Abstract
Objectives
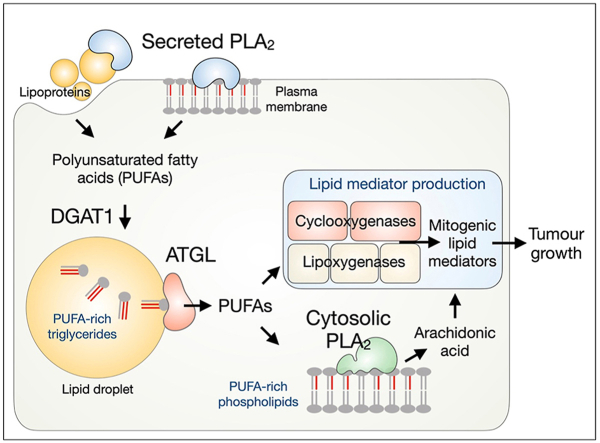
Polyunsaturated fatty acids (PUFAs) are structural components of membrane phospholipids and precursors of oxygenated lipid mediators with diverse functions, including the control of cell growth, inflammation and tumourigenesis. However, the molecular pathways that control the availability of PUFAs for lipid mediator production are not well understood. Here, we investigated the crosstalk of three pathways in the provision of PUFAs for lipid mediator production: (i) secreted group X phospholipase A2 (GX sPLA2) and (ii) cytosolic group IVA PLA2 (cPLA2α), both mobilizing PUFAs from membrane phospholipids, and (iii) adipose triglyceride lipase (ATGL), which mediates the degradation of triacylglycerols (TAGs) stored in cytosolic lipid droplets (LDs).
Methods
We combined lipidomic and functional analyses in cancer cell line models to dissect the trafficking of PUFAs between membrane phospholipids and LDs and determine the role of these pathways in lipid mediator production, cancer cell proliferation and tumour growth in vivo.
Results
We demonstrate that lipid mediator production strongly depends on TAG turnover. GX sPLA2 directs ω-3 and ω-6 PUFAs from membrane phospholipids into TAG stores, whereas ATGL is required for their entry into lipid mediator biosynthetic pathways. ATGL controls the release of PUFAs from LD stores and their conversion into cyclooxygenase- and lipoxygenase-derived lipid mediators under conditions of nutrient sufficiency and during serum starvation. In starving cells, ATGL also promotes the incorporation of LD-derived PUFAs into phospholipids, representing substrates for cPLA2α. Furthermore, we demonstrate that the built-up of TAG stores by acyl-CoA:diacylglycerol acyltransferase 1 (DGAT1) is required for the production of mitogenic lipid signals that promote cancer cell proliferation and tumour growth.
Conclusion
This study shifts the paradigm of PLA2-driven lipid mediator signalling and identifies LDs as central lipid mediator production hubs. Targeting DGAT1-mediated LD biogenesis is a promising strategy to restrict lipid mediator production and tumour growth.
Keywords: Lipid droplets, Diacylglycerol acyltransferase, Adipose triglyceride lipase, Phospholipase A2, Lipid mediators, Cancer
Graphical abstract
Highlights
-
•
Phospholipase A2-induced lipid mediator production depends on lipid droplet turnover.
-
•
Group X sPLA2-released polyunsaturated fatty acids are incorporated into triglycerides.
-
•
Triglyceride lipolysis by ATGL drives lipid mediator production.
-
•
ATGL also delivers polyunsaturated fatty acids into phospholipids to be targeted by cPLA2α.
-
•
Inhibition of DGAT1 reduces cancer cell proliferation and tumour growth in vivo.
1. Introduction
Fatty acids (FAs) are universal cellular energy sources and membrane building blocks. FAs are also involved in signalling pathways that control cell growth, inflammation and tumorigenesis [1]. The ensemble of signalling FAs is vastly expanded by the oxygenation of polyunsaturated FAs (PUFAs) by cyclooxygenase (COX), lipoxygenase (LOXs) and CYP450 monooxygenase enzymes into several families of bioactive lipid mediators, including the eicosanoids [2]. These short-lived autocrine and paracrine signalling molecules are released from cells to collectively modulate various processes in their microenvironment, e.g., to orchestrate a shift into pro-inflammatory or anti-inflammatory states. Pathways that control the availability of different PUFAs for oxygenation determine the types of lipid mediator species produced and the dynamics of their production [[3], [4]]. Our current understanding of the control of PUFA supply for lipid mediator production is limited, particularly as this is intrinsically dependent on complex (PU)FA metabolism, including PUFA biosynthesis, uptake, storage, breakdown, lipid remodelling and trafficking [[5], [6], [7]].
The canonical pathway that supplies arachidonic acid (C20:4n–6; AA) for eicosanoid production depends on the group IVA cytosolic phospholipase A2 (cPLA2α), whose role in stimulus-induced eicosanoid production has been demonstrated in various pathophysiological settings [[8], [9], [10], [11]]. Upon cell activation, cPLA2α binds to perinuclear membranes of the ER and Golgi complex and selectively hydrolyses phospholipids containing AA at the sn-2 position [12]. Numerous other members of the PLA2 superfamily promote lipid mediator production, either through activation of cPLA2α or by acting independently on their respective phospholipid pools, thereby releasing not only AA but also other PUFAs [6,13,14]. In particular, several secreted PLA2s (sPLA2s) have been implicated in lipid mediator production [[15], [16], [17]]. The group X sPLA2 is the most potent among mammalian sPLA2s at hydrolysing the phosphatidylcholine (PC)-rich plasma membrane, lipoproteins and extracellular vesicles [[18], [19], [20], [21]]. It releases various unsaturated FAs, including ω-3 and ω-6 PUFAs, and it is involved in inflammation, immunity and cancer [15,[21], [22], [23], [24], [25]].
Recent studies suggest that besides membrane phospholipids, other cellular lipid pools, including neutral lipids stored in lipid droplets (LDs) or derived from lipoprotein uptake, are also sources of PUFAs for lipid mediator production [[7], [26], [27], [28], [29]]. LDs are specialized organelles that store FAs and other lipids in their esterified forms, primarily as triglycerides (TAGs) and sterol esters [[30], [31], [32], [33]]. A hallmark role of LDs is taking up excess FAs to prevent lipotoxicity and fine-tune FA release via lipolysis to match various cellular demands [[34], [35], [36], [37], [38]]. Adipose triglyceride lipase (ATGL), the major mammalian TAG lipase, provides FAs for mitochondrial energy production, but it also controls FA-induced signalling pathways that coordinate metabolism and inflammation [[39], [40], [41], [42], [43]]. In immune and cancer cells, LDs have long been implicated in AA storage, trafficking and eicosanoid production [[44], [45], [46], [47], [48]]. It was only recently shown that ATGL provides precursors for lipid mediator production and modulates neutrophil immune responses in vivo [26,27,49]. However, it is not yet clear how LDs manage PUFA trafficking for lipid mediator production in mammalian cells and how they cooperate with the canonical PLA2-mediated pathways [7,50].
The trafficking of PUFAs between membranes and LDs is an emerging mechanism controlling PUFA oxygenation and lipid-induced oxidative damage, including ferroptotic cell death. On the one hand, the sequestration of PUFAs into LDs limits their availability for oxidation [[20], [51], [52], [53]]. For instance, in fly embryos exposed to hypoxia, membrane lipid peroxidation and neuronal damage are prevented via a mechanism mediated by phospholipase D and diacylglycerol acyltransferase (DGAT) that diverts PUFAs from phospholipids into TAGs stored within LDs [51]. On the other hand, the proportion of (poly)unsaturated FAs released from LDs may determine membrane saturation, oxidative stress and cell fate [20,54]. Accordingly, in cancer cells exposed to exogenous PUFAs, LDs are enriched with PUFA-TAGs and their hydrolysis by ATGL promotes oxidative stress-dependent cell death [20]. Under these conditions, the transfer of unsaturated FAs from membrane phospholipids into LDs mediated by the human group X (hGX) sPLA2 balances phospholipid and TAG acyl-chain composition and prevents PUFA lipotoxicity [20,24]. Based on these findings, we hypothesize that LD turnover modulates membrane PUFA content and affects PLA2-mediated lipid mediator production.
Here, we investigated the crosstalk between different molecular pathways providing PUFAs for lipid mediator production in human cancer cells. We show that the incorporation of PUFAs into TAGs and their subsequent release via lipolysis are essential for production of mitogenic lipid mediators. We demonstrate that LDs integrate several PUFA trafficking pathways and act as central hubs that control PLA2-driven lipid mediator production. Furthermore, we provide evidence for a pathophysiological relevance of these findings by showing that inhibition of TAG synthesis impairs the production of mitogenic lipid mediators and reduces cancer cell proliferation in vitro and tumour growth in vivo.
2. Results
2.1. Membrane phospholipid hydrolysis by hGX sPLA2 leads to enrichment of LDs with long-chain PUFA-TAGs
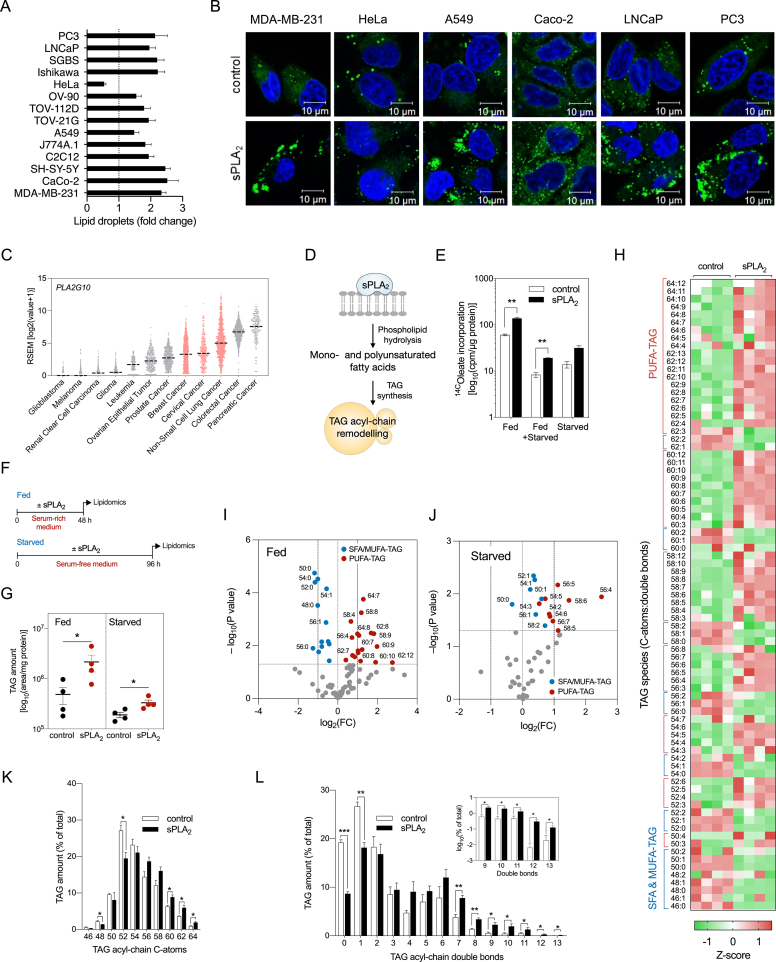
We have shown previously that hGX sPLA2 releases various unsaturated FAs from adherent cells, including oleic acid (C18:1n–9; OA) and PUFAs [20], and induces LD accumulation in several breast cancer cell lines [20,24]. Recombinant hGX sPLA2 also induced LD accumulation in other cancer and immortalised non-tumorigenic cell lines (Figure 1A, B), suggesting that its effects on LD metabolism are not limited to specific cell types. A comparative analysis of PLA2G10 mRNA levels and gene alterations in different cancer tissues (TCGA PanCancer Atlas) revealed that the PLA2G10 gene is amplified and its mRNA highly expressed in several types of cancer, such as colorectal, breast, cervical and lung cancer (Figure 1C; Supp. Figure 1A). Moreover, we observed a positive correlation between PLA2G10 mRNA levels and the expression of several genes involved in TAG and LD metabolism, including PNPLA2, DGAT1, AGPAT2 and PLIN5 (Supp. Figure 1B). We then asked whether hGX sPLA2 alters TAG acyl chain composition in LDs of MDA-MB-231 cells (Figure 1D), an invasive and metastatic breast cancer cell line showing enhanced proliferation and resistance to starvation upon treatment with hGX sPLA2 [24]. Cells pre-incubated with [14C]-OA readily incorporated the radiolabelled FA into phospholipids and TAGs, while hGX sPLA2 treatment specifically increased the abundance of TAGs containing [14C]-OA in various growth conditions (Figure 1E; Supp. Figure 1C and D). Lipidomic liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of TAG species extracted from hGX sPLA2-treated serum-fed (Figure 1F–I, K, L; Supp. Figure 1E and F) and serum-starved cells (Figure 1G, J) revealed an unexpected enrichment of TAG species with long-chain and highly unsaturated FAs (Figure 1K, L; Supp. Figure 1E). Interestingly, hGX sPLA2 treatments under fed conditions enriched cells with TAGs containing 7–12 double bonds, whereas starved cells were enriched with TAGs with a lower level of unsaturation (Figure 1I, J). These results suggested that among the unsaturated FAs released by hGX sPLA2, the polyunsaturated species are preferentially redistributed from membrane phospholipids into LDs. Experiments with serum-starved cells (Figure 1E, G, J) demonstrated that serum lipoproteins, which are major targets for the enzyme [20,55], are not required for hGX sPLA2-induced enrichment of TAGs with PUFAs. Although we did not assess the involvement of extracellular vesicles [21] as substrates for the enzyme under these conditions, our observations suggest a direct action of hGX sPLA2 on the plasma membrane leading to enrichment of LDs with PUFA-containing TAGs.
Figure 1.
hGX sPLA2promotes enrichment of LDs with long-chain PUFA-containing triglycerides. (A, B) LD levels in control cells and cells treated with 10 nM recombinant hGX sPLA2 for 48 h in serum-rich medium. (A) Neutral lipid content was quantified by Nile Red staining and flow cytometry (n = 3 independent experiments; 20000 cells per treatment). (B) Representative live-cell confocal microscopy images of LDs stained with BODIPY 493/503 (green) and nuclei with Hoechst 33342 (blue). (C) PLA2G10 mRNA expression in samples from cancer patients (TCGA PanCancer Atlas Studies, cBioportal). mRNA levels were batch normalized from Illumina HiSeq_RNASeqV2. Dashed line indicates median. (D) Diagram illustrating the hypothesis that unsaturated fatty acids (FAs) released through hGX sPLA2 membrane hydrolysis are incorporated into triacylglycerols (TAGs) and lead to lasting changes in LD TAG acyl-chain composition. (E) hGX-sPLA2-induced changes in the incorporation of radiolabelled oleate into cellular TAGs in MDA-MB-231 cells grown in complete medium for 24 h (Fed), in complete medium for 24 h followed by 96 h of serum starvation (Fed + Starved), and in serum-free medium for 96 h (Starved) (n = 4 independent experiments). (F) Diagram illustrating the experimental treatments used for lipidomic analysis in (G)–(L). (G–L) TAG lipidomic analyses of MDA-MB-231 cells treated with recombinant hGX sPLA2 in complete medium for 48 h (Fed; G–I, K, L) or in the absence of serum for 96 h (Starved; G, J). Cell lysates were collected and analysed by UPLC/qTOF-MS (n = 4 independent experiments). (H–J) hGX sPLA2-induced changes in the levels of individual TAG species presented as a representative z-score heat-map (H) and volcano plots (I, J) prepared by log2 data transformation and multiple t-test analyses (n = 4 independent experiments). Statistically significant changes (–log10(P value) > 1.30) in TAGs containing mostly saturated and mono-unsaturated FAs (SFA/MUFA-TAGs with 0–2 double bonds; blue) and those containing polyunsaturated FAs (PUFA-TAGs with 3–12 double bonds; red) are shown. (K, L) hGX sPLA2-induced changes in relative levels of TAG species grouped by number of acyl-chain C-atoms (chain length) and double bonds (chain unsaturation). (A–L) Data are means ± SEM of at least three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (unpaired t-tests).
