Abstract
Microbial nitrite oxidation is the primary pathway that generates nitrate in wastewater treatment systems and can be performed by a variety of microbes: namely, nitrite-oxidizing bacteria (NOB). Since NOB were first isolated 130 years ago, the understanding of the phylogenetical and physiological diversities of NOB has been gradually deepened. In recent endeavors of advanced biological nitrogen removal, NOB have been more considered as a troublesome disruptor, and strategies on NOB suppression often fail in practice after long-term operation due to the growth of specific NOB that are able to adapt to even harsh conditions. In line with a review of the history of currently known NOB genera, a phylogenetic tree is constructed to exhibit a wide range of NOB in different phyla. In addition, the growth behavior and metabolic performance of different NOB strains are summarized. These specific features of various NOB (e.g., high oxygen affinity of Nitrospira, tolerance to chemical inhibitors of Nitrobacter and Candidatus Nitrotoga, and preference to high temperature of Nitrolancea) highlight the differentiation of the NOB ecological niche in biological nitrogen processes and potentially support their adaptation to different suppression strategies (e.g., low dissolved oxygen, chemical treatment, and high temperature). This review implicates the acquired physiological characteristics of NOB to their emergence from a genomic and ecological perspective and emphasizes the importance of understanding physiological characterization and genomic information in future wastewater treatment studies.
Keywords: microbial nitrification, nitrite oxidation, nitrite-oxidizing bacteria (NOB), suppression, kinetics; short-cut nitrogen removal
1. Introduction
Microbial nitrite oxidation to nitrate is performed by a group of microorganisms, named after their major function as nitrite-oxidizing bacteria (NOB). As one of the primary producers of nitrate, NOB are widespread in natural environments, such as in soil,1,2 ocean,3,4 freshwater,5 and hot springs.6−8 Members of NOB are diverse, which have been found spanning 4 phyla and 12 genera (including candidate genera) with considerable functional and physiological diversities.
NOB also play an essential role in biological nitrogen removal processes within modern wastewater treatment plants (WWTPs).9−11 Biological nitrogen removal was initially proposed in the middle of the twentieth century in response to the growing issue of eutrophication. Until now, nitrification–denitrification has still been the most widely adopted nitrogen removal process. In this process, nitrite generated from ammonia oxidation is subsequently oxidized to nitrate by NOB, which is then anoxically reduced back to nitrite and further to dinitrogen gas by denitrifying microorganisms. An obvious issue associated with the conventional nitrification–denitrification is the transformation between nitrite and nitrate, which leads to significant dissipation of energy (from nitrite to nitrate) and organic carbon (from nitrate to nitrite).12
In recent years, increasing attention has been paid to upgrading traditional WWTPs with the aim of reducing operational costs and carbon footprints. These advanced biological nitrogen removal processes are usually associated with the fate of nitrite. Unlike the traditional nitrification–denitrification pattern, researchers are endeavoring to develop a more energy-efficient and more sustainable flow of nitrogen in WWTPs, that is, to directly reduce nitrite to nitrogen gas, bypassing nitrate. For example, partial nitritation and anammox (PN/A, or deammonification) and nitrite shunt are the two most widely known state-of-the-art shortcut nitrogen removal techniques, which could significantly save energy and organic carbon sources compared to the traditional nitrification–denitrification process.13,14 With that, nitrite accumulation becomes a critical prerequisite, while as an oxidizer of nitrite, NOB are considered as the enemy to the shortcut nitrogen removal systems. Indeed, the NOB suppression can be achieved easily in high-strength wastewater due to the in situ formed free ammonia (FA) and free nitrous acid (FNA). However, in mainstream systems where the nitrogen is low (40–60 mg N/L), stable NOB suppression is a known challenge, which is a hot research topic and also the focus of the present review. To suppress the NOB in low-strength wastewater, various control strategies have been proposed. For example, lowering the dissolved oxygen (DO) concentration is a very typical approach, as it manipulates the competition between AOB and NOB.15 Shortening the solid retention time (SRT) is another strategy to wash out NOB due to the higher growth rates of AOB compared to those of NOB.16 Moreover, the ex situ treatment of mainstream sludge has been proven effective as well. About 20–30% of mainstream sludge is centrifuged and treated by different methods on a daily basis, e.g., FA and FNA, which is shown capable of selectively suppressing NOB in the mainstream treatment.17 However, most of these strategies have only been shown to inhibit NOB in a relatively short period, and the long-term operation under practically relevant conditions usually results in the emergence and adaptation of specific nitrite oxidizers.18 This is intrinsically because of distinct physiological characteristics of diverse nitrite oxidizers, which may pose a fundamental challenge to next-generation shortcut nitrogen removal processes.
Previous reviews focused either more on the fundamentals of NOB or summarizing approaches to suppress NOB from practical engineering aspects. For instance, Daims et al.19 delved into the biological mechanisms and ecological interaction of NOB, while Cao et al.20 discussed key microbes and interactions in PN/A and suggested critical factors for the process control, including operational parameters and bioreactor selection. Additionally, Wang et al.12 models from a kinetic perspective for sidestream and mainstream PN/A processes and identifies suitable operational windows. This review aims to build a bridge between these two aspects. Specifically, the integration of perspectives from microbiologists and wastewater engineers will be beneficial to shed light on the question of why NOB suppression in engineered wastewater systems is difficult. Moreover, the review will facilitate the development of more practical solutions for NOB suppression in municipal WWTPs based on the most recent fundamental knowledge, thus underpinning the paradigm shift to carbon-neutral and energy-positive wastewater management in the future.
2. History of NOB Discovery
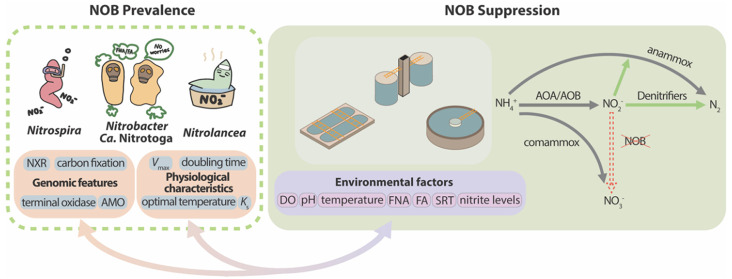
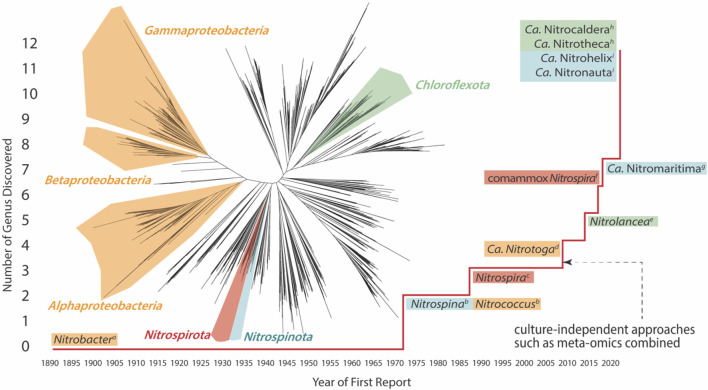
The advancement in cultivation technology, molecular methods, and bioinformatics has broadened the diversity of NOB. To date, all known chemolithoautotrophic nitrite oxidizers are bacteria spanning 4 phyla including Pseudomonadota (also known as Proteobacteria), Nitrospirota (also known as Nitrospirae), Nitrospinota (also known as Nitrospinae), and Chloroflexota (also known as Chloroflexi), which consist of 7 genera and 5 candidate genera (without isolates obtained) (Figure 1).
Figure 1.
Historic discovery of NOB and complete ammonia oxidation (comammox) genera. The red line indicates the increase in the total number of NOB/comammox genera identified. The colored background of the genera names represents the phylum to which each NOB/comammox belongs to. The phylogenetic tree is a pruned tree from a previous “tree of life” study,21 and branches with colored contour depict the phylum. The discovery of NOB genera was first reported in the following studies: (a) ref (22), (b) ref (23), (c) ref (24), (d) ref (1), (e) ref (25), (f) refs (26 and 27), (g) ref (28) (h) ref (29), and (i) ref (30).
Nitrobacter (Nb.) winogradsky was the first NOB isolate obtained by serial dilution, which is a significant milestone in the history of NOB discovery.31Nb. winogradsky grows aerobically with nitrite as an electron donor and can also alternatively live on simple organic compounds (e.g., pyruvate, formate, acetate, and yeast extract-peptone).32−34 After about a century, Nitrococcus and Nitrospina were identified in marine ecosystems in 1971, with two strains isolated from surface samples of South Pacific waters and South Atlantic waters, respectively.23 Unlike Nitrobacter, they were both obligate chemoautotrophic NOB, as no growth was observed in organic media. Until 1986, a new genus of NOB Nitrospira was discovered, and Nitrospira (Ns.) marina was isolated from a water sample at a depth of 206 m from the Gulf of Maine in the Atlantic Ocean.24 In the following more than a century, until now, the genus Nitrospira has been found to be the most ubiquitous NOB in natural and engineered ecosystems. A phylogenetic analysis of 16S rRNA genes indicates that Nitrospira can be classified into at least six sublineages, exhibiting a greater phylogenetic diversity than other NOB genera.19 Within sublineage II, a number of Nitrospira species including the representative isolate Ns. inopinata were shown to be able to perform complete ammonia oxidation (comammox).26,27 This discovery is a breakthrough that fundamentally changed the previous perception by demonstrating that complete nitrification can be carried out by a single microorganism rather than multiple microorganisms in concert. Additionally, Nitrospina belonging to a distinct novel phylum Nitrospinota was recently discovered based on an in-depth evaluation of the draft genome of Nitrospina (Nn.) gracilis.35 It is considered to be an important NOB population in marine environments.30,36,37
The knowledge of NOB diversity keeps evolving, and more novel NOB have come to light based on the improvement of culture-independent approaches (e.g., meta-omics). The novel NOB genus Candidatus (Ca.) Nitrotoga belonging to β-Proteobacteria was discovered in 2007 based on the enrichment from permafrost-affected soil in Siberian Arctic.1 Members of the genus Ca. Nitrotoga can generally adapt to the cold environment (<10 °C) except for Ca. Nitrotoga (Nt.) fabula.38 In contrast, the genus Nitrolancea positioning in the Chloroflexota phylum seems to be thermophilic, which was initially discovered in a nitrifying bioreactor at an elevated temperature of 37 °C.25 More recently, two draft genomes of putative thermophilic NOB were retrieved from a metagenome yielded from the Yellowstone Hot Spring enrichment, provisionally named “Ca. Nitrocaldera robusta” and “Ca. Nitrotheca patiens”.8 Both candidate genera belong to the phylum Chloroflexota and expectedly prefer a thermophilic lifestyle. In the Nitrospinae phylum, two new marine genera, “Ca. Nitrohelix vancouverensis” and “Ca. Nitronauta litoralis” were recovered by using cell sorting, activity screening, and incubation.30 These studies exemplify an efficient protocol enabling physiological investigation rather than traditional cultivation. A recent work using metagenome-assembled genomes and single-cell amplified genomes also proposed a provisional genus “Candidatus Nitromaritima” in a deep-branching lineage under Nitrospinota.28 These results collectively indicate that applications of culture-independent approaches will likely further enlarge the pool of NOB than we expected before.
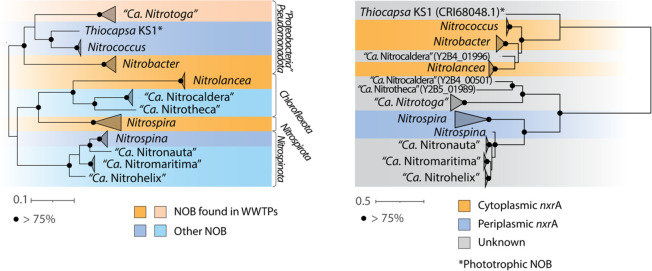
According to the 16S rRNA phylogeny (Figure 2a), NOB is distributed in at least four deep branches. The phylogenetic distance between NOB groups is large, with a 16S rRNA sequence identity of 62.4–92.2% between genera. The phylogeny of the key marker gene nxrA (alpha subunit of nitrite oxidoreductase) of NOB shows a congruent pattern (Figure 2b), which can be divided into three main clusters and likely correlated with the different features, for example, energy efficiency (see more discussions in Section 4.1) and the locale in the cell membrane. Specifically, the cytoplasmic nitrite oxidoreductase (NXR) is represented by Nitrobacter and Nitrolancea, and the periplasmic NXR is represented by Nitrospira and Nitrospina. The recent identification of Ca. Nitrotoga has revealed novel sub-branches of periplasm NXR (Figure 2b), which is associated with a unique soluble NXR periplasmic holoenzyme.39 Analogous to the 16S rRNA phylogeny, the three branches of nxrA are divergent from each other, indicating at least three evolution origins.39,40
Figure 2.
