Abstract
Cancer stem cells (CSCs) are major drivers of metastasis, drug resistance and recurrence in numerous cancers. However, critical factors that can modulate CSC stemness have not been clearly identified. Nuclear receptor subfamily 2 group E member 3 (nr2e3) expression has been previously reported to be positively associated with drug sensitivity and favorable clinical outcomes in patients with estrogen receptor (ER)+ breast cancer. This suggests that nr2e3 expression may be inversely associated with CSC stemness in this type of tumor cells. The present study aimed to investigate the regulatory roles of NR2E3 in the stem-like properties of ER+ breast cancer cells and to identify the underlying mechanisms. Bioinformatics analysis was performed using the data derived from the Cancer Genome Atlas database. Nr2e3-specific shRNA and nuclear receptor subfamily 2 group C member 2 (nr2c2) overexpressed plasmids were constructed to silence and enhance the expression of nr2e3 and nr2c2, respectively. Transwell and wound healing experiments were conducted to evaluate the migration and invasion ability of MCF7 cells, while colony formation tests were used to evaluate the clonality. Flow cytometry was used to detect the percentage of CD44+CD24-/low cells. Reverse transcription-quantitative PCR and western blotting were performed to detect expression at the mRNA and protein levels. The results showed that compared with normal breast tissues and MCF10A cells, the expression of nr2e3 was increased in ER+ breast tumor tissues and cell lines. Nr2e3 silencing promoted the migration, invasion and colony-forming ability of the ER+ MCF7 cells. It also increased the expression of epithelial-mesenchymal transition markers and stem cell-related transcription factors, in addition to the percentage of CD44+CD24-/low cells. The expression of nr2e3 and nr2c2 was found to be positively correlated. Nr2e3 knockdown decreased the mRNA and protein expression levels of nr2c2, whereas nr2c2 overexpression reversed the elevated CD44+CD24-/low cell ratio and the increased migratory activity caused by nr2e3 silencing. The results of the present study suggest that NR2E3 may serve an important role in modulating the stem-like properties of ER+ breast cancer cells, where NR2E3/NR2C2 signaling may be a therapeutic target in ER+ breast cancer.
Keywords: estrogen receptor-positive breast cancer, stem-like properties, nuclear receptor, nuclear receptor subfamily 2 group E member 3, nuclear receptor subfamily 2 group C member 2
Introduction
Nuclear receptor subfamily 2, group E member 3 (NR2E3) serves an important function in retinal photoreceptor cell development and maintenance (1). Mutations in the human nr2e3 gene have been reported to cause several retinal degenerative diseases, such as enhanced S-cone syndrome and retinitis pigmentosa (2). In recent years, research on the molecular function of NR2E3 in other tissues and its role in various diseases, such as liver injury and breast cancer, has been attracting attention (3,4). In estrogen receptor (ER)+ breast cancer, the level of nr2e3 expression has been found to be positively associated with recurrence-free survival. In addition, patients with higher nr2e3 expression tended to be more sensitive to tamoxifen treatment, which in turn confers more positive clinical outcomes compared with those with lower nr2e3 expression (5). Nr2e3 is typically expressed at low levels in ER- breast cancer tissues and its overexpression induces cancer cell growth, invasion and metastasis (6). In addition, elevated levels of nr2e3 expression have been associated with improved clinical prognosis in patients with hepatic carcinoma, and with the occurrence and progression of lung carcinoma and pancreatic cancer (7-9). These findings suggest that outside the retina, NR2E3 can serve biological functions in cancers.
The cancer stem cell (CSC) theory hypothesizes that a subgroup of malignant cancer cells is endowed with stem-like properties that are undifferentiated, express self-renewing capacities and can replenish other differentiated bulk tumor cells (10). Clinically, conventional radiotherapy and drug treatments can terminate the majority of differentiated tumor cells, but they have limited efficacy against CSCs. CSCs also appear to form the basis of tumor heterogeneity and the pathological cause of tumor growth, drug resistance, metastasis and recurrence (11,12). In this regard, promoting the differentiation of CSCs into drug-sensitive cancer cells, known as differentiation therapy, has been proposed to be a promising treatment strategy for eradicating cancer cells (13-15). Therefore, identifying novel biomarkers that match the specific molecular signatures of cancer cells to broaden the target spectrum may prove beneficial.
The present study hypothesized that nr2e3 is expressed at higher levels in differentiated tumor cells in ER+ breast cancer compared with breast cancer stem cells based on the following considerations: i) CSCs form a fraction of the tumor cell population that can differentiate into the majority of the bulk tumor cell types, which can then contribute to poor prognosis and drug resistance (16); ii) patients with breast cancer with higher nr2e3 expression levels tend to have superior clinical outcomes and exhibit favorable responses to tamoxifen treatment (5); and iii) Nr2e3 expression was previously found to be increased in ER+ breast tumor tissues compared with that in normal and ER- breast tissues (17). In addition, it was hypothesized that NR2E3 may facilitate the differentiation of breast CSCs into bulk tumor cells, not too dissimilar to its activity in promoting the differentiation of rod photoreceptors from retinal pluripotent cells in the retina (18). Therefore, the present study aimed to investigate the relationship between nr2e3 expression and the stem-like characteristics of ER+ breast cancer cells to evaluate the suitability of NR2E3 as a diagnostic and therapeutic biomarker for ER+ breast cancer.
Materials and methods
Cell culture
Human normal mammary epithelial cells MCF10A (cat. no. CL-0525) and the ER+ cell line MCF7 (cat. no. CL-0149) were purchased from Procell Life Science & Technology Co., Ltd. The two cell lines were cultured in their cell-specific complete medium (cat. nos. CM-0525 for MCF10A and CM-0149 for MCF7; Procell Life Science & Technology Co., Ltd.,). When MCF10A cells were cultured, 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 µg/ml hydrocortisone, 1% non-essential amino acid (NEAA), 10 µg/ml insulin and 1% penicillin-streptomycin solution (P/S) were added to DMEM/F12 medium. When MCF7 cells were cultured, 10% fetal bovine serum (FBS), 1% NEAA, 10 µg/ml insulin and 1% P/S were added to the MEM. According to a previous study (19), paclitaxel-resistant MCF7 cells were cultured with 10 µg/ml paclitaxel (cat. no. ab120143; Abcam) over 6 months. All cells were cultured at 37˚C with 5% CO2.
Plasmids and transfection
The negative control (NC) and nr2e3 short-hairpin RNAs (shRNAs) were designed and purchased from RiboBio Co., Ltd. The target sequences (Table I) were cloned into the pRNAT-U6.1/Neo plasmid (RiboBio Co., Ltd.). The full-length cDNA sequence of human nuclear receptor subfamily 2 group C member 2 (nr2c2) was cloned into the pEGFP-N1 vector (Miaoling Biotechnology Co., Ltd.). Lipofectamine™ 3000 reagents (cat. no. L300000; Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect the shRNA-carrying plasmids and/or the nr2c2-overexpression plasmids into MCF7 cells. Plasmids were incubated with Lipofectamine™ 3000 reagents and Opti-MEM™ (cat. no. 11058021; Invitrogen; Thermo Fisher Scientific, Inc.) for 20 min at room temperature. Dishes with a diameter of 35 mm and 15 mm were transfected with 6 and 4 µg of plasmids, respectively. For the nr2c2 overexpression experiment, MCF7 cells were co-transfected with nr2e3 shRNA plasmids and the nr2c2-overexpression vector. 36 h after transfection, cells were harvested for subsequent experiments.
