Summary
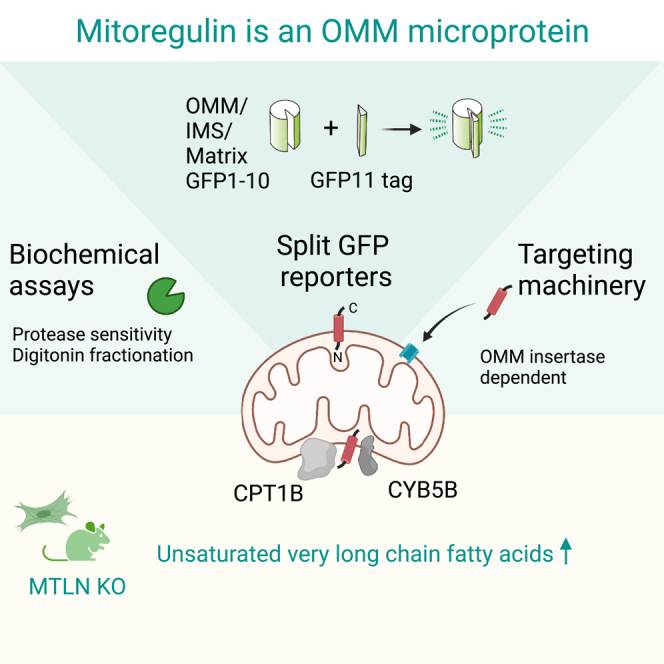
LINC00116 encodes a microprotein first identified as Mitoregulin (MTLN), where it was reported to localize to the inner membrane of mitochondria to regulate fatty acid oxidation and oxidative phosphorylation. These initial discoveries were followed by reports with differing findings about its molecular functions and submitochondrial localization. To clarify the apparent discrepancies, we constructed multiple orthogonal methods of determining the localization of MTLN, including split GFP-based reporters that enable efficient and reliable topology analyses for microproteins. These methods unequivocally demonstrate MTLN primarily localizes to the outer membrane of mitochondria, where it interacts with enzymes of fatty acid metabolism including CPT1B and CYB5B. Loss of MTLN causes the accumulation of very long-chain fatty acids (VLCFAs), especially docosahexaenoic acid (DHA). Intriguingly, loss of MTLN protects mice against western diet/fructose-induced insulin-resistance, suggests a protective effect of VLCFAs in this context. MTLN thus serves as an attractive target to control the catabolism of VLCFAs.
Subject areas: Biological sciences, Biochemistry, Molecular biology, Cell biology
Graphical abstract
Highlights
-
•
Split GFP is devised to report submitochondrial localizations of proteins in vivo
-
•
Microprotein Mitoregulin primarily localizes to the outer membrane of mitochondria
-
•
Mitoregulin modulates lipid metabolisms and levels of very long-chain fatty acids
Biological sciences; Biochemistry; Molecular biology; Cell biology
Introduction
The advent of ribosome sequencing (Ribo-seq) technology allows comprehensive annotations of protein translation at single nucleotide resolution with unprecedented sensitivity.1 This has led to the discovery of protein translations from many noncanonical open reading frames (ORFs) in diverse samples and species.2,3 These ORFs often nest in long non-coding RNAs (LncRNAs), 5′ untranslated regions (UTRs), and 3′UTRs of classical transcripts.4,5,6 As most noncanonical ORFs are shorter than 100 codons, they were also referred to as small ORFs (sORFs). The detection of protein translations from sORFs significantly expanded the protein-coding capability of our genome. For instance, sORF databases OpenProt and sORFs.org registered more than half a million and more than two million novel ORFs by analyzing more than 100 public available datasets, respectively.7,8 Even after applying stringent criteria to remove redundant annotations, nearly 200,000 entries remain.9 Deep sequencing of ribosome footprints in human tissues and primary cell cultures also identified nearly 8,000 novel ORFs (Chothani et al., 2022).
Despite the rapid expansion of sORF annotations, efforts to characterize sORF-encoded proteins (SEPs) have not kept pace, even as growing evidence point to their requirements in diverse biological processes. Notably, many SEPs are imported into mitochondria and play diverse functions as shown by us and others. For instance, mitochondrial-localized SEPs (mito-SEPs) BRAWNIN and SMIM4 form a complex to safeguard the early assembly of electron transport chain complex III (CIII) in response to metabolic cues.10,11 Mito-SEP MOCCI and Mm47 were found to regulate innate immunity.12,13,14 At the outer membrane, microprotein PIGBOS interacts with ER proteins to regulate ER stress response.15
Among these mito-SEPs, a peptide encoded by LINC00116 was discovered and characterized in parallel by multiple research groups and considered as a prototypical mito-SEPs. Indeed, the 56 a.a. peptide was first discovered by mass spectrometry and identified as one of the most highly expressed SEPs by Ribo-seq.16,17 It has since been named Mitoregulin (MTLN), MOXI, MPM, and LEMP by independent groups, with MTLN now being accepted as the standard name.18,19,20,21,22,23 MTLN is most highly expressed in brown fat, cardiac and skeletal muscles.19
Despite in depth analysis by multiple groups, the molecular function of MTLN is still obscure and remains contested. Knock-out of MTLN in mice does not affect the viability and growth of animals.19,20 Reported interaction partners of MTLN range from the outer membrane protein CYB5R3, inner membrane protein NUDFA7, and matrix protein HADHA.20,21,24 The plethora of phenotypes associated with MTLN deficiency include defects in fatty acid oxidation,20 calcium handling,19 and smaller muscle fibers.22 More intriguingly, both impaired and enhanced mitochondrial respiration were reported in MTLN mutants.21,22,24 In view of these discrepant findings, further investigation is required to clarify the function of MTLN.
Here, by applying multiple orthogonal methods, we concluded that MTLN is primarily localized to the mitochondrial outer membrane and has a cytosolic tail. At the outer membrane, MTLN interacts with CPT1 and CYB5B to regulate fatty acid metabolism. We propose that this is the bona fide molecular function of MTLN from which stems its associated phenotypes in lipid metabolism.
Results
LINC00116 encodes a microprotein that is localized to the outer membrane of mitochondria
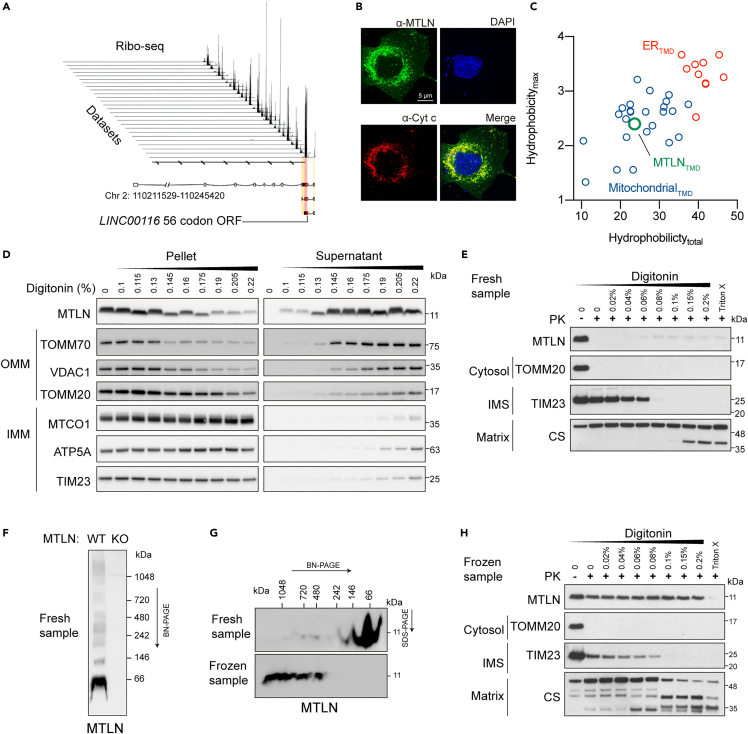
Using a customized proteogenomic pipeline for the identification and validation of SEPs that localize to the mitochondria,10 we independently detected MTLN as a bona fide mito-SEP with strong evidence of translation in a wide range of cell lines as ascertained by ribosome profiling (Figure 1A). Using a polyclonal antibody raised against the C-terminus of the predicted protein product, we detected endogenous MTLN in U2OS cells that was diminished by siRNAs against LINC00116 (Figure S1A). Consistent with previous reports, both endogenous and hemagglutinin (HA) epitope-tagged MTLN is localized to the mitochondria (Figures 1B and S1B).
Figure 1.
Microprotein MTLN is localized to the outer membrane of mitochondria
(A) LINC00116-associated ribosome footprints in multiple ribosome profiling datasets.
(B) Immunostaining of MTLN in U2OS cell using an antibody against its C-terminus. Mitochondria were marked by cytochrome c (Cyt c).
(C) Hydropathic scales of the transmembrane segments of mitochondrial and ER single pass transmembrane microproteins calculated by Kyte & Doolittle’s method.
(D)Solubilization of mitochondrial proteins isolated from freshly harvested HEK293T cells by a gradient of digitonin. Marker proteins of the outer membrane of mitochondria (OMM) and the inner membrane of mitochondria (IMM) were indicated.
(E) Proteinase K digestion of mitochondrial proteins isolated from freshly harvested HEK293T cell after digitonin permeabilization. TOMM20, TIM23, and citrate synthase (CS) possess cytosolic, inter membrane space (IMS) and matrix domains, respectively.
(F) Mobility of MTLN analyzed by blue native-PAGE (BN-PAGE) after digitonin solubilization. Mitochondria were isolated from freshly dissected mouse skeletal muscle.
(G) Mobility of MTLN analyzed by BN-PAGE and second-dimensional SDS-PAGE. Mitochondria were isolated from freshly dissected mouse skeletal muscle or mouse skeletal muscle stored at −80°C after flash freezing with liquid nitrogen.
(H) Protease sensitivity assay similar to (E), except that mitochondria were prepared from frozen HEK293T cell pellets.
The protein sequence of MTLN is well conserved in vertebrates and has a single pass transmembrane domain (spTMD) near its N-terminus (Figure S1C). We previously found the presence of a spTMD to be a good predictor of mitochondrial localized SEPs.10 Compared to spTMDs used for ER targeting, we found spTMDs of mitochondrial-localized microproteins have a significantly lower hydrophobicity index (Figure 1C). Consistent with its mitochondrial localization, the N-terminal spTMD of MTLN clustered with mitochondrial spTMDs (Figure 1C). Mitochondria are formed by the two separated layers of membranes, the outer membrane of mitochondria (OMM) and the inner membrane of mitochondria (IMM). Owing to their distinct lipid composition, the OMM can be solubilized by lower concentrations of digitonin, a non-ionic steroidal detergent, compared to the IMM (Figure 1D). Importantly, the solubility of MTLN in digitonin solutions followed the pattern of multiple OMM proteins in both HEK293T (Figure 1D) and mouse skeletal muscle mitochondria (Figure S1D). Protease sensitivity assay was then used to interrogate the membrane topology of MTLN. MTLN was fully degraded by proteinase K in the absence of detergent, a unique pattern presented only by OMM proteins with a cytosolic epitope, like TOM20 (Figures 1E and S1E). In contrast, proteins with IMS and matrix domains were only digested by proteinase K when the OMM and IMM were sequentially solubilized by digitonin at different concentrations (Figures 1E and S1E). These results jointly argue that MTLN is an OMM protein, and the C-terminus of MTLN faces the cytosol.