Phylogenetic tree of currently known NOB and their nxrA. (a, left) 16S rRNA phylogenomic tree of NOB. Colored backgrounds indicate the NOB found in WWTPs. Light blue and light orange indicate not validly published nomenclature, or no isolates were reported so far. (b, right) Phylogenetic tree of NOB nxrA. Colored backgrounds indicate the types of nxrA. Black dots at the branch nodes indicate bootstrap values (based on 1000 iterations) >75%. On both sides, an asterisk indicates phototrophic NOB.
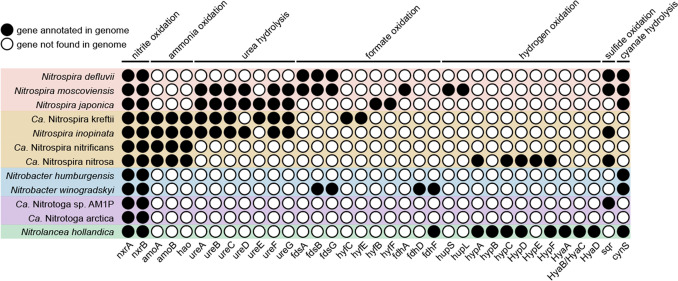
The metabolic versatility of NOB has been revealed previously, showing their additional capability of performing for example urea hydrolysis and using formate,41 hydrogen,42 sulfide,4 and cyanate43,44 as substrates for their growth (Figure 3). Such highly functional diversity supports the survival of NOB in different natural and engineered systems but adds difficulty to suppressing the nitrite oxidation in wastewater treatment using a single strategy. Moreover, these diverse functions are widely possessed by different NOB, while the species within the same genus may show different functions. As the nitrifying community in wastewater-engineered systems is mostly revealed by 16S rRNA amplicon sequencing, the resolution is inadequate to show the microbial community structure at the species level. In specific circumstances, especially when the NOB control failed and the adaptation of NOB is observed, using advanced metagenomic sequencing will be conducive to providing more accurate information in high resolution.
Figure 3.
Functional genes in NOB Nitrospira, comammox Nitrospira, Nitrobacter, Ca. Nitrotoga, and Nitrolancea based on mapped reads to the annotated ORFs. Twelve representative genomes that contained at least one of the related genes are displayed. Metagenemarks45 was used to predict the genes of each genome, and functional annotation referred to KEGG,46 COG,47 and Uniprot48 databases. Notably, due to differences in MAG quality and limitations of databases we used, it is possible that some genomes missed functional genes. As gene expression is regulated by various complex factors, such as environmental conditions, nutritional status, and intra- and extracellular signals, gene annotation only represents the potential for functional expression of these genes and cannot ensure that they will be expressed in the actual environment, which requires further investigation.
Both phylogenetic trees based on 16S rRNA and the nxrA gene are rooted at midpoints. In the 16S rRNA gene tree (left), there are 4 major branches in accordance to phyla level, namely Pseudomonadota (Proteobacteria), Chloroflexota, Nitrospiraota, and Nitrospinota. In contrast, the nxrA phylogenetic tree is mainly divided into three major branches. These include cytoplasmic nxrA represented by Nitrococcus, Nitrobacter, and Nitrolancea, periplasmic nxrA represented by Nitrospira and Nitrospina, and a middle branch mainly represented by Ca. Nitrotoga yet to be clarified. The major branches of the trees are composed of highly divergent sequences, suggesting significant evolutionary divergence between lineages. Upon closer inspection, the topology of the branching patterns has similarities between the trees. For example, in both trees, Nitrospira, Nitrospina, and other Nitrospinota-affiliated genera still cluster together (although Nitrospira seems more independent), with only minor differences in their branching patterns or sequence divergence. Moreover, Nitrococcus, Nitrobacter, and Thiocapsa also remain relatively close to each other. These clades likely represent closely related lineages that have undergone relatively recent evolutionary diversification. However, the overall patterns of evolutionary relationships among the 16S rRNA and nxrA genes are not necessarily alike; therefore, it is necessary to choose the appropriate gene when using genetic analysis to interpret NOB evolutionary relationships.
3. Primary NOB in Global Full-Scale WWTPs
Nitrification converts ammonia (NH3) to nitrite (NO2–) and then to nitrate (NO3–), which is generally carried out by ammonia-oxidizing bacteria (AOB)/ammonia-oxidizing archaea (AOA), NOB, and comammox bacteria in wastewater treatment systems. NOB Nitrobacter cells were frequently detected together with Nitrospira, which were believed to dominate nitrification in wastewater treatment systems.49,50 In recent years, with more accessible sequencing and quantification technologies, there has been an increasing amount of evidence highlighting the prevalence of Nitrospira in activated sludge of WWTPs.51 A systematic global-sampling effort of ∼1200 activated sludge samples from 269 WWTPs demonstrated that Nitrospira were the dominant NOB in most examined WWTPs.52 Similarly, other studies also showed that sublineages I and II of the genus Nitrospira were ubiquitously detected in WWTPs.50,53 It is therefore tempting to assume that the operational conditions in conventional activated sludge processes are favorable to Nitrospira in comparison to those of other NOB genera. The dominance of Nitrospira is likely due to the physiological features and metabolic versatility of this genus, as elaborated below.
3.1. Physiological Features
Table 1 summarizes the key physiological features of the most kinetically characterized NOB, including the optimum temperature, half-saturation nitrite constant (Km), and maximum nitrite oxidation rate (Vmax). These characterization studies unanimously reveal that the nitrite affinity constant of Nitrospira (Km = 6–54 μM NO2–, excluding comammox Nitrospira) is much lower than the counterpart of other NOBs found in wastewater treatment. Since the in situ nitrite concentration is generally below 50 μM NO2– in widely installed continuous flow activated sludge processes, Nitrospira may have a greater resilience and adaptive ability over other NOB genera. For example, Nitrospira outcompeted Nitrobacter when the nitrite concentration was controlled below 3 mg NO2–-N/L.54 However, it should be noted that the sequencing batch reactor (SBR), a configuration widely applied in small-/medium-scale WWTPs, often transiently accumulates nitrite in the aerobic period. The increased nitrite concentration can provide opportunities for the growth of other NOB such as Ca. Nitrotoga, which have a slightly higher Km value in the range 24–86.5 μM NO2–.38,55,56 By using 16S rRNA gene specific PCR and FISH, Lücker et al. verified the presence of Nitrotoga-like bacteria in 11 of 15 full-scale SBRs surveyed, in which the nitrite level could reach as high as 0.48 mg/L NO2– within a typical cycle.10
Table 1. Physiological Features of 22 Isolated/Enriched NOB Cultures, Including Optimum Temperature, Mean (±SD) Half-Saturation Constants (Km), and Maximum Nitrite Oxidation Activities (Vmax)a.
| organism name | sample source | optimum temp (°C) | Km (μM NO2–) | Vmax (μmol NO2–/(mg protein h)) | source |
|---|---|---|---|---|---|
| Nitrospira moscoviensis | corroded iron pipe of a heating system | 39 | 9 ± 3 | 18 ± 1 | (56, 85) |
| Nitrospira defluvii | enrichment from activated sludge sample | 32 | 9 ± 3 | 48 ± 2 | (56, 84, 86) |
| Nitrospira sp. NJ1 | WWTP | 31 | 10 ± 2 | 31 ± 5 | (87) |
| Nitrospira sp. ND1 | WWTP | ND | 6 ± 1 | 45 ± 7 | (88) |
| Nitrospira sp. Ecomares 2.1 | moving-bed filter of an aquaculture system | 28–30 | 54.0 ± 11.9 | 21.4 ± 1.2 | (89) |
| Nitrospira sp. BS10 | WWTP | 28 | 27 ± 11 | 20 ± 2 | (56) |
| Nitrospira inopinata | hot water pipe from an oil exploration well | 37 | 372 ± 55 | 16.9 | (56, 69) |
| Nitrobacter hamburgensis X14 | soil | ND | 544 ± 55 | 64 ± 1 | (56, 90, 91) |
| Nitrobacter winogradskyi | soil | 32 | 309 ± 92 | 78 ± 5 | (34, 56, 91, 92) |
| Nitrobacter vulgaris | sewage | ND | 49 ± 11 | 164 ± 9 | (84, 93) |
| Nitrobacter sp. Nb-311A | surface waters of the Atlantic Ocean | ND | 27.6 ± 6.7 | 95.2 ± 7.0 | (89) |
| Ca. Nitrotoga sp. AM1 | sand of eelgrass zone | 16 | 24.7 ± 9.8 | ND | (55) |
| Ca. Nitrotoga arctica | permafrost soil | 10/13–19 | 58 ± 28 | 26 ± 3 | (56, 80) |
| Ca. Nitrotoga sp. HAM-1 | WWTP | ND | ND | ND | (83) |
| Ca. Nitrotoga fabula KNB | WWTP | 24–28 | 89.3 ± 3.9 | 27.6 ± 8.4 | (38) |
| Ca. Nitrotoga sp. HW29 | recirculation aquaculture system | 22 | ND | ND | (94) |
| Nitrospina watsonii 347 | Black Sea, 100m depth | 28 | 18.7 ± 2.1 | 36.9 ± 2.2 | (23, 89, 95) |
| Nitrococcus mobiliz | surface water of the South Pacific Ocean | 25–30 | 119.7 ± 34.0 | 141.0 ± 10.6 | (23, 89) |
| Nitrolancea hollandica | laboratory-scale bioreactor | 40 | 1000 | NA | (25,96) |
| Ca. Nitrohelix vancouverensis | sandy coastal surface sediment samples | ND | 8.7 ± 2.5 | ND | (30) |
| Ca. Nitronauta litoralis | sandy coastal surface sediment samples | ND | 16.2 ± 1.6 | ND | (30) |
| Nitrospina gracilis 3/211 | surface water of the Atlantic Ocean | 25–30 | 20.1 ± 2.1 | 41.4 ± 9.4 | (23) |
ND means not determined.
Nitrospira generally has a slow growth rate. For example, the generation times for two Nitrospira representatives, Ns. moscoviensis (sublineage II) and Ns. defluvii (sublineage I), were determined to be 32 and 37 h, respectively,56 which were longer than the doubling time of 13 h for Nb. vulgaris and 26 h for Nb. winogradskyi. This indicates that the SRT applied in many WWTPs (∼15 days) is important to the retention of Nitrospira. In agreement with this hypothesis, a study adopting different SRTs reported that Nitrospira greatly outnumbered Nitrobacter at a longer SRT of 40 days.57 Attributed to the slow growth rate, biofilms provide a more suitable niche for the growth of Nitrospira. In a full-scale hybrid biofilm and activated sludge reactor, the metagenomic approach revealed that the biofilm had significantly higher abundances of Nitrospira compared to the suspended sludge.58 In addition to the canonical Nitrospira, researchers highlighted that the comammox Nitrospira also prefer to grow in biofilms with long SRT. For example, comammox Nitrospira were found dominating the biofilm in a rotating biological contactor in WWTP,59 and a survey on 14 full-scale nitrogen removal systems also revealed that the long SRT (>10 days) and attached growth phase were significantly correlated to the prevailing comammox Nitrospira.60 Our recent studies also showed the prevalence of comammox Nitrospira in nitrification biofilms attached to sponge and plastic carriers.61,62
Although oxygen is also essential to NOB growth, the oxygen affinity was rarely quantified due to the mass transfer resistance among cell aggregates. In engineered systems, researchers usually use apparent half-saturation constants for oxygen (KO) to describe the oxygen affinity of NOB, which typically ranges from 0.06 to 1.0 mg O2/L for NOB in WWTPs.63−67 Since DO concentrations in nitrifying tanks are usually in the range of 1–3 mg O2/L, which is higher than most of the apparent KO values reported, the oxygen may not be the factor affecting the competition between Nitrospira and other NOB. However, this is only applicable to the NOB community in flocs, while in biofilms, the DO level may play an important role because of the higher apparent KO due to mass transfer resistance.68 From the above discussion, the low in situ nitrite concentration and the long SRT are likely to jointly contribute to the dominance of Nitrospira in full-scale WWTPs.
3.2. Metabolic Versatility
Recent findings have suggested that the prevalence of Nitrospira in wastewater treatment systems could also be related to their metabolic versatility. First, some Nitrospira members are capable of utilizing ammonia as an energy source (i.e., comammox Nitrospira), which is the main form of nitrogen in raw wastewater. In particular, comammox Nitrospira exhibit a higher affinity for ammonia compared to most other canonical ammonia oxidizers.69 Field investigations have shown that comammox Nitrospira are present in nearly all wastewater treatment processes.70−72 Due to the high affinity for ammonia, the systems with comammox Nitrospira may have higher ammonia removal efficiencies compared to nitrogen removal systems where they are absent.60 Still, more studies should be dedicated to further quantifying the contribution of comammox Nitrospira to ammonia and nitrite removal in WWTPs.
