ABSTRACT
Regular high-intensity exercise can cause changes in athletes’ gut microbiota, and the extent and nature of these changes may be affected by the athletes’ exercise patterns. However, it is still unclear to what extent different types of athletes have distinct gut microbiome profiles and whether we can effectively monitor an athlete’s inflammatory risk based on their microbiota. To address these questions, we conducted a multi-cohort study of 543 fecal samples from athletes in three different sports: aerobics (n = 316), wrestling (n = 53), and rowing (n = 174). We sought to investigate how athletes’ gut microbiota was specialized for different types of sports, and its associations with inflammation, diet, anthropometrics, and anaerobic measurements. We established a microbiota catalog of multi-cohort athletes and found that athletes have specialized gut microbiota specific to the type of sport they engaged in. Using latent Dirichlet allocation, we identified 10 microbial subgroups of athletes’ gut microbiota, each of which had specific correlations with inflammation, diet, and anaerobic performance in different types of athletes. Notably, most inflammation indicators were associated with Prevotella-driven subgroup 7. Finally, we found that the effects of sport types and exercise intensity on the gut microbiota were sex-dependent. These findings shed light on the complex associations between physical factors, gut microbiota, and inflammation in athletes of different sports types and could have significant implications for monitoring potential inflammation risk and developing personalized exercise programs.
IMPORTANCE
This study is the first multi-cohort investigation of athletes across a range of sports, including aerobics, wrestling, and rowing, with the goal of establishing a multi-sport microbiota catalog. Our findings highlight that athletes’ gut microbiota is sport-specific, indicating that exercise patterns may play a significant role in shaping the microbiome. Additionally, we observed distinct associations between gut microbiota and markers of inflammation, diet, and anaerobic performance in athletes of different sports. Moreover, we expanded our analysis to include a non-athlete cohort and found that exercise intensity had varying effects on the gut microbiota of participants, depending on sex.
KEYWORDS: multi-sport, gut microbiota, inflammation, latent Dirichlet allocation, aerobics, wrestling, rowing
INTRODUCTION
Regular physical exercise is beneficial for human health, such as optimizing cardiorespiratory fitness, immunity, insulin sensitivity, and body composition (1, 2). This is especially true for athletes, who focus on fitness-enhancing training to improve their athletic performance. Previous studies have found significant changes in the composition and function of the gut microbiome in athletes under the influence of prolonged high-intensity physical exercise (2 - 4). For instance, compared to normal people, rugby players had a higher abundance of Firmicutes and lower levels of Bacteroidetes (5). Marathon runners had an increased presence of Veillonella (6). Professional cyclists had an increased abundance of Methanobrevibacter smithii and Prevotella (7). These suggested that different types of athletes with specialized training might have different gut microbial variations compared to healthy individuals. Specialized gut microbial changes can affect athletic performance through multiple avenues, such as regulating excitatory and inhibitory neurotransmitters to alleviate psychological stress (8, 9), and converting exercised-induced lactate into propionate to improve endurance (6). However, it is unclear how the gut microbiota profiles of athletes differ across different types of sports and whether specialized microbial patterns are associated with specific training models. A better understanding of these links could deepen our understanding of the specialized microbial profiles under specific training patterns, thereby facilitating the development of personalized exercise modulation for individuals.
However, when physical activity is too intense, athletes may experience negative effects of gut microbiota changes during exercise (10), such as gut inflammation. Gut inflammation was correlated with an increased risk of viral or bacterial infection, partially caused by excessive physical activity (11). Previous studies have found that physical activity resulted in profound differences in inflammatory and metabolic markers between professional athletes and controls (12). For example, rugby players exhibit lower levels of proinflammatory cytokines (12), and endurance athletes show increased production of butyrate (a modulator of proper immune function) (13). Changes in inflammation-related factors in athletes might imply different degrees of potential gut inflammation risk. However, few studies have focused on microbes’ correlations with the gut inflammation that athletes may experience during exercise. Exploring the potential of gut microbiota in adjusting the inflammation of athletes might help them to alleviate the effects of sports injuries. Therefore, we constructed a comprehensive multi-cohort study including information about physical fitness, diet, blood measurements, and gut microbial profiles for different types of athletes. We aimed to grasp the specialization of gut microbiota across multiple sports, as well as their associations with inflammation. This information could prove highly significant for monitoring the potential inflammation risk and further achieving personalized exercise modulation.
In this study, we built a microbiome catalog for athletes across different types of sports and answer key questions including how gut microbiota differed in athletes from different types of sports; which microbes were associated with and their potential effects on inflammation, diet, and anaerobic performance; and what were the differences in gut microbiota among participants with different exercise intensities.
RESULTS
Overview of the multi-cohort study across different types of sports
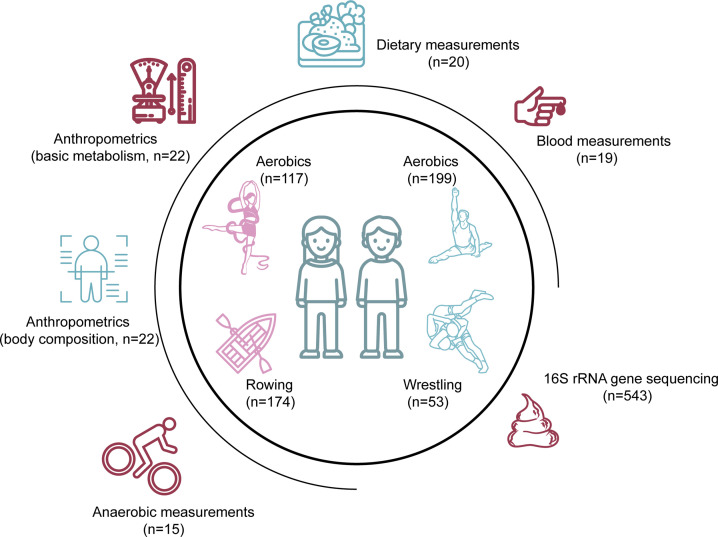
We collected a total of 543 fecal samples from athletes in three types of sports, including aerobics (AER), wrestling (WRE), and rowing (ROW), and designated these samples as the multi-sport meta-cohort (MS cohort, Fig. 1). These various sports have specialized training patterns with AER focusing on flexibility, ROW on endurance, and WRE on both endurance and explosive power. Owing to the genetic, physical, and environmental differences between females and males, we divided the MS cohort into two sub-cohorts for the following analysis, comprising the MS-female and MS-male cohorts. The MS-female cohort was composed of 117 AER female athletes and 174 ROW female athletes, while the MS-male cohort was composed of 199 AER male athletes and 53 WRE male athletes. In addition, we collected 19 blood measurements, 20 dietary measurements, 22 anthropometrics (basic metabolism and body composition), and 15 anaerobic measurements to be correlated with athletes' gut microbial profiles (Table S1). Furthermore, to investigate the impact of exercise intensities on gut microbiota, we additionally recruited and examined 178 fecal samples from non-athletes.