Interestingly, we observed a minor population of protease resistant MTLN under digitonin treatments (Figure 1E). To investigate this phenomenon, we separated MTLN on a native PAGE and observed high molecular weight smears (Figure 1F). When using mitochondria isolated from frozen cell and tissue samples for blue native PAGE analysis, we serendipitously found that freezing samples before mitochondria isolation, a common procedure in mitochondrial isolation protocols to increase yield, promoted the formation of high molecular weight smears and made MTLN resistant to protease digestion (Figures 1G, 1H, and S1F). These data suggest that extra cautiousness is needed when performing biochemical assays for MTLN.
Assaying protein submitochondrial localization in vivo by split GFP
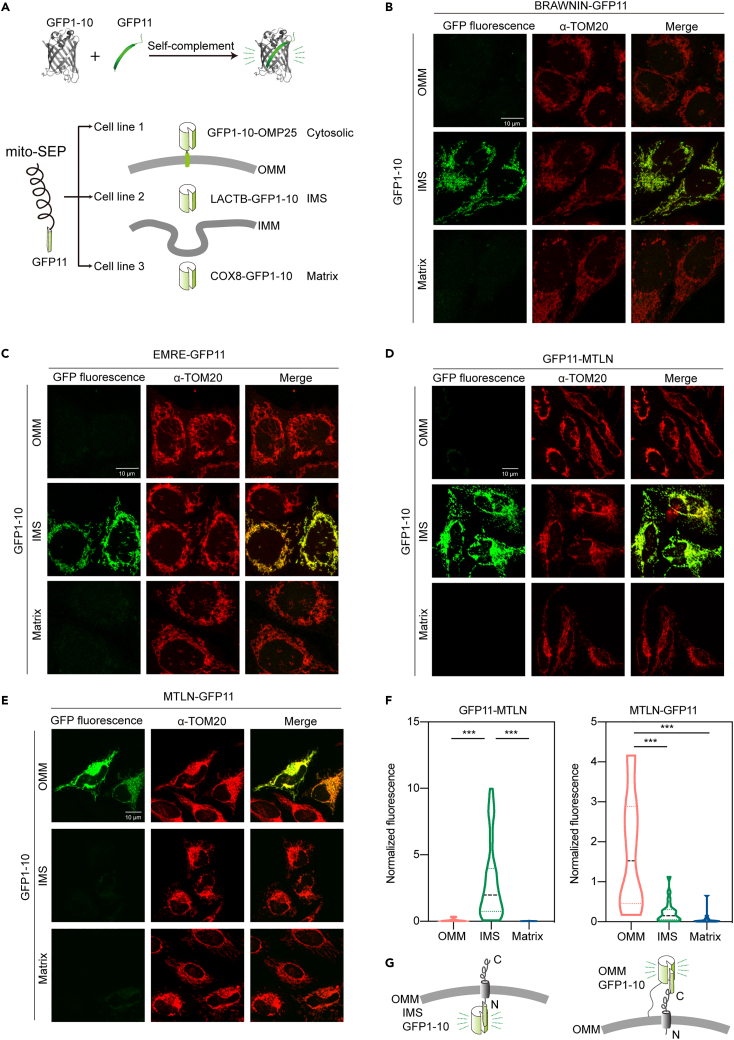
Since our results are in contradiction to previous reports, we sought to rule out potential artifacts that are intrinsically associated with cell lysis in biochemical assays. We achieved this goal by devising a split GFP-based orthogonal method to analyze the membrane topology of mitochondrial proteins in vivo. GFP1-10 and GFP11, the two components of split GFP system, self-complement and become fluorescent when present in the same cellular compartment.25 A SEP of interest can be tagged either on the N- or C-terminus with GFP11, a minimal 16 amino acids tag that is free of bulky side chain and strong charges residues (Figure 2A). On the other end, to report the 3 submitochondrial spaces, we developed cell lines with GFP1-10 localized to the matrix (COX8A-GFP1-10), IMS (LACTB-GFP1-10), and the cytosolic side of OMM (GFP1-10-OMP25) (Figure 2A). For each reporter, we validated their localizations with protease sensitivity assays (Figures S2A–S2C). We first tested this system with examples of known localization and topology by tagging GFP11 to the C-terminus of mitochondrial microprotein BRAWNIN. When co-expressed with GFP1-10s of different submitochondrial localizations, BRAWNIN-GFP11 complemented most efficiently with the IMS GFP1-10, indicating a C-terIMS topology, in agreement with former biochemical assays (Figure 2B).10 We next asked whether this tool can be applied to resolve conflicts of protein topology analyses in literature. EMRE, an accessory protein of mitochondrial calcium uniporter MCU, is a single pass transmembrane protein of the IMM. Previously, different orientations have been proposed for EMRE.26,27 In our analysis, EMRE-GFP11 and GFP11-EMRE interacted strongly with IMS GFP1-10 and matrix GFP1-10, respectively, suggesting a Nmatrix-CIMS topology (Figures 2C and S2D).28 This topology was corroborated by structural analysis and consistent with our protease sensitivity assay29 (Figure S2E). We then tagged MTLN with GFP11 on its C and N terminus, validated their mitochondrial localization, and introduced them into cells expressing mitochondrial GFP1-10s (Figures S2F, 2D, and 2E). The N terminal tagged GFP11-MTLN complemented exclusively with IMS GFP1-10 (Figures 2D and 2F). By contrast, C-terminal tagged MTLN-GFP11 fluoresced most efficiently with OMM GFP1-10 (Figures 2E and 2F). These data confirmed the OMM localization of MTLN and furthermore clarified its topology: the N terminus of MTLN faces the IMS while the C terminus of MTLN resides in the cytosol (i.e., NIMS/Ccyto orientation) (Figure 2G).
Figure 2.
In vivo protein topology analysis confirms the OMM localization of MTLN
(A) Schematic presentation of fluorescence-based mitochondrial protein topology reporters. The localization and topology of peptide of interest can be deduced by identifying the most efficient complemented GFP1-10 reporter.
(B-E) GFP fluorescence and TOM20 staining of representative U2OS cells co-transfected with BRAWNIN-GFP11 (panel B), EMRE-GFP11 (panel C), GFP11-MTLN (panel D), and MTLN-GFP11 (panel E) together with OMM, IMS, and matrix GFP1-10. Images were acquired and presented using identical settings. Scale bar = 10 μM.
(F) GFP fluorescence of U2OS cells was normalized with the immunofluorescence of GFP1-10 stained by a GFP antibody in the same cell. Data from 3 biological replicates were pooled and shown in violin plots. p values from two-sided unpaired t-test. ∗∗∗p values < 0.001.
(G) Cartoons showing the topology of MTLN as indicted by the most efficient complementation between specific GFP1-10 and the GFP11 tag.
Lastly, we sought the third orthogonal method for determining the subcellular localization of MTLN, testing the hypothesis that the MTLN C-terminus resides in the cytosol. In this method, we tested the ability of anti-MTLN antibody (raised against the C-terminus) to pull down intact mitochondria (Figure S2G). Consistent with the finding that MTLN C-terminus resides in the cytosol, we found that anti-MTLN antibody coated magnetic beads successfully enriched intact mitochondria as efficiently as an antibody against bona fide OMM protein TOM22 (Figure S2H). In contrast, an antibody against an IMM protein TIM23 only partially pulled down damaged mitochondria devoid of outer membrane as shown by the western of IMM protein ATP5A and OMM protein TOM20, respectively (Figure S2H).
Deficiency of OMM helical protein insertase impairs the mitochondrial targeting of MTLN
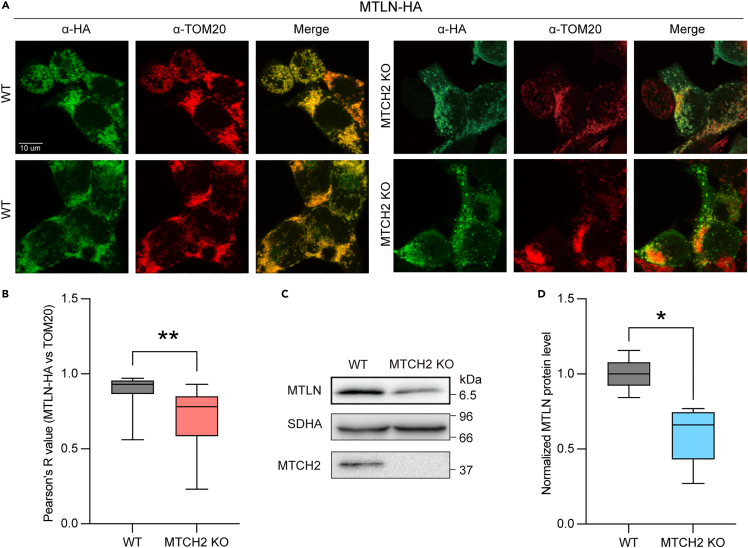
Non-canonical mitochondrial solute carriers MTCH2 and MTCH3 were recently identified as the insertases of OMM α-helical proteins.30 MTCH2 and MTCH3 are synthetically lethal and work in a partially redundant manner.30 As an α-helical protein in the OMM, MTLN should be a bona fide substrate of these insertases, and the mitochondrial targeting of MTLN might be impaired when the level MTCH2/MTCH3 is limited. To test this hypothesis, we knocked out MTCH2 in HEK293T cells and analyzed the localization of MTLN by confocal microscopy (Figures 3A and S3A). We found that MTLN is partially localized to the mitochondria and apparent non-mitochondrial punctate stains were observed in MTCH2 KO cells (Figure 3A). Correlation analysis between MTLN-HA and TOM20 signals confirmed a partial mislocalization of MTLN in MTCH2 KO cells (Figure 3B). By contrast, the targeting of an IMM α-helical protein, BRAWNIN, remains unaffected (Figures S3B and S3C). In addition, the loss MTCH2 decreases the protein level of endogenous MTLN (Figures 3C and 3D). These genetic data further support the OMM localization of MTLN.
Figure 3.
Mitochondrial targeting of MTLN is impaired by the deficiency of OMM α-helical protein insertase
(A) Representative image of WT and MTCH2 KO HEK293T cells that were transfected with MTLN-HA and stained with HA and TOM20 antibodies.
(B) Quantitative analysis of mitochondrial localization of MTLN-HA in WT and MTCH2 KO HEK293T cells. Pearson’s correlated values between HA and TOM20 signals were calculated for individual cells using ImageJ and plotted. Error bars of the boxplot shows the minimum and maximum values with boxes extend from the 25th to 75th percentiles, and the p value was calculated by unpaired t-test. ∗∗p values < 0.01.
(C) Western analysis of endogenous MTLN in WT and MTCH2 KO HEK293T cells.
(D) Quantification of MTLN protein level in WT and MTCH2 KO HEK293T cells. SDHA signal was used as the loading control. Data from 5 replicates were plotted. Error bars of the boxplot shows the minimum and maximum values with boxes extend from the 25th to 75th percentiles, and p values from two-sided paired t-test. ∗p values < 0.05.
Loss of MTLN does not impact mitochondria oxidative phosphorylation
Multiple studies linked the function of MTLN to mitochondrial respiration but conclusions remain controversial.19,21,22,24 To clarify this issue, we generated both MTLN KO cultured cells and mice (Figures S4A–S4E) and examined the assembly, activity, and function of the mitochondrial respiratory chain (RC).