Second, Nitrospira members have versatile metabolic pathways, such as the metabolism of urea, cyanate, and hydrogen. Among all, the degradation of urea is more relevant to wastewater treatment, as over 80% of ammonia nitrogen in domestic wastewater is from urine.73 It has been demonstrated that Ns. moscoviensis belonging to Nitrospira sublineage II can cleave urea to ammonia and CO2,41 and the produced ammonia can be further supplied to ammonia oxidizers. Indeed, the operon encoding the functional urease (ure) exists in the genomes of many other species of Nitrospira such as Ns. sp. BS1074 and Ns. sp. ND1,75 illustrating the ecological importance of urea to Nitrospira. Additionally and intriguingly, three novel comammox species were selectively enriched in a urine-fed reactor.76 Similar to other comammox, Ns. inopinata, Ca. Ns. nitrosa, and Ca. Ns. nitrificans, all these comammox Nitrospira genomes contained a complete urea utilization pathway. The slow release of ammonia from urea hydrolysis might contribute to the prevalence of comammox Nitrospira in the urine-treatment reactor. Although some AOB and AOA also have ureolytic activities,77−79 the successful enrichment from the urine-fed reactor indicates that urea is an important factor to the selection of comammox Nitrospira, which requires further investigations.
3.3. Impact of Seasonal Temperature Variation
From a global perspective, seasonal temperature variation is an important factor shaping the microbial communities in full-scale WWTPs. In particular, some full-scale WWTPs are located in the temperate zone, which generally exhibits annual average temperatures lower than those of tropical and subtropical regions. Various Ca. Nitrotoga strains were discovered from the surface layer of permafrost soil with an extreme temperature range (−48 to 18 °C) and permanently frozen sediments.80 Despite the extremely low temperature where they were found, the optimal temperatures of seven Ca. Nitrotoga strains were above 10 °C, and five among them were above 20 °C. The result suggested that Ca. Nitrotoga are psychrotolerant rather than psychrophilic, which might adapt to the low temperature when the soil froze more than 3000 years ago and survive as a frozen “living fossil”.80 Because of the psychrotolerant feature, Ca. Nitrotoga may become the primary NOB in wastewater-engineered systems with low temperatures. For example, Ca. Nitrotoga were observed to be the transiently dominant NOB in cold seasons at some WWTPs.11,81 An investigation recognized Ca. Nitrotoga as the key NOB in full-scale WWTPs, with Ca. Nitrotoga detected in WWTPs between 7 and 16 °C.10 In this study, Ca. Nitrotoga was the only detectable NOB in two full-scale WWTPs from Germany at temperatures of 16 and 9 °C, respectively. A recent study showed Ca. Nitrotoga co-occurred with Nitrospira only in an SBR reactor operated at low temperatures (4–14 °C), while it was almost not detected in a similar reactor but at elevated temperatures (22–34 °C). This together indicates that Ca. Nitrotoga may have a specific temperature preference and highlights the importance of operating temperature as a factor in the selection of NOB communities in wastewater treatment processes.82 Indeed, the optimal temperature for the growth in the Ca. Nitrotoga genus is obviously lower than that of Nitrospira (Table 1), which supports the competitive advantages of Ca. Nitrotoga over Nitrospira at low temperatures.83 In a coculture of Ns. defluvii and Ca. Nt. sp. BS, the microbial community shifted at 17 °C, where Ca. Nt. sp. BS became the dominant NOB.84 Laboratory bioreactor and coincubation studies also confirmed that temperature is a deciding factor affecting niche occupation of Nitrotoga-like bacteria in activated sludge.1,82
4. NOB Suppression and Adaptation in Advanced Nitrogen Removal Processes
In traditional wastewater treatment, aeration for nitrification consumes significant energy for the oxidation of nitrite to nitrate, while the reduction of nitrate to nitrite requires organics for heterotrophic denitrification or potentially through endogenous denitrification.97 Therefore, increasing attention has been drawn to advanced nitrogen removal via the nitrite pathway, which is a cost-effective alternative and also known as shortcut biological nitrogen removal. The shortcut nitrogen removal technologies mainly include the nitrite shunt and PN/A (or deammonification), which can be achieved in both two-stage (i.e., oxidation and reduction processes in two bioreactors) and one-stage (i.e., oxidation and reduction processes in one bioreactor) configurations. In theory, ammonia is first converted to nitrite, which will be removed by heterotrophic denitrifiers in the nitrite shunt and by anammox bacteria in the PN/A process. Compared with conventional nitrification and denitrification, the nitrite shunt process reduces the aeration requirement by 25%, organic carbon consumption by 40%, and sludge production by up to 55%,98 while these benefits are further enhanced in the PN/A process achieving the reduction of aeration, organic carbon consumption, and sludge production by nearly 60%, 100%, and 80%, respectively.99,100
Despite these remarkable benefits, a critical challenge associated with the success of nitrite shunt and PN/A is the suppression of NOB. Generally, strategies to minimize NOB activity and achieve PN are developed based on three principles: (1) different kinetics of AOB and NOB, (2) different resilience of AOB and NOB to harsh treatment, and (3) the competition for nitrite by other microorganisms (e.g., anammox bacteria) (Table 2). Accordingly, the suppression of NOB can be realized by in situ control and ex situ treatment. For the in situ control, strategies adopting low DO, high residual ammonium, short SRT, and combining anammox bacteria to compete for nitrite are proposed as favorable strategies to selectively inhibit NOB. Apart from that, recently, a novel approach was developed by forming an in situ acidic condition in promoting the stable nitrite accumulation, or partial nitritation.101,102 Regarding the ex situ control, biocidal treatments using such as FA,103 FNA,104 sulfide,105 ultrasound,106,107 and light irradiation108 were found to be efficient for the selective inhibition of NOB. Both in situ control and ex situ treatment can be practical. While the in situ strategies are generally easier to implement, the ex situ treatment most times requires additional capital costs for a separate treatment tank. However, the previous economic analysis showed that the capital cost of the ex situ treatment by FNA was only approximately 5% of the total reduced cost by replacing nitrification-denitrification with the PN/A process.109
Table 2. Dominant NOB under Different Control Strategies.
| strategy | reactor type | NOB | DO (mg O2/L) | SRT (days) | harsh treatment | ref |
|---|---|---|---|---|---|---|
| oxygen-based control | UMABR | Nitrospira | 0.6 ± 0.1 | infinite | (110) | |
| oxygen-based control | MBBR | Nitrospira | 0.15–0.18 | uncontrolled | (111) | |
| oxygen-based control | MBBR | Nitrospira and Nitrobacter | <0.03 | uncontrolled | (112) | |
| oxygen-based control | SBR | Nitrospira | 0.2–6.5 | uncontrolled to 14.4 ± 2 | (64) | |
| FNA-based sidestream treatment | SBR | Ca. Nitrotoga | >1 mg HNO2-N/L FNA | (113) | ||
| FA-based sludge treatment | SBR | Ca. Nitrotoga | 1.5–2.0 | 15 | 354 ± 58 mg N/L FA | (114) |
| FNA-based sidestream treatment | anoxic/oxic (An/O) reactor | Ca. Nitrotoga | 15 | 1.87 mg N/L FNA | (115) | |
| ex situ control with harsh treatments | SBR | Nitrospira and Nitrobacter | 1.5–2.0 | 15 | 4.23 mg N/L FNA and sequentially 210 mg N/L FA | (103) |
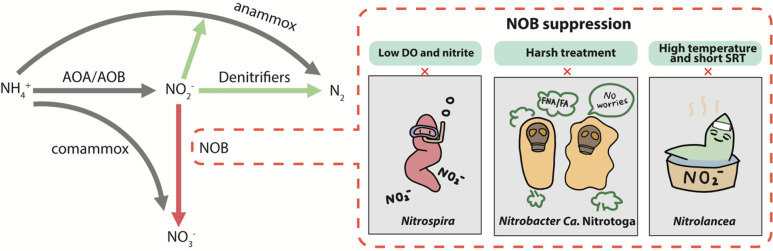
Nonetheless, the stable NOB suppression in long-term operation remains challenging under mainstream conditions with low ammonium concentration and seasonally varying temperatures. From a kinetic point of view, the development of NOB control strategies can be found in a recent review.12 Herein, a microbial perspective of NOB suppression and adaptation to different treatment processes is detailed, since different types of NOB (i.e., Nitrospira, Nitrobacter, Nitrolancea, and Ca. Nitrotoga) appear to be adapted to different control strategies, leading to the failure of nitrogen removal via the nitrite pathway (Figure 4).
Figure 4.
Conceptual diagram describing the adaptation of NOB to different treatment processes in the shortcut nitrogen removal processes.
4.1. Nitrospira Surviving in Low-DO and Low-Nitrite Systems
Low-DO control is undoubtedly the most recognized strategy for NOB inhibition and is widely applied in shortcut nitrogen removal processes. In comparison to the two-stage configuration, the one-stage shortcut nitrogen removal processes require more strict DO control strategies because the residual oxygen may pose inhibitory effects to anaerobic microbes such as anammox bacteria and denitrifiers growing in the same niche. Therefore, the supplied oxygen in theory should be sufficient only to support the ammonia oxidation while limiting the subsequent nitrite oxidation by NOB and any potential inhibitory impacts on anammox bacteria or denitrifiers. In previous reports, measures such as transient anoxia, DO-based aeration, and intermittent aeration have been proven to be effective in achieving shortcut nitrogen removal performance.116−118
However, Nitrospira has stubbornly appeared in many one-stage shortcut nitrogen removal processes, likely because the Nitrospira sublineage I, as one of the most abundant Nitrospira branches in WWTPs, has an even higher affinity for oxygen (0.09 ± 0.02 mg O2/L) than most of AOB.64 For example, Liu and Wang57 showed that Nitrospira gradually became predominant, leading to the failure of nitritation in a long-term reactor operated at low DO (≤0.5 mg O2/L). The complete nitrification was achieved at a low DO of 0.3 ± 0.14 mg O2/L with Nitrospira abundance of up to 2.64 × 106 cells/mL, Liu et al.119 also detected Nitrospira under extremely hypoxic conditions (0.02–0.10 O2 mg/L) in a one-stage PN/A process.
The different oxygen affinities of the NOB are likely related to electron transfer and terminal oxidases. Specifically, aerobic respiration relies on the respiratory chain, where electrons generated during nitrite oxidation flow from NXR to cytochrome c and then to the terminal oxidase.38 Three distinct types of terminal oxidase have been well-studied and present in the respiratory chain of different NOB.120−122 The aa3-type heme-copper oxidase (A-class HCO) is commonly found in Nitrobacter,123,124 which shows a lower affinity for oxygen. Nitrospira contain a putative cytochrome bd-like terminal oxidase, which could also receive electrons derived from nitrite or low-potential donors like organic carbon.125,126 The cbb3-type terminal oxidase, a member of the C-class HCO possessing an extremely high affinity for oxygen,38 is used by NOB species to adapt to O2 concentrations at nanomolar concentration, e.g., Nn. gracilis35 and Ca. Nt. fabula.39 This thus supports the occurrence of Ca. Nitrotoga and Nitrospira in a low-oxygen nitrifying bioreactor.127
Previous studies also reported the dominance of comammox Nitrospira in a mainstream nitrifying moving bed biofilm reactor (MBBR)128 and nitrogen removal system based on an anammox process,129 both of which were under low-DO conditions. However, a recent full-scale survey suggested that the presence of comammox is not significantly related to DO levels.60 In addition, comammox Nitrospira were successfully enriched from an MBBR with a sufficient DO supply,62 and the apparent KO value of this comammox-dominated culture was determined to be 2.8 ± 0.4 mg O2/L, indicating that this cluster of comammox Nitrospira may not have a strong affinity for oxygen. However, the study has its limitations on the mixed culture. Moreover, the efficiency of oxygen mass transfer in biofilms can also affect the measurement of the apparent affinity for oxygen.
