ABSTRACT
The cellular processes that support human coronavirus replication and contribute to the pathogenesis of severe disease remain incompletely understood. Many viruses, including coronaviruses, cause endoplasmic reticulum (ER) stress during infection. IRE1α is a component of the cellular response to ER stress that initiates non-conventional splicing of XBP1 mRNA. Spliced XBP1 encodes a transcription factor that induces the expression of ER-related targets. Activation of the IRE1α–XBP1 pathway occurs in association with risk factors for severe human coronavirus infection. In this study, we found that the human coronaviruses HCoV-OC43 (human coronavirus OC43) and SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) both robustly activate the IRE1α–XBP1 branch of the unfolded protein response in cultured cells. Using IRE1α nuclease inhibitors and genetic knockdown of IRE1α and XBP1, we found that these host factors are required for optimal replication of both viruses. Our data suggest that IRE1α supports infection downstream of initial viral attachment and entry. In addition, we found that ER stress–inducing conditions are sufficient to enhance human coronavirus replication. Furthermore, we found markedly increased XBP1 in circulation in human patients with severe coronavirus disease 2019 (COVID-19). Together, these results demonstrate the importance of IRE1α and XBP1 for human coronavirus infection.
IMPORTANCE
There is a critical need to understand the cellular processes co-opted during human coronavirus replication, with an emphasis on identifying mechanisms underlying severe disease and potential therapeutic targets. Here, we demonstrate that the host proteins IRE1α and XBP1 are required for robust infection by the human coronaviruses, SARS-CoV-2 and HCoV-OC43. IRE1α and XBP1 participate in the cellular response to ER stress and are activated during conditions that predispose to severe COVID-19. We found enhanced viral replication with exogenous IRE1α activation, and evidence that this pathway is activated in humans during severe COVID-19. Together, these results demonstrate the importance of IRE1α and XBP1 for human coronavirus infection.
KEYWORDS: coronavirus, unfolded protein response, ER stress, SARS-CoV-2, HCoV-OC43, endoplasmic reticulum, IRE1α, XBP1
INTRODUCTION
Coronaviruses are a family of enveloped, positive-stranded RNA viruses, including the recently emergent, currently pandemic, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Coronavirus replication utilizes incompletely understood host cell pathways and relies on intracellular membranes derived from the endoplasmic reticulum (ER) (1). Disruption of the normal ER environment causes a state termed ER stress, which is detected by the cellular unfolded protein response (2). Inositol-requiring enzyme 1α (IRE1α) is a component of the unfolded protein response and ER-resident transmembrane protein that oligomerizes and autophosphorylates during ER stress (3, 4). This activates its cytosolic RNase domain to initiate non-conventional splicing of the mRNA encoding X-box binding protein-1 (XBP1). Spliced XBP1 mRNA is a specific product of activated IRE1α and, when translated, encodes a transcription factor that upregulates genes, including those involved in ER function (5, 6). IRE1α also targets other specific RNAs, leading to their degradation in a process termed regulated IRE1-dependent decay (RIDD) (7). IRE1α activation occurs in association with risk factors for severe SARS-CoV-2 infection, including diabetes, hypertension, aging, and obesity (8 - 10).
Some coronaviruses manipulate IRE1α for their own benefit. Transmissible gastroenteritis virus (TGEV) is a porcine alphacoronavirus that activates IRE1α, leading to the induction of XBP1 targets (11). In TGEV-infected cells, IRE1α also cleaves a microRNA that regulates type I interferon receptor signaling, leading to the evasion of interferon responses. IRE1α activation and XBP1 target gene induction also occur during infection with the avian gammacoronavirus and infectious bronchitis virus (IBV) (12). Similar to our findings with hepatitis C virus (13), IRE1α plays an anti-apoptotic role during IBV infection (12). Murine hepatitis virus is a betacoronavirus that activates IRE1α (14, 15). However, XBP1 target genes are not induced (14) or only slightly elevated (16). Unlike these non-human pathogens, SARS coronavirus, a human betacoronavirus closely related to SARS-CoV-2, limits IRE1α activation and XBP1 splicing in infected cells (17, 18).
In this study, we examined the role of IRE1α and XBP1 during infection with the human betacoronaviruses, HCoV-OC43 and SARS-CoV-2. We found that IRE1α is activated and promotes infection via XBP1 during infection of cultured cells with both viruses. We demonstrated that pre-existing ER stress and activation of IRE1α resulted in heightened human coronavirus infection, and elevated XBP1(S) protein is present in samples from patients with severe COVID-19. Together, these findings reveal that IRE1α and XBP1 are cellular host factors that promote the replication of the human coronaviruses HCoV-OC43 and SARS-CoV-2.
RESULTS
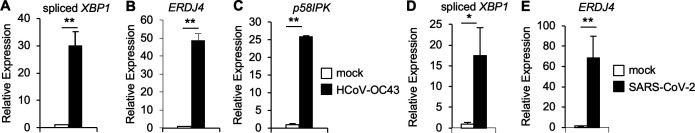
Human coronaviruses activate IRE1α and induce XBP1 targets
The human betacoronavirus SARS-CoV inhibits IRE1α activation resulting in minimal XBP1 splicing during infection (17, 18). To determine whether other human betacoronaviruses similarly limit the activation of the IRE1α branch of the unfolded protein response, we infected human HCT-8 epithelial cells with HCoV-OC43 and assessed XBP1 mRNA splicing using qRT-PCR with the primers specific for spliced XBP1 mRNA. We found that HCoV-OC43 infection stimulated robust XBP1 splicing in this system (Fig. 1A), consistent with findings in infected neurons (19). The model betacoronavirus murine hepatitis virus activates IRE1α, but XBP1 target genes are not induced (14) or only slightly elevated (16). Induction of ERDJ4 and P58IPK requires XBP1 (20, 21), and we found strong expression of these XBP1-responsive genes in HCoV-OC43-infected cells (Fig. 1B and C).