2.2. ATGL-mediated LD breakdown is required for PGE2 production
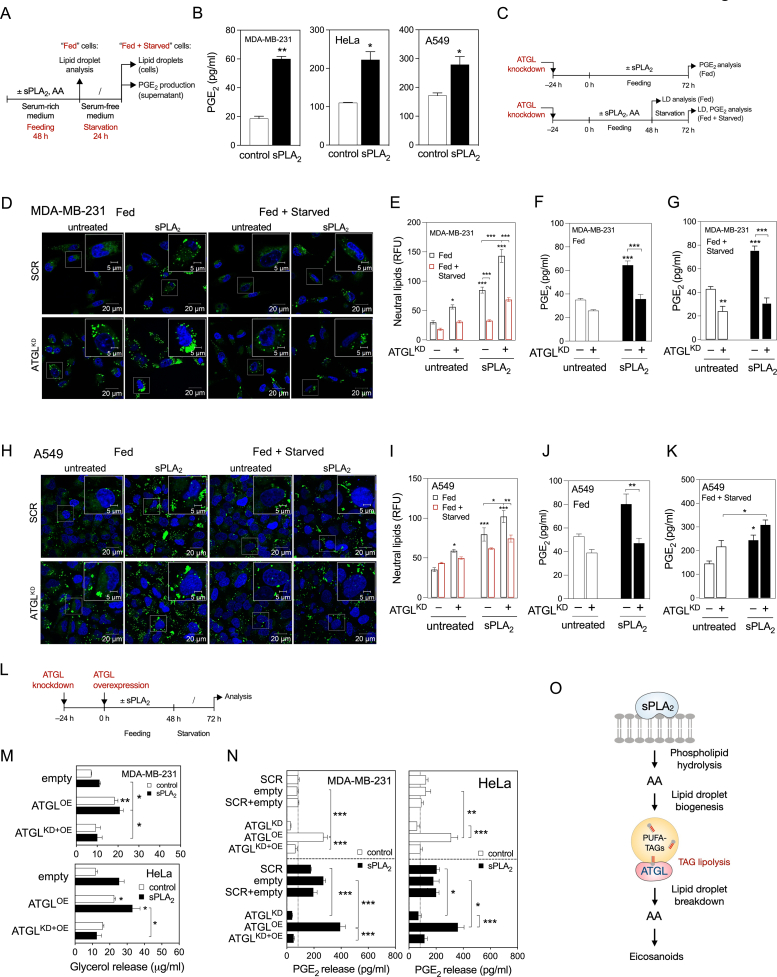
Serum withdrawal induces LD breakdown in most cell types [56]. Given the hGX-sPLA2-induced enrichment of LDs with PUFAs, we next asked if starvation-induced breakdown of PUFA-rich LDs is sufficient to stimulate eicosanoid production. LD biogenesis was first stimulated with hGX sPLA2 (or with exogenous AA) in cells grown in serum-rich medium. Then the cells were serum-starved in the absence of these stimuli to induce LD breakdown (Figure 2A; Supp. Figure 2A). In comparison with untreated cells, serum-starved MDA-MB-231, HeLa and A549 cells pre-treated with hGX sPLA2 released more glycerol (Supp. Figure 2C), an indicator of TAG lipolysis, and produced more prostaglandin (PG)E2 (Figure 2B), a major AA-derived eicosanoid. Although HeLa cells were unique among cell lines in showing a net reduction in neutral lipid levels upon treatment with hGX sPLA2 during feeding (Figure 1A, B; Supp. Figure 2B), there was an increase in lipolysis (Supp. Figure 2C) and PGE2 production (Figure 2B) during serum starvation. This is indicative of an accelerated LD turnover in hGX-sPLA2-treated HeLa cells that leads to net reduction in LD levels and drives lipid mediator production. Similarly, pre-treatment with exogenous AA led to increased PGE2 production in starving MDA-MB-231 and HeLa cells (Supp. Figure 2D). These observations indicate that starvation-induced LD breakdown drives PGE2 synthesis.
Figure 2.
ATGL-mediated LD breakdown is required for PGE2production. (A) Diagram illustrating the experimental set-up used to load cells with LDs (Feeding) and then to induce their breakdown (Starvation). (B) PGE2 levels in cell supernatants of control and hGX-sPLA2-treated cells quantified by ELISA at the end of the starvation period. (C) Diagram illustrating the experimental conditions used in (D)–(K) and Supp. Figure 2(E)–(L). (D–F, H–J) LD levels and PGE2 production in ATGL-silenced control and sPLA2-treated MDA-MB-231 and A549 cells grown as shown in (C) and analysed after 72 h of feeding (Fed) or after 48 h feeding plus 24 h starvation (Fed + Starved). Neutral lipids were quantified by Nile Red staining and flow cytometry, PGE2 was quantified by ELISA. (G, K) Representative confocal microscopy images showing effects of ATGL depletion on cellular LD content in control and hGX-sPLA2-treated MDA-MB-231 and A549 cells, under serum-rich (Fed) and serum-free (Fed + Starved) conditions. LDs and nuclei were stained using BODIPY 493/503 and Hoechst 33342, respectively. (L) Diagram illustrating the experimental set-up used in (M), (N) and Supp. Figure 2M, N. (M, N) Glycerol release and PGE2 production in ATGL-overexpressing serum-starved cells (both untreated and hGX sPLA2 pretreated), in comparison with cells co-transfected with ATGL-specific siRNAs (ATGLKD + OE), non-targeting siRNA (scrambled) and control plasmid (empty), grown as illustrated in (L). (O) Diagram illustrating the proposed model of LD-mediated eicosanoid production in cancer cells. Data are means ± SEM of two (M) or three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-way ANOVA with Tukey (N) or Bonferroni (E–G, I–K, M) adjustment; unpaired t-tests (B)).
We next investigated if ATGL, the rate-limiting enzyme in cytosolic TAG lipolysis, is responsible for LD breakdown and supports PGE2 production during feeding as well as during starvation. ATGL silencing (Figure 2C; Supp. Figure 2E) reduced lipolytic glycerol release (Supp. Figure 2F and G) and increased LD abundance (Figure 2D, E) in both fed and starved MDA-MB-231 cells. Importantly, ATGL knockdown also reduced basal and hGX-sPLA2-stimulated PGE2 production under conditions of nutrient sufficiency and during serum starvation (Figure 2F, G). The finding that ATGL is essential for LD breakdown and PGE2 production during serum starvation were reproduced in hGX-sPLA2-treated HeLa cells (Supp. Figure 2E–J) and in MDA-MB-231 cells pre-treated with exogenous AA (Supp. Figure 2K and L). However, experiments with ATGL silencing in A549 cells (Supp. Figure 2E) indicated that whereas ATGL participates in LD breakdown during both feeding and starvation (Figure 2H, I; Supp. Figure 2F and G), it is involved in PGE2 production only under nutrient-rich conditions in these cells (Figure 2J, K). Interestingly, ATGL expression was elevated in starving A549 cells pre-treated with hGX-sPLA2 (Supp. Figure 2E). This effect aligns with the correlation observed between PLA2G10 and PNPLA2 expression in human tumour samples (Supp. Figure 1B).
To corroborate our findings on the role of ATGL in LD turnover and eicosanoid production, we asked whether ATGL overexpressing cells would have increased levels of lipolysis and PGE2 production. ATGL overexpression in the breast and cervical cancer cells (Figure 2L; Supp. Figure 2M) enhanced glycerol release (Figure 2M) and PGE2 production (Figure 2N), albeit without significantly altering total neutral lipid levels (Supp. Figure 2N). The effects of ATGL overexpression were fully reversed in the presence of ATGL-targeting siRNA (Figure 2M, N). Collectively, these data demonstrate that LD breakdown via ATGL is required for basal, AA-induced and hGX-sPLA2-stimulated PGE2 production (Figure 2O) in both fed and starved cancer cells.
2.3. ATGL-mediated release of ω-3 and ω-6 PUFAs from LDs drives lipid mediator production
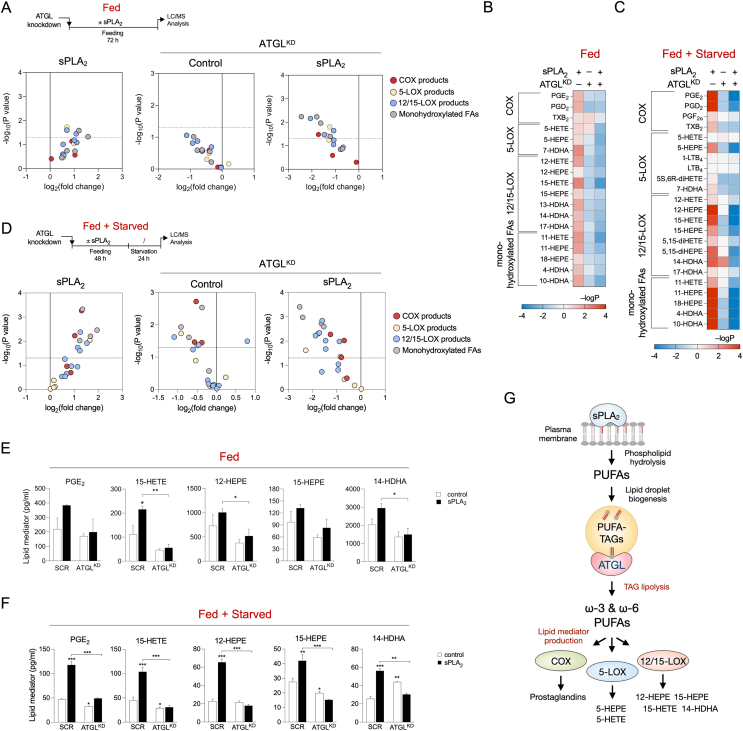
The data presented above demonstrate that transient storage of AA within LD TAGs followed by AA release from TAGs by ATGL are intermediate steps in basal and hGX-sPLA2-stimulated production of PGE2. To find out if ATGL-mediated TAG lipolysis promotes the production of a wider range of eicosanoids and related oxylipins (which might also derive from other PUFAs), we performed targeted LC-MS/MS-based metabololipidomics to examine the effects of ATGL silencing on the lipid mediator profiles of untreated and hGX-sPLA2-treated cells. Here, treatment of breast cancer cells with hGX sPLA2 stimulated the production of numerous lipid mediators, biosynthesised from different PUFAs by various enzymatic pathways (Figure 3, Supp. Figure 3). hGX sPLA2 promoted the synthesis of eicosapentaenoic acid (C20:5n–3; EPA)-derived lipid mediators, as well as the production of AA-derived and docosahexaenoic acid (C22:6n–3; DHA)-derived products. These lipid mediators were produced during the treatment with hGX sPLA2 in nutrient-rich conditions (Figure 3A, B) and during serum starvation (Figure 3C, D), in the absence of hGX sPLA2. Notably, ATGL depletion suppressed this hGX-sPLA2-primed release of lipid mediators under both conditions (Figure 3A–F, Supp. Figure 3). Furthermore, ATGL silencing reduced the basal, hGX-sPLA2-independent production of lipid mediators. These results suggested that ATGL-mediated TAG lipolysis provides LD-derived PUFAs for lipid mediator production during both nutrient sufficiency and during starvation. Additionally, the hGX sPLA2-stimulated release of unesterified PUFAs from cells was suppressed by ATGL silencing under both nutrient-rich and starvation conditions (Supp. Figure 3A and B). On the contrary, in starving A549 cells, ATGL silencing did not supress hGX-sPLA2-stimulated PUFA release or lipid mediator production (Supp. Figure 3C), which is in line with the results of the PGE2 ELISA analyses in these cells (Figure 2J). Overall, our results in MDA-MB-231 cells indicated that PUFAs released from cells by hGX sPLA2 cell treatment were not directly derived from sPLA2 phospholipid hydrolysis, but were rather released from TAG stores through lipolysis by ATGL. The enrichment of LDs with PUFAs in nutrient-rich conditions is thus coupled with an increased immediate release of PUFAs via basal ATGL-mediated lipolysis during nutrient sufficiency or with a delayed PUFA release occurring during serum starvation-induced lipolysis. These results demonstrate that ATGL controls the availability of PUFAs for lipid mediator synthesis under both nutrient sufficiency and starvation conditions (Figure 3G).
Figure 3.
ATGL-mediated lipolysis drives the production of a wide spectrum of bioactive lipid mediators during nutrient sufficiency and during serum starvation. (A–F) UPLC-MS-MS analysis of lipid mediators released from fed (A, B, E) and serum-starved (C, D, F) MDA-MB-231 cells pre-treated with hGX sPLA2, and from ATGL-depleted cells without and with hGX sPLA2 pre-treatment, presented as volcano plots (A, D), heat maps (B, C), and graphs for selected individual species (E, F). Volcano plots were prepared using log2-transformed fold-change values and multiple t-test analysis, and the heat map by –logP data transformation and two-way ANOVA with Sidak adjustment (n = 3 independent experiments). (G) Diagram illustrating the involvement of hGX sPLA2 and ATGL in the production of a wide range of PUFA-derived cyclooxygenase (COX) and lipoxygenase (LOX) signalling molecules. Data are means ± SEM of three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-way ANOVA with Sidak adjustment).
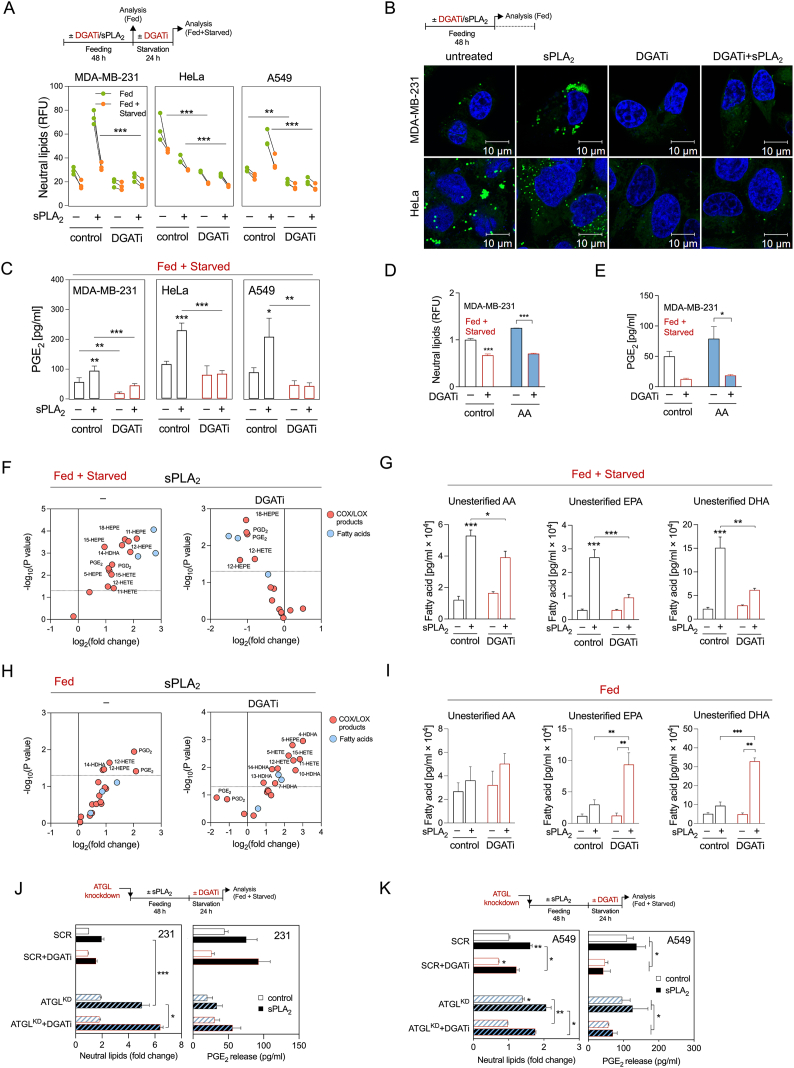
2.4. Inhibition of DGAT-mediated LD biogenesis impairs PGE2 production
To examine whether the incorporation of PUFAs into TAGs is a prerequisite for the conversion of PUFAs into lipid mediators, we inhibited TAG synthesis using a combination of specific inhibitors of DGAT1 and DGAT2 (DGATi), which mediate the final committed step in TAG synthesis [33], and followed changes in LD turnover and lipid mediator production. Combined inhibition of both DGATs was necessary because individual inhibition of DGAT1 or DGAT2 did not reduce neutral lipid levels in hGX-sPLA2-treated cells (Supp. Figure 4A). DGAT inhibition during serum feeding strongly suppressed basal and hGX-sPLA2-induced LD accumulation (Figure 4A, B). There was an almost complete depletion of LDs in serum-fed DGATi-treated cells (Figure 4B), which allowed only a minimal breakdown of residual LDs during the subsequent serum starvation (Figure 4A). Notably, cells depleted of LDs displayed low basal PGE2 release during serum starvation and did not increase PGE2 production upon stimulation with either hGX sPLA2 or exogenous AA (Figure 4C–E). DGATi treatment of A549 cells during feeding abolished PGE2 production in starving A549 cells (Figure 4C), whereas ATGL depletion failed to do so (Figure 2K; Supp. Figure 3C), which indicated that the build-up of TAGs drives eicosanoid production in these cells as well. Clearly, the deficiency of LDs at the start of the starvation phase diminishes the capacity of the different types of cancer cells to produce PGE2. These results were corroborated by lipidomic analyses in starving MDA-MB-231 cells, which showed that inhibition of DGAT1 and DGAT2 suppresses hGX sPLA2-primed production of several AA- and EPA-derived lipid mediators (Figure 4F; Supp. Figure 4B), reduces exogenous AA-primed production of prostaglandins (Supp. Figure 4C) and blunts the release of unesterified PUFAs during the starvation (Figure 4G). On the contrary, under nutrient-rich conditions, DGAT inhibition enhanced the release of EPA and DHA and their oxygenated products (Figure 4H, I). Interestingly, the release of AA was not significantly affected. In addition, the AA-derived PGE2 and PGD2 were the only lipid mediators among those detected that were decreased by DGAT inhibition during nutrient sufficiency (Figure 4H; Supp. Figure 4D). This could be an indication that LD depletion interferes specifically with the production of prostaglandins, possibly by affecting the function of cPLA2α, COXs and prostaglandin synthetases, which have been previously found to be localised to LDs [46,[57], [58], [59]]. Thus, interfering with TAG synthesis during nutrient sufficiency disrupts the production of AA-derived eicosanoids and redirects EPA and DHA into mediator production pathways that are independent of LDs. This also promotes the release of excess EPA and DHA from cells. In summary, DGAT-mediated TAG biosynthesis under serum-rich conditions controls the pool of PUFAs available for immediate lipid mediator production and is a prerequisite for lipid mediator biosynthesis during subsequent periods of serum starvation.