Table I.
shNC and NR2E3-specific shRNA sequences.
| shRNA used | shRNA sequence (5'-3') |
|---|---|
| shNC | Sense: CAACAAGATGAAGAGCACCAA |
| Antisense: TTGGTGCTCTTCATCTTGTTG | |
| shRNA1 | Sense: GAAGGATCCTGAGCACGTA |
| Antisense: TACGTGCTCAGGATCCTTC | |
| shRNA2 | Sense: GGGAAGCACTATGGCATCT |
| Antisense: AGATGCCATAGTGCTTCCC | |
| shRNA3 | Sense: CATGGCCAGCCTTATAACA |
| Antisense: TGTTATAAGGCTGGCCATG |
NC, negative control; nr2e3, nuclear receptor subfamily 2 group E member 3; sh, short hairpin RNA.
The Cancer Genome Atlas (TCGA) data analysis
According to our previous study (19), the expression data of nr2e3 and nr2c2 in 808 ER+ breast tumor and 113 normal tissues were downloaded from the TCGA database (https://portal.gdc.cancer.gov/; version 32.0) for bioinformatics analysis. Furthermore, the expression profile of nr2e3 in 1,109 breast tumor samples that were not categorized according to ER content was also downloaded from the TCGA database. BRB-Arra/Tools (http://brb.nci.nih.gov/BRB-ArrayTools/download.html, version 4.6.2) was used to analyze these data. Briefly, data collation and gene labeling modules were used, differentially expressed genes in the dataset were screened with P<0.05 and logFC >1 as criteria, and hierarchical cluster analysis was performed.
MTT assay
The MTT cytotoxicity assay was conducted using an MTT Cell Proliferation and Cytotoxicity Detection kit (cat. no. C0009S; Beyotime Institute of Biotechnology). Briefly, ~2x103 MCF7 cells transfected with either NR2E3 shRNAs or shNC were seeded into 96-well plates, before 10 µl MTT reagent was added. After incubation at 37˚C for 4 h, 100 µl formazan solvent (DMSO) was added. After incubation for another 4 h (at 37˚C), absorbance at 570 nm was measured.
Wound healing assay
MCF7 cells were cultured in complete medium. shRNA-transfected MCF7 cell monolayers were scratched when the confluence reached ~80%. Scratched cells were serum starved, and incubated at 37˚C with 5% CO2. Images were captured at 0 and 24 h post-scratching using an inverted fluorescent microscope (cat. no. CKX53; Olympus Corporation) under the same magnification.
Two-dimensional colony formation assay
A total of 1,000 transfected MCF7 cells were seeded into 6-cm dishes, maintained in complete medium, and incubated for 2 weeks at 37˚C with 5% CO2. Colonies were fixed with 4% paraformaldehyde (cat. no. P0099; Beyotime Institute of Biotechnology) for 30 min and stained with crystal violet (cat. no. C0121; Beyotime Institute of Biotechnology) for 10 min, all at room temperature. Images were taken using an Ordinary camera (cat. no. VlogR7; Canon Corporation). Colonies that contain more than 50 cells were counted manually.
Transwell assays
Transwell chambers (cat. no. 3422; BD Biosciences) were used to investigate cell migration capacities. In every well, a total of ~2x103 cells were resuspended in 200 µl serum-free DMEM and placed into the upper chamber, whereas 600 µl complete medium with 10% FBS was added to the lower chamber. After incubation for 24 h at 37˚C, the cells were fixed in 4% formaldehyde for 30 min and stained with crystal violet for 10 min, both at room temperature. The number of cells on the underside of the membrane was counted under an Olympus CKX53 inverted microscope at x200 magnification, before cells in three randomly selected fields were counted.
Quantitative PCR (qPCR)
MCF7 cells that were transfected with nr2e3 shRNAs were harvested when the confluence reached ~90%. RNAiso Plus Reagent (cat. no. 9108; Takara Bio Inc.) was used to extract the total RNA. cDNA was synthesized through reverse transcription using the BeyoRT™ II kit (cat. no. D7168M; Beyotime Institute of Biotechnology). Briefly, total RNA, oligo(dT)18 primer, reaction buffer, RNase inhibitor, dNTP Mix, BeyoRT™ II M-MLV and DEPC-treated water formed a reaction system. Following the manufacturer's instruction, this system was incubated at 42˚C for 60 min, and then for 10 min at 80˚C. For the qPCR experiment, AceQ™ qPCR SYBR® Green Master Mix (cat. no. Q111-02; Vazyme Biotechnology Co., Ltd.) was used. According to the manufacturer's instructions, thermocycling conditions for PCR were 95˚C for 10 sec and then 60˚C for 30 sec. The relative mRNA expression was calculated using the 2-ΔΔCq method. The primer sequences used for qPCR are listed in Table II. GAPDH was used as the normalization control.
Table II.
Primers used for reverse transcription-quantitative PCR.
| Gene (accession number) | Primer sequence (5' to 3') |
|---|---|
| Nr2e3 | F: GATCCTGAGCACGTAGAGGC |
| (NM_016346.4) | R: GCAATTTCCCAAACCTCACGG |
| Nr2c2 | F: GGCGCCAAATCCTGAGGTAA |
| (NM_003298.5) | R: GGTGAGGCTACAGCAGAGTC |
| Nanog | F: TCCTCCTCTTCCTCTATACTAAC |
| (NM_024865.4) | R: CCCACAAATCACAGGCATAG |
| Klf4 | F: ATCTCGGCCAATTTGGGGTT |
| (NM_004235.6) | R: CCAGGTGGCTGCCTCATTA |
| Oct4 | F: ATCGAGAACCGAGTGAGA |
| (NM_002701.6) | R: ACACTCGGACCACATCCTT |
| Sox2 | F: GGGAAATGGGAGGGGTGCAAAAGAGG |
| (NM_003106.4) | R: TTGCGTGAGTGTGGATGGGATTGGTGT |
| E-cadherin | F: CCTCCAGAGTTTACTGCCATGAC |
| (NM_001792.5) | R: GTAGGATCTCCGCCACTGATTC |
| N-cadherin | F: GGCGCCACCTGGAGAGA |
| (NM_004360.5) | R: TGTCGACCGGTGCAATCTT |
| Vimentin | F: TACAGGAAGCTGCTGGAAGG |
| (NM_003380.5) | R: ACCAGAGGGAGTGAATCCAG |
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT |
| (NM_002046.7) | R: GGCTGTTGTCATACTTCTCATGG |
Nr2e3, nuclear receptor subfamily 2 group E member 3; Nr2c2, nuclear receptor subfamily 2 group C member 2; Nanog, nanog homeobox; Klf4, Krüppel-like factor 4; Oct4, POU class 5 homeobox 1; Sox2, SRY-box transcription factor 2; E-cadherin, cadherin 1; N-cadherin, cadherin 2. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward primer sequence; R, Reverse primer sequence.