Fig 1.
Human coronavirus infection activates IRE1α and induces XBP1 targets. (A–C) HCT-8 cells were infected with HCoV-OC43 for 48 hours. (D+E) Calu-3 cells were infected with SARS-CoV-2 for 48 hours. The relative abundance of spliced XBP1 (A+D), ERDJ4 (B+E), and p58IPK (C) were determined by quantitative RT-PCR. Data are means ± SD of four replicates and are representative of three independent experiments. *P < 0.05, **P < 0.01 by unpaired t-test.
In some settings, IRE1α’s RNase domain also degrades host RNAs including BLOC1S1, SCARA3, COL6A1, and HGSNAT through the process known as RIDD (22). Consistent with observations in other cell types (23), these RIDD targets were reduced in HCT-8 cells treated with the ER stress–inducing agent, tunicamycin (Fig. S1A). However, BLOC1S1, SCARA3, COL6A1, and HGSNAT were unaffected by HCoV-OC43 (Fig. S1B), suggesting that IRE1α activity is limited to XBP1 splicing during infection.
To determine whether SARS-CoV-2 similarly activates IRE1α and induces XBP1-dependent gene expression, we infected human Calu-3 lung epithelial cells with the clinically derived USA-WA1/2020 SARS-CoV-2 strain. SARS-CoV-2 infection stimulated XBP1 splicing (Fig. 1D) and expression of the XBP1-responsive gene, ERDJ4 (Fig. 1E). However, the RIDD targets BLOC1S1, SCARA3, COL6A1, and HGSNAT were not reduced (Fig. S1C), similar to our findings with HCoV-OC43 infection. Together these results indicate that the human betacoronaviruses SARS-CoV-2 and HCoV-OC43 both activate IRE1α to splice XBP1 mRNA, leading to the expression of XBP1-responsive genes, but RIDD targets are not degraded.
IRE1α is required for optimal HCoV-OC43 infection
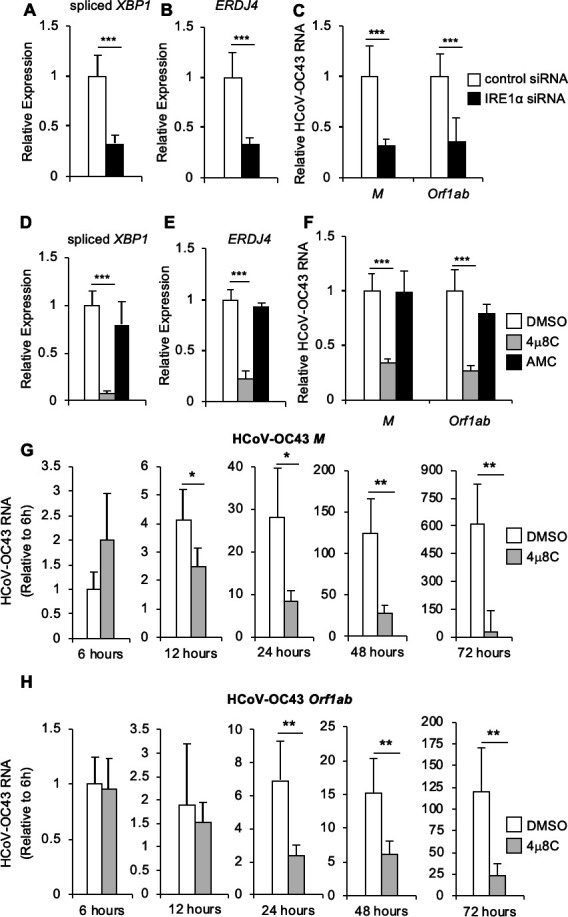
IRE1α promotes infection by a number of RNA viruses, including hepatitis C (13), Zika (20), and influenza A (24). To determine the role of this host factor during HCoV-OC43 infection, we used small-interfering RNA (siRNA) to knock down IRE1α in HCT-8 cells. As a functional control for IRE1α inactivation, we assessed XBP1 mRNA splicing (Fig. 2A) and sXBP1 protein (Fig. S2A and B) and found both were reduced in cells treated with siRNA targeting IRE1α. Consistently, XBP1-dependent ERDJ4 expression in HCoV-OC43-infected cells was also reduced by siRNA targeting IRE1α (Fig. 2B). We then assessed the effect of IRE1α knockdown on viral replication and found reduced HCoV-OC43 RNA, using qRT-PCR with two different primer sets to quantify viral RNA (Fig. 2C). These results suggest that HCoV-OC43 requires IRE1α for efficient viral infection.
