Summary
Background
Hip minimum joint space width (mJSW) provides a proxy for cartilage thickness. This study aimed to conduct a genome-wide association study (GWAS) of mJSW to (i) identify new genetic determinants of mJSW and (ii) identify which mJSW loci convey hip osteoarthritis (HOA) risk and would therefore be of therapeutic interest.
Methods
GWAS meta-analysis of hip mJSW derived from plain X-rays and DXA was performed, stratified by sex and adjusted for age and ancestry principal components. Mendelian randomisation (MR) and cluster analyses were used to examine causal effect of mJSW on HOA.
Findings
50,745 individuals were included in the meta-analysis. 42 SNPs, which mapped to 39 loci, were identified. Mendelian randomisation (MR) revealed little evidence of a causal effect of mJSW on HOA ( 0.98 [95% CI 0.82–1.18]). However, MR-Clust analysis suggested the null MR estimates reflected the net effect of two distinct causal mechanisms cancelling each other out, one of which was protective, whereas the other increased HOA susceptibility. For the latter mechanism, all loci were positively associated with height, suggesting mechanisms leading to greater height and mJSW increase the risk of HOA in later life.
Interpretations
One group of mJSW loci reduce HOA risk via increased mJSW, suggesting possible utility as targets for chondroprotective therapies. The second group of mJSW loci increased HOA risk, despite increasing mJSW, but were also positively related to height, suggesting they contribute to HOA risk via a growth-related mechanism.
Funding
Primarily funded by the Medical Research Council and Wellcome Trust.
Keywords: Osteoarthritis, Genome-wide association study, Mendelian randomisation, Cartilage
Research in context.
Evidence before this study
Osteoarthritis is the most common cause of hip pain worldwide. One previous study found 4 genetic loci associated with cartilage thickness at the hip, some of which showed nominal associations with hip osteoarthritis suggesting that a portion of hip osteoarthritis (HOA) heritability is conveyed through cartilage thickness.
Added value of this study
This study presents a genome-wide association meta-analysis of minimum joint space width (mJSW), a proxy of cartilage thickness. It identified 39 loci that contain mJSW associated single nucleotide polymorphisms (SNPs). Interestingly, initial Mendelian randomisation (MR) results showed no causal effect of decreasing mJSW on hip osteoarthritis risk. Using MR clustering analyses, 3 groups of mJSW loci were revealed based on associations with HOA risk: Cluster one was associated with larger mJSW and lower HOA risk; Cluster two was associated with larger mJSW, increased HOA risk and increased height; Cluster three was unrelated to HOA risk. The equivalent opposing effects of Cluster one and two loci explained the initial null MR results. Subsequent, fine mapping techniques revealed the likely causal genes implicated by these genetic associations.
Implications of all the available evidence
The evidence suggests mJSW associated loci can affect HOA risk in two distinct clusters; those that decrease HOA risk with increasing mJSW and those that increase HOA risk via increasing mJSW. The first group of SNPs most likely act via cartilage mediated pathways, suggesting possible utility as targets for chondroprotective therapeutics. In contrast, the latter group of SNPs are associated with greater height and likely act through growth-related mechanisms which might have less therapeutic utility.
Introduction
Hip osteoarthritis (HOA) is the commonest cause of pain and loss of function of the hip worldwide.1 It is a disease of the whole joint with multiple biological pathways implicated in its pathogenesis, involving cartilage, bone and synovium.2 The prevalence of HOA is approximately 10% and is predicted to increase.1,3 Currently there are no known drugs to prevent disease and/or symptomatic progression, leaving total hip replacement (THR) as the treatment of choice for those with end-stage disease. As a result, HOA costs European countries over €400 billion/year in both direct and indirect healthcare costs illustrating its substantial health and economic burden.4 A better understanding of the pathogenesis of HOA may uncover new opportunities for treatment, prevention and early diagnosis.
A key component of HOA pathogenesis is the loss of cartilage, and this is often seen as a narrowing of the joint space on imaging.5 A standardised measure of joint space is minimum joint space width (mJSW), which serves as a proxy of cartilage thickness in large epidemiological studies.5,6 A well-powered systematic review found hip mJSW to have little association with hip pain which counters the idea of it being a useful predictor of disease.7 One reason for this could be the heterogeneity of pathways that affect cartilage thickness. For example, a tall individual has a higher risk of HOA8 and would be expected to have a wider joint space. Whereas other individuals might have altered cartilage metabolism and homeostasis that predisposes them to early cartilage loss, a smaller mJSW and with this an increased HOA risk.9 These opposing disease pathways are difficult to contextualise and understand using conventional epidemiological approaches.
Genome-wide association studies (GWAS) offer the opportunity to identify genes and their biological pathways that predispose an individual to disease and which might offer potential therapeutic targets.10,11 To date, 45 independent genetic loci have been associated with HOA, but the underlying genetic pathways causing disease remain largely unclear.12 A more focused GWAS of mJSW might help to identify pathways involved in cartilage metabolism that would be seen as a priority for therapeutic development.13 In addition, post-GWAS methods such as Mendelian randomisation (MR) can test if observed associations are causal, rather than being confounded, and using newer techniques cluster genetic loci into effect groups,14,15 potentially discovering previously unseen and unknown opposing genetic effects.16
An earlier GWAS found four independent loci associated with mJSW obtained from antero-posterior (AP) radiographs, many of which showed nominal associations with HOA.17 Larger sample sizes and updated genotype reference panels provide the opportunity for a more comprehensive characterization of mJSW genetic architecture. The UK Biobank study (UKB) has recently conducted over 40,000 high-resolution dual-energy X-ray absorptiometry (DXA) scans of the hip that have been automatically annotated for mJSW.3 The present study aimed to conduct a GWAS meta-analysis of hip mJSW combining X-ray and DXA cohorts to maximize the study power, and then explore the genetic architecture of mJSW and its relationship with HOA. Subsequently, we aimed to evaluate causal effects of mJSW on HOA risk using MR and cluster analyses, to allow for the possibility that distinct sets of SNPs associated with mJSW map to directionally opposite causal pathways.15
Methods
Cohort descriptions
GWAS cohorts comprised the UKB, The Rotterdam Study (RS) I&II, Osteoporotic Fractures in Men (MrOS) Study and Study of Osteoporotic Fractures (SOF). mJSW was measured automatically in UKB and manually in RS, MrOS and SOF (see Supplementary Methods).
Genome-wide association study
GWAS for mJSW were conducted separately in UKB, RS I&II, SOF and MrOS. In each cohort, mJSW was stratified by sex and adjusted for age, ancestry principal components, and in addition study site in the case of MrOS and SOF. Given potential relationships between mJSW and height, a further GWAS was performed including height adjustment for each cohort. Residuals resulting from female and male analysis were standardised to mean = 0, SD = 1, and then combined into a single outcome for GWAS. UKB used a linear mixed model for GWAS, implemented in BOLT-LMM (v2.3),18 SOF, MrOS used an OLS linear regression model implemented in PLINK19 and RS1 and RS2 used RVtests.5 RS I&II were imputed to Haplotype Reference Consortium (HRC v.1.1), UK Biobank release V3 was imputed to 3 reference panels (UK10K, 1000 Genomes and HRC) and SOF and MrOS were imputed to 1000 Genomes. All cohorts used the hg19 build.
Statistics and meta-analysis
Before meta-analysis, quality control of summary statistics was performed using EasyQC.20 Briefly, missing data, mono-allelic SNVs, implausible values (Linear regression: P > 1, infinite SE, beta >10, EAF>1.) and duplicates were removed from the data. We excluded variants with poor imputation quality (INFO <0.4) and minor allele frequency ≤0.005. Allele coding was harmonized across cohorts (A/T/C/G or I/D) and allele frequency checked against HRC imputed reference (http://www.haplotype-reference-consortium.org/) to identify possible allele coding errors. P-Z scatter plots were inspected for problems with beta estimates, standard errors and P values. Cleaned files were used to perform an inverse variance weighted fixed effects meta-analysis was performed with METAL.21 Following the meta-analysis SNPs were only considered if they were in more than one cohort and a SNP heterogeneity below a prespecified threshold (I2 ≤ 30). A separate GWAS meta-analysis was conducted excluding UKB so that mJSWDXA and mJSWX-ray could be compared. Genome-wide statistical significance threshold was set at a P-value less than 5 × 10−8 (linear regression).
