Abstract
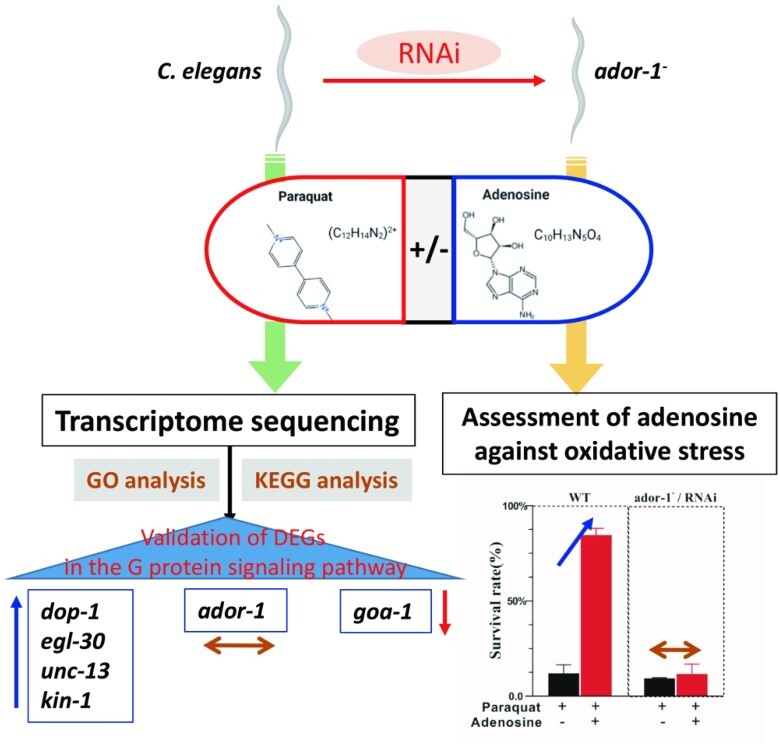
This study sought to identify the genes associated with adenosine’s protective action against paraquat (PQ)-induced oxidative stress via the adenosine receptor (ADOR-1) in Caenorhabditis elegans (C. elegans). The C. elegans was divided into 3 groups—2 groups exposed to PQ, one in presence, and one in absence of adenosine—and a control group that was not treated. Each group’s total RNA was extracted and sequenced. When the transcriptomes of these groups were analyzed, several genes were found to be differently expressed. These differentially expressed genes were significantly enriched in adenosine-response biological processes and pathways, including gene ontology terms related to neuropeptide and kyoto encyclopedia of genes and genomes pathways associated to cAMP pathway regulator activity. Quantitative reverse-transcription PCR confirmed that G-protein-coupled receptors signaling pathway involving dop-1, egl-30, unc-13, kin-1, and goa-1 genes may play crucial roles in modulating adenosine’s protective action. Interestingly, there are no significant variations in the expression of the ador-1 gene across the 3 treatments, thereby indicating that adenosine receptor exerts a consistent and stable influence on its related pathways irrespective of the presence or absence of PQ. Furthermore, the wild-type group with ador-1 gene has higher survival rate than that of the ador-1−/RNA interference group while treated with PQ in the presence of adenosine. Conclusively, our study uncovered a number of novel PQ-response genes and adenosine receptor-related genes in C. elegans, which may function as major regulators of PQ-induced oxidative stress and indicate the possible protective effects of adenosine.
Keywords: GO analysis, KEGG, GPCRs, qPCR, RNAi
Graphical Abstract
Graphical Abstract.
Introduction
Presently, oxidative stress has been linked to the emergence and progression of a variety of diseases, including cancers, Parkinson’s disease, atherosclerosis, and Alzheimer’s disease.1,2 Oxidative stress typically results from the pathological conditions of particular diseases or external causes, such as toxins.3 Paraquat (PQ; also known as 1,1′-dimethyl-4,4′-bipyridinium dichloride) undergoes an in vivo, NADPH-dependent reduction, producing a stable PQ radical that reacts with oxygen to generate a superoxide anion, a reactive oxygen species, allowing it to cause immediate damage via direct contact, ingestion, or inhalation.4,5 Therefore, PQ has been as an effective chemical for establishing a model of oxidative stress in the laboratory.
Adenosine (ADO, adenine nucleoside) plays an essential function in the physiological and pathological conditions of the body (such as oxidant injury in neural cells) through combining several types of adenosine receptors (ADORs) that are widely distributed throughout the body’s tissues.6 ADORs, which belong to the superfamily of G-protein-coupled receptors (GPCRs), have 4 distinct ADOR isoforms, including A1, A2A, A2B, and A3 in mammals.7 It is well documented that ADO functions via signaling pathways regulated by the adenosine receptor.8
Recently, Caenorhabditis elegans has been used as an alternative in the in vivo animal model for toxicological analyses of several chemicals.9,10 Recent research from our laboratory indicates that a certain dosage of adenosine has protective effects on C. elegans exposed to PQ by reducing its incidence of mortality.11 However, little is known about its protective mechanism against PQ-induced oxidative damage. Notably, ADOR-1, which is encoded by the ador-1 gene, is the only homolog of the adenosine receptor in C. elegans and is known to be an ortholog of human adenosine A2b receptor. Although ADOR-1 has not been characterized in terms of function, adenosine has been identified to have a protective effect against oxidative stress as a neurotransmitter.12 RNA interference (RNAi) is a valuable technique to determine gene function. In C. elegans, RNAi can be achieved by feeding nematodes bacteria carrying a plasmid expressing double-stranded RNA targeting a gene of interest.13 Therefore, in the present study, transcriptome and RNAi data from C. elegans groups treated with PQ in the presence or absence of adenosine were examined to identify PQ-response genes and ADOR-1-related genes for adenosine’s potential protective action.
