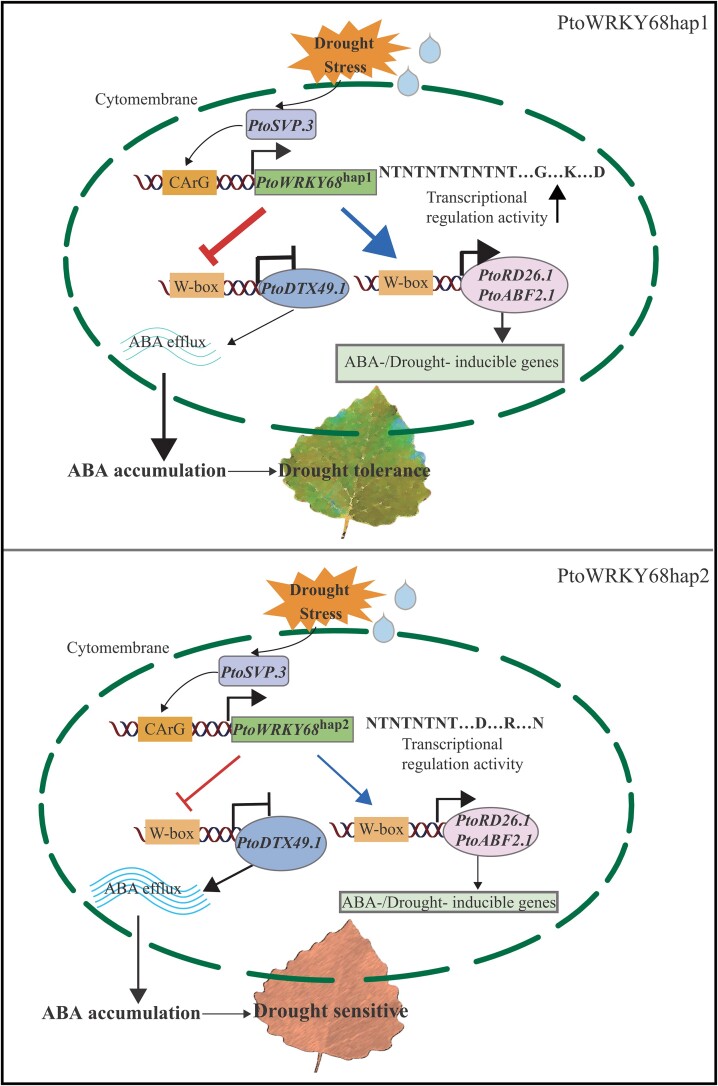
In light of the outcomes of the system genetics strategy, we propose an ABA-dependent drought-responsive network of PtoWRKY68 alleles that are positively regulated by PtoSVP.3 in respond to drought stress. The allelic variation in the CDS region of PtoWRKY68 differentially activated PtoRD26.1 and PtoABF2.1 and repressed PtoDTX49.1 to confer drought tolerance by modulating ABA signaling and accumulation in Populus (Fig. 6).
Figure 6.
Proposed molecular model of the regulation of the Populus drought tolerance regulatory module. Under drought stress, PtoWRKY68 alleles are positively regulated by PtoSVP.3, and the allelic variation in the CDS region of PtoWRKY68hap1 (above) enhances its binding and activation of PtoRD26.1 and PtoABF2.1 and represses PtoDTX49.1 to confer drought tolerance by modulating ABA efflux and signaling transduction in Populus tomentosa. PtoWRKY68hap2 (below) has lower binding affinity and activation of downstream targets than PtoWRKY68hap1. Thus, PtoWRKY68hap1 accessions show better drought tolerance than PtoWRKY68hap2 accessions of Populus. The arrows indicate promotion, the blunt indicate inhibition. The bold arrow indicates the significantly enhanced expression levels of PtoRD26.1 and PtoABF2.1 and the bold blunt indicates the significantly repressed expression levels of PtoDTX49.1.
The PtoSVP.3-PtoWRKY68-PtoDTX49.1/PtoABF2.1/PtoRD26.1 cascade forms a drought-response regulatory network
AtWRKY3 in Arabidopsis is induced by pathogen infection or salicylic acid treatment (Lai et al. 2008) but its role in drought tolerance is unclear. Here we report that PtoWRKY68 is a homolog of AtWRKY3 which functions as an activator of PtoRD26.1 and PtoABF2.1 but as a repressor of PtoDTX49.1 under drought stress. PtoDTX49.1 is a MATE family member which is a homolog of AtDTX50 in Arabidopsis, participates in ABA efflux and thus negatively regulates drought tolerance. However, how AtDTX50 and its homologs in other plants are involved in the regulatory cascade of the ABA pathway to response to drought was unclear. We verified the interaction between PtoWRKY68 and the promoter of PtoDTX49.1, and functionally characterized the role of PtoWRKY68 in modulation of PtoDTX49.1 expression and ABA accumulation for improving drought tolerance.
The ABFs are core components of the canonical PYR/PYL/RCAR-PP2Cs-SnRK2s-ABFs/ABI5 module of the ABA signaling pathway, and a variety of stress-responsive genes are regulated by ABF2 in various plants. For example, overexpression of AtABF2 in Arabidopsis activated the expression of the ABA-inducible gene AtHIS1–3 (Fujita et al. 2005). In this study, PtoABF2.1, a homolog of AtABF2, which was specifically bound and activated by PtoWRKY68, implicating PtoABF2.1 involve in PtoWRKY68-mediated ABA signaling transduction in response to drought stress. In Arabidopsis, AtRD26 is involved in ABA-dependent stress signaling, and the ABA signaling transduction gene AtRD20 is upregulated by overexpression of AtRD26 (Fujita et al. 2004). A homolog in poplar (P. tomentosa Carr. clone 741), PtoRD26.1, is induced by SRMT and the PtoNF-YC9 complex to respond to salt stress (Tong et al. 2022). The co-expression network of PtoRD26 in P. tomentosa Carr. clone 741 demonstrated that the homologs of PtoWRKY68 (P.x_tomentosa21811), PtoRD26.1 (P.x_tomentosa47952), and PtoABF2.1 (P.x_tomentosa01515) have similar patterns of expression and are involved in drought, salt stress, and the ABA pathway (Tong et al. 2022). We identified and validated the PtoWRKY68-PtoRD26.1/PtoABF2.1 regulatory pathway in our constructed co-expression network via systems genetics. Therefore, we illustrated the drought-stress-responsive mechanisms, which are composed of PtoWRKY68, PtoABF2.1, PtoRD26.1, and PtoDTX49.1 together provoke and amplify the ABA-dependent drought response in P. tomentosa.