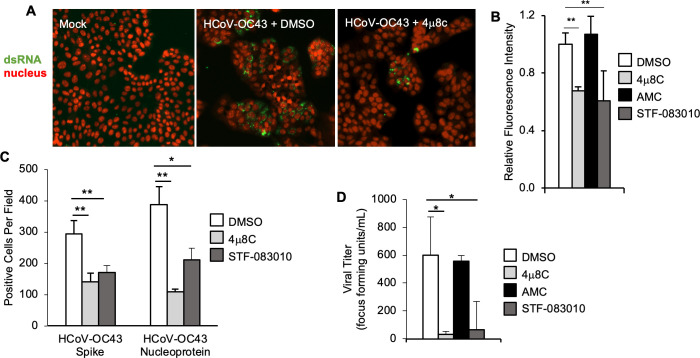
Fig 2.
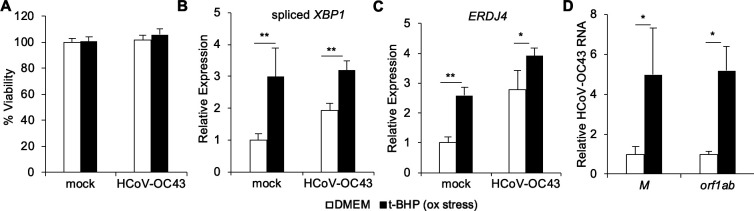
IRE1α is required for optimal HCoV-OC43 infection. (A–C) HCT-8 cells were transfected with siRNA targeting IRE1α or non-targeting control siRNA and then infected with HCoV-OC43 at an MOI of 0.01. RNA was harvested 48-hour post-infection, and the relative abundance of spliced XBP1 (A), ERDJ4 (B), and HCoV-OC43 viral RNA (C) were determined by quantitative RT-PCR. (D–H) HCT-8 cells were treated with IRE1α nuclease inhibitor 4μ8C, structurally similar negative control AMC or DMSO solvent control prior to infection with HCoV-OC43 at an MOI of 0.01 (D–F) or an MOI of 1 (G–H). RNA was harvested 48-hour post-infection (D–F) or at the indicated time point (G and H), and the relative abundance of spliced XBP1 (D), ERDJ4 (E), and HCoV-OC43 viral RNA (F–H) was determined by quantitative RT-PCR. HCoV-OC43 viral RNA abundance at the indicated time points (G and H) is calculated relative to the first time point. Data are means ± SD of six (A–C) or four (D–H) replicates and are representative of three (A–F) and two (G and H) independent experiments, respectively. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t-test.
IRE1α affects multiple cellular pathways via its kinase and nuclease functions (2). To determine whether IRE1α’s RNase activity promotes HCoV-OC43 infection, we treated cells with the selective IRE1α nuclease inhibitor, 4μ8C (25) and structurally similar inactive control molecule AMC (13, 20). We found that 4μ8C, but not the inactive control AMC, prevented XBP1 mRNA splicing (Fig. 2D), sXBP1 protein expression (Fig. S2C), and induction of the XBP1 target ERDJ4 (Fig. 2E) verifying its effect on IRE1α. Inhibiting IRE1α’s nuclease activity with 4μ8C significantly reduced the abundance of HCoV-OC43 viral RNA (Fig. 2F), indicating that the RNase activity is required for optimal infection. We then tested a structurally distinct IRE1α nuclease inhibitor, STF-083010 (26), which also inhibited XBP1 mRNA splicing (Fig. S3A), sXBP1 protein expression (Fig. S2C), ERDJ4 induction (Fig. S3B), and reduced HCoV-OC43 viral RNA (Fig. S3C).
These experiments used a low multiplicity of infection (MOI) of 0.01, so to further characterize the requirement for IRE1α throughout a high MOI infection, we performed a time course experiment with an MOI of 1. We found that at early time points during HCoV-OC43 infection, there was no significant decrease in viral RNA in the presence of IRE1α inhibitor compared to dimethylsulfoxide (DMSO) control (Fig. 2G and H). However, at 24-hour post-infection and later time points, viral RNA was significantly reduced with IRE1α inhibition (Fig. 2G and H), similar to our findings from the low MOI infection model.
To assess viral RNA using an orthogonal approach, we performed immunostaining for the viral replication intermediate, double-stranded RNA (dsRNA) (Fig. 3A). We found that the selective IRE1α nuclease inhibitors 4μ8C and STF-083010 both reduced dsRNA staining (Fig. 3A and B). We next used immunostaining to assess viral spike and nucleoprotein production (Fig. S4A). IRE1α nuclease inhibition with 4μ8C and STF-083010 also significantly reduced viral protein, compared to DMSO solvent control (Fig. 3C; Fig. S4A). Finally, we collected supernatant from infected cells and determined infectious viral titer using an endpoint focus-forming assay. We found that both IRE1α inhibitors, but not the inactive AMC control, resulted in a significant reduction in infectious HCoV-OC43 viral titer (Fig. 3D). Together, these findings demonstrate that HCoV-OC43 infection requires IRE1α RNase activity for optimal replication.
Fig 3.
IRE1α is required for optimal HCoV-OC43 infection assessed by immunofluorescence and titer. (A–D) HCT-8 cells were treated with IRE1α nuclease inhibitor 4μ8C, structurally similar negative control AMC, IRE1α nuclease inhibitor STF-083010 or DMSO solvent control prior to infection with HCoV-OC43. (A–C) Cells were fixed 48-hour post-infection. (A) Double-stranded RNA (dsRNA, green) was visualized by immunostaining, and nuclei were counterstained with TO-PRO-3 (red). (B) Relative total fluorescence intensity was calculated for dsRNA. (C) HCoV-OC43 viral proteins spike and nucleoprotein were visualized by immunostaining, and positive cells were quantified per field at 10× magnification. (D) Viral supernatant was harvested 48-hour post-infection, then diluted serially and plated on HCT-8 cells for an endpoint focus-forming assay. Data are means ± SD of six (C) or four (C+D) replicates and are representative of three (A+B) and two (C+D) independent experiments, respectively. *P < 0.05, **P < 0.01 by unpaired t-test.