Materials and methods
Culture and treatment of C. elegans
Wild-type (WT) Bristol (N2) C. elegans used in the present study was originally obtained from the Caenorhabditis Genetic Center (Minneapolis, United States). They were maintained at 20 °C on nematode growth medium (NGM) plates inoculated with Escherichia coli (E. coli) OP50 according to protocol.14
As previously described,15 age-synchronized populations of L4-larval nematodes were obtained at 20 °C for ~48 h. Individually, approximate 1,000 age-synchronized young adults were treated individually with PQ (150 mM) and PQ (150 mM) in the presence of adenosine (3,000 μM), which were abbreviated as PQ group and PQ_AD group. Nematodes treated with sterilized M9 buffer solution (Na2HPO4 6 g, KH2PO4 3 g, NaCl 5 g, MgSO4∙7H2O .25 g) was used as the control (CK group). PQ and adenosine (provided by Sigma-Aldrich; Merck KGaA) were, respectively, dissolved into 150 mM and 3,000 μM with 10 mL of sterilized M9 buffer according to the previous study.11
Three groups of nematodes were exposed to the respective conditions for 5 min before being washed 3 times with M9 buffer solution to eliminate the bulk of the residual bacteria. Total RNA was extracted immediately using TRIzol (Invitrogen) according to the manufacturer’s instructions.
RNA transcriptome sequencing analysis
The RNA transcriptome sequencing analysis was performed by BGI Genomics Corporation (Beijing, China) using HiSeq technology. Reference genome and gene model annotation files were downloaded from WormBase (WS269). To evaluate alterations of molecular signaling pathway after PQ treatment in the presence or absence of adenosine in C. elegans, the data were divided into 2 comparisons, containing CK versus PQ (comparison A) and PQ versus PQ_AD (comparison B). An index of the reference genome was created using Hisat2 v2..5, and paired-end clean reads were aligned with the reference genome using Hisat2 v2..5.16–18 FeatureCounts v1.5.0-p3 was used to count the read numbers mapped to each gene.19 The FPKM of each gene was then computed based on the length of the gene and the number of mapped reads. Differential expression analysis of 2 groups was performed via the R package DESeq2 (1.16.1) based on the negative binomial distribution.20
The method of Benjamini and Hochberg was utilized to modulate the obtained P-values in order to control the false discovery rate. As a criterion for significantly differential expression, a corrected value of FDR ≤ .001 and fold change ≥ 2 was chosen. The clusterProfiler R package was used to perform the gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) with an adjusted value of P < 0.05.21 Fisher’s exact test was also applied to these DEGs lists to identify gene sets that significantly overlapped with the kyoto encyclopedia of genes and genomes (KEGG) pathway gene sets.
Quantitative reverse-transcription PCR
On the basis of comparison of major differential genes between CK versus PQ and PQ versus PQ_AD in the transcriptome, quantitative reverse-transcription PCR (qRT-PCR) assay was utilized to validate the differential genes among the mRNA expression of G-protein signaling pathway genes. Primers for each genes are listed in Table 1. PrimeScript RT reagent Kit with gDNA Eraser was used to generate cDNA from total RNA (TaKaRa Bio, Japan). Comparative qRT-PCR was performed using Takara TB GREEN Premix Ex Taq II reagent (TaKaRa Bio, Japan) on Bio-Rad CFX 384 to amplify single-stranded cDNA. The PCR cycles were as follows: 95 °C for 60 s; 40 cycles of 95 °C for 5 s; 40 cycles of 60 °C for 30 s. Relative gene expression levels were calculated using the 2−ΔΔCt method.22 The ΔCt value was computed for each sample using act-1 as an endogenous control gene. The qRT-PCR gene expression was statistically compared using the Student’s t-test.
Table 1.
qRT-PCR primers for G-protein signaling pathway-related genes.
| Symbol | WormBase ID | KOG (other description)a | Primer sequence(5′-3′) | Size (bp) |
|---|---|---|---|---|
| dop-1 | WBGene00001052 | 7 transmembrane receptor, enables dopamine neurotransmitter receptor activity, coupled via Gs | F:CCTTGTTTCTTTGGCCGTGT R:GAACGGCCAGTATCCCAAGA |
91 |
| egl-30 | WBGene00001196 | G-protein subunit Galphaq/Galphay, small G-protein superfamily | F:ACTCGCATCTCGCTGACTAC R:CGCGCACGTAAAATGAGAGT |
140 |
| unc-13 | WBGene00006752 | Neurotransmitter release regulator, UNC-13 | F:TGTGACCACTTTGGAACCCC R:TAGAACTGCATCGGCTTCCG |
121 |
| ador-1 | WBGene00011878 | 7 transmembrane receptor | F:GCGCTCACGGATTTTCTAGC R:CACAAAGAGGCACCCGTAGA |
99 |
| goa-1 | WBGene00001648 | G-protein alpha subunit (small G-protein superfamily) | F:TTTTGCGAGCCATGAGCAAC R:TCTGTGTCCTCCATTCGTGC |
103 |
| kin-1 | WBGene00002189 | cAMP-dependent protein kinase types I and II, regulatory subunit | F:AAGCATAAGCAGTCGGGCAA R:GATAGCCTGGAGAATGCGCT |
114 |
| act-1 | WBGene00000063 | Actin | F:CTTCCCTCTCCACCTTCCAAC R:GCTGGTGGTGACGATGGTTT |
144 |
aGenes are categorized based on their molecular function (KOG or other description, http://www.wormbase.org).
The ador-1 RNAi and assessment of adenosine against oxidative stress
The nematodes grown on ador-1 RNAi bacteria were prepared exactly as described previously.23 Briefly, E. coli HT115 RNAi clones were grown overnight in LB with 50 μg/mL carbenicillin and 12.5 μg/mL tetracycline hydrochloride, then seeded into the NGM agar plates containing 2 mM isopropylthiogalactoside and 25 μg/mL carbenicillin. Plates were kept for 7 days before feeding and then L4-stage larvae were transferred to the NGM plates seeded with RNAi-induced bacteria. Nematodes which were feed with HT115 E. coli bacteria, transformed with the L4440 vector (empty vector or carrying specific fragments of cDNA), were divided into WT and ador-1−/RNAi type. For the oxidative stress assay, approximately, 1,080 age-synchronized young adults were transferred to 96-well plates. Three groups of nematodes (CK, PQ, and PQ_AD) were treated with M9 buffer solution, PQ (150 mM) and PQ (150 mM) in the presence of adenosine (3,000 μM), respectively. The survival rate of each cohort was recorded individually at 6-hour intervals. All experiments were repeated for 3 times.