SVP is a temperature-dependent transcriptional repressor of flowering (Andres et al. 2014; Sureshkumar et al. 2016). In addition, it confers drought tolerance by regulating ABA catabolism rather than ABA signaling in Arabidopsis (Bechtold et al. 2016; Wang et al. 2018). Our findings suggest a molecular mechanism by which a homolog of AtSVP, PtoSVP.3, indirectly regulates ABA signaling by interacting with the promoter of PtoWRKY68 in P. tomentosa. These data suggest that PtoWRKY68 is a master regulator in the PtoSVP.3-PtoWRKY68-PtoDTX49.1/PtoABF2.1/PtoRD26.1 cascade regulatory network of the ABA-dependent drought-responsive pathway.
Allelic variation in PtoWRKY68 affects drought tolerance by modulating ABA signaling
The identification of functional SNPs or indels in genes coupled with analyses of their phenotypic impacts has recently been used as an effective approach to elucidate gene function for genetic improvement (Mao et al. 2015; Wang et al. 2016; Han et al. 2018). In this study, a 12 bp indel and three nonsynonymous variants in PtoWRKY68 were found to indirectly regulate ABA signaling and accumulation by affecting its efficacy in regulating downstream genes. The data are in line with the fact that drought tolerance is a complex trait controlled by multiple QTLs. These variants divided a natural population of 300 P. tomentosa accessions into two haplotype groups representing drought-tolerant (PtoWRKY68hap1) and drought-sensitive (PtoWRKY68hap2) alleles. Conceivably, the natural distribution of P. tomentosa, in terms of water availability, broadly reflects its ability to tolerate drought stress. Populations in water-scarce locations (NE and NW climate regions) are more frequently exposed to drought stress than those in well-hydrated environments (S climate region) (Supplemental Fig. S2). Hence, the drought tolerance conferred by PtoWRKY68hap1 is likely to be an adaptative mechanism formed by long-term functional differentiation and natural selection.
Similar allelic variation exists in homologs in other Populus species (e.g. PtrWRKY68 and PagWRKY68) but the number of Asn-Thr repeats varies by species (Supplemental Fig. S8). These data suggest that the number of Asn-Thr repeats is related to drought tolerance in Populus species; however, the drought response mechanisms of different species are diverse and complex and warrant further investigation. Stress-induced phosphorylation of TFs occurs mainly at Ser/Thr sites (Zhao et al. 2021). For example, SnRK2 kinases phosphorylate Ser/Thr residues in an ABA-dependent manner (Nakashima et al. 2009). The question arises as to whether the Thr site of the Asn-Thr repeat unit is a phosphorylation site in PtoWRKY68 under drought stress.
A potential regulatory pathway and a set of loci implicated in the response to drought stress
Sessile plants have multifaceted strategies to prevent water loss, maintain cellular homeostasis, and persevere through periods of drought. Among seven drought-related physiological and photosynthetic traits, Gs, Ci, Tr, and Chl were reduced by drought stress, indicating decreased photosynthesis efficiency and transpiration to prevent water loss (Xiong et al. 2002). ABA, PRO, and WUE were increased by drought stress, mitigating plant cellular damage and ensuring water supply for plants growth (Pinheiro and Chaves 2011). These drought-related measurements showed substantial variability and a high level of repeatability (Fig. 1), suggesting that these physiological changes have utility as indices of Populus drought tolerance.
Numerous genetic factors have been implicated in drought responses and to a lesser extent in drought tolerance, which is a multigenic trait governed by complex regulatory pathways. System genetics has been used to characterize the genetic mechanisms underpinning key regulators of complex traits. In this study, to understand the biological functions of causal genes that are identified by GWAS, we performed co-expression network analysis under well-watered and drought-stress conditions revealed that both well-watered and drought networks contained PtoWRKY68, which was significantly correlated with drought-related traits. By integration of co-expression, DAP-seq and RNA-seq approaches to identify the direct transcriptional targets of PtoWRKY68, we found evidence that PtoWRKY68 plays a prominent role in modulating ABA efflux and multiple ABA signaling core components, including PtoDTX49.1, PtoABF2.1, and PtoRD26.1. These results suggest that PtoWRKY68 is a key regulator of drought tolerance in Populus and may function in an ABA regulatory pathway. The sequence variation in the PtoWRKY68 CDS region influences its binding ability to these downstream genes, suggestive of an imperative role of co-expression networks in characterizing causal genes. Hence, regulatory network analysis facilitates the explanatory power of GWAS by leveraging prior biological information on gene function (Wang et al. 2010). eQTLs affect complex traits by regulating gene expression and allowing the construction of regulatory networks (Porcu et al. 2019). Therefore, incorporating eQTL information into GWAS analyses will unleash the potential of increasing the power of GWAS by identifying pathways associated with complex traits (Wu and Pan 2018). eQTL analysis showed that the expression of PtoWRKY68 was regulated by PtoSVP.3 under both well-watered and drought conditions, highlighting the ability of eQTLs to evaluate the regulatory networks associated with complex traits.