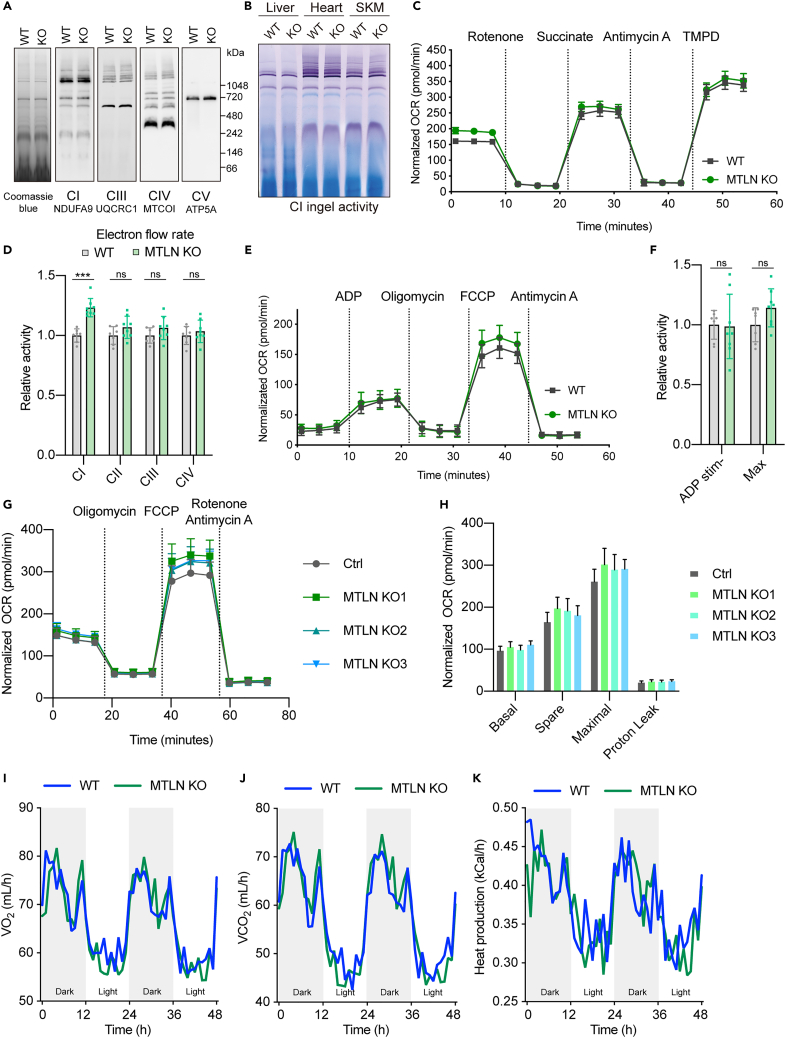
First, protein abundances of the core subunits of RC complexes remain unchanged in the heart of MTLN KO mouse (Figure S4F). Native PAGE analysis of RC complexes of the MTLN KO mouse also showed normal abundance and assemblies (Figure 4A). The enzymatic activity of respiratory chain complexes remains unchanged in mitochondria isolated from hearts of MTLN KO mice (Figure S4G). Considering that both enhanced and reduced complex I activity were reported for MTLN KO in literature, we performed complex I in-gel assay but failed to discern visible defects in the MTLN KO (Figures 4B and S4H). We then investigated the electron transport activity of individual complexes in isolated mouse heart mitochondria using pyruvate, malate, and succinate as substrates (Figure 4C). Complex I-dependent electron flow was mildly increased while electron flow through other complexes remained unchanged in MTLN KO (Figures 4C and 4D). Likewise, the electron transport chain of MTLN KO was well-coupled with ATP production and displayed normal respiratory capacity (Figures 4E and 4F).
Figure 4.
Loss of MTLN does not impair mitochondrial respiratory chain
(A) BN-PAGE and immunoblotting analysis of mitochondrial respiratory chain (RC) complex in mouse heart mitochondria. Staining by Coomassie blue G250 served as a loading reference.
(B) Complex I in-gel assay of BN-PAGE separated mitochondrial RC complexes that were isolated from mouse liver, heart, and skeletal muscle (SKM).
(C) Oxygen consumption rate (OCR) of purified mouse heart mitochondria was monitored along the sequential injection of indicated inhibitors and substrates. Rotenone and antimycin A are inhibitors of complex I and III, respectively. Succinate and TMPD are electron donors of complex II and IV, respectively. Data are represented as mean ± standard error of the mean (SEM), n = 8 technical replicates. Representative data of 3 biological replicates are shown.
(D) Related to panel C, rates of electron flow through the RC complexes in isolated mouse heart mitochondria were measured in Seahorse using oxygen consumption rate as the readout. Mitochondria were pretreated FCCP to uncouple electron flow with ATP production. Electron flows through the individual complexes were defined as: CI, rotenone sensitive rate; CII, succinate stimulated rate; CIII, antimycin A sensitive rate; CIV, TMPD stimulated rate. Data are represented as mean ± SEM, n = 8 technical replicates. p values are from unpaired, two-tailed t test. ∗∗∗p values < 0.001. ns p values ≥ 0.05.
(E) OCR of mouse heart mitochondria was measured along the sequential injection of indicated reagents. Oligomycin inhibits ATP synthase while FCCP uncouples ATP synthesis with electron transport. Data are represented as mean ± SEM, n = 8 technical replicates.
(F) Related to panel E, ADP stimulated (coupled) and maximal (uncoupled) oxygen consumption rate of isolated mouse heart mitochondria were measured by Seahorse. Pyruvate and malate were used as the fuel. Data are represented as mean ± SD, n = 8 technical replicates. ns p values ≥ 0.05.
(G) Representative seahorse Mito Stress test using multiple MTLN KO C2C12 myoblast cell lines. Data are represented as mean ± SD, n = 8 technical replicates.
(H)Basal, spare, maximal respiration, and electron leak of the mitochondria of MTLN KO C2C12 cell measured by a Mito Stress test. Data are represented as mean ± SD, n = 8 technical replicates.
(I–K) Rates of mouse whole body oxygen consumption (VO2), CO2 production (VCO2), and heat production measured by indirect calorimetry.
We then assayed the respiration at the cellular and whole-body levels using MTLN KO cell and mouse models, respectively. All three MTLN deficient C2C12 myoblast lines showed normal basal, spare, and maximum respiration in the Mito Stress test (Figures 4G and 4H). To further ascertain this observation in animals, we housed wild type (WT) and MTLN KO mice in metabolic cages and tracked their activity, food intake and energy expenditure. No significant difference was detected in water and food intake while rates of oxygen consumption and carbon dioxide production stayed unchanged (Figures 4I–4K). Meanwhile, abundances of TCA cycle metabolites, lactate and amino acids were not disturbed in MTLN KO mice (Figures S4I–S4K). Altogether, both cellular and animal models of MTLN KO do not present any biochemical, functional, or physiological defect in mitochondrial oxidative phosphorylation.
Loss of MTLN perturbs lipid metabolism but does not grossly impair FAO
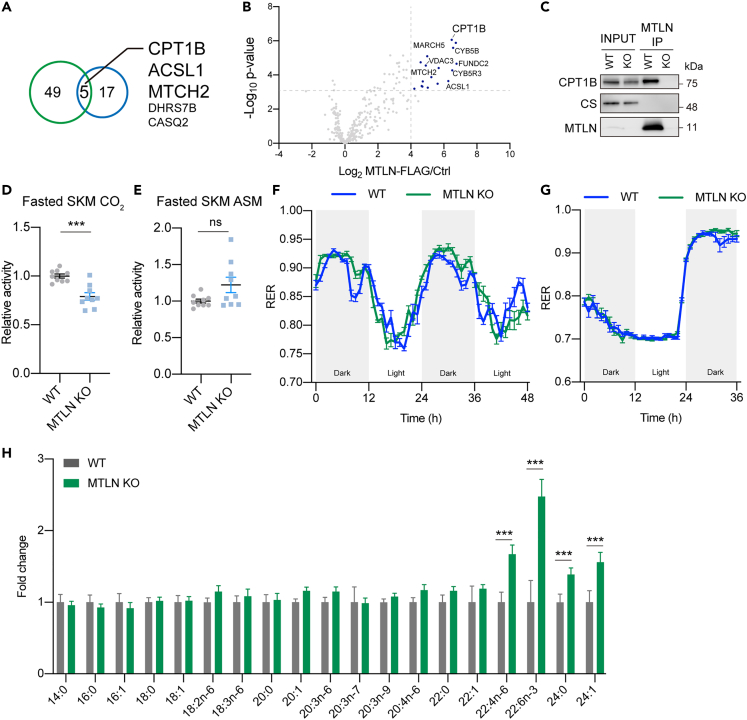
To directly interrogate the molecular function of MTLN, we determined the protein interactome of MTLN. We purified endogenous MTLN from DDM-solubilized mouse skeletal muscle mitochondria and analyzed its interactome with mass spectrometry (Table S1). In parallel, FLAG-tagged MTLN was expressed in mouse skeletal muscle using adeno-associated virus (AAV) and purified following digitonin solubilization for interactome analysis. 5 proteins were highly enriched in both set-ups, including 3 OMM proteins (Figure 5A). Among these co-purified proteins, MTCH2 was recently identified as the insertase of α-helical OMM proteins.30 Importantly, multiple proteins related to fatty acid metabolism are highly enriched in the MTLN interactome (Figure 5B). One of the strongest enriched proteins is CPT1B, the rate-limiting enzyme of fatty acid oxidation responsible for the formation of acyl-carnitine. We confirmed that CPT1B, but not unrelated protein CS, specifically interacts with endogenous MTLN by IP and western analysis (Figures 5C and S5A). Interestingly, a fraction of MTLN migrates as a ∼150 kDa complex in native PAGE, and the majority of CPT1B migrates in this range (Figure S5B). Notably, several CPT1-associated proteins are also detected in the MTLN interactome, including ACSL1, an acyl-CoA synthase,31 and VDAC3, a voltage-dependent anion channel that assists acyl-carnitine import (Figure 5B).32 CYB5B and CYB5R3, a pair of NADH-dependent enzymes required for lipid reduction and remodeling, also interact with MTLN (Figure S5C). These observations suggest that MTLN is part of the lipid metabolism network at the OMM.
Figure 5.
MTLN is required for cellular fatty acid metabolism
(A) Venn diagram showing the highly enriched proteins (Log2 enriched value > 4) detected in MTLN IPs that were reproducibly identified by two independent IPs strategies. Endogenous MTLN and FLAG tagged MTLN were purified by MTLN and FLAG antibodies from freshly isolated mitochondria solubilized with DDM and digitonin, respectively.
(B) Volcano plot showing enriched mitochondrial proteins that were enriched by MTLN-FLAG IP. Highly enriched proteins (Log2MTLN-FLAG/Ctrl>4 and -Log10p value>3) were marked in blue.
(C) Western validation of the physical interaction between endogenous MTLN and CPT1B. Endogenous MTLN was immuno-isolated from mouse skeletal muscles.
(D) Oxidation of radioactive palmitic acid as measured by the production of 14C labeled CO2. Skeletal muscle (SKM) homogenates were prepared from animals fasted for 12h. Mean and SEM from 9 biological replicates are shown. p values from two-sided unpaired t-test. ∗∗∗p values < 0.001.
(E) Similar to panel D, oxidation of radioactive palmitic acid as measured by the production of 14C labeled by short chain acid solute metabolites (ASM). Mean and SEM from 9 biological replicates are shown. ns p values ≥ 0.05.
(F) Respiratory exchange ratios (RERs) of indicated WT and MTLN KO mice measured in metabolic home cages under the Chow diet. Mean and SEM from 4 biological replicates are shown.
(G) RERs of indicated animals in fasting and refeeding. The fasting started together with a night cycle. Mean and SEM from 4 biological replicates are shown.