Another strategy to suppress NOB in one-stage nitrogen removal processes is a low in situ nitrite concentration. Ideally, nitrite produced by AOB in a one-stage nitrite shunt or PN/A is immediately consumed by denitrifiers and anammox bacteria, leading to a gradual washout of NOB due to failure in the competition for nitrite. However, the nitrite uptake competency depends on the nitrite affinities of NOB and other nitrite scavengers, which can vary between genera or even species levels. As shown in Figure 3b, the nxr genes can be divided into at least two major types, based on their subcellular locus at periplasm or cytoplasm.19 In particular, Nitrospira and Nitrospina harbor the periplasmic NXR,130 while cytoplasmic NXR mainly occurs in Nitrobacter, Nitrolancea, and Nitrococcus. Theoretically, the periplasmic NXR energetically prevails over cytoplasmic NXR,131,132 as nitrite oxidation takes place outside the cytoplasm and the liberated protons directly contribute to the proton motive force (PMF). This could become the greater energic advantage of NOB using periplasmic NXR than cytoplasmic NXR, as nitrite oxidation only yields low energy (△G°′ = −74 kJ/mol NO2–).19,130 In agreement with this fundamental knowledge, previous studies observed that the cytoplasmic group usually dominates over the periplasmic group in environments with a relatively higher level of nitrite, and vice versa for low concentration of nitrite.25,56 These two different types of NXR are likely associated with different affinities for nitrite, which therefore differentiate the growing niche of different NOB. Members of the Nitrospira with periplasmic NXR have a lower Km for nitrite than other NOB populations or even anammox bacteria (Table 1). This may explain why Nitrospira have a stronger endurance to low levels of nitrite and can compete for nitrite against denitrifiers and anammox bacteria in the one-stage shortcut nitrogen removal process.
4.2. Nitrobacter and Ca. Nitrotoga Tolerating Ex Situ Harsh Treatment
In recent years, a number of studies have achieved NOB suppression based on FNA (HNO2)/FA (NH3) inhibitory and biocidal effects,133 by sidestream sludge treatment,134−136 or ex situ treatment. Under acidic conditions, nitrite can form FNA (HNO2 ⇌ H+ + NO2–), which is an inhibitor of nitrite oxidizers,17,137 and free ammonia is the un-ionized form of ammonium that can form under alkaline conditions.138 Generally, FNA causes biocidal effects likely due to the oxidative damage by various reactive nitrogen and oxygen species dissociated from FNA, which can lead to oxidative damage to cellular proteins, cell membrane, and nucleic acids.133 Differently, the inhibitory effects of FA are attributed to the passive diffusion of FA molecules into cells, causing proton imbalance or potassium deficiency.103,104 All of these strategies assume that NOB are more sensitive to harsh conditions than AOB, thus being selectively inhibited; Nitrospira is especially sensitive to these harsh conditions.
However, several strains of NOB show a certain robustness against these harsh treatments. As observed in the alternating treatment by FNA and FA, a clear shift in microbial community indicated that Nitrobacter may be tolerant to the FNA treatment.103 FNA treatment often comes with a low pH, while to date, only a Nitrobacter strain has been reported to be acidophilic NOB that can oxidize nitrite at a pH as low as 3.5.2 In the natural environment, Nitrobacter have been also detected in large numbers of acidic habitats such as acidic soil.2 However, the mechanism of acidophilic Nitrobacter tolerating the acidic environment remains unknown.
Another recently recognized NOB, Ca. Nitrotoga, have been shown to have a high tolerance to both FNA and FA treatment as well.113 It was reported that FNA ranging from 0 to 1.37 mg HNO2-N/L was unable to inactivate nitrite oxidation due to the emergence of Ca. Nitrotoga (from 0% to 4.51%).115 Likewise, in an activated sludge system regularly exposed to ∼220 mg NH3-N/L FA, the dominant NOB shifted from Nitrospira (from 61.12% to 2.18% of the nitrifiers) to Ca. Nitrotoga (from 4.6% to 85.25% of the nitrifiers).134 The proliferation of Ca. Nitrotoga was also observed in a recent study, in which alternating FNA and FA treatment and low DO control were both applied.113 All of these results imply that Ca. Nitrotoga could be a critical challenge to the shortcut nitrogen removal that is achieved based on sidestream inactivation using FNA/FA. The mechanism for the tolerance of Ca. Nitrotoga to FNA (weakly acidic conditions) and FA (weakly alkaline conditions) is a fundamental question worth more exploration in the future.
As is known, the NOB suppression is easy in treating high-strength wastewater during which the entire system is continuously exposed to the in situ high FNA/FA. However, in the ex situ sludge treatment by FNA/FA to achieve NOB suppression in low-strength wastewater, only 20–30% of mainstream sludge is generally treated every day, which will provide a certain feasibility for NOB to adapt to the treatment in long-term operation, as observed in many aforementioned studies.102,109 Based on the learning from the NOB suppression in high-strength wastewater, researchers hypothesized that an in situ harsh treatment of mainstream sludge (e.g., in situ high FNA) might enable robust NOB suppression as the whole bulk of the sludge is continuously exposed to the harsh condition continuously. Drawing upon this hypothesis, a robust nitritation process and stable NOB suppression have been reported recently in an in situ acidic reactor, where the operating pH was controlled at 5–6.101 Of note, the low pH was not achieved by adding acids but was self-sustained by the microbial ammonia oxidation (NH4+ + 1.5 O2 → NO2– + 2 H+ + H2O), which produces protons. Operation using real sewage demonstrated stable suppression of NOB in the long term because AOB can produce nitrite and protons to form in situ FNA (NO2– + H+ ↔ HNO2) at a ppm level, as an inhibitor to NOB.139,140 As the entire system was exposed to high FNA continuously, the NOB suppression was very stable, similar to the principle of NOB suppression in high-strength wastewater treatment. Interestingly, the novel AOB Ca. Nitrosoglobus were enriched within these acidic systems, which exhibited extreme tolerance to acid and FNA,141 while no known NOB was able to survive under such harsh conditions (i.e., FNA > 1 mg HNO2-N/L).142
It is reasonable to speculate that Nitrobacter and Ca. Nitrotoga can be as phylogenetically complex as Nitrospira, with some branches being tolerant to the harsh treatment and some not. However, in comparison to Nitrospira, understanding the diversities of Nitrobacter and Ca. Nitrotoga is to be advanced, and the mechanisms behind their tolerance against FNA or FA are to be further explored. The above experimental evidence also suggests that it would be extremely challenging to suppress NOB growth using a single strategy.132,143
4.3. Nitrolancea Proliferating with High Temperature and Short SRT
Due to the slow growth rate of NOB and different responses to temperature variation, short SRT and higher temperatures are proposed to suppress NOB activity. These strategies have been used in the classical SHARON (single reactor system for high activity ammonium removal over nitrite) process, one of the earliest processes designed to treat high-strength wastewater.133 The SHARON reactors employed high temperatures (30–40 °C) and short SRT (∼1.5 days). In spite of many successful demonstrations, the new NOB genus Nitrolancea has been identified in engineered systems with elevated ammonium or nitrite concentrations. Nitrolancea (Nl.) hollandica was first isolated from a bioreactor with 428 mM of ammonia to achieve partial nitrification but ended up with nitrite oxidation. In this case, only a small amount of Nitrobacter cells were detected, without other known NOB, indicating the major contribution from an unknown NOB. The obtained isolate Nitrolancea (Nl.) hollandica can tolerate a broad temperature range of 25–63 °C and nitrite up to 75 mM, with an optimum temperature of 40 °C.25 More importantly, a maximal specific growth rate of 0.019 h–1 was observed for Nl. hollandica,25 which is several times higher than that of other known NOB, suggesting that to wash out Nl. Hollandica, an even shorter SRT may be needed. Recently, Spieck et al. obtained four more Nitrolancea strains from a centrate treatment reactor, all of which have similar physiological features including thermotolerance, the capability to grow at high nitrite concentrations, and a fast-growing pattern.96 To date, limited studies have reported the presence of Nitrolancea in reactors; similar 16S rRNA sequences were found in a partial nitrification reactor144 and FNA-treated wastewater.145
5. Conclusions and Future Perspectives
The discovery of a novel NOB is a key path to broaden our knowledge for understanding microbial nitrite oxidation in wastewater treatment. In recent years, novel NOB have come to light with differentiated physiology features and metabolic versatility, resulting in robust adaptability of the NOB to different environments. It can be foreseen that more unknown NOB species will be discovered in the future. A basic approach to study novel NOB in previous studies is based on cultivation/isolation and characterization, which is time-consuming and labor-intensive. In many studies focusing on wastewater treatment, 16S rRNA gene amplicon sequencing has been applied, but the resolution is too low to distinguish species or reveal their functions. Considering that activated sludge systems are significantly complex, in situ investigation of NOB in wastewater treatment processes can be an eclectic solution. Given the emergence of advanced detection methods, there should be a tendency for researchers to incorporate advanced detection techniques with traditional experimental approaches. For example, metagenomic sequencing is now intensely used to unravel functional NOB communities in environmental samples, and this can be more explored in wastewater samples to identify new NOB. In particular, this approach can also be used in advanced nitrogen removal processes that require NOB suppression yet face challenges. Genomic features of these adapted NOB in shortcut nitrogen removal can be an important indicator of their physiological effects, like the nitrite and oxygen affinities, as mentioned above. In parallel, metatranscriptomics can be used to identify gene expressions in the coculture of NOB and other nitrifiers, which helps to reveal the synergy and other physiological effects with their partners compared to single culture and the NOB physiology, such as the mechanism of the tolerance to FNA and affinity for oxygen. Moreover, many previous studies have focused on the development of new control strategies while relatively missing the characterization of abundant NOB when the process performance fails. To better suppress NOB in such systems, more attention should be paid to profiling the physiological characteristics of the NOB, especially at species levels.
The discovery of a novel NOB also provides opportunities to suppress or promote nitrite oxidation in wastewater treatment. Taking the genus Nitrospira as an example, it is now gradually realized that the functional diversity within genera is non-negligible, exhibiting diverse physiological features. Among this genus, comammox Nitrospira feature an extremely high affinity to ammonia and may outcompete other ammonia-oxidizers under oligotrophic conditions.69 While being detected in various wastewater treatment units, comammox Nitrospira was believed to damage the shortcut nitrogen removal via the nitrite pathway, as ammonia is oxidized straight away to nitrate by this unique single microorganism. However, more recent studies have indicated that ammonia oxidation can be interrupted at nitrite by comammox bacteria, which have been successfully coupled with anammox bacteria in different configurations.134,135 In two very recent reports, comammox Nitrospira were found to be the main ammonia-oxidizing community (accounting for 89.2 ± 7.9% in total prokaryotic amoA copies) in a PN/A process at low ammonium loading,135 and in a symbiotic association of coincubating comammox Nitrospira and anammox bacteria enabled a sustained nitrogen loss.134 The new knowledge highlights a potentially positive role of comammox Nitrospira in shortcut nitrogen removal processes, in addition to producing less greenhouse gas N2O and leading to low residual ammonium in the final effluent.136,146,147 More specifically, this review also elaborates that some members in the genera of Ca. Nitrotoga, Nitrobacter, and Nitrolancea can become important nitrite oxidizers under specific conditions, such as cold temperature, acidic pH, and thermophilic processes. With new insights into subdivided NOB species, there is thus a need to re-evaluate the regarding contribution to nitrite oxidation especially when corresponding strategies are being designed to suppress a specific NOB in advanced nitrogen removal processes. To resolve the puzzle of mechanisms that are not yet clear, cultivating and characterization of these new NOB would be the prerequisites, and new information needs to be bridged to detailed operation of engineering systems.
Finally, the review also highlighted the challenge of NOB suppression using a single strategy. This is primarily because while using a single strategy to suppress NOB, some specific NOB that are resistant to this applied strategy may gain a competitive edge over others, leading to the failure of suppression in long-term operation. Due to the high diversity of NOB, it appears that there is no “one-size-fits-all” solution. However, it is also unlikely that a specific NOB strain can survive under all of the different strategies. For example, although Nitrospira can survive under low-DO and low-nitrite conditions, they are very sensitive to harsh treatment by FNA and FA. Adversely, Nitrobacter are relatively tolerant to FNA and FA treatment, while they do not usually survive under low-oxygen conditions. This knowledge suggests that the integration of multiple strategies may be necessary to effectively inhibit NOB in long-term operation, which can be applied either alternatively or concurrently. For instance, the alternation of ex situ FNA and FNA treatment on a monthly basis has shown to be more effective in maintaining NOB suppression than applying each of them individually;103 a combined strategy including low DO, ex situ treatment by FA, and in situ competition by anammox bacteria has been applied to stably inhibit NOB growth, while without any of the three factors, NOB adaptation was observed.148 In addition to the combined strategies, another potential option to suppress NOB is to achieve in situ harsh treatment, inspired by the learning from high-strength wastewater treatment. Due to the low nitrogen concentration, achieving high in situ FNA as that in high-strength wastewater needs to lower the pH to 5–6. Fortunately, this can be achieved by the newly discovered acidic-tolerant AOB “Ca. Nitrosoglobus”, which generates protons and nitrite forming in situ high FNA without any chemical input.139 As the entire system is continuously exposed to the in situ high FNA, NOB adaptation is unlikely to be achieved. Notably, partial denitrification and anammox (PD/A) could be also a feasible alternative to skirt NOB suppression, which has been the challenging step in PN/A.149 PD/A includes nitrification (NH4+ → NO3–), partial denitrification (NO3– → NO2–), and anammox and exhibits higher stability compared to PN/A,150,151 which can be a compromise if PN/A fails.