In the post-GWAS era, how to enhance the value of GWAS datasets and reveal the genomic underpinnings of complex phenotypes is a challenge (Leiserson et al. 2013; Jia and Zhao 2014). Systems genetics enables the identification of genetic pathways modulated by master regulators under drought stress in Populus. Systems genetics also allows analysis of the molecular regulatory mechanisms of complex traits. Our findings highlight the role of systems genetics, providing insights into how to search for drought-tolerant individuals in a drier atmosphere in the future. Genome editing using the CRISPR-Cas9 system enables the generation of alleles that can improve plant performance under abiotic stresses (Gupta et al. 2020). Allelic variation and the drought tolerance allele (PtoWRKY68hap1) are common in Populus germplasm adapted to water-scarce conditions, which impact developmental and regulatory pathways to modulate plant growth and production performance under the rapidly changing global climate environment. The identification of the PtoWRKY68 regulatory module provides insight into the molecular mechanisms that underpin drought tolerance in Populus and provides potential targets for engineering drought-tolerant varieties.
Materials and methods
Association population of Chinese white poplar (Populus tomentosa)
The population used for association mapping included 300 accessions selected from a clonal arboretum of 1,047 accessions in Guan Xian County, Shandong Province, China (36°23′N, 115°47′E), representing almost the entire natural distribution of P. tomentosa (30–40°N, 105–125°E) (Du et al. 2012). The geographic distribution of these 300 accessions was divided into the southern (S, n = 84), NW(n = 108), and NE (n = 108) geographic regions (Huang 1992). In the past 5 yr, the annual average precipitation in the S region was higher than in the NW and NE regions (Supplemental Fig. S2) (http://www.cma.gov.cn/). An experimental population of 300 1-yr-old accessions was asexually propagated via grafting with three duplicated cultivations per accession. In the initial growth stage, the water content in the soil was kept at 75% to 80%. Water supply was stopped when the plants were 4-mo-old until the water content in the soil dropped from 20% to 25%. During drought treatment, the soil moisture content was maintained at ≤25% for 1 mo before trait evaluation.
Phenotypic data of the P. tomentosa association population
To estimate plant responses to drought stress in the P. tomentosa population, seven drought-related (photosynthetic and physiological) traits were measured using functional leaves (fourth to sixth leaves from the top of the main stem). Measurements of the P. tomentosa population were performed first under well-watered conditions. Subsequently, the same plants were subjected to drought treatment and the drought-related measurements were repeated. Photosynthetic characteristics were measured using the LI-6400 portable photosynthesis system (LI-COR Inc., Lincoln, NE). To reduce the influence of circadian rhythms on photosynthesis, these traits were measured in functional leaves from 9:00 to 11:00 Am on a sunny day. All measurements were performed in triplicate per plant and three biological replications were performed for each accession. WUE was defined as the ratio between CO2 assimilation and Tr.
To quantify PRO content, endogenous PRO was extracted from 0.2 g fresh leaf tissue using 5 mL 6 mol/L hydrochloric acid and hydrolyzed at 110°C for 24 h, neutralized using 5 mL 6 mol/L Na(OH)2, and centrifuged at 5000 rpm for 10 min. The supernatant (0.5 mL) was transferred to a new tube, and 0.5 mL NaHCO3 (0.5 mol/L; pH 9.0) and 0.5 mL DNFB reaction solution were added. The mixture was incubated at 60°C for 60 min, cooled to room temperature, and phosphate buffer (pH 7.0) was added to a 5 mL volume. The PRO content was determined using HPLC-1100 (Agilent) with a Symmetry C18 column. To quantify ABA content, ∼0.3 g fresh leaf tissue was extracted with 5 mL extraction buffer (80% isopropyl alcohol, 1% formic acid; v/v), at 4°C for 1 h using ultrasound, and centrifuged at 10,000 rpm for 10 min. The supernatant was transferred to a new tube, ultrasonically extracted with 1 mL dichloromethane at 4°C for 30 min, and centrifuged as above. The ABA content was determined via UPLC (Agilent) with a Symmetry C18 column. To quantify Chl content, ∼1.0 g fresh leaf tissue was extracted with 4 mL 0.1% butylated hydroxytoluene-ethanol solution (v/v), at 25°C with shaking in darkness for 4 h. The Chl content was determined via HPLC (Agilent) with a Symmetry C18 column. The percentage reduction or increase of these drought-related traits was calculated using the formula: (measurement of drought-stressed—measurement of well-watered)/measurement of well-watered. Phenotype repeatability was estimated according to broad-sense heritability (h2) as described previously (Yang et al. 2011; Speed and Balding 2014).
SNP calling following resequencing of the association population
The methods and clean data for resequencing the association population with 300 accessions were previously described (Xiao et al. 2021). In brief, clean reads were aligned to the P. tomentosa reference genome by the Burrows–Wheeler Aligner (v. 0.7.5a–r405) using default parameters (Li and Durbin 2009). Low mapping quality (MQ < 20) reads were filtered using SAMtools (v. 1.1) and removed. Genome variant calling was performed using Genome Analysis Toolkit v. 4.0 (https://gatk.broadinstitute.org/hc/en-us) with conservative parameters. SNPs with only two alleles were pruned using Vcftools_0.1.13 (Danecek et al. 2011). Following the removal of those with a MAF < 0.05, heterozygous genotype frequency <0.25, and missing genotype >0.2, we identified 3,002,432 SNPs across all individuals.
RNA-sequencing analysis
Leaf tissues for RNA-sequencing (RNA-seq) were collected from well-watered and drought-stress-treated plants. The duration of sampling was minimized to reduce changes in gene expression. Total RNA was extracted from the mature leaves of 300 unrelated individuals, using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. RNA libraries were constructed and sequenced by Novogene (Beijing, China). Paired-end sequencing was performed on the Illumina Hiseq 2005 platform (Illumina, San Diego, CA). Next, clean data were uniquely mapped to the P. tomentosa reference genome using TopHat v. 2.1.1 with default options (Trapnell et al. 2009). The isoform levels and gene-level counts of the assembled transcripts were computed and normalized based on FPKM units using Cufflinks v. 2.1.1 with default options (Trapnell et al. 2012). Genes with q < 0.05 were identified as DEGs using Cufflinks v. 2.1.1.