Western blotting
MCF7 cells that were transfected with nr2e3 shRNAs were harvested when the confluence reached ~90%. Total proteins were extracted using cell lysis buffer for western blotting (cat. no. P0013; Beyotime Institute of Biotechnology). The protein concentration was determined with a bicinchoninic acid kit (cat. no. P0010; Beyotime Institute of Biotechnology). A 10% gel was used for electrophoresis and each lane was loaded with 20 µg protein. Proteins were then transferred to a polyvinylidene fluoride membrane (cat. no. FFP78; Beyotime Institute of Biotechnology), and incubated in blocking buffer (cat. no. P0023B; Beyotime Institute of Biotechnology) for 2 h at room temperature. After washing with TBST buffer (containing 20% Tween), the membranes were then incubated with the primary and secondary antibodies (primary antibodies, 1:500; secondary antibodies, 1;10,000). A visualization reagent (cat. no. KF8005; Affinity Biosciences) was the applied. The primary and secondary antibodies used for western blotting are listed in Table III. GAPDH was used as the normalization control.
Table III.
Antibodies used for WB and FCM.
| Protein | Experiment | Company | Cat. no. |
|---|---|---|---|
| NR2E3 | WB | Proteintech Group, Inc. | 14246-1-AP |
| NR2E3 | WB | Santa Cruz Biotechnology, Inc. | sc-374513 |
| E-cadherin | WB | ProteinTech Group, Inc. | 20874-1-AP |
| N-cadherin | WB | ProteinTech Group, Inc. | 22018-1-AP |
| VIMENTIN | WB | Affinity Biosciences, Ltd. | BF8006 |
| SLUG | WB | Affinity Biosciences, Ltd. | AF4002 |
| NR2C2 | WB | ABclonal Biotech Co., Ltd. | A6422 |
| LSD1 | WB | ABclonal Biotech Co., Ltd. | A1156 |
| H3K4me2 | WB | ABclonal Biotech Co., Ltd. | A2356 |
| GAPDH | WB | ProteinTech Group, Inc. | 60004-1-Ig |
| Goat anti-rabbit IgG | WB | Proteintech Group, Inc. | SA00001-2 |
| Goat anti-mouse IgG | WB | Proteintech Group, Inc. | SA00001-1 |
| CD44-FITC | FCM | Invitrogen; Thermo Fisher Scientific, Inc. | 11-0441-82 |
| CD24-PE | FCM | Invitrogen; Thermo Fisher Scientific, Inc. | 12-0247-42 |
NR2E3, nuclear receptor subfamily 2 group E member 3; E-cadherin, cadherin 1; N-cadherin, cadherin 2; NR2C2, nuclear receptor subfamily 2 group C member 2; LSD1, Lysine-specific histone demethylase 1A; H3K4me2, Histone H3 lysine 4 dimethylation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WB, western blotting; FCM, flow cytometry.
JASPAR prediction
The online prediction software JASPAR (https://jaspar.genereg.net) was used to predict the NR2E3 binding site on the nr2c2 gene promoter. The human nr2c2 gene promoter sequence (https://www.ncbi.nlm.nih.gov/nuccore/NC_000003.12?from=14947583&to=15049273&report=fasta) with FASTA format was scanned at 2 kbp length.
Flow cytometry
A total of ~1x106 nr2e3-silenced MCF7 cells and the negative control cells, as well as the nr2c2-overexpressed MCF7 cells were incubated with 5 µl FC receptor blocker (cat. no. abs9476; Absin Bioscience, Inc.) at 4˚C for 10 min, and then incubated on ice with 0.25 µg FITC-conjugated CD44 and 0.25 µg PE-conjugated CD24 antibodies for 30 min. The percentages of CD44+CD24-/low subgroup cells were detected using flow cytometry with the Beckman CytoFlex system (Beckman Coulter, Inc.) and analyzed using FlowJo software (version 10.8.1). The antibodies used for flow cytometry are listed in Table III.
Statistical analysis
All data were analyzed using GraphPad Prism (version 6; Dotmatics). One-way ANOVA followed by Tukey's post hoc test was used for all comparisons. Data are presented as the mean ± SD. All data were obtained from at least three independent experiments. P<0.05 was used to indicate a statistically significant difference.
Results
Expression of nr2e3 is increased in ER+ breast cancer tissues and cell lines
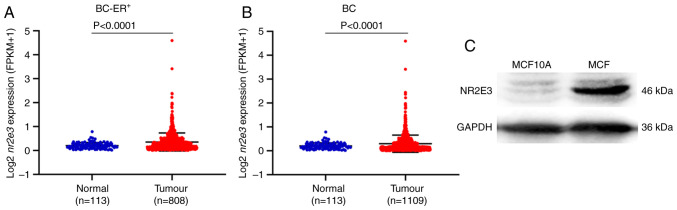
To investigate the biological roles of NR2E3 in ER+ breast cancer, its expression was first investigated in ER+ breast cancer tissues compared with that in normal breast tissue samples. Data from 808 ER+ breast tumors and 113 normal samples were downloaded from TCGA database, where the subsequent analysis revealed that the expression of nr2e3 was significantly higher in the tumor samples (Fig. 1A). When the tumor types were not categorized according to the ER content, the expression of nr2e3 remained significantly higher in breast cancer tissues (Fig. 1B). The protein expression level of NR2E3 was found to be elevated in the ER+ cell line MCF7 compared with that in the MCF10A normal human breast epithelial cell line (Fig. 1C). These results suggest that nr2e3 expression is increased in ER+ breast cancer and tumor types not categorized according to the ER content.
Figure 1.
Expression of nr2e3 is upregulated in ER+ breast adenoma tissue cells. Nr2e3 expression was analyzed in (A) ER+ and (B) total breast adenoma tissues compared with normal tissue samples. Data were downloaded from The Cancer Genome Atlas database. (C) Western blotting showing the protein expression levels of NR2E3 in MCF10A and MCF7 cells, with GAPDH used as a loading control. BC, breast cancer; ER, estrogen receptor; NR2E3, nuclear receptor subfamily 2 group E member 3; FPKM, fragments per kilobase of exon model per million mapped fragments.