Linkage disequilibrium score regression and genome-wide conditional and joint complex trait analysis (GCTA-COJO).
Linkage disequilibrium (LD) score regression (LDSC) v1.0.1 was used to estimate SNP heritability, and the genetic correlation between mJSW and several other traits, including HOA, height, and body mass index (BMI) (see Supplementary Methods).12,22 In addition, the genetic correlation between mJSWDXA and mJSWX-ray was examined. A European based LD reference panel was used, and analysis was limited to HapMap3 SNPs (therefore excluding major histocompatibility regions).22 Conditional and joint analysis (GCTA-COJO) was performed in conjunction with a UKB reference panel to identify statistically independent mJSW associated signals.23
Mendelian randomisation and MR cluster analysis
The conditionally independent mJSW lead SNPs were used as genetic instruments for MR analyses to investigate the causal effect of mJSW on HOA, using the TwoSampleMR package v0.5.6 in R.24 The HOA GWAS was a meta-analysis combining the latest genetics of osteoarthritis consortium HOA GWAS without UKB12 and an updated UKB HOA GWAS removing those individuals with mJSW measures to avoid sample overlap (see Supplementary Methods). Steiger filtering was applied to demonstrate the exposure instruments were upstream of the outcome. Inverse variance weighted (IVW) analysis was used as the primary method, with MR Egger, weighted median, simple mode and weighted mode approaches as sensitivity analyses.14 MR-Clust was applied in relation to HOA to group variants into distinct groups with similar causal estimates.15 This method, which may help to identify different causal mechanisms underlying HOA, is used when heterogeneity in causal effect estimates for a complex trait is observed, and different biological mechanisms are suspected. Two sample MR was then used to quantify cluster specific effects on HOA and height. A previous GWAS of height (GWAS ID: ukb-b-10787), available via the IEU open GWAS project,25 was used.
Gene prioritisation and downstream analyses
Initially, the independent mJSW lead SNPs were looked-up in a previous GWAS of height (GWAS ID: ukb-b-10787) and BMI (GWAS ID: ukb-b-19953) in UKB25 and HOA. SNPs were prioritised based on MR-Clust results and a look-up in previous height and HOA summary statistics. In these fine mapping analyses that used the coloc R package, we compared 100 kb regions on either side of the lead mJSW SNP in the mJSW and HOA GWAS to look for shared signals.26 Then generalised gene-set analysis of GWAS data (MAGMA v1.08)27 was implemented in Functional Mapping and annotation of GWAS (FUMA) tool.28 Briefly, SNPs were mapped to the protein coding genes using default settings (SNP-wise (mean) model for gene test) and gene-set analysis was performed using 10,894 gene sets obtained from MsigDB v5.2. In addition, the list of mapped genes was annotated for overlapping gene ontology biological processes genes using PANTHER.29 Subsequently, the expression quantitative trait loci (eQTL) database GTEx was searched for each leading SNP to identify cis-acting effects, with cultured fibroblasts considered the most relevant tissue. LocusFocus was used to conduct Bayesian colocalisation with all expressed genes over 100 kb either side of the sentinel SNP.26,30,31 To further identify which cis-genes share the same causal variants, we used colocalisation to look at eQTL data assessed on highly degraded (diseased) and less degraded (healthy) cartilage, and synovial tissue retrieved following knee and hip joint replacements.32 When referring to the posterior probability (PP) obtained from colocalisation analyses we are referring to the fourth PP indicating a shared causal signal. We considered a SNP to colocalise with an eQTL if the PP was >80%. In addition, regulatory elements of non-coding human genome were identified using RegulomeDB.33
Ethics
All participants provided informed consent for this study and ethical approval was gained from UK Biobank (application number 17295) which is overseen by the Ethics Advisory Committee and received approval from the National Information Governance Board for Health and Social Care and Northwest Multi-Centre Research Ethics Committee (11/NW/0382).
Role of funders
None of the funders had any role in study design, data collection, data analyses, interpretation, or the writing of this manuscript.
Results
Genome wide association analysis
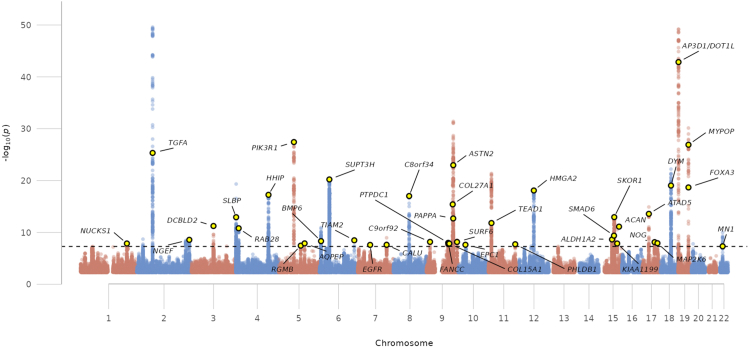
We conducted a GWAS meta-analysis of hip mJSW in 50,745 participants from 5 cohorts, of whom 24,429 were males and 26,316 females with a mean age of 65.1 years (range 45–97 years), height of 169.7 cm (135–204 cm), weight of 75.5 kg (34–171 kg) and mJSW of 3.05 mm (0.0–7.4 mm) (Supplementary Table S1). Following conditional analyses, 42 independent SNPs were identified at genome-wide significance (Linear regression: P ≤ 5·0 × 10−8) (Supplementary Fig. S1.1–S1.42), together accounting for 4.6% of mJSW variance (Table 1, Fig. 1).17 The identified SNPs mapped to 39 loci, of which 35 had not previously been associated with mJSW (defined as >1 MB from previously reported variants17). mJSW SNP heritability (h2) was 0.20 (95% CI 0.16, 0.25), and there was moderate genomic inflation (λ = 1.11; UKB λ = 1.10, MrOS 1.02, SOF 1.00, RS1 1.01, RS2 1.00). However, the intercept from LDSC, and the ratio attenuation statistic (Intercept = 1.01 [Standard error = 0.01]/RPS = 0.28 [0.15]) suggested that most of the inflation reflected polygenicity rather than confounding due to population stratification or relatedness (Supplementary Fig. S2). Equivalent results were obtained in a further GWAS following height adjustment (Supplementary Fig. S3).
Table 1.