(H) Cellular total lipid acyl profile was determined by targeted mass spectrometry in WT and MTLN KO C2C12. Mean and SEM from WT C2C12 samples and MTLN KO cells separately generated using 3 different gRNAs are shown. p values from two-sided unpaired t-test. ∗∗∗p values < 0.001.
We next tested whether the loss of MTLN imposed a general effect on the rate of fatty acid oxidation (FAO) using 14C labeled palmitic acid as the substrate. In heart and skeletal muscles, oxidation of palmitic acid as marked by CO2 emission trended lower in the MTLN KO although acid soluble metabolites remained unchanged (Figures 5D, 5E, and S5D–S5I). The rate of respiration trended lower in MTLN KO mitochondria when using palmitoylcarnitine, an FAO substrate that bypasses CPT1 (Figure S5J). These observations suggest that the metabolism of short chain acyl-CoAs is mildly defective under MTLN deficiency. Consistent with a lack of overt defects in FAO, the acylcarnitine profile of MTLN KO heart mitochondria was unchanged compared to WT (Figure S5K). Likewise, MTLN KO did not affect CPT1B enzymatic activity in vitro (Figure S5L).
To further clarify this issue at whole body level, respiratory exchange ratio (RER) of mice were measured by metabolic cages. The RER ratio of MTLN KO oscillated normally in the day and night cycles and dropped to 0.7 during fasting, suggesting that fatty acid oxidation is not grossly impaired (Figures 5F and 5G). Intriguingly, we observed modest but reproducible dips in RER readings 3 h before day cycles in WT animals, which was attenuated in the MTLN KOs, suggesting a lipid catabolism defect specific to these periods in the circadian rhythm of the mouse.
To discover alternate links between MTLN and lipid metabolism, we profiled fatty acid methyl esters of cellular lipids by targeted metabolomics (Figure 5H). We observed a significant accumulation of multiple very long-chain fatty acids (VLCFA) upon MTLN depletion, including polyunsaturated VLCFA docosahexaenoic acid (DHA, C22:6 n-3) (Figure 5H). By contrast, no significant difference was observed for all the other molecules. Taken together, our analysis clearly ruled out a gross fatty acid oxidation defect in the MTLN KO. Instead, MTLN, by virtue of its interaction with CPT1B and other lipid metabolizing enzymes in the OMM, is likely to be involved in the catabolism of VLCFA and the loss of which led to a perturbation in the lipid metabolism.
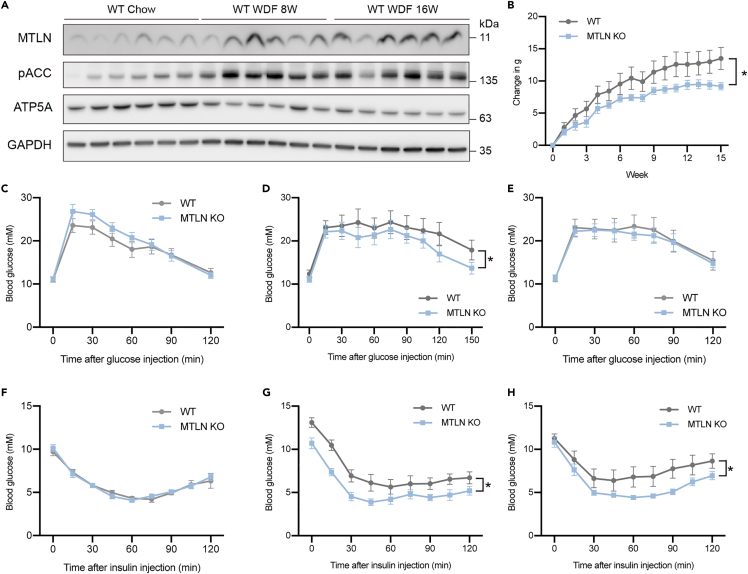
MTLN deficiency protects animals from diet-induced insulin resistance
Alternations of fatty acid metabolism are widely implicated in metabolic disorders. Additionally, we observed that the Western diet and fructose feeding induced the protein level of MTLN (Figure 6A). To investigate the physiological relevance of MTLN KO in the context of diet-induced insulin resistance, we fed mice with a Western diet supplemented with 15% fructose w/v in drinking water (WDF) for 16W. Before switching to the WDF diet from the standard Chow diet, MTLN KO mice have normal body weight and fat mass (Figures S6A and S6B). After switching to WDF diet, MTLN KO mice gained significantly less body weight, as both fat and lean mass were accumulated in MTLN KO mice at a slower rate (Figures 6B and S6A–S6C). As the WDF diet is known to impair the glucose tolerance and insulin sensitivity of animals, we performed glucose and insulin tolerance test in animals (Figures 6C–6H). After an 8-week WDF feeding, WT animals showed an elevated level of basal blood glucose and blunt insulin response (Figures 6D and 6G). By contrast, the insulin sensitivity remained unchanged in MTLN KO animals after both 8-week and 16-week WDF feeding, compared with age-matched animals on the Chow diet (Figures 6F–6H). These observations suggest that the metabolic remodeling in the MTLN KO protected the animal from diet-induced insulin resistance.
Figure 6.
Loss of MTLN protects mouse from diet-induced insulin resistance
(A) Western analysis of protein expression after feeding with the Western diet supplemented with 15% fructose in drinking water (WDF).
(B) The gain of body weight after WDF feeding. Mean and SEM of 6 biological replicates are shown. p value from two-way ANOVA test. ∗p values < 0.05.
(C) Glucose tolerance test before WDF feeding. Mean and SEM of 6 biological replicates are shown.
(D) Glucose tolerance test after 8 weeks of WDF feeding. Mean and SEM of 6 biological replicates are shown. p value from two-way ANOVA test. ∗p values < 0.05.
(E) Glucose tolerance test after 16 weeks of WDF feeding. Mean and SEM of 6 biological replicates are shown.
(F) Insulin tolerance test before WDF feeding. Mean and SEM of 6 biological replicates are shown.
(G) Insulin tolerance test after 8 weeks of WDF feeding. Mean and SEM of 6 biological replicates are shown. p value from two-way ANOVA test. ∗p values < 0.05.
(H) Insulin tolerance test after 16 weeks of WDF feeding. Mean and SEM of 6 biological replicates are shown. p value from two-way ANOVA test. ∗p values < 0.05.
Discussion
Recent discoveries of protein translations from many sORFs have expanded our knowledge of the functional proteome of eukaryotes.3 Many of these sORF-encoded peptides (SEPs) are localized to mitochondria, pointing to the critical role of SEPs in metabolism regulation.10,33 As one of the first characterized mito-SEPs, Mitoregulin (MTLN) encoded by LINC00116 is considered a prototypic mito-SEP and was analyzed in parallel by multiple research groups.18,19,20,21,22,23 However, the submitochondrial localization and basic biological functions of MTLN remain debated. Here, using multiple orthogonal methods, we showed that MTLN is localized to the outer membrane of mitochondria. At the outer membrane, MTLN interacts with a network of enzymes involved in fatty acid metabolism. Loss of MTLN mildly impaired fatty acid oxidation and led to an accumulation of very long-chain fatty acids (VLCFA), especially docosahexaenoic acid (DHA). When fed under obesity/diabetes-inducing diet, MTLN-deficient animals accumulated less body fat and showed better insulin sensitivity. Taken together, our data demonstrate that MTLN is a key regulator of fatty acid metabolism at the outer membrane of mitochondria.
Our report provides a comprehensive analysis on the submitochondrial localization of MTLN. In literature, MTLN was first assigned as an IMM protein based on protease sensitivity assays.19,20 However, the OMM localization of MTLN is implicated in published data as well. First, immunogold staining of MTLN primarily marked the OMM in adipocytes.18 Second, MTLN was found to interact with OMM protein CYB5R3.21 In our study, using multiple orthogonal methods, we concluded that MTLN primarily is localized to the OMM instead of the IMM. We also showed that MTLN forms high molecular weight smears in detergent and phase transition. In line with our observation, MTLN was shown to form high molecular weight smears in SDS-PAGE.19
The sub-organelle localization is integral to the biological function of a mitochondrial protein. Classical topology analysis methods require mitochondrial purification and tedious biochemical assays, and the result of which can be distorted by factors including the mitochondria integrity, protein to detergent ratios, and the efficiency of protease digestion and termination. In contrast, the split GFP-based reporters we constructed in this study enable efficient and reliable topology analysis in vivo. Albeit the tagging of protein with the GFP11 tag can potentially change the targeting of the tagged protein, high-throughput analysis of protein subcellular localizations using a similar tag showed this type of tag is largely benign in protein sorting.34 This robust and convenient method is readily applicable in the topology analysis of many uncharacterized mito-SEPs. Although MTLN is identified primarily as an OMM protein in all cell lines and conditions we have examined, we cannot formally rule out that MTLN can enter the IMM in certain cell types and physiological conditions. New techniques that can trace submitochondrial traffickings of protein in real-time and in vivo may help to resolve this issue.
MTLN was reported to involve both mitochondrial respiration and lipid metabolism.19,21 Our data demonstrated that MTLN primarily functions in fatty acid metabolism. We observed a reduced rate of fatty acid oxidation and accumulation of polyunsaturated VLCFA in MTLN deficient models. Mechanistically, MTLN is likely to function through associations with key enzymes of fatty acid oxidation, CPT1, and fatty reduction and remodeling, CYB5B and CYB5R3. By contrast, we showed that the assembly and function of respiratory chain complexes remain unchanged at cellular and animal levels.
In line with the role of MTLN in regulating lipid metabolism, we showed that MTLN deficiency protected mice from Western diet and fructose-induced insulin insensitivity. We chose to challenge MTLN KO mice with the WDF diet as we found this diet induces the protein expression of MTLN (Figure 6A). Other groups have fed MTLN KO mice with high-fat diet and reported both increased and decreased serum-free fatty acid, an indicator of body fat content, in MTLN KO mice.18,35 Aside from these inconclusive observations, one phenotype that was repeatedly identified by different researchers is the accumulation of DHA.21,35 The accumulation of DHA and several other VLCFA species might be due to either a defect in their oxidation or an enhancement in biogenesis. We prefer the former explanation for two reasons. First, MTLN is highly expressed in catabolic tissues such as muscle and brown fat but low in anabolic tissues like liver. Second, multiple species of VLCFA are enriched in the MTLN KO and different pathways were involved in their biogenesis.36 The beta-oxidation of VLCFA is initiated in the peroxisome and continued in the mitochondria. Although we did not detect an interaction between MTLN and peroxisomal proteins using an affinity purification approach, a microprotein-compatible proximity labeling technique should be used to formally test this hypothesis in the future. Dietary supplement of ω-3 fatty acid such as DHA is well known to improve insulin sensitivity even though the molecular mechanism is less understood.37 Our observation suggests that a change in DHA metabolism and storage may directly impact the insulin sensitivity of animals. Future investigations on the molecular link between the regulation of DHA oxidation and insulin sensitivity will be required to unravel the underlying causal effect.