Figure 4.
DGAT-mediated LD biogenesis is required for lipid mediator production in serum-starved cancer cells. (A) Diagram illustrating the experimental conditions used (top) and cellular neutral lipid content before (Fed) and after (Fed + Starved) serum starvation in cells treated with DGAT1 (T863) and DGAT2 (PF-06427878) inhibitors (DGATi), without and with stimulation of LD biogenesis by hGX sPLA2 during serum sufficiency. (B) Representative confocal microscopy images of live cells under nutrient-replete conditions and treated with DGAT inhibitors in the absence and presence of hGX sPLA2. LDs and nuclei were visualised using BODIPY 493/503 and Hoechst 33342 staining, respectively, and confocal microscopy. (C) DGATi-induced changes in PGE2 production in serum-starved cancer cells (Fed + Starved), without and with additional stimulation of LD biogenesis by hGX sPLA2 pre-treatment. Cells were treated according to (A). (D, E) DGATi-induced changes in neutral lipids (D) and PGE2 production (E) in serum-starved MDA-MB-231 cancer cells (Fed + Starved), without and with additional stimulation of LD biogenesis by arachidonic acid (AA) pre-treatment. Cells were treated according to (A). (F–I) DGATi-induced changes in the profiles of lipid mediators and fatty acids released from MDA-MB-231 cells serum-starved for 24 h after a 48-h feeding phase (F, G) or from cells fed for 72 h (H, I), treated with hGX sPLA2 during growth in complete media, without and with treatment with an equimolar mix of DGAT1 and DGAT2 inhibitors according to (A). Volcano plots (F, H) show significant changes (–log10(P value) > 1.30) in individual lipids between hGX sPLA2-treated versus control cells (left) and hGX sPLA2-treated versus hGX sPLA2- and DGATi-treated (right) cells and were prepared using log2-transformed fold-change values and multiple t-test analysis (n = 4 independent experiments). (J, K) Diagrams illustrating the experimental conditions used (top) and changes in PGE2 production induced by DGATi treatments during serum starvation in control (SCR) and ATGL-depleted (ATGLKD) MDA-MB-231 (J) and A549 (K) cells, without and with hGX sPLA2 pre-treatment. (A, D, J, K) Neutral lipid content was quantified by Nile Red staining and flow cytometry. (C, E, J, K) PGE2 levels were determined in cell supernatants as described in Methods. (F–I) UPLC-MS/MS analysis was performed as described in Methods. Data are means ± SEM of at least three or four (F, G) independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-way ANOVA with Tukey (A, G, I, J, K) or Bonferroni (C, D, E) adjustment).
Besides promoting LD breakdown, serum removal in MDA-MB-231 cells stimulates the transcription of lipogenic genes, including the sterol-regulatory element binding 1 protein (SREBP-1) transcription factor and key enzymes involved in FA synthesis [24]. In accordance with ongoing lipogenesis, treatment of serum-starved cells with hGX sPLA2 resulted in a net increase in TAG levels (Figure 1G). To examine whether TAG synthesis occurring during serum starvation might also contribute to eicosanoid production, control and ATGL-depleted cells were treated with DGAT1 and DGAT2 inhibitors during serum feeding or during serum starvation. As expected, DGAT inhibition during serum feeding fully depleted MDA-MB-231 and HeLa cells of LDs, thereby abolishing the effects of hGX sPLA2 and ATGL silencing on LD abundance (Supp. Figure 4E). In contrast, DGAT inhibition during serum starvation did not affect LD abundance in these two cell lines (Figure 4J; Supp. Figure 4F), nor did it affect PGE2 release in MDA-MB-231 cells (Figure 4J). However, DGAT inhibition in serum-starved A549 cells reduced LD abundance and suppressed PGE2 production (Figure 4K). This was observed in both control and in ATGL-deficient cells indicating that the build-up of TAG stores during starvation drives eicosanoid biosynthesis in A549 cells via an ATGL-independent mechanism. Accordingly, DGAT inhibition during both serum feeding and starvation was necessary for full suppression of PGE2 production in A549 cells (Supp. Figure 4G).
Collectively, these results demonstrate that DGAT-mediated TAG synthesis and LD turnover are required for lipid mediator production. ATGL-mediated breakdown of pre-existing DGAT-induced LDs is the predominant mechanism of LD-driven eicosanoid production in the serum-starved MDA-MB-231 breast and HeLa cervical cancer cells. On the contrary, A549 lung cancer cells employ TAG synthesis during starvation and ATGL-independent mechanisms to support eicosanoid production.
2.5. cPLA2α cooperates with ATGL and depends on LD turnover to drive eicosanoid production
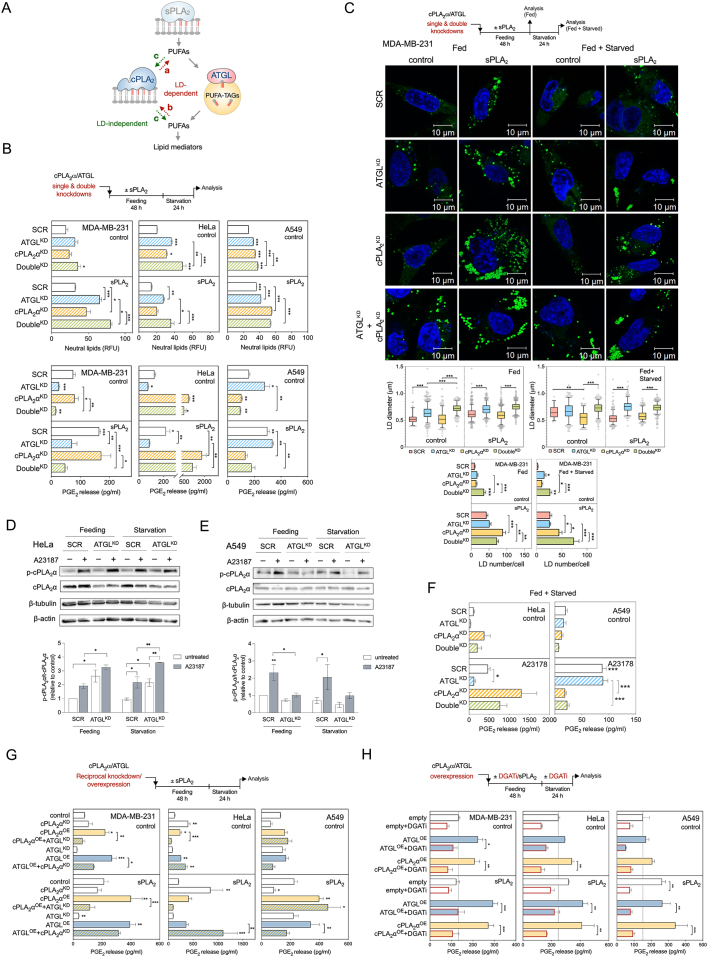
Given the well-accepted role of cPLA2α in providing AA for eicosanoid biosynthesis, we next asked whether cPLA2α participates in LD-driven lipid mediator production. We speculated that at least three main scenarios are possible (Figure 5A): (a) cPLA2α-induced incorporation of phospholipid-derived AA into TAGs, followed by AA release from LDs by ATGL; (b) ATGL-dependent transfer of TAG-derived AA into phospholipid pools, which are then accessed by cPLA2α; and (c) an LD-independent action of cPLA2α on membrane phospholipids.
Figure 5.
cPLA2α depends on LD turnover to drive lipid mediator production. (A) Scheme illustrating hypothetical models of interplay between cPLA2α and LDs in providing PUFAs for lipid mediator production. (B) Changes in neutral lipid content and PGE2 production induced by ATGL (ATGLKD) and cPLA2α (cPLA2αKD) single and double (DoubleKD) knockdowns, in comparison with control siRNA-treated cells (SCR), without and with stimulation of LD biogenesis by hGX sPLA2 pre-treatment. (C) Diagram illustrating the experimental conditions used (top), representative live-cell confocal microscopy images (middle) and image analysis (bottom) of LDs in ATGL (ATGLKD) and cPLA2α (cPLA2αKD) single and double (DoubleKD) knockdown MDA-MB-231 cells, in comparison with non-targeting control siRNA-treated cells (SCR), without and with stimulation of LD biogenesis by hGX sPLA2 pre-treatment. LDs were stained with BODIPY 493/503 (green) and nuclei with Hoechst 33342 (blue) and images analysed using ImageJ and the LD Counter Plugin. Box plots are showing changes in LD diameters and bar plots changes in LD numbers per cell in serum-fed (Fed) and serum-starved (Starved) cells. Data are geometric means (diameter analysis) or means (number analysis) ±SEM (n > 40 cells/sample) of two independent experiments. (D, E) Representative western blots and densitometry analyses showing phosphorylated and total cPLA2α protein levels in fed (48 h) and starved cells (48 h feeding followed by 24 h starvation for A549 or 1.5 h starvation for HeLa cells) treated for 1 h with 1 μM A23187. (F) Changes in PGE2 production induced by ATGL (ATGLKD) and cPLA2α (cPLA2αKD) single and double (DoubleKD) knockdowns in A549 and HeLa cells grown for 48 h in complete media followed by 1.5 h of starvation and treated in the last hour with 1 μM A23187. (G) PGE2 production in cells with reciprocal knockdown/overexpression of cPLA2α and ATGL. Cells were reverse transfected with siRNAs specific for ATGL (ATGLKD) and/or cPLA2α (cPLA2αKD), then forward transfected with ATGL-encoding (ATGLOE) and/or cPLA2α-encoding (cPLA2αOE) plasmids, without and with pre-treated with hGX sPLA2, as illustrated in the scheme (top). In controls (control), non-targeting siRNA reverse transfections were combined with backbone (‘empty’) vector forward transfections. (H) DGAT inhibition (DGATi)-induced changes in PGE2 production in serum-starved control cells (empty) and in cells overexpressing ATGL (ATGLOE) or cPLA2α (cPLA2αOE), without and with additional stimulation of LD biogenesis by hGX sPLA2 pre-treatment. Neutral lipid content was quantified by Nile Red staining and flow cytometry (B), PGE2 levels were determined in cell supernatants using ELISA (B, F, G, H). Data are means ± SEM of two (B, A549 cells) or at least three independent experiments ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (nested one-way ANOVA with Sidak adjustment (C, box plots), two-way ANOVA with Tukey (B, C, H, F), Holm-Sidak (D, E) or Dunnet (G) adjustment).
To examine these possibilities, we first used siRNA to deplete cells of cPLA2α alone or in combination with ATGL (Figure 5B; Supp. Figure 5A and B). ATGL deficient cells had elevated total neutral lipid levels (Figure 5B; Supp. Figure 5C) and displayed an increase in the diameter of cellular LDs, with only minor changes in the number of LDs per cell (Figure 5C; Supp. Figure 5E and F). In contrast, cPLA2α silencing resulted in a significant increase in the number of cellular LDs in all three cell lines (Figure 5C; Supp. Figure 5E and F), which was accompanied by only a slight increase in total neutral lipid levels (Figure 5B; Supp. Figure 5C). This finding was particularly evident in ATGL-deficient and hGX sPLA2 pre-treated starving cells and suggested a modulatory role of cPLA2α in LD turnover. Most notably, cPLA2α depletion in A549 cells, but not ATGL silencing, abolished basal and hGX-sPLA2-primed eicosanoid release during starvation (Figure 5B). On the contrary, while ATGL knockdown lowered PGE2 production in MDA-MB-231 cells, cPLA2α silencing did not alter PGE2 levels. Of note, the knockdown of cPLA2α in HeLa cells led to a marked increase in PGE2 release, which was partially reduced upon depletion of both ATGL and cPLA2α (Figure 5B). These data suggested that cPLA2α modulates LD turnover and that its contribution to PGE2 production during serum starvation is cell type-specific.
Given that the cellular activity of cPLA2α in promoting eicosanoid production depends on Ca2+-induced phosphorylation and membrane translocation of the enzyme [11], which typically occurs during cell activation, we next asked whether cPLA2α is phosphorylated under our experimental feeding/starvation conditions and whether this is affected by ATGL. Indeed, basal cPLA2α phosphorylation levels were observed in both fed and starved A549 and HeLa cells (Figure 5D, E). Treating cells with the calcium ionophore A23187 stimulated cPLA2α phosphorylation and promoted PGE2 production in both cell lines (Figure 5D–F). Importantly, ATGL silencing suppressed PGE2 production in A23187-treated HeLa cells (Figure 5F), even though the ATGL deficiency in these cells was accompanied by a slight increase of cPLA2α phosphorylation (Figure 5D). This suggested that ATGL is capable of promoting PGE2 production even when cPLA2α is optimally activated. In A549 cells, ATGL silencing reduced cPLA2α phosphorylation during feeding, but not during starvation (Figure 5E), which could explain the suppressive effect of ATGL deficiency on PGE2 production observed only in fed A549 cells (Figure 2J). Our efforts to analyse the effects of ATGL on cPLA2α activation in MDA-MB-231 cells were hampered by the very low protein levels of the enzyme (Supp. Figure 5A and B) and the poor capacity of A23187 to stimulate PGE2 production in these cells (data not shown). Of note, the lower cPLA2α expression in the breast cancer cell lines relative to the cervical and lung cancer cell lines (Supp. Figure 5A and B) approximately corresponds to the expression ratios observed in breast tumours relatively to cervical and lung tumours (Supp. Figure 5G).