Genome-wide association analysis
GWAS was performed in TASSEL v. 5.0 using high-quality data for the 3,002,432 SNPs (Bradbury et al. 2007). The standard mixed linear model was applied, in which the population structure (Q) and kinship (K) were estimated as described previously (Du et al. 2019). For the degree of replication, we performed ten replicate runs with random seed at each value of K from one to ten with 10-fold cross-validation (CV). We calculated the average CV error and standard deviation for every K-value. K = 3 was used because it separated the S, NE, and NW geographic subpopulations. The SNP set was used to construct a neighbor-joining (NJ) phylogenetic tree. The phylogenetic tree encompassed three clades, consistent with the admixture results. Collectively, K = 3 was the optimal K-value for the separation of subpopulations. The percentage reduction or increase of the seven drought-related traits was normalized by Z-score. The compromised significance threshold for GWAS was set as P < 3.3 × 10−7 (1/n, n = effective number of independent SNPs) based on Bonferroni-adjusted correction for multiple testing. LD analysis using the R package, LD heatmap was used to define LD blocks surrounding significant SNPs by intervals (Shin et al. 2006).
Sequence alignment and phylogenetic analysis
To annotate the functions of candidate genes, we performed BLASTP (v. 2.9.0+; 1e−5) searches of the Swiss-Prot database and the NCBI nonredundant protein database (NR). Sequence alignments were performed using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle). Using complete protein sequences, the phylogenetic tree was constructed by rooting at the midpoint using the NJ method in MEGA v. 7 (Kumar et al. 2016). The reliability of the tree was estimated by bootstrapping using 2,000 replications. Evolutionary distances were computed using the Poisson correction method. Alignments used for phylogenetic analysis are provided in Supplemental Files S1 to S12.
Vector construction and genetic transformation of Arabidopsis
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 were surface-sterilized and cold-stratified in sterile water at 4°C for 48 h, followed by germination on agar-solidified 1/2 MS medium (pH 5.8) supplemented with 1% sucrose at 22°C. Seven-day-old seedlings were transplanted in soil and grown in a growth room at 22°C with a 16/8 h diurnal cycle.
The CDSs of PtoWRKY68hap1 and PtoWRKY68hap2 were PCR-amplified from P. tomentosa clones LM50 and 3169, respectively, which were cloned into the pCXSN vector under the transcriptional control of the CaMv 35S promoter. The resultant overexpression vector was introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform WT A. thaliana (Col-0) plants using the floral-dip method (Clough and Bent 1998). Transformants were selected on hygromycin-containing (50 mmol/L) medium and progressed to the T3 generation when the transgene became homozygous. The drought-related traits and relative expression levels of PtoWRKY68hap1 and PtoWRKY68hap2 were measured in two independent homozygous transformed lines.
Drought tolerance assay
Individual A. thaliana plants were grown in pots under short-day conditions (10/14 h, light/dark) in a growth room. Twenty-eight-day-old plants were saturated in water to 95% relative gravimetric soil water content (rSWC), measured daily. For drought treatment, watering was withheld until a 17% rSWC was reached. Control plants were maintained under well-watered conditions at a 95% rSWC. The experiments were performed three times, and each experiment contained 12 plants per line. The gross fresh weight of the detached leaves of five plants was measured to assess water loss. The water loss rate was calculated using the formula: water loss rate (%) = (FW − DW)/FW × 100, where FW is the leaves the fresh weight of harvest, and DW is the fresh weight of leaves detached at 10 min intervals.
RT-qPCR analysis
Transgenic and WT A. thaliana plants were maintained in a greenhouse under well-watered or drought-stress conditions, with three independent biological replicates. Total RNA was extracted from the mature leaves of seedlings using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. Total RNA was reverse transcribed into first-strand cDNA using the Reverse Transcription System (Promega Corporation, Madison, WI). After the inactivation of enzymes by heating, a 0.5 μL aliquot was used for RT-qPCR on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) using SYBR Premix Ex Taq (TaKaRa, Dalian, China) according to the manufacturer's protocol. 18S rDNA and ACTIN2 (AT3G18780) were used as the internal controls. PCR involved preincubation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s, and dissociation at 60°C to 95°C at 0.05°C s−1. The relative transcript abundance of candidate genes was calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). The transcript levels of candidate genes in transgenic lines were relative to the expression in WT plants. Three biological replicates were performed, each of which consisted of three technical repeats. The primers used are listed in Supplemental Table S3.
ABA-sensitivity assay in transgenic Arabidopsis and P. tomentosa accessions
Leaves of six P. tomentosa accessions and both transgenic and WT plants were immersed in stomatal opening buffer (10 mM MES, 5 mM KCl, 50 mM CaCl2, pH 6.15) for 3 h before being transferred to opening buffer with 1 μM ABA. The stomatal aperture was measured every 30 min for 90 min. Epidermal strips were peeled and photographed using a DMi8 microscope (Leica Biosystems, Nussloch, Germany). Stomatal apertures were measured using Image J software (http://rsb.info.nih.gov/ij).
To assess the impact of ABA on seed germination, surface-sterilized seeds from transgenic and WT A. thaliana were plated on a solid 1/2 MS medium with ABA (0, 0.5, and 1 μM). The rates of seed germination were recorded daily and photographed on days 4 and 7. To measure root length, sterilized seeds of the transgenic lines and WT were germinated on 1/2 MS medium, and 5-d-old seedlings were transplanted to fresh medium with ABA (0, 1, and 3 μM) for 7 d; subsequently, root length and the number of lateral roots were analyzed (Shang et al. 2010; Seo et al. 2012; Zhang et al. 2014; Ma et al. 2019). Root elongation of A. thaliana plants was analyzed using Image J.