Acknowledgments
This study was supported by the Australian Research Council (ARC) Discovery Project DP230101340. The authors thank Dr. Yue Zheng, Dr. Zhiyao Wang, and Professor Zhiguo Yuan for a review and helpful discussions. M.Z. is the recipient of an Advance Queensland Industry Research Fellowship and an ARC Industry Fellowship (IE230100245). T.L. is an ARC Discovery Early Career Researcher Award (DECRA) Fellow (DE220101310). Z.S. acknowledges the China Scholarship Council (CSC) and the University of Queensland (UQ) for scholarship support.
The authors declare no competing financial interest.
References
- Alawi M.; Lipski A.; Sanders T.; Eva-Maria-Pfeiffer; Spieck E. Cultivation of a Novel Cold-Adapted Nitrite Oxidizing Betaproteobacterium from the Siberian Arctic. Isme J. 2007, 1 (3), 256–264. 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- Hankinson T.; Schmidt E. An Acidophilic and a Neutrophilic Nitrobacter Strain Isolated from the Numerically Predominant Nitrite-Oxidizing Population of an Acid Forest Soil. Appl. Environ. Microbiol. 1988, 54 (6), 1536–1540. 10.1128/aem.54.6.1536-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuessel J.; Lam P.; Lavik G.; Jensen M. M.; Holtappels M.; Guenter M.; Kuypers M. M. M. Nitrite Oxidation in the Namibian Oxygen Minimum Zone. Isme J. 2012, 6 (6), 1200–1209. 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuessel J.; Lücker S.; Yilmaz P.; Nowka B.; van Kessel M. A. H. J.; Bourceau P.; Hach P. F.; Littmann S.; Berg J.; Spieck E.; Daims H.; Kuypers M. M. M.; Lam P. Adaptability as the Key to Success for the Ubiquitous Marine Nitrite Oxidizer Nitrococcus. Sci. Adv. 2017, 3 (11), e1700807 10.1126/sciadv.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black E. M.; Just C. L. The Genomic Potentials of NOB and Comammox Nitrospira in River Sediment Are Impacted by Native Freshwater Mussels. Front. Microbiol. 2018, 9, 2061. 10.3389/fmicb.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T. A.; Calica N. A.; Huang D. A.; Manoharan N.; Hou W.; Huang L.; Panosyan H.; Dong H.; Hedlund B. P. Cultivation and Characterization of Thermophilic Nitrospira Species from Geothermal Springs in the US Great Basin, China, and Armenia. Fems Microbiol. Ecol. 2013, 85 (2), 283–292. 10.1111/1574-6941.12117. [DOI] [PubMed] [Google Scholar]
- Lebedeva E. V.; Off S.; Zumbraegel S.; Kruse M.; Shagzhina A.; Lücker S.; Maixner F.; Lipski A.; Daims H.; Spieck E. Isolation and Characterization of a Moderately Thermophilic Nitrite-Oxidizing Bacterium from a Geothermal Spring. Fems Microbiol. Ecol. 2011, 75 (2), 195–204. 10.1111/j.1574-6941.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- Spieck E.; Spohn M.; Wendt K.; Bock E.; Shively J.; Frank J.; Indenbirken D.; Alawi M.; Lücker S.; Hüpeden J. Extremophilic Nitrite-Oxidizing Chloroflexi from Yellowstone Hot Springs. ISME J. 2020, 14 (2), 364–379. 10.1038/s41396-019-0530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretschko S.; Timmermann G.; Schmid M.; Schleifer K.-H.; Pommerening-Röser A.; Koops H.-P.; Wagner M. Combined Molecular and Conventional Analyses of Nitrifying Bacterium Diversity in Activated Sludge: Nitrosococcus Mobilis and Nitrospira-Like Bacteria as Dominant Populations. Appl. Environ. Microbiol. 1998, 64 (8), 3042–3051. 10.1128/AEM.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S.; Schwarz J.; Gruber-Dorninger C.; Spieck E.; Wagner M.; Daims H. Nitrotoga -like Bacteria Are Previously Unrecognized Key Nitrite Oxidizers in Full-Scale Wastewater Treatment Plants. ISME J. 2015, 9 (3), 708–720. 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A. M.; Albertsen M.; Vollertsen J.; Nielsen P. H. The Activated Sludge Ecosystem Contains a Core Community of Abundant Organisms. ISME J. 2016, 10 (1), 11–20. 10.1038/ismej.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Duan H.; Yuan Z.; Hu S. A 20-Year Journey of Partial Nitritation and Anammox (PN/A): From Sidestream toward Mainstream. Environ. Sci. Technol. 2022, 56, 7522. 10.1021/acs.est.1c06107. [DOI] [PubMed] [Google Scholar]
- Turk O.; Mavinic D. S. Preliminary Assessment of a Shortcut in Nitrogen Removal from Wastewater. Can. J. Civ. Eng. 1986, 13 (6), 600–605. 10.1139/l86-094. [DOI] [Google Scholar]
- Lackner S.; Gilbert E. M.; Vlaeminck S. E.; Joss A.; Horn H.; van Loosdrecht M. C. M. Full-Scale Partial Nitritation/Anammox Experiences – An Application Survey. Water Res. 2014, 55, 292–303. 10.1016/j.watres.2014.02.032. [DOI] [PubMed] [Google Scholar]
- Zheng M.; Li H.; Duan H.; Liu T.; Wang Z.; Zhao J.; Hu Z.; Watts S.; Meng J.; Liu P.; Rattier M.; Larsen E.; Guo J.; Dwyer J.; Van Den Akker B.; Lloyd J.; Hu S.; Yuan Z. One-year stable pilot-scale operation demonstrates high flexibility of mainstream anammox application. Water Res. X 2023, 19, 100166. 10.1016/j.wroa.2023.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinga C.; Schellen A. A. J. C.; Mulder J. W.; van Loosdrecht M. C. M.; Heijnen J. J. The sharon process: an innovative method for nitrogen removal from ammonium-rich waste water. Water Sci. Technol. 1998, 37 (9), 135–142. 10.2166/wst.1998.0350. [DOI] [Google Scholar]
- Hu Z.; Liu T.; Wang Z.; Meng J.; Zheng M. 2023. Toward energy neutrality: novel wastewater treatment incorporating acidophilic ammonia oxidation. Environ. Sci. Technol. 2023, 57 (11), 4522–4532. 10.1021/acs.est.2c06444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S.; Seuntjens D.; De Cocker P.; Lackner S.; Vlaeminck S. E. Success of Mainstream Partial Nitritation/Anammox Demands Integration of Engineering, Microbiome and Modeling Insights. Curr. Opin. Biotechnol. 2018, 50, 214–221. 10.1016/j.copbio.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Daims H.; Luecker S.; Wagner M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016, 24 (9), 699–712. 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; van Loosdrecht M. C. M.; Daigger G. T. Mainstream Partial Nitritation–Anammox in Municipal Wastewater Treatment: Status, Bottlenecks, and Further Studies. Appl. Microbiol. Biotechnol. 2017, 101 (4), 1365–1383. 10.1007/s00253-016-8058-7. [DOI] [PubMed] [Google Scholar]
- Hug L. A.; Baker B. J.; Anantharaman K.; Brown C. T.; Probst A. J.; Castelle C. J.; Butterfield C. N.; Hernsdorf A. W.; Amano Y.; Ise K.; Suzuki Y.; Dudek N.; Relman D. A.; Finstad K. M.; Amundson R.; Thomas B. C.; Banfield J. F. A New View of the Tree of Life. Nat. Microbiol. 2016, 1 (5), 1–6. 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- Vinogradskij S. N.Recherches sur les organismes de la nitrification; Impr Charaire: 1889. [Google Scholar]
- Watson S. W.; Waterbury J. B. Characteristics of Two Marine Nitrite Oxidizing Bacteria, Nitrospina Gracilis Nov. Gen. Nov. Sp. and Nitrococcus Mobilis Nov. Gen. Nov. Sp.. Arch. Für Mikrobiol. 1971, 77 (3), 203–230. 10.1007/BF00408114. [DOI] [Google Scholar]
- Watson S.; Bock E.; Valois F.; Waterbury J.; Schlosser U. Nitrospira-Marina Gen-Nov Sp-Nov - a Chemolithotrophic Nitrite-Oxidizing Bacterium. Arch. Microbiol. 1986, 144 (1), 1–7. 10.1007/BF00454947. [DOI] [Google Scholar]
- Sorokin D. Y.; Luecker S.; Vejmelkova D.; Kostrikina N. A.; Kleerebezem R.; Rijpstra W. I. C.; Damste J. S. S.; Le Paslier D.; Muyzer G.; Wagner M.; van Loosdrecht M. C. M.; Daims H. Nitrification Expanded: Discovery, Physiology and Genomics of a Nitrite-Oxidizing Bacterium from the Phylum Chloroflexi. Isme J. 2012, 6 (12), 2245–2256. 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel M. A. H. J.; Speth D. R.; Albertsen M.; Nielsen P. H.; Op den Camp H. J. M.; Kartal B.; Jetten M. S. M.; Lücker S. Complete Nitrification by a Single Microorganism. Nature 2015, 528 (7583), 555–559. 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H.; Lebedeva E. V.; Pjevac P.; Han P.; Herbold C.; Albertsen M.; Jehmlich N.; Palatinszky M.; Vierheilig J.; Bulaev A.; Kirkegaard R. H.; von Bergen M.; Rattei T.; Bendinger B.; Nielsen P. H.; Wagner M. Complete Nitrification by Nitrospira Bacteria. Nature 2015, 528 (7583), 504–509. 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi D. K.; Blom J.; Stepanauskas R.; Stingl U. Diversification and Niche Adaptations of Nitrospina-like Bacteria in the Polyextreme Interfaces of Red Sea Brines. Isme J. 2016, 10 (6), 1383–1399. 10.1038/ismej.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieck E.; Spohn M.; Wendt K.; Bock E.; Shively J.; Frank J.; Indenbirken D.; Alawi M.; Lücker S.; Hüpeden J. Extremophilic Nitrite-Oxidizing Chloroflexi from Yellowstone Hot Springs. ISME J. 2020, 14 (2), 364–379. 10.1038/s41396-019-0530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A. J.; Jung M.-Y.; Strachan C. R.; Herbold C. W.; Kirkegaard R. H.; Wagner M.; Daims H. Genomic and Kinetic Analysis of Novel Nitrospinae Enriched by Cell Sorting. ISME J. 2021, 15 (3), 732–745. 10.1038/s41396-020-00809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winogradsky S. Contributions a La Morphologie Des Organisms de La Nitrification. Arkhiv Biol. Nauk St Petersbourg 1892, 1, 87–137. [Google Scholar]
- Bock E. Growth of Nitrobacter in Presence of Organic-Matter 0.2. Chemo-Organotrophic Growth of Nitrobacter-Agilis. Arch. Microbiol. 1976, 108 (3), 305–312. 10.1007/BF00454857. [DOI] [PubMed] [Google Scholar]
- Steinmuller W.; Bock E. Enzymatic Studies on Autotrophically, Mixotrophically and Heterotrophically Grown Nitrobacter-Agilis with Special Reference to Nitrite Oxidase. Arch. Microbiol. 1977, 115 (1), 51–54. 10.1007/BF00427844. [DOI] [PubMed] [Google Scholar]
- Sundermeyer H.; Bock E. Energy-Metabolism of Autotrophically and Heterotrophically Grown Cells of Nitrobacter-Winogradskyi. Arch. Microbiol. 1981, 130 (3), 250–254. 10.1007/BF00459529. [DOI] [Google Scholar]