Limitations of the study
Future study is needed to pinpoint the exact molecular function of MTLN in fatty acid metabolism. We show that MTLN can interact with both CPT1 and CYB5B in the OMM. However, it is unclear whether MTLN needs to interact with both proteins simultaneously to fulfill its function. Moreover, it remains possible that CPT1 and CYB5B are not direct interactors of MTLN since co-IP/MS under native conditions can potentially detect second or third degree interactors residing in the same neighborhood. As such, we could refer to these as MTLN “associated proteins” instead of direct interactors. Besides, CPT1 is currently known as the rate-limiting carnitine acyltransferase of the fatty acid β-oxidation; whether it plays other moonlighting functions in fatty acid metabolism will need to be explored in the future. Additionally, we showed that the loss of MTLN protects mice from diet-induced insulin insensitivity in a relatively limited cohort size in C57BL/6J animals. Future animal studies with larger cohort sizes in diversified genetic backgrounds and large-scale phenotype-genotype association studies in human populations will help to validate our findings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ATP5A | Santa Cruz | Cat#SC-136178 RRID: AB_2061764 |
| Rabbit polyclonal anti-DYKDDDDK | ProteinTech | Cat#20543-1-AP RRID: AB_11232216 |
| Rabbit monoclonal anti-GAPDH | Cell signaling Technology | Cat#2118 RRID: AB_561053 |
| Mouse monoclonal anti-NDUFA9 | Invitrogen | Cat#459100 RRID: AB_2532223 |
| Rabbit polyclonal anti-SDHA | AB Clonal | Cat#A2594 RRID: AB_2764479 |
| Rabbit polyclonal anti-TIM23 | ProteinTech | Cat#11123-1-AP RRID: AB_615045 |
| Rabbit polyclonal anti-TOMM20 | ProteinTech | Cat#11802-1-AP RRID: AB_2207530 |
| Rabbit polyclonal anti-TOMM70 | AB Clonal | Cat#A4349 RRID: AB_2765636 |
| Rabbit polyclonal anti-UQCRC1 | AB Clonal | Cat#A3339 RRID: RRID: AB_2765058 |
| Mouse monoclonal anti-GFP | ProteinTech | Cat#66002-1-Ig RRID: AB_11182611 |
| Mouse monoclonal anti-Cytochrome C | Santa Cruz | Cat#SC-13561 RRID: AB_627381 |
| Rabbit polyclonal anti-VDAC1 | AB Clonal | Cat#A0810 RRID: AB_2757406 |
| Mouse monoclonal anti-MTCO1 | Abcam | Cat#Ab14705 RRID: AB_2084810 |
| Mouse monoclonal anti-Citrate Synthase | Santa Cruz | Cat#SC-390693 RRID: AB_2813783 |
| Mouse monoclonal anti-HA | BioLegend | Cat# 901502 RRID: AB_2565007 |
| Rabbit polyclonal anti-CPT1B | ABclonal | Cat# A6796 RRID: AB_2767379 |
| Mouse monoclonal anti-GRP 78 | Santa Cruz | Cat# sc-376768 RRID: AB_2819145 |
| Mouse monoclonal anti-HSP 60 | Santa Cruz | Cat# sc-13115 RRID: AB_627758 |
| Rabbit polyclonal anti-CYB5R3 | ProteinTech | Cat# 10894-1-AP RRID: AB_2292715 |
| Alexa Fluor 594 donkey anti−rabbit IgG (H + L) | Life Technology | Cat# A21207 |
| Alexa Fluor 488 donkey anti−goat IgG (H + L) | Life Technology | Cat# A11055 |
| HRP-conj Donkey Anti-Mouse IgG (H + L) | Jackson ImmunoResearch | Cat#715-035-150 |
| HRP-conj Goat Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch | Cat# 111-035-003 |
| Rabbit polyclonal anti-phospho-ACC (Ser79) | Cell Signaling Technology | Cat#3661; RRID: AB_330337 |
| Rabbit polyclonal anti-MTCH2 | ProteinTech | Cat#16888-1-AP RRID: AB_2266733 |
| Rabbit polyclonal anti-MITOREGULIN | This paper | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Sucrose | Sigma-Aldrich | Cat#S3089 |
| Ethylenediaminetetraacetic acid, ACS reagent, 99.4–100.6%, powder | Sigma-Aldrich | Cat#E9884 |
| Trizma® base, anhydrous, free-flowing, Redi-Dri™, ≥99.9% | Sigma-Aldrich | Cat#RDD008 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid, ≥97.0% | Sigma-Aldrich | Cat#E4378 |
| n-Dodecyl β-D-maltoside, ≥98% (GC) | Sigma-Aldrich | Cat#D4641 |
| Digitonin | Sigma-Aldrich | Cat#D141 |
| D-Sorbitol, ≥98% | Sigma-Aldrich | Cat#S1876 |
| ANTI-FLAG® M2 Affinity Gel, purified immunoglobulin, buffered aqueous glycerol solution | Sigma-Aldrich | Cat#A2220 |
| 3X FLAG® Peptide, lyophilized powder | Sigma-Aldrich | Cat#F4799 |
| Protein A/G agarose | Thermo Fisher Scientific | Cat#20421 |
| Sodium chloride, ReagentPlus®, ≥99% | Sigma-Aldrich | Cat#S9625 |
| Potassium chloride, powder, BioReagent, suitable for cell culture, suitable for insect cell culture, ≥99.0% | Sigma-Aldrich | Cat#P5405 |
| Lipofectamine 3000 | Invitrogen | Cat#L3000015 |
| Puromycin | Invivogen | Cat#ant-pr |
| G418 | Invivogen | Cat#ant-gn |
| Blotting-Grade Blocker | Bio-Rad | Cat#1706404 |
| 20X Tris-Buffered Saline (TBS) Buffer, Ultra Pure Grade | 1st base | Cat#BUF-3030-20X4L |
| TWEEN® 20, viscous liquid | Sigma-Aldrich | Cat#P1379 |
| NativePAGE™ Running Buffer (20X) | Invitrogen | Cat#BN2001 |
| NativePAGE™ Cathode Buffer Additive (20X) | Invitrogen | Cat#BN2002 |
| NativePAGE™ 5% G-250 Sample Additive | Invitrogen | Cat#BN2004 |
| NativePAGE™ Sample Buffer (4X) | Invitrogen | Cat#BN2003 |
| 20X MES SDS Running Buffer | Invitrogen | Cat#B0002 |
| cOmplete™, Mini Protease Inhibitor Cocktail | Roche | Cat#11836153001 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat#11836170001 |
| Methanol, for analysis EMSURE® ACS,ISO,Reag. Ph Eur | Merck | Cat#1.06009.2511 |
| Acetic acid (glacial) 100%, anhydrous for analysis EMSURE® ACS,ISO,Reag. Ph Eur | Merck | Cat#1.00063.2500 |
| DMEM/High glucose (4500 mg/mL) with L-glutamine; without sodium pyruvate | HyClone | Cat#SH30022.01 |
| DMEM/Low glucose (1000 mg/mL) with L-glutamine, sodium pyruvate | HyClone | Cat#SH30021.01 |
| Sodium pyruvate solution, 100 mM, sterile-filtered, BioReagent, suitable for cell culture | Sigma-Aldrich | Cat#S8636 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140-122 |
| Fetal Bovine Serum, South American Origin, Heat Inactivated | HyClone | Cat#SV30160.03 |
| Fetal Bovine Serum, dialyzed, US origin | Gibco | Cat#26400-044 |
| Triton™ X-100, laboratory grade | Sigma-Aldrich | Cat#X100 |
| Bovine Serum Albumin, heat shock fraction, protease free, fatty acid free, essentially globulin free, pH 7, ≥98% | Sigma-Aldrich | Cat#A7030 |
| Sodium deoxycholate, ≥97% (titration) | Sigma-Aldrich | Cat#D6750 |
| Sodium dodecyl sulfate, BioReagent, suitable for electrophoresis, for molecular biology, ≥98.5% (GC) | Sigma-Aldrich | Cat#L3771 |
| Glycerol, ACS reagent, ≥99.5% | Sigma-Aldrich | Cat#G7893 |
| HEPES, ≥99.5% (titration) | Sigma-Aldrich | Cat#H3375 |
| Magnesium chloride, anhydrous, ≥98% | Sigma-Aldrich | Cat#M8266 |
| D-(+)-Glucose solution, 100 g/L in H2O, sterile-filtered, BioXtra, suitable for cell culture | Sigma-Aldrich | Cat#G8644 |
| D-(+)-Galactose, powder, anhydrous, BioReagent, suitable for cell culture, suitable for insect cell culture | Sigma-Aldrich | Cat#G5388 |
| Oligomycin A, ≥99% (HPLC) | Sigma-Aldrich | Cat#75351 |
| Rotenone, ≥95% | Sigma-Aldrich | Cat#R8875 |
| Antimycin A from Streptomyces sp. | Sigma-Aldrich | Cat#A8674 |
| Acetyl coenzyme A, trilithium salt, trihydrate | MP Biomedicals | Cat#100490 |
| Oxaloacetic acid, ≥97% (HPLC) | Sigma-Aldrich | Cat#O4126 |
| Trypsin-EDTA (0.25%), phenol red | Gibco | Cat#25200-056 |
| DPBS without calcium, magnesium | HyClone | Cat#SH30028.02 |
| Sodium hydroxide, reagent grade, 97%, powder | Sigma-Aldrich | Cat#655104 |
| Agarose | 1st base | Cat#BIO-1000 |
| Pierce™ Trypsin Protease, MS Grade | Thermo Fisher Scientific | Cat#90057 |
| Proteinase K | Thermo Fisher Scientific | EO0491 |
| Trifluoroacetic acid, eluent additive for LC-MS, LiChropur, ≥99.0% (GC) | Sigma-Aldrich | Cat#80457 |
| Acetonitrile, anhydrous, 99.8% | Sigma-Aldrich | Cat#271004 |
| Ammonium hydroxide solution, puriss., 30–33% NH3 in H2O | Sigma-Aldrich | Cat#05002 |
| Pierce™ Formic Acid, LC-MS Grade | Thermo Fisher Scientific | Cat#TS-28905 |
| Acetone, ACS reagent, ≥99.5% | Sigma-Aldrich | Cat#179124 |
| Urea AR | Chem-Supply | Cat#UA001 |
| Rabbit IgG Isotype Control | Invitrogen | Cat#02-6102 |
| Pierce™ Protein A/G Agarose | Thermo Scientific | Cat#20422 |
| DharmaFECT1 | Horizon | Cat# T-2001-01 |
| Formaldehyde | Sigma-Aldrich | Cat#47608 |
| L-(−)-Malic acid | Sigma-Aldrich | Cat#M6413 |
| N,N,N′,N′-Tetramethylp-phenylenediamine | Sigma-Aldrich | Cat#T7394 |
| Adenosine 5′-diphosphate | Sigma-Aldrich | Cat#A5285 |
| L-carnitine | Sigma-Aldrich | Cat#C0283 |
| Palmitoyl-l-carnitine chloride | Sigma-Aldrich | Cat# P1645 |
| Palmitic acid, [1-14C]-40-60mCi(1.48–2.22GBq)/mmol | PerkinElmer | Cat#NEC075H250UC |
| Palmitic acid | Sigma-Aldrich | Cat#P5585 |
| Western diet | Research Diets | Cat# D12079B |
| D-(−)-Fructose | Sigma-Aldrich | Cat# F0127 |
| Acetonitrile | Sigma-Aldrich | Cat# 34851 |
| Pyruvic acid | Sigma-Aldrich | Cat#107360 |
| REDiant PCR master mix | 1st BASE | Cat# BIO-5185-200 |
| Seahorse XF DMEM medium | Agilent | Cat#103575-100 |
| FCCP | MedChenExpress | Cat#HY-100410 |
| Coomassie blue G250 | Sangon Biotech | Cat#A600038-0025 |
| Nitrotetrazolium blue chloride | Sigma-Aldrich | Cat#N6876 |
| Succinate | Sigma-Aldrich | Cat#S2378 |
| Fatty acid methyl esters i(FAMEs) mix | Anpel | Cat#CDAA-252795-MIX |
| 19:0 FAME | Anpel | Cat#CDAA-251019M |
| Critical commercial assays | ||
| Pierce BCA Protein Quantification Kit | Thermo Scientific | Cat#23225 |
| Mitochondria isolation kit for cultured cells | Abcam | Cat#ab110170 |
| Mitochondria isolation kit | Miltenyi Biotec | Cat#130-094-532 |
| Deposited data | ||
| Mass spectrometry data | PRIDE | PXD030258 |
| Experimental models: Cell lines | ||
| Human HEK293T | ATCC | Cat#CRL-3216 |
| Human HeLa | ATCC | Cat#CCL2 |
| Human U2OS | ATCC | Cat#HTB-96 |
| Mouse C2C12 | ATCC | Cat#CRL-1772 |
| Experimental models: Organisms/strains | ||
| Mouse: WT (C57BL/6J) | The Jackson Laboratory | Cat#000664 |
| Mouse: MTLN−/− (C57BL/6J) | This paper | NA |
| Oligonucleotides | ||
| Lincode Human LINC00116 (205251) siRNA | Horizon | Cat# RU-017831-00-0002 |
| MTLN KO mice gRNA1 (5′CAATGGCGGACGTGTCTGAG3′) | This study | NA |
| MTLN KO mice gRNA2 (5′ TCGGGCTGCAGCGCGTGCTC 3′) | This study | NA |
| HsMTCH2 gRNA (5′ TTTCACGTACATGAGCGGCT 3′) | This study | NA |
| MTLN KO mice genotyping forward (5′ACTTTGTGCTGAGTGGTTGC3′) | This study | NA |
| MTLN KO mice genotyping reverse (5′ AAGCCATGTTCTTGCAAGCC 3′) | This study | NA |
| MTLN KO C2C12 gRNA1 (5′CAATGGCGGACGTGTCTGAG3′) | This study | NA |
| MTLN KO C2C12 gRNA2 (5′GCTGGCAAGCGAATCGGCTG3′) | This study | NA |
| MTLN KO C2C12 gRNA3 (5′GCTGCAGGTGTCCGTGCTAG3′) | This study | NA |
| Recombinant DNA | ||
| lentiCRISPRv2 puro | Gift from Shang Li Lab | NA |
| pCS2-MTLN-HA | This study | NA |
| pcDNA3.1/Hygro(+)-GFP1-10-OMP25 | This study | NA |
| pcDNA3.1/Hygro(+)-LACTB-GFP1-10 | This study | NA |
| pcDNA3.1/Hygro(+)-Cox8a-GFP1-10 | This study | NA |
| pcDNA3.1/Hygro(+)-EMRE-GFP11 | This study | NA |
| pcDNA3.1/Hygro(+)-MTLN-GFP11 | This study | NA |
| pcDNA3.1/Hygro(+)-GFP11- MTLN | This study | NA |
| pcDNA3.1/Hygro(+)-Brawnin-GFP11 | This study | NA |
| pscAAV-CMV-MsMTLN-Flag | This study | NA |
| pCDH-CMV-MTLN-HA | This study | NA |
| pCDH- CMV-FLAG-CYB5B | This study | NA |
| pLV3-CMV-EMREMTS-GFP11-EMRE(54–107) | This study | NA |
| Software and algorithms | ||
| ImageJ | Schneider et al.38 | https://imagej.nih.gov/ij/ |
| GraphPad-Prism 9 | https://www.graphpad.com | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lena Ho (lena@ho-lab.org).
