Abstract
A sensitive and specific nested PCR assay was developed for the detection of granulocytic ehrlichiae. The assay amplifies the 16S rRNA gene and was used to examine acute-phase EDTA-blood and serum samples obtained from seven humans with clinical presentations compatible with human granulocytic ehrlichiosis. Five of the seven suspected cases were positive by the PCR assay using DNA extracted from whole blood as the template, compared with a serologic assay that identified only one positive sample. The PCR assay using DNA extracted from the corresponding serum samples as the template identified three positive samples. The sensitivity of the assay on human samples was examined, and the limit of detection was shown to be fewer than 2 copies of the 16S rRNA gene. The application of the assay to nonhuman samples demonstrated products amplified from template DNA extracted from Ixodes scapularis ticks collected in Rhode Island and from EDTA-blood specimens obtained from white-tailed deer in Maryland. All PCR products were sequenced and identified as specific to granulocytic ehrlichiae. A putative variant granulocytic ehrlichia 16S rRNA gene sequence was detected among products amplified from both the ticks and the deer blood specimens.
The ehrlichiae are obligately intracellular bacteria and have been characterized as pathogens of medical and veterinary importance. Two forms of human ehrlichiosis have been reported in the United States: human monocytic ehrlichiosis (HME) and human granulocytic ehrlichiosis (HGE). The first cases of HME were described in 1987 (32), and the etiologic agent was subsequently identified as Ehrlichia chaffeensis (2, 15). HGE was first described in 1994 and differs from HME in that granulocytic leukocytes serve as the primary target cells (13). The Ehrlichia species responsible for HGE has not been defined, although the nucleotide sequence of the 16S rRNA gene (16S rDNA) of the HGE agent suggests a close relationship to Ehrlichia equi, the agent of equine granulocytic ehrlichiosis, and Ehrlichia phagocytophila, which is responsible for infections of ruminants in Europe (13). Studies supporting the close relationship of the HGE agent to E. equi and E. phagocytophila have included descriptions of the serologic cross-reactivity of these organisms (3, 19) and a report that a horse experimentally infected with the HGE agent was resistant to an E. equi challenge (7).
The initial reports of HGE were in Minnesota and Wisconsin, but recent studies have shown that the disease also occurs in the northeastern United States and northern California (3, 11, 13, 21, 25, 28, 42). Human infections with a granulocytic Ehrlichia species have also been described in Europe (4, 10, 22, 36, 38). The HGE agent has been associated with the deer tick, Ixodes scapularis (3, 34, 35), which might serve as the primary vector. Additional vectors, and a mammalian reservoir, have not been determined, although dogs living in areas where HGE is endemic can be naturally infected with granulocytic ehrlichiae (GE) (27, 31, 37). Rodents, particularly white-footed mice (Peromyscus leucopus), are competent experimental hosts and may serve as a natural reservoir (39, 40, 42).
HGE presents clinically as an acute febrile illness, although the nonspecific symptoms of infection, most commonly fever, headache, and myalgia, make the diagnosis problematic. Likewise, HGE-associated laboratory findings, including leukopenia, thrombocytopenia, and elevated liver enzymes, are relatively nonspecific. A history of tick bite or tick exposure, while suggestive, is not diagnostic. Although several recent reports have described successful cultivation of the HGE agent (26, 29), the procedure is lengthy and the sensitivity has not been determined. Serologic tests, particularly indirect immunofluorescence antibody (IFA) assays, are commonly used but are often negative during the acute phase of infection. The recognition of cytoplasmic inclusions, or morulae, in infected cells during the examination of blood smears can be diagnostic; however, morulae have not been observed in numerous cases subsequently confirmed by other criteria (1, 5). PCR assays have been used often, mostly in research-oriented settings, and have the advantage of being able to yield positive results during the early acute phase of illness. PCR-based assays have proven critical for the identification and subsequent characterization of the ehrlichiae and have been developed for use with various clinical samples, but whole blood is the most commonly used source (2, 9, 12, 30). A PCR assay has been shown to be sensitive for identifying E. chaffeensis in acute-phase human blood samples (2, 24), and a nested PCR assay was used in the initial study that identified an Ehrlichia species as being the etiologic agent of HGE (13). More recent HGE studies have mainly used the primer set originally described by Chen et al. (13) for identification of the agent (23). To date, several alternative PCR assays have been described (6, 35), but these assays have not been thoroughly tested on human and veterinary specimens. The goal of this study was to develop a sensitive and specific PCR assay for the diagnosis of HGE and for the identification of GE in potential vector and host species.
MATERIALS AND METHODS
Samples.
EDTA–whole-blood and serum samples were collected in June 1995 from seven human patients with suspected ehrlichiosis in Westchester County, N.Y. I. scapularis ticks were collected from Trustom Pond, R.I. Blood specimens from white-tailed deer were collected from Prince Georges County, Md. Human blood samples were stored at 4°C prior to DNA extraction. DNA was extracted from each sample within 4 weeks of the date the blood samples were drawn. White-tailed deer blood samples and ticks were stored at −20°C prior to DNA extraction.
Controls.