Weighted gene co-expression network analysis
WGCNA was applied to analyze the RNA-seq data of 55 accessions randomly selected from the natural population of P. tomentosa. The expression of 34,106 genes in well-watered and drought-stress samples of these 55 accessions was evaluated from the RNA-seq dataset. On average, 24,381 genes were detected in 80% of the samples, and 9,714 genes were significantly differentially expressed between well-watered and drought-stress samples (P < 0.001, t-test). Log10 (FPKM + 1) was used to normalize FPKM values, which were used to generate co-expression networks with the WGCNA package in R (Langfelder and Horvath 2012). Independent signed networks were constructed from well-watered and drought-stress time-course samples. An adjacency matrix was constructed using soft threshold powers of four and nine under well-watered and drought-stress conditions, respectively. Network interconnectedness was measured by calculating the topological overlap using the TOMdist function with a signed TOMType. Average hierarchical clustering using the hclust function was performed to group the genes based on the topological overlap dissimilarity measure (1-TOM) of their connection strengths. Network modules were identified using a dynamic tree-cut algorithm with a minimum cluster size of 30 and a merging threshold function of 0.25. To identify hub genes within the modules, module membership for each gene was calculated based on the Pearson correlation between the expression level and the module eigengene. Genes within the module with the highest module membership were considered highly connected within that module. To test the correlations between each module and the photosynthetic and physiological traits, the module eigengenes were correlated with the physiological data.
DAP-seq sampling and data analysis
DAP-seq was carried out as described previously (Bartlett et al. 2017) and performed at Bluescape Hebei Biotech to purify gDNA from the leaves of P. tomentosa clone LM50. A genomic DNA library was prepared using the NGS0602-MICH TLX DNA-Seq Kit (MICH, Hebei, China). The coding sequences of PtoWRKY68hap1 and PtoWRKY68hap2 were cloned into pFN19K HaloTag T7 SP6 Flexi vector and expressed using the TNT SP6 Coupled Wheat Germ Extract System (Promega, Madison, WI). PtoWRKY68hap1 and PtoWRKY68hap1 bead mixtures were incubated with the genomic DNA library. Eluted DNA was sequenced on an Illumina NavoSeq6000 with two technical duplicates. DAP-seq reads were aligned to the reference P. tomentosa genome (CRA000903) using Bowtie2 (Langmead and Salzberg 2012). The conserved motifs in peaks were identified via MEME-ChIP (Machanick and Bailey 2011). We performed KEGG analysis using the KOBAS v. 2.0 database (http://kobas.cbi.pku.edu.cn/). PtoWRKY68 target genes were defined as peaks located 2 kb upstream of ATG.
Expression quantitative trait loci analysis
eQTL analysis was conducted using the normalized relative transcript abundances of PtoWRKY68 from 300 accessions under each condition as the phenotypic variable in the GWAS analysis. The standard mixed linear model and the compromised significance threshold were consistent with the above GWAS.
Prediction of cis-elements
A 2 kb DNA fragment spanning upstream of the start codon ATG of PtoWRKY68 was extracted as its promoter sequence, from which cis-regulatory elements were predicted in silico using PlantTFDB v. 5.0 software (http://planttfdb.gao-lab.org/index.php) (Tian et al. 2020). CArG motifs in the PtoWRKY68 promoter region were identified as described previously (Tang and Perry 2003; Li et al. 2008). The primers used for PCR of the promoter sequence are shown in Supplemental Table S3.
Electrophoretic mobility shift assay
The full-length CDSs of PtoWRKY68hap1, PtoWRKY68hap2, and PtoSVP.3 were amplified by PCR, and individually cloned into the BamHI/SalI sites of the expression vector pET-32a-HIS using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). Following expression in Escherichia coli BL21 (DE3), the recombinant PtoWRKY68hap1, PtoWRKY68hap2, and PtoSVP.3 proteins were purified using the His-Tagged Protein Purification Kit (Cwbio, Taizhou, China), diluted in PBS buffer (135 mM NaCl, 4.7 mM KCl, 10 mM Na2HPO4, and 2 mM NaH2PO4; pH 7.4), and quantified on a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Motif DNA representing target gene promoters was synthesized by annealing the forward and reverse complementary oligos containing W1-box, W2-box, and CArG elements (Supplemental Table S3). EMSA was performed using an EMSA Kit (Beyotime, Shanghai, China). DNA–protein complexes were fractionated on a non-denaturing 6% polyacrylamide gel, transferred to a positive nylon membrane, and UV cross-linked. Complexes were detected using streptavidin-HRP conjugate with an Enhanced Chemiluminescence (ECL) Kit (Beyotime).
Dual-luciferase reporter assay
DLRA was conducted as described previously (Hellens et al. 2005). Briefly, PtoWRKY68hap1, PtoWRKY68hap2, and PtoSVP.3 cDNAs were individually cloned into the BamHI/SalI sites of the effector plasmid pGreenII 62-SK using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The promoters of PtoABF2.1, PtoDTX49.1, PtoRD26.1, and PtoWRKY68 have cloned into the SalI/HindIII sites of the reporter plasmid pGreenII 0800-LUC with the 35S promoter-driven Renilla luciferase (REN) gene. These constructs were introduced to A. tumefaciens GV3101 (pSoup-p19). Equal concentrations and volumes of A. tumefaciens constructs, promoter-LUC, pGreenII 0800-LUC, PtoSVP.3/PtoWRKY68–62-SK, and pGreenII62-SK were co-infiltrated into abaxial mesophyll cells in fully expanded Nicotiana benthamiana leaves using needle-free syringes, followed by incubation at room temperature for 48 to 60 h (Chen et al. 2008). The infiltrated N. benthamiana leaves were sprayed with 1 mM d-fluorescein and photographed using an LB983 Night Owl II fluorescence imaging system (Berthold Technologies, Bad Wildbad, Germany). Quantification was performed by normalizing firefly luciferase (LUC) activity to that of REN, using 35S:REN as the internal control. Luciferase activity was assayed using the DLRA system (Beyotime) according to the manufacturer's instructions. DLRAs were performed in at least triplicate. Relative luciferase activities using PtoWRKY68/PtoSVP.3-pGreenII62-SK as the effector was compared to the control effector (pGreenII62-SK empty vector).