We previously found that IRE1α promotes the replication of hepatitis C virus by blocking apoptosis in infected cells (13). Inhibition of IRE1α sensitized HCV-infected cells to death, limiting viral replication at the later stages of infection. IRE1α also plays an anti-apoptotic role during infection with IBV (12). Based on these findings, we hypothesized that inhibiting IRE1α may sensitize HCoV-OC43-infected cells to die, thus terminating viral replication. To test this hypothesis, we assessed viability by measuring cellular ATP (Fig. S4B). Neither 4μ8C nor STF-083010 was cytotoxic alone. In contrast to our prediction, we found that neither IRE1α inhibitor sensitized HCoV-OC43-infected cells to die. These data suggest that this virus does not require IRE1α to maintain host cell viability and uses IRE1α for another aspect of its life cycle.
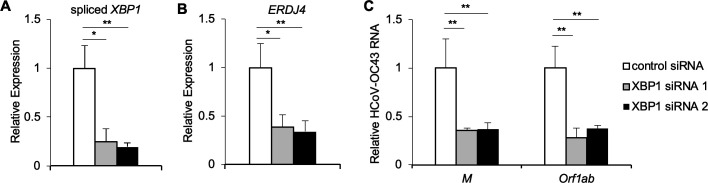
XBP1 is required for optimal HCoV-OC43 infection
The inhibitors 4μ8c and STF-083010 are selective for the IRE1α nuclease domain and have no effect on the kinase activity of IRE1α (27). Our results demonstrate XBP1 target induction during HCoV-OC43 infection (Fig. 1B and C), suggesting that the requirement for IRE1α could be via XBP1 splicing and transcriptional induction of XBP1 targets. To test the hypothesis that IRE1α promotes HCoV-OC43 infection via XBP1, we knocked down XBP1 using siRNA. We found a significant reduction in spliced XBP1 mRNA (Fig. 4A), sXBP1 protein (Fig. S2A and B), and expression of the XBP1-induced target ERDJ4 (Fig. 4B) in cells treated with two independent siRNAs targeting XBP1. We additionally found that knocking down XBP1 reduced HCoV-OC43 RNA abundance (Fig. 4C), indicating that the requirement for IRE1α in HCoV-OC43 infection is XBP1-dependent.
Fig 4.
XBP1 is required for optimal HCoV-OC43 infection. HCT-8 cells were transfected with siRNA targeting XBP1 or non-targeting control siRNA and then infected with HCoV-OC43. RNA was harvested 48-hour post-infection, and the relative abundance of spliced XBP1 (A), ERDJ4 (B), and HCoV-OC43 viral RNA (C) was determined by quantitative RT-PCR. Data are means ± SD of six replicates and are representative of three independent experiments. *P < 0.01, **P < 0.005 by unpaired t-test.
IRE1α and XBP1 promote SARS-CoV-2 infection and inflammatory cytokine responses independently of viral entry
In addition to HCoV-OC43, we found that SARS-CoV-2 stimulates robust IRE1α activation with XBP1 splicing and induction of XBP1 target genes (Fig. 1D and E). To determine whether the requirement for IRE1α is shared by this highly pathogenic human coronavirus, we tested the IRE1α nuclease inhibitor, 4μ8C. Inhibiting IRE1α with 4μ8C reduced the abundance of viral RNA by almost 2-logs at 48-hour post-infection, whereas the inactive control had no effect (Fig. 5A). Similarly, to our results with HCoV-OC43, we found that the structurally distinct IRE1α nuclease inhibitor, STF-083010 also reduced viral RNA at this time point, but viral RNA was unaffected by either inhibitor at an earlier time point (Fig. S5A) in a low MOI infection model. To determine whether IRE1α also supports viral replication during a high MOI infection, we infected cells with an MOI of 1. We again observed a significant reduction of viral RNA in cells treated with the IRE1α nuclease inhibitor, 4μ8C, but not inactive control AMC (Fig. S5B). Together, these results indicate that efficient SARS-CoV-2 replication requires the activity of this host protein.
Fig 5.