Fig 1.
Overview of the multi-sport cohort. Diagram summarizes data cohort and available metadata (n = number of variables collected).
Establishment and comparison of multi-sport gut microbial profiles
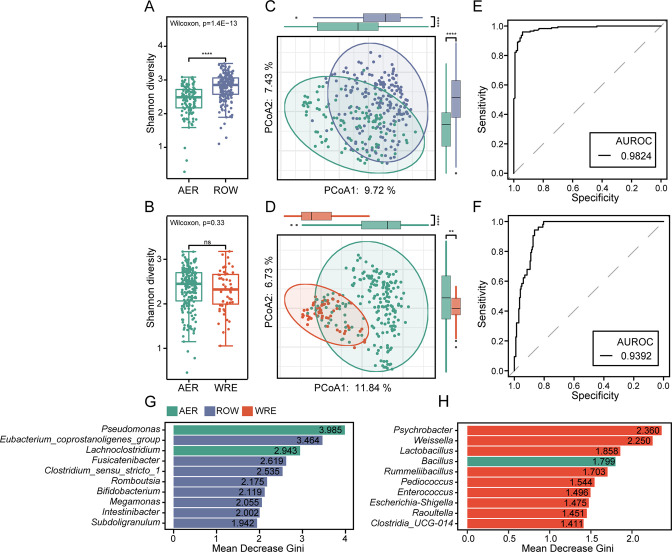
We first established a multi-sport gut microbial profile and found that aerobics, wrestling, and rowing athletes have their specific gut microbial profiles. In the MS cohort, we identified gut microbes at phylum (n = 9), class (n = 13), order (n = 27), family (n = 52), and genus (n = 111) levels that were presented in more than 10% of all the 543 fecal samples, as the MS microbial catalog. We then compared these gut microbes at each taxonomic level between different types of sports in the MS-female and MS-male cohorts, respectively (Table S2). Phyla Actinobacteriota (P = 2.09E−13) and Proteobacteria (P = 1.74E−14) were most differently distributed in the MS-female and MS-male cohorts, respectively. We found that the differentiation in gut microbes was notable, with 77% of genera differently distributed between AER and ROW in the MS-female cohort and 64% of genera between AER and WRE in the MS-male cohort (q < 0.05). We then investigated the differentiation in microbial diversity. In the MS-female cohort, ROW athletes’ gut microbiota had higher Shannon diversity than AER athletes’ gut microbiota (P = 1.4E−13, Fig. 2A), and their overall profiles were obviously separated against the PCoA1 axis (P = 1.09E−12) and PCoA2 axis (P = 1.42E−14) of principal coordinates analysis (PCoA) based on Bray-Curtis dissimilarities at the genus level (Fig. 2C). The random forest algorithm was applied to these samples and arrived at a receiver operating characteristic (AUROC) of 0.9824 (Fig. 2E), further validating the differentiation of their gut microbial profiles. In the MS-male cohort, the Shannon diversity showed no significant difference between WRE athletes’ gut microbiota and AER athletes’ gut microbiota (Fig. 2B). However, the differentiation in their overall profiles was notable, as demonstrated by their obvious separation in PCoA and a high AUROC of 0.9392 in random forest algorithm (Fig. 2D and F). In addition, we introduced sedentary (SD) populations to the MS cohort as a control to construct a Random Forest model (Fig. S1A through D). Compared with the MS cohort, the area under curve receiver operating characteristic (AUROC) values were reduced in both male and female groups (0.9464 and 0.6846, respectively).
Fig 2.
Establishment and comparisons of multi-sport gut microbiota profiles. (A–D) Comparison of the microbial Shannon diversity and microbial overall profiles between different types of sports in the MS-female cohort (A and C), as well as in the MS-male cohort (B and D). Microbial overall profiles were determined by the PCoA using Bray-Curtis differences at genus level (E–F). The ROC curves of the random forest classification in types of sports using microbial genera as features, with AUROC displayed in the MS-female cohort (E) and the MS-male cohort (F). The top 10 genus contributed to the random forest classification in the MS-female cohort (G) and the MS-male cohort (H). **P < 0.05; ****P < 0.001; and ns, not significant. ROC, receiver operating characteristic.
Moreover, Fig. 2G and H showed the top 10 genera with the largest contribution to random forest classification in the MS-female and MS-male cohorts, respectively. In the MS-female cohort, Pseudomonas and Eubacterium_coprostanoligenes_group were the most discriminant features for female AER and ROW samples. In the MS-male cohort, Psychrobacter and Bacillus were the most discriminant features for male WRE and AER samples. These results suggested that both female and male athletes had specialized gut microbiota for their respective sports.