Results
RNA sequencing
RNA-seq data quality assessment results are summarized in Supplementary Table S1. Three groups of samples were processed, and each group’s clean reads were >40 M. On average, the number of bases with a sequencing error rate of ≤0.1% (Q30) comprised >85% of the total number of bases. These results indicated that the sequences and their respective alignments were of high quality. Sequencing information was submitted to BioProject database as PRJNA809866, and the website is ‘http://www.ncbi.nlm.nih.gov/bioproject/809866’.
Differential gene expression in RNA-seq data
Using statistical inference based on DESeq, fold changes of C. elegans RNA-seq data involved in CK group, PQ group, and PQ_AD group is shown in Fig. 1. In pair-wise comparisons of the 2 groups, a total of 2,269 genes were found to be differentially expressed, including 1,878 upregulated and 391 downregulated genes in CK versus PQ group (comparison A, Fig. 1A); and 213 upregulated and 975 downregulated genes in PQ versus PQ_AD group (comparison B, Fig. 1B); 922 genes were discovered to be coregulated between CK versus PQ group and PQ versus PQ_AD group.
Fig. 1.
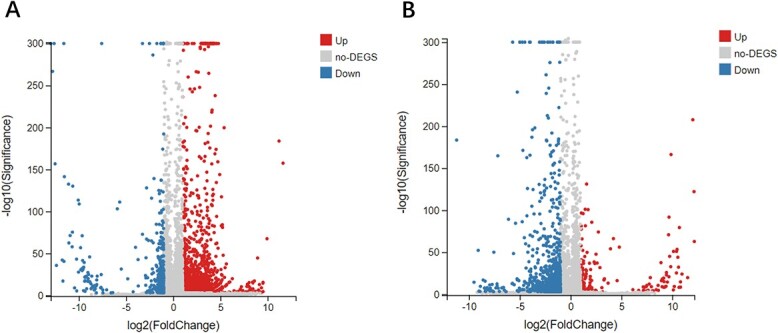
Volcano plot of fold changes and P-values of C. elegans RNA-seq data following treatment with sterile M9 buffer solution (CK group), 150 mM PQ group, and 150 mM PQ in present of 3,000 μM adenosine(PQ_AD group). A) CK versus PQ group; B) PQ versus PQ_AD group. Notes: The X-axis represents the fold of difference value after log2 transformation, and the Y-axis represents the significance value after −log10 transformation. Red color represents upregulated DEGs, blue color represents downregulated DEGs, and gray color represents non-DEGs.
GO-based enrichment analysis
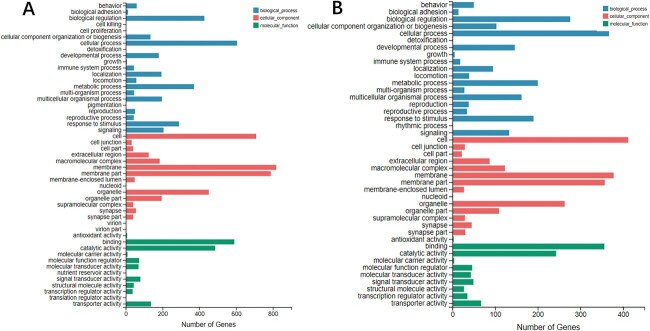
The DEGs from the RNA-seq data in the comparison A (CK vs. PQ) and comparison B (PQ vs. PQ_AD) clustered into 3 categories, including biological process, cellular component, and molecular function. There are a total of 1,566 and 830 DEGs in comparison A and comparison B, respectively (Fig. 2A and B).
Fig. 2.
GO functional annotation classification statistics of DEGs in comparison A (CK vs. PQ) and B (PQ vs. PQ_AD). Note: The X-axis represents the number of genes annotated to GO entries, and the Y-axis represents the GO functional classification.
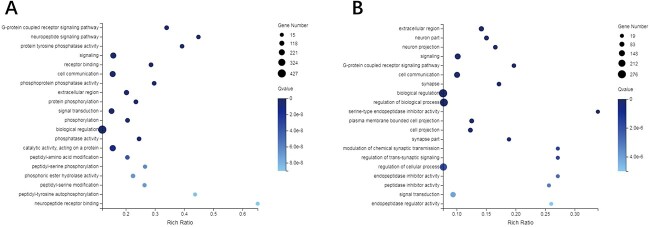
Figure 3A and 3B display the top 20 GO terms resulting from the GO enrichment of 2 comparisons. Additionally, it was discovered that the GO terms shared by both comparisons comprised five categories, namely signaling (GO:0023052), extracellular region (GO:0005576), G-protein-coupled receptor signaling pathway (GO:0007186), cell communication (GO:0007154), and cell signal transduction (GO:0007165) (Table 2).
Fig. 3.
Top 20 GO terms in GO enrichment of (A) CK versus PQ and (B) PQ versus PQ_AD.
Table 2.
GO terms coexisting in CK versus PQ versus PQ_AD.
| GO term ID | GO term | (CK vs. PQ) rich ratio | (CK vs. PQ) Q value | (PQ vs. PQ_AD) rich ratio | (PQ vs. PQ_AD) Q value |
|---|---|---|---|---|---|
| GO:0023052 | Signaling | 0.157 | 1.16E-12 | 0.102 | 1.60E-09 |
| GO:0005576 | Extracellular region | 0.202 | 3.34E-11 | 0.142 | 1.83E-13 |
| GO:0007186 | GPCR signaling pathway | 0.340 | 3.52E-19 | 0.197 | 1.80E-09 |
| GO:0007154 | Cell communication | 0.155 | 2.82E-12 | 0.101 | 1.90E-09 |
| GO:0007165 | Signal transduction | 0.151 | 1.79E-09 | 0.094 | 4.02E-06 |
According to the enrichment results, the gene classes containing >3 genes in the GO terms are considered to have a high frequency. The obtained high-frequency gene families and their functions in the 5 GO classifications are displayed in Supplementary Table S2 (all data are based on Wormbase). Additionally, it was discovered that certain gene families may be associated with the protective effect of adenosine, mainly clustering in neural-related function. These gene families include the dop genes family, neuropeptide-related genes family (such as flp, ins, and nlp), peptidase activity regulation-related genes family (such as sbt), neuropeptide receptor genes family (such as npr), and acetylcholine release-related unc genes family. Figure 4 depicts DEGs of the neural-related function in 3 groups.