Materials availability
Unique reagents generated in this study will be made available upon reasonable request to the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and study participant details
Mice
C57BL/6J wild type (WT) and MTLN knock-out (KO) mice were housed in 12 h :12 h light : dark cycles at 21°C-24°C with relative humidity maintained at 40–60%. Twelve-week-old male mice were used in the WDF diet feeding experiment, and 4-month-old male mice were used in the mouse indirect calorimetry analysis. All the animal handling protocols were approved by the Duke-NUS, SingHealth Institutional Animal Care and Use Committee.
Cell lines
HEK293T cells (human, female), HeLa cells (human, female), U2OS cells (human, female), and C2C12 myoblast cells (mouse, female) were obtained from ATCC with no additional authentication performed as described in the key resources table.
Method details
Cell culture
HEK293T, HeLa, U2OS, and C2C12 myoblast were cultured at sub-confluency in DMEM high glucose (4500 mg/mL) medium supplemented with 10% FBS, 1 mM pyruvate and 100 U/mL penicillin-streptomycin. Cell culture incubators were maintained at 37°C with 5% CO2. To knock-down the MTLN expression by RNA interference, 25 nM of non-targeting and human MTLN-targeting siRNA were transfected into U2OS cells using the Dharmafect 1 reagent. To transiently express proteins of interest, plasmids were transfected into HEK293T and U2OS cells using Lipofectamine 2000. To generate MTLN KO cells, C2C12 cells were transduced with lentiviruses expressing guide RNAs and Cas9 protein. Briefly, 4.4 million of HEK293T cells were seeded one day before and transfected with 3 μg of transfer vector (lentiCRISPR v2 carries gRNAs as specified in the key resources table) and the third generation packaging constructs (1 μg of pMDL, 1 μg of pREV, and 0.5 μg of pVSV-G) in antibiotics free DMEM high glucose medium supplemented with 2% FBS. The transfection medium was replaced with normal DMEM medium (DMEM high glucose medium supplemented with 10% FBS, 1 mM pyruvate and 100 U/mL penicillin-streptomycin) 6 h after the transfection. Lentivirus-containing media were collected 48 h after the transfection and concentrated to 1 mL using Vivaspin 100,000 MWCO columns. 25, 000 C2C12 cells were transduced with crude viruses that 1:1 (vol:vol) mixed with fresh medium using polybrene at 8 μg/mL. Cells were selected with 1 μg/mL of puromycin 72 h post transduction.
Immunofluorescence analysis of cultured cells
Cells seeded in glass bottom dishes were washed with PBS and fixed with 4% paraformaldehyde for 15 min at 37°C. After PBS washes, cells were permeabilized and blocked with the blocking buffer (0.3% Triton X, 3% BSA, 10% FBS in PBS) for 1 h at room temperature (RT). Samples were stained with specific primary antibodies (1:500 diluted) in the staining buffer (0.1% Triton X, 3% BSA, 10% FBS in PBS) overnight at 4°C and washed with PBS+0.1% Tween20 (PBST) for 3 times. Samples were then stained with Alexa fluor 488, 594 antibodies (1:500 diluted) in the staining buffer and washed with PBST 3 times before imaging. Images were acquired using an Olympus FV3000 confocal laser scanning microscope.
Hydrophobicity for transmembrane microproteins
The list of curated microproteins (<100 a.a.) was downloaded from Uniprot with annotations of subcellular localization. Transmembrane domains of ER and mitochondrial microproteins were predicted by TMHMM-2.0. Hydropathic scores of the transmembrane domains of single-pass transmembrane proteins were calculated using Kyte & Doolittle’s method.39 The maximum and total hydropathic scores of the transmembrane domains were calculated and plotted.
Protein electrophoresis and immunoblotting
Proteins and protein complexes were analyzed by denaturing and native polyacrylamide gel electrophoresis (PAGE). In denaturing PAGE, samples were lysed in RIPA buffer supplemented with cOmplete protein inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl fluorid (PMSF). Protein concentrations were measured by bicinchoninic acid (BCA) assay and normalized. Protein lysates were incubated at 95°C for 5 min in 1× sample buffer (50 mM Tris-HCl pH6.8, 2% SDS, 10% glycerol, 12.5 mM EDTA, 0.02% bromophenol blue, 50 mM DTT) and separated by SDS-PAGE in NuPAGE MES SDS running buffer with 4-12% Bis-Tris precast gels (Thermo). Proteins were transferred to 0.2 μM PVDF membranes (Bio-Rad) using a Trans-Blot Turbo system (Bio-Rad). Membranes were incubated in the blocking buffer (TBS with 0.1% Tween20 and 5% skim milk powder) for 1 h at RT and incubated with diluted primary antibodies in the blocking buffer overnight at 4°C. Primary antibodies were diluted for 1:2000 diluted or 1:5000 diluted in the blocking buffer. After extensive washes in TBST, membranes were probed with HRP-conjugated secondary antibodies against species-specific primary antibodies and washed. Chemiluminescence was recorded by X-ray films or a ChemiDoc imaging system (Bio-Rad).
In native-PAGE, 50 μg of mitochondrial proteins were solubilized in 20 μL of 1× Native PAGE sample buffer (Thermo) containing 400 μg of digitonin and incubated on ice for 20 min. Samples were spun at 20, 000 ×g for 10 min at 4°C, and the supernatants were collected and stained with 0.05% of Coomassie blue G250. Protein lysates were resolved by 3-12% Native PAGE Bis-Tris gels (Thermo) in the dark blue cathode running buffer (Thermo) at 150 V for 30 min and in the light cathode running buffer at 250 V for 60 min in an ice bath. Protein complexes were transferred to 0.2 μM PVDF membranes (Bio-Rad) using a Trans-Blot Turbo system (Bio-Rad). Membranes were fixed in 8% acetic acid for 15 min at RT, washed with water, air dried, detained with methanol, and washed with water again. Membranes were blocked and probed as described above. In experiments where 2nd dimensional SDS-PAGE was performed, gel slices of lanes of interest were excised by a microtome blade and slotted horizontally on top of a comb-free SDS gel and subjected to electrophoresis. When complex I in-gel assay was performed, native PAGE gels were washed with water and incubated with 20 mL of complex I activity buffer (2 mM Tris-HCl pH7.4, 0.1 mg/mL NADH, 2.5 mg/mL nitrotetrazolium blue chloride) and fixed with 8% of acetic acid until complex I containing bands are obvious.
Mitochondria isolation
Mitochondria were isolated cultured cells using the mitochondria isolation kit (Abcam) following the user manual. Mitochondria were isolated from mouse tissue as described. Briefly, freshly dissected mouse skeletal muscle and heart were minced and resuspended in 10 volumes of skeletal muscle mitochondria isolation buffer (67 mM sucrose, 50 mM KCl, 10 mM EDTA, 0.1% fatty acid-free BSA, 50 mM Tris-HCl, pH7.4) or heart mitochondria isolation buffer (75 mM sucrose, 225 mM sorbitol, 1mM EGTA, 0.1% fatty acid-free BSA, 10 mM Tris-HCl, pH7.4) supplemented with protease inhibitors cocktail and 1mM PMSF. Tissues were homogenized with a motor-driven Potter-Elvehjem tissue grinders operated at 1300 rpm with 10-15 strokes performed. Tissue homogenates were spun at 600 ×g for 10 min and the supernatant was collected. Mitochondria were precipitated by centrifugation at 7,000 ×g for 15 min and resuspended in Buffer C (320 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH7.4). Mitochondria protein concentration was measured by BCA assay and used for downstream experiments.
Mitochondrial membrane fractionation
Fractionation of mitochondria membranes was performed as described.10 Briefly, mitochondria were isolated freshly from HEK293T cells and resuspended to 1 mg of mitochondrial protein per 1 mL of isotonic buffer (250 mM sucrose, 1 mM EDTA, 10 mM HEPES-KOH pH 7.4, protease inhibitor cocktail and 1 mM PMSF). Mitochondria were aliquoted and solubilized by adding digitonin to different concentrations (0%, 0.1%, 0.115%, 0.13%, 0.145%, 0.16%, 0.175%, 0.19%, 0.205%, and 0.22%) and incubated at 4°C for 1 h with nutation. Aliquots were centrifuged at 20, 000 ×g for 20 min at 4°C to separate the soluble and insoluble fractions. Insoluble pellets were resuspended in equal volumes of isotonic buffer and analysed together with the soluble fraction by SDS-PAGE and immunoblotting.