Co-occurrence networks for multi-sport gut microbiota
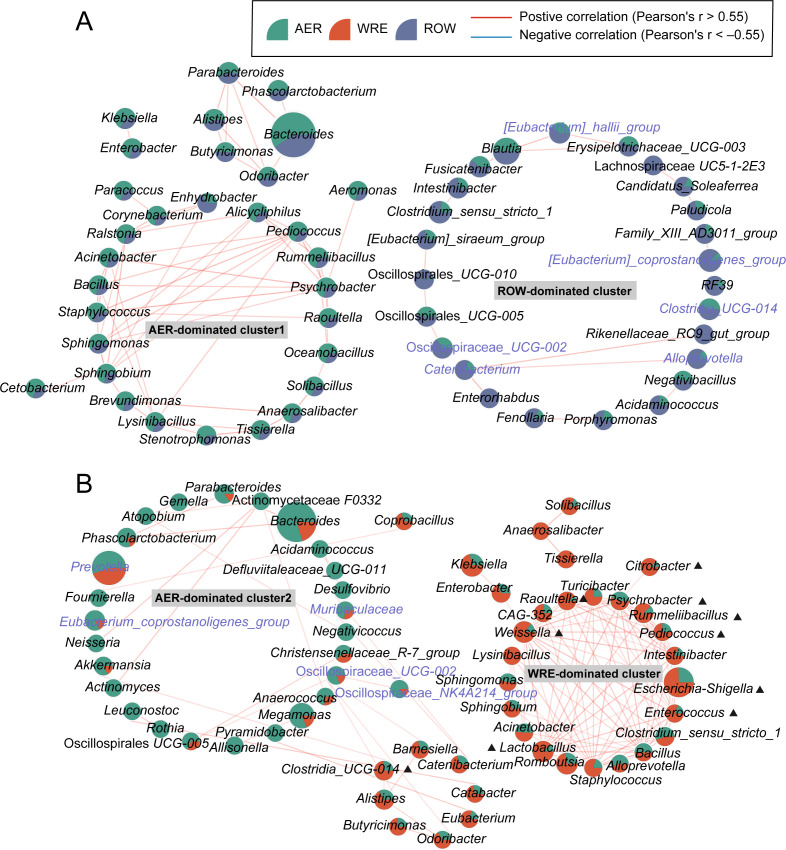
We next investigated the interaction patterns in the gut microbiota of different types of athletes. In the MS-female cohort (Fig. 3A), we identified two network clusters based on Spearman correlations at the genus level, one of which contained bacteria enriched in the AER group (AER-dominated cluster 1), while the other contained bacteria enriched in the ROW group (ROW-dominated cluster), such as Eubacterium_hallii_group and Clostridia_UCG-014. In the MS-male cohort, we also identified two network clusters including AER-dominated cluster 2 and WRE-dominated cluster (Fig. 3B). Moreover, when SD populations were recruited in the analysis, we identified an SD-dominated cluster, differentiated from those MS clusters (Fig. S1E and F). These findings suggested that different types of populations shared different interaction patterns of the gut microbiota.
Fig 3.
Co-occurrence networks of multi-sport gut microbiota. (A and B) The networks of genera were constructed in MS-female (A) and MS-male cohorts (B), respectively. Each circle represents a genus with the size representing relative abundance and the color representing its enrichment in the group. The edges indicate significant Spearman correlations between genus (P < 0.05 and absolute value of SCC > 0.55). Red edges indicate positive correlations, and blue edges indicate negative correlations. SCC, Spearman correlation coefficient.
Gut microbial composition patterns resolved by latent Dirichlet allocation
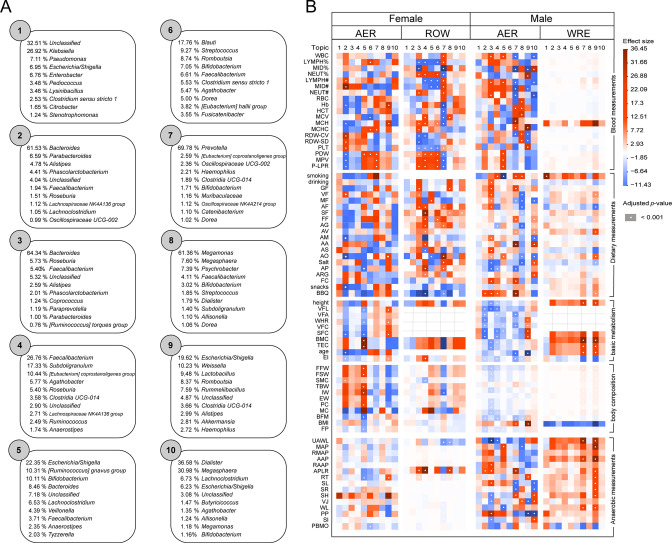
Further, we introduced latent Dirichlet allocation (LDA) to resolve the latent structure of athletes' gut microbiota. LDA, a Bayesian probabilistic generative model (14 - 16), could reduce the dimensionality of the microbial data into subgroups (topics), where microbes within the same topic were modulated by shared latent factors. We identified a total of 10 microbial topics at the genus level to describe each participant’s gut microbiota composition (Table S3). Each topic represented a specific probability distribution of certain genera (Fig. 4A). We found that the topic proportions were different across multi-sport gut microbiota (Fig. S2A and B). Hierarchical clustering of the 10 topics (Fig. S2C) revealed that topics containing the same dominant bacteria (e.g., topics 2 and 3) or containing certain overlapped genera (e.g., topics 8 and 10) were clustered into the same group.
Fig 4.
Associations of athletes gut microbial subgroups with blood measurements, dietary measurements, anthropometrics, and anaerobic measurements. (A) Gut microbial subgroups (topics) were identified by LDA. The top 10 genera with the highest probability per topic were displayed. NA represents operational taxonomic units (OTUs) not classified to a genus. The topic number is random and serves only as an identifier. (B) The heatmap shows the associations of athletes gut microbial subgroups with blood measurements, dietary measurements, anthropometrics, and anaerobic measurements. The associations were calculated by Dirichlet regression models. Each association between a topic and measurement was represented by a square, where the intensity of the color indicates the effect size. Red indicates the positive association and blue indicates the negative association. A white dot in the center of a circle indicates that the association remained significant after FDR correction (q < 0.001). Gray grid means no data. WBC, white blood cell count; LYMPH%, lymphocyte ratio; MID%, intermediate cell ratio; NEUT%, neutrophil ratio; LYMPH#, lymphocyte count; MID#, intermediate cell count; NEUT#, neutrophil count; RBC, red blood cells; Hb, hemoglobin; HCT, hematocrit; MCV, mean red blood cell volume; MCH, mean erythrocyte hemoglobin; MCHC, mean hemoglobin concentration; RDW-CV, red blood cell distribution concentration; RDW-SD, red blood cell distribution width; PLT, platelet; PDW, platelet distribution; MPV, mean platelet volume; P-LPR, large platelet ratio; GF, grain frequency; VF, vegetables frequency; MF, meat frequency; AF, aquatic frequency; SF, soy frequency; FF, fruit frequency; AG, amount of grain; AV, amount of vegetables; AM, amount of meat; AA, amount of aquatic; AS, amount of soy; AO, amount of oil; AP, amount of pepper; ARG, amount of raw garlic; FC, fruit count; VFL, visceral fat levels; VFA, visceral fat area; WHR, waist to hip ratio; VFC, visceral fat content; SFC, subcutaneous fat content; BMC, basal metabolic capacity; TEC, total energy consumption; EI, electrical impedance; FFW, fat free weight; FSW, fat free soft weight; SMC, skeletal muscle content; TBW, total body water; IW, intracellular water; EW, extracellular water; PC, protein capacity; MC, minerals capacity; BFM, body fat mass; BMI, body mass index; FP, fat percentage; UAWL, upper extremity anaerobic work load; MAP, maximum anaerobic power; RMAP, relative maximum anaerobic power; AAP, average anaerobic power; RAAP, relative average anaerobic power; APLR, anaerobic power lapse rate; RT, reaction time; SL, standing on left leg with eyes closed; SR, standing on right leg with eyes closed; SH, sitting height; VJ, vertical jump; WL, wing length; PP, push-ups 1 min; SI, step index; PBMO, power bike max oxygen uptake; FDR, false discovery rate.