Fig. 4.
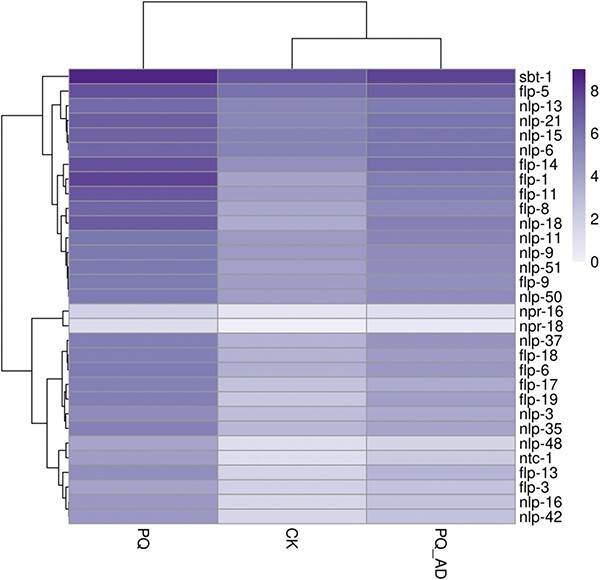
Heatmap of DEGs of neural-related function for adenosine’s protective effects.
KEGG pathway-based enrichment analysis
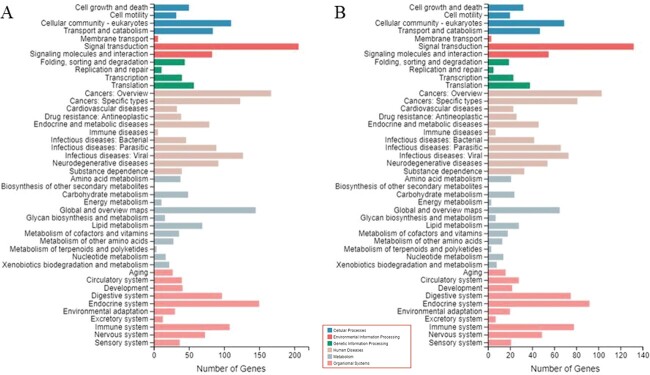
The pathway types and trend of the DEGs were similar between comparison A (CK vs. PQ) and comparison B (PQ vs. PQ_AD), but the number of genes differed (Fig. 5A and B). It appears that exposure to PQ altered the pathways of DEGs, which were partially repaired or responded with adding adenosine due to its protective effects.
Fig. 5.
KEGG pathway analysis of A) CK versus PQ and B) PQ versus PQ_AD.
It demonstrated that amyotrophic lateral sclerosis, amoebiasis, and complement and coagulation cascade pathways have the largest difference in comparison A (CK vs. PQ) (Fig. 6A). As for PQ treatment in the presence of adenosine in comparison B (PQ vs. PQ_AD), the complement and coagulation cascade pathways and amoebiasis were the most significantly altered (Fig. 6B).
Fig. 6.
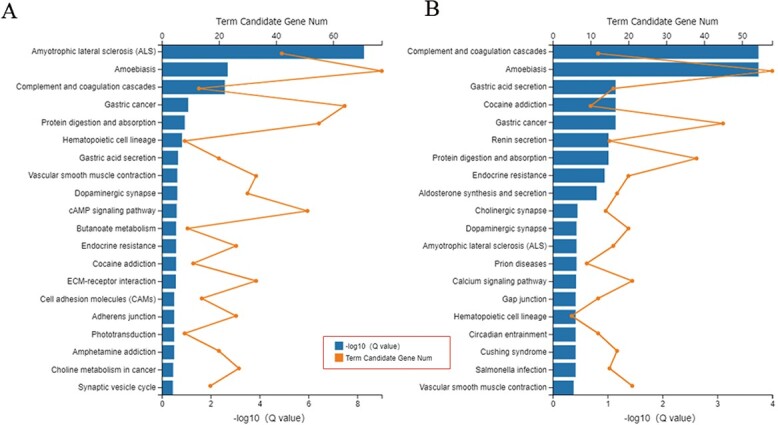
KEGG pathway enrichments of A) CK versus PQ and B) PQ versus PQ_AD.
Two pathways, namely the complement and coagulation cascade pathways and amoebiasis pathway, were identified as the most significant by pathway enrichment of common differential genes (Fig. 7). Additional investigation revealed that 11 of 12 differential complement and coagulation cascade pathways genes encoded the homologs of human tissue factor pathway inhibitor (TFPI) (Supplementary Table S3). Moreover, among the 43 differential genes involved in the amoebiasis pathway, 34 genes encode homologs of the mucin family member mucin-2 (MUC2) (Supplementary Table S3). It was postulated that these 2 components may play a crucial function in the injury and recovery of nematodes.
Fig. 7.
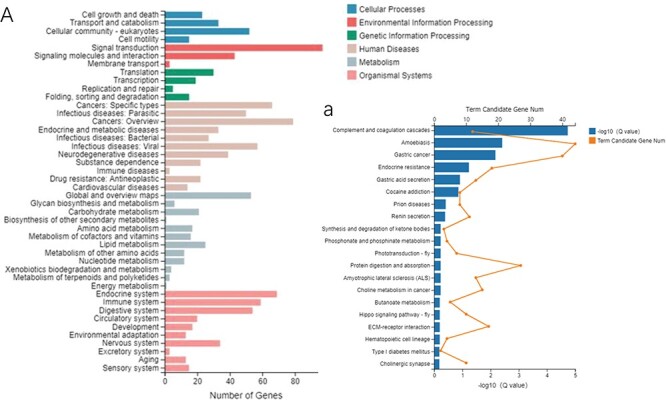
(A) KEGG pathway analysis and (a) enrichment of common DEGs.