A PCR-amplified region of the 16S rDNA was cloned and used as a positive control for subsequent amplifications. The cloned fragment was amplified from DNA extracted from the blood of one of the PCR-positive patients from Westchester County, N.Y., by using primers ec12 and ec9 as previously described (2). The amplified product was purified by using the manufacturer’s recommended protocol with the Wizard PCR Preps DNA Purification System (Promega Corporation, Madison, Wis.). The purified product was ligated into plasmid vector pGEM-T, by using the pGEM-T Vector System kit (Promega). The ligation product was transformed into Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) by electroporation, and individual white colonies were selected after growth on Luria-Bertani agarose with ampicillin (50 μg/ml). Plasmid DNA was purified from overnight cultures by using the Qiagen (Chatsworth, Calif.) Plasmid Kit, and the insertion was confirmed as the HGE agent 16S rRNA gene by DNA sequencing. This construct, named pFC4, was used as a positive control where indicated in the text.
Serologic testing.
An IFA assay was used to screen human sera for anti-HGE agent antibodies by using E. equi antigen slides as a surrogate antigen (Fuller Laboratories, Fullerton, Calif.). Sera were initially screened at dilutions of 1:64 and 1:128. Fluorescein isothiocyanate (FITC)-conjugated goat anti-human immunoglobulin G (IgG) (heavy and light chain; Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was used to identify antibodies reactive to E. equi-infected horse neutrophils. Positive- and negative-control sera were included in each assay. Titers of positive serum samples were subsequently determined to end point. The IgM-specific IFA assay utilized FITC-conjugated goat anti-human IgM (Kirkegaard & Perry Laboratories). Sera for the IgM assay were first depleted of IgG by use of a protein G affinity method (Quik-Sep IgM; Isolab, Akron, Ohio).
DNA purification.
Total DNA was purified from 200 μl of EDTA-whole blood (from humans or white-tailed deer) with the QIAamp blood kit (Qiagen). The protocol used was that suggested by the manufacturer. Briefly, detergent lysis was carried out in the presence of proteinase K for 10 min at 70°C. The lysed material was applied to a spin column containing a silica gel-based membrane and was washed twice. Purified DNA was eluted from the columns in 200 μl of Tris (10 mM; pH 8.0) and was stored at 4°C until it was used as the template for PCR amplification.
DNA was purified from serum samples by using the QIAamp blood kit essentially as described above. The only modification from the EDTA-blood protocol was an initial concentration step that involved the centrifugation of approximately 1 ml of each serum sample for 2 min at 12,000 × g. The supernatant was removed except for approximately 200 μl that was used to resuspend the serum sediment. The resuspended material was then used for DNA extraction.
DNA was purified from adult I. scapularis ticks by using a modification of the QIAamp tissue kit protocol (Qiagen). Individual ticks were placed in 1.5-ml microcentrifuge tubes containing 50 μl of phosphate-buffered saline (10 mM; pH 7.4) and were crushed with a disposable micropestle (Kontes Scientific Glassware/Instruments, Vineland, N.J.). After the addition of 150 μl of phosphate-buffered saline (10 mM; pH 7.4), the QIAamp tissue extraction protocol was followed as described by the manufacturer (Qiagen).
PCR amplification.
PCR amplifications were performed in a Perkin-Elmer 9600 thermal cycler, and reagents were from the GeneAmp PCR Kit with AmpliTaq DNA polymerase (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Primary reactions used 5 μl of purified DNA as the template in a total volume of 50 μl. Amplifications contained 200 μM each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 1.25 U of Taq polymerase, and 0.5 μM each primer. Primers were ge3a (5′ CACATGCAAGTCGAACGGATTATTC) and ge10r (5′ TTCCGTTAAGAAGGATCTAATCTCC). Cycling conditions involved an initial 2-min denaturation at 95°C, followed by 40 cycles, each consisting of a 30-s denaturation at 94°C, a 30-s annealing at 55°C, and a 1-min extension at 72°C. These 40 cycles were followed by a 5-min extension at 72°C. Reaction products were subsequently maintained at 4°C until they were analyzed by agarose gel electrophoresis or used as templates for nested reactions.
Nested amplifications used 1 μl of the primary PCR product as the template in a total volume of 50 μl. Each nested amplification contained 200 μM each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 1.25 U of Taq polymerase, and 0.2 μM each primer: ge9f (5′ AACGGATTATTCTTTATAGCTTGCT) and ge2 (5′ GGCAGTATTAAAAGCAGCTCCAGG). Nested cycling conditions were as described for the primary amplification, except that 30 cycles were used. Reaction products were subsequently maintained at 4°C until they were analyzed by agarose gel electrophoresis or purified for DNA sequencing.
Quality control included both positive and negative controls that were extracted and PCR amplified in parallel with all specimens. Human whole-blood DNA extractions and PCR amplifications were done in duplicate. In order to minimize the potential for contamination, DNA extractions, PCR setup, and agarose gel electrophoresis were performed in three separate rooms.
DNA sequencing and data analysis.
DNA sequencing reactions used fluorescence-labeled dideoxynucleotide technology (Dye Terminator Cycle Sequencing Ready Reaction Kit; Perkin-Elmer, Applied Biosystems Division). Sequencing reaction products were separated, and data were collected with an ABI 377 automated DNA sequencer (Perkin-Elmer, Applied Biosystems Division). Sequences were edited and assembled with the Staden software programs (17) and were analyzed with the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.) (18). The 16S rDNA sequences used for comparison were obtained from the GenBank database and included the HGE agent (accession no. U02521), E. equi (M73223), and E. phagocytophila (M73220).