- Lücker S.; Nowka B.; Rattei T.; Spieck E.; Daims H. The Genome of Nitrospina Gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer. Front. Microbiol. 2013, 4, 27. 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman J. M.; Leilei Shih J.; Popp B. N. Nitrite Oxidation in the Upper Water Column and Oxygen Minimum Zone of the Eastern Tropical North Pacific Ocean. ISME J. 2013, 7 (11), 2192–2205. 10.1038/ismej.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Kop L. F. M.; Lau M. C. Y.; Frank J.; Jayakumar A.; Lücker S.; Ward B. B. Uncultured Nitrospina -like Species Are Major Nitrite Oxidizing Bacteria in Oxygen Minimum Zones. ISME J. 2019, 13 (10), 2391–2402. 10.1038/s41396-019-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzinger K.; Koch H.; Lücker S.; Sedlacek C. J.; Herbold C.; Schwarz J.; Daebeler A.; Mueller A. J.; Lukumbuzya M.; Romano S.; Leisch N.; Karst S. M.; Kirkegaard R.; Albertsen M.; Nielsen P. H.; Wagner M.; Daims H. Characterization of the First “Candidatus Nitrotoga” Isolate Reveals Metabolic Versatility and Separate Evolution of Widespread Nitrite-Oxidizing Bacteria. mBio 2018, 9 (4), e01186–18. 10.1128/mBio.01186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker A. M.; Mosier A. C. Genomic Profiling of Four Cultivated Candidatus Nitrotoga Spp. Predicts Broad Metabolic Potential and Environmental Distribution. Isme J. 2018, 12 (12), 2864–2882. 10.1038/s41396-018-0240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemp J.; Lücker S.; Schott J.; Pace L. A.; Johnson J. E.; Schink B.; Daims H.; Fischer W. W. Genomics of a Phototrophic Nitrite Oxidizer: Insights into the Evolution of Photosynthesis and Nitrification. ISME J. 2016, 10 (11), 2669–2678. 10.1038/ismej.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H.; Lücker S.; Albertsen M.; Kitzinger K.; Herbold C.; Spieck E.; Nielsen P. H.; Wagner M.; Daims H. Expanded Metabolic Versatility of Ubiquitous Nitrite-Oxidizing Bacteria from the Genus Nitrospira. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (36), 11371–11376. 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H.; Galushko A.; Albertsen M.; Schintlmeister A.; Gruber-Dorninger C.; Lücker S.; Pelletier E.; Le Paslier D.; Spieck E.; Richter A.; Nielsen P. H.; Wagner M.; Daims H. Growth of Nitrite-Oxidizing Bacteria by Aerobic Hydrogen Oxidation. Science 2014, 345, 1052–1054. 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- Kitzinger K.; Marchant H. K.; Bristow L. A.; Herbold C. W.; Padilla C. C.; Kidane A. T.; Littmann S.; Daims H.; Pjevac P.; Stewart F. J.; Wagner M.; Kuypers M. M. M. Single Cell Analyses Reveal Contrasting Life Strategies of the Two Main Nitrifiers in the Ocean. Nat. Commun. 2020, 11 (1), 767. 10.1038/s41467-020-14542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatinszky M.; Herbold C.; Jehmlich N.; Pogoda M.; Han P.; von Bergen M.; Lagkouvardos I.; Karst S. M.; Galushko A.; Koch H.; Berry D.; Daims H.; Wagner M. Cyanate as an Energy Source for Nitrifiers. Nature 2015, 524 (7563), 105–108. 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Lomsadze A.; Borodovsky M. Ab Initio Gene Identification in Metagenomic Sequences. Nucleic Acids Res. 2010, 38 (12), e132 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Sato Y.; Kawashima M.; Furumichi M.; Tanabe M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44 (D1), D457–462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y.; Wolf Y. I.; Makarova K. S.; Vera Alvarez R.; Landsman D.; Koonin E. V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49 (D1), D274–D281. 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51 (D1), D523–D531. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M.; Koops H.-P.; Flood J.; Amann R.; Rath G. In Situ Analysis of Nitrifying Bacteria in Sewage Treatment Plants. Water Sci. Technol. 1996, 34 (1-2), 237. 10.2166/wst.1996.0377. [DOI] [Google Scholar]
- Daims H.; Nielsen J. L.; Nielsen P. H.; Schleifer K. H.; Wagner M. In Situ Characterization of Nitrospira-like Nitrite Oxidizing Bacteria Active in Wastewater Treatment Plants. Appl. Environ. Microbiol. 2001, 67 (11), 5273–5284. 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.; Shao M.-F.; Ye L. 454 Pyrosequencing Reveals Bacterial Diversity of Activated Sludge from 14 Sewage Treatment Plants. ISME J. 2012, 6 (6), 1137–1147. 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Ning D.; Zhang B.; Li Y.; Zhang P.; Shan X.; Zhang Q.; Brown M. R.; Li Z.; Van Nostrand J. D.; Ling F.; Xiao N.; Zhang Y.; Vierheilig J.; Wells G. F.; Yang Y.; Deng Y.; Tu Q.; Wang A.; Zhang T.; He Z.; Keller J.; Nielsen P. H.; Alvarez P. J. J.; Criddle C. S.; Wagner M.; Tiedje J. M.; He Q.; Curtis T. P.; Stahl D. A.; Alvarez-Cohen L.; Rittmann B. E.; Wen X.; Zhou J. Global Diversity and Biogeography of Bacterial Communities in Wastewater Treatment Plants. Nat. Microbiol. 2019, 4 (7), 1183–1195. 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- Gruber-Dorninger C.; Pester M.; Kitzinger K.; Savio D. F.; Loy A.; Rattei T.; Wagner M.; Daims H. Functionally Relevant Diversity of Closely Related Nitrospira in Activated Sludge. Isme J. 2015, 9 (3), 643–655. 10.1038/ismej.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangkitjawisut W.; Limpiyakorn T.; Powtongsook S.; Pornkulwat P.; Suwannasilp B. B. Differences in Nitrite-Oxidizing Communities and Kinetics in a Brackish Environment after Enrichment at Low and High Nitrite Concentrations. J. Environ. Sci. 2016, 42, 41–49. 10.1016/j.jes.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Ishii K.; Fujitani H.; Soh K.; Nakagawa T.; Takahashi R.; Tsuneda S. Enrichment and Physiological Characterization of a Cold-Adapted Nitrite-Oxidizing Nitrotoga Sp from an Eelgrass Sediment. Appl. Environ. Microbiol. 2017, 83 (14), e00549–17. 10.1128/AEM.00549-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowka B.; Daims H.; Spieck E. Comparison of Oxidation Kinetics of Nitrite-Oxidizing Bacteria: Nitrite Availability as a Key Factor in Niche Differentiation. Appl. Environ. Microbiol. 2015, 81 (2), 745–753. 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Wang J. Long-Term Low DO Enriches and Shifts Nitrifier Community in Activated Sludge. Environ. Sci. Technol. 2013, 47 (10), 5109–5117. 10.1021/es304647y. [DOI] [PubMed] [Google Scholar]
- Chao Y.; Mao Y.; Yu K.; Zhang T. Novel Nitrifiers and Comammox in a Full-Scale Hybrid Biofilm and Activated Sludge Reactor Revealed by Metagenomic Approach. Appl. Microbiol. Biotechnol. 2016, 100 (18), 8225–8237. 10.1007/s00253-016-7655-9. [DOI] [PubMed] [Google Scholar]
- Spasov E.; Tsuji J. M.; Hug L. A.; Doxey A. C.; Sauder L. A.; Parker W. J.; Neufeld J. D. High Functional Diversity among Nitrospira Populations That Dominate Rotating Biological Contactor Microbial Communities in a Municipal Wastewater Treatment Plant. Isme J. 2020, 14 (7), 1857–1872. 10.1038/s41396-020-0650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto I.; Dai Z.; Huo L.; Anderson C. L.; Vilardi K. J.; Ijaz U.; Khunjar W.; Wilson C.; De Clippeleir H.; Gilmore K.; Bailey E.; Pinto A. J. Long Solids Retention Times and Attached Growth Phase Favor Prevalence of Comammox Bacteria in Nitrogen Removal Systems. Water Res. 2020, 169, 115268 10.1016/j.watres.2019.115268. [DOI] [PubMed] [Google Scholar]
- Huang T.; Xia J.; Liu T.; Su Z.; Guan Y.; Guo J.; Wang C.; Zheng M. Comammox Nitrospira Bacteria Are Dominant Ammonia Oxidizers in Mainstream Nitrification Bioreactors Emended with Sponge Carriers. Environ. Sci. Technol. 2022, 56 (17), 12584–12591. 10.1021/acs.est.2c03641. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Zheng M.; Su Z.; Liu T.; Li J.; Guo J.; Yuan Z.; Hu S. Selective Enrichment of Comammox Nitrospira in a Moving Bed Biofilm Reactor with Sufficient Oxygen Supply. Environ. Sci. Technol. 2022, 56 (18), 13338–13346. 10.1021/acs.est.2c03299. [DOI] [PubMed] [Google Scholar]
- Laanbroek H. J.; Gerards S. Competition for Limiting Amounts of Oxygen between Nitrosomonas Europaea and Nitrobacter Winogradskyi Grown in Mixed Continuous Cultures. Arch. Microbiol. 1993, 159 (5), 453–459. 10.1007/BF00288593. [DOI] [Google Scholar]
- Law Y.; Matysik A.; Chen X.; Thi S. S.; Nguyen T. Q. N.; Qiu G.; Natarajan G.; Williams R. B. H.; Ni B.-J.; Seviour T. W.; Wuertz S. High Dissolved Oxygen Selection against Nitrospira Sublineage I in Full-Scale Activated Sludge. Environ. Sci. Technol. 2019, 53 (14), 8157–8166. 10.1021/acs.est.9b00955. [DOI] [PubMed] [Google Scholar]
- Manser R.; Gujer W.; Siegrist H. Consequences of Mass Transfer Effects on the Kinetics of Nitrifiers. Water Res. 2005, 39 (19), 4633–4642. 10.1016/j.watres.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Picioreanu C.; Pérez J.; van Loosdrecht M. C. M. Impact of Cell Cluster Size on Apparent Half-Saturation Coefficients for Oxygen in Nitrifying Sludge and Biofilms. Water Res. 2016, 106, 371–382. 10.1016/j.watres.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Moussa M. S.; Hooijmans C. M.; Lubberding H. J.; Gijzen H. J.; van Loosdrecht M. C. M. Modelling Nitrification, Heterotrophic Growth and Predation in Activated Sludge. Water Res. 2005, 39 (20), 5080–5098. 10.1016/j.watres.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Lackner S.; Terada A.; Horn H.; Henze M.; Smets B. F. Nitritation Performance in Membrane-Aerated Biofilm Reactors Differs from Conventional Biofilm Systems. Water Res. 2010, 44 (20), 6073–6084. 10.1016/j.watres.2010.07.074. [DOI] [PubMed] [Google Scholar]
- Kits K. D.; Sedlacek C. J.; Lebedeva E. V.; Han P.; Bulaev A.; Pjevac P.; Daebeler A.; Romano S.; Albertsen M.; Stein L. Y.; Daims H.; Wagner M. Kinetic Analysis of a Complete Nitrifier Reveals an Oligotrophic Lifestyle. Nature 2017, 549 (7671), 269–272. 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M. K.; Kapoor V.; Santo-Domingo J.; Chandran K. Comammox Functionality Identified in Diverse Engineered Biological Wastewater Treatment Systems. Environ. Sci. Technol. Lett. 2018, 5 (2), 110–116. 10.1021/acs.estlett.7b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Ma L.; Mao Y.; Jiang X.; Xia Y.; Yu K.; Li B.; Zhang T. Comammox in Drinking Water Systems. Water Res. 2017, 116, 332–341. 10.1016/j.watres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Daims H.; Liu Y.; Herbold C. W.; Pjevac P.; Lin J.-G.; Li M.; Gu J.-D. Activity and Metabolic Versatility of Complete Ammonia Oxidizers in Full-Scale Wastewater Treatment Systems. mBio 2020, 11 (2), 1. 10.1128/mBio.03175-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udert K. M.; Larsen T. A.; Biebow M.; Gujer W. Urea Hydrolysis and Precipitation Dynamics in a Urine-Collecting System. Water Res. 2003, 37 (11), 2571–2582. 10.1016/S0043-1354(03)00065-4. [DOI] [PubMed] [Google Scholar]