DEGs associated with the adenosine receptor signal transduction pathway
On the basis of pathway analysis of the common DEGs between 2 comparisons, the genes clustering in the signal transduction pathway are most affected by exposure to PQ in the presence or absence of adenosine. The ador-1 is the homologous gene of the adenosine receptor in C. elegans. It is well-documented that ador-1 mainly functions via the cAMP signaling pathway. Consequently, clustering of DEGs in the cAMP pathway were calculated based on KEGG pathway analysis (Fig. 8, Supplementary Table S4). PQ groups have higher expression levels of GPCR (C24A8.6, F10D7.1, ador-1, dop-1, gcy-9, gcy-11, gcy-19, gcy-22/daf-37, dmsr-8, dop-2, dop-3, dop-5, frpr-17, npr-16, npr-18, npr-25, npr-29, and npr-32), AC (acy-2), cyclic nucleotide gated channel (CNGC) (K05F1.9, T06E4.10, T22B3.3, ZK354.7, and cec-1), calmodulin (CaM) (C50C3.5, K03A1. 4, cal-3, cal-4, cal-8, and tnc-2), PDE (ZC443.4), phosphatidylinositol phospholipase C, epsilon (PLCε) (frm-9), PP1 (C06A1.3, C24H11.2, C27B7.6, F20D6.6, F40E3.5, and F42G8.8), and MYPT1 (K02F6.3) and have lower expression levels of Gi (gpa-16), GLI3 (sup-35), ACO (acox-1.4, acox-1.5, and acox-3), and ATP (catp-2) than the CK group. In addition, PQ_AD group have lower expression levels of GPCR (C24A8.6, F10D7.1, dop-6, gcy-11, gcy-18, gcy-19, gcy-22/ C24A8.6, daf-37, dmsr-7, dop-2, dop-3, dop-5, dop-6, frpr-5, gbb-1, and npr-18), Gs (gsa-1), AC (acy-2), CNGC (T06E4.10), CaM (C50C3.5, cal-3, cal-4, and tnc-2), VDCC (unc-2), and PDE (ZC443.4), JNK (jnk-1) but have higher expression levels of Gi (gpa-16) than the PQ group.
Fig. 8.
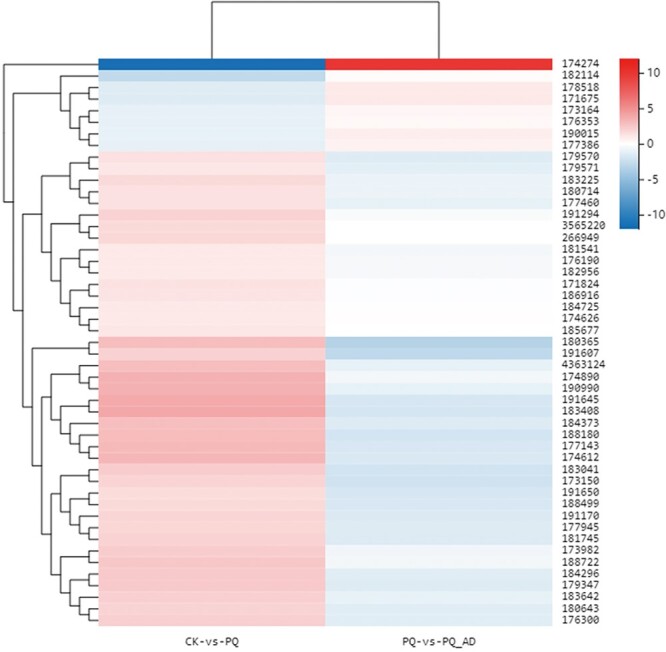
Differential expression clustering of DEGs in cAMP signal pathway between 2 comparisons (CK vs. PQ and PQ vs. PQ_AD).
Validation of DEGs in the G-protein signaling pathway
The implicated genes, which were known to play a role in C. elegans motor neuron G-protein signaling pathway, were screened out via transcriptome analysis, and these are demonstrated in Table 1. All the qPCR validation results matched the transcriptome results. In the qPCR validation results, the genes related to promote acetylcholine release pathway (such as dop-1, egl-30, and unc-13) and the gene related to promote neuropeptide release pathway (such as kin-1) exhibited a similar trend in that their expression were upregulation during PQ-treated but downregulation during PQ_AD-treated (treatments with PQ in present of adenosine) (Fig. 9). In addition, the expression of these 4 genes was significantly increased when treated with PQ compared to CK and PQ_AD (P < 0.01). Interestingly, although the expression of ador-1 gene follows the same pattern as that of the dop-1, egl-30, unc-13, and kin-1 genes, there was no significant change among the 3 treatments CK, PQ, and PQ_AD (P > 0.05). It indicated that adenosine receptor has a consistent and steady action on its respective pathways regardless of whether or not PQ is present. Furthermore, qPCR was used to access the dominance of GPCR signaling pathway for adenosine protection effect by validating the gene (goa-1) that suppressed the mentioned pathways, including acetylcholine release pathway and neuropeptide release pathway. The expression of the gene goa-1 followed the exact opposite pattern, decreasing by 65.3% after the PQ-treating and increasing to (74.5 ± 8)% following adenosine addition.
Fig. 9.
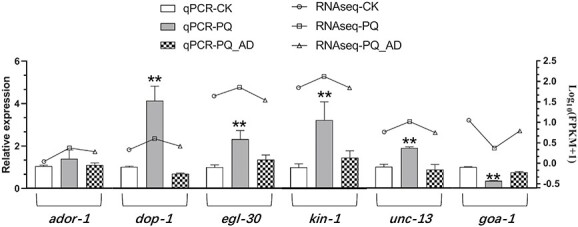
qPCR validation of G-protein signaling pathway genes in C. elegans following treatment with sterile M9 buffer solution (CK group), 150 mM PQ group, and 150 mM PQ in presence of 3,000 μM adenosine(PQ_AD group).
Assessment of adenosine against oxidative stress
The knockdown of ador-1 changed the oxidative-stress resistance of C. elegans in the presence of adenosine. The survival rates of WT and ador-1−/RNAi nematodes treated with CK, PQ, and PQ_AD are counted in Fig. 10. There was no significant difference between WT and ador-1−/RNAi while treated with M9 buffer solution and PQ. By contrast, RNAi of genes encoding ADOR-1 proteins exhibited significant negative effects on survival under oxidative stress conditions in PQ_AD treatment (P < 0.01). Comparing PQ treatment with PQ_AD treatment, there was no significant difference in the survival rate among the RNAi groups, whereas there was a significant increase among the WT groups (P < 0.01). Thus, it indicated that the presence of ador-1 gene played an important role in improving the survival rate when nematodes were treated with adenosine under oxidative stress conditions.