Statistical analysis
Statistical significance was determined by t-test or one-way ANOVA with Tukey's test analysis using the IBM SPSS Statistics 25.0 software (IBM SPSS, Chicago, USA). Significant differences were indicated by *P < 0.05, **P < 0.01, or ***P < 0.001, ns, with no significant difference.
Accession numbers
Sequence data in this study has been submitted to GenBank: accession numbers OQ789667–OQ789672. The RNA-seq of P. tomentosa under well-water and water-deficient phases, DAP-seq of PtoWRKY68hap1 and PtoWRKY68hap2, and genome resequencing of P. tomentosa are available at the BIGD Genome Sequence Archive (https://bigd.big.ac.cn) under accession number CRA007408, CRA009302, CRA009303, and CRA000903, respectively. Sequence data used in this article can be found in the Arabidopsis Information Resource (https://www.arabidopsis.org/index.jsp) under the following accession numbers: AtNCED3 (AT3G14440), AtRD20 (AT2G33380), AtRD29B (AT5G52300), AtHIS1–3 (AT2G18050), AtSnRK2.6 (AT4G33950), and AtABI2 (AT5G57050).
Supplementary Material
Acknowledgments
We thank Xueqing Ma (College of Resources and Environmental Sciences, China Agricultural University) for assisting us to analyze average precipitation in P. tomentosa distribution regions.
Contributor Information
Yuanyuan Fang, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Dan Wang, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Liang Xiao, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Mingyang Quan, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Weina Qi, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Fangyuan Song, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Jiaxuan Zhou, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Xin Liu, Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing 100093, People’s Republic of China.
Shitong Qin, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Qingzhang Du, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Qing Liu, The Institute of Agriculture and Food Research, CSIRO Agriculture and Food, Black Mountain, Canberra ACT 2601, Australia.
Yousry A El-Kassaby, Department of Forest and Conservation Sciences, Faculty of Forestry, Forest Sciences Centre, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Deqiang Zhang, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China; Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, No. 35, Qinghua East Road, Beijing 100083, People’s Republic of China.
Author contributions
D.Z. designed the experiment and conception; Y.F., D.W., and X.L. performed the experiments; Y.F. wrote the manuscript; W.Q., F.S., and S.Q. helped to collect and analyze the data; L.X., M.Q., and J.Z. provided the valuable suggestions on the manuscript; Q.D., Q.L., and Y.E.K. helped to revise the manuscript; D.Z. obtained the funding and were responsible for this manuscript. All authors read and approved the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Seven drought-related traits of P. tomentosa under well-watered and drought-stress conditions in three climate regions.
Supplemental Figure S2. Average precipitation in the past 5 yr in P. tomentosa distribution regions.
Supplemental Figure S3. Manhattan plots of GWAS with six drought-related traits.
Supplemental Figure S4. Phylogenetic consensus trees of PtoDUF3598 and PtoWRKY68 in different species.
Supplemental Figure S5. Drought tolerance of transgenic Arabidopsis thaliana overexpressing PtoWRKY68.
Supplemental Figure S6. Sequence alignment of PtoWRKY68hap1 and PtoWRKY68hap2 proteins.
Supplemental Figure S7. Drought-stress phenotype of WT, PtoWRKY68hap1, and PtoWRKY68hap1 transgenic plants.
Supplemental Figure S8. Sequence alignment of PtoWRKY68 homologs in other Populus species.
Supplemental Figure S9. Dynamic changes in drought-related traits are correlated with co-expression networks.
Supplemental Figure S10. Phylogenetic consensus tree of PtoABF2.1 in different species.
Supplemental Figure S11. Phylogenetic consensus tree of PtoRD26.1 in different species.
Supplemental Figure S12. Phylogenetic consensus tree of PtoDTX49.1 in different species.
Supplemental Figure S13. PtoWRKY68 alleles positively regulated ABA-/drought-inducible genes and were associated with ABA-related phenotypes.
Supplemental Figure S14. Verification and RT-qPCR analysis of svp mutants.
Supplemental Table S1. Details of significant SNPs associated with drought-related traits in the association population of P. tomentosa.
Supplemental Table S2. Conserved hub genes (intramodular connectivity > 20) were detected using WGCNA.
Supplemental Table S3. Oligonucleotide sequences of the primers used in this study.
Supplemental Data Set 1. Variations in a genomic region of PtoWRKY68.
Supplemental Data Set 2. Well-watered network gene lists with module membership and gene significance for each drought-related trait.
Supplemental Data Set 3. Drought network gene lists with module membership and gene significance for each drought-related trait.
Supplemental Data Set 4. DEGs in PtoWRKY68hap1 vs. PtoWRKY68hap2 haplogroups with a significant difference (P < 0.05) and at least 2-fold change.
Supplemental File 1. DUF3598 homologs sequence alignment.
Supplemental File 2. DUF3598 homologs phylogenetic consensus tree.
Supplemental File 3. WRKY I genes sequence alignment.
Supplemental File 4. WRKY I genes phylogenetic consensus tree.
Supplemental File 5. ABF2 homologs sequence alignment.
Supplemental File 6. ABF2 homologs phylogenetic consensus tree.
Supplemental File 7. RD26 homologs sequence alignment.
Supplemental File 8. RD26 homologs phylogenetic consensus tree.
Supplemental File 9. MATE efflux family protein sequence alignment.
Supplemental File 10. MATE efflux family protein phylogenetic consensus tree.
Supplemental File 11. MADS-box protein SVP homologs sequence alignment.
Supplemental File 12. MADS-box protein SVP homologs phylogenetic consensus tree.