Conditionally independent minimum joint space width single nucleotide polymorphisms, and their associations with height, BMI, and HOA risk.
| RSID | CHR | BP | C.GENE | EA | NEA | EAF | Cluster | Cluster prob | mJSW beta | mJSW P | HOA Beta | HOA P | Height Beta | Height P | BMI Beta | BMI P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7571789 | 2 | 70,714,793 | TGFA | C | T | 0.48 | 1 | 1 | 0.09 | 2.62 × 10−50 | −0.06 | 5.32 × 10−18 | 0.00 | 0.11 | −0.01 | 3.00 × 10−03 |
| rs2236996 | 4 | 1,703,646 | SLBP | A | G | 0.48 | 1 | 0.99 | 0.05 | 1.15 × 10−13 | −0.03 | 6.74 × 10−04 | −0.01 | 6.00 × 10−07 | 0.00 | 0.11 |
| rs10948155 | 6 | 44,687,957 | SUPT3H | T | C | 0.65 | 1 | 0.99 | 0.06 | 6.07 × 10−21 | −0.05 | 9.17 × 10−11 | 0.00 | 0.78 | 0.01 | 1.70 × 10−03 |
| rs35199713 | 6 | 155,415,593 | TIAM2 | G | A | 0.03 | 1 | 0.82 | 0.11 | 3.25 × 10−09 | −0.05 | 0.03 | 0.01 | 0.02 | −0.02 | 6.50 10−03 |
| rs17172430 | 7 | 55,122,650 | EGFR | A | G | 0.12 | 1 | 0.56 | 0.05 | 2.51 × 10−08 | −0.02 | 0.1 | 0.00 | 0.93 | 0.00 | 0.67 |
| rs7846438 | 8 | 69,578,824 | C8orf34 | A | G | 0.77 | 1 | 1 | 0.06 | 9.65 × 10−18 | −0.04 | 1.48 × 10−06 | 0.00 | 0.82 | 0.01 | 0.006 |
| rs4979342 | 9 | 116,905,618 | COL27A1 | C | T | 0.27 | 1 | 1 | 0.06 | 3.88 × 10−16 | −0.03 | 9.40 × 10−05 | 0.00 | 0.24 | 0.00 | 0.52 |
| rs76164690 | 10 | 32,590,362 | EPC1 | T | G | 0.86 | 1 | 0.93 | 0.05 | 2.37 × 10−08 | −0.06 | 1.59 × 10−07 | −0.01 | 5.90 × 10−06 | 0.01 | 9.10 × 10−03 |
| rs11857461 | 15 | 58,319,690 | ALDH1A2 | C | T | 0.49 | 1 | 0.92 | 0.04 | 2.26 × 10−09 | −0.02 | 0.01 | 0.00 | 0.47 | 0.01 | 9.70 × 10−03 |
| rs34656141 | 19 | 2,158,228 | AP3D1 | T | C | 0.4 | 1 | 0.96 | 0.09 | 1.42 × 10−43 | −0.03 | 2.34 × 10−05 | 0.02 | 2.00 × 10−76 | 0.00 | 0.11 |
| rs2106973 | 22 | 28,055,460 | MN1 | G | A | 0.48 | 1 | 0.95 | 0.03 | 4.63 × 10−08 | −0.02 | 6.95 × 10−03 | 0.01 | 1.70 × 10−13 | 0.00 | 0.81 |
| rs981269 | 4 | 12,897,698 | RAB28 | T | C | 0.77 | 2 | 1 | 0.05 | 1.55 × 10−11 | 0.05 | 2.71 × 10−07 | 0.02 | 5.10 × 10−30 | 0.00 | 0.46 |
| rs7711053 | 5 | 67,822,620 | PIK3R1 | G | A | 0.38 | 2 | 1 | 0.07 | 3.74 × 10−28 | 0.05 | 1.57 × 10−12 | 0.01 | 1.90 × 10−04 | −0.01 | 4.70 × 10−03 |
| rs270417 | 6 | 7,729,614 | BMP6 | T | C | 0.72 | 2 | 0.7 | 0.04 | 4.69 × 10−09 | 0.02 | 0.01 | 0.03 | 7.10 × 10−87 | −0.01 | 4.40 × 10−04 |
| rs7869550 | 9 | 119,134,796 | PAPPAd | A | G | 0.8 | 2 | 1 | 0.06 | 2.01 × 10−13 | 0.06 | 2.15 × 10−09 | 0.03 | 1.40 × 10−74 | 0.00 | 0.27 |
| rs76248879 | 9 | 119,325,659 | ASTN2d | A | T | 0.87 | 2 | 1 | 0.1 | 1.07 × 10−23 | 0.10 | 2.18 × 10−11 | Proxy NA | |||
| rs597974a | 9 | 136,144,297 | SURF6 | A | G | 0.68 | 2 | 0.92 | 0.04 | 6.82 × 10−09 | 0.03 | 5.44 × 10−03 | 0.00 | 4.1 × 10−03 | 0.00 | 0.15 |
| rs34651525 | 11 | 12,846,729 | TEAD1 | T | A | 0.69 | 2 | 1 | 0.05 | 1.53 × 10−12 | 0.04 | 1.28 × 10−07 | 0.01 | 5.40 × 10−14 | 0.00 | 0.73 |
| rs34949187 | 15 | 89,386,652 | ACAN | G | A | 0.18 | 2 | 0.87 | 0.06 | 7.65 × 10−12 | 0.03 | 2.69 × 10−03 | 0.02 | 1.40 × 10−37 | 0.00 | 0.42 |
| rs2716212 | 17 | 67,503,653 | MAP2K6 | G | A | 0.61 | 2 | 0.67 | 0.04 | 1.18 × 10−08 | 0.06 | 2.89 × 10−13 | 0.01 | 3.40 × 10−14 | 0.00 | 0.27 |
| rs227734 | 17 | 54,767,470 | NOG | T | C | 0.3 | 2 | 0.98 | 0.04 | 7.44 × 10−09 | 0.05 | 2.66 × 10−09 | 0.02 | 2.80 × 10−37 | 0.00 | 0.09 |
| rs823097 | 1 | 205,681,370 | NUCKS1 | G | A | 0.43 | 3 | 0.99 | 0.04 | 1.35 × 10−08 | 0.01 | 0.34 | 0.01 | 8.20 × 10−23 | 0.01 | 5.2 × 10−03 |
| rs10933424 | 2 | 233,872,408 | NGEF | T | C | 0.89 | 3 | 0.91 | 0.06 | 2.65 × 10−09 | −0.01 | 0.36 | 0.02 | 1.20 × 10−14 | 0.00 | 0.77 |
| rs7633464 | 3 | 98,715,823 | DCBLD2 | A | G | 0.48 | 3 | 1 | 0.04 | 5.71 × 10−12 | 0.00 | 0.85 | 0.01 | 4.90 × 10−10 | 0.00 | 0.29 |
| rs12511230 | 4 | 145,471,245 | HHIP | A | T | 0.6 | 3 | 1 | 0.05 | 5.66 × 10−18 | 0.00 | 0.96 | −0.01 | 2.70 × 10−18 | 0.00 | 0.14 |
| rs2545730 | 5 | 98,109,985 | RGMB | G | A | 0.52 | 3 | 0.98 | 0.03 | 3.52 × 10−08 | 0.01 | 0.26 | 0.00 | 0.11 | 0.00 | 0.34 |
| rs17138646 | 5 | 115,346,245 | AQPEP | T | G | 0.88 | 3 | 0.84 | 0.05 | 1.28 × 10−08 | −0.01 | 0.32 | 0.00 | 0.097 | 0.00 | 0.99 |
| rs62479589b | 7 | 128,406,506 | CALU | G | A | 0.38 | 3 | 0.95 | 0.04 | 2.42 × 10−08 | 0.01 | 0.17 | 0.00 | 7.4 × 10−04 | 0.00 | 0.69 |
| rs4744313 | 9 | 96,846,061 | PTPDC1 | T | C | 0.63 | 3 | 0.79 | 0.04 | 1.01 × 10−08 | −0.01 | 0.25 | 0.00 | 0.22 | 0.01 | 0.01 |
| rs10962293 | 9 | 16,136,648 | C9orf92 | C | T | 0.29 | 3 | 0.99 | 0.04 | 6.58 × 10−09 | 0.00 | 0.95 | 0.00 | 0.82 | 0.00 | 0.93 |
| rs1413299 | 9 | 101,761,241 | COL15A1 | T | G | 0.37 | 3 | 0.56 | 0.04 | 1.39 × 10−08 | −0.01 | 0.13 | −0.01 | 5.30 × 10−17 | 0.01 | 0.004 |