- Sakoula D.; Nowka B.; Spieck E.; Daims H.; Lücker S. The Draft Genome Sequence of “Nitrospira Lenta” Strain BS10, a Nitrite Oxidizing Bacterium Isolated from Activated Sludge. Stand. Genomic Sci. 2018, 13 (1), 32. 10.1186/s40793-018-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki N.; Fujitani H.; Shimada Y.; Morohoshi T.; Sekiguchi Y.; Tsuneda S. Genomic Analysis of Two Phylogenetically Distinct Nitrospira Species Reveals Their Genomic Plasticity and Functional Diversity. Front. Microbiol. 2018, 8, 2637. 10.3389/fmicb.2017.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Hua Z.-S.; Liu T.; Wang C.; Li J.; Bai G.; Lücker S.; Jetten M. S. M.; Zheng M.; Guo J. Selective Enrichment and Metagenomic Analysis of Three Novel Comammox Nitrospira in a Urine-Fed Membrane Bioreactor. ISME Commun. 2021, 1 (1), 1–8. 10.1038/s43705-021-00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Sáez L.; Waller A. S.; Mende D. R.; Bakker K.; Farnelid H.; Yager P. L.; Lovejoy C.; Tremblay J.-É.; Potvin M.; Heinrich F.; Estrada M.; Riemann L.; Bork P.; Pedrós-Alió C.; Bertilsson S. Role for Urea in Nitrification by Polar Marine Archaea. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (44), 17989–17994. 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzinger K.; Padilla C. C.; Marchant H. K.; Hach P. F.; Herbold C. W.; Kidane A. T.; Könneke M.; Littmann S.; Mooshammer M.; Niggemann J.; Petrov S.; Richter A.; Stewart F. J.; Wagner M.; Kuypers M. M. M.; Bristow L. A. Cyanate and Urea Are Substrates for Nitrification by Thaumarchaeota in the Marine Environment. Nat. Microbiol. 2019, 4 (2), 234–243. 10.1038/s41564-018-0316-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M.; Stieglmeier M.; Spang A.; Könneke M.; Schintlmeister A.; Urich T.; Engel M.; Schloter M.; Wagner M.; Richter A.; Schleper C. Nitrososphaera Viennensis, an Ammonia Oxidizing Archaeon from Soil. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (20), 8420–8425. 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuter S.; Koch H.; Sass K.; Wegen S.; Lee N.; Lücker S.; Spieck E. Some like It Cold: The Cellular Organization and Physiological Limits of Cold-Tolerant Nitrite-Oxidizing Nitrotoga. Environ. Microbiol. 2022, 24 (4), 2059–2077. 10.1111/1462-2920.15958. [DOI] [PubMed] [Google Scholar]
- Keene N. A.; Reusser S. R.; Scarborough M. J.; Grooms A. L.; Seib M.; Domingo J. S.; Noguera D. R. Pilot Plant Demonstration of Stable and Efficient High Rate Biological Nutrient Removal with Low Dissolved Oxygen Conditions. Water Res. 2017, 121, 72–85. 10.1016/j.watres.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Li S.; Ni G.; Duan H.; Huang X.; Yuan Z.; Zheng M. Temperature Variations Shape Niche Occupation of Nitrotoga-like Bacteria in Activated Sludge. ACS EST Water 2021, 1 (1), 167–174. 10.1021/acsestwater.0c00060. [DOI] [Google Scholar]
- Alawi M.; Off S.; Kaya M.; Spieck E. Temperature Influences the Population Structure of Nitrite-Oxidizing Bacteria in Activated Sludge. Environ. Microbiol. Rep. 2009, 1 (3), 184–190. 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- Wegen S.; Nowka B.; Spieck E. Low Temperature and Neutral PH Define “Candidatus Nitrotoga Sp.” as a Competitive Nitrite Oxidizer in Coculture with Nitrospira Defluvii. Appl. Environ. Microbiol. 2019, 85 (9), e02569-18. 10.1128/AEM.02569-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich S.; Behrens D.; Lebedeva E.; Ludwig W.; Bock E. A New Obligately Chemolithoautotrophic, Nitrite-Oxidizing Bacterium,Nitrospira Moscoviensis Sp. Nov. and Its Phylogenetic Relationship. Arch. Microbiol. 1995, 164 (1), 16–23. 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- Spieck E.; Hartwig C.; McCormack I.; Maixner F.; Wagner M.; Lipski A.; Daims H. Selective Enrichment and Molecular Characterization of a Previously Uncultured Nitrospira-like Bacterium from Activated Sludge. Environ. Microbiol. 2006, 8 (3), 405–415. 10.1111/j.1462-2920.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- Ushiki N.; Fujitani H.; Aoi Y.; Tsuneda S. Isolation of Nitrospira Belonging to Sublineage II from a Wastewater Treatment Plant. Microbes Environ. 2013, 28 (3), 346–353. 10.1264/jsme2.ME13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani H.; Ushiki N.; Tsuneda S.; Aoi Y. Isolation of Sublineage I Nitrospira by a Novel Cultivation Strategy. Environ. Microbiol. 2014, 16 (10), 3030–3040. 10.1111/1462-2920.12248. [DOI] [PubMed] [Google Scholar]
- Jacob J.; Nowka B.; Merten V.; Sanders T.; Spieck E.; Daehnke K. Oxidation Kinetics and Inverse Isotope Effect of Marine Nitrite-Oxidizing Isolates. Aquat. Microb. Ecol. 2017, 80 (3), 289–300. 10.3354/ame01859. [DOI] [Google Scholar]
- Laanbroek H.; Bodelier P.; Gerards S. Oxygen-Consumption Kinetics of Nitrosomonas-Europaea and Nitrobacter-Hamburgensis Grown in Mixed Continuous Cultures at Different Oxygen Concentrations. Arch. Microbiol. 1994, 161 (2), 156–162. 10.1007/BF00276477. [DOI] [Google Scholar]
- Both G. J.; Gerards S.; Laanbroek H. J. Kinetics of Nitrite Oxidation in Two Nitrobacter Species Grown in Nitrite-Limited Chemostats. Arch. Microbiol. 1992, 157 (5), 436–441. 10.1007/BF00249101. [DOI] [Google Scholar]
- Laudelout H.; Vantichelen L. Kinetics of the Nitrite Oxidation by Nitrobacter-Winogradskyi. J. Bacteriol. 1960, 79 (1), 39–42. 10.1128/jb.79.1.39-42.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock E.; Koops H.-P.; Möller U. C.; Rudert M. A New Facultatively Nitrite Oxidizing Bacterium, Nitrobacter Vulgaris Sp. Nov. Arch. Microbiol. 1990, 153 (2), 105–110. 10.1007/BF00247805. [DOI] [Google Scholar]
- Hüpeden J.; Wegen S.; Off S.; Luecker S.; Bedarf Y.; Daims H.; Kuehn C.; Spieck E. Relative Abundance of Nitrotoga Spp. in a Biofilter of a Cold-Freshwater Aquaculture Plant Appears To Be Stimulated by Slightly Acidic PH. Appl. Environ. Microbiol. 2016, 82 (6), 1838–1845. 10.1128/AEM.03163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieck E.; Keuter S.; Wenzel T.; Bock E.; Ludwig W. Characterization of a New Marine Nitrite Oxidizing Bacterium, Nitrospina Watsonii Sp Nov., a Member of the Newly Proposed Phylum “Nitrospinae.. Syst. Appl. Microbiol. 2014, 37 (3), 170–176. 10.1016/j.syapm.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Spieck E.; Sass K.; Keuter S.; Hirschmann S.; Spohn M.; Indenbirken D.; Kop L. F. M.; Luecker S.; Giaveno A. Defining Culture Conditions for the Hidden Nitrite-Oxidizing Bacterium Nitrolancea. Front. Microbiol. 2020, 11, 1522. 10.3389/fmicb.2020.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J.; Peng Y.; Wang B.; Wang S. Achievement of High Nitrite Accumulation via Endogenous Partial Denitrification (EPD). Bioresour. Technol. 2017, 224, 140–146. 10.1016/j.biortech.2016.11.070. [DOI] [PubMed] [Google Scholar]
- Antileo C.; Medina H.; Bornhardt C.; Muñoz C.; Jaramillo Montoya F.; Proal Najera J. B. Actuators Monitoring System for Real-Time Control of Nitrification–Denitrification via Nitrite on Long Term Operation. Chem. Eng. J. 2013, 223, 467–478. 10.1016/j.cej.2013.02.079. [DOI] [Google Scholar]
- Jetten M. S. M.; Horn S. J.; van Loosdrecht M. C. M. Towards a More Sustainable Municipal Wastewater Treatment System. Water Sci. Technol. 1997, 35 (9), 171–180. 10.2166/wst.1997.0341. [DOI] [Google Scholar]
- Wett B. Development and Implementation of a Robust Deammonification Process.. Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 2007, 56 (7), 81–88. 10.2166/wst.2007.611. [DOI] [PubMed] [Google Scholar]
- Li J.; Xu K.; Liu T.; Bai G.; Liu Y.; Wang C.; Zheng M. Achieving Stable Partial Nitritation in an Acidic Nitrifying Bioreactor. Environ. Sci. Technol. 2020, 54 (1), 456–463. 10.1021/acs.est.9b04400. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Ye L.; Jiang G.; Hu S.; Yuan Z. Side-Stream Sludge Treatment Using Free Nitrous Acid Selectively Eliminates Nitrite Oxidizing Bacteria and Achieves the Nitrite Pathway. Water Res. 2014, 55, 245–255. 10.1016/j.watres.2014.02.029. [DOI] [PubMed] [Google Scholar]
- Duan H.; Ye L.; Lu X.; Yuan Z. Overcoming Nitrite Oxidizing Bacteria Adaptation through Alternating Sludge Treatment with Free Nitrous Acid and Free Ammonia. Environ. Sci. Technol. 2019, 53 (4), 1937–1946. 10.1021/acs.est.8b06148. [DOI] [PubMed] [Google Scholar]
- Wang D.; Wang Q.; Laloo A.; Xu Y.; Bond P. L.; Yuan Z. Achieving Stable Nitritation for Mainstream Deammonification by Combining Free Nitrous Acid-Based Sludge Treatment and Oxygen Limitation. Sci. Rep. 2016, 6, 25547. 10.1038/srep25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado Vela J.; Dick G. J.; Love N. G. Sulfide Inhibition of Nitrite Oxidation in Activated Sludge Depends on Microbial Community Composition. Water Res. 2018, 138, 241–249. 10.1016/j.watres.2018.03.047. [DOI] [PubMed] [Google Scholar]
- Huang S.; Zhu Y.; Lian J.; Liu Z.; Zhang L.; Tian S. Enhancement in the Partial Nitrification of Wastewater Sludge via Lowintensity Ultrasound: Effects on Rapid Start-up and Temperature Resilience. Bioresour. Technol. 2019, 294, 122196 10.1016/j.biortech.2019.122196. [DOI] [PubMed] [Google Scholar]
- Huang S.; Zhu Y.; Zhang G.; Lian J.; Liu Z.; Zhang L.; Tian S. Effects of Low-Intensity Ultrasound on Nitrite Accumulation and Microbial Characteristics during Partial Nitrification. Sci. Total Environ. 2020, 705, 135985 10.1016/j.scitotenv.2019.135985. [DOI] [PubMed] [Google Scholar]
- Wang L.; Qiu S.; Guo J.; Ge S. Light Irradiation Enables Rapid Start-Up of Nitritation through Suppressing NxrB Gene Expression and Stimulating Ammonia-Oxidizing Bacteria. Environ. Sci. Technol. 2021, 55 (19), 13297–13305. 10.1021/acs.est.1c04174. [DOI] [PubMed] [Google Scholar]
- Duan H.; Wang Q.; Erler D. V.; Ye L.; Yuan Z. Effects of Free Nitrous Acid Treatment Conditions on the Nitrite Pathway Performance in Mainstream Wastewater Treatment. Sci. Total Environ. 2018, 644, 360–370. 10.1016/j.scitotenv.2018.06.346. [DOI] [PubMed] [Google Scholar]
- Li X.; Sun S.; Badgley B. D.; Sung S.; Zhang H.; He Z. Nitrogen Removal by Granular Nitritation–Anammox in an Upflow Membrane-Aerated Biofilm Reactor. Water Res. 2016, 94, 23–31. 10.1016/j.watres.2016.02.031. [DOI] [PubMed] [Google Scholar]
- Laureni M.; Falås P.; Robin O.; Wick A.; Weissbrodt D. G.; Nielsen J. L.; Ternes T. A.; Morgenroth E.; Joss A. Mainstream Partial Nitritation and Anammox: Long-Term Process Stability and Effluent Quality at Low Temperatures. Water Res. 2016, 101, 628–639. 10.1016/j.watres.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Peng Y.; Zhang L.; Liu J.; Wang X.; Gao R.; Pang L.; Zhou Y. Quantify the Contribution of Anammox for Enhanced Nitrogen Removal through Metagenomic Analysis and Mass Balance in an Anoxic Moving Bed Biofilm Reactor. Water Res. 2019, 160, 178–187. 10.1016/j.watres.2019.05.070. [DOI] [PubMed] [Google Scholar]
- Zheng M.; Li S.; Ni G.; Xia J.; Hu S.; Yuan Z.; Liu Y.; Huang X. Critical Factors Facilitating Candidatus Nitrotoga To Be Prevalent Nitrite-Oxidizing Bacteria in Activated Sludge. Environ. Sci. Technol. 2020, 54, 15414. 10.1021/acs.est.0c04192. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Hu Z.; Duan H.; De Clippeleir H.; Al-Omari A.; Hu S.; Yuan Z. Unravelling Adaptation of Nitrite-Oxidizing Bacteria in Mainstream PN/A Process: Mechanisms and Counter-Strategies. Water Res. 2021, 200, 117239 10.1016/j.watres.2021.117239. [DOI] [PubMed] [Google Scholar]