Therefore, to find out more about the interplay between cPLA2α and ATGL, we asked how reciprocal overexpression of one and silencing of the other might affect LD turnover and PGE2 production (Figure 5G). In all three cell lines, the overexpression of ATGL or cPLA2α had a minimal effect on neutral lipid levels (Supp. Figure 5D, H, I), but both significantly augmented PGE2 production (Figure 5G). ATGL silencing blocked PGE2 production induced by overexpression of cPLA2α in MDA-MB-231 and HeLa cells, whereas cPLA2α silencing only partially reduced ATGL-overexpression-driven eicosanoid production in MDA-MB-231 cells, and even increased it in HeLa cells. On the contrary, in starving A549 cells, ATGL knockdown did not affect cPLA2α-induced PGE2 production, but silencing of cPLA2α reduced PGE2 release from ATGL-overexpressing cells (Figure 5G). Taken together, these data suggested that ATGL is the main enzyme in the provision of PUFAs for eicosanoid production in MDA-MB-231 and HeLa cells, whereby cPLA2α stimulation of eicosanoid production depends on ATGL. In contrast, in A549 cells, cPLA2α has a dominant role in PGE2 production, which is independent of ATGL, but is still associated with changes in LD turnover.
To examine if TAG synthesis is involved in cPLA2α-driven lipid mediator production, cPLA2α and ATGL were overexpressed in cells depleted of LDs using DGAT inhibitors (Figure 5H; Supp. Figure 5I). As expected, TAG synthesis was necessary for ATGL stimulation of eicosanoid production (Figure 5H). However, in all three cell lines, cPLA2α-overexpression-induced eicosanoid production was also completely abolished upon DGAT inhibition (Figure. 5H). Therefore, LDs are required for cPLA2α-driven lipid mediator production. cPLA2α and ATGL appear to have cell-type-specific roles that are either cooperative or complementary for both LD turnover and eicosanoid production. In the breast and cervical cancer cells, deficiency of ATGL impairs cPLA2α-induced PGE2 production, which suggests the possibility that the transfer of AA from TAGs to phospholipids is a prerequisite for the action of cPLA2α.
2.6. ATGL and cPLA2α have complementary roles in TAG and phospholipid acyl-chain remodelling
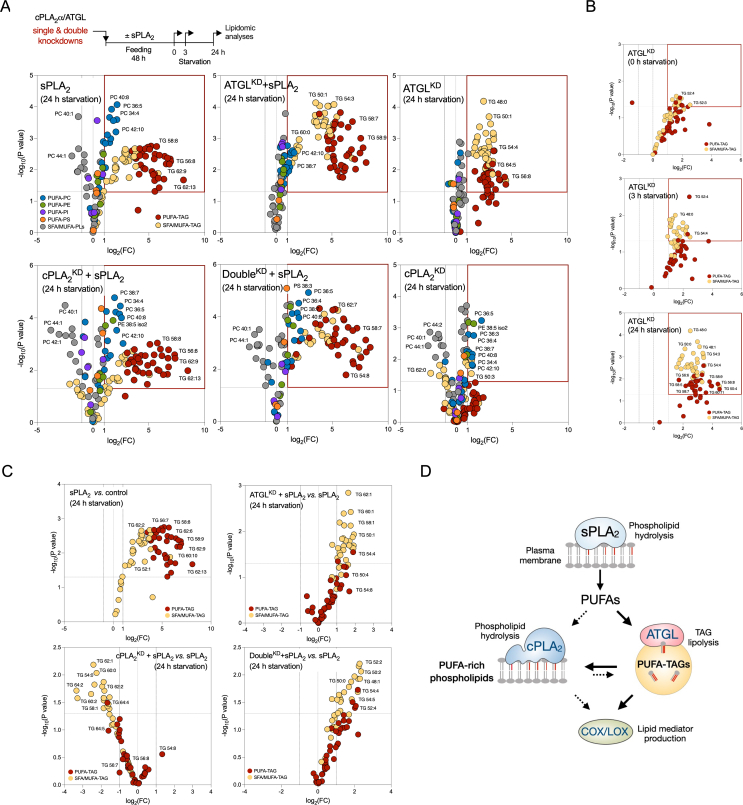
To examine how hGX sPLA2, ATGL and cPLA2α affect PUFA trafficking between the phospholipid and TAG pools in MDA-MB-231 cells during serum starvation, we performed single and double knockdowns of ATGL and cPLA2α followed by lipidomic analyses comparing the lipid compositions of cells first pre-treated with hGX sPLA2 during nutrient sufficiency and subsequently serum-starved for 0 h, 3 h or 24 h (Figure 6A).
Figure 6.
ATGL and cPLA2α cooperatively modulate PUFA trafficking between triglycerides and membrane phospholipids. (A–C) Untargeted lipidomic analysis of phospholipids and triglycerides (TAGs) in serum-starved MDA-MB-231 cells depleted of ATGL (ATGLKD), cPLA2α (cPLA2αKD), or both (DoubleKD), without and with hGX sPLA2 pre-treatment under serum-rich conditions, and grown as shown in the diagram (A). Volcano plots show significant changes (–log10(P value) > 1.30) in individual lipids between each treatment condition versus control cells (unless otherwise indicated), and were prepared by log2 fold-change (FC) data transformation and multiple t-test analysis (n = 3 independent experiments). TAGs and phospholipids (PLs) containing saturated and mono-unsaturated acyl chains (SFA/MUFA-TAGs, SFA/MUFA-PLs, with 0–3 and 0–2 double bonds, respectively) and those containing polyunsaturated FAs (PUFA-TAGs, PUFA-PLs, with at least 4 and 3 double bonds, respectively) are colour-coded as indicated. TG, triglyceride; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol, PS, phosphatidylserine. (D) Schematic illustration of the predominant pathways involved in LD-mediated PUFA trafficking between the membrane phospholipid and TAG pools in serum-starved cancer cells.
First, hGX sPLA2 treatment led to enrichment of both TAGs and phospholipids with PUFAs (Figure 6A; Supp. Figure 6A; Supp. Figure 7A). hGX-sPLA2-induced PUFA-TAG enrichment was seen for serum-fed MDA-MB-231 cells, and it persisted upon hGX sPLA2 and serum withdrawal. Importantly, the levels of PUFA-containing phospholipids (PUFA-PLs) progressively increased during the course of serum starvation (Supp. Figure 6A). These data suggest that a significant portion of hGX sPLA2-released PUFAs that are incorporated into TAGs in serum-fed cells are gradually released via lipolysis during serum starvation and are re-esterified into phospholipids.
Second, ATGL silencing led to an increased abundance of most TAG species, but also altered phospholipid composition (Figure 6A; Supp. Figure 6A). The retention of various TAG species within LDs due to ATGL deficiency is in agreement with the general lack of TAG acyl-chain specificity of ATGL [60]. During serum starvation, ATGL depletion caused a gradual increase of numerous PUFA-TAGs (Figure 6B), which confirmed that ATGL hydrolyses PUFA-TAGs in starving cells. Importantly, ATGL deficiency reverted the elevation of PUFA-PLs induced by hGX sPLA2 pre-treatment (Supp. Figure 6A). This is consistent with a reduced flux of PUFAs from TAGs into phospholipids due to ATGL depletion. Thus, ATGL hydrolyses various TAG species, including numerous PUFA-TAGs, and provides PUFAs for esterification into phospholipids.
Third, depletion of cPLA2α predominantly affected phospholipid (Figure 6A; Supp. Figure 6A), but also altered TAG composition (Figure 6C). Numerous PUFA-PC and PUFA-phosphatidylethanolamine (PUFA-PE) species were progressively elevated in cPLA2α-deficient cells over the course of the serum starvation (Supp. Figure 6A), which suggested that these lipids are targeted by cPLA2α. The decreased abundance of PUFA-PLs observed in ATGL-deficient cells was reversed in double-knockdown cells (Figure 6A; Supp. Figure 6B), which suggested that ATGL-derived PUFAs are incorporated into a phospholipid pool that is targeted by cPLA2α. A LION/web lipid ontology analysis suggested that the increased content of PUFA-PLs in cPLA2α-deficient cells leads to significant changes in membrane biophysical properties, including increased lateral diffusion, reduced bilayer thickness and a lower transition temperature (Supp. Figure 7B and C). Furthermore, cPLA2α silencing did not affect hGX-sPLA2-induced PUFA-TAG enrichment (Figure 6A), but it reduced the content of SFA/MUFA-TAGs (Figure 6C). This effect was fully reversed in double-knockdown cells, suggesting that cPLA2α depletion alters TAG composition in an ATGL-dependent manner. Therefore, ATGL promotes the transfer of PUFAs from TAGs into a phospholipid pool that is targeted by cPLA2α, which in turn reciprocally modulates the TAG pool.
Collectively, our results suggest that hGX-sPLA2-liberated PUFAs are first and predominantly incorporated into TAGs of growing LDs, and are then redistributed into phospholipids upon TAG lipolysis by ATGL, particularly during serum starvation (Figure 6D). Some of the ATGL-released PUFAs are directly used for eicosanoid production (i.e., independent of cPLA2α), while some are re-esterified into phospholipids and can be then mobilised by cPLA2α for the production of eicosanoids.
2.7. LDs drive cancer cell proliferation by promoting eicosanoid production
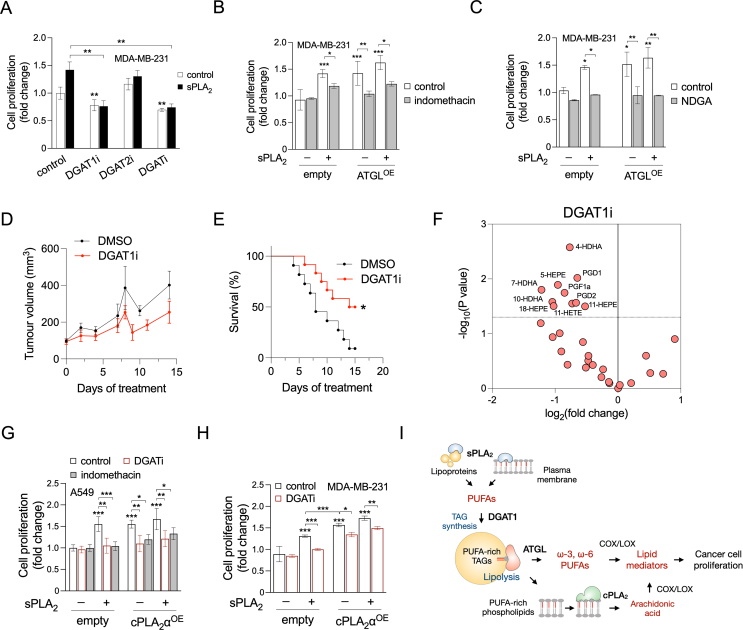
Given that PGE2 and other eicosanoids are mitogenic factors that promote tumour growth [3], we next asked whether LD turnover by DGAT and ATGL affects cancer cell proliferation and possibly mediates the proliferative effect of hGX sPLA2 [22,24]. The induction of MDA-MB-231 cell proliferation by hGX sPLA2 was fully blocked by DGAT1 inhibition and by concurrent inhibition of both DGAT1 and DGAT2; however, DGAT2 inhibition alone had no effects (Figure 7A). Inhibition of DGAT1, but not DGAT2, also reduced the basal, hGX-sPLA2-independent rate of breast cancer cell proliferation (Figure 7A) and colony formation (Supp. Figure 8A and B).
Figure 7.
LD-mediated lipid mediator production promotes cancer-cell proliferation and tumour growth. (A) Proliferation of MDA-MB-231 cells treated with T863 (DGAT1i) and PF-06427878 (DGAT2i), or an equimolar mix of both DGAT inhibitors (DGATi), grown under nutrient-rich conditions in the absence and presence of hGX sPLA2. (B, C) Proliferation of MDA-MB-231 cells overexpressing ATGL (ATGLOE), treated with indomethacin (B) or nordihydroguaiaretic acid (NDGA) (C), in the absence or presence of hGX sPLA2. (D, E) Tumour growth (D) and corresponding Kaplan–Meier survival curves (E) of mice bearing MDA-MB-231 xenografts and treated daily with the DGAT1 inhibitor T863 or with 0.2% DMSO vehicle. Significance was determined by log-rank (Mantel–Cox) test. (F) DGAT1-inhibitor-induced changes in the abundance of lipid mediators and PUFAs in tumour samples isolated from mice bearing MDA-MB-231 xenografts and treated daily with DGAT1 inhibitor T863 or with 0.2% DMSO vehicle. The volcano plot depicts significant changes (–log10(P value) > 1.30) in individual lipids detected by UPLC-MS/MS in DGAT1i-treated versus vehicle-treated mice and was prepared using log2-transformed fold-change values and multiple t-test analysis (n = 4 independent experiments). (G) Proliferation of A549 cells overexpressing cPLA2α (cPLA2αOE) and grown in the absence and presence of DGATi, indomethacin and recombinant hGX sPLA2. (H) Proliferation of MDA-MB-231 cells overexpressing cPLA2α (cPLA2αOE) and grown in the absence and presence of DGATi and recombinant hGX sPLA2. Data are means ± SEM of three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-way ANOVA with Tukey (A), Bonferroni (B, C) or Sidak (G, H) adjustments).
As ATGL promotes lipid mediator production, we hypothesized that ATGL overexpression should induce cell proliferation in a COX/LOX-dependent manner. Indeed, alone or in combination with hGX sPLA2 treatment, ATGL overexpression stimulated cancer cell proliferation (Figure 7B, C), which was suppressed by the non-selective COX and LOX inhibitors indomethacin and nordihydroguaiaretic acid, respectively (Figure 7B, C). Exogenous PGE2-stimulated MDA-MB-231 cell proliferation, confirming its mitogenic potency under the present conditions (Supp. Figure 8C). The effect of PGE2 was suppressed by inhibition of DGAT1, but not by inhibition of DGAT2, which suggested a positive-feedback loop between eicosanoids and DGAT1-mediated TAG turnover. After conducting a comparative analysis of DGAT1 mRNA levels and genetic alterations in various cancer tissues (TCGA PanCancer Atlas Studies), we discovered that the DGAT1 gene is amplified and its mRNA significantly expressed in multiple cancer types, including breast, cervical and lung cancer (Supp. Figure 8D and E). Interestingly, we observed a strong correlation across the different cancer types between the mRNA levels of DGAT1 and PTGES2, which encodes prostaglandin E synthase 2 (Supp. Figure 8F).
To examine the relevance of DGAT1-driven lipid mediator production for cancer cell proliferation in vivo, we treated mice bearing aggressive MDA-MB-231 xenografts with DGAT1 inhibitors, followed tumour growth and determined lipid mediator content in tumours. DGAT1 inhibitor treatments reduced tumour volume and prolonged the survival of mice (Figure 7D, E). Furthermore, tumour samples of DGAT1 inhibitor-treated mice contained reduced levels of lipid mediators (Figure 7F). Thus, DGAT1 inhibition suppressed lipid mediator production and tumour growth leading to prolonged survival of mice bearing breast cancer xenografts. Altogether, these results with MDA-MB-231 cancer cells demonstrate that DGAT1-mediated PUFA incorporation into LDs and PUFA release via ATGL drive basal and hGX-sPLA2-primed production of COX- and LOX-derived lipid mediators that in turn stimulate cancer cell proliferation and tumour growth.
cPLA2α-induced eicosanoid production promotes cancer cell proliferation and tumour growth [11,61], although this activity has not yet been associated with LDs. As cPLA2α has a major role in LD-dependent eicosanoid production in A549 cells (Figure 5B), we asked whether cPLA2α stimulates A549 cell proliferation in a DGAT-and COX-dependent manner. Indeed, cPLA2α-overexpression led to increased A549 cell proliferation, which was blocked by either DGAT or COX inhibitors (Figure 7G), demonstrating that DGAT-mediated TAG synthesis and COX-mediated conversion of cPLA2α-released AA into lipid mediators are required for cPLA2α-stimulated A549 lung cancer cell proliferation. Exogenous hGX sPLA2 also stimulated A549 cell proliferation, which was suppressed by the inhibition of DGAT and COX (Figure 7G). Finally, in MDA-MB-231 cells, where DGAT-mediated TAG synthesis was required for cPLA2α-stimulated PGE2 production (Figure 5H), cPLA2α overexpression resulted in higher proliferation rates, which were reduced by DGAT inhibition (Figure 7H). Together, these data demonstrate that LD biogenesis controls both hGX-sPLA2-and cPLA2α-mediated cancer cell proliferation, which is driven by lipid mediators produced by COX and LOX enzymes.