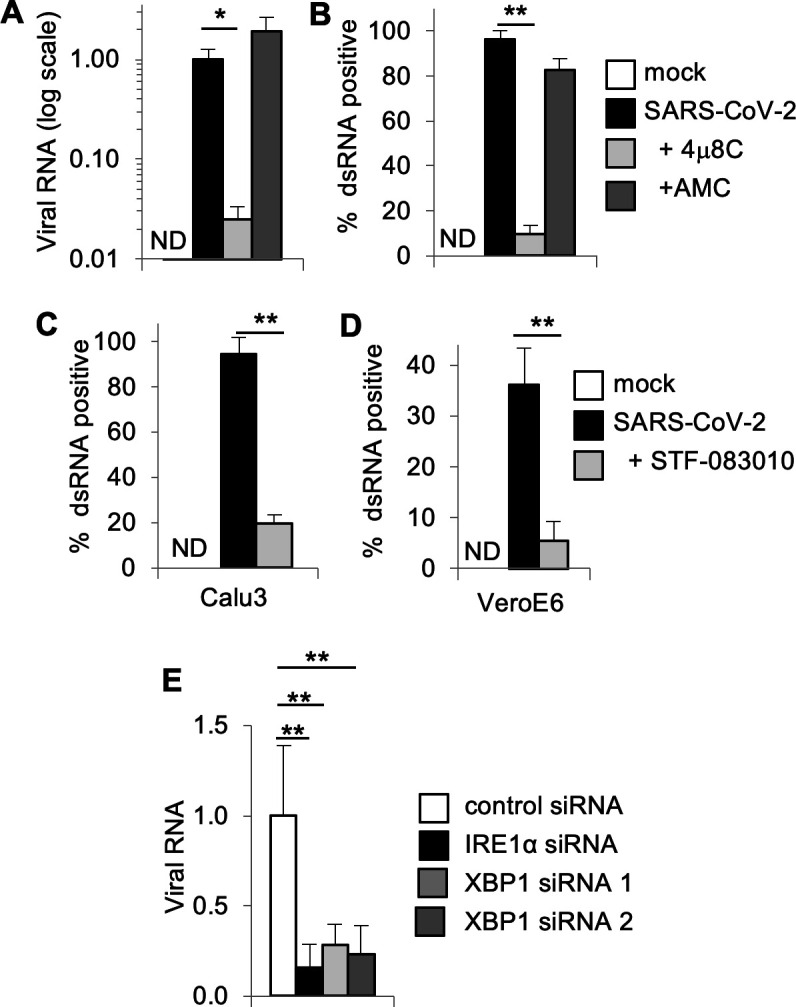
IRE1α and XBP1 are required for optimal SARS-CoV-2 infection. (A+B) Calu-3 cells were treated with IRE1α nuclease inhibitor 4μ8C, structurally similar negative control AMC or DMSO solvent control prior to infection with SARS-CoV-2. (A) RNA was harvested 48-hour post-infection, and the relative abundance of viral RNA was determined by quantitative RT-PCR. (B) The viral replication intermediate, dsRNA, was visualized by immunostaining and positive cells were quantified. (C) Calu-3 or (D) VeroE6 cells were treated with IRE1α nuclease inhibitor STF-083010 or DMSO solvent control prior to infection with SARS-CoV-2 and dsRNA immunostaining. (E) Calu-3 cells were transfected with siRNA targeting IRE1α, XBP1, or non-targeting control siRNA prior to SARS-CoV-2 infection and measurement of viral RNA by quantitative RT-PCR. Data are means ± SD of three (A, B, C), five (D), or six (E) replicates and are representative of two independent experiments. *P < 0.05, **P < 0.01 by unpaired t-test. ND, not detected.
To verify these results, we performed immunostaining for dsRNA and observed that 4μ8C, but not the inactive control, robustly inhibited viral infection (Fig. 5B). The structurally distinct IRE1α nuclease inhibitor STF-083010 also strongly limited viral infection as assessed by dsRNA immunostaining (Fig. 5C). Neither 4μ8C nor STF-083010 were cytotoxic to Calu-3 cells, nor did they reduce the viability of SARS-CoV-2-infected cells (Fig. S5C). To determine whether this effect would extend to other cell types, we infected VeroE6 cells and confirmed the inhibition of SARS-CoV-2 infection with the IRE1α inhibitor, STF-083010 (Fig. 5D) without an effect on cell viability (Fig. S5D).
To test the hypothesis that IRE1α promotes SARS-CoV-2 infection via XBP1, we knocked down XBP1 using siRNA in Calu-3 cells. We verified the reduction in spliced XBP1 mRNA with XBP1 siRNA (Fig. S5E). We found a significant reduction in SARS-CoV-2 viral RNA in cells treated with two independent siRNAs targeting XBP1 (Fig. 5E). We also genetically confirmed the requirement for IRE1α, as IRE1α siRNA knockdown also suppressed viral replication (Fig. 5E). Together, these data demonstrate that SARS-CoV-2 requires host IRE1α RNase activity and XBP1 for optimal infection.
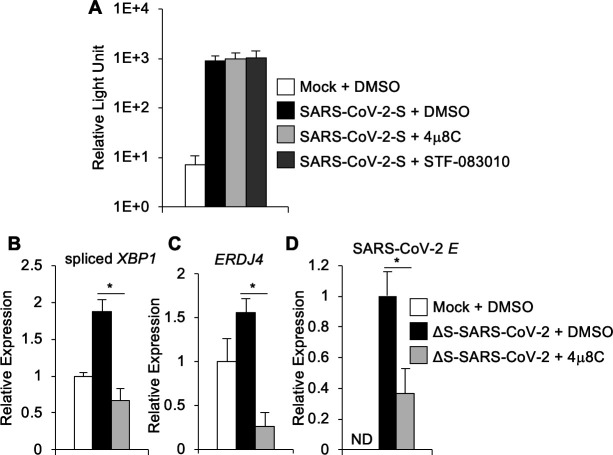
Coronaviruses, including SARS-CoV-2, interact with cell surface receptors via their spike envelope glycoprotein to initiate receptor-mediated entry. To determine whether IRE1α supports the initial viral binding and entry processes, we used SARS-CoV-2 spike pseudotyped lentivirus carrying a luciferase reporter (28). We infected HEK293T cells by overexpressing the ACE2 receptor and assessed reporter virus infection by measuring luciferase activity. We found that the IRE1α inhibitors 4μ8C and STF-083010 did not prevent pseudovirus infection (Fig. 6A). Together with our earlier observation that IRE1α inhibitors had no significant effect on viral RNA abundance at an early time point post-infection, these results suggest that receptor binding and spike-dependent entry are not IRE1α-dependent.