RESULTS
Nested PCR assay design and use with human whole blood.
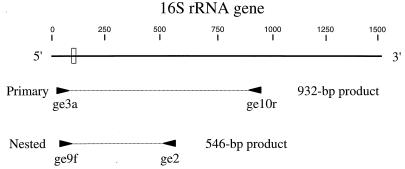
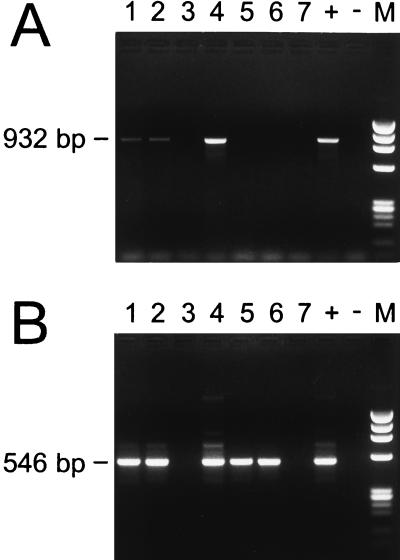
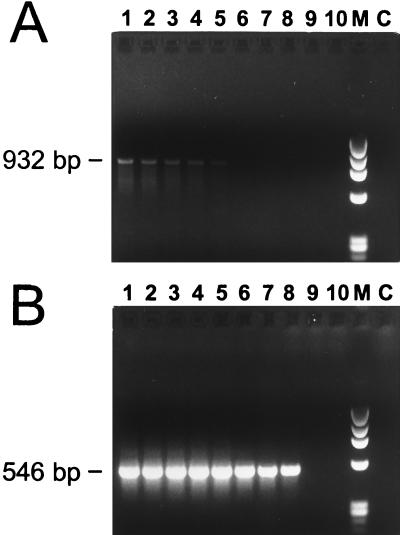
The initial testing of PCR assays used numerous primers and primer combinations and included both direct and nested PCR protocols (data not shown). A nested assay using primers ge3a and ge10r in the primary reaction, followed by ge9f and ge2 in a nested reaction, appeared to provide excellent sensitivity. The locations of these primers within the 16S rRNA gene of the HGE agent are illustrated in Fig. 1. Primers ge9f and ge10r were previously described by Chen et al. (13). Samples were collected from seven humans in Westchester County, N.Y., an area within the geographic range of I. scapularis ticks. These patients were suspected of having HGE on the basis of clinical presentation, and both EDTA–whole-blood and serum samples from the acute phase were available. Results of the nested assay using template DNA extracted from EDTA–whole-blood specimens are shown in Fig. 2. Samples 1, 2, and 4 were positive in the primary amplification (Fig. 2A, lanes 1, 2, and 4), as indicated by the 932-bp product. Two of the three positives showed low levels of product (Fig. 2A, lanes 1 and 2). Sample 4 (Fig. 2A, lane 4) showed an amount of product fairly comparable to that of the positive control. The results of the nested PCR assay (Fig. 2B) demonstrated that samples 1, 2, 4, 5, and 6 (lanes 1, 2, 4, 5, and 6) were positive, as indicated by the 546-bp product. Products from each of the five positive samples show bands clearly evident on the ethidium bromide-stained agarose gel, and the intensity of each band is comparable to that of the positive control. Duplicate aliquots of each human blood sample were independently extracted and PCR amplified, and these produced results identical to those shown in Fig. 2 (data not shown).
FIG. 1.
Schematic representation of the GE 16S rDNA, showing the relative locations of PCR primers used for the primary (ge3a and ge10r) and nested (ge9f and ge2) amplification reactions. The orientation of each primer is indicated by an arrowhead, and the sizes of the primary and nested products are shown. Scale, 16S rDNA base numbering. Small rectangle, region where sequence differences between the HGE agent, E. equi, and the GE variant were noted.
FIG. 2.
PCR products amplified from DNA purified from human EDTA-blood samples (lanes 1 to 7) and from positive (+) and negative (−) controls. (A) Primary amplification products. The expected size (932 bp) is indicated. (B) Nested amplification products. The expected size (546 bp) is noted. Lanes M are size standards and show the HaeIII digest of phage φX174 DNA.
PCR assay using human serum.
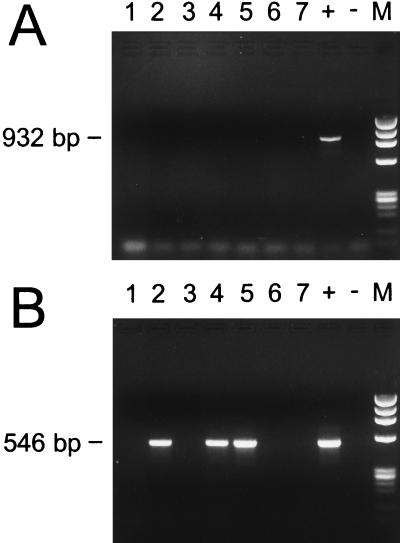
PCR was performed on DNA extracted from serum samples obtained from the same seven patients whose whole-blood specimens were tested by PCR as described above. The products of the amplifications on the serum-derived templates are shown in Fig. 3. Whereas the primary amplification produced no detectable products for any of the serum samples (Fig. 3A, lanes 1 to 7), the nested PCR assays showed products of the appropriate size for samples 2, 4, and 5 (Fig. 3B, lanes 2, 4, and 5).