Nr2e3 silencing promotes the migration, invasion and colony-formation by MCF7 cells
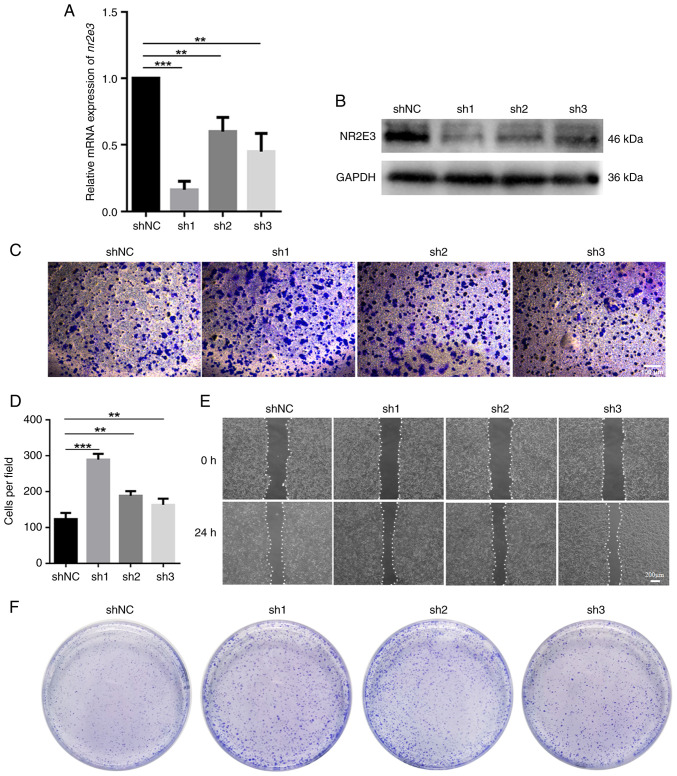
The association between nr2e3 expression and the migration, invasion and colony-formation of MCF7 cells was next assessed as an indication of the stemness property of CSCs in vitro. In total, three shRNAs targeting nr2e3 were designed and transfected into MCF7 cells. None of the three nr2e3-specific shRNAs, sh1, sh2 and sh3, nor the shNC, exhibited cytotoxicity towards MCF7 cells according to the MTT assay (Fig. S1A). In addition, mRNA and protein expression levels of nr2e3 were found to be significantly decreased in shRNA-transfected cells compared with those in the shNC group (Fig. 2A and B). Among them, sh1 exerted the highest silencing effect and so was selected for use in further experiments. The results of the Transwell and wound healing assays showed that nr2e3 knockdown markedly increased both their migratory and invasive capabilities (Figs. 2C-E and S1B). Two-dimensional colony formation tests showed that silencing nr2e3 expression markedly promoted colony-formation (Figs. 2F and S2A). Considering that CSCs contribute to the migration and invasion ability of tumor cells (12), while nr2e3 knockdown increases the migration and invasion ability of MCF7 cells and promotes the colony formation of MCF7 cells, it can be inferred that nr2e3 knockdown enhances the stem cell-like properties of ER+ tumor cells.
Figure 2.
Efficiency of nr2e3 shRNA transfection on the migration, invasion and colony-formation of MCF7 cells. Evaluation of the effectiveness of nr2e3-targetting shRNAs using (A) reverse transcription-quantitative PCR and (B) western blotting, with GAPDH used as a control. (C) Representative images and (D) evaluation of the migratory ability using Transwell assays. The migratory cells were observed using an Olympus CKX53 inverted microscope at x200 magnification. Scale bar, 100 µm. (E) Evaluation of the migratory ability was examined by wound healing experiments. (F) Evaluation of the colony-formation ability. **P<0.01 and ***P<0.001. C, control; NR2E3, nuclear receptor subfamily 2 group E member 3; ns, no significance; shRNA or sh, short hairpin RNA.
In addition, paclitaxel treatment terminated most differentiated tumor cells, whereas the ratio of stem-like cells was elevated in paclitaxel-resistant (PR) breast tumor cells (20). In the present study, it was found that nr2e3 was expressed at lower levels in MCF7-PR cells (Fig. S2B). This finding supported the notion that nr2e3 is mainly expressed in differentiated tumor cells, whereas in stem-like breast tumor cells nr2e3 expression is low.
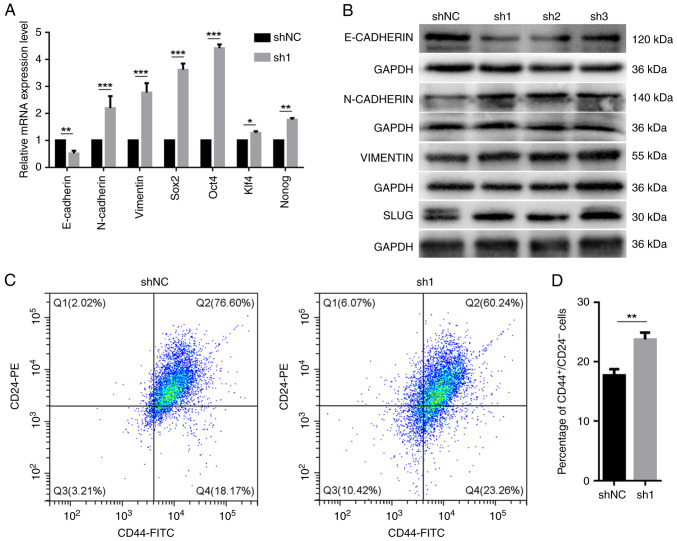
nr2e3 silencing promotes epithelial-mesenchymal transition (EMT), enhances the expression of stem cell-related transcription factors and increases the proportion of CD44+CD24-/low cells
Tumor cells that undergo epithelial-mesenchymal transition (EMT) usually have enhanced migration, invasion and drug resistance. Additionally, EMT gives carcinoma cells the capacity to renew themselves and increases the ratio of tumor stem cells (21). Thus, the relationship between nr2e3 expression and the expression of EMT-related marker genes was next evaluated. It was shown that nr2e3 knockdown significantly reduced the mRNA expression levels of E-cadherin, but increased the expression of N-cadherin and vimentin mRNA (Fig. 3A). Similar directions of changes in the protein expression levels of E-cadherin, N-cadherin and vimentin were found using western blotting (Figs. 3B and S3). The protein expression of slug, a transcription factor that can promote EMT (22), was also shown to be increased when nr2e3 expression was knocked down (Figs. 3B and S3). These results support the hypothesis that nr2e3 knockdown can promote the EMT process.
Figure 3.
Effects of nr2e3 silencing on the expression of EMT markers, stem cell-related transcription factors and the proportion of CD44+CD24-/low cells. Detection of the mRNA and protein expression levels of EMT markers and stem cell-related transcription factors using (A) reverse transcription-quantitative PCR and (B) western blotting, respectively, with GAPDH used as the endogenous control. (C) Proportion of CD44+CD24-/low subgroup cells was analyzed using flow cytometry. (D) Semi-quantification of the expression ratio from (C). *P<0.05, **P<0.01 and ***P<0.001. C, control; EMT, epithelial-mesenchymal transition; NR2E3, nuclear receptor subfamily 2 group E member 3; sh, short hairpin RNA; klf4, Krüppel-like factor 4.