3. Discussion
In this study, we provide evidence that TAG turnover controls the production of a wide range of ω-3 and ω-6 PUFA-derived oxygenated lipid mediators. We show that the esterification of PUFAs into TAGs and their lipolytic release from LDs are necessary for PUFA entry into lipid mediator biosynthetic pathways. We demonstrate that ATGL liberates ω-3 and ω-6 PUFAs from TAGs, alters membrane phospholipid composition and drives lipid mediator production via the COX and LOX pathways (Figure 7I). Our data further suggest that LDs control canonical PLA2-stimulated lipid mediator biosynthesis pathways through 1) a DGAT-dependent sequestration of exogenous and membrane-derived PUFAs and 2) by modulation of membrane PUFA content through the delivery of LD-derived PUFAs into phospholipids via ATGL. We demonstrate that through the initial release of PUFAs from the plasma membrane and serum lipoproteins, hGX sPLA2 primes cells for lipid mediator production by DGAT-driven enrichment of TAGs with PUFAs, followed by their delayed release by ATGL-mediated lipolysis. Lipid mediator production induced by cPLA2α, which acts on perinuclear membranes to release AA for eicosanoid production, also depends on intact TAG turnover. These findings change the paradigm of PLA2-mediated inflammatory and mitogenic signalling by integrating membrane hydrolysis with LD metabolism. Furthermore, our results demonstrate that the inhibition of DGAT1-mediated TAG synthesis compromises lipid mediator production in cancer cells, thereby suppressing basal, hGX sPLA2-, and cPLA2α-induced cancer cell proliferation in vitro and reducing tumour growth in vivo. This study identifies the LD organelle as a central lipid trafficking hub that controls major PUFA supply routes for lipid mediator biosynthesis.
One of the most striking findings of the present study is that LD turnover regulates eicosanoid production from both exogenously-added PUFAs and membrane-hydrolysis-derived PUFAs, suggesting that the build-up of PUFA-rich TAG stores is a required step in the control of lipid mediator signalling. A similar mechanism has been described in cardiomyocytes, whereby exogenous FAs have to be esterified into TAGs and then released by ATGL to activate peroxisome-proliferator-activated receptor (PPAR) signalling [40,62]. Furthermore, as we found DGAT activity to be a prerequisite for both ATGL-induced and sPLA2/cPLA2α-primed eicosanoid production, it can be assumed that LDs control PUFA availability for eicosanoid production through the regulation of both TAG and membrane phospholipid pools.
Our previous data showed that breast cancer cells challenged with exogenous PUFAs depend on the balance between DGAT1-mediated sequestration of PUFAs into LDs and their release via ATGL-mediated lipolysis to survive lethal oxidative damage [20]. In support of this, inhibition of DGAT activity diverts dietary PUFAs towards esterification into membrane phospholipids, thereby increasing their peroxidation and leading to ferroptosis in acidic tumours [52]. Accordingly, DGAT1 increases the resilience of cancer cells against the stress of increased FA acquisition, which is a hallmark of transformed cells [63]. It was recently shown that DGAT1-mediated TAG synthesis is required for prostaglandin formation during macrophage activation [64] and Drosophila oogenesis (preprint [65]). Our results suggest that DGAT activity controls cancer cell growth and proliferation by generating PUFA stores in LDs, which can be used for the production of potent, albeit short-lived, pro-inflammatory and anti-inflammatory lipid signalling molecules. As these signalling molecules are released from cells to act in autocrine and paracrine manners, they can modulate tumour growth by altering the function of cancer cells, immune cells and other cells in the tumour microenvironment [3,4]. Thus, in addition to their protective role against nutrient deficiency, lipotoxicity and oxidative stress [38], LDs modulate tumour growth also through the control of lipid mediator-dependent cell-autonomous and non-cell-autonomous mitogenic and inflammatory signalling pathways, and thus emerge as targets for therapeutic intervention. Targeting the DGAT enzymes might improve cancer treatments, particularly under conditions of elevated lipid influx (e.g., abundance of dietary fats, dyslipidemia, oncogene-driven elevated endogenous FA synthesis, high autophagic flux) [36,52,66].
sPLA2 enzymes display tissue- and cell-specific expression patterns and differ in their membrane binding affinities and hydrolytic activities depending on the phospholipid composition and structural features of their target lipid assemblies [67,68]. Their ability to promote lipid mediator production has been attributed either to direct PUFA release from phospholipids (e.g., in the case of hGX sPLA2) or cooperative action with cPLA2α (e.g., in the case of the group IIA sPLA2) [13,18,69]. Regardless of the particular PLA2(s) involved, the release of PUFAs from phospholipids has been considered the final step in the control of PUFA entry into oxygenation pathways. Here, we show that lipid mediator production by two of the most potent mammalian PLA2s, hGX sPLA2 and cPLA2α, depends on LD turnover. Notably, hGX-sPLA2-treated MDA-MB-231 breast cancer cells showed an enrichment of both TAGs and phospholipids with PUFAs, but their release from TAGs by ATGL was essential for their conversion into lipid mediators and preceded the action of cPLA2α. Intriguingly, in A549 lung cancer cells, cPLA2α rather than ATGL was required for hGX-sPLA2-primed PGE2 production, but intact TAG synthesis was still a prerequisite for both hGX-sPLA2-and cPLA2α-dependent eicosanoid synthesis and cell proliferation. This suggests that lipid mediator production could be mediated in different cell types by various combinations of PLA2s and (TAG) lipases, but the LD emerges as a central hub that controls the trafficking and final destination of PUFAs.
This study provides a novel view of PLA2-mediated inflammatory and mitogenic signalling, which integrates membrane hydrolysis with cellular LD metabolism. LDs could be the missing link that will help explain the elusive cross-talk between sPLA2s and cPLA2α in lipid mediator biosynthesis [13,19,67]. It will be interesting to investigate in future studies whether LD metabolism governs PLA2-dependent lipid mediator production in various pathophysiological settings. For example, PUFA-enriched TAGs have been observed in visceral adipose tissue of patients with colorectal cancer, along with elevated expression of hGX sPLA2 and prostaglandin biosynthetic enzymes [70]. As shown in Figure 1C and Supp. Figure 1B and C, PLA2G10 gene expression is amplified in several cancer types and correlates with the expression of several LD metabolism and membrane remodelling genes, including ATGL and DGAT1, which suggests that the pathophysiological role of hGX sPLA2 in cancer might well be linked with LD metabolism. However, more work will be needed to identify the type of cells within tumours that express and release the group X sPLA2 and demonstrate that the enzyme enriches LDs with PUFAs in vivo, leading to enhanced eicosanoid production and tumour growth. Given that MDA-MB-231 cells express very low levels of hGX sPLA2 [71], we could not test the capacity of sPLA2 inhibitors to limit xenograft cancer growth in our model system. However, a recent study has shown that the group X sPLA2 released from macrophages hydrolyzes tumour-cell-derived extracellular vesicles and accelerates B cell lymphoma formation [21]. These effects were inhibited by the sPLA2 inhibitor varespladib, identifying the enzyme as a potential therapeutic target in cancer. Furthermore, the hGX sPLA2-induced incorporation of ω-3 and ω-6 PUFAs into TAGs, which is a novel mechanism of enzymatically-induced enrichment of LDs with phospholipid-derived PUFAs, could help clarify the various proposed roles of the group X sPLA2 that also extend beyond cancer and lipid mediator production [15,23,68,72,73].
Our results indicate that different cell-type-specific mechanisms might explain the dependence of cPLA2α-dependent eicosanoid production on LDs. In the MDA-MB-231 and HeLa cells, cPLA2α depends on TAG synthesis and ATGL-mediated TAG lipolysis to drive the incorporation of PUFAs into phospholipids, which are then targeted by cPLA2α. In A549 cells, cPLA2α controls basal and sPLA2-primed PGE2 production independently of ATGL, although its activity still depends on intact LD biogenesis. In both cases, our data suggest that cPLA2α-mediated AA release for eicosanoid production occurs downstream of, and is controlled by, LD turnover. Altogether, our findings suggest that LD turnover regulates the supply of PUFAs for lipid mediator production via at least two pathways: direct delivery of LD-derived PUFAs, which is independent of cPLA2α; and an indirect route, which requires cPLA2α but depends on LDs for the control of PUFA availability in membrane phospholipids. Notably, our data indicate that other lipases and LD breakdown mechanisms (e.g., lipophagy) besides (or instead of) ATGL might contribute to LD-dependent lipid mediator biosynthesis.
Interestingly, cPLA2α has been shown to drive mitochondrial β-oxidation of both FAs and eicosanoids for energy production [74], which is another indication that ATGL and cPLA2α share common metabolic and signalling pathways. Indeed, LD metabolism, and ATGL in particular, are tightly coupled with mitochondrial β-oxidation for energy production and other purposes [[62], [75]]. Notably, our previous research has demonstrated that hGX-sPLA2-induced LD accumulation and enhanced MDA-MB-231 cell proliferation are associated with the activation of 5' AMP-activated protein kinase (AMPK) and increased expression of genes involved in mitochondrial β-oxidation under nutrient-rich conditions [20,24]. These effects were further augmented and contributed to cell survival during serum starvation, which suggested that ATGL-mediated lipolysis is coupled with AMPK-driven increase in β-oxidation in controlling cellular fate in both fed and starved MDA-MB-231 cells. Nevertheless, the involvement of β-oxidation in LD-induced cell proliferation observed in this study and its potential contribution to eicosanoid metabolism remains unknown.
One of the intriguing questions that remains to be addressed in future studies is whether PUFA trafficking between the TAG core and the LD phospholipid monolayer is relevant for lipid mediator production. In principle, TAG lipolysis-derived PUFAs can be re-esterified in monolayer phospholipids and targeted by cPLA2α or other PLA2s. In agreement with this, cPLA2α, COXs, several prostaglandin synthetases and de novo produced eicosanoids have been localised to LDs [[46], [57], [58], [59], [76], [77]]. Our findings in cells treated with DGAT inhibitors under nutrient sufficiency, showing that the AA-derived PGE2 and PGD2 are the only mediators decreased upon LD depletion, are in accordance with a mechanism whereby the localization of cPLA2α and the cyclooxygenase/prostaglandin synthase machinery on LDs is required for the production of AA-derived prostaglandins. The strong correlation between the expression of DGAT1 and PTGES2 in human tumour samples derived from multiple cancer types is also indicative of a relationship between LDs and prostaglandin production. In addition, cPLA2α is involved in remodelling of membrane shape and it might affect LD biogenesis and lipolysis by altering membrane composition and its biophysical properties [[6], [7], [50], [77], [78], [79], [80], [81]]. In agreement with this, our results indicate that cPLA2α modulates membrane PUFA-phospholipid content and LD metabolism, including LD abundance and TAG acyl-chain composition. The molecular basis and functional relevance of these findings are currently unclear. We speculate that eicosanoid production does not necessarily occur on LDs as isolated cytosolic platforms, but at specific LD–ER contact sites that enable rapid lipid and protein transfer between the structures involved (i.e., the ER membrane bilayer, the LD phospholipid monolayer and TAG core) [82]. Such compartmentalisation would support the interplay among cPLA2α, ATGL and other enzymes in the control of LD turnover, membrane remodelling and the eicosanoid synthesis machinery.
In conclusion, our study reveals that LDs are essential for lipid mediator production and cancer cell proliferation. Esterification of PUFAs into TAGs by the DGAT enzymes and PUFA release from LDs (which depends on TAG lipolysis by ATGL, and/or other lipases depending on the cell type) drives lipid mediator production either directly, by feeding PUFAs into the COX/LOX machinery, or by redirecting some of the PUFAs into membrane phospholipids first, whereby they are then targeted by cPLA2α. Notably, both exogenous and sPLA2-membrane-hydrolysis-derived PUFAs have to cycle through LDs to be converted into lipid mediators, which reveals that LDs are optimal storage reservoirs for excess PUFAs that actively control PUFA release. LDs thus have an important regulatory role in lipid mediator production, thereby potentially affecting numerous downstream signalling pathways that are involved in inflammation, immunity and cancer.
4. Materials and methods
4.1. Materials
MDA-MB-231 human breast adenocarcinoma cells, A549 human lung carcinoma cells, HeLa human cervical adenocarcinoma cells, C2C12 mouse myoblasts cells, and Caco-2 human colorectal adenocarcinoma cells were obtained from American Type Culture Collection (ATCC, USA). J774A.1 mouse reticulum cell sarcoma macrophages were from the European Collection of Authenticated Cell Cultures (ECACC, UK), and MDA-MB-231 human breast adenocarcinoma cells with stable luciferase 2A and RFP expression (MDA-MB-231/Luciferase-2A-RFP) were from GeneTarget (USA). PC-3 human prostate adenocarcinoma cells were a kind gift from Dr. Mojca Pavlin (University of Ljubljana, Slovenia). OV-90 human ovarian papillary serous adenocarcinoma cells, TOV-112D human ovarian endometrioid carcinoma cells, TOV-21G human ovarian clear cell carcinoma cells and Ishikawa human endometrial adenocarcinoma cells were a kind gift from Dr. Brett McKinnon (Berne University Hospital, Switzerland). SGBS human Simpson-Golabi-Behmel syndrome preadipocytes were a kind gift from Dr. Merce Miranda (Joan XXIII University Hospital Tarragona, Spain), and SH-SY5Y human neuroblastoma cells were a kind gift from Dr. Boris Rogelj (Jožef Stefan Institute, Slovenia). RPMI-1640 culture medium was from ATCC (USA), and Dulbecco's modified Eagle's medium nutrient mixture F-12 (DMEM/F12), DMEM with high glucose and GlutaMAX supplement (DMEM-GlutaMax), fetal bovine serum, Dulbecco's phosphate-buffered saline (DPBS), TrypLE Select and Opti-MEM were from Life Technologies (USA). AA and PGE2 standards were from Cayman Chemical (USA), and BODIPY 493/503, Lipofectamine RNAiMAX, Lipofectamine 3000, High-capacity cDNA reverse transformation kits were from Thermo Fisher Scientific (USA). Hoechst 33342 nuclear stain was from Enzo Life Sciences (USA). Human ATGL-targeting and cPLA2α-targeting siRNAs and the AllStars Negative Control siRNA were from Qiagen (Germany). T863 (DGAT1 inhibitor), PF-06424439 (DGAT2 inhibitor), indomethacin (COX inhibitor), nordihydroguaiaretic acid (LOX inhibitor), essentially fatty acid-free (EFAF) bovine serum albumin (BSA) (cat. no. A7511), FAF-BSA (cat. no. A8806) and Nile red were from Sigma–Aldrich (USA). A23187 was from Alomone Labs, (Israel). High Pure RNA isolation kits were from Roche (Germany), horseradish-peroxidase-labelled secondary antibodies were from Jackson ImmunoResearch Laboratories (USA), ATGL (cat. no. 2138) and phospho-cPLA2α (Ser505) (cat. no. 2831) antibodies were from Cell Signaling Technology (USA), cPLA2α antibodies (cat. no. sc-454) were from Santa Cruz (USA), β-tubulin antibodies (cat. no. T52O1) were from Sigma and β-actin antibodies (cat. no. NB600-532) were from Novus Biologicals (UK). Recombinant wild-type hGX sPLA2 was prepared as described previously [24]. The full-length cDNAs coding for human cPLA2α (NCBI RefSeq NM.024420.3) and ATGL (NCBI RefSeq NM.020376.4) were cloned into the pcDNA 4/HisMaxC vector (Thermo Fisher Scientific, USA) using Gibson assembly cloning kits (New England Biolabs, USA), after removal of the N-terminal His-tag region. All of the other chemicals were of at least analytical grade, and were purchased from Sigma-Aldrich (USA) or Serva (Germany).