Funding
The study was supported by the Major Project of Agricultural Biological Breeding (No. 2022ZD0401502), the Project of National Natural Science Foundation of China (No. 32170370), the Project funded by China Postdoctoral Science Foundation (No. 2022M710406) and the 111 Project (No. B20050). The funding body had no involvement in designing the study or collecting, analyzing, or interpreting data, or in writing the manuscript.
References
- Andres F, Porri A, Torti S, Mateos J, Romera-Branchat M, Garcia-Martinez JL, Fornara F, Gregis V, Kater MM, Coupland G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci U S A. 2014:111(26):E2760–E2769. 10.1073/pnas.1409567111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS. A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol. 1997:34(4):629–641. 10.1023/A:1005886011722 [DOI] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP. RD20, A stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2010:51(12):1975–1987. 10.1093/pcp/pcq155 [DOI] [PubMed] [Google Scholar]
- Barber VA, Juday GP, Finney BP. Reduced growth of Alaskan white spruce in the twentieth century from temperature-induced drought stress. Nature. 2000:405(6787):668–673. 10.1038/35015049 [DOI] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017:12(8):1659–1672. 10.1038/nprot.2017.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Penfold CA, Jenkins DJ, Legaie R, Moore JD, Lawson T, Matthews JSA, Vialet-Chabrand SRM, Baxter L, Subramaniam S, et al. Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. Plant Cell. 2016:28(2):345–366. 10.1105/tpc.15.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert HJ, Gong Q, Li P, Ma S. Unraveling abiotic stress tolerance mechanisms–getting genomics going. Curr Opin Plant Biol. 2006:9(2):180–188. 10.1016/j.pbi.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007:23(19):2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol. 2020:62(1):25–54. 10.1111/jipb.12899 [DOI] [PubMed] [Google Scholar]
- Chen J, Nolan T, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell. 2017:29(6):1425–1439. 10.1105/tpc.17.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yin Y. WRKY Transcription factors are involved in brassinosteroid signaling and mediate the crosstalk between plant growth and drought tolerance. Plant Signal Behav. 2017:12(11):e1365212. 10.1080/15592324.2017.1365212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):323–324. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014:15(1):34–48. 10.1038/nrg3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Coustham V, Loudet O. Lessons from eQTL mapping studies: non-coding regions and their role behind natural phenotypic variation in plants. Curr Opin Plant Biol. 2012:15(2):192–198. 10.1016/j.pbi.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Wang B, Wei Z, Zhang D, Li B. Genetic diversity and population structure of Chinese white poplar (Populus tomentosa) revealed by SSR markers. J Hered. 2012:103(6):853–862. 10.1093/jhered/ess061 [DOI] [PubMed] [Google Scholar]
- Du Q, Yang X, Xie J, Quan M, Xiao L, Lu W, Tian J, Gong C, Chen J, Li B, et al. Time-specific and pleiotropic quantitative trait loci coordinately modulate stem growth in Populus. Plant Biotechnol J. 2019:17(3):608–624. 10.1111/pbi.13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007:10(4):366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LP, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004:39(6):863–876. 10.1111/j.1365-313X.2004.02171.x [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 Is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005:17(12):3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Rico-Medina A, Cano-Delgado AI. The physiology of plant responses to drought. Science. 2020:368(6488):266–269. 10.1126/science.aaz7614 [DOI] [PubMed] [Google Scholar]
- Han Z, Hu Y, Lv Y, Rose JKC, Sun Y, Shen F, Wang Y, Zhang X, Xu X, Wu T, et al. Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol. 2018:176(3):2292–2304. 10.1104/pp.17.01320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):1–14. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. The study on the climatic regionalization of the distributional region of Populus tomentosa. J Beijing For Univ. 1992:14(S3):26–32. [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001:27(4):325–333. 10.1046/j.1365-313x.2001.01096.x [DOI] [PubMed] [Google Scholar]
- Jia P, Zhao Z. Network assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet. 2014:133(2):125–138. 10.1007/s00439-013-1377-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Duan Y, Yin J, Ye S, Zhu J, Zhang F, Lu W, Fan D, Luo K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot. 2014:65(22):6629–6644. 10.1093/jxb/eru381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Tong S, Chen N, Liu B, Bai Q, Chen Y, Bi H, Zhang Z, Lou S, Tang H, et al. The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant J. 2021:105(5):1258–1273. 10.1111/tpj.15109 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lingeman J, Colon AM, Coruzzi G, Shasha D. Gene regulatory networks in plants: learning causality from time and perturbation. Genome Biol. 2013:14(6):123. 10.1186/gb-2013-14-6-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher C, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:33(7):1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Vinod K, Zheng Z, Fan B, Chen Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008:8(1):68. 10.1186/1471-2229-8-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012:46(11):1–17. 10.18637/jss.v046.i11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011:7(1):e1001057. 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Eldridge JV, Ramachandran S, Raphael BJ. Network analysis of GWAS data. Curr Opin Genet Dev. 2013:23(6):602–610. 10.1016/j.gde.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009:25(14):1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Li X, Jiang M. CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol Biol Rep. 2021:48(8):5821–5832. 10.1007/s11033-021-06541-4 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008:15(1):110–120. 10.1016/j.devcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Lim C, Kang K, Shim Y, Yoo SC, Paek NC. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022:188(4):1900–1916. 10.1093/plphys/kiab492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gu HY, Zhu J, Niu YM, Zhang C, Guo GL. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 2019:10:1081. 10.3389/fphys.2019.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009:324(5930):1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Ma Q, Xia Z, Cai Z, Li L, Cheng Y, Liu J, Nian H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front Plant Sci. 2019:9:1979. 