Furthermore, nr2e3 knockdown was found to significantly increase the mRNA expression levels of stem cell-associated transcription factors sox2, oct4, nanog and Kruppel-like factor 4 (Fig. 3A). The association between NR2E3 and the proportion of the CD44+CD24-/low population was investigated, where it was found that nr2e3 knockdown significantly enhanced the proportion of CD44+CD24-/low in MCF7 tumor cells (Fig. 3C and D).
NR2C2 is a potential downstream target of NR2E3
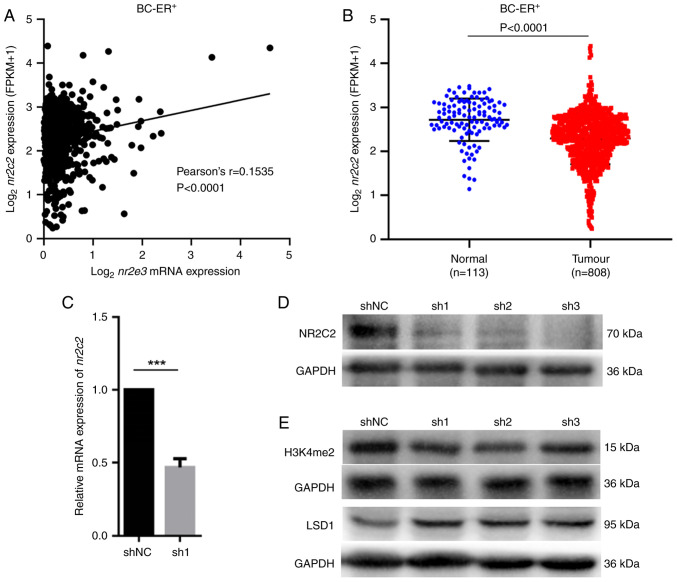
Using genome-wide chromatin immunoprecipitation assays and publicly available database analysis (NCI-60 database), Park et al (5) previously showed that NR2E3 can directly regulate the expression of the ERα (esr1) gene. In addition to ESR1, NR2E3 also has the highest number of correlated genes encoding nuclear receptors, such as peroxisome proliferator activated receptor α (PPARA), thyroid hormone receptor α (THRA), estrogen related receptor α (ESRRA), hepatocyte nuclear factor 4α (HNF4A) and NR2C2, suggesting that there may be further cross-talk between NR2E3 and these nuclear receptors (5). In the present study, using TCGA database, the correlation between mRNA expression of nr2e3 and ppara, thra, esrra, hnf4a and nr2c2 in ER+ breast tumor cells were evaluated, where it was found that there was little or no correlation between nr2e3 expression and the expression of ppara, thra, esrra and hnf4a (Fig. S4). By contrast, a significant positive correlation between the mRNA expression of nr2e3 and nr2c2 was found (Fig. 4A). Compared with that in the normal breast samples, nr2c2 expression was significantly lower in ER+ breast cancer tissues compared with that in normal tissues (Fig. 4B).
Figure 4.
Nr2e3 knockdown decreases the expression level of nr2c2. (A) Correlation analysis between the mRNA expression levels between nr2e3 and nr2c2 in ER+ breast cancer tissues. Data were downloaded from TCGA database. (B) Nr2c2 expression was investigated in ER+ breast tumor samples compared with normal specimens from TCGA database. Expression levels of nr2c2 (C) mRNA and (D) protein were detected in nr2e3-silenced MCF7 and shNC-transfected cells using reverse transcription-quantitative PCR and western blotting, respectively, with GAPDH used as the control. ***P<0.001. (E) Protein expression levels of LSD1 and H3K4me2 were detected in NR2E3-depleted MCF7 and shNC cells using western blotting. GAPDH served as the endogenous control. BC, breast cancer; ER, estrogen receptor; LSD1, lysine-specific histone demethylase 1A; NR2C2, nuclear receptor subfamily 2 group C member 2; NR2E3, nuclear receptor subfamily 2 group E member 3; TCGA, The Cancer Genome Atlas.
To determine if NR2E3 can directly regulate nr2c2 expression, the mRNA and protein levels of nr2c2 were detected in nr2e3-silenced MCF7 cells. It was shown that nr2e3 silencing markedly reduced nr2c2 expression (Fig. 4C and D). JASPAR (https://jaspar.genereg.net) was used to scan the 2 kb promoter sequence upstream from the transcriptional start site of the nr2c2 gene to identify an NR2E3 binding site. In total, two predicted NR2E3 binding sites were identified on the proximal promoter of the nr2c2 gene (-1748 to -1742 bp, and -1516 to -1510 bp), with the predicted binding sequence being xAAGCTT (x represents nucleotide A, T, C or G; Fig. S5). This suggested that NR2E3 can directly bind to the nr2c2 promoter to regulate its transcription. Furthermore, nr2e3 knockdown was found to decrease the expression levels of the active histone marker histone H3 lysine 4 dimethylation (H3K4me2), in addition to markedly increasing the protein expression level of lysine-specific histone demethylase 1A (lsd1) (Fig. 4E), which usually retains the suppressive histone status (9). These results suggest that NR2E3 may also serve a role as an epigenetic modification factor that can sustain nr2c2 promoter chromatin accessibility. Taken together, these findings suggest that the orphan nuclear receptor NR2C2 may be implicated in the NR2E3 signaling pathway upstream of the regulation of ER+ breast cancer cell physiology.
Nr2c2 overexpression decreases the proportion of CD44+CD2-/low cells and suppresses migratory activity
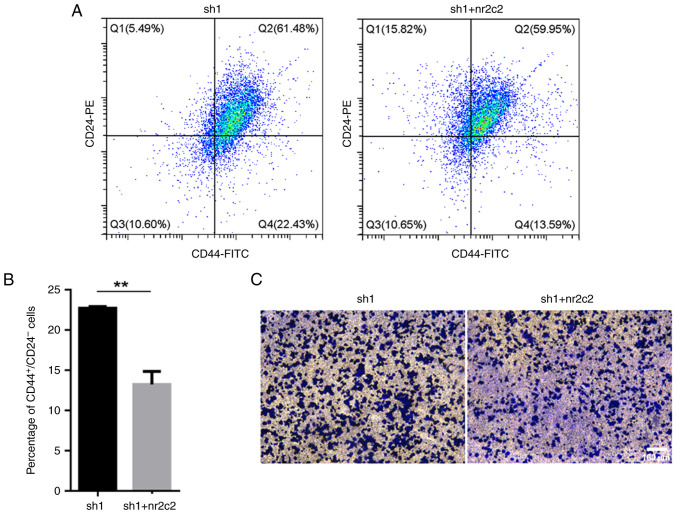
To determine if NR2E3 can modulate the stem-like characteristics of ER+ breast carcinoma cells through NR2C2, vectors containing the full-length coding sequence of the human nr2c2 cDNA were transfected into the nr2e3-silenced MCF7 cells. The protein expression levels of nr2c2 were found to be increased following nr2c2 overexpression (Fig. S6). It was then observed that nr2c2 overexpression reversed the elevated ratio of both CD44+CD24-/low cells and the increased number of migratory cells caused by nr2e3 silencing (Fig. 5A-C). These findings suggest that an NR2E3/NR2C2 network can modulate the stem-like activities of ER+ breast tumor cells.