Associations between gut microbial subgroups and inflammation related to intensive exercise
Next, we investigated the gut microbial subgroups (topics) associated with inflammation. The Dirichlet regression model was performed to test the associations between gut microbial subgroups (topics) and blood measurements (Fig. 4B), and these phenotypic factors had a highly explained variance for microbial diversity and function (Fig. S3). In the ROW female group, topics 3, 4, 5, 6, 7, and 8 were associated with one or more blood measurement factors (q < 0.001), and topic 7 exhibited the strongest positive associations. As a result, ROW female athletes with a high probability of topic 7 tended to have higher leukocyte (P = 1.99E−13), lymphocyte count (P = 9.81E−12), intermediate cell count (P = 2.42E−13), and neutrophil count (P = 2.76E−08). These factors were common indicators of inflammation in the blood test and varied with different types of sports (Fig. S4A and B). Moreover, we noticed that most genera of topic 7 were reported to be associated with sports inflammation, including Prevotella, Oscillospira, Coprococcus, and Haemophilus (Table S4). Prevotella (69.78% of topic 7) colonization in the gut resulted in metabolic changes in the microbiota, reduced interleukin 18 (IL-18) production that exacerbated intestinal inflammation, and might lead to systemic autoimmunity. Additionally, Prevotella contained enzymes that played an important role in mucus degradation, possibly leading to increased intestinal permeability (17). Previous research has suggested that increased intestinal permeability might favor colonic bacterial translocation, with a consequent risk of gastrointestinal problems (18, 19). In mouse models, exhaustive exercise promoted intestinal inflammation and increased the growth of Oscillospira, and Coprococcus (20), which also had a high percentage in topic 7. In addition, when evaluated in a post-exercise phase, exhaustive exercise can cause immune function depression, thereby increasing the risk of viral and bacterial infections (11), with Haemophilus (2.21% of topic 7) hosting various pathogenic species (21). In general, these genera might play a crucial role in enhancing gut permeability and the risk of infection, promoting inflammatory responses in athletes.
Interestingly, the significant associations between topic 7 and those inflammation factors were also observed in the AER male group but they were negative (Fig. 4B). The AER male group with a high probability for topic 7 was characterized by a low leukocyte (P = 1.06E−09), lymphocyte count (P = 1.74E−05), intermediate cell count (P = 7.33E−37), and neutrophil count (P = 5.18E−12). The correlation between topic 7 and inflammation may reflect the collective response of the microbes it encompasses, and we found that Eubacterium_hallii_group (P = 4.1E−07), Clostridia_UCG-014 (P = 0.015), Alloprevotella (P = 1.6E−05), and Oscillospiraceae_UCG-002 (P = 7.0E−08) were more enriched in the ROW female group, while Prevotella (P = 0.006) was more enriched in the AER male group (Fig. 3 and Fig. S4C). Therefore, the difference in correlation orientation might result from the different enriched genera of topic 7 between the female ROW and male AER groups. We further performed a regression analysis of these bacteria and inflammatory indicators (Fig. S4D and E). For the ROW female group, Clostridia_UCG-014 and Eubacterium_coprostanoligenes_group were consistently negatively correlated with leukocyte (P = 0.00023, P = 9.6E−07) and neutrophil count (P = 3E−05, P = 5.1E−07). In AER male group, it can be observed that Alloprevotella and Prevotella were consistently negatively correlated with leukocyte (P = 0.012, P = 0.0052) and lymphocyte count (P = 0.0012, P = 0.047), while AER female did not observe a significant correlation with inflammation. Overall, these results revealed that certain microbes enriched in topic 7 were associated with inflammation, and this association was influenced by sports’ types. Differences in the abundance of microbes in hosts with different sport types or sex may lead to specialized inflammation patterns, such as Eubacterium_coprostanoligenes_group, Clostridia_UCG-014 were correlated with the inflammation of female rowing athletes.
Associations of gut microbial subgroups with dietary information and anaerobic performance
Moreover, the Dirichlet regression model showed that most associations of gut microbial subgroups (topics) with dietary measurements and anthropometrics measurements were observed in the AER male group (Fig. 4B). Topic 3 was positively associated with food intake including drinking, vegetable frequency, meat frequency, aquatic frequency, and barbecue (BBQ; q < 0.0005). Meanwhile, topic 3 was negatively correlated with fat free weight, skeletal muscle content, and body fat mass (q < 0.0001). The results suggested that topic 3 might be the key microbial subgroup in inhibiting obesity under high caloric intake. Topic 2 was found to be strongly negatively correlated with anthropometrics (basic metabolism) factors such as visceral fat levels, waist to hip ratio, and subcutaneous fat content (q < 0.0001), indicating a potential role in weight control. Notably, topics 2 and 3 belonged to the same topic cluster and shared similar genera probability distributions (Fig. S2D and E). Phascolarctobacterium has a high proportion in both topics, and numerous studies reported that it was inversely associated with obesity (22). In general, both topics 2 and 3 might play a crucial role in controlling the weight of athletes.
We then explored the associations between gut microbial subgroups and anaerobic measurements. Topic 9 had the strongest positive association with anaerobic measurements such as upper extremity anaerobic work load, maximum anaerobic power, and average anaerobic power (q < 2.48E−5) in the WRE male group. This indicated that topic 9 might have a potential role in enhancing anaerobic performance, which warranted further investigations.