| rs10739993c | 9 | 97,982,669 | FANCC | C | T | 0.59 | 3 | 0.98 | 0.04 | 1.79 × 10−08 | 0.00 | 0.94 | 0.00 | 7.4 × 10−04 | 0.00 | 0.15 |
| rs45540840 | 11 | 118,486,110 | PHLDB1 | G | A | 0.22 | 3 | 0.99 | 0.04 | 1.87 × 10−08 | 0.00 | 0.96 | 0.01 | 3.60 × 10−04 | 0.00 | 0.07 |
| rs2260671 | 12 | 66,174,909 | HMGA2 | A | G | 0.08 | 3 | 1 | 0.1 | 8.19 × 10−19 | 0.00 | 0.98 | 0.01 | 3.30 × 10−04 | 0.00 | 0.58 |
| rs1809360 | 15 | 68,189,737 | SKOR1 | C | T | 0.57 | 3 | 1 | 0.05 | 1.12 × 10−13 | 0.00 | 0.65 | 0.00 | 0.84 | −0.02 | 2.90 × 10−14 |
| rs117564279 | 15 | 81,224,038 | CEMIP | A | G | 0.02 | 3 | 0.85 | 0.15 | 1.35 × 10−08 | 0.06 | 0.07 | 0.00 | 0.67 | −0.01 | 0.14 |
| rs7179372 | 15 | 67,036,441 | SMAD6 | G | A | 0.2 | 3 | 1 | 0.05 | 4.12 × 10−10 | 0.01 | 0.44 | 0.01 | 2.70 × 10−11 | −0.01 | 0.02 |
| rs62070652 | 17 | 29,221,277 | ATAD5 | C | T | 0.27 | 3 | 0.98 | 0.05 | 2.62 × 10−14 | 0.02 | 0.05 | 0.04 | 1.60 × 10−166 | 0.00 | 0.4 |
| rs8097746 | 18 | 46,640,782 | DYM | T | C | 0.59 | 3 | 1 | 0.06 | 9.12 × 10−20 | 0.01 | 0.14 | 0.02 | 4.80 × 10−51 | 0.00 | 0.02 |
| rs34717890 | 19 | 46,400,443 | MYPOPd | T | C | 0.12 | 3 | 1 | 0.1 | 1.24 × 10−27 | −0.01 | 0.25 | 0.00 | 0.18 | 0.00 | 0.39 |
| rs61648765 | 19 | 46,381,864 | FOXA3d | C | G | 0.78 | 3 | 1 | 0.07 | 2.11 × 10−19 | 0.00 | 0.9 | 0.01 | 4.40 × 10−04 | 0.00 | 0.17 |
| rs34687269 | 9 | 119,484,132 | ASTN2d | A | T | 0.52 | Pal | Pal | 0.07 | 3.96 × 10−32 | 0.07 | 2.32 × 10−19 | 0.01 | 6.20 × 10−23 | 0.00 | 0.71 |
Each conditionally independent mJSW SNP is assigned to a cluster according to HOA effect by MR-Clust. The probability for it being a member of that cluster is given. Each SNP effect and P-value is given for a GWAS of mJSW, HOA, standing height and BMI.
C.Gene – closest gene, mJSW – minimum joint space width, Pal – Palindromic SNP, SNP – single nucleotide polymorphism, HOA – hip osteoarthritis, P – P-value.
Proxy SNP rs687621 (r2 = 0.98).
rs6954748 (r2 = 0.97).
rs7854570 (r2 = 0.99).
rs7869550, rs76248879, rs34687269 mapped to PAPPA-ASTN2 locus, and rs61648765 and rs34717890 mapped to FOXA3-MYPOP locus based on a 1 mb sliding window approach.
Fig. 1.
Manhattan plot showing mJSW genome-wide meta-analysis results. The dashed black line denotes the threshold for declaring genome-wide significance (Linear regression: P ≤ 5.0 × 10−8). Yellow circles represent not previously reported mJSW loci (defined as > 1 MB from previously known genome-wide significant mJSW variants).
Genetic correlation
LDSC provided estimates of genetic correlation. A strong genetic correlation was seen between mJSWDXA (N = 38,175) versus mJSWX-ray (N = 12,570) (rg 0.87 [95% CI 0.59, 1.14]). While the SNP heritability z-score for mJSWDXA was 10.8, the SNP heritability z-score for mJSWX-ray was 3.3, which is below the threshold of 4 that is suggested for reliable LDSC estimates.22 There was a moderate correlation between mJSWcombined (mJSWDXA and mJSWX-ray combined) versus height (rg 0.28 [0.22, 0.33]) and between mJSWDXA versus height (rg 0.34 [0.28, 0.39]). There was weak genetic correlation between mJSW and BMI and HOA, confidence intervals excluding zero in the case of mJSWDXA and BMI (rg 0.08 [0.03, 0.14]) and HOA (rg 0.14 [0.04, 0.25]), but including zero for mJSWcombined versus BMI (rg 0.06 [0.01, 0.12]) and versus HOA (rg 0.10 [−0.01, 0.21]) (Supplementary Table S3).
Mendelian randomisation and MR-cluster
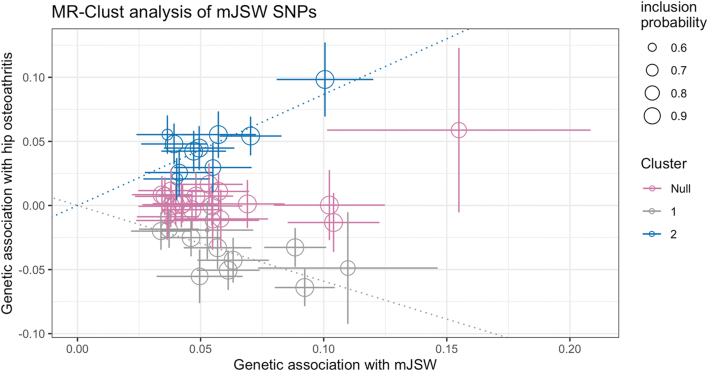
To examine the causal relationship between mJSW and osteoarthritis, we performed a two sample MR. 41 of the 42 independent mJSW lead SNPs were used as genetic instruments (mean F-statistic = 59, range 30–222, Supplementary Table S4). Rs34687269 was not included in the MR analyses because its alleles are palindromic. Despite good instrument strength, two sample MR showed no causal effect of mJSW on HOA (IVW: Odds Ratio (OR) 0.98 [95% CI 0.82–1.18], MR Egger: OR 0.69 [0.40–1.18] and Weighted Median: OR 0.98 [0.88–1.09]) (Supplementary Fig. S4). Subsequent cluster analysis of the mJSW genetic instruments displayed three distinct clusters, with two sample MR used to quantify each cluster's effects: (i) Cluster one SNPs (n = 11) were associated with a higher mJSW and a decreased risk of HOA (IVW: OR 0.55 [95% CI 0.49–0.62]); (ii) Cluster two SNPs (n = 10) were associated with both greater mJSW and an increased risk of HOA (IVW: OR 2.40 [2.04–2.82]); and (iii) Cluster three SNPs (n = 20) had no clear association with HOA (IVW: OR 1.03 [0.95–1.11]) (Table 2, Fig. 2, and Supplementary Figs. S5–S7). Heterogeneity of SNP effects between mJSW and HOA identified by cluster analysis illustrated why no net causal effect between these traits was detected. To further understand these SNP clusters, SNP associations with other traits were investigated.
Table 2.