Fig 6.
IRE1α promotes SARS-CoV-2 viral RNA replication independently of viral entry. (A) Hek293 + ACE2 cells were pre-treated with small molecule IRE1α inhibitors or DMSO solvent control prior to infection with SARS-CoV-2 spike pseudotyped lentivirus or mock infection. Reporter virus infection was assessed 72 hours later by measuring luciferase activity, which is shown on a log scale. (B–D) Calu-3 cells were treated with IRE1α nuclease inhibitor 4μ8C or DMSO solvent control prior to infection with ΔS-SARS-CoV-2 single-cycle virus replicon particles. RNA was harvested 24 hours post-infection, and the relative abundance of spliced XBP1 (B), ERDJ4 (C), or SARS-CoV-2 E gene (D) was determined by quantitative RT-PCR. Data are means ± SD of eight (A) or five (B) replicates and are representative of two (A) or three (B–D) independent experiments, respectively. *P < 0.05 by unpaired t-test. ND, not detected.
To further examine post-entry aspects of viral replication, we used a single-cycle infectious SARS-CoV-2 virus replicon particle system (29). This approach utilizes co-transfection of a bacterial artificial chromosome–encoded viral genome in which spike is deleted, together with plasmid-encoded VSV-G. Infection with the resulting virus replicon particles is mediated by VSV-G and independent of ACE2. Infection is limited to a single cycle, as viral RNA replication, but not subsequent virion packaging, can occur. We infected cells with single-cycle virus replicon particles and found that 4μ8C prevented XBP1 mRNA splicing (Fig. 6B) and subsequent ERDJ4 induction (Fig. 6C) verifying its effect on IRE1α. We observed that 4μ8C significantly reduced viral RNA in this system (Fig. 6D). Together, these data suggest that IRE1α supports post-entry SARS-CoV-2 viral RNA replication.
ER stress enhances human coronavirus infection via IRE1α
Risk factors for severe COVID-19 include advanced age, hypertension, diabetes, and obesity, which are all associated with ER stress (8 - 30 - 31 - 31). Although there are likely multiple mechanisms contributing to disease severity, we hypothesized that conditions associated with pre-existing ER stress may prime exuberant viral replication via IRE1α. Oxidative stress is associated with aging and metabolic diseases, causes ER stress, and activates IRE1α (32 - 34). To determine if exogenous oxidative stress enhances coronavirus infection, we pre-treated cells with the oxidative stress inducer tBHP (35, 36) at non-cytotoxic concentrations (Fig. 7A). We verified that tBHP triggered XBP1 splicing (Fig. 7B) and expression of the XBP1 target gene ERDJ4 (Fig. 7C), consistent with prior studies (36). Oxidative stress induced by tBHP enhanced HCoV-OC43 infection, as indicated by increased viral RNA (Fig. 7D). To understand whether the effects on increased viral RNA were due to IRE1α activity, we pre-treated cells with tBHP paired with IRE1α nuclease inhibitor prior to HCoV-OC43 infection. We found that cells treated with tBHP and either 4μ8C or STF-083010 had viral RNA levels (Fig. S6A) and XBP1 splicing (Fig. S6B) similar to DMSO-only control samples. These results overall indicate that oxidative stress activates IRE1α and promotes HCoV-OC43 infection.
Fig 7.
Pre-treatment with tert-butyl hydroperoxide enhances HCoV-OC43 viral infection. HCT-8 cells were treated with the oxidative stress inducer, tert-butyl hydroperoxide (t-BHP) or medium control 2 hours prior to infection with HCoV-OC43. Viability was measured by quantifying dehydrogenase activity in metabolically active cells (A). RNA was harvested 48 hours post-infection, and the relative abundance of spliced XBP1 (B), ERDJ4 (C), and HCoV-OC43 viral RNA (D) was determined by quantitative RT-PCR. Data are means ± SD of four replicates and are representative of two independent experiments.*P < 0.05, **P < 0.001 by unpaired t-test.
XBP1 activation occurs during human COVID-19
We hypothesized that SARS-CoV-2 infection activates IRE1α not only in cultured cells but also in patients with COVID-19 and may represent a prognostic marker for disease severity. To test this hypothesis, we measured the protein product of spliced XBP1 in serum from patients with severe COVID-19 and normal healthy controls using western blot (Fig. 8A). We found a strong and consistent increase in XBP1(S) protein in samples from patients hospitalized in the intensive care unit with acute, severe COVID-19 compared to normal controls (Fig. 8B). This finding suggests that XBP1 activation occurs not only in cultured cells but also in humans suffering from severe SARS-CoV-2 infection.
Fig 8.
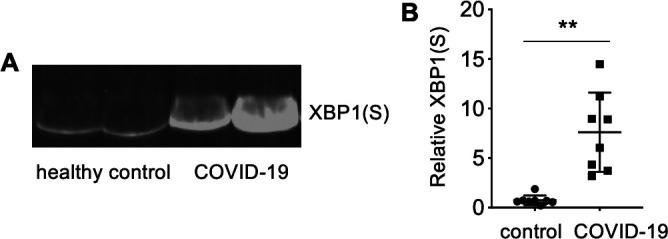
Elevated serum XBP1(S) in human SARS-CoV-2 infection. (A+B) XBP1(S) was measured in serum from normal healthy controls or patients with COVID- 19 by western blot. (A) The image shows representative samples. (B) Relative quantification. Means + SD, n = 8. **P < 0.001 by Mann–Whitney U test.