FIG. 3.
PCR products amplified from DNA purified from human serum samples (lanes 1 to 7) and from positive (+) and negative (−) controls. (A) Primary amplification products. The expected size (932 bp) is indicated. (B) Nested amplification products. The expected size (546 bp) is noted. Lanes M are size standards and show the HaeIII digest of phage φX174 DNA.
Serologic testing of human samples.
The serum samples from the seven human patients were tested for antibodies by IFA assay, and six of the seven were negative (IFA titer, <64). The only IFA-positive serum was from patient 1, with a reciprocal titer of 512. Furthermore, an isotype-specific IFA assay showed that the response of this patient was restricted to the IgM class, indicative of an acute infection. The results of the PCR and IFA assays for the human blood and serum samples are summarized in Table 1.
TABLE 1.
Results of serologic and PCR assays for acute-phase samples collected from suspected HGE patients residing in Westchester County, N.Y.
| Patient | Serologic assaya result | PCR result for:
|
|
|---|---|---|---|
| EDTA-blood | Serum | ||
| 1 | + | + | − |
| 2 | − | + | + |
| 3 | − | − | − |
| 4 | − | + | + |
| 5 | − | + | + |
| 6 | − | + | − |
| 7 | − | − | − |
IFA assay using E. equi antigen slides and FITC-conjugated goat anti-human IgG (heavy and light chain).
PCR detection of ehrlichial DNA in ticks.
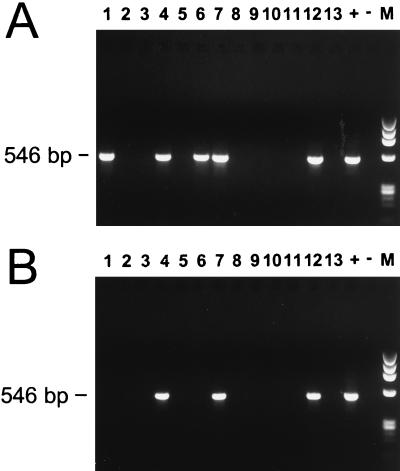
Application of the assay to potential vector species was tested with DNA extracted from 31 flat, unengorged female I. scapularis ticks and 30 flat, unengorged female Dermacentor variabilis ticks collected from Trustom Pond, R.I. Whereas none of the amplifications using the DNA extracted from D. variabilis ticks showed any products (data not shown), five of the Ixodes extracts (16%) showed products of the correct size. Figure 4A shows the results of the nested amplification using template DNA extracted from 13 of the I. scapularis ticks and includes the five positive products (lanes 1, 4, 6, 7, and 12). All the tick extracts were negative when tested by PCR assay for E. chaffeensis (data not shown).
FIG. 4.
Nested PCR products amplified from DNA purified from I. scapularis ticks (A) and white-tailed deer (B). Positive (+) and negative (−) controls are shown, and the expected size of the nested products (546 bp) is indicated. Lanes M are size standards and show the HaeIII digest of phage φX174 DNA.
PCR of ehrlichial DNA from white-tailed deer.
DNA was also extracted from EDTA–whole-blood samples obtained from white-tailed deer in Prince Georges County, Md. Thirty-two deer blood samples were tested with the HGE-specific nested PCR assay, and products of the correct size were amplified from three of these (9.4%). Figure 4B shows the nested PCR products for 13 of the deer templates, including the three positives (lanes 4, 7, and 12). All the deer blood extracts were negative when tested by PCR assay for E. chaffeensis (data not shown).
Sensitivity of the nested PCR assay.
The sensitivity of the assay was assessed by a spiking experiment that involved the addition of a known quantity of a plasmid (pFC4) containing the HGE 16S rRNA gene sequence to DNA extracted from ehrlichia PCR-negative human EDTA-blood samples. Each primary reaction mixture contained the same amount of background human genome DNA (5 μl) and 1 μl from a series of 10-fold serial dilutions of pFC4. The nested reactions were performed as described in Materials and Methods and used 1 μl of the primary reaction product as the template. By using this approach, 10-fold serial dilutions of the purified, quantitated plasmid DNA were tested, and the nested PCR amplification products are shown in Fig. 5. The limit of detection was determined to be approximately 1.93 copies (Fig. 5, lane 8). These data were reproduced by using three different serial dilutions of pFC4 and by using the DNA purified from three different noninfected humans as a nonspecific competitor (data not shown).
FIG. 5.
Sensitivity testing of the GE PCR assays. (A) Products of the primary assay using 10-fold serial dilutions of the positive control (pFC4) as templates. Lanes 1 through 10 correspond to template copy numbers ranging from 1.93 × 107 (lane 1) to 0.0193 (lane 10). The expected size of the primary product (932 bp) is indicated. (B) Products of nested PCR using 1 μl of the corresponding primary product as the template, with the expected size of the nested PCR product (546 bp) indicated. Lanes M are molecular weight standards and show the HaeIII digest of phage φX174 DNA. Lanes C, negative controls representing the background human genome DNA as the template without the addition of the plasmid-cloned ehrlichia 16S rDNA.