Two sample Mendelian randomisation results.
| Exposure | Outcome | SNPs | IVW |
MR egger |
Weighted median |
Simple mode |
Weighted mode |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||
| mJSW | HOA | 41 | 0.98 (0.82–1.18) | 0.87 | 0.69 (0.40–1.18) | 0.18 | 0.98 (0.88–1.09) | 0.72 | 0.97 (0.79–1.20) | 0.80 | 0.93 (0.76–1.13) | 0.49 |
| Cluster 1 | HOA | 11 | 0.55 (0.49–0.62) | 2.47 × 10−25 | 0.59 (0.42–0.85) | 0.02 | 0.56 (0.49–0.64) | 1.61 × 10−16 | 0.58 (0.45–0.73) | 1.94 × 10−03 | 0.52 (0.40–0.67) | 3.64 × 10−04 |
| Cluster 2 | HOA | 10 | 2.40 (2.04–2.82) | 2.98 × 10−26 | 1.97 (1.07–3.63) | 0.06 | 2.36 (1.98–2.81) | 8.62 × 10−21 | 2.40 (1.82–3.17) | 1.99 × 10−04 | 2.33 (1.90–2.84) | 3.71 × 10−05 |
| Cluster 3 | HOA | 20 | 1.03 (0.95–1.11) | 0.47 | 1.01 (0.79–1.29) | 0.91 | 1.01 (0.91–1.12) | 0.86 | 1.00 (0.81–1.23) | 0.98 | 0.99 (0.83–1.18) | 0.90 |
| Exposure | Outcome | SNPs | Beta (95% CI) | P | Beta (95% CI) | P | Beta (95% CI) | P | Beta (95% CI) | P | Beta (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Height | 11 | 0.06 (−0.02 to 0.14) | 0.16 | 0.20 (−0.05 to 0.45) | 0.16 | 0.01 (−0.01 to 0.03) | 0.17 | 0.01 (−0.01 to 0.03) | 0.45 | 0.01 (0.00–0.03) | 0.16 |
| Cluster 2 | Height | 10 | 0.25 (0.14–0.36) | 1.36 × 10−05 | −0.07 (−0.40 to 0.26) | 0.69 | 0.10 (0.07–0.13) | 2.75 × 10−11 | 0.30 (0.11–0.50) | 0.01 | 0.11 (0.08–0.13) | 1.01 × 10−05 |
| Cluster 3 | Height | 20 | 0.09 (−0.01 to 0.19) | 0.07 | 0.09 (−0.22 to 0.40) | 0.58 | 0.02 (−0.01 to 0.04) | 0.22 | 0.02 (−0.01 to 0.06) | 0.25 | 0.01 (−0.01 to 0.03) | 0.22 |
IVW – inverse variance weighted, MR – Mendelian randomisation, OR – odds ratio, mJSW – minimum joint space width, SNP – single nucleotide polymorphism, CI – confidence interval, P – P-value, HOA – hip osteoarthritis.
Fig. 2.
MR-Clust results. Each independent minimum joint space width (mJSW) single nucleotide polymorphism (SNP) is plotted comparing their mJSW and hip osteoarthritis (HOA) effects. Three clusters are identified: Cluster one SNPs show a protective effect on HOA with increasing mJSW, Cluster two SNPs show an increasing risk of HOA with increasing mJSW and the null cluster SNPs show no effect on HOA.
Trait look-ups and SNP prioritisation
The 42 independent mJSW-associated SNPs were examined in previous GWAS of HOA, height, and BMI (Table 1). SNPs in Cluster one (n = 11), which were associated with a decreased risk of HOA with increasing mJSW, showed mixed associations with height; MR showed limited evidence of a small causal effect on height overall (IVW: β 0.06 [95% CI −0.02, 0.14]) (Table 2). SNPs in Cluster two (n = 10), which were associated with a higher HOA risk with increasing mJSW, all (except one for which a proxy SNP was not found) showed strong and consistent positive associations with height and some evidence of association with BMI, and MR showed a strong causal effect of these SNPs on height (IVW: β 0.25 [95% CI 0.14, 0.36]) (Table 2 and Supplementary Figs. S8 and S9). Colocalisation analysis revealed that two SNPs in Cluster two (near BMP6 and MAP2K6) shared common signals with height (Bayesian colocalisation: PP 99% and 97%, respectively) (Supplementary Table S5).
The palindromic SNP (rs34687269) showed strong positive associations with mJSW, HOA and height, in keeping with rs76248879, a Cluster two SNP that was also situated close to the ASTN2 locus (Table 1). There is a clear null cluster outlier (rs117564279) in Fig. 2, which shows a strong mJSW effect (Linear regression: β 0.15, P 1.35 × 10−8) and weaker HOA (Linear regression: β 0.06, P 0.07) effect. Interestingly, it has no association with height (Linear regression: β 0.002, P 0.67). Rs117564279 is a rare allele with a MAF 0.02 (Table 1).
Identification of candidate osteoarthritis pathogenesis genes
SNPs in Cluster one, which were thought to increase HOA risk through reduced mJSW and hence are potential targets for chondro-protective therapies, were assessed further. Colocalisation was used to compare GWAS signals between mJSW and HOA. Loci closest to TGFA, COL27A1, C8orf34 and SLBP showed strong evidence of a shared signal (Bayesian colocalisation: PP 100%, 100%, 99% & 97%, respectively). No other loci showed such evidence (Table 3).
Table 3.
Cluster one candidate gene identification.
| C.Gene | RSID | HOA Beta | HOA P | HOA Coloc (PP) | MAGMA–top genes (P-value) |
|---|---|---|---|---|---|
| TGFA | rs7571789 | −0.06 | 5.32 × 10−18 | 1.00 | TGFA (4.83 × 10−17) |
| SUPT3H | rs10948155 | −0.05 | 9.17 × 10−11 | 0.03 | SUPT3H (6.93 × 10−15), RUNX2 (1.55 × 10−12) |
| EPC1 | rs76164690 | −0.06 | 1.59 × 10−07 | 0.00 | EPC1 (3.41 × 10−06) |
| C8orf34 | rs7846438 | −0.04 | 1.48 × 10−06 | 0.99 | C8orf34 (3.83 × 10−09) |
| AP3D1 | rs34656141 | −0.03 | 2.34 × 10−05 | 0.40 | DOT1L (3.11 × 10−15) |
| COL27A1 | rs4979342 | −0.03 | 9.40 × 10−05 | 1.00 | COL27A1 (2.21 × 10−12) |
| SLBP | rs2236996 | −0.03 | 6.74 × 10−04 | 0.97 | TMEM129 (3.45 × 10−12), TACC3 (9.78 × 10−11), SLBP (1.05 × 10−14) |
| MN1 | rs2106973 | −0.02 | 6.95 × 10−03 | 0.16 | N/A |
| ALDH1A2 | rs11857461 | −0.02 | 0.01 | 0.25 | ALDH1A2 (8.45 × 10−09) |
| TIAM2 | rs35199713 | −0.05 | 0.03 | 0.00 | N/A |
| EGFR | rs17172430 | −0.02 | 0.10 | 0.05 | N/A |
Cluster one SNPs were labelled with the closest gene. They were looked up in a HOA GWAS and their beta and P-value is given in the columns “HOA Beta” and “HOA” P respectively. Colocalisation was used to assess whether mJSW and HOA GWAS signals share common genetic causal variant in a given region with the posterior probability (PP) reported (H4: both traits are associated and share a single causal variant). Gene set analysis (MAGMA) was used to identify further candidate genes. The P-value threshold was 2.65 × 10−06 (linear regression). eQTL signals were assessed in GTEx using LocusFocus to conduct colocalisation with posterior probabilities reported. Cultured fibroblasts or if not present the tissue with the largest evidence of expression are reported.
C.Gene – closest gene, HOA – hip osteoarthritis, PP – posterior probability, Coloc – colocalisation, GTEx – genotyping expression project, N/A – not applicable, MAGMA – generalised gene-set analysis of GWAS data.