Fig. 10.
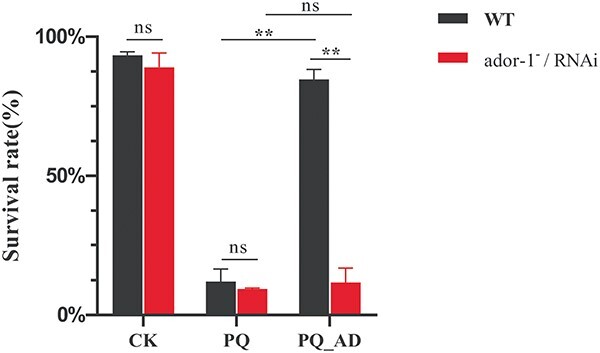
The survival rate of C. elegans, including WT and ador-1−/RNAi type. CK represents treatment with sterile M9 buffer solution, PQ represents treatment with 150 mM PQ, and PQ_AD represents treatment with 150 mM PQ in the presence of 3,000 μM adenosine.
Discussion
In the present study, we performed RNA-seq analysis of C. elegans on PQ exposed in the presence/absence of adenosine and performed the GO and KEGG pathway analyses on the DEGs in order to investigate the mechanism of adenosine’s protective effects.
In comparison A (CK vs. PQ), 82.8% of the DEGs of PQ group were upregulated relative to the CK group. By contrast, in comparison B (PQ vs. PQ_AD), 82.1% of the DEGs of PQ_AD group were downregulated relative to the PQ group. It appeared that the protective impact of adenosine on nematodes exposed to PQ has an expression mechanism that is inverse to that of PQ exposure.
Intriguingly, in the comparison of CK versus PQ and PQ versus PQ_AD, the DEGs were clustered in 3 identical GO categories: biological process, cellular component, and molecular function (Fig. 2). Considering our prior finding,11 it is possible to predict that the survival rate was improving in C. elegans exposed to PQ in the presence of adenosine due to a significant portion of the gene expression being recovered by the addition of adenosine. Surely, the extent to which genes play a critical role in this process requires more investigation.
GO enrichment analysis identified several gene families associated with the protective effects of adenosine, including abu, dop, daf, flp, nlp, npr, gcy, nhr, etc. Furthermore, the neuropeptide gene family comprised the majority. Although previous reports on neuropeptides are less about resistance to oxidative stress and are directly affecting the survival of nematodes, most of the neuropeptide genes and its signaling pathway discovered this study may play a crucial role in the survival of nematodes. As the largest neuropeptide family in C. elegans, the flp gene family expressed FMRF-like amide peptides that serve as sensory, motor, and interneurons in >50% of the nervous system.24,25 The nematode flp gene has neuropeptide receptor binding activity and functions in the neuropeptide signaling pathway to regulate sleep, movement, mating regulation, action potential regulation, oviposition regulation, etc.26 In the signal transduction system, FLP can function as a neurotransmitter, bind to the transmembrane receptor on the cell membrane, and activate the adjacent G-protein; and the activated G-protein can activate or inhibit a specific enzyme (such as cAMP, cGMP, etc.), which as a second messenger, activates protein kinase, which in turn activates phosphokinase, converts ATP into ADP, and mediates related reactions; activated G-protein have the ability to activate ion channels, such as Ca2+ channels, thereby causing changes in the membrane potential, ultimately leading to the regulation of various physiological activities.
Notably, the nlp gene family initially recorded as being resistant to oxidative stress is now predicted to have neuropeptide hormone activity in the Wormbase database. Similar to the flp gene family, nlp genes are widely expressed in neuronal and nonneuronal tissues.27 The nlp-7 has been reported to affect lifespan extension through dietary restriction.28 The expression of nlp-29, 31 can induce defense responses.29,30 The nlp-24, 25, 27, 28, 29, 30, 31, and 33 can function as antimicrobial peptides,31 and antimicrobial peptides are widely used in innate immunity,29 while nlp-27, 29, and 31 may also function as neuromodulators.27 In addition, the expression product of the npr gene family is the neuropeptide receptor family, a G-protein-coupled receptor.
KEGG pathway-based enrichment analysis of common differential genes revealed that TFPI in the complement and coagulation cascade pathway and MUC2 in the amebiasis pathway may play a significant role in the process of nematode’s injury and recovery. TFPI is an endogenous serine protease inhibitor produced and released by endothelial cells, as is well known. TFPI inhibits a variety of serine proteases, such as trypsin, α-chymotrypsin, plasmin, and cathepsin G, with quite wide inhibitory effects.32 TFPI inhibits the progression of atherosclerosis in humans via its effects on endothelial cell activation, vascular smooth muscle cell proliferation and migration, inflammatory cell recruitment, and extracellular matrix.33 TFPI also plays a significant role in the development and treatment of hemophilia.34 In C. elegans, the homologous gene of TFPI has not been thoroughly explored, but we hypothesize that its function is strongly related to oxidative stress in C. elegans and adenosine’s protective impact.
MUC2 is a member of the mucin family, which are high molecular weight glycoproteins produced by numerous epithelial tissues and are the major proteins in the intestinal mucus. MUC2 functions as the intestinal mucus barrier, inhibiting the entry of intestinal pathogens in mice35 and preserving cell viability to reduce oxidative stress in the intestinal epithelial cells.36
cAMP pathway analysis of comparison A (CK vs. PQ) revealed that the expression of GPCRs, including adenosine receptors and dopamine receptors, rose in PQ group, but Gi protein dropped. Together, their expression encouraged the upregulation of cAMP. As the second messenger in the G-protein-coupled signaling pathway, cAMP regulates the expression of CNGC on the cell membrane, increases the influx of Ca2+, and passes CaM forms a positive feedback regulation on AC. On the other hand, it transmits the signal to the downstream Epac (RAPGEF3, Rap guanine nucleotide exchange factor) through PLCε-DAG/IP3 pathway further regulates Ca2+.