Specificity of the nested PCR assay.
Positive and negative controls were included with each PCR amplification to ensure the specificity of the assay. Negative controls consisted of human EDTA-blood or serum samples collected from noninfected individuals and were extracted in parallel with the test samples. No products were amplified from any of the negative-control samples (Fig. 2 to 4). All PCR amplifications were repeated in order to confirm the initial results and assess the reproducibility of the assay; the results of the repeat amplifications were identical to the results shown in Fig. 2 to 4 (data not shown). The specificity of the assay was further assessed by attempting to amplify products from non-GE bacteria by using DNA templates purified from infected human EDTA-blood samples or tissue culture-grown bacteria. No products were amplified from other Ehrlichia species that were tested, including E. chaffeensis, E. canis, E. sennetsu, and E. risticii. Also negative were DNAs from the phylogenetically related bacteria Bartonella henselae and Rickettsia rickettsii and from purified DNA products containing Rhizobium sp., Agrobacterium sp., and Mycoplana sp. (data not shown).
DNA sequencing.
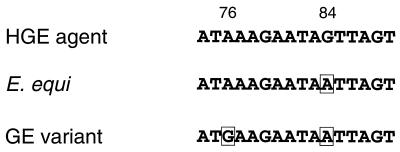
The nested PCR assays amplified a 546-bp portion of the GE 16S rRNA gene (Fig. 1). Each PCR product was sequenced, and the sequences determined for the products from the human blood and serum samples, and from three of the five positive ticks, were identical to each other and to the 16S rRNA gene sequence of the HGE agent previously described by Chen et al. (13). Two of the tick-derived products and all three of the deer PCR products showed sequences that differed from the 16S rDNA sequence of the HGE agent by 2 bases and from the corresponding E. equi sequence by 1 nucleotide. The region near the 5′ end of the 16S rRNA gene is where the GE variant differs from the HGE agent and E. equi. This region of variation is indicated in Fig. 1, and the nucleotides that differ are shown in Fig. 6.
FIG. 6.
GE sequence differences. The region of the 16S rRNA gene where sequence differences were noted is shown for the HGE agent, E. equi, and a novel GE variant. The differing nucleotides are boxed, and the numbers above these are the corresponding base number designations for the HGE agent 16S rDNA sequence reported by Chen et al. (13) (GenBank accession no. U02521). The E. equi sequence was obtained from GenBank (accession no. M73223).
DISCUSSION
HGE produces a wide range of clinical manifestations, from a relatively benign febrile illness to severe and sometimes fatal infections. Tetracycline and doxycycline have been reported to be therapeutically efficacious, and patients generally respond within 48 h of treatment (20). If administered promptly, these antibiotics can effectively prevent severe forms of the illness. Therefore, the prompt diagnosis and treatment of HGE is imperative. Diagnosis of HGE has been based primarily on IFA and PCR-based assays. The nested PCR assay described in this report is both sensitive and specific. The assay confirmed infection with the HGE agent in five of the seven human samples examined. Of these five PCR-positive cases, only one was positive by serologic (IFA) assay, and an isotype-specific IFA assay showed that the response was restricted to the IgM class (titer, 1:512). Follow-up studies by IFA assay on convalescent-phase samples showed that two patients who were PCR positive and IFA negative in this study subsequently seroconverted (41). Convalescent-phase samples were not available for the other two PCR-confirmed cases. These data suggest that PCR-based assays represent a preferable method for evaluating suspected HGE cases during the acute phase of infection. Additionally, some sera from confirmed HGE cases are cross-reactive with antigens of the agent of HME, E. chaffeensis, when assayed by IFA assay (16, 33). PCR-based assays are capable of discriminating between these two closely related organisms that produce infections with very similar clinical presentations.
The nested PCR assay was tested with template DNA purified from human serum samples and resulted in positive results for samples collected from three of the five patients whose whole-blood specimens were positive by PCR. These results support the studies of Pancholi et al. (35) and of Dumler and Bakken (21) by demonstrating that PCR amplification of the HGE agent DNA from human serum is feasible, although not as sensitive as PCR using EDTA-blood. However, PCR represents a valuable adjunct to serologic testing when only an acute-phase serum sample is available, and it will facilitate the confirmation of cases that would otherwise be missed.
The nested assay as described, from DNA extraction through agarose gel electrophoretic analysis, can be completed by an experienced laboratorian within an 8-h workday. A simple, commercially available kit was used for DNA extraction, further simplifying the procedure and making it more amenable to routine use in diagnostic laboratories. Modifications of the assay using colorimetric, fluorescent, and luminescent detection are currently being evaluated and have the potential to further enhance the applicability of the assay. The assay can be used for the routine diagnosis of HGE in human clinical samples and is being used in this manner in the Centers for Disease Control and Prevention (CDC) Rickettsial Laboratory, but it was designed primarily as a research tool to provide optimal sensitivity and specificity with a variety of samples. The sensitivity of the assay requires the use of positive and negative controls for both DNA extraction and PCR amplification, and laboratorians must be trained in proper PCR techniques in order to avoid contamination.