Subsequently, attempts were made to identify the underlying causal gene responsible for the SNP association. MAGMA assigned TGFA, SUPT3H-RUNX2, C8orf34, EPC1, COL27A1, SLBP-TMEM129-TACC3, ALDH1A2 and DOT1L as candidate genes (Table 3). TGFA and SUPT3H mJSW association signals colocalised with GTEx expression in amygdala and basal ganglia respectively, but not in fibroblasts (Supplementary Table S6). The outlier SNP (rs117564279) with the largest effect size near CEMIP was also examined in GTEx and colocalised with eQTL SNPs in skeletal muscle (PP 0.98). RegulomeDB suggested the SNPs nearest to TGFA, AP3D1, EGFR and TIAM2 were non-coding regulatory regions with probability scores > 0.5 (Supplementary Table S7). Colocalisation between mJSW SNPs and human cartilage eQTL data provided no further gene-SNP evidence for our prioritised SNPs (Supplementary Table S8). However, it did show evidence of colocalisation for two null cluster SNPs; rs62479589 with OPN1SW in both highly and less degraded cartilage (Bayesian colocalisation: PP 97% & 90% respectively) and rs823097 with RAB7L1 in highly degraded cartilage (Bayesian colocalisation: PP 96%) (Supplementary Table S8).
Gene ontology biological process annotations
PANTHER and FUMA GeNE2FUNC analyses showed that three Cluster two SNPs (which mapped to ACAN, NOG, BMP6) overlapped with skeletal system morphogenesis and development (Supplementary Tables S9 and S10). However, MAGMA gene-set analysis returned no results suggesting little evidence for overlap with any of the gene sets tested.
Discussion
In the largest GWAS of hip mJSW to date, we identified 42 conditionally independent SNPs, mapping to 39 loci. Overall MR analysis revealed little evidence for a causal effect of mJSW on HOA risk. However, cluster analysis identified three groups of SNPs with distinct effects. One cluster comprised 11 SNPs which increase mJSW leading to a decrease in HOA risk. In contrast, a second cluster comprised 10 SNPs which increased mJSW but led to an increase in HOA risk. The latter set of SNPs was also related to height, a known risk factor for HOA. A null cluster comprised 20 SNPs with no association with HOA risk. Taken together, these findings suggest that SNPs associated with mJSW may exert distinct effects on HOA risk according to whether this is instrumented by SNPs which are also related to height.
Of the 11 loci in Cluster one, which were protective for HOA with increasing mJSW, TGFA, C8orf34, COL27A1 and SLBP-TMEM129-TACC3 colocalised with the same causal signal for HOA in a large-scale HOA GWAS. The present findings suggest that these previously identified loci cause HOA through reduced cartilage thickness, suggesting potential utility as therapeutic targets for chondro-protective therapy. TGFA was implicated in mJSW by a previous much smaller GWAS and is known to be involved in endochondral bone formation.17,34, 35, 36 Likewise, COL27A1 is established in cartilage regulation and formation, and mutations are associated with osteochondrodysplasias in humans such as Steel syndrome which feature early hip dislocations and OA.37,38 There is little known about C8orf34 regulation of joint tissues such as cartilage but it has been implicated in vertebral disc disease.39 In addition, MAGMA suggested TMEM129, SLBP and TACC3 might be the genes responsible for the association with mJSW at rs2236996 locus but this was not supported by eQTL findings. TMEM129 mutations can lead to facial dysmorphias such as Wolf-Hirschhorn syndrome and has been suggested to be a genetic risk factor for OA through disrupted protein degradation in the endoplasmic reticulum.40, 41, 42
The other loci identified in Cluster one, SUPT3H-RUNX2, AP3D1, EPC1, MN1, ALDH1A2, TIAM2 and EGFR did not colocalise with corresponding HOA GWAS signals but nonetheless showed at least a nominal HOA association. The SUPT3H-RUNX2 locus was identified in the previous mJSW GWAS and has been implicated in chondrocyte and osteoblast differentiation respectively.17,36 MAGMA suggested EPC1 as a candidate for rs76164690, however this signal did not colocalise with eQTL expression in fibroblasts. Pigment epithelium derived factor (PEDF) is the product of the EPC1 gene and is known to be anti-angiogenic. Previously PEDF has been shown to be preferentially expressed in OA cartilage contributing to OA pathogenesis by upregulating matrix degrading factors.43,44 Whilst there was evidence of an association between rs34656141 with eQTL expression for AP3D1, DOT1L and AMH in fibroblasts these signals did not colocalise. DOT1L was previously implicated in mJSW in a smaller GWAS and is known to regulate cartilage homeostasis and protect against OA.17,45 Anti-Mullerian Hormone, the product of AMH, is associated with knee OA in women.46 ALDH1A2, TIAM2 and EGFR showed less evidence of an association with HOA, that said, ALDH1A2 and EGFR have previously been identified as potential treatment targets for OA.47,48 Less is known about AP3D1, TIAM2 and MN1 in the context of cartilage and HOA.
One locus showed a SNP effect of increased mJSW and HOA risk that was not associated with height; rs117564279 (CEMIP) is a rare variant with a MAF 0.02 and large effect size for both mJSW and HOA (β 0.15 & β 0.06 respectively). CEMIP is the closest gene and showed colocalisation between eQTL expression (skeletal muscle) and the mJSW GWAS signal. CEMIP has recently been shown to be expressed in cartilage from osteoarthritic joints, and to induce a fibrosis type response within chondrocytes.49 Therefore, CEMIP warrants further investigation to understand if altered expression leads to thicker more fibrous cartilage which in turn could lead to a wider joint space and a higher risk of HOA.
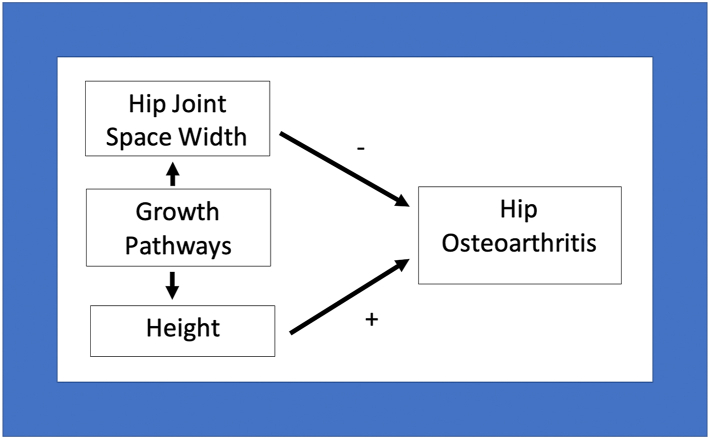
The opposing effects of SNPs in clusters one and two, as shown by the MR analyses of each cluster, presumably lead to a net null effect of mJSW on HOA. This may help to explain why mJSW, when examined observationally, displays little or no associations with HOA and symptoms, yet a decreased mJSW is often seen clinically in severely symptomatic individuals.7 Our observation that Cluster two SNPs are related to both height and HOA is consistent with previous findings that height GWAS signals overlap with OA.50,51 This also corresponds with findings from observational studies that taller individuals are at an increased risk of HOA.8,52 Whereas Cluster two SNPs are related to height, mJSW GWAS results showed little attenuation following height adjustment. Therefore, Cluster two SNPs appear to increase HOA risk through co-association with greater height, although height itself does not appear to be on the causal pathway for mJSW, suggesting the role of an intermediary growth-related mechanism (Fig. 3). Consistent with this suggestion, gene ontology annotation suggested that three Cluster two SNPs (ACAN, NOG, BMP6) have a role in skeletal development. Extra-skeletal endocrine actions that influence growth might also play a role, given two loci, PAPPA and PIK3R1, are involved in the action of IGF-1 and insulin.53,54
Fig. 3.
A directed acyclic graph to represent the proposed relationships between hip minimum joint space width, height and hip osteoarthritis.
The strengths of this study include its large sample size which has afforded the power to identify 35 loci not previously known to be associated with mJSW. In addition, by combining a GWAS meta-analysis with other genetic analyses such as LDSC, MR and MR-Clust we have been able to tease out different causal pathways related to mJSW. Arguably, the main limitation was our combination of DXA and X-ray based measures for deriving mJSW. Though DXA-derived mJSW represents a different method, the finding of an inverse relationship with rHOA (see Supplementary Methods) provides face validity. However certain differences exist in mJSW measurements using these methods. For example, unlike DXA scans where only the superior joint space can be evaluated, mJSW can also be measured on X-rays at other sites. That said, X-ray based mJSW measurements were based solely on the superior joint space in SOF and MrOS. In contrast, in RS, mJSW measurements were also obtained laterally, axially and medially, with the smallest value used. Despite these differences, genetic correlation between mJSW obtained using these two methods was relatively high, albeit the mJSWX-ray GWAS was underpowered for LDSC analysis.