cAMP pathway analysis of comparison B (PQ vs. PQ_AD) revealed that the expression of several of the aforementioned pathways is reversed in PQ_AD group, particularly the expression of GPCR. In the protective impact of adenosine, the expressions of Gs, Gi, AC, CNGC, CaM, and PDE were reversed, indicating that the regulation of cAMP by GPCR, CaM, and PDE represented the restoration of cell activity and the function of mitochondrial cells. The maintenance of intracellular Ca2+ homeostasis is important, which is associated with opening of the mitochondrial permeability transition pore, loss of membrane potential, and impairment of energy metabolism and cellular function.37,38
PLCε, MYPT1, ATP, and ADOR-1 were among those whose expressions did not reverse in comparison B, indicating that these genes remained to function or could not be restored from the cell damage to recovery. The regulation of PLCε on Ca2+ obviously continues in the protective effect of adenosine, whereas the continuous inhibition of myosin regulatory light chain (MLC) by MYPT1 leads to the tension of nematode endothelial cells and smooth muscle cells; MLC phosphorylation is essential for muscle contraction function, and its decrease in phosphorylation is 1 of the key reasons for the impairment of contraction function.39 This might be due to the tight state of the nematode’s body when it is exposed to PQ, and it appears that the protective action of adenosine does not properly ameliorate this condition. The decrease in Na+/K+-transporting ATPase subunit expression in PQ-treated cells may be attributable to a malfunction in ion control caused by a decrease in cell activity. In rats with cerebral ischemia, the activity of Na+/K+-transporting ATPase decreases with the increase of oxidative stress, and the ability of antioxidant protection is destroyed at the same time.40 Since powerful antioxidants can protect its activity, we hypothesized that nonantioxidant adenosine cannot directly restore its activity. The elevated expression level of ADOR-1 in the PQ group indicates its antistress action in an environment of oxidative stress. Among all GPCR genes in this pathway, only ador-1 did not undergo callback in the PQ_AD group (P = 0.264). It demonstrated that it continued to contribute to adenosine’s protective action on the nematode cells.
Meanwhile, qPCR validation yielded identical results. The C. elegans neuron GPCR signaling pathway involving dop-1, egl-30, unc-13, kin-1, and goa-1 genes may play crucial roles in modulating adenosine’s protective action (Table 1, Fig. 9). Intriguingly, there are no significant expression differences between the PQ group and PQ_AD group for ador-1 gene despite a large number of genes in the PQ group being downregulated by the addition of adenosine (Fig. 9). It suggested that the adenosine receptor homolog ADOR-1 may continue to play a consistent role in the protective action of adenosine regardless of whether or not PQ is present. Furthermore, the WT group with ador-1 gene has a higher survival rate than that of the ador-1−/RNAi group when they were treated with PQ in the presence of adenosine (Fig. 10). It further demonstrated that ador-1-relative genes were of great significance for improving the survival rate of nematodes under oxidative stress. Cytotoxicity induced by PQ has been shown to lead to an increase in mitochondrial membrane permeability, which is manifested as a decrease in mitochondrial membrane potential (ΔΨ).41 Furthermore, PQ also has a strong oxidizing ability, which can lead to a decrease in antioxidant enzymes (such as superoxide dismutase, glutathione peroxidase, and total superoxide dismutase). Therefore, the mitochondrial function and antioxidant system should be explored further in C. elegans, which were treated with PQ in the presence or absence of adenosine.
Conclusion
Our findings indicated that the adenosine’s protective action against PQ-induced oxidative stress in C. elegans is mediated via adenosine receptor (ADOR-1)-related genes. Several novel GO and KEGG pathway analyses were carried out to investigate the DEGs in 2 comparisons (CK vs. PQ and PQ vs. PQ_AD). Furthermore, GO enrichment analysis revealed several neuropeptide gene families associated with the protective effect of adenosine, such as abu, dop, daf, flp, nlp, npr, gcy, nhr, etc. cAMP pathway analysis and qPCR demonstrated that the expression of GPCRs, including adenosine receptors and dopamine receptors, may play a role in the protective effect of adenosine on nematode cells, and this needs further investigation. RNAi of ador-1 gens indicate that ADOR-1 plays an important role during adenosine against PQ-exposed condition. Overall, our study provides a new insight into the biological basis of the adenosine’s protective effect against PQ-induced oxidative stress via RNA-seq and may lead to a greater understanding of the molecular mechanism of the adenosine receptor (ADOR-1).
Supplementary Material
Acknowledgments
The study was supported by the Natural Science Foundation of China (31461143030), National Key Research and Development Plan (2018YFA0108403), and the Zhejiang Natural Science Foundation (Y18C140002, LGN18C200026, and 21SBYB08) and the National Innovation and Entrepreneurship Training Program for College Students (202210356046).
Contributor Information
Lingmei Ma, College of Life Sciences, Engineering Training Centre/College of Innovation, China Jiliang University, Hangzhou 310018, China.
Chunyan Ling, College of Life Sciences, Engineering Training Centre/College of Innovation, China Jiliang University, Hangzhou 310018, China.
Shuning Hu, College of Life Sciences, Engineering Training Centre/College of Innovation, China Jiliang University, Hangzhou 310018, China.
Sudan Ye, College of Applied Engineering, Zhejiang Institute of Economics and Trade, Hangzhou 310018, China.
Chun Chen, College of Life Sciences, Engineering Training Centre/College of Innovation, China Jiliang University, Hangzhou 310018, China.
Authors’ contributions
Lingmei Ma, Chunyan Ling, and Shuning Hu: Methodology, experiment, data analysis, writing; Sudan Ye: conceptualization, review, and editing; Chun Chen: supervision, conceptualization, review, and editing.
Conflict of interest statement: None declared.