DISCUSSION
In this study, we found that HCoV-OC43 and SARS-CoV-2 both robustly activate the IRE1α–XBP1 branch of the unfolded protein response in cultured cells. The abundance of the protein product of spliced XBP1 in patients with COVID-19 indicates activation of this pathway also occurs during human infection. Although we focused on the IRE1α–XBP1 branch of the unfolded protein response, Protein kinase RNA (PKR)-like ER kinase (PERK) and activating transcription factor-6 (ATF6) have also been found to be activated during infection with SARS-CoV-2, but interestingly not HCoV-OC43 (16, 19, 37, 38). These responses may be influenced by cell type and infection conditions, as partial IRE1α activation was found in SARS-CoV-2-infected A549 cells and during infection with a higher MOI of 5 (39). Compared to other cell types, A549 cells also demonstrate reduced XBP1 splicing in response to the chemical ER stress inducers thapsigargin and tunicamycin (40). SARS-coronavirus limits IRE1α activation via the envelope (E) gene (18). The E gene is conserved, but not identical between SARS-CoV and SARS-CoV-2 (41), and there are multiple other genetic differences between these two viruses, which contribute to differences in biological behavior via poorly understood mechanisms. Future studies may reveal specific determinants of differential IRE1α regulation between human coronaviruses and during different infection conditions.
There may be multiple pathways contributing to IRE1α activation during HCoV-OC43 and SARS-CoV-2 infection. For example, overexpression of either SARS-CoV-2 ORF3a, ORF8, or spike proteins is sufficient to activate all three branches of the unfolded protein response, including IRE1α activation and XBP1 mRNA splicing (16, 37, 42, 43). In addition, SARS-CoV-2 NSP6 interacts with the sigma-1 receptor (44), an ER-resident transmembrane protein that contributes to IRE1α activation (45). Although XBP1 splicing occurs during infection, we found no evidence of degradation of other IRE1α targets via RIDD in HCoV-OC43- or SARS-CoV-2-infected cells. XBP1 splicing and RIDD are distinct outputs of the IRE1α RNase activity that are not always present simultaneously (46) and may be differentially regulated by the oligomerization state of IRE1α (47). Thus, viral infection may be sufficient to trigger XBP1 splicing, but limit broader RNA degradation.
COVID-19 is characterized by a curiously broad spectrum of disease severity, with some patients having mild to no symptoms and others succumbing to lethal disease (48, 49). Risk factors for severe disease include advanced age, diabetes, hypertension, obesity, and increased viral load (50, 51). HCoV-OC43 is also associated with a spectrum of clinical symptoms and has been associated with lethal infections (52). Although the mechanisms contributing to severe coronavirus disease are likely multifactorial, all of these risk factors are well-associated with ER stress and IRE1α activation (2).
IRE1α promotes infection by a number of different viruses via distinct mechanisms. XBP1-independent activities of IRE1α limit apoptosis of cells infected with the avian coronavirus (IBV) (12), as well as hepatitis C virus (13). Unlike these studies, we found that IRE1α inhibition did not result in the death of HCoV-OC43- or SARS-CoV-2-infected cells, suggesting that IRE1α does not play an anti-apoptotic role for these viruses. IRE1α promotes replication of the porcine coronavirus TGEV via XBP1-independent evasion of antiviral interferon responses (11). We found the activity of IRE1α inhibition against SARS-CoV-2 in interferon-deficient (53) VeroE6 cells, and a requirement for XBP1, suggesting a mechanism distinct from interferon evasion. IRE1α inhibition had no effect on SARS-CoV-2 spike pseudotyped viral entry, suggesting that receptor binding and spike-dependent entry are not IRE1α-dependent. Instead, we hypothesize that IRE1α leads to XBP1-dependent transcriptional changes that promote post-entry aspects of viral replication. In support of this, we found that IRE1α supports optimal viral RNA replication in a single-cycle ACE2-independent SARS-CoV-2 virus replicon particle system. Future work is necessary to further define the specific step(s) of the viral life cycle that are supported by IRE1α and determine if this host factor impacts initial translation, polyprotein processing, or viral RNA replication. Spliced XBP1 is required and sufficient to expand the ER in specialized secretory cells and induce morphological alterations including perinuclear vesicles that appear similar to changes observed with SARS-CoV-2 infection (6, 54 - 58). These observations suggest the hypothesis that IRE1α may support the biogenesis of ER-derived viral replication platforms. Further work is needed to understand whether IRE1α differentially affects specific aspects of viral RNA replication, including synthesis of negative RNA or positive genomic or subgenomic RNA species. Overall, this warrants future study.