Protease sensitivity assay
Protein topology was analysed by protease sensitivity assay as described before. Mitochondria were isolated from fresh HEK293T cells and resuspended to 1 mg of mitochondrial protein per 1 mL of the isotonic buffer without protease inhibitors (250 mM sucrose, 1 mM EDTA, 10 mM HEPES-KOH pH 7.4). Mitochondria membranes were solubilized by adding digitonin to different concentrations (0%, 0.02%, 0.04%, 0.06%, 0.08%, 0.1%, 0.15%, 0.2%) or Triton X to 1%. Samples were incubated on ice for 10 min and treated with 100 μg/mL of proteinase K for 30 min on ice. Proteinase K was omitted in one of the detergent free samples to serve as the total control. Digestion was terminated by 8 mM of freshly prepared PMSF. Samples were analysed by SDS-PAGE and immunoblotting. To test whether a freeze-thaw cycle would affect the protease sensitivity of MTLN, HEK293T cell pellets were snap frozen and thawed on ice before mitochondrial isolation where appropriate.
Analysis of membrane topology and sub-mitochondrial localization by split GFP
The N terminus (68 a.a.) of mouse LACTB N and the presequence (29 a.a.) of human COX8A were fused to the N terminus of GFP1-10 to target it to the inter-membrane space and matrix of mitochondria, respectively. The C terminus (39 a.a.) of human OMP25 was fused to the C terminus GFP1-10 to target it to the outer membrane of mitochondria. The correct targeting of these fusion proteins was confirmed by protease sensitivity assays as described above. The GFP11 tag was fused to the N or C terminus of proteins to be assayed as described in figure legends. GFP11-tagged proteins were co-transfected with the three mitochondrial GFP1-10s in pairs in U2OS cells. The GFP fluorescence was recorded by confocal microscopy.
Immunoisolation of mitochondria
Immunoisolation of mitochondria was performed using magnetic mitochondria isolation kit (Miltenyi Biotec) following the user manual. Protein A magnetic beads (Miltenyi Biotec) were coated with MTLN antibody or TIM23 antibody and used together with the TOM22 antibody pre-coated magnetic beads (Miltenyi Biotec). Cells were lysed by Dounce homogenization in buffer A and incubated with magnetic beads. The labelled mitochondria was captured by a magnetic separator, washed and eluted.
Colocalization analysis
Pearson correlation coefficients between HA-tagged microproteins and TOM20 immunofluorescence signals were quantified using the Coloc 2 plugin of ImageJ. Images were processed to yield individual cells for the colocalization analysis.
Mitochondrial respiratory assay with isolated mitochondria
Mitochondria were isolated from mouse heart as described above. In the coupled assay, mitochondria were resuspended in activity assay buffer (70 mM sucrose, 220 mM mannitol, 5 mM KH2PO4, 5 mM MgCl2, 1 mM EGTA, 0.2% fatty acid free BSA, 2 mM HEPES pH7.4) supplemented with 10 mM of pyruvate and 5 mM of malate. In the electron flow assay, mitochondria were resuspended in activity assay buffer supplemented with 5 mM pyruvate, 1 mM malate and 4 μM FCCP. 96 well Seahorse cell culture plates were coated with purified mitochondria (1.4 μg of mitochondrial protein per well) by centrifugation at 2, 000 g for 20 min at 4°C. Pre-warmed assay buffer and substrates were added to 180 μL, and the Seahorse plate was loaded. In coupled assays, 3.2 mM of ADP (concentrations in wells are indicated), 3 μM of oligomycin, 4 μM of FCCP, and 4 μM of antimycin A diluted in the activity assay buffer without BSA were sequentially injected. In the electron flow assay, 2 μM of rotenone, 5 mM of succinate, 4 μM of antimycin A, and 100 μM of TMPD were sequentially injected. Oxygen consumption rates were recorded on an Agilent XFe96 analyzer.
Generation and genotyping of MTLN KO mice
The C57BL6/J MTLN KO line was generated by pronuclear microinjection of 2 gRNA as described in the key resources table and Cas9 protein at Walter and Eliza Hall Institute of Medical Research. The Cas9 cleavage led to a severe truncation in the coding sequence of MTLN. To genotype the animals, mouse tail clips were treated in lysis buffer (25 mM NaOH, 0.2 mM EDTA) at 95°C for 2 h and neutralized with an equal volume of neutralization buffer (80 mM Tris-HCl pH 3.0). Crude DNA extracts were amplified with REDiant PCR master mix (1st Base) with primers described in the key resources table. Heterozygous KOs was backcrossed with WT C57BL6/J mice for 8 generations before inbreeding to generate WT and MTLN KO that were used in experiments.
Seahorse mitostress test
12, 000 C2C12 myoblast were seeded on Seahorse 96 well cell culture plate with normal DMEM high glucose media 1 day before the assay. The sensor cartridge was hydrated with water overnight and incubated with the calibration buffer on the day of assay. Right before the assay, the original media was replaced with XF basal DMEM media supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose. The cell culture plates were incubated at 37°C at ambient CO2 level for 45 min and loaded on a Agilent XFe96 analyzer. 2 μM of oligomycin, 1 μM of FCCP, and 1 μM of rotenone/antimycin were sequentially injected following a standard to Seahorse MitoStress assay protocol (Agilent). Citrate synthase activity was performed to normalize cell number as described.11
Mouse indirect calorimetry
4-month-old male mice (4 animals each phenotype) were housed inside the calorimetric chamber (Phenomaster) for a total of 6 days at a constant temperature of 23°C and humidity of 65%. The in-and-out O2 and CO2 concentrations were measured for each home cage to calculate O2 consumption and CO2 production. O2 and CO2 sensors were calibrated with calibration gas mixtures (Calibration report: Good air equivalent: CO2: 0.050 ± 0.0005%, O2: 20.900 ± 0.1% in N2; CO2 span: CO2: 1.000 ± 0.001%, O2: 20.000 ± 0.1% in N2). The sample air flow was adjusted to 0.37 L/min. High precision weighing stations in combination with leak- and spill-proof containers recorded accurately the body weight, food and water intake (in grams). Spontaneous activity was recorded with two levels of infrared light beam frames surrounding each cage. Recording began from the first entry in the chamber but measures were considered only from the 2nd day in the chamber. All measurements were taken every 15 min and are presented here most of the time hourly or as means per hour.
Protein immunoprecipitation and mass spectrometry
Endogenous MTLN and FLAG-tagged were purified from mouse skeletal muscles and AAV infection mouse hearts, respectively. In the purification of endogenous MTLN, skeletal muscle mitochondria were isolated from WT and MTLN KO mice as described above. 2 mg of mitochondrial proteins were solubilized in 800 μL of IP buffer (100 mM NaCl, 10% glycerol, 1 mM EDTA, 25 mM Tris- HCl, pH7.4) supplemented with 1% DDM and protease inhibitor cocktail. After nutation at 4°C for 1 h, lysates were centrifuged at 20, 000 ×g for 15 min at 4°C to remove the insoluble fraction. The supernatant was incubated with 5 μg of MTLN antibody and nutated at 4°C for 2 h and centrifuged again at 20, 000 ×g for 10 min at 4°C. The supernatant was then incubated with 20 μL of protein A/G beads and nutated for 1 h at 4°C. Beads were washed with 1mL of IP buffer supplemented with 0.5% DDM 5 times, and bound protein were eluted with 2% SDS. To purify FLAG-tagged MTLN from mouse hearts, MTLN with a C-terminal FLAG tag was cloned into the psAAV construct under a CMV promoter. Adeno-associated viruses were produced by Vector Core @ GIS (A∗STAR, Singapore). 1×1010 vg of viruses were injected into the thoracic cavity of P6 pups. AAV expressing GFP was used as a control. Hearts were collected 6 weeks after the injection, and heart mitochondria were isolated. 600 μg of mitochondrial proteins were solubilized in 1200 μL of IP buffer (100 mM NaCl, 10% glycerol, 1 mM EDTA, 25 mM Tris-HCl, pH7.4) supplemented with 0.2% digitonin and protease inhibitor cocktail. Solubilized protein was incubated with anti-FLAG M2 beads for 3 h at 4°C. Beads were extensively washed and eluted with 3×FLAG peptides. Eluted proteins were analyzed by mass spectrometry as described.11
Fatty acid oxidation assay
Rates of fatty acid oxidation were measured in mouse tissue homogenates using 14C labelled palmitic acid. First, 50 μL of 0.1mCi/mL of 14C palmitic acid was dry down under nitrogen gas and incubated with 500 μL of 7% BSA containing 5mM palmitate at 37°C for 30 min with frequent vortexing. Solubilized 14C palmitic acid was diluted 10 times in oxidation reaction mixture (100 mM sucrose, 5 mM KH2PO4, 0.2 mM EDTA, 80 mM KCl, 1 mM MgCl2, 2 mM L-carnitine, 0.1 mM malate, 0.05 mM coenzyme A, 10 mM Tris-HCl, pH8). Freshly digested mouse tissues were homogenized with 5 volumes of STE buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH7.4) using a Potter-Elvehjem homogenizer. Tissue homogenates were centrifuged at 420 ×g for 10 min at 4°C. To start the assay, 30 μL of tissue homogenate was incubated with 370 μL of the oxidation reaction mixture containing 14C palmitic acid at 37°C for 30 min. After the reaction, reaction mix was incubated with 200 μL of 1 M perchloric acid, and radioactive CO2 oxidized from 14C palmitate was trapped by a wet paper disk containing 20 μL of 1M NaOH. Radioactivity in the acid-soluble fraction and trapped CO2 was measured by liquid scintillation.
WDF diet feeding
Twelve-week-old male mice were fed with Western diet (40%) supplemented with 15% (w/v) fructose in drinking water for 16 weeks. Body weight and body content were measured by quantitative magnetic resonance imaging (EchoMRI) every 7 days.
Glucose tolerance test and insulin tolerance test
Mice were fasted for 6 h before the test. 2 g of glucose per kg of lean body mass and 0.5 U of insulin per kg of body mass were intraperitoneally injected. Blood glucose levels were measured before and after the injection using an Accu-Chek blood glucose meter via tail snip.
Targeted metabolomics analysis
Mouse heart and gastrocnemius muscle were homogenized in 50% acetonitrile/50% H2O (300 μL per 50 mg of tissue) with two 20 s cycles of 5000 rpm bead beating in a liquid nitrogen cooled tissue homogenizer (Cayman Chem). Tissue homogenates were extracted by ethylacetate and analysed by liquid chromatography–mass spectrometer as described.10
Quantification of fatty acid methyl esters by GC-MS
C2C12 cells in 6-well plates were washed, trypsinized, and resuspended in PBS for cell counting. Samples were prepared alongside standard curve samples made up of Fatty acid methyl esters (FAMEs) mix (Anpel). Fresh cell and mitochondria pellets were subjected to downstream derivatization and lipid extraction procedures without freezing. Samples were incubated at 45°C overnight in an acetyl chloride/toluene/methanol mixture supplemented with 19:0 FAME (Anpel) as an internal standard. After incubation, tubes were cooled to room temperature. 0.66 M aqueous sodium carbonate was added to yield a final aqueous layer of approximately 0.1 M sodium carbonate. Resultant FAMEs were extracted by adding 2 mL of n-hexane, and then the tubes were shaken and centrifuged at 2,000 ×g for 5 min. The organic layer was extracted to a new tube, and the 2 mL n-hexane extraction was repeated. The combined organic layers were evaporated to dryness under nitrogen flow. The dried lipids were redissolved in a small amount of n-hexane appropriate for the sample and analyzed by gas chromatography-mass spectrometry (GC-MS/MS) using an Agilent 7890B/7000C with HP-5 column (Agilent, 19091J-413). Complete GC-MS/MS configurations and method programs are available upon request. FAMEs were identified by molecular ion m/z (unit mass) spectrum and retention time compared to known standards. Integration and quantification of selected ions were performed on MassHunter Quantitative Analysis Program (Agilent Technologies, B.06.00).