Figure 5.
Nr2c2 overexpression reverses the elevated proportion of CD44+CD24-/low cells and the increased migratory ability originally induced by nr2e3 silencing. (A) Flow cytometry was used to analyze the proportion of CD44+CD24-/low subgroups in nr2c2-overexpressing MCF7 tumor cells with nr2e3 expression knocked down. (B) Semi-quantification of the CD44+CD24-/low ratio in (A). (C) Migratory activity of MCF7 cells following nr2c2 overexpression and nr2e3 silencing was analyzed using Transwell assay. The migratory cells were observed using an Olympus CKX53 inverted microscope at x200 magnification. Scale bar, 100 µm. **P<0.01. sh, short hairpin RNA; NR2C2, nuclear receptor subfamily 2 group C member 2; NR2E3, nuclear receptor subfamily 2 group E member 3.
Discussion
Nr2e3 was initially thought to be uniquely expressed in the retinal photoreceptor cells, it was therefore also called the photoreceptor-specific nuclear receptor (23). However, nr2e3 has also been reported to be expressed in other tissues, such as liver, mammary-glands, adrenal gland, thyroid gland, prostate, testis, uterus, trachea, digestive tract and salivary glands (4,24,25). In addition, its expression has been reported in several cancer cell lines, including the Y79, HepG2, MCF7, T47D, HeLa and HCT116. Nr2e3 expression has been associated with the occurrence, progression and drug sensitivity depending on the cancer type (5-9,26,27).
Proteins specifically designed for the development of photoreceptors in the retina are employed to direct the proliferation of tumor cells. Neuroretinal leucine zipper protein and cone-rod homeobox transcription factor, two pivotal transcription factors that can form functional complexes with NR2E3 to regulate photoreceptor differentiation, are closely associated with the growth of the medulloblastoma (28). In patients with liver cancer and ER+ breast carcinoma, high levels of nr2e3 expression are associated with favorable clinical outcomes and higher sensitivity to tamoxifen treatment (5,9). In ER- breast cancer, nr2e3 overexpression has been previously found to induce migration and metastasis (6), suggesting that NR2E3 serves a tumorigenic and antineoplastic function influenced by the molecular environment.
In the present study, nr2e3 expression was found to be increased in ER+ breast cancer tissues and cell lines, which is consistent with previously reported data from TaqMan PCR assays and data re-elaboration (4,17). In the present study, nr2e3 silencing promoted EMT progression, increased the ratio of CD44+CD24-/low cells and promoted the expression of stem cell-related transcription factors. By contrast, knocking down nr2e3 expression enhanced the ability of migration, invasion and colony formation of ER+ MCF7 cells. Data in the present study also verified that nr2e3 expression is inversely associated with the stem-like properties of ER+ breast tumor cells. In addition, changes to the stem-like properties of the MCF7 cells appeared to be in part mediated by the regulation of nr2c2 expression. Therefore, the present study provided a novel finding that the NR2E3/NR2C2 nuclear receptor network can modulate the physiological behaviors of breast cancer cells.
In retinal cells, NR2E3 mediates the expression of photoreceptor genes such as rhodopsin and gnat1 on the transcriptional level (29). In tumor cells, NR2E3 can function as an epigenetic modulator to regulate the chromatin accessibility of target genes, such as esr1, aryl hydrocarbon receptor and long non-coding RNA damage-induced noncoding (3,9,30). NR2E3 can also regulate protein activity through post-translational modifications. NR2E3 has been reported to enhance the stability of p53 proteins by increasing acetylation, thereby strengthening p53 signaling (27), These results suggest that NR2E3 can modulate signal transduction on pre-transcriptional, transcriptional and post-translational levels.
The expression of nr2e3 and nr2c2 mRNA was positively correlated in ER+ breast carcinoma, although not as high as the correlation between nr2e3 and esr1 (5). In the present study, nr2e3 knockdown markedly downregulated the mRNA and protein expression of nr2c2. Mechanistically, several predicted binding sequences (for example, xAAGCTT) of NR2E3 were predicted at the proximal promoter of the nr2c2 gene (-1748 to -1742 bp, and -1516 to -1510 bp), suggesting that NR2E3 may directly activate nr2c2 transcription. nr2c1, a homologous gene that is associated with nr2c2 and with high degrees of sequence homology (the overall structural identity is 65%, and the DNA binding domain is 82%), was previously identified as a direct target of NR2E3 (31,32). The consensus sequence AAGTCA recognized by NR2E3 proteins in retinal photoreceptors is also present on the nr2c2 promoter (33). Further studies to determine the binding sequences of NR2E3 in ER+ breast cancer cells are warranted using specific antibodies in chromatin immunoprecipitation experiments. Another potential mechanism by which NR2E3 can regulates nr2c2 expression could be by the modulation of chromatin accessibility, since nr2e3 knockdown was found to increase the expression of LSD1 whilst decreasing that of the active histone marker H3K4me2, consistent with previous findings (3,9,30).
Nr2c2 is ubiquitously expressed in the human brain, lung, kidney, skeletal muscle, prostate, ovary and testis, where they serve as a factor in neuronal development, glucose metabolism, hematogenesis and spermatogenesis (34). Since it is abundantly expressed in testicular tissues, it is also called testicular orphan nuclear receptor 4(35). Depending on the tumor type, NR2C2 may function as a tumorigenic or tumor-suppressive factor. In prostatic carcinoma, non-small-cell lung carcinoma and malignant neuroglioma, NR2C2 was found to enhance the migratory and infiltrative capabilities of tumor cells (35-37). In hepatocellular carcinoma and bladder cancer, the opposite effect is observed (38,39). Consistent with a previously reported TaqMan array analysis (17), the present study showed that nr2c2 was expressed at lower levels in ER+ breast tissues. In ER+ breast carcinoma, NR2C2 breaks the ER homodimers by binding to monomeric ESR1, thereby reducing cell proliferation (40). In addition, NR2C2 can alter the oxygen state of MCF7 cells by decreasing the expression of oncogenic microRNAs (miR)-526b and miR-655, which then suppresses tumor migration and invasion (41). These data suggest that NR2C2 may inhibit the tumorigenicity of ER+ breast cancer cells.