The sensitivity of the PCR amplification was assessed by using a plasmid control and was shown to be fewer than 2 copies of the 16S rRNA gene. Infected granulocytes appear to contain many bacteria when examined by light or electron microscopy, and ehrlichia DNA can often be detected by less-sensitive PCR assays. However, for samples where few bacteria are present, the sensitivity provided by this assay should prove useful. As evidence, single-step PCR assays were negative for each of the seven human serum samples tested in this report, while the nested assay produced positive results from three of these samples. In our laboratory, the assay has not produced equivocal plus/minus results; all positives have been distinguished by products easily visible on ethidium bromide-stained agarose gels. In addition to the HGE cases described in this study, the assay has been used to identify GE DNA and confirm infections in 4 cases when only acute-phase EDTA-blood samples were available and 11 cases when only acute-phase serum samples were available.
The ability of the assay to detect ehrlichia DNA in veterinary samples was demonstrated. The amplification of DNA from I. scapularis ticks collected from Trustom Pond, R.I., showed that 16% (5 of 31) were PCR positive for GE. PCR of DNA from deer blood specimens collected in Prince Georges County, Md., showed that 9.4% (3 of 32) were positive. Recently, it was reported that 50% of I. scapularis ticks collected in Connecticut were PCR positive, although it was not indicated if these positives were confirmed by DNA sequencing, and geographic variations in prevalence were noted (34). The evidence accumulated to date suggests that, like that of other tick-transmitted pathogens, the prevalence of GE in tick populations may vary significantly based on location and seasonal parameters.
The specificity of the assay was examined, and the assay will not amplify products from DNA purified from the other Ehrlichia species that have been tested or from B. henselae or R. rickettsii. The assay will amplify the 16S rDNA of a white-tailed deer agent determined to be an Ehrlichia species that was recently described from Georgia and Oklahoma (reference 14 and data not shown). Although the white-tailed deer agent was not observed in the current study involving deer from Maryland, positive PCR products from deer-derived templates need to be further analyzed by DNA sequencing, particularly because there may be some overlap in the geographic distribution of these agents.
The specificity of the assay was also assessed by sequencing the PCR amplicons. The DNA sequence determined for each of these products was strongly homologous (≥99.4% identity in 497 bp) to previously reported 16S rDNA sequences of GE. The sequences for the products amplified from each of the human blood and serum samples were identical to the published nucleotide sequence of the HGE agent (13). In addition to the samples described in this report, the nested assay has been used by the CDC Rickettsial Laboratory to test more than 500 human samples (EDTA-blood, citrated blood, and serum), and the only products ever amplified (20 total positives, including the 5 in this study) were identical in sequence to the HGE agent 16S rDNA. The sequences for three of the tick-derived amplicons were also identical to each other and to the HGE agent sequence. These data support the correspondence between HGE and I. scapularis as a potential vector of the disease, as previously suggested (35, 39). However, the 16S rRNA gene sequences determined for the PCR products from two of the ticks, and from all three positive white-tailed deer, showed a variant GE sequence that differs from the HGE agent sequence by 2 bases. The sequence of the GE variant we found in Maryland white-tailed deer and Rhode Island ticks was identical to the sequence recently described for an Ehrlichia species from white-tailed deer in Wisconsin (8). The identification of this variant in three diverse locations, and in populations of both white-tailed deer and ticks, suggests that it may be relatively widespread and constant in nature. Whether the GE variant identified in this report causes human or veterinary disease remains to be determined. An isolate of this variant would be valuable and would allow experimental infection studies to address this issue.
A recent report described the PCR amplification of an ehrlichia 16S rDNA sequence from white-footed mice in Minnesota (40). The only difference between this sequence and the 16S rDNA sequence of the HGE agent (GenBank accession no. U02521) is a single deletion of a thymidine residue at position 153 of the HGE agent. This thymidine residue is present in the GE variant described in the present study. It remains to be determined whether the GE variant reported here (found in ticks and deer), the HGE agent, the agent found in Minnesota white-footed mice, E. equi, and E. phagocytophila represent natural variants of a single species. Certainly, the 16S rDNA sequences that have been amplified and sequenced for these organisms are very well conserved, with 1 to 3 nucleotide differences. The identification and DNA sequencing of genetic elements that are more variable than the 16S rDNA will be instrumental in resolving the phylogenetic relationship between the HGE agent and the increasing number of GE variants that are being found in nature.
ACKNOWLEDGMENTS
We are extremely grateful to John Sumner and Dana Jones for DNA sequencing assistance and to the Biotechnology Core Facility of the National Center for Infectious Diseases for oligonucleotide synthesis. We are grateful to Burt Anderson for the design of primers ge2 and ge3, which were modified for this study.
V.B.S. acknowledges support from the American Wildlife Research Foundation.