In terms of other limitations, as this is a GWAS of individuals with European ancestry this limits generalisability to other ancestries. In addition, there were some methodological differences in how GWAS was performed in the different cohorts, reflecting the fact that these had been initially undertaken as part of separate studies. Finally, there was limited evidence of colocalisation between GWAS and eQTL data which hinders the identification of effector genes. However, it is increasingly recognised that many true GWAS signals fail to colocalise with eQTL signals.55,56
In conclusion, we present findings from a GWAS meta-analysis of hip mJSW which identified 39 loci. Subsequently, we showed that mJSW SNPs act on HOA in two distinct clusters; those that decrease HOA risk with increasing mJSW and those that increase HOA risk via increasing mJSW. We postulate the first group of SNPs may act via cartilage mediated pathways, suggesting possible utility as targets for chondroprotective therapies. In contrast, the latter group of SNPs are associated with greater height and likely act through growth-related mechanisms which require further clarification.
Contributors
BGF and MF have verified the underlying data and take responsibility for the findings in the manuscript.
Study design: MF, BGF; Data Analysis: MF, BGF, RE, KAF, ML, AH, FRS.; Interpretation of results: all authors; Replication data: CGB, DSE.; Manuscript drafting: MF, BGF, JPK, JT.; Manuscript reviewing and editing: all authors. In addition, all authors have read and approved the final version of the manuscript.
Data sharing statement
The summary minimum joint space width summary statistics have been uploaded to the GWAS Catalog (https://www.ebi.ac.uk/gwas/). The UK Biobank mJSW data from this study will be available in a forthcoming data release. Users must be registered with UK Biobank to access their resources (https://bbams.ndph.ox.ac.uk/ams/).
Declaration of interests
BGF is a member of the Versus Arthritis Research Advisory Group. DE reports grants income from NIH/NIA U24AG051129. FRS reports Wellcome Trust payments to fund meetings and travel. ML reports a CSIRO PhD Top Up scholarship. CL have a patent Image processing apparatus and method for fitting a deformable shape model to an image using random forest regression voting. This is licensed with royalties to Optasia Medical. NH reports consultancy fees and honoraria from UCB, Amgen, Kyowa Kirin, Thornton Ross, Consilient. AEH started working for Novo Nordisk A/S after drafting this manuscript. GDS reports funding from the MRC for the MRC Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/1). JHT reports funding from the MRC, a payment from Liverpool Medical Institution and he is a member of the Royal Osteoporosis Society grants panel.
Acknowledgements
This work has been conducted using the UK Biobank resource (application number 17295). The authors would like to thank the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. The generation and management of GWAS genotype data for the Rotterdam Study (RSI & RSII) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The authors would like to thank Pascal Arp, Mila Jhamai, Marijn Verkerk, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Linda Broer PhD, for the creation of the imputed data.
No funders had any role in study design or manuscript writing. BGF is a National Institute of Health and Care Research Academic Clinical Lecturer and was previously supported by a Medical Research Council (MRC) clinical research training fellowship (MR/S021280/1). MF, RE, FS are supported, and this work is funded by a Wellcome Trust collaborative award (209233/Z/17/Z). ML is supported by a University of Queensland Research Training Scholarship from The University of Queensland (UQ). ML thanks the Commonwealth Scientific and Industrial Research Organisation for the support through a Postgraduate Top-Up Scholarship. CL was funded by the MRC (MR/S00405X/1) as well as a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (223267/Z/21/Z). This research was funded in whole, or in part, by the Wellcome Trust [Grant numbers 080280/Z/06/Z, 20378/Z/16/Z, 223267/Z/21/Z]. For the purpose of open access, the authors have applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. NCH acknowledges support from the MRC (MC_PC_21003; MC_PC_21001) and NIHR Southampton Biomedical Research Centre, University of Southampton and University Hospital Southampton. BGF, MF, AEH, GDS, JHT work in the MRC Integrative Epidemiology Unit at the University of Bristol, which is supported by the MRC (MC_UU_00011/1). JPK is funded by a National Health and Medical Research Council (Australia) Investigator grant (GNT1177938), and by the Lions Medical Research Foundation (2020 Lions Dunning-Orlich Investigator Award). DSE acknowledges funding from NIH/NIA U24AG051129. The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institute of Health funding from the following institutes: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: R01 AR052000, K24 AR048841, U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. The Study of Osteoporotic Fractures (SOF) is supported by National Institutes of Health funding. The National Institute on Aging (NIA) provides support under the following grant numbers: R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, and R01 AG027576.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104759.
Appendix A. Supplementary data
References
- 1.Aresti N., Kassam J., Nicholas N., Achan P. Hip osteoarthritis. BMJ. 2016;354:i3405. doi: 10.1136/bmj.i3405. [DOI] [PubMed] [Google Scholar]
- 2.Hunter D.J., Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 3.Faber B.G., Ebsim R., Saunders F.R., et al. A novel semi-automated classifier of hip osteoarthritis on DXA images shows expected relationships with clinical outcomes in UK Biobank. Rheumatology (Oxford) 2021;61(9):3586–3595. doi: 10.1093/rheumatology/keab927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmon J.H., Rat A.C., Sellam J., et al. Economic impact of lower-limb osteoarthritis worldwide: a systematic review of cost-of-illness studies. Osteoarthritis Cartilage. 2016;24(9):1500–1508. doi: 10.1016/j.joca.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Croft P., Cooper C., Wickham C., Coggon D. Defining osteoarthritis of the hip for epidemiologic studies. Am J Epidemiol. 1990;132(3):514–522. doi: 10.1093/oxfordjournals.aje.a115687. [DOI] [PubMed] [Google Scholar]
- 6.Faber B.G., Ebsim R., Saunders F.R., et al. Osteophyte size and location on hip DXA scans are associated with hip pain: findings from a cross sectional study in UK Biobank. Bone. 2021;153 doi: 10.1016/j.bone.2021.116146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu Miow Lin D., Reichmann W.M., Gossec L., Losina E., Conaghan P.G., Maillefert J.F. Validity and responsiveness of radiographic joint space width metric measurement in hip osteoarthritis: a systematic review. Osteoarthritis Cartilage. 2011;19(5):543–549. doi: 10.1016/j.joca.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croft P., Coggon D., Cruddas M., Cooper C. Osteoarthritis of the hip: an occupational disease in farmers. BMJ. 1992;304(6837):1269–1272. doi: 10.1136/bmj.304.6837.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holyoak D.T., Otero M., Armar N.S., et al. Collagen XI mutation lowers susceptibility to load-induced cartilage damage in mice. J Orthop Res. 2018;36(2):711–720. doi: 10.1002/jor.23731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachmazidou I., Hatzikotoulas K., Southam L., et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019;51(2):230–236. doi: 10.1038/s41588-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zengini E., Hatzikotoulas K., Tachmazidou I., et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet. 2018;50(4):549–558. doi: 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boer C.G., Hatzikotoulas K., Southam L., et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184(18):4784–4818 e17. doi: 10.1016/j.cell.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oo W.M., Hunter D.J. Disease modification in osteoarthritis: are we there yet? Clin Exp Rheumatol. 2019;37 Suppl 120(5):135–140. [PubMed] [Google Scholar]