References
- 1. Barnham K, Masters C, Bush A. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004:3:205–214. [DOI] [PubMed] [Google Scholar]
- 2. Liu J, Qu W, Kadiiska MB. Role of oxidative stress in cadmium toxicity and carcinogenesis. Toxicol Appl Pharmacol. 2009:238:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993:90:7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pronczuk GJ. Epidemiology of paraquat poisoning. In: Bismuth C, Hall AH, editors. Paraquat poisoning: mechanisms, prevention, treatment. New York: Marcel Dekker; 1995. pp. 37–51. [Google Scholar]
- 5. Bus JS, Gibson JE. Paraquat: model for oxidant-initiated toxicity. Environ Health Perspect. 1984:55:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linden J. Molecular approach to adenosine receptors: receptor-mediated mechanisms of tissue protection. Annu Rev Pharmacol Toxicol. 2001:1:775–787. [DOI] [PubMed] [Google Scholar]
- 7. Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998:50:413–492. [PubMed] [Google Scholar]
- 8. Chen JF, Eltzschig HK, Fredholm BB. Adenosine receptors as drug targets—What are the challenges? Nat Rev Drug Discov. 2013:12(4):265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salgueiro WG, Xavier MC, Duarte LF. Direct synthesis of 4-organylsulfenyl-7-chloro quinolines and their toxicological and pharmacological activities in Caenorhabditis elegans. Eur J Med Chem. 2014:75:448–459. [DOI] [PubMed] [Google Scholar]
- 10. Mariele C, Caroline S, Brucker N, Anelise B, Denise J, Daiandra F. Caenorhabditis elegans as an alternative in vivo model to determine oral uptake, nanotoxicity, and efficacy of melatonin-loaded lipid-core nanocapsules on paraquat damage. Int J Nanomedicine. 2015:10:5093–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie X, Shang L, Ye S, Chen C. The protective effect of adenosine-preconditioning on paraquat-induced damage in Caenorhabditis elegans. Dose Response. 2020:18(2):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013:13:1–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De-Souza EA, Camara H, Salgueiro WG, Moro RP, Knittel TL, Tonon G, Pinto S, Pinca APF, Antebi A, Pasquinelli AE, et al. RNA interference may result in unexpected phenotypes in Caenorhabditis elegans. Nucleic Acids Res. 2019:47(8):3957–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974:77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amrit FR, Ratnappan R, Keith SA, Ghazi A. The C. elegans lifespan assay toolkit. Methods. 2014:68:465–475. [DOI] [PubMed] [Google Scholar]
- 16. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015:12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat Protoc. 2016:11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013:30:923–930. [DOI] [PubMed] [Google Scholar]
- 20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15:550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012:16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cappelli K, Felicetti M, Capomaccio S, Spinsanti G, Silvestrelli M, Verini Supplizi A. Exercise induced stress in horses: selection of the most stable reference genes for quantitative RT-PCR normalization. BMC Mol Biol. 2008:9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shang LC. Study on the adenosine receptor ador-1 gene of Caenorhabditis elegans. PhD thesis. Hangzhou: China Jiliang University; 2022. [Google Scholar]
- 24. Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol. 2010:475(4):540. [DOI] [PubMed] [Google Scholar]
- 25. Schinkmann K, Li C. Localization of FMRF amide-like peptides in Caenorhabditis elegans. J Comp Neurol. 1992:316(2):251–260. [DOI] [PubMed] [Google Scholar]
- 26. Li C. The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology. 2005:131:109–127. [DOI] [PubMed] [Google Scholar]
- 27. Nathoo AN, Moeller RA, Westlund BA, Anne CH. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci U S A. 2001:98(24):14000–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park SK, Link CD, Johnson TE. Life-span extension by dietary restriction is mediated by NLP-7 signaling and coelomocyte endocytosis in C. elegans. FASEB J. 2010:24(2):383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008:18(7):481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muir RE, Tan MW. Virulence of Leucobacter chromiireducens subsp. solipictus to Caenorhabditis elegans: characterization of a novel host-pathogen interaction. Appl Environ Microbiol. 2008:74(13):4185–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004:5(5):488–494. [DOI] [PubMed] [Google Scholar]
- 32. Petersen LC, Bjørn SE, Olsen OH, Nordfang O, Norris F, Norris K. Inhibitory properties of separate recombinant Kunitz-type-protease-inhibitor domains from tissue-factor-pathway inhibitor. Eur J Biochem. 1996:235(1–2):310–316. [DOI] [PubMed] [Google Scholar]
- 33. Yuan HQ, Hao YM, Ren Z, Gu HF, Liu FT, Yan BJ, Qu SL, Tang ZH, Liu LS, Chen DX, et al. Tissue factor pathway inhibitor in atherosclerosis. Clin Chim Acta. 2019:491:97–102. [DOI] [PubMed] [Google Scholar]
- 34. Chowdary P. Anti-tissue factor pathway inhibitor (TFPI) therapy: a novel approach to the treatment of haemophilia. Int J Hematol. 2020:111(1):42–50. [DOI] [PubMed] [Google Scholar]
- 35. Wei X, Yang Z, Rey FE, Ridaura VK, Davidson NO, Gordon JI, Semenkovich CF. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe. 2012:11(2):140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao ZG, Qu W, Wang K, Chen S, Zhang LJ, Wu DL, Chen ZG. Bisphenol A inhibits mucin 2 secretion in intestinal goblet cells through mitochondrial dysfunction and oxidative stress. Biomed Pharmacother. 2019:111:901–908. [DOI] [PubMed] [Google Scholar]
- 37. Mallilankaraman K, Cárdenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenár T, Csordás G, Madireddi P, Yang J, Müller M, et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012:14(12):1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaufman RJ, Malhotra JD. Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochim Biophys Acta. 2014:1843(10):2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toepfer C, Caorsi V, Kampourakis T, Sikkel MB, West TG, Leung M, Al-Saud SA, MacLeod KT, Lyon AR, Marston SB, et al. Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. J Biol Chem. 2013:288(19):13446–13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simão F, Matté A, Matté C, Soares FMS, Wyse ATS, Netto CA, Salbego CG. Resveratrol prevents oxidative stress and inhibition of Na+ K+-ATPase activity induced by transient global cerebral ischemia in rats. J Nutr Biochem. 2011:22(10):921–928. [DOI] [PubMed] [Google Scholar]
- 41. Huang CL, Chao CC, Lee YC, Lu MK, Cheng JJ, Yang YC, Wang VC, Chang WC, Huang NK. Paraquat induces cell death through impairing mitochondrial membrane permeability. Mol Neurobiol. 2016:53(4):2169–2188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.