REFERENCES
- 1.Aguero-Rosenfeld M E, Horowitz H W, Wormser G P, McKenna D F, Nowakowski J, Munoz J, Dumler J S. Human granulocytic ehrlichiosis: a case series from a medical center in New York State. Ann Intern Med. 1996;125:904–908. doi: 10.7326/0003-4819-125-11-199612010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B E, Dawson J E, Jones D C, Wilson K H. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken J S, Dumler J S, Chen S-M, Eckman M R, Van Etta L L, Walker D H. Human granulocytic ehrlichiosis in the upper Midwest United States. A new species emerging? JAMA. 1994;272:212–218. [PubMed] [Google Scholar]
- 4.Bakken J S, Krueth J, Tilden R L, Dumler J S, Kristiansen B E. Serological evidence of human granulocytic ehrlichiosis in Norway. Eur J Clin Microbiol Infect Dis. 1997;15:829–832. doi: 10.1007/BF01701530. [DOI] [PubMed] [Google Scholar]
- 5.Bakken J S, Krueth J, Wilson-Nordskog C, Tilden R L, Asanovich K, Dumler J S. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA. 1996;275:199–205. [PubMed] [Google Scholar]
- 6.Barlough J E, Madigan J E, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genome DNA in horses and ticks (Ixodes pacificus) Vet Parasitol. 1996;63:319–329. doi: 10.1016/0304-4017(95)00904-3. [DOI] [PubMed] [Google Scholar]
- 7.Barlough J E, Madigan J E, DeRock E, Dumler J S, Bakken J S. Protection against Ehrlichia equi is conferred by prior infection with the human granulocytotropic ehrlichia (HGE agent) J Clin Microbiol. 1995;33:3333–3334. doi: 10.1128/jcm.33.12.3333-3334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belongia E A, Reed K D, Mitchell P D, Kolbert C P, Persing D H, Gill J S, Kazmierczak J J. Prevalence of granulocytic Ehrlichia infection among white-tailed deer in Wisconsin. J Clin Microbiol. 1997;35:1465–1468. doi: 10.1128/jcm.35.6.1465-1468.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas B, Mukherjee D, Mattingly-Napier B L, Dutta S K. Diagnostic application of polymerase chain reaction for detection of Ehrlichia risticii in equine monocytic ehrlichiosis (Potomac horse fever) J Clin Microbiol. 1991;29:2228–2233. doi: 10.1128/jcm.29.10.2228-2233.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouqui P, Dumler J S, Lienhard R, Brossard M, Raoult D. Human granulocytic ehrlichiosis in Europe. Lancet. 1995;346:782–783. doi: 10.1016/s0140-6736(95)91544-3. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Human granulocytic ehrlichiosis—New York, 1995. Morbid Mortal Weekly Rep. 1995;44:593–595. [PubMed] [Google Scholar]
- 12.Chang W-L, Pan M-J. Specific amplification of Ehrlichia platys DNA from blood specimens by two-step PCR. J Clin Microbiol. 1996;34:3142–3146. doi: 10.1128/jcm.34.12.3142-3146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S-M, Dumler J S, Bakken J S, Walker D H. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–595. doi: 10.1128/jcm.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson J E, Warner C K, Baker V, Ewing S A, Stallknecht D E, Davidson W D, Kocan A A, Lockhart J M, Olson J G. Ehrlichia-like 16S rDNA sequence from wild white-tailed deer (Odocoileus virginianus) J Parasitol. 1996;82:52–58. [PubMed] [Google Scholar]
- 15.Dawson J E, Anderson B E, Fishbein D B, Sanchez J L, Goldsmith C S, Wilson K H, Duntley C W. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol. 1991;29:2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson J E, Fuller L, Telford S R., III . Abstracts of the 12th Sesqui-Annual Meeting of ASRRD. Pacific Grove, Calif: American Society for Rickettsiology and Rickettsial Diseases; 1996. Serologic cross-reactivity between Ehrlichia chaffeensis and the agent of human granulocytic ehrlichiosis, abstr. 17; p. 17. [Google Scholar]
- 17.Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumler J S, Asanovich K M, Bakken J S, Richter P, Kimsey R, Madigan J E. Serologic cross-reactions among Ehrlichia equi, Ehrlichia phagocytophila, and human granulocytic ehrlichia. J Clin Microbiol. 1995;33:1098–1103. doi: 10.1128/jcm.33.5.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumler J S, Bakken J S. Ehrlichial diseases of humans: emerging tick-borne infections. Clin Infect Dis. 1995;20:1102–1110. doi: 10.1093/clinids/20.5.1102. [DOI] [PubMed] [Google Scholar]
- 21.Dumler J S, Bakken J S. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J Infect Dis. 1996;173:1027–1030. doi: 10.1093/infdis/173.4.1027. [DOI] [PubMed] [Google Scholar]
- 22.Dumler J S, Dotevall L, Gustafson R, Granström M. A population-based seroepidemiologic study of human granulocytic ehrlichiosis and Lyme disease on the west coast of Sweden. J Infect Dis. 1997;175:720–722. doi: 10.1093/infdis/175.3.720. [DOI] [PubMed] [Google Scholar]
- 23.Edelman D C, Dumler J S. Evaluation of an improved PCR diagnostic assay for human granulocytic ehrlichiosis. Mol Diagn. 1996;1:41–49. doi: 10.1054/MODI00100041. [DOI] [PubMed] [Google Scholar]