10.3389/fpls.2018.01979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011:27(12):1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler N, Schiffthaler B, Robinson KM, Terebieniec BK, Vucak M, Mannapperuma C, Bailey M, Jansson S, Hvidsten TR, Street NR. Leaf shape in Populus tremula is a complex, omnigenic trait. Ecol Evol. 2020:10(21):11922–11940. 10.1002/ece3.6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Pastinen T. The study of eQTL variations by RNA-Seq: from SNPs to phenotypes. Trends Genet. 2011:27(2):72–79. 10.1016/j.tig.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LP, Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun. 2015:6(1):8326. 10.1038/ncomms9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell NG. Mechanisms linking drought, hydraulics, carbon metabolism, and vegetation mortality. Plant Physiol. 2011:155(3):1051–1059. 10.1104/pp.110.170704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukarram M, Choudhary S, Kurjak D, Petek A, Khan M. Drought: sensing, signalling, effects and tolerance in higher plants. Physiol Plant. 2021:172(2):1291–1300. 10.1111/ppl.13423 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009:50(7):1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol. 2006:60(1):51–68. 10.1007/s11103-005-2418-5 [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Chaves MM. Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot. 2011:62(3):869–882. 10.1093/jxb/erq340 [DOI] [PubMed] [Google Scholar]
- Porcu E, Rueger S, Lepik K, Santoni FA, Reymond A, Kutalik Z. Mendelian Randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat Commun. 2019:10(1):3300. 10.1038/s41467-019-10936-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallett JP, Leak DJ, Liotta CL, et al. The path forward for biofuels and biomaterials. Science. 2006:311(5760):484–489. 10.1126/science.1114736 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010:15(7):395–401. 10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotech J. 2012:10(1):2–11. 10.1111/j.1467-7652.2011.00634.x [DOI] [PubMed] [Google Scholar]
- Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT. Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 2012:160(1):556–568. 10.1104/pp.112.202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010:22(6):1909–1935. 10.1105/tpc.110.073874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw. 2006:16(Code Snippet 3):1–9. 10.18637/jss.v016.c03 [DOI] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003:6(5):410–417. 10.1016/S1369-5266(03)00092-X [DOI] [PubMed] [Google Scholar]
- Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res. 2014:24(9):1550–1557. 10.1101/gr.169375.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants. 2016:2(5):16055. 10.1038/nplants.2016.55 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003:35(1):44–56. 10.1046/j.1365-313X.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Tang W, Perry SE. Binding site selection for the plant MADS domain protein AGL15. J Biol Chem. 2003:278(30):28154–28159. 10.1074/jbc.M212976200 [DOI] [PubMed] [Google Scholar]
- Tang S, Zhao H, Lu S, Yu L, Zhang G, Zhang Y, Yang QY, Zhou Y, Wang X, Ma W, et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant. 2021:14(3):470–487. 10.1016/j.molp.2020.12.003 [DOI] [PubMed] [Google Scholar]
- Tian F, Yang DC, Meng YQ, Jin J, Gao G. Plantregmap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020:48(D1):D1104–D1113. 10.1093/nar/gkz1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Wang Y, Chen N, Wang D, Liu B, Wang W, Chen Y, Liu J, Ma T, Jiang Y. PtoNF-YC9-SRMT-PtoRD26 module regulates the high saline tolerance of a triploid poplar. Genome Biol. 2022:23(1):148. 10.1186/s13059-022-02718-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009:25(9):1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012:7(3):562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006:45(4):523–539. 10.1111/j.1365-313X.2005.02593.x [DOI] [PubMed] [Google Scholar]
- Wang K, Bu T, Cheng Q, Dong L, Su T, Chen Z, Kong F, Gong Z, Liu B, Li M. Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 2021:229(5):2660–2675. 10.1111/nph.17019 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010:11(12):843–854. 10.1038/nrg288 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang F, Hong Y, Yao J, Ren Z, Shi H, Zhu J. The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol Plant. 2018:11(9):1184–1197. 10.1016/j.molp.2018.06.009 [DOI] [Google Scholar]
- Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Yang X, Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet. 2016:48(10):1233–1241. 10.1038/ng.3636 [DOI] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei DFN, Leung J. An update on abscisic acid signaling in plants and more. Mol Plant. 2008:1(2):198–217. 10.1093/mp/ssm022 [DOI] [PubMed] [Google Scholar]
- Wu C, Pan W. Integrating eQTL data with GWAS summary statistics in pathway-based analysis with application to schizophrenia. Genet Epidemiol. 2018:42(3):303–316. 10.1002/gepi.22110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Du Q, Fang Y, Quan M, Lu W, Wang D, Si J, El-Kassaby YA, Zhang D. Genetic architecture of the metabolic pathway of salicylic acid biosynthesis in Populus. Tree Physiol. 2021:41(11):2198–2215. 10.1093/treephys/tpab068 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002:14(suppl 1):S165–S183. 10.1105/tpc.000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011:88(1):76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013:162(3):1378–1391. 10.1104/pp.113.217596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TF, Liu Y, Fu JD, Ma J, Fang ZW, Chen J, Zheng L, Lu ZW, Zhou YB, Chen M, et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol J. 2021:19(12):2589–2605. 10.1111/pbi.13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pu H, Duan Z, Li Y, Liu B, Zhang Q, Li W, Rochaix J, Liu L, Peng L. Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell. 2018:30(8):1770–1788. 10.1105/tpc.18.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu J, Gong Z, Zhu JK. Abiotic stress responses in plants. Nat Rev Genet. 2022:23(2):104–119. 10.1038/s41576-021-00413-0 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu H, Pan Y, Yu Y, Luan S, Li L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant. 2014:7(10):1522–1532. 10.1093/mp/ssu063 [DOI] [PubMed] [Google Scholar]
- Zhao L, Yan J, Xiang Y, Sun Y, Zhang A. ZmWRKY104 transcription factor phosphorylated by ZmMPK6 functioning in ABA-induced antioxidant defense and enhance drought tolerance in maize. Biology (Basel). 2021:10(9):893. 10.3390/biology10090893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang P, Guo P, Chong L, Yu G, Sun X, Hu T, Li Y, Hsu CC, Tang K, et al. CDK8 Is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020:228(5):1573–1590. 10.1111/nph.16787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.