- 14.Sanderson E., Glymour M.M., Holmes M.V., et al. Mendelian randomization. Nat Rev Methods Primers. 2022;2(1):6. doi: 10.1038/s43586-021-00092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foley C.N., Mason A.M., Kirk P.D.W., Burgess S. MR-Clust: clustering of genetic variants in Mendelian randomization with similar causal estimates. Bioinformatics. 2021;37(4):531–541. doi: 10.1093/bioinformatics/btaa778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S., Mason A.M., Grant A.J., et al. Using genetic association data to guide drug discovery and development: review of methods and applications. Am J Hum Genet. 2023;110(2):195–214. doi: 10.1016/j.ajhg.2022.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castano-Betancourt M.C., Evans D.S., Ramos Y.F., et al. Novel genetic variants for cartilage thickness and hip osteoarthritis. PLoS Genet. 2016;12(10) doi: 10.1371/journal.pgen.1006260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh P.R., Tucker G., Bulik-Sullivan B.K., et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangamaran V.R., Uppili B., Gopal D., Ramalingam K. EasyQC: tool with interactive user interface for efficient next-generation sequencing data quality control. J Comput Biol. 2018;25(12):1301–1311. doi: 10.1089/cmb.2017.0186. [DOI] [PubMed] [Google Scholar]
- 21.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulik-Sullivan B., Finucane H.K., Anttila V., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., Ferreira T., Morris A.P., Medland S.E., et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–375. doi: 10.1038/ng.2213. S1-S375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11) doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon M.S., Andrews S.J., Elsworth B., Gaunt T.R., Hemani G., Marcora E. The variant call format provides efficient and robust storage of GWAS summary statistics. Genome Biol. 2021;22(1):32. doi: 10.1186/s13059-020-02248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giambartolomei C., Vukcevic D., Schadt E.E., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4) doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas P.D., Ebert D., Muruganujan A., Mushayahama T., Albou L.P., Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31(1):8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panjwani N., Wang F., Mastromatteo S., et al. LocusFocus: web-based colocalization for the annotation and functional follow-up of GWAS. PLoS Comput Biol. 2020;16(10) doi: 10.1371/journal.pcbi.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberg J., Southam L., Roumeliotis T.I., et al. A molecular quantitative trait locus map for osteoarthritis. Nat Commun. 2021;12(1):1309. doi: 10.1038/s41467-021-21593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle A.P., Hong E.L., Hariharan M., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Usmani S.E., Ulici V., Pest M.A., Hill T.L., Welch I.D., Beier F. Context-specific protection of TGFalpha null mice from osteoarthritis. Sci Rep. 2016;6 doi: 10.1038/srep30434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usmani S.E., Pest M.A., Kim G., Ohora S.N., Qin L., Beier F. Transforming growth factor alpha controls the transition from hypertrophic cartilage to bone during endochondral bone growth. Bone. 2012;51(1):131–141. doi: 10.1016/j.bone.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 36.van Meurs J.B., Boer C.G., Lopez-Delgado L., Riancho J.A. Role of epigenomics in bone and cartilage disease. J Bone Miner Res. 2019;34(2):215–230. doi: 10.1002/jbmr.3662. [DOI] [PubMed] [Google Scholar]
- 37.Swingler T.E., Wheeler G., Carmont V., et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64(6):1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 38.Gonzaga-Jauregui C., Yesil G., Nistala H., et al. Functional biology of the steel syndrome founder allele and evidence for clan genomics derivation of COL27A1 pathogenic alleles worldwide. Eur J Hum Genet. 2020;28(9):1243–1264. doi: 10.1038/s41431-020-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjornsdottir G., Stefansdottir L., Thorleifsson G., et al. Rare SLC13A1 variants associate with intervertebral disc disorder highlighting role of sulfate in disc pathology. Nat Commun. 2022;13(1):634. doi: 10.1038/s41467-022-28167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice S.J., Cheung K., Reynard L.N., Loughlin J. Discovery and analysis of methylation quantitative trait loci (mQTLs) mapping to novel osteoarthritis genetic risk signals. Osteoarthritis Cartilage. 2019;27(10):1545–1556. doi: 10.1016/j.joca.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 41.Engbers H., van der Smagt J.J., van 't Slot R., Vermeesch J.R., Hochstenbach R., Poot M. Wolf-Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet. 2009;17(1):129–132. doi: 10.1038/ejhg.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brumwell A., Aubourg G., Hussain J., et al. Identification of TMEM129, encoding a ubiquitin-protein ligase, as an effector gene of osteoarthritis genetic risk. Arthritis Res Ther. 2022;24(1):189. doi: 10.1186/s13075-022-02882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfander D., Grimmer C., Aigner T., Swoboda B., Schmidt R., Cramer T. Pigment epithelium derived factor--the product of the EPC-1 gene--is expressed by articular chondrocytes and up regulated in osteoarthritis. Ann Rheum Dis. 2006;65(7):965–967. doi: 10.1136/ard.2005.047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klinger P., Lukassen S., Ferrazzi F., et al. PEDF is associated with the termination of chondrocyte phenotype and catabolism of cartilage tissue. BioMed Res Int. 2017;2017 doi: 10.1155/2017/7183516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monteagudo S., Cornelis F.M.F., Aznar-Lopez C., et al. DOT1L safeguards cartilage homeostasis and protects against osteoarthritis. Nat Commun. 2017;8 doi: 10.1038/ncomms15889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki E., Chiba D., Ota S., et al. Reduced serum levels of anti-Mullerian hormone is a putative biomarker of early knee osteoarthritis in middle-aged females at menopausal transition. Sci Rep. 2021;11(1):4931. doi: 10.1038/s41598-021-84584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shepherd C., Zhu D., Skelton A.J., et al. Functional characterization of the osteoarthritis genetic risk residing at ALDH1A2 identifies rs12915901 as a key target variant. Arthritis Rheumatol. 2018;70(10):1577–1587. doi: 10.1002/art.40545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Y., Luo L., Gui T., et al. Targeting cartilage EGFR pathway for osteoarthritis treatment. Sci Transl Med. 2021;13(576) doi: 10.1126/scitranslmed.abb3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deroyer C., Charlier E., Neuville S., et al. CEMIP (KIAA1199) induces a fibrosis-like process in osteoarthritic chondrocytes. Cell Death Dis. 2019;10(2):103. doi: 10.1038/s41419-019-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott K.S., Chapman K., Day-Williams A., et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Ann Rheum Dis. 2013;72(6):935. doi: 10.1136/annrheumdis-2012-202081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lettre G. The osteoarthritis and height GDF5 locus yields its secrets. Nat Genet. 2017;49(8):1165–1166. doi: 10.1038/ng.3924. [DOI] [PubMed] [Google Scholar]
- 52.Liu B., Balkwill A., Banks E., Cooper C., Green J., Beral V. Relationship of height, weight and body mass index to the risk of hip and knee replacements in middle-aged women. Rheumatology (Oxford) 2007;46(5):861–867. doi: 10.1093/rheumatology/kel434. [DOI] [PubMed] [Google Scholar]
- 53.Rojas-Rodriguez R., Ziegler R., DeSouza T., et al. PAPPA-mediated adipose tissue remodeling mitigates insulin resistance and protects against gestational diabetes in mice and humans. Sci Transl Med. 2020;12(571) doi: 10.1126/scitranslmed.aay4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuentes E.N., Bjornsson B.T., Valdes J.A., et al. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. Am J Physiol Regul Integr Comp Physiol. 2011;300(6):R1532–R1542. doi: 10.1152/ajpregu.00535.2010. [DOI] [PubMed] [Google Scholar]
- 55.Nieuwenhuis T.O., Yang S.Y., Verma R.X., et al. Consistent RNA sequencing contamination in GTEx and other data sets. Nat Commun. 2020;11(1):1933. doi: 10.1038/s41467-020-15821-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mostafavi H., Spence J.P., Naqvi S., Pritchard J.K. Limited overlap of eQTLs and GWAS hits due to systematic differences in discovery. bioRxiv. 2022 doi: 10.1101/2022.05.07.491045v1. 2022.05.07.491045. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.