Sex-dependent different intensities of exercise affect the human gut microbiota
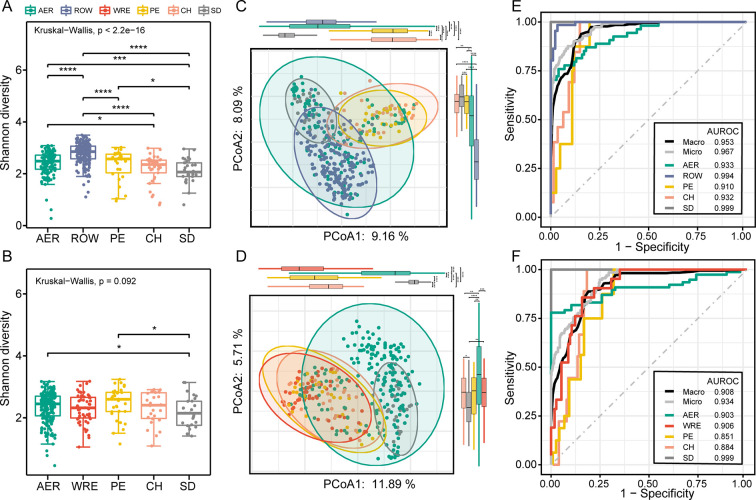
Finally, we sought to investigate the effects of different exercise intensities on gut microbiota. To this end, we introduced three additional non-athlete cohorts, including physical major students (PE), children (CH), and sedentary people (SD). We found Shannon diversity was also affected by exercise intensity. For females, we found that athletes had higher microbial Shannon diversity than that non-athletes (P < 0.05). Especially, the ROW group had the highest Shannon diversity of all the other four groups (P = 2.86E−22, Fig. 5A). However, for males, the differences in Shannon diversity were not significant between athletes and non-athletes groups (P = 0.092, Fig. 5B). And the gut microbial compositions could be differentiated by exercise intensity for both males and females (P < 0.05, Fig. 5C and D). Moreover, the Random Forest model showed that female groups had higher classification accuracy when discriminating one group out of the five groups (AUROCAER = 0.933, AUROCROW = 0.994, AUROCPE = 0.910, AUROCCH = 0.932, AUROCSD = 0.999, Fig. 5E), as compared with that of the male groups (AUROCAER = 0.903, AUROCWRE = 0.906, AUROCPE = 0.851, AUROCCH = 0.884, AUROCSD = 0.999, Fig. 5F). These results suggested that exercise intensity had effects on the gut microbiota, and these effects were sex-dependent.
Fig 5.
Effects of different degrees of exercise on human gut microbiota. (A) The boxplots show the microbial Shannon diversity among five female groups including AER, ROW, PE, CH, and SD. (B) The boxplots show the microbial Shannon diversity among five male groups including AER, WRE, PE, CH, and SD. (C and D) PCoA of female participants (C) and male participants (D) based on the Bray-Curtis differences. (E and F) The ROC curves of the random forest classification in five groups using microbial genera as features, with AUROC displayed in the female groups (E) and the male groups (F). *P < 0.1; **P < 0.05; ***P < 0.01; ****P < 0.001; ROC, receiver operating characteristic.
To further assess the effect of sex-dependent exercise intensity on gut microbiota, we then identified the top 5 genera with the highest importance in the Random Forest model (quantified by the mean decrease accuracy) for each group (Fig. S5). We found that the abundance of Clostridium_sensu_stricto_1 decreased as the female participants’ exercise intensity decreased, especially in athletes compared to SD, while a similar trend was observed for Intestinibacter in male participants (Fig. S5). In addition, we investigated the distribution of inflammation-associated microbiota (the microbiota in topic 7) in each cohort. Prevotella as the dominant microbe in topic 7 (69.78% of topic 7) was found to decrease as the intensity of exercise decreased, especially in male participants (Fig. S6). Taken together, certain gut microbial alterations could respond to the different intensities of exercise, and sex was a non-negligible factor in both microbial changes and the potential inflammation risk.
DISCUSSION
Athletes’ gut microbial profiles were influenced by the types of sports they participated in. Previous studies have demonstrated that exercise could increase microbial diversity in the gut microbiome (12, 23). In the present study, the microbial diversity of the ROW female group was profoundly higher than AER female group, suggesting that different types of exercise have different effects on microbial diversity in female athletes, but no significant differences were observed between male athletes. The analysis of microbial composition and diversity of different types of athletes revealed that the gut microbial community of different types of athletes differs profoundly. These results confirmed that the pattern, intensity, and frequency of physical exercise cannot be ignored when studying the effects of physical activity on the microbial community. In other words, to monitor the potential risk of inflammation and further realize individualized exercise modulation, it is necessary to consider different types of athletes when studying the association between microbiota and inflammatory factors.
The associations between gut microbiota and blood measurements confirmed that inflammatory factors were affected by the types of sports. Blood measurements showed the strongest and most consistent associations with topic 7 in ROW female and AER male, respectively. However, topic 7 was positively correlated with inflammatory indicators in the female ROW group but negatively correlated in the AER male group. This intriguing result suggested that gut microbiota and inflammation might have a sport-specific relationship. We need to investigate further to determine the causes of this sport-specific correlation. Specifically, we observed a strong positive association between topic 7 and inflammatory markers, but Prevotella (69.78% of topic 7) was not significantly correlated with the inflammatory markers in ROW female group. These results suggested that the correlation between topic 7 and inflammation may reflect the collective response of the microbes it encompasses, rather than the response of a single microbe. For the AER male group, a negative association was observed between topic 7 and inflammatory markers. Certain Prevotella species have been linked to inflammation (24), but not all of them are harmful. For example, Prevotella copri CB7 has been reported to have both beneficial and detrimental effects depending on the context (25). Our data cannot identify which Prevotella species are enriched in the AER male group, and the negative correlation observed may be a unique characteristic of the gut microbiota of athletes that requires further investigation. Overall, gut microbiota can reflect specific inflammatory patterns in different types of athletes, and the association information between the microbial community and inflammatory factors could prove highly significant for monitoring the potential inflammation risk and further achieving personalized exercise modulation. In particular, ROW female athletes should pay more attention to the changes in topic 7 and conduct gut microbial modulation and training monitoring, to avoid the risk of inflammation caused by training as much as possible.
Moreover, by combining athletes and non-athletes cohorts, we found that differences in exercise intensity could lead to specific microbial changes, some of which were associated with inflammation. Broadly speaking, different types and intensities of exercise may have distinct impacts on the gut microbes of general people. In addition to exercising regularly, regulating the types and intensities of exercises is equally important for normal people to gain the benefit of exercise.
Our work has focused on gut microbial community profiling and inflammation pattern mining in multi-sport cohorts. Firstly, the results of this cohort study would lay the foundation for future experiments on the modulation of exercise intensity for inflammation control, or to explore probiotics for athletes to adjust their inflammation levels rather than using antibiotics. Further investigations into mechanisms are warranted to validate the findings of this study. Secondly, conducting multi-omic studies, such as metagenomic and metabolomic analysis may further enhance our understanding of how gut microbial communities adjust inflammation when athletes take high-intensity exercise. Finally, samples from a wider variety of sports in the future will considerably expand our understanding of the athlete microbiota.