4.2. Cell culture and treatments
MDA-MB-231 cells were cultured in RPMI-1640 medium, A549 cells in DMEM/F12 medium containing 2 mM l-glutamine (Gibco, USA), and HeLa cells in DMEM-Glutamax medium, all supplemented with 10% fetal bovine serum. Adherent cells were detached using TrypLE Select. Unless otherwise indicated, the cells were seeded in 24-well plates at a density of 3 × 104 (MDA-MB-231), 2.5 × 104 (A549) or 1.5 × 104 (HeLa) cells/well and grown for 48 h in complete medium, followed by 24 h of serum deprivation in their respective media containing 0.02% EFAF-BSA. In experiments with the A23187 calcium ionophore, cells were treated with 10 nM hGX sPLA2 for 48 h in complete medium, washed with DPBS and serum-starved for 1.5 h or for 24 h in serum-free medium containing 0.02% EFAF-BSA and treated with 1 μM A23187 during the last 60 min. Aliquots of stock solutions of AA and PGE2 in absolute ethanol were stored under argon at −80 °C. Prior to addition to cell cultures, AA was resuspended in the relevant complete medium and incubated for 1 h at room temperature. Unless otherwise indicated, T863 and PF-06424439 were added to cells 2 h before treatments with recombinant hGX sPLA2 (1–10 nM) or AA (10 μM), and were present in the medium for the duration of the treatments. Prior to addition to cell culture, radiolabelled [14C]-OA was saponified by removing ethanol from the stock aliquot of [14C]-OA and resuspending in 50 μL 0.1 mM NaOH. Saponified [14C]-OA was incubated in the relevant complete medium for 30 min at room temperature and stored at −20 °C.
4.3. Silencing of ATGL and cPLA2α expression using small-interfering RNAs
Reverse transfection was performed in 24-well plates at cell densities of 6 × 104 (MDA-MB-231), 5 × 104 (A549) or 3 × 104 (HeLa) cells/well, or in 6-well plates at 3 × 105 (MDA-MB-231), 2.5 × 105 (A549) or 1.5 × 105 (HeLa) cells/well. Gene expression silencing was performed with a 20 nM mixture of two ATGL-specific siRNAs (10 nM each) or a 40 nM mixture of four siRNAs specific for cPLA2α. Non-targeting siRNA controls contained 20 nM (for ATGL) or 40 nM (for cPLA2α) AllStars Negative Control siRNA (Qiagen). Transfection complexes were generated using 1 μL/well Lipofectamine RNAiMAX in 24-well plates, or 7.5 μL/well in 6-well plates, with Opti-MEM medium, according to manufacturer instructions.
4.4. Transient overexpression of ATGL and cPLA2α
For transient overexpression experiments, the cells were seeded in complete medium in 24-well plates at a density of 9 × 104 (MDA-MB-231), 6 × 104 (A549) and 4.5 × 104 (HeLa) cells/well, or in 6-well plates at 4.5 × 105 (MDA-MB-231), 3 × 105 (A549) and 2.5 × 105 (HeLa) cells/well. Cells were then incubated for 24 h in complete medium, washed, and transfected with 0.5 μg/well plasmid DNA in 24-well plates, or 2.5 μg/well in 6-well plates, using Lipofectamine 3000 and Opti-MEM medium, according to manufacturer instructions; they were then left for 6 h in serum-depleted medium containing 0.02% EFAF-BSA (serum starvation). After 6 h, the cells were washed and treated according to the experimental set-up.
4.5. Real-time quantitative PCR
Real-time quantitative (q)PCR analysis was performed as described previously [24,71]. Briefly, the cells were seeded in complete medium in 6-well plates at 3 × 105 (MDA-MB-231), 2.5 × 105 (A549) and 1.5 × 105 (HeLa cells) cells/well, grown 48 h in complete medium, followed by 24 h serum deprivation in medium containing 0.02% EFAF-BSA. Total RNA was isolated from cell lysates and first-strand cDNA was generated using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, USA), according to the manufacturer instructions. qPCR analysis was performed on a StepOnePlus real-time PCR system (Thermo Scientific, USA) using FastStart Universal SYBR Green Master (Rox; Roche, Switzerland). Calibrator cDNA was transcribed from Quantitative PCR Human Reference Total RNA (Agilent Technologies, USA). Relative gene expression was calculated upon normalisation to two reference genes, considering primer-specific PCR efficiency and error propagation.
4.6. Western blotting
Cells were seeded in complete medium in 6-well plates and reverse transfected with siRNAs and/or transiently transfected with pDNA, as described above. Cell lysates were prepared by scrapping adherent cells in Tris-glycine sodium dodecyl sulphate (SDS) sample buffer (Novex, Life Technologies, USA) that contained 800 mM dithiothreitol (Sigma-Aldrich, USA), with the addition of Halt protease inhibitor cocktail (Thermo Scientific, USA). Lysates were incubated at 95 °C for 10 min and stored on ice. Total protein concentrations were determined using Pierce 660 nm protein assays (Thermo Scientific, USA). Proteins (10–40 μg) were separated on 10% SDS-PAGE gels and then transferred to nitrocellulose membranes (Serva, Germany). The membranes were blocked for 1 h (for ATGL) or 2 h (for cPLA2α) in 5% non-fat dry milk in TBS/0.1% Tween-20 (TBST) or in 1% Western blocking reagent (WBR) (Roche Applied Science, Germany) in TBS (for β-actin). For the detection of phosphorylated cPLA2α (p-cPLA2α) membranes were blocked for 1 h in 5% BSA in TBST and 1 h in 1% WBR in TBS for β-tubulin. Membranes were then incubated overnight at 4 °C in the presence of rabbit anti-human primary antibodies for ATGL (1:1000 dilution), rabbit anti-human primary p-cPLA2α antibodies (1:1000 dilution), mouse anti-human primary β-tubulin antibodies (1:5000 dilution) or mouse anti-human primary antibodies for cPLA2α (1:250 dilution), all in 5% non-fat dry milk in TBST, or rabbit anti-human primary antibodies for β-actin (1:5000 dilution) in 0.5% WBR in TBS. After washing with TBST, the membranes were incubated for 1 h with horseradish-peroxidase-conjugated secondary antibodies (1:10,000 dilution) in 5% non-fat dry milk in TBST for ATGL, in 0.5% WBR in TBST for cPLA2α and p-cPLA2α, and in 0.5% WBR in TBS for β-actin and β-tubulin. The signals were visualised using Lumi-Light Western Blotting Substrate (Roche Applied Science, Germany) on a Gel Doc XR system (Bio-Rad, USA). Images were analysed using densitometry with ImageJ (National Institutes of Health, USA).
4.7. Neutral lipid quantification by flow cytometry
Cellular neutral lipid levels were quantified by flow cytometry as described previously [24]. Floating and adherent cells were harvested and centrifuged at 300×g for 10 min, and the pellets were resuspended in 500 μL 1 μg/mL Nile Red solution in DPBS. After a 10-min incubation in the dark, cell analysis was performed by flow cytometry on a FACSCalibur system, equipped with a 488-nm Ar-ion laser, and using the CellQuest software (Becton Dickinson, USA) and an FL-1 (530/30) filter, for at least 2 × 104 events per sample.
4.8. Triglyceride assays
Cellular TAG contents were determined using a TAG quantification assay (Abcam, USA). MDA-MB-231 and HeLa cells were seeded in 6-well plates at 1.5 × 105 and 7 × 104 cells/well, respectively. After 24 h, MDA-MB-231 cells were treated with 10 nM hGX sPLA2 and HeLa cells with 1 nM hGX sPLA2 in complete medium for 48 h. Cell lysates were prepared and used for TAG quantification according to the manufacturer instructions.
4.9. Glycerol release assays
Cellular lipolytic activity was assessed by measuring glycerol release in cell supernatants. Briefly, the cells were reverse transfected and seeded in 48-well plates at a density of 3 × 104 (MDA-MB-231 cells) or 1.5 × 104 cells/well (HeLa cells). For reverse transfection, 0.5 μL Lipofectamine RNAiMAX, 20 nM siRNA and 40 μL OPTI-MEM medium were used per well. After 24 h, the cells were washed, placed in serum-free medium containing 0.02% EFAF-BSA, and transfected with 0.25 μg plasmid DNA for protein overexpression using Lipofectamine 3000, according to the manufacturer instructions. After 6 h, the cells were washed and treated with 10 nM hGX sPLA2 for 48 h in complete medium. Cell supernatants were collected in low-binding microcentrifuge tubes and centrifuged for 10 min (4 °C, 16,000×g), and the glycerol concentrations were determined using the Glycerol Cell-Based assay kits (Cayman Chemicals, USA), according to the manufacturer instructions.
4.10. Untargeted lipidomic analysis of phospholipids and triglycerides
For untargeted lipidomic analysis of hGX sPLA2-induced changes in TAG acyl-chain composition, MDA-MB-231 cells were seeded in complete medium on 10-cm plates at 1 × 106 cells/plate. After 24 h, the cells were treated with 1 nM hGX sPLA2 in complete medium for 48 h. Cell lysates were prepared by washing the cells twice with DPBS and scraping in 1 mL lysis buffer (20 mM Tris–HCl, pH 7.4, 2 mM EDTA, 2 μL Halt protease inhibitor cocktail), followed by centrifugation for 10 min (1000×g, 4 °C). Cell pellets were resuspended in 150 μL lysis buffer and sonicated on ice. Total lipids were extracted in chloroform/methanol (2/1, v/v) containing 1% acetic acid, 500 nM butylated hydroxytoluene (BHT) and internal standards (IS; 100 pmol 17:0/17:0/17:0 triacylglycerol, Larodan, Solna, Sweden) under constant shaking for 1 h (30 rpm/min, 4 °C). After centrifugation at 3300 rpm for 20 min at room temperature, the upper aqueous layer was removed, and the organic solvents were evaporated using a sample concentrator (Techne, UK) equipped with the Dri-Block DB-3 heater (Techne, UK). Lipids were resolved in 200 μL chloroform and stored at −20 °C. Prior to mass spectrometry, the samples were placed at room temperature and dried and resuspended in 1 mL chloroform/methanol (2/1, v/v). An aliquot of each sample (20 μL) was mixed with 180 μL isopropanol, and 5 μL was used for chromatographic separation on an Acquity-UPLC system (Waters Corporation, Milford, MA, USA), equipped with an ACQUITY BEH C18 column (2.1 × 50 mm, 1.7 μm; Waters Corporation, Milford, MA, USA). A SYNAPTTMG1 qTOF HD mass spectrometer (Waters Corporation, Milford, MA, USA) equipped with an ESI source was used for detection. Data acquisition was carried out using the MassLynx 4.1 software (Waters), and the lipid classes were analysed with the Lipid Data Analyser 1.6.2 software. The data were normalised for recovery, extraction and ionisation efficacy by calculating analyte/internal standard ratios (AU) and expressed as percentage composition.
To determine the changes in the phospholipid and TAG profiles in ATGL-depleted and cPLA2α-depleted cells, MDA-MB-231 cells were seeded in complete medium on 6-well plates at 3 × 105 cells/well and reverse transfected with ATGL-specific and/or cPLA2α-specific siRNAs, as described above. After 24 h, the cells were washed and grown for 48 h in complete medium in the presence or absence of 10 nM sPLA2. The cells were then washed twice with DPBS and serum starved for the following 24 h in RPMI-1640 medium containing 0.02% EFAF-BSA. Samples were collected (at 0, 3, 24 h of serum starvation) by placing the plates on ice, washing the cells twice with ice-cold DPBS, and scraping the cells in 300 μL lysis buffer, followed by centrifugation for 10 min (1000×g, 4 °C). The cell pellets were resuspended in 150 μL lysis buffer and sonicated on ice. Then, 10 μL of each sample was used for protein determination (Pierce 660). Total lipids were extracted by transferring the pellets into 2 ml tubes followed by homogenisation in 700 μL of a 3:1 (v/v) mixture of methyl tert-butyl ether and methanol, containing 1% acetic acid, 500 nM BHT and IS (8 pmol 18:3/18:3/18:3 triacylglycerol, 14:0/14:0 phosphatidylcholine, Larodan, Solna, Sweden; 50 pmol 17:0/17:0 phosphatidylethanolamine, 12 pmol 17:0/17:0 phosphatidylserine, Avanti Polar Lipids, Alabaster, AL, USA), with two steel beads on a mixer mill (30 Hz, Retsch, Germany) at 4 °C. After homogenisation, the samples were mixed at room temperature under constant shaking for 30 min. Then 140 μL distilled H2O was added, and the samples were thoroughly mixed and centrifugated (14,000 rpm, 10 min), to establish phase separation. The organic phase (500 μL) was transferred into new tubes and the organic solvent was evaporated off under a stream of nitrogen. The residual protein slurry was dried and used for protein determination after lysis in 400 μL NaOH/SDS (0.3 M/0.1%). Prior to mass spectrometry analysis, the lipids were resolved in 200 μL isopropanol/methanol/H2O (70/25/10, v/v/v). Chromatographic separation was performed on a 1290 Infinity II LC system (Agilent, Santa Clara, CA, USA) equipped with a C18 column (Zorbax RRHD Extend; 2.1 × 50 mm, 1.8 μm; Agilent, Santa Clara, CA, USA), using a 16 min linear gradient from 60% solvent A (H2O; 10 mM ammonium acetate, 0.1% formic acid, 8 μM phosphoric acid) to 100% solvent B (2-propanol; 10 mM ammonium acetate, 0.1% formic acid, 8 μM phosphoric acid). The column compartment was kept at 50 °C. A Q-TOF mass spectrometer (6560 Ion Mobility; Agilent, Santa Clara, CA, USA) equipped with electrospray ionisation source (Dual AJS) was used for detection of the lipids in positive and negative Q-TOF mode. Lipid data were acquired at MS1 level using the MassHunter Data Acquisition software (B.09; Agilent, Santa Clara, CA, USA) and initial feature analysis was performed in MassHunter Mass Profiler (V10.0, Agilent). A manually curated, in-house lipid library of naturally occurring lipids, containing ∼1000 MS2-identified/characterized lipid species, was used for lipid-feature identification based on accurate precursor mass (< 5 ppm) and retention time (< 0.2 min). All identifications were inspected manually and RT and m/z of identified lipid species were used as target list for peak integration in MassHunter Quantitative Analysis (B.09.00). Peak areas were blank subtracted, single-point quantified using the respective internal standard, normalized using the determined protein amount of the respective sample and expressed as fmol/μg protein. Lipidomic data were analysed by multiple t-test analysis of log-transformed data to compare two conditions at a time, using GraphPad Prism 9.0.2 (GraphPad Software, USA). Lipid ontology enrichment analysis was performed using the LION/web enrichment and principal component analysis modules [83]. Lipidomics data were deposited in the MetaboLights database under study ID MTBLS7786.
4.11. Thin layer chromatography of radiolabelled cellular lipids
MDA-MB-231 cells were seeded in complete medium on T-25 flasks at a density of 5 × 105 cells/flask. After 24 h, the cells were treated with 1 μCi/sample [14C]-OA for 18 h, washed twice with DPBS, and treated with 10 nM hGX sPLA2 under three different conditions: (a) 24 h in complete medium; (b) 24 h in complete medium followed by 96 h serum deprivation in 0.02% EFAF-BSA in RPMI-1640; and (c) 24 h in 0.02% FAF-BSA in RPMI-1640 followed by 96 h in 0.02% EFAF-BSA in RPMI-1640. Lipid extraction was performed with 1 mL hexane/isopropanol (3:2, v/v) under constant shaking for 10 min at room temperature, and repeated twice. The samples were stored in microcentrifuge tubes at −20 °C. Total proteins were isolated from cell remnants in 2 mL lysis buffer (0.3 M NaOH, 0.1% SDS) under constant shaking for 2 h at room temperature. Protein concentrations were determined using BSA standard solutions (Thermo Scientific, USA) and the BCA protein assay reagent (Thermo Scientific, USA). Samples of cellular lipids were dried, resuspended in 20 μL chloroform (repeated three times), and loaded onto the stationary phase of silica TLC plates immobilised on a polymeric binder (Merck, Germany). Dry TLC plates were developed using chloroform/methanol/acetone/acetic acid/H2O (50/10/20/12/5, v/v/v/v/v) for phospholipid separation, and hexane/diethyl ether/acetic acid (70:29:1, v/v/v) for neutral lipid separation. Radiolabelled lipids were detected with autoradiography using an imager (PhosphorImager SI; Amersham Biosciences, UK). To quantify the TAG content, the TLC plates were incubated in an iodine steam, the TAG patches were cut out and placed in vials with scintillation cocktail, and the radioactivity was measured using a liquid scintillation counter (Tri-Carb 1600CA; PerkinElmer, USA).