- Ma B.; Yang L.; Wang Q.; Yuan Z.; Wang Y.; Peng Y. Inactivation and Adaptation of Ammonia-Oxidizing Bacteria and Nitrite-Oxidizing Bacteria When Exposed to Free Nitrous Acid. Bioresour. Technol. 2017, 245, 1266–1270. 10.1016/j.biortech.2017.08.074. [DOI] [PubMed] [Google Scholar]
- Ma Y.; Domingo-Félez C.; Plósz B. Gy.; Smets B. F. Intermittent Aeration Suppresses Nitrite-Oxidizing Bacteria in Membrane-Aerated Biofilms: A Model-Based Explanation. Environ. Sci. Technol. 2017, 51 (11), 6146–6155. 10.1021/acs.est.7b00463. [DOI] [PubMed] [Google Scholar]
- Ge S.; Peng Y.; Qiu S.; Zhu A.; Ren N. Complete Nitrogen Removal from Municipal Wastewater via Partial Nitrification by Appropriately Alternating Anoxic/Aerobic Conditions in a Continuous Plug-Flow Step Feed Process. Water Res. 2014, 55, 95–105. 10.1016/j.watres.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Pellicer-Nàcher C.; Sun S.; Lackner S.; Terada A.; Schreiber F.; Zhou Q.; Smets B. F. Sequential Aeration of Membrane-Aerated Biofilm Reactors for High-Rate Autotrophic Nitrogen Removal: Experimental Demonstration. Environ. Sci. Technol. 2010, 44 (19), 7628–7634. 10.1021/es1013467. [DOI] [PubMed] [Google Scholar]
- Liu T.; Hu S.; Yuan Z.; Guo J. High-Level Nitrogen Removal by Simultaneous Partial Nitritation, Anammox and Nitrite/Nitrate-Dependent Anaerobic Methane Oxidation. Water Res. 2019, 166, 115057 10.1016/j.watres.2019.115057. [DOI] [PubMed] [Google Scholar]
- Gong X.; Garcia-Robledo E.; Schramm A.; Revsbech N. P. Respiratory Kinetics of Marine Bacteria Exposed to Decreasing Oxygen Concentrations. Appl. Environ. Microbiol. 2016, 82 (5), 1412–1422. 10.1128/AEM.03669-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher R. S.; Watmough N. J. The Bacterial Cytochrome Cbb3 Oxidases. Biochim. Biophys. Acta BBA - Bioenerg. 2004, 1655, 388–399. 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Ushiki N.; Jinno M.; Fujitani H.; Suenaga T.; Terada A.; Tsuneda S. Nitrite Oxidation Kinetics of Two Nitrospira Strains: The Quest for Competition and Ecological Niche Differentiation. J. Biosci. Bioeng. 2017, 123 (5), 581–589. 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- Calhoun M. W.; Thomas J. W.; Gennis R. B. The Cytochrome Oxidase Superfamily of Redox-Driven Proton Pumps. Trends Biochem. Sci. 1994, 19 (8), 325–330. 10.1016/0968-0004(94)90071-X. [DOI] [PubMed] [Google Scholar]
- Starkenburg S. R.; Larimer F. W.; Stein L. Y.; Klotz M. G.; Chain P. S. G.; Sayavedra-Soto L. A.; Poret-Peterson A. T.; Gentry M. E.; Arp D. J.; Ward B.; Bottomley P. J. Complete Genome Sequence of Nitrobacter Hamburgensis X14 and Comparative Genomic Analysis of Species within the Genus Nitrobactei. Appl. Environ. Microbiol. 2008, 74 (9), 2852–2863. 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov V. B.; Gennis R. B.; Hemp J.; Verkhovsky M. I. The Cytochrome Bd Respiratory Oxygen Reductases. Biochim. Biophys. Acta-Bioenerg. 2011, 1807 (11), 1398–1413. 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundinger A. B.; Lawson C. E.; Jetten M. S. M.; Koch H.; Lücker S. Cultivation and Transcriptional Analysis of a Canonical Nitrospira Under Stable Growth Conditions. Front. Microbiol. 2019, 10, 1. 10.3389/fmicb.2019.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-D.; Noguera D. R. Nitrospira Community Composition in Nitrifying Reactors Operated with Two Different Dissolved Oxygen Levels. J. Microbiol. Biotechnol. 2008, 18 (8), 1470–1474. [PubMed] [Google Scholar]
- Roots P.; Wang Y.; Rosenthal A. F.; Griffin J. S.; Sabba F.; Petrovich M.; Yang F.; Kozak J. A.; Zhang H.; Wells G. F. Comammox Nitrospira Are the Dominant Ammonia Oxidizers in a Mainstream Low Dissolved Oxygen Nitrification Reactor. Water Res. 2019, 157, 396–405. 10.1016/j.watres.2019.03.060. [DOI] [PubMed] [Google Scholar]
- Shao Y.-H.; Wu J.-H. Comammox Nitrospira Species Dominate in an Efficient Partial Nitrification–Anammox Bioreactor for Treating Ammonium at Low Loadings. Environ. Sci. Technol. 2021, 55 (3), 2087–2098. 10.1021/acs.est.0c05777. [DOI] [PubMed] [Google Scholar]
- Lücker S.; Wagner M.; Maixner F.; Pelletier E.; Koch H.; Vacherie B.; Rattei T.; Damste J. S. S.; Spieck E.; Le Paslier D.; Daims H. A Nitrospira Metagenome Illuminates the Physiology and Evolution of Globally Important Nitrite-Oxidizing Bacteria. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (30), 13479–13484. 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincke M.; Bock E.; Kastrau D.; Kroneck P. Nitrite Oxidoreductase from Nitrobacter-Hamburgensis - Redox Centers and Their Catalytic Role. Arch. Microbiol. 1992, 158 (2), 127–131. 10.1007/BF00245215. [DOI] [Google Scholar]
- Spieck E.; Aamand J.; Bartosch S.; Bock E. Immunocytochemical Detection and Location of the Membrane-Bound Nitrite Oxidoreductase in Cells of Nitrobacter and Nitrospira. Fems Microbiol. Lett. 1996, 139 (1), 71–76. 10.1111/j.1574-6968.1996.tb08181.x. [DOI] [Google Scholar]
- Duan H.; Gao S.; Li X.; Ab Hamid N. H.; Jiang G.; Zheng M.; Bai X.; Bond P. L.; Lu X.; Chislett M. M.; Hu S.; Ye L.; Yuan Z. Improving Wastewater Management Using Free Nitrous Acid (FNA). Water Res. 2020, 171, 115382 10.1016/j.watres.2019.115382. [DOI] [PubMed] [Google Scholar]
- Li S.; Duan H.; Zhang Y.; Huang X.; Yuan Z.; Liu Y.; Zheng M. Adaptation of Nitrifying Community in Activated Sludge to Free Ammonia Inhibition and Inactivation. Sci. Total Environ. 2020, 728, 138713 10.1016/j.scitotenv.2020.138713. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Xue Y.; Xia J.; Zhong H.; Ni G.; Liu Y.; Yuan Z.; Hu S. Free Ammonia Shock Treatment Eliminates Nitrite-Oxidizing Bacterial Activity for Mainstream Biofilm Nitritation Process. Chem. Eng. J. 2020, 393, 124682 10.1016/j.cej.2020.124682. [DOI] [Google Scholar]
- Zheng M.; Wang Z.; Meng J.; Hu Z.; Liu Y.; Yuan Z.; Hu S. Inactivation Kinetics of Nitrite-Oxidizing Bacteria by Free Nitrous Acid. Sci. Total Environ. 2021, 752, 141876 10.1016/j.scitotenv.2020.141876. [DOI] [PubMed] [Google Scholar]
- Anthonisen A. C.; Loehr R. C.; Prakasam T. B. S.; Srinath E. G. Inhibition of Nitrification by Ammonia and Nitrous Acid. J. Water Pollut. Control Fed. 1976, 48 (5), 835–852. [PubMed] [Google Scholar]
- Liu Y.; Ngo H. H.; Guo W.; Peng L.; Wang D.; Ni B. The Roles of Free Ammonia (FA) in Biological Wastewater Treatment Processes: A Review. Environ. Int. 2019, 123, 10–19. 10.1016/j.envint.2018.11.039. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Meng J.; Hu Z.; Ni G.; Guerrero Calderon A.; Li H.; De Clippeleir H.; Al-Omari A.; Hu S.; Yuan Z. Robust Nitritation Sustained by Acid-Tolerant Ammonia-Oxidizing Bacteria. Environ. Sci. Technol. 2021, 55 (3), 2048–2056. 10.1021/acs.est.0c05181. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Duan H.; Ni G.; Yu W.; Liu Y.; Yuan Z.; Hu S. Acidic Aerobic Digestion of Anaerobically-Digested Sludge Enabled by a Novel Ammonia-Oxidizing Bacterium. Water Res. 2021, 194, 116962 10.1016/j.watres.2021.116962. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Ni G.; Maulani N.; Xia J.; De Clippeleir H.; Hu S.; Yuan Z.; Zheng M. Stoichiometric and Kinetic Characterization of an Acid-Tolerant Ammonia Oxidizer ‘Candidatus Nitrosoglobus.’. Water Res. 2021, 196, 117026 10.1016/j.watres.2021.117026. [DOI] [PubMed] [Google Scholar]
- Meng J.; Hu Z.; Wang Z.; Hu S.; Liu Y.; Guo H.; Li J.; Yuan Z.; Zheng M. Determining Factors for Nitrite Accumulation in an Acidic Nitrifying System: Influent Ammonium Concentration, Operational PH, and Ammonia-Oxidizing Community. Environ. Sci. Technol. 2022, 56 (16), 11578–11588. 10.1021/acs.est.1c07522. [DOI] [PubMed] [Google Scholar]
- Duan H.; Watts S.; Zheng M.; Wang Z.; Zhao J.; Li H.; Liu P.; Dwyer J.; McPhee P.; Rattier M.; Larsen E.; Yuan Z.; Hu S. Achieving Robust Mainstream Nitrite Shunt at Pilot-Scale with Integrated Sidestream Sludge Treatment and Step-Feed. Water Res. 2022, 223, 119034 10.1016/j.watres.2022.119034. [DOI] [PubMed] [Google Scholar]
- Yu H.; Tian Z.; Zuo J.; Song Y. Enhanced Nitrite Accumulation under Mainstream Conditions by a Combination of Free Ammonia-Based Sludge Treatment and Low Dissolved Oxygen: Reactor Performance and Microbiome Analysis. RSC Adv. 2020, 10 (4), 2049–2059. 10.1039/C9RA07628J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Wang Z.; Wang S.; Qiao X.; Gong X.; Gong Q.; Liu X.; Peng Y. Recovering Partial Nitritation in a PN/A System during Mainstream Wastewater Treatment by Reviving AOB Activity after Thoroughly Inhibiting AOB and NOB with Free Nitrous Acid. Environ. Int. 2020, 139, 105684 10.1016/j.envint.2020.105684. [DOI] [PubMed] [Google Scholar]
- Gottshall E. Y.; Bryson S. J.; Cogert K. I.; Landreau M.; Sedlacek C. J.; Stahl D. A.; Daims H.; Winkler M. Sustained Nitrogen Loss in a Symbiotic Association of Comammox Nitrospira and Anammox Bacteria. Water Res. 2021, 202, 117426 10.1016/j.watres.2021.117426. [DOI] [PubMed] [Google Scholar]
- Kits K. D.; Jung M.-Y.; Vierheilig J.; Pjevac P.; Sedlacek C. J.; Liu S.; Herbold C.; Stein L. Y.; Richter A.; Wissel H.; Brueggemann N.; Wagner M.; Daims H. Low Yield and Abiotic Origin of N2O Formed by the Complete Nitrifier Nitrospira Inopinata. Nat. Commun. 2019, 10, 1836. 10.1038/s41467-019-09790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Zheng M.; Hu Z.; Duan H.; De Clippeleir H.; Al-Omari A.; Hu S.; Yuan Z. Unravelling Adaptation of Nitrite-Oxidizing Bacteria in Mainstream PN/A Process: Mechanisms and Counter-Strategies. Water Res. 2021, 200, 117239 10.1016/j.watres.2021.117239. [DOI] [PubMed] [Google Scholar]
- Du R.; Peng Y.; Cao S.; Wang S.; Wu C. Advanced Nitrogen Removal from Wastewater by Combining Anammox with Partial Denitrification. Bioresour. Technol. 2015, 179, 497–504. 10.1016/j.biortech.2014.12.043. [DOI] [PubMed] [Google Scholar]
- Cao S.; Du R.; Peng Y.; Li B.; Wang S. Novel Two Stage Partial Denitrification (PD)-Anammox Process for Tertiary Nitrogen Removal from Low Carbon/Nitrogen (C/N) Municipal Sewage. Chem. Eng. J. 2019, 362, 107–115. 10.1016/j.cej.2018.12.160. [DOI] [Google Scholar]
- Du R.; Peng Y.; Ji J.; Shi L.; Gao R.; Li X. Partial Denitrification Providing Nitrite: Opportunities of Extending Application for Anammox. Environ. Int. 2019, 131, 105001 10.1016/j.envint.2019.105001. [DOI] [PubMed] [Google Scholar]