Conclusions
In this study, we collected one of the largest sets of gut microbiota samples from athletes across different sports types and examined their specialized gut microbial profiles. We determined the association between microbes and inflammation in athletes, compared the different strengths of association among different types of sports, and explained their possible roles in regulating inflammation. We found that the differences in microbial abundance in hosts with different sports types may lead to specialized inflammation patterns. Additionally, certain microbial subgroups were also found to be associated with dietary information and anaerobic performance, which were also influenced by the types of sports. Furthermore, changes in the abundance of inflammation-related microbiota in healthy individuals were influenced by different exercise intensities. Finally, we found that sex was a non-negligible factor in both microbial changes and the potential risk of inflammation. Collectively, our work has elucidated the complicated association of physical factors, gut microbiota, and inflammations across different types of sports, which could prove highly significant for monitoring the potential gut inflammation risk and further achieving personalized exercise modulation.
MATERIALS AND METHODS
Study design and sample collection
Professional aerobics athletes (AER, n = 316, female = 117, male = 199, 18 ± 7 years, 24.80 ± 25.46 kg/m2), male wrestling athletes (WRE, n = 53, 16 ± 4 years, 22.73 ± 10.60 kg/m2), and female rowing athletes (ROW, n = 174, 15 ± 3 years, 36.03 ± 19.49 kg/m2) were selected for fecal sample collection. According to the grade in athletic competition and information regarding technical level obtained from General Administration of Sport of China, we found that these athletes have participated in at least one competition above the province-level. These athletes differ in terms of training patterns, with aerobics focusing on flexibility, rowing on endurance, and wrestling on both endurance and explosive power. Therefore, we can explore the differences in gut microbiota between athletes under different exercise patterns. Additionally, to further evaluate the effects of exercise with different intensities on the gut microbiota, we constructed a non-athlete cohort. The non-athlete cohort consisted of 58 sports college students (PE, female = 26, male = 32, 21 ± 2 years, 26.19 ± 40.67 kg/m2), 63 children (CH, female = 37, male = 26, 9 ± 2 years, 15.80 ± 6.78 kg/m2), and 57 sedentary individuals (SD, female = 30, male = 27, 21 ± 2 years). The sports college students come from Wuhan Sports University, and their training intensity and athletic performance are lower than those of professional athletes. The children ranged in age from 7 to 11 years, and they started exercising from early childhood (<6 years). The sedentary individuals are computer-related workers who spend all their working hours sitting (>8 h). All the cohorts had no medical issues or received antibiotic treatment in the past 4 months. Fecal samples were collected and stored in sterilized 50 mL tubes, immediately placed on freezer packs, and stored at −80°C.
Dietary factors, physical characteristics, and sports-related indicators were recorded and examined by the questionnaire and professional measurements for the athlete cohort. These factors were divided into five groups: blood measurements, dietary measurements, anthropometrics (basic metabolism), anthropometrics (body composition), and anaerobic measurements. Blood measurements such as leukocyte, lymphocyte ratio, neutrophil count, hematocrit, and hemoglobin were measured by routine blood tests. Dietary measurements such as the level of smoking, drinking, grain, vegetables, fruit, and soy were recorded by the questionnaires. Anthropometrics referred to the combination of various complex factors such as height, weight, age, basal metabolic capacity, fat free weight, protein capacity, body fat mass, and age obtained by the questionnaire; the height and weight were measured using an electronic height tester; others such as fat free weight, protein capacity, and body fat mass were measured with the professional body composition analyzer (X-SCAN PLUS II, Jawon Medical Co., Ltd, South Korea) (26). We divided these indicators into anthropometrics (basic metabolism) and anthropometrics (body composition), the first of which mainly focused on basal metabolism-related indicators and the latter of which described the composition of the body. Anaerobic measurements were measured using MetaLyzer II (Cortex, Leipzig, Germany) (27) and Technogym multipower system D4773L (Technogym, Italy) (28).
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from fecal samples using the PowerSoil DNA Isolation Kit (MoBio, USA) according to the manufacturer’s instructions. All extracted DNA was dissolved in Tris-EDTA buffer and stored at −20°C. DNA concentration quantification was performed with a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), and DNA quality assessment was performed with 0.8% agarose gels. The V3–V4 hypervariable region of the 16S rRNA gene was sequenced for each sample and we used 5–50 ng of DNA as a template for amplifying the V3–V4 amplicon using the forward primer (5ʹ-CCTACGGRRBGCASCAGKVRVGAAT-3ʹ) and reverse primer (5ʹ-GGACTACNVGGGTWTCTAATCC-3ʹ). The sequencing library was constructed using the MetaVxTM Library Preparation kit (Genewiz, Inc., South Plainfield, NJ, USA) via adding indexed adapters to the ends of 16S rDNA amplicons in limited cycle PCR. DNA libraries were verified and quantified by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and Qubit 2.0 (Applied Biosystems, Carlsbad, CA, USA). All sequencing reactions were performed on the Illumina MiSeq (San Diego, CA, USA) platform using a paired-end sequencing strategy.
16S rRNA gene sequence data process
The raw multiplexed-paired-end sequences were firstly input to QIIME2 (version 2020.11.0) (29) and were demultiplexed using “qiime cutadapt demux-paired” with “--p-error-rate 0.” The primers of demultiplexed sequences were then trimmed using “qiime cutadapt trim-paired” with “--p-match-adapter-wildcards--p-match-read-wildcards--p-discard-untrimmed.” The trimmed sequences were quality-controlled using “qiime dada2 denoise-paired” with “--p-trunc-len-f 270--p-trunc-len-r 230--p-n-threads 20--p-min-fold-parent-over-abundance 4.” The dada2-produced representative sequences were taxonomically annotated using “qiime feature-classifier classify-sklearn” against the V3–V4 region of the Silva 138 database (30). The taxonomic annotations were integrated into the feature table using “qiime taxa collapse.” For beta diversity analysis, the dada2-produced feature table was rarefied to 8,000 reads per sample using “qiime feature-table rarefy” with “--p-sampling-depth 8,000” based on the curve plateaus of the alpha diversity.