4.12. Lipid mediator analysis using UPLC-MS-MS
MDA-MB-231 cells were seeded in complete medium in 6-well plates at 3 × 105 cells/well. In case of ATGL silencing, cells were reverse transfected with ATGL-specific siRNAs as described above. After 24 h, the cells were treated with 10 nM hGX sPLA2 and/or a mixture of 20 μM DGAT1 and 20 μM DGAT2 inhibitors for 72 h (fed cells) or for 48 h in complete medium, then washed with DPBS and starved for 24 h in RPMI-1640 medium containing 0.02% EFAF-BSA (fed + starved cells). Cells with DGAT inhibition were treated with the inhibitors also during the serum starvation phase. The cells were lysed and total protein concentrations were determined as described above. Supernatants were collected in low-binding microcentrifuge tubes, centrifuged at 4 °C (16,000 × g, 10 min) and stored at −80 °C before shipping on dry ice. 1 mL of the supernatants (MDA-MB-231 experiment) were first mixed with 2 mL of ice-cold methanol containing deuterium-labelled internal standards (200 nM d8-5S-HETE, d4-LTB4, d5-LXA4, d5-RvD2, d4-PGE2 and 10 μM d8-AA; Cayman Chemical/Biomol GmbH, Hamburg, Germany) to facilitate quantification and sample recovery. Samples were kept at −20 °C for 60 min to allow protein precipitation.
Tumour samples for UPLC-MS-MS analysis were collected from mice xenografts. After cancer cell injection and visible tumour growth, mice were subcutaneously treated daily with T863 at a dose of 9.6 mg/kg or corresponding vehicle (0.2% DMSO) for two weeks before being sacrificed and sampled for tumours. Tumour samples (30–50 mg) were weighed and 25 μL methanol of each mg tumour was added and then homogenized. After that, samples were centrifuged (16,000 × g, 4 °C, 5 min), and 800 μL (corresponding to 32 mg tumour) were filled up to 2 mL methanol and 1 mL PBS was added containing deuterium-labelled internal standards (200 nM d8-5S-HETE, d4-LTB4, d5-LXA4, d5-RvD2, d4-PGE2 and 10 μM d8-AA; Cayman Chemical/Biomol GmbH) to facilitate quantification and sample recovery. Lipid mediator analysis using UPLC-MS-MS was performed as described previously [84] with some minor modifications [85]. Briefly, after centrifugation (1200 × g, 4 °C, 10 min) 8 mL of acidified H2O was added (final pH = 3.5) and the samples were subjected to solid phase extraction. The solid phase cartridges (Sep-Pak® Vac 6cc 500 mg/6 mL C18; Waters, Milford, MA) were equilibrated with 6 mL methanol and then with 2 mL H2O prior sample loading onto the columns. After washing with 6 mL H2O and additional 6 mL n-hexane, lipid mediators were eluted with 6 mL methyl formate. The eluates were brought to dryness using a TurboVap LV evaporation system (Biotage, Uppsala, Sweden) and resuspended in 200 μL methanol/water (50/50, v/v) for UPLC-MS-MS analysis. Lipid mediator profiling was analyzed with an Acquity™ UPLC system (Waters, Milford, MA, USA) and a QTRAP 5500 Mass Spectrometer (ABSciex, Darmstadt, Germany), equipped with a Turbo V™ Source and electrospray ionization. Lipid mediators were separated using an ACQUITY UPLC® BEH C18 column (1.7 μm, 2.1 × 100 mm; Waters, Eschborn, Germany) at 50 °C with a flow rate of 0.3 mL/min and a mobile phase consisting of methanol/water/acetic acid of 42/58/0.01 (v/v/v) that was ramped to 86/14/0.01 (v/v/v) over 12.5 min and then to 98/2/0.01 (v/v/v) for 3 min [85]. The QTrap 5500 was operated in negative ionization mode using scheduled multiple reaction monitoring (MRM) coupled with information-dependent acquisition. The scheduled MRM window was 60 s, optimized lipid mediator parameters were adopted [85], and the curtain gas pressure was set to 35 psi. The retention time and at least six diagnostic ions for each lipid mediator were confirmed by means of external standards (Cayman Chemical/Biomol GmbH). Quantification was achieved by calibration curves for each lipid mediator. Linear calibration curves were obtained for each lipid mediator and gave r2 values of 0.998 or higher (for fatty acids 0.95 or higher). Additionally, the limit of detection for each targeted lipid mediator was determined [85].
4.13. Quantification of PGE2 in cell supernatants
The amounts of PGE2 released into culture medium were routinely determined using a commercial enzyme immunoassay (Prostaglandin E2 Express ELISA kits; Cayman Chemicals, USA). For quantification of PGE2 under serum-free conditions, the cells were seeded in 24-well plates and reverse transfected with siRNAs as described above. After 24 h, the cells were transfected with plasmid DNA as described above and treated with 10 nM hGX sPLA2 and/or 10 μM AA and/or a mixture of 20 μM T863 and 20 μM PF-06424439 (DGAT inhibitors) for 48 h in complete medium. The cells were then washed and serum starved for 24 h in the appropriate culture medium containing 0.02% EFAF-BSA. In some experiments, DGAT inhibitors were included in the serum-starvation phase. For PGE2 quantification in serum-containing supernatants, MDA-MB-231 cells were seeded at 5 × 104 cells/well and A549 cells at 4 × 104 cells/well, reverse transfected as described above and treated with 10 nM hGX sPLA2 for 72 h in complete medium. Cell supernatants were collected in low-binding microcentrifuge tubes, centrifuged at 4 °C (16,000 × g, 10 min) and immediately used for analysis or stored at −80 °C for up to 7 days. For analysis, 50 μL undiluted samples were used, and standard curves prepared by diluting the PGE2 standard in culture medium.
4.14. Confocal microscopy
Cytosolic LDs were visualised using BODIPY 493/503 neutral lipid staining. Cells were reverse transfected and seeded on glass-bottomed culture plates at 6 × 104 (MDA-MB-231 cells), 3 × 104 (HeLa cells) and 5 × 104 (A549 cells) cells/well. Twenty-four hours later, the media were replaced and the cells were treated with 10 nM hGX sPLA2 and the DGAT inhibitors in complete medium for 48 h, followed by 24-h starvation in serum-free medium containing 0.02% EFAF-BSA. For confocal microscopy, the cells were washed twice with DPBS, stained with 1 μg/mL BODIPY 493/503 and 1 μg/mL Hoechst stain solution in RPMI-1640 medium for 15 min in a CO2 incubator, washed twice with DPBS, and left in fresh RPMI-1640 medium. Live imaging was carried out at the beginning of and after the 24-h serum starvation using a confocal laser scanning microscope (LSM 710; Carl Zeiss, Germany) and a stage-top microscope CO2 incubation system (Tokai Hit, Japan). Images were processed using the Zen software (Carl Zeiss, Germany).
4.15. LD counting and diameter analysis
Confocal microscopy images of BODIPY 493/503 stained LDs were used for computer image analysis with the ImageJ software (National Institutes of Health, USA) and the LD Counter Plugin (https://doi.org/10.5281/zenodo.2581434). Analysis was performed on 32-bit two-dimensional images, whereby the numbers and sizes of the LDs were determined according to the plugin instructions. LD diameters were calculated from the LD surface areas. Analyses were performed on at least 40 cells/sample. Data were loaded into the Prism 9.0.2 software (GraphPad Software, USA), with the geometric means of the LD diameters and numbers calculated per individual cell in the samples, followed by the statistical analysis.
4.16. Cell proliferation
MDA-MB-231/Luciferase-2A-RFP cells were seeded in complete medium in 96-well plates at 5 × 103 cells/well in at least triplicates. After 24 h, the cells were treated with 20 μM DGAT1 and 20 μM DGAT2 inhibitors and/or 1 μM PGE2 and/or 10 nM hGX sPLA2 as described above, and left in complete medium for 72 h. PGE2 was replenished every 24 h by direct addition to the culture medium. In transient transfection experiments, the cells were seeded in complete medium on 48-well plates at 3 × 104 (MDA-MB-231/Luciferase-2A-RFP cells) or 1.5 × 104 (A549 cells) cells/well. After 24 h, the cells were washed and transfected with 0.250 μg plasmid DNA/well using 0.5 μL P3000 reagent and 1 μL Lipofectamine 3000 in OptiMEM medium, and incubated in RPMI-1640 (MDA-MB-231) or DMEM/F12 with 2 mM l-glutamine (A549) containing 0.02% EFAF-BSA. After 6 h, the cells were treated with 10 nM recombinant hGX sPLA2 in complete medium, with 25 μM (A549) or 50 μM (MDA-MB-231) indomethacin, 25 μM nordihydroguaiaretic acid, and/or 20 μM each of the T863 and PF-06424439 DGAT1 and DGAT2 inhibitors, respectively. A549 cell proliferation was determined after 48 h using the direct cell proliferation assay kits (CyQUANT; Invitrogen, USA). MDA-MB-231/Luciferase-2A-RFP cell proliferation was determined by measuring RFP fluorescence emission (excitation, 558 nm; emission, 583 nm) on a microplate reader (Infinite M1000; Tecan, Austria). The final data were obtained after subtracting the background signal of the blank samples that contained culture medium without cells.
4.17. Clonogenic assay
MDA-MB-231 cells were seeded in 24-well plates in complete medium as described above. After 24 h, the cells were treated with 20 μM T863 and/or 20 μM PF-06424439 and left for 24 h. Cells were then washed and harvested for cell counting. 500 cells/well were seeded in 6-well plates and colony formation followed for nine days in complete medium containing 20 μM T863 and/or 20 μM PF-06424439. Medium was replaced every 48–72 h. For analysis, colonies were washed with DPBS, fixed in 100% cold methanol at 4 °C for 15 min and stained with 0.5% crystal violet in DBPS. Images were taken for colony counting with ImageJ software (National Institutes of Health, USA).
4.18. Mouse studies
All animal experiments were performed according to the directives of the EU 2010/63 and were approved by the Administration of the Republic of Slovenia for Food Safety, Veterinary and Plant Protection of the Ministry of Agriculture, Forestry and Foods, Republic of Slovenia (Permit Number U34401-9/2020/9). Laboratory animals were housed in IVC cages (Techniplast), fed standard chow (Mucedola) and tap water was provided ab libitum. Mice were maintained in 12-12 h dark–light cycle at approximately 40–60% relative humidity and an ambient temperature of 22 °C. All animals used in the study were healthy and accompanied with health certificate from the animal vendor. Health status was confirmed by FELASA recommended Mouse Vivum immunocompetent panel (QM Diagnostics). To test the anti-tumour effect of DGAT1 inhibition, female 8–10 weeks old SCID C.B-17/IcrHsd-Prkdcscid mice (Envigo, Italy) were used for xenograft cancer studies. 3 × 106 MDA-MB-231/Luciferase-2A-RFP cells were implanted into the right flank of the mouse. When a certain tumour dimension reached 5 mm, subcutaneous daily treatments were started with T863 at a dose of 9.6 mg/kg or with corresponding 0.2% DMSO vehicle. Body weight and tumour size via calliper measurements were performed every couple of days. Tumour burden was calculated using formula V=(a∗b∗c∗Π)/6. Animals were humanely euthanized either when the observed tumour dimension reached 12 mm, the tumour ulcerated or when animals lost over 20% of their own body mass. Tumour samples at the end of the experiment were collected and snap frozen in liquid nitrogen for further UPLC-MS-MS analysis.
4.19. Data and statistical analysis
Please refer to figure legends for sample size and statistical tests performed. Publicly available cancer genomics data from the Cancer Genome Atlas (TCGA) consortium, the Pan-Cancer Atlas Studies [86], were analysed using cBioPortal [87,88]. Data were plotted and statistical analyses performed using Prism 9.0.2 (GraphPad Software, USA). Unless otherwise indicated, the data are presented as means ± SEM of at least three independent experiments. Statistical significance was determined using t-tests, one-way or two-way ANOVA, followed by Bonferroni, Sidak or Tukey's multiple comparison tests. P values < 0.05 were considered as statistically significant.
Author contributions
E.J.J. performed most of the experiments, analysed the data, and prepared manuscript draft; A.P.J. and V.B. performed initial lipidomic experiments; Š.K. prepared samples for lipid mediator mass spectrometry analysis and performed LD diameter analysis; T.O.E. performed mass spectrometry analyses, designed experiments, contributed ideas and revised the paper; R.Z. provided materials, designed experiments, contributed ideas and revised the paper; G.L. contributed materials, ideas and revised the paper; P.M.J., J.G. and O.W. performed lipid mediator mass spectrometry analyses and revised the paper; D.L. and A.G.U. performed in vivo studies, and R.J. revised the paper; T.P. conceptualised the study, analysed the data, prepared the figures and illustrations, and wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Mauro Danielli, Leja Perne, Ana Temprano Lopez, Petra Hruševar, Ana Kump, Ema Guštin and Belen Vilanova Baeza for their technical help, Dr. Mojca Pavlin (University of Ljubljana, Slovenia), Dr. Brett McKinnon (Berne University Hospital, Switzerland), Dr. Merce Miranda (Joan XXIII University Hospital Tarragona, Spain) and Dr. Boris Rogelj (Jožef Stefan Institute, Slovenia) for kindly providing cell lines. We thank Dr. Chris Berrie for critical reading of the manuscript. The results shown here are in part based upon data generated by the TCGA Research Network:https://www.cancer.gov/tcga. This work was supported by the Slovenian Research Agency young researcher (1000-15-106) and postdoctoral grants (Z3-2650) to E.J.J., the P1-0207 Programme grant, J7-1818 Project grant to T.P., P4-0176 Programme grant to R.J. and J4-4563 Project grant to D.L.; by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Project-ID 239748522 - SFB 1127 ChemBioSys, and the EU COST Action CA19105 EpiLipidNet.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101791.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Lipidomics data were deposited in the MetaboLights database under study ID MTBLS7786.