Acknowledgments
This work was funded by NRF-NRFF2017-05, HHMI IRSP 55008732 awarded to L.H. S.Z. was supported by a KTP postdoctoral fellowship from Duke-NUS Medical School and is supported by start-up funds from the Zhejiang University. D.S. is supported by grants from the Australian National Health and Medical Research Council (NHMRC Project Grant 1140906 and NHMRC Fellowships 1140851). Q.Z. is supported by the National Natural Science Foundation of China (32171275). We acknowledge the support of the Mito Foundation (Australia) for the provision of mass spectrometry instrumentation through the large equipment grant support scheme. We thank the Bio21 Mass Spectrometry and Proteomics Facility (MMSPF) for provision of instrumentation, training, and technical support. We thank Zhaoxiaonan Lin from the Core Facilities, Zhejiang University School of Medicine for their technical support. We thank Zhiwei Ge and Ya’er Zhu (Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University) for their assistance of GC-MS/MS data collection. D.K. is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative.
Author contributions
S.Z. and L.H. conceived this work, designed and interpreted all experiments. S.Z. performed experiments with collaborative help from Y.G., G.F., and R.G. (biochemical assays, protein topology analysis and split-GFP), D.R, D.K., and D.S. (mass spectrometry), C.L. (Crispr KO cell), B.K., and P.Y. (WDF feeding and genotyping). Z.B. and R.L. performed indirect calorimetry. G.W., H.X., and, Q.Z. performed fatty acid methyl esters quantification. S.Z. and L.H. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107558.
Contributor Information
Shan Zhang, Email: shanzhang@zju.edu.cn.
Lena Ho, Email: lena@ho-lab.org.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. Mass spectrometry data have been deposited to PRIDE: PXD030258. No new code has been generated in this study. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J.S., Jackson S.E., Wills M.R., Weissman J.S. Ribosome Profiling Reveals Pervasive Translation Outside of Annotated Protein-Coding Genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright B.W., Yi Z., Weissman J.S., Chen J. The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol. 2022;32:243–258. doi: 10.1016/j.tcb.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez T.F., Chu Q., Donaldson C., Tan D., Shokhirev M.N., Saghatelian A. Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 2020;16:458–468. doi: 10.1038/s41589-019-0425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., Brunner A.D., Cogan J.Z., Nuñez J.K., Fields A.P., Adamson B., Itzhak D.N., Li J.Y., Mann M., Leonetti M.D., Weissman J.S. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367:1140–1146. doi: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mudge J.M., Ruiz-Orera J., Prensner J.R., Brunet M.A., Gonzalez J.M., Magrane M., Martinez T., Schulz J.F., Yang Y.T., Albà M.M., et al. A community-driven roadmap to advance research on translated open reading frames detected by Ribo-seq. bioRxiv. 2021 doi: 10.1101/2021.06.10.447896. [DOI] [Google Scholar]
- 7.Olexiouk V., Van Criekinge W., Menschaert G. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018;46:D497–D502. doi: 10.1093/nar/gkx1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunet M.A., Lucier J.F., Levesque M., Leblanc S., Jacques J.F., Al-Saedi H.R.H., Guilloy N., Grenier F., Avino M., Fournier I., et al. OpenProt 2021: Deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 2021;49:D380–D388. doi: 10.1093/nar/gkaa1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neville M.D.C., Kohze R., Erady C., Meena N., Hayden M., Cooper D.N., Mort M., Prabakaran S. A platform for curated products from novel open reading frames prompts reinterpretation of disease variants. Genome Res. 2021;31:327–336. doi: 10.1101/gr.263202.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S., Reljić B., Liang C., Kerouanton B., Francisco J.C., Peh J.H., Mary C., Jagannathan N.S., Olexiouk V., Tang C., et al. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 2020;11:1312. doi: 10.1038/s41467-020-14999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C., Zhang S., Robinson D., Ploeg M.V., Wilson R., Nah J., Taylor D., Beh S., Lim R., Sun L., et al. Mitochondrial microproteins link metabolic cues to respiratory chain biogenesis. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111204. [DOI] [PubMed] [Google Scholar]
- 12.Lee C.Q.E., Kerouanton B., Chothani S., Zhang S., Chen Y., Mantri C.K., Hock D.H., Lim R., Nadkarni R., Huynh V.T., et al. Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat. Commun. 2021;12:2130. doi: 10.1038/s41467-021-22397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatta A., Atianand M., Jiang Z., Crabtree J., Blin J., Fitzgerald K.A. A Mitochondrial Micropeptide Is Required for Activation of the Nlrp3 Inflammasome. J. Immunol. 2020;204:428–437. doi: 10.4049/jimmunol.1900791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarewich C.A., Munir A.Z., Bezprozvannaya S., Gibson A.M., Young Kim S., Martin-Sandoval M.S., Mathews T.P., Szweda L.I., Bassel-Duby R., Olson E.N. The cardiac-enriched microprotein mitolamban regulates mitochondrial respiratory complex assembly and function in mice. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2120476119. e2120476119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu Q., Martinez T.F., Novak S.W., Donaldson C.J., Tan D., Vaughan J.M., Chang T., Diedrich J.K., Andrade L., Kim A., et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019;10:4883. doi: 10.1038/s41467-019-12816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catherman A.D., Li M., Tran J.C., Durbin K.R., Compton P.D., Early B.P., Thomas P.M., Kelleher N.L. Top down proteomics of human membrane proteins from enriched mitochondrial fractions. Anal. Chem. 2013;85:1880–1888. doi: 10.1021/ac3031527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C., Giraldez A.J. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesen M., Warren C.R., Yu H., Toyohara T., Ding Q., Florido M.H.C., Sayre C., Pope B.D., Goff L.A., Rinn J.L., Cowan C.A. Mitoregulin Controls β-Oxidation in Human and Mouse Adipocytes. Stem Cell Rep. 2020;14:590–602. doi: 10.1016/j.stemcr.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein C.S., Jadiya P., Zhang X., McLendon J.M., Abouassaly G.M., Witmer N.H., Anderson E.J., Elrod J.W., Boudreau R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018;23:3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarewich C.A., Baskin K.K., Munir A.Z., Bezprozvannaya S., Sharma G., Khemtong C., Shah A.M., McAnally J.R., Malloy C.R., Szweda L.I., et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chugunova A., Loseva E., Mazin P., Mitina A., Navalayeu T., Bilan D., Vishnyakova P., Marey M., Golovina A., Serebryakova M., et al. LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. USA. 2018;116:4940–4945. doi: 10.1073/pnas.1809105116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y.F., Xiao M.H., Chen H.X., Meng Y., Zhao N., Yang L., Tang H., Wang J.L., Liu X., Zhu Y., Zhuang S.M. A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 2019;10:528. doi: 10.1038/s41419-019-1767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Fan J., Han L., Qi H., Wang Y., Wang H., Chen S., Du L., Li S., Zhang Y., et al. The micropeptide LEMP plays an evolutionarily conserved role in myogenesis. Cell Death Dis. 2020;11:357. doi: 10.1038/s41419-020-2570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao M.H., Lin Y.F., Xie P.P., Chen H.X., Deng J.W., Zhang W., Zhao N., Xie C., Meng Y., Liu X., et al. Downregulation of a mitochondrial micropeptide, MPM, promotes hepatoma metastasis by enhancing mitochondrial complex I activity. Mol. Ther. 2022;30:714–725. doi: 10.1016/j.ymthe.2021.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamiyama D., Sekine S., Barsi-Rhyne B., Hu J., Chen B., Gilbert L.A., Ishikawa H., Leonetti M.D., Marshall W.F., Weissman J.S., Huang B. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 2016;7:11046–11049. doi: 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vais H., Mallilankaraman K., Mak D.O.D., Hoff H., Payne R., Tanis J.E., Foskett J.K. EMRE Is a Matrix Ca2+ Sensor that Governs Gatekeeping of the Mitochondrial Ca2+ Uniporter. Cell Rep. 2016;14:403–410. doi: 10.1016/j.celrep.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto T., Yamagoshi R., Harada K., Kawano M., Minami N., Ido Y., Kuwahara K., Fujita A., Ozono M., Watanabe A., et al. Analysis of the structure and function of EMRE in a yeast expression system. Biochim. Biophys. Acta. 2016;1857:831–839. doi: 10.1016/j.bbabio.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Tsai M.-F., Phillips C.B., Ranaghan M., Tsai C.W., Wu Y., Willliams C., Miller C. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. Elife. 2016;5:e155455. doi: 10.7554/eLife.15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Nguyen N.X., She J., Zeng W., Yang Y., Bai X.C., Jiang Y. Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter. Cell. 2019;177:1252–1261.e13. doi: 10.1016/j.cell.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guna A., Stevens T.A., Inglis A.J., Replogle J.M., Esantsi T.K., Muthukumar G., Shaffer K.C.L., Wang M.L., Pogson A.N., Jones J.J., et al. MTCH2 is a mitochondrial outer membrane protein insertase. Science. 2022;378:317–322. doi: 10.1126/science.add1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L.O., Grevengoed T.J., Paul D.S., Ilkayeva O., Koves T.R., Pascual F., Newgard C.B., Muoio D.M., Coleman R.A. Compartmentalized Acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes. 2015;64:23–35. doi: 10.2337/db13-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K., Kerner J., Hoppel C.L. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J. Biol. Chem. 2011;286:25655–25662. doi: 10.1074/jbc.M111.228692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L., et al. The Translational Landscape of the Human Heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Cho N.H., Cheveralls K.C., Brunner A.D., Kim K., Michaelis A.C., Raghavan P., Kobayashi H., Savy L., Li J.Y., Canaj H., et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science. 2022;375:eabi6983. doi: 10.1126/science.abi6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Averina O.A., Permyakov O.A., Emelianova M.A., Grigoryeva O.O., Gulyaev M.V., Pavlova O.S., Mariasina S.S., Frolova O.Y., Kurkina M.V., Baydakova G.V., et al. Mitochondrial peptide Mtln contributes to oxidative metabolism in mice. Biochimie. 2023;204:136–139. doi: 10.1016/j.biochi.2022.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Sassa T., Kihara A. Metabolism of very long-chain fatty acids: Genes and pathophysiology. Biomol. Ther. 2014;22:83–92. doi: 10.4062/biomolther.2014.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lepretti M., Martucciello S., Aceves M.A.B., Putti R., Lionetti L. Omega-3 fatty acids and insulin resistance: focus on the regulation of mitochondria and endoplasmic reticulum stress. Nutrients. 2018;10:1–20. doi: 10.3390/nu10030350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider, Caroline A., Wayne S. Rasband, Kevin W. Eliceiri. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012; 9: 671-675. [DOI] [PMC free article] [PubMed]
- 39.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. Mass spectrometry data have been deposited to PRIDE: PXD030258. No new code has been generated in this study. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.