The molecular mechanism underlying the NR2E3-mediated regulation of the characteristics of ER+ breast cancer cells can be complex. In addition to the aforementioned NR2C2, NR2E3 can enhances esr1 transcription by interacting with protein inhibitor of activated STAT protein 3 (PIAS3), a representative inhibitor of STAT3 (5,42). Although ESR1 functions in cancer progression (43,44), its high expression has been associated with superior recurrence-free survival in ER+ breast cancer (5). Furthermore, patients with higher levels of expression of both nr2e3 and esr1 tended to show the optimal recurrence-free survival (5). Esr1 expression was no longer associated with prognosis when patients were treated with tamoxifen. However, nr2e3 expression was still relevant (5), suggesting that NR2E3 can modulates the characteristics of breast tumor cells through distinct pathways in patients who received hormonal therapy. PIAS3 acts as an essential protein that recruits NR2E3 to the esr1 promoter (5). Although PIAS3 is an inhibitor of STAT3, a transcription factor that facilitates self-renewal and metastasis of breast cancer cells (13,14), ectopic expression of PIAS3 was shown to enhance the proliferation of MCF7 cells, attenuate the cytotoxicity of tamoxifen and decrease the survival time of patients with ER+ breast cancer (42). Therefore, according to the ER content, further studies are needed to investigate the molecular association of NR2E3 with these factors.
In conclusion, results from the present study suggest that nr2e3 expression is inversely associated with the migratory and invasive capability of ER+ breast cancer cells. Nr2e3 silencing reinforced the EMT process, enhanced the expression of stem cell-related transcription factors and elevated the proportion of CD44+CD24-/low cells. In addition, NR2E3 may perform its function by targeting NR2C2. Therefore, NR2E3/NR2C2 signaling may represent a target to eliminate stem-like cells in this type of breast cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by The Key Project of Anhui Educational Committee (grant nos. KJ2020A0573 and KJ2021A0748).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SX was responsible for the study conceptualization, investigation, and writing and editing of the manuscript. YH was responsible for performing experiments, writing and reviewing of the manuscript. JJ was responsible for performing experiments and methodology carried out. LF was responsible for performing experiments. CZ was responsible for the methodology and data analysis. QY was responsible for the project administration and data interpretation. YN was responsible for the methodology. ZS was responsible for the conceptualization, funding acquisition and reviewing of the manuscript. SX and YH confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Bengbu Medical College (approval no. 2022-138).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Aísa-Marín I, López-Iniesta MJ, Milla S, Lillo J, Navarro G, de la Villa P, Marfany G. Nr2e3 functional domain ablation by CRISPR-Cas9D10A identifies a new isoform and generates retinitis pigmentosa and enhanced S-cone syndrome models. Neurobiol Dis. 2020;146(105122) doi: 10.1016/j.nbd.2020.105122. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Datta S, Brabbit E, Love Z, Woytowicz V, Flattery K, Capri J, Yao K, Wu S, Imboden M, et al. Nr2e3 is a genetic modifier that rescues retinal degeneration and promotes homeostasis in multiple models of retinitis pigmentosa. Gene Ther. 2021;28:223–241. doi: 10.1038/s41434-020-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanal T, Leung YK, Jiang W, Timchenko N, Ho SM, Kim K. NR2E3 is a key component in p53 activation by regulating a long noncoding RNA DINO in acute liver injuries. FASEB J. 2019;33:8335–8348. doi: 10.1096/fj.201801881RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garattini E, Bolis M, Paroni G, Fratelli M, Terao M. Lipid-sensors, enigmatic-orphan and orphan nuclear receptors as therapeutic targets in breast-cancer. Oncotarget. 2016;7:42661–42682. doi: 10.18632/oncotarget.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YY, Kim K, Kim SB, Hennessy BT, Kim SM, Park ES, Lim JY, Li J, Lu YL, Gonzalez-Angulo AM, et al. Reconstruction of nuclear receptor network reveals that NR2E3 is a novel upstream regulator of ESR1 in breast cancer. EMBO Mol Med. 2012;4:52–67. doi: 10.1002/emmm.201100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z, Wang L, Xu W. IL-13Rα2 mediates PNR-induced migration and metastasis in ERα-negative breast cancer. Oncogene. 2015;34:1596–1607. doi: 10.1038/onc.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracci PM, Zhou M, Young S, Wiemels J. Serum autoantibodies to pancreatic cancer antigens as biomarkers of pancreatic cancer in a San Francisco Bay Area case-control study. Cancer. 2012;118:5384–5394. doi: 10.1002/cncr.27538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichen JG, Dalmau J, Demopoulos A, Wade D, Posner JB, Rosenfeld M. The photoreceptor cell-specific nuclear receptor is an autoantigen of paraneoplastic retinopathy. J Neuroophthalmol. 2001;21:168–172. doi: 10.1097/00041327-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Khanal T, Choi K, Leung YK, Wang J, Kim D, Janakiram V, Cho SG, Puga A, Ho SM, Kim K. Loss of NR2E3 represses AHR by LSD1 reprogramming, is associated with poor prognosis in liver cancer. Sci Rep. 2017;7(10662) doi: 10.1038/s41598-017-11106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Chen W, Liu S, Chen C. Targeting breast cancer stem cells. Int J Biol Sci. 2023;19:552–570. doi: 10.7150/ijbs.76187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taurin S, Alkhalifa H. Breast cancers, mammary stem cells, and cancer stem cells, characteristics, and hypotheses. Neoplasia. 2020;22:663–678. doi: 10.1016/j.neo.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim JR, Mouawad J, Gorton OK, Bubb WA, Kwan AH. Cancer stem cell characteristics and their potential as therapeutic targets. Med Oncol. 2021;38(76) doi: 10.1007/s12032-021-01524-8. [DOI] [PubMed] [Google Scholar]
- 13.Rahmati M, Johari B, Kadivar M, Rismani E, Mortazavi Y. Suppressing the metastatic properties of the breast cancer cells using STAT3 decoy oligodeoxynucleotides: A promising approach for eradication of cancer cells by differentiation therapy. J Cell Physiol. 2020;235:5429–5444. doi: 10.1002/jcp.29431. [DOI] [PubMed] [Google Scholar]
- 14.Johari B, Rahmati M, Nasehi L, Mortazavi Y, Faghfoori MH, Rezaeejam H. Evaluation of STAT3 decoy oligodeoxynucleotides' synergistic effects on radiation and/or chemotherapy in metastatic breast cancer cell line. Cell Biol Int. 2020;44:2499–2511. doi: 10.1002/cbin.11456. [DOI] [PubMed] [Google Scholar]
- 15.Johari B, Moradi M. Application of transcription factor decoy oligodeoxynucleotides (ODNs) for cancer therapy. Methods Mol Biol. 2022;2521:207–230. doi: 10.1007/978-1-0716-2441-8_11. [DOI] [PubMed] [Google Scholar]