Statistical analysis
Microbial diversity
Statistical analysis was conducted mainly using the R platform (http://www.r-project.org/). Alpha diversity was quantified by the Shannon diversity that was calculated using the function “diversity” of the R package “vegan” (version 2.6-2) (31). Mann-Whitney-Wilcoxon test was used to calculate the statistical significance (P values) of the differences in alpha diversity between groups. Beta diversity was quantified by the Bray-Curtis dissimilarity that was calculated using the function “vegdist” of the R package “vegan” (version 2.6-2) (31). Principal coordinates analysis (PCoA) based on Bray-Curtis distances was applied to the samples and visualized by the R package “ggplot2” (version 3.3.6) (32). Mann-Whitney-Wilcoxon test was used to calculate the statistical significance of the sample separation between groups against the PCo1 and PCo2 axes. Permutational analysis of variance (PERMANOVA) on the Bray-Curtis distances with 9,999 permutations was used to test the associations of the phenotype with both microbiome composition and function. PERMANOVA (33) was carried out using the “adonis” function in the R package “vegan” package (version 2.6-2) (31).
Prediction model based on gut microbiota to distinguish different athlete types
Random Forest models were generated based on microbial compositions to differentiate three athlete types using the R package “randomForest” (version 4.7-1.1) (34). The data set was randomly divided into the training set (40%) and the testing set (60%). Function “trainControl” in R package “caret” was used to perform 10 repeats of 10-fold cross-validation. Function “train” in R package “caret” was used to fit models over different tuning parameters to determine the “mtry” for Random Forest algorithm. Gini coefficients were used to measure how each variable contributed to the homogeneity of the nodes and leaves in the resulting Random Forest. The receiver operating characteristic (ROC) curve was generated to evaluate the performance of the prediction model.
Co-occurrence network analysis
Co-occurrence network analysis was based on the Spearman correlations between genera. The co-occurrence relationship between the two genera was accepted if the Spearman correlation coefficient was greater than 0.55 or less than −0.55 (calculated by R function “cor”) and P < 0.05 (calculated by R function "cor.test”). Cytoscape (version 3.9.1) (35) was used to visualize the microbial networks.
Latent Dirichlet allocation and phenotype-subgroup associations
Latent Dirichlet allocation (LDA), a Bayesian probabilistic generative model, was used to reveal latent structure present in gut microbial data, as previously described (16). Genera subgroups were identified by LDA and their correlations with phenotype including blood measurements, dietary measurements, anthropometrics (basic metabolism and body composition), and anaerobic measurements were assessed separately using Dirichlet regression models by R package “DirichletReg” (version 0.7.1) (36). To remove the sex effects on gut microbiota or the phenotypic data, we grouped the samples based on sex and the athlete type into four comparison groups including AER males versus WRE males and AER females versus ROW females. The covariates age and body mass index were adjusted in the regression analysis. P values for all associations were adjusted using a false discovery rate (FDR), and a significant threshold was FDR < 0.001.
Prediction of functional composition and regression analysis
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, version 1.0.0-dev) (37) was applied to profile the functional composition of microbial communities based on the high-quality of 16S rRNA gene according to the manual of PICRUSt. The functional trait abundances were determined using the KEGG database (version 66.1, 1 May 2013) (38). Regression analysis of microbiota in topic 7 and inflammatory indicators was performed using R function “cor.test.”
ACKNOWLEDGMENTS
The authors thank all the volunteers who made this research possible.
This work was partially supported by the National Natural Science Foundation of China grant no. 32071465, 31871334, and 31671374 and the China Ministry of Science and Technology’s National Key R&D Program grant (no. 2018YFC0910502). Numerical computations were performed on the Hefei Advanced Computing Center.
Y.L., M.C., S.W., Q.L., and K.N. designed the study, reviewed, and verified the data. Y.L., M.C., Y.Z., K.Y., and Y.T. collected samples and conducted experiments. Y.L., M.C., and K.N. conducted data analysis and produced the figures and tables. Y.L., M.C., and K.N. wrote the manuscript. All authors revised the manuscript. Y.L., M.C., S.W., Q.L., and K.N. supervised the study. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
The authors declare no competing interests.
Contributor Information
Song Wang, Email: wangsong@whsu.edu.cn.
Qunwei Lu, Email: luqw@hust.edu.cn.
Kang Ning, Email: ningkang@hust.edu.cn.
Daniel Garrido, Pontificia Universidad Catolica de Chile, Santiago, Chile .
DATA AVAILABILITY
Sequencing data are available in the Genome Sequence Archive (GSA) section of National Genomics Data Center (project accession number CRA007901).
ETHICS STATEMENT
All procedures involving human participants were approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (No. IEC-S102). Informed consent was obtained from all individual participants.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00259-23.
Comparisons of gut microbiota profiles in multi-sport and sedentary populations.
Distribution of topics across the MS cohorts.
Identification of gut microbial covariates.
Distribution of inflammatory indicators and microbiota related with inflammation.
Taxonomical biomarkers in athletes and non-athletes.
The distribution of microbiota in topic 7 in athletes and non-athletes.
Phenotype statistics for athletes.
Comparisons of taxonomic abundances between different types of sport.
The each sample's gut microbiota described by a unique composition of 10 topics.