References
- 1.Röhrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 2.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., DuBois R.N. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greene E.R., Huang S., Serhan C.N., Panigrahy D. Regulation of inflammation in cancer by eicosanoids. Prostag Other Lipid Mediat. 2011;96:27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Chacón G., Astudillo A.M., Balgoma D., Balboa M.A., Balsinde J. Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim Biophys Acta, Mol Cell Biol Lipids. 2009;1791:1103–1113. doi: 10.1016/j.bbalip.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Astudillo A.M., Balboa M.A., Balsinde J. Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:772–783. doi: 10.1016/j.bbalip.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Jarc E., Petan T. A twist of FATe: lipid droplets and inflammatory lipid mediators. Biochimie. 2020;169:69–87. doi: 10.1016/j.biochi.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Bonventre J. Cytosolic phospholipase A2alpha reigns supreme in arthritis and bone resorption. Trends Immunol. 2004;25:116–119. doi: 10.1016/j.it.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu Rev Pharmacol. 2009;49:123–150. doi: 10.1146/annurev.pharmtox.011008.145616. [DOI] [PubMed] [Google Scholar]
- 10.Murakami M., Taketomi Y., Miki Y., Sato H., Hirabayashi T., Yamamoto K. Recent progress in phospholipase A₂ research: from cells to animals to humans. Prog Lipid Res. 2011;50:152–192. doi: 10.1016/j.plipres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Leslie C.C. Cytosolic phospholipase A2: physiological function and role in disease. J Lipid Res. 2015;56:1386–1402. doi: 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi D., Mouchlis V.D., Dennis E.A. Omega-3 versus omega-6 fatty acid availability is controlled by hydrophobic site geometries of phospholipase A2s. J Lipid Res. 2021;62 doi: 10.1016/j.jlr.2021.100113. [paper in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saiga A., Uozumi N., Ono T., Seno K., Ishimoto Y., Arita H., et al. Group X secretory phospholipase A2 can induce arachidonic acid release and eicosanoid production without activation of cytosolic phospholipase A2 alpha. Prostag Other Lipid Mediat. 2005;75:79–89. doi: 10.1016/j.prostaglandins.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Duchez A.-C., Boudreau L.H., Naika G.S., Rousseau M., Cloutier N., Levesque T., et al. Respective contribution of cytosolic phospholipase A2α and secreted phospholipase A2 IIA to inflammation and eicosanoid production in arthritis. Prostag Other Lipid Mediat. 2019;143 doi: 10.1016/j.prostaglandins.2019.106340. [DOI] [PubMed] [Google Scholar]
- 15.Murase R., Sato H., Yamamoto K., Ushida A., Nishito Y., Ikeda K., et al. Group X secreted phospholipase A2 releases ω3 polyunsaturated fatty acids, suppresses colitis, and promotes sperm fertility. J Biol Chem. 2016;291:6895–6911. doi: 10.1074/jbc.M116.715672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato H., Taketomi Y., Miki Y., Murase R., Yamamoto K., Murakami M. Secreted phospholipase PLA2G2D contributes to metabolic health by mobilizing ω3 polyunsaturated fatty acids in WAT. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107579. [DOI] [PubMed] [Google Scholar]
- 17.Brglez V., Lambeau G., Petan T. Secreted phospholipases A2 in cancer: diverse mechanisms of action. Biochimie. 2014;107 Pt:114–123. doi: 10.1016/j.biochi.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Bezzine S., Bollinger J.G., Singer A.G., Veatch S.L., Keller S.L., Gelb M.H. On the binding preference of human groups IIA and X phospholipases A2 for membranes with anionic phospholipids. J Biol Chem. 2002;277:48523–48534. doi: 10.1074/jbc.M203137200. [DOI] [PubMed] [Google Scholar]
- 19.Mounier C.M., Ghomashchi F., Lindsay M.R., James S., Singer A.G., Parton R.G., et al. Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A2 occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A2-α. J Biol Chem. 2004;279:25024–25038. doi: 10.1074/jbc.M313019200. [DOI] [PubMed] [Google Scholar]
- 20.Jarc E., Kump A., Malavašič P., Eichmann T.O., Zimmermann R., Petan T. Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:247–265. doi: 10.1016/j.bbalip.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Kudo K., Miki Y., Carreras J., Nakayama S., Nakamoto Y., Ito M., et al. Secreted phospholipase A2 modifies extracellular vesicles and accelerates B cell lymphoma. Cell Metabol. 2022;34:615–633.e8. doi: 10.1016/j.cmet.2022.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Surrel F., Jemel I., Boilard E., Bollinger J.G., Payré C., Mounier C.M., et al. Group X phospholipase A2 stimulates the proliferation of colon cancer cells by producing various lipid mediators. Mol Pharmacol. 2009;76:778–790. doi: 10.1124/mol.108.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schewe M., Franken P.F., Sacchetti A., Schmitt M., Joosten R., Böttcher R., et al. Secreted phospholipases A2 are intestinal stem cell niche factors with distinct roles in homeostasis, inflammation, and cancer. Cell Stem Cell. 2016;19:38–51. doi: 10.1016/j.stem.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Pucer A., Brglez V., Payré C., Pungerčar J., Lambeau G., Petan T. Group X secreted phospholipase A2 induces lipid droplet formation and prolongs breast cancer cell survival. Mol Cancer. 2013;12:111. doi: 10.1186/1476-4598-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogden H.L., Lai Y., Nolin J.D., An D., Frevert C.W., Gelb M.H., et al. Secreted phospholipase A2 group X acts as an adjuvant for type 2 inflammation, leading to an allergen-specific immune response in the lung. J Immunol. 2020;204:3097–3107. doi: 10.4049/jimmunol.2000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dichlberger A., Schlager S., Maaninka K., Schneider W.J., Kovanen P.T. Adipose triglyceride lipase regulates eicosanoid production in activated human mast cells. J Lipid Res. 2014;55:2471–2478. doi: 10.1194/jlr.M048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlager S., Goeritzer M., Jandl K., Frei R., Vujic N., Kolb D., et al. Adipose triglyceride lipase acts on neutrophil lipid droplets to regulate substrate availability for lipid mediator synthesis. J Leukoc Biol. 2015;98:837–850. doi: 10.1189/jlb.3A0515-206R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlager S., Vujic N., Korbelius M., Duta-Mare M., Dorow J., Leopold C., et al. Lysosomal lipid hydrolysis provides substrates for lipid mediator synthesis in murine macrophages. Oncotarget. 2017;8:40037–40051. doi: 10.18632/oncotarget.16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gartung A., Zhao J., Chen S., Mottillo E., VanHecke G.C., Ahn Y.-H., et al. Characterization of eicosanoids produced by adipocyte lipolysis: implication of cyclooxygenase-2 in adipose inflammation. J Biol Chem. 2016;291:16001–16010. doi: 10.1074/jbc.M116.725937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welte M.A., Gould A.P. Lipid droplet functions beyond energy storage. Biochim Biophys Acta 1862. 2017:1260–1272. doi: 10.1016/j.bbalip.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiam A.R., Ikonen E. Lipid droplet nucleation. Trends Cell Biol. 2020;31:108–118. doi: 10.1016/j.tcb.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Walther T.C., Chung J., Farese R.V. Lipid droplet biogenesis. Annu Rev Cell Dev Biol. 2017;33:491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V., Ory D.S., et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty-acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell. 2017;42:9–21.e5. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henne W.M., Reese M.L., Goodman J.M. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 2018;37 doi: 10.15252/embj.201898947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petan T. Rev Physiol Biochem Pharmacol. Springer; Berlin, Heidelberg: 2020. Lipid droplets in cancer. [paper in press] [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 40.Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt A., van de Weijer T., et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweiger M., Romauch M., Schreiber R., Grabner G.F., Hütter S., Kotzbeck P., et al. Pharmacological inhibition of adipose triglyceride lipase corrects high-fat diet-induced insulin resistance and hepatosteatosis in mice. Nat Commun. 2017;8 doi: 10.1038/ncomms14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 43.Smirnova E., Goldberg E.B., Makarova K.S., Lin L., Brown W.J., Jackson C.L. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2005;7:106–113. doi: 10.1038/sj.embor.7400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dvorak A.M., Dvorak H.F., Peters S.P., Shulman E.S., MacGlashan D.W., Pyne K., et al. Lipid bodies: cytoplasmic organelles important to arachidonate metabolism in macrophages and mast cells. J Immunol. 1983;131:2965–2976. [PubMed] [Google Scholar]
- 45.Triggiani M., Oriente A., Marone G. Differential roles for triglyceride and phospholipid pools of arachidonic acid in human lung macrophages. J Immunol. 1994;152:1394–1403. [PubMed] [Google Scholar]
- 46.Bozza P.T., Bakker-Abreu I., Navarro-Xavier R.A., Bandeira-Melo C. Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot Essent Fatty Acids. 2011;85:205–213. doi: 10.1016/j.plefa.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Accioly M.T., Pacheco P., Maya-Monteiro C.M., Carrossini N., Robbs B.K., Oliveira S.S., et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin E2 synthesis in colon cancer cells. Cancer Res. 2008;68:1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 48.Pereira-Dutra F.S., Teixeira L., Costa M.F.S., Bozza P.T. Fat, fight, and beyond: the multiple roles of lipid droplets in infections and inflammation. J Leukoc Biol. 2019;106:563–580. doi: 10.1002/JLB.4MR0119-035R. [DOI] [PubMed] [Google Scholar]
- 49.Riederer M., Lechleitner M., Köfeler H., Frank S. Reduced expression of adipose triglyceride lipase decreases arachidonic acid release and prostacyclin secretion in human aortic endothelial cells. Arch Physiol Biochem. 2017;123:249–253. doi: 10.1080/13813455.2017.1309052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guijas C., Rodríguez J.P., Rubio J.M., Balboa M.A., Balsinde J. Phospholipase A2 regulation of lipid droplet formation. Biochim Biophys Acta. 2014;1841:1661–1671. doi: 10.1016/j.bbalip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Bailey A.P., Koster G., Guillermier C., Hirst E.M.A., MacRae J.I., Lechene C.P., et al. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell. 2015;163:340–353. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dierge E., Debock E., Guilbaud C., Corbet C., Mignolet E., Mignard L., et al. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metabol. 2021;33:1701–1715.e5. doi: 10.1016/j.cmet.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 53.Cheng X., Geng F., Pan M., Wu X., Zhong Y., Wang C., et al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metabol. 2020;32:229–242.e8. doi: 10.1016/j.cmet.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackerman D., Tumanov S., Qiu B., Michalopoulou E., Spata M., Azzam A., et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 2018;24:2596–2605.e5. doi: 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillaume C., Payré C., Jemel I., Jeammet L., Bezzine S., Naika G.S., et al. In vitro anti-Plasmodium falciparum properties of the full set of human secreted phospholipases A2. Infect Immun. 2015;83:2453–2465. doi: 10.1128/IAI.02474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosch M., Parton R.G., Pol A. Lipid droplets, bioenergetic fluxes, and metabolic flexibility. Semin Cell Dev Biol. 2020;108:33–46. doi: 10.1016/j.semcdb.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Wooten R.E., Willingham M.C., Daniel L.W., Leslie C.C., Rogers L.C., Sergeant S., et al. Novel translocation responses of cytosolic phospholipase A2α fluorescent proteins. Biochim Biophys Acta. 2008;1783:1544–1550. doi: 10.1016/j.bbamcr.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreira L.S., Piva B., Gentile L.B., Mesquita-Santos F.P., D'Avila H., Maya-Monteiro C.M., et al. Cytosolic phospholipase A2-driven PGE2 synthesis within unsaturated fatty acids-induced lipid bodies of epithelial cells. Biochim Biophys Acta, Mol Cell Biol Lipids. 2009;1791:156–165. doi: 10.1016/j.bbalip.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Cruz A.L.S., Barreto E. de A., Npb Fazolini, Viola J.P.B., Bozza P.T. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020;11:105. doi: 10.1038/s41419-020-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eichmann T.O., Kumari M., Haas J.T., Farese R.V., Zimmermann R., Lass A., et al. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J Biol Chem. 2012;287:41446–41457. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koundouros N., Karali E., Tripp A., Valle A., Inglese P., Perry N.J.S., et al. Metabolic fingerprinting links oncogenic PIK3CA with enhanced arachidonic-acid-derived eicosanoids. Cell. 2020;181:1596–1611.e27. doi: 10.1016/j.cell.2020.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., et al. Fat SIGNALS - lipases and lipolysis in lipid metabolism and signaling. Cell Metabol. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilcock D.J., Badrock A.P., Wong C.W., Owen R., Guerin M., Southam A.D., et al. Oxidative stress from DGAT1 oncoprotein inhibition in melanoma suppresses tumor growth when ROS defenses are also breached. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castoldi A., Monteiro L.B., Bakker N. van T., Sanin D.E., Rana N., Corrado M., et al. Triacylglycerol synthesis enhances macrophage inflammatory function. Nat Commun. 2020;11:4107. doi: 10.1038/s41467-020-17881-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giedt M.S., Thomalla J.M., Johnson M.R., Lai Z.W., Tootle T.L., Welte M.A. Adipose triglyceride lipase promotes prostaglandin-dependent actin remodeling by regulating substrate release from lipid droplets. Preprint at bioRxiv 2021. 2021. 8.2. [DOI] [PMC free article] [PubMed]
- 66.Chitraju C., Mejhert N., Haas J.T., Diaz-Ramirez L.G., Grueter C.A., Imbriglio J.E., et al. Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metabol. 2017;26:407–418.e3. doi: 10.1016/j.cmet.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lambeau G., Gelb M.H. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem. 2008;77:495–520. doi: 10.1146/annurev.biochem.76.062405.154007. [DOI] [PubMed] [Google Scholar]
- 68.Murakami M., Sato H., Taketomi Y. Updating phospholipase A2 biology. Biomolecules. 2020;10:1457. doi: 10.3390/biom10101457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanasaki K., Ono T., Saiga A., Morioka Y., Ikeda M., Kawamoto K., et al. Purified group X secretory phospholipase A2 induced prominent release of arachidonic acid from human myeloid leukemia cells. J Biol Chem. 1999;274:34203–34211. doi: 10.1074/jbc.274.48.34203. [DOI] [PubMed] [Google Scholar]
- 70.Liesenfeld D.B., Grapov D., Fahrmann J.F., Salou M., Scherer D., Toth R., et al. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare study. Am J Clin Nutrition. 2015;102:433–443. doi: 10.3945/ajcn.114.103804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brglez V., Pucer A., Pungerčar J., Lambeau G., Petan T. Secreted phospholipases A2 are differentially expressed and epigenetically silenced in human breast cancer cells. Biochem Biophys Res Commun. 2014;445:230–235. doi: 10.1016/j.bbrc.2014.01.182. [DOI] [PubMed] [Google Scholar]
- 72.Li X., Shridas P., Forrest K., Bailey W., Webb N.R. Group X secretory phospholipase A2 negatively regulates adipogenesis in murine models. Faseb J. 2010;24:4313–4324. doi: 10.1096/fj.10-154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato H., Isogai Y., Masuda S., Taketomi Y., Miki Y., Kamei D., et al. Physiological roles of group X-secreted phospholipase A2 in reproduction, gastrointestinal phospholipid digestion, and neuronal function. J Biol Chem. 2011;286:11632–11648. doi: 10.1074/jbc.M110.206755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slatter D.A., Aldrovandi M., O'Connor A., Allen S.M., Brasher C.J., Murphy R.C., et al. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metabol. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 76.Ward K.E., Sengupta R., Ropa J.P., Amiar S., Stahelin A.R.V. The cytosolic phospholipase A2α N-terminal C2 domain binds and oligomerizes on membranes with positive curvature. Biomolecules. 2020;10:647. doi: 10.3390/biom10040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward K.E., Ropa J.P., Adu-Gyamfi E., Stahelin R.V. C2 domain membrane penetration by group IVA cytosolic phospholipase A2 induces membrane curvature changes. J Lipid Res. 2012;53:2656–2666. doi: 10.1194/jlr.M030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gubern A., Casas J., Barceló-Torns M., Barneda D., Rosa X de la, Masgrau R., et al. Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J Biol Chem. 2008;283:27369–27382. doi: 10.1074/jbc.M800696200. [DOI] [PubMed] [Google Scholar]
- 79.Antonny B., Vanni S., Shindou H., Ferreira T. From zero to six double bonds: phospholipid unsaturation and organelle function. Trends Cell Biol. 2015;25:427–436. doi: 10.1016/j.tcb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 80.Cao Z., Hao Y., Fung C.W., Lee Y.Y., Wang P., Li X., et al. Dietary fatty acids promote lipid droplet diversity through seipin enrichment in an ER subdomain. Nat Commun. 2019;10:2902. doi: 10.1038/s41467-019-10835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zoni V., Khaddaj R., Campomanes P., Thiam A.R., Schneiter R., Vanni S. Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. Elife. 2021;10 doi: 10.7554/eLife.62886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schuldiner M., Bohnert M. A different kind of love - lipid droplet contact sites. Biochim Biophys Acta 1862. 2017:1188–1196. doi: 10.1016/j.bbalip.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Molenaar M.R., Jeucken A., Wassenaar T.A., van de Lest C.H.A., Brouwers J.F., Helms J.B. LION/web: a web-based ontology enrichment tool for lipidomic data analysis. GigaScience. 2019;8 doi: 10.1093/gigascience/giz061. giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Werz O., Gerstmeier J., Libreros S., Rosa XD la, Werner M., Norris P.C., et al. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun. 2018;9:59. doi: 10.1038/s41467-017-02538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Werner M., Jordan P.M., Romp E., Czapka A., Rao Z., Kretzer C., et al. Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome. Faseb J. 2019;33:6140–6153. doi: 10.1096/fj.201802509R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., et al. Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Lipidomics data were deposited in the MetaboLights database under study ID MTBLS7786.