- 16.Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152–163. doi: 10.1016/j.ctrv.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Muscat GEO, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, Davis MJ, Clyne C, Funder JW, Simpson ER, et al. Research resource: Nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie S, Han S, Qu Z, Liu F, Li J, Yu S, Reilly J, Tu J, Liu X, Lu Z, et al. Knockout of Nr2e3 prevents rod photoreceptor differentiation and leads to selective L-/M-cone photoreceptor degeneration in zebrafish. Biochim Biophys Acta Mol Basis Dis. 2019;1865:1273–1283. doi: 10.1016/j.bbadis.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Zhang L, Wang Y, Ding Y, Chen T, Wang Y, Wang H, Li Y, Duan K, Chen S, et al. Involvement of miR-451 in resistance to paclitaxel by regulating YWHAZ in breast cancer. Cell Death Dis. 2017;8(e3071) doi: 10.1038/cddis.2017.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OL, He M, Wei M. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J Exp Clin Cancer Res. 2018;37(256) doi: 10.1186/s13046-018-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gooding AJ, Schiemann WP. Epithelial-mesenchymal transition programs and cancer stem cell phenotypes: Mediators of breast cancer therapy resistance. Mol Cancer Res. 2020;18:1257–1270. doi: 10.1158/1541-7786.MCR-20-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Recouvreux MV, Moldenhauer MR, Galenkamp KMO, Jung M, James B, Zhang Y, Lowy A, Bagchi A, Commisso C. Glutamine depletion regulates Slug to promote EMT and metastasis in pancreatic cancer. J Exp Med. 2020;217(e20200388) doi: 10.1084/jem.20200388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venturini G, Kokona D, Steiner BL, Bulla EG, Jovanovic J, Zinkernagel MS, Escher P. In vivo analysis of onset and progression of retinal degeneration in the Nr2e3rd7/rd7 mouse model of enhanced S-cone sensitivity syndrome. Sci Rep. 2021;11(19032) doi: 10.1038/s41598-021-98271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi MY, Romer AI, Hu M, Lepourcelet M, Mechoor A, Yesilaltay A, Krieger M, Gray PA, Shivdasaniet RA. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura M, Naito S, Yokoi T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab Pharmacokinet. 2004;19:135–149. doi: 10.2133/dmpk.19.135. [DOI] [PubMed] [Google Scholar]
- 26.Cai X, Conley SM, Cheng T, Al-Ubaidi MR, Naash MI. A 350 bp region of the proximal promoter of Rds drives cell-type specific gene expression. Exp Eye Res. 2010;91:186–194. doi: 10.1016/j.exer.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Z, Pyeon D, Wang Y, Lambert P, Xu W, Ahlquist P. Orphan nuclear receptor PNR/NR2E3 stimulates p53 functions by enhancing p53 acetylation. Mol Cell Biol. 2012;32:26–35. doi: 10.1128/MCB.05513-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garancher A, Lin CY, Morabito M, Richer W, Rocques N, Larcher M, Bihannic L, Smith K, Miquel C, Leboucher S, et al. NRL and CRX define photoreceptor identity and reveal subgroup-specific dependencies in medulloblastoma. Cancer Cell. 2018;33:435–449.e6. doi: 10.1016/j.ccell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Khuzaei S, Broadgate S, Halford S, Jolly JK, Shanks M, Clouston P, Downes SM. Novel pathogenic sequence variants in NR2E3 and clinical findings in three patients. Genes (Basel) 2020;11(1288) doi: 10.3390/genes11111288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khanal T, Kim D, Johnson A, Choubey D, Kim K. Deregulation of NR2E3, an orphan nuclear receptor, by benzo(a)pyrene-induced oxidative stress is associated with histone modification status change of the estrogen receptor gene promoter. Toxicol Lett. 2015;237:228–236. doi: 10.1016/j.toxlet.2015.06.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin SJ, Yang DR, Yang G, Lin CY, Chang HC, Li G, Chang C. TR2 and TR4 orphan nuclear receptors: An overview. Curr Top Dev Biol. 2017;125:357–373. doi: 10.1016/bs.ctdb.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Haider NB, Mollema N, Gaule M, Yuan Y, Sachs AJ, Nystuen AM, Naggert JK, Nishina PM. Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res. 2009;89:365–372. doi: 10.1016/j.exer.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roduit R, Escher P, Schorderet DF. Mutations in the DNA-binding domain of NR2E3 affect in vivo dimerization and interaction with CRX. PLoS One. 2009;4(e7379) doi: 10.1371/journal.pone.0007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao X, Zhang Y, Wu L, Cheng R, Li C, Qu C, Ji H. Immunohistochemical study of NR2C2, BTG2, TBX19, and CDK2 expression in 31 paired primary/recurrent nonfunctioning pituitary adenomas. Int J Endocrinol. 2019;2019(5731639) doi: 10.1155/2019/5731639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Zhang J, Ma Y, Chen J, Dong B, Zhao W, Wang X, Zheng Q, Fang F, Yang Y. Testicular orphan receptor 4 (TR4) is a marker for metastasis and poor prognosis in non-small cell lung cancer that drives the EMT phenotype. Lung Cancer. 2015;89:320–328. doi: 10.1016/j.lungcan.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Qiu X, Zhu J, Sun Y, Fan K, Yang DR, Li G, Yang G, Chang C. TR4 nuclear receptor increases prostate cancer invasion via decreasing the miR-373-3p expression to alter TGFβR2/p-Smad3 signals. Oncotarget. 2015;6:15397–15409. doi: 10.18632/oncotarget.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Z, Zheng J, Xue Y, Liu X, Wang D, Yang C, Ma J, Liu L, Ruan X, Wang Z, Liu Y. NR2C2-uORF targeting UCA1-miR-627-5p-NR2C2 feedback loop to regulate the malignant behaviors of glioma cells. Cell Death Dis. 2018;9(1165) doi: 10.1038/s41419-018-1149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren W, Hu J, Li H, Chen J, Ding J, Zu X, Fan B. miR-616-5p promotes invasion and migration of bladder cancer via downregulating NR2C2 expression. Front Oncol. 2021;11(762946) doi: 10.3389/fonc.2021.762946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin R, Lin H, Li G, Xu J, Shi L, Chang C, Cai X. TR4 nuclear receptor suppresses HCC cell invasion via downregulating the EphA2 expression. Cell Death Dis. 2018;9(283) doi: 10.1038/s41419-018-0287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shyr CR, Hu YC, Kim E, Chang C. Modulation of estrogen receptor-mediated transactivation by orphan receptor TR4 in MCF-7 cells. J Biol Chem. 2002;277:14622–14628. doi: 10.1074/jbc.M110051200. [DOI] [PubMed] [Google Scholar]
- 41.Gervin E, Shin B, Opperman R, Cullen M, Feser R, Maiti S, Majumder M. Chemically induced hypoxia enhances miRNA functions in breast cancer. Cancers (Basel) 2020;12(2008) doi: 10.3390/cancers12082008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang SF, Hou MF, Chen FM, Ou-Yang F, Wu YC, Chai CY, Yeh YT. Prognostic value of protein inhibitor of activated STAT3 in breast cancer patients receiving hormone therapy. BMC Cancer. 2016;16(20) doi: 10.1186/s12885-016-2063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei JT, Gou X, Seker S, Ellis MJ. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J Cancer Metastasis Treat. 2019;5(38) doi: 10.20517/2394-4722.2019.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matutino A, Joy AA, Brezden-Masley C, Chia S, Verma S. Hormone receptor-positive, HER2-negative metastatic breast cancer: Redrawing the lines. Curr Oncol. 2018;25 (Suppl 1):S131–S141. doi: 10.3747/co.25.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.