The probability composition of the genus for each topic.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, Ahmadi-Vand Z, Marsden KR, Gibson DL. 2016. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 4:42. doi: 10.1186/s40168-016-0189-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, Matsubara N, Hara H. 2008. Voluntary running exercise alters microbiota composition and increases N-butyrate concentration in the rat cecum. Bioscience, Biotechnology, and Biochemistry 72:572–576. doi: 10.1271/bbb.70474 [DOI] [PubMed] [Google Scholar]
- 3. Liu Y, Wang Y, Ni Y, Cheung CKY, Lam KSL, Wang Y, Xia Z, Ye D, Guo J, Tse MA, Panagiotou G, Xu A. 2020. Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab 31:77–91. doi: 10.1016/j.cmet.2019.11.001 [DOI] [PubMed] [Google Scholar]
- 4. Camilleri M. 2019. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68:1516–1526. doi: 10.1136/gutjnl-2019-318427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, Shanahan F, Cotter PD, O’Sullivan O. 2018. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67:625–633. doi: 10.1136/gutjnl-2016-313627 [DOI] [PubMed] [Google Scholar]
- 6. Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, Yang Z, Hattab MW, Avila-Pacheco J, Clish CB, Lessard S, Church GM, Kostic AD. 2019. Meta-Omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med 25:1104–1109. doi: 10.1038/s41591-019-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, Sodergren E, Weinstock GM. 2017. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 5:98. doi: 10.1186/s40168-017-0320-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moloney RD, Desbonnet L, Clarke G, Dinan TG, Cryan JF. 2014. The microbiome: stress, health and disease. Mamm Genome 25:49–74. doi: 10.1007/s00335-013-9488-5 [DOI] [PubMed] [Google Scholar]
- 9. Holzer P, Farzi A.. 2014. Neuropeptides and the Microbiota-Gut-Brain Axis, p 195–219. In Lyte M, Cryan JF (ed), Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease doi:10.1007/978-1-4939-0897-4_9. Springer New York, New York, NY. [Google Scholar]
- 10. Wegierska AE, Charitos IA, Topi S, Potenza MA, Montagnani M, Santacroce L. 2022. The connection between physical exercise and gut microbiota: implications for competitive sports athletes. Sports Med 52:2355–2369. doi: 10.1007/s40279-022-01696-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, Nimmo MA. 2011. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol 11:607–615. doi: 10.1038/nri3041 [DOI] [PubMed] [Google Scholar]
- 12. Shanahan F, Clarke SF, Murphy EF, O’Sullivan O, Cotter P. 2015. Author response: linking lifestyle and microbes. Gut 64:520. doi: 10.1136/gutjnl-2014-308107 [DOI] [PubMed] [Google Scholar]
- 13. Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, Wells SD, Arent SM, Kreider RB, Campbell BI, Bannock L, Scheiman J, Wissent CJ, Pane M, Kalman DS, Pugh JN, Ortega-Santos CP, ter Haar JA, Arciero PJ, Antonio J. 2020. The athletic gut microbiota. Journal of the International Society of Sports Nutrition 17:24. doi: 10.1186/s12970-020-00353-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blei DM, Ng AY, Jordan MI.. 2003. Latent Dirichlet allocation. J Mach Learn Res 3:993–1022. [Google Scholar]
- 15. Sankaran K, Holmes SP. 2019. Latent variable modeling for the microbiome. Biostatistics 20:599–614. doi: 10.1093/biostatistics/kxy018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Breuninger TA, Wawro N, Breuninger J, Reitmeier S, Clavel T, Six-Merker J, Pestoni G, Rohrmann S, Rathmann W, Peters A, Grallert H, Meisinger C, Haller D, Linseisen J. 2021. Associations between habitual diet, metabolic disease, and the gut microbiota using latent Dirichlet allocation. Microbiome 9:61. doi: 10.1186/s40168-020-00969-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iljazovic A, Roy U, Gálvez EJC, Lesker TR, Zhao B, Gronow A, Amend L, Will SE, Hofmann JD, Pils MC, Schmidt-Hohagen K, Neumann-Schaal M, Strowig T. 2021. Perturbation of the gut microbiome by Prevotella Spp. enhances host susceptibility to Mucosal inflammation. Mucosal Immunol 14:113–124. doi: 10.1038/s41385-020-0296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monda V, Villano I, Messina A, Valenzano A, Esposito T, Moscatelli F, Viggiano A, Cibelli G, Chieffi S, Monda M, Messina G. 2017. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev 2017:3831972. doi: 10.1155/2017/3831972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA.. 2019. Exercise and the gut microbiome: a review of the evidence, Potential Mechanisms, and Implications for Human Health. Exerc Sport Sci Rev 47. [DOI] [PubMed] [Google Scholar]
- 20. Clark A, Mach N. 2016. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: A systematic review for athletes. Journal of the International Society of Sports Nutrition 13:43. doi: 10.1186/s12970-016-0155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kakanis MW, Peake J, Brenu EW, Simmonds M, Gray B, Hooper SL, Marshall-Gradisnik SM. 2010. The open window of susceptibility to infection after acute exercise in healthy young male elite athletes. Exerc Immunol Rev 16:119–137. [PubMed] [Google Scholar]
- 22. Watanabe Y, Nagai F, Morotomi M. 2012. Characterization of phascolarctobacterium succinatutens sp. Nov., an Asaccharolytic, Succinate-utilizing bacterium isolated from human Feces. Appl Environ Microbiol 78:511–518. doi: 10.1128/AEM.06035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. 2013. Exercise attenuates PCB-induced changes in the Mouse gut Microbiome. Environ Health Perspect 121:725–730. doi: 10.1289/ehp.1306534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen JM. 2017. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151:363–374. doi: 10.1111/imm.12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley RE. 2016. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 13:69–70. doi: 10.1038/nrgastro.2016.4 [DOI] [PubMed] [Google Scholar]
- 26. Piepiora P, Superson M, Witkowski K. 2017. Personality and the body composition of athletes using the example of the Polish national youth female wrestling team. Journal of Combat Sports and Martial Arts 2:107–109. doi: 10.5604/01.3001.0010.8692 [DOI] [Google Scholar]
- 27. Yuktasir B. 2008. Warm-up: a case study on maximal oxygen consumption as it relates to acute stretching. J Hum Kinet 19:165–176. doi: 10.2478/v10078-008-0013-y [DOI] [Google Scholar]
- 28. Capodaglio P, Capodaglio Edda M, Facioli M, Saibene F. 2007. Long-term strength training for community-dwelling people over 75: impact on muscle function, functional ability and life style. Eur J Appl Physiol 100:535–542. doi: 10.1007/s00421-006-0195-8 [DOI] [PubMed] [Google Scholar]
- 29. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS II, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oksanen JAI. 2016. Vegan: ecological diversity. Available from: https://github.com/vegandevs/vegan
- 32. Ito K, Murphy D.. 2013. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol 2:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- 34. Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 35. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maier M. 2014. DirichletReg: dirichlet regression for compositional data in R. Research Report Series / Department of Statistics and Mathematics 125. [Google Scholar]
- 37. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang C, Zhang Y, Zhu X, Liu K, Wang X, Chen M, Wang J, Chen H, Hui S, Huang L, Zhang Q, Zhu J, Wang B, Mi M. 2016. Healthy subjects differentially respond to dietary capsaicin correlating with specific gut enterotypes. J Clin Endocrinol Metab 101:4681–4689. doi: 10.1210/jc.2016-2786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparisons of gut microbiota profiles in multi-sport and sedentary populations.
Distribution of topics across the MS cohorts.
Identification of gut microbial covariates.
Distribution of inflammatory indicators and microbiota related with inflammation.
Taxonomical biomarkers in athletes and non-athletes.
The distribution of microbiota in topic 7 in athletes and non-athletes.
Phenotype statistics for athletes.
Comparisons of taxonomic abundances between different types of sport.
The each sample's gut microbiota described by a unique composition of 10 topics.
The probability composition of the genus for each topic.
Data Availability Statement
Sequencing data are available in the Genome Sequence Archive (GSA) section of National Genomics Data Center (project accession number CRA007901).