- 24.Everett E D, Evans K A, Henry R B, McDonald G. Human ehrlichiosis in adults after tick exposure: diagnosis using polymerase chain reaction. Ann Intern Med. 1994;120:730–735. doi: 10.7326/0003-4819-120-9-199405010-00002. [DOI] [PubMed] [Google Scholar]
- 25.Gerwirtz A S, Cornbleet P J, Vugia D J, Traver C, Niederhuber J, Kolbert C P, Persing D H. Human granulocytic ehrlichiosis: report of a case in northern California. Clin Infect Dis. 1996;23:653–654. doi: 10.1093/clinids/23.3.653. [DOI] [PubMed] [Google Scholar]
- 26.Goodman J L, Nelson C, Vitale B, Madigan J E, Dumler J S, Kurtti T J, Munderloh U G. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med. 1996;334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 27.Greig B, Asanovich K M, Armstrong P J, Dumler J S. Geographic, clinical, serologic, and molecular evidence of granulocytic ehrlichiosis, a likely zoonotic disease, in Minnesota and Wisconsin dogs. J Clin Microbiol. 1996;34:44–48. doi: 10.1128/jcm.34.1.44-48.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardalo C J, Quagliarello V, Dumler J S. Human granulocytic ehrlichiosis in Connecticut: report of a fatal case. Clin Infect Dis. 1995;21:910–914. doi: 10.1093/clinids/21.4.910. [DOI] [PubMed] [Google Scholar]
- 29.Heimer R, Van Andel A, Wormser G P, Wilson M L. Propagation of granulocytic Ehrlichia spp. from human and equine sources in HL-60 cells induced to differentiate into functional granulocytes. J Clin Microbiol. 1997;35:923–927. doi: 10.1128/jcm.35.4.923-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iqbal Z, Chaichanasiriwithaya W, Rikihisa Y. Comparison of PCR with other tests for early diagnosis of canine ehrlichiosis. J Clin Microbiol. 1994;32:1658–1662. doi: 10.1128/jcm.32.7.1658-1662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madewell B R, Gribble D H. Infection of two dogs with an agent resembling Ehrlichia equi. J Am Vet Med Assoc. 1982;180:512–514. [PubMed] [Google Scholar]
- 32.Maeda K, Markowitz N, Hawley R C, Ristic M, Cox D, McDade J E. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med. 1987;316:853–856. doi: 10.1056/NEJM198704023161406. [DOI] [PubMed] [Google Scholar]
- 33.Magnarelli L A, Dumler J S, Anderson J F, Johnson R C, Fikrig E. Coexistence of antibodies to tick-borne pathogens of babesiosis, ehrlichiosis, and Lyme borreliosis in human sera. J Clin Microbiol. 1995;33:3054–3057. doi: 10.1128/jcm.33.11.3054-3057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnarelli L A, Stafford III K C, Mather T N, Yeh M-T, Horn K D, Dumler J S. Hemocytic rickettsia-like organisms in ticks: serologic reactivity with antisera to ehrlichiae and detection of DNA of agent of human granulocytic ehrlichiosis by PCR. J Clin Microbiol. 1995;33:2710–2714. doi: 10.1128/jcm.33.10.2710-2714.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pancholi P, Kolbert C P, Mitchell P D, Reed K D, Dumler J S, Bakken J S, Telford S R, Persing D H. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1007–1012. doi: 10.1093/infdis/172.4.1007. [DOI] [PubMed] [Google Scholar]
- 36.Petrovic M, Furlan S L, Zupanc T A, Strle F, Brouqui P, Roux V, Dumler J S. Human disease in Europe caused by a granulocytic Ehrlichia species. J Clin Microbiol. 1997;35:1556–1559. doi: 10.1128/jcm.35.6.1556-1559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodgers S J, Morton R J, Baldwin C A. A serological survey of Ehrlichia canis, Ehrlichia equi, Rickettsia rickettsii and Borrelia burgdorferi in dogs in Oklahoma. J Vet Diagn Invest. 1989;1:154–159. doi: 10.1177/104063878900100212. [DOI] [PubMed] [Google Scholar]
- 38.Sumption K J, Wright D J M, Cutler S J, Dale B A S. Human ehrlichiosis in the UK. Lancet. 1995;346:1487–1488. doi: 10.1016/s0140-6736(95)92502-3. [DOI] [PubMed] [Google Scholar]
- 39.Telford S R, III, Dawson J E, Katavolos P, Warner C K, Kolbert C P, Persing D H. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls J J, Greig B, Neitzel D F, Dumler J S. Natural infection of small mammal species in Minnesota with the agent of human granulocytic ehrlichiosis. J Clin Microbiol. 1997;35:853–855. doi: 10.1128/jcm.35.4.853-855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong S J, Brady G S, Dumler J S. Serological responses to Ehrlichia equi, Ehrlichia chaffeensis, and Borrelia burgdorferi in patients from New York State. J Clin Microbiol. 1997;35:2198–2205. doi: 10.1128/jcm.35.9.2198-2205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh M-T, Mather T N, Coughlin R T, Gingrich-Baker C, Summer J W, Massung R F. Serologic and molecular detection of granulocytic ehrlichiosis in Rhode Island. J Clin Microbiol. 1997;35:944–947. doi: 10.1128/jcm.35.4.944-947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]