In addition to regulating ER-associated genes, XBP1 binds directly to promoters for cytokines including interleukin 6 (IL-6) (59), which is strongly induced in severe COVID-19 and associated with the risk of respiratory failure and death (60 - 62). However, coronaviruses disrupt many pathways of pattern recognition receptor-mediated innate immune sensing (63), and XBP1-mediated cytokine production could provide a means of inciting inflammation via the detection of pathogen-induced cellular stress, even in the setting of viral innate immune antagonism. In addition to promoting acute inflammation, IRE1α is associated with chronic lung injury and pulmonary fibrosis (64, 65). We are only beginning to understand the long-term consequences faced by patients after acute COVID-19, which include symptoms from pulmonary fibrosis (66, 67) that may be a consequence of viral IRE1α activation. Based on emerging evidence for the role of IRE1α in multiple diseases, IRE1α inhibitors have been developed and evaluated as potential therapeutics. These drugs have provided robust in vivo inhibition of IRE1α and have been well-tolerated in both pre-clinical studies and early clinical trials in humans (27, 68 - 70). Ultimately, based on the cellular findings presented here, further animal studies are warranted to determine the impact of IRE1α on viral pathogenesis and replication in vivo.
MATERIALS AND METHODS
Reagents
Cells were treated with 1 µg/mL tunicamycin (Sigma-Aldrich), 50 µM 4μ8c (8-formyl-7-hydroxy-4-methylcoumarin, MilliporeSigma), 50 µM AMC (7-amino-4-methylcoumarin, VWR), 50 µM STF-083010 (MilliporeSigma), or 50 µM tBHP (tert-butyl hydroperoxide; Sigma-Aldrich). Viability of HCoV-OC43 infected cells was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Viability of SARS-CoV-2-infected cells was assessed using the CellTiter-Glo 2.0 Assay (Promega).
Cells and viruses
Cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Serum Plus II, MilliporeSigma), 10 mM HEPES, 50 U/mL penicillin-streptomycin (Gibco), and 0.05 mM β-mercaptoethanol. HCT-8, Calu-3, and VeroE6 cells were obtained from the American Type Culture Collection. 293T-ACE2 cells that overexpress ACE2 (28) were a kind gift from Dr. Jesse D. Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). HCoV-OC43 (NR-52725) was obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) and propagated in HCT-8 cells using medium containing 2% FBS. HCoV-OC43 was titered in HCT-8 cells using a focus-forming assay and used for infections at an MOI of 0.01, unless otherwise indicated. SARS-CoV-2 Isolate USA-WA1/2020 (NR-52281) was obtained through BEI Resources, NIAID, NIH; propagated in VeroE6 cells; titered in VeroE6 cells using a plaque-forming assay; and used for infections at an MOI of 0.01, unless otherwise indicated. Cells were treated with small-molecule IRE1α inhibitors for 2.5 hours prior or tBHP for 2 hours prior to infection.
Single-cycle infectious SARS-CoV-2 virus replicon particles
The generation of SARS-CoV-2 virus replicon particles (VRP(G)) was performed using the delta spike dual reporter bacmid and vesicular stomatitis virus glycoprotein G (VSV-G) plasmid (Addgene; 138479) as described previously (29) with some modifications. Briefly, a mixture of 293T/Huh7.5 cells (−1 × 106 cells of each type) was transfected with 3.5 µg of ΔS-Luc-GFP (green fluorescent protein) bacmid and 0.5 µg of VSV-G plasmid using polyethyleneimine (PEI) transfection reagent (Polysciences; DNA/PEI ratio, 1:4). At 5–6 hours, the transfection mixture containing media was replaced with DMEM/2% and incubated at 37°C. At 48–72 hours post media change, the supernatants were collected and used seed stocks for producing ΔS-VRP(G) working stocks. To generate working stocks, approximately 1.5–2 × 107 Huh7.5 cells seeded in 15-cm plates a day prior were transfected with 20 μg of VSV-G plasmid using PEI reagent (DNA/PEI ratio, 1:4), and the media was changed to DMEM/10% FBS at 5–6 hours post-transfection. The following day, VSV-G-transfected Huh7.5 cells were placed in 10-mL DMEM/2% FBS and infected with 1 mL of ΔS-VRP(G) seed stock. After 2–3 hours of incubation, the inoculum mixture was replaced with 25 mL of DMEM/2%FBS and GFP expression in the cells as well as the luciferase activity in the supernatant. At the peak of GFP expression or luciferase activity (~24–48 hours post media change), the supernatants were collected, clarified of cellular debris by centrifugation at 3,000 rpm for 10 minutes, and stored at –80°C until use. Calu-3 cells were seeded in 96-well plates and treated with small molecule IRE1α inhibitor 4u8C or DMSO for 2.5 hours. Cells were then incubated for 3 hours with 50 μL of ΔS-VRP(G) seed stock, and plates were rocked back and forth every 15 minutes during the incubation period. The supernatant was removed and replaced with a fresh medium containing IRE1α inhibitor or DMSO.
Expression analysis
RNA isolated from cell lysates using the SingleShot Cell Lysis Kit (Bio-Rad) was used to synthesize cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR (qRT-PCR) was performed on a Bio-Rad CFX Connect or Roche Lightcycler 480 using SYBR Green (Bio-Rad) with the following primers (all primers listed in the 5′–3′ orientation):
human spliced XBP1: TGCTGAGTCCGCAGCAGGTG (forward) and GCTGGCAGGCTCTGGGGAAG (reverse)
human ERDJ4: TAGTCGGAGGGTGCAGGATA (forward) and CGCTCTGATGCCGATTTTGG (reverse)
human P58IPK: (forward) TGTGTTTGGGATGCAGAACTAC and (reverse) TCTTCAACTTTGACGCAGCTT
human BLOC1S1: (forward) CAGACAGGCCAGTGGATCG and (reverse) TCTCCACATCCCCAATTTCCTTG
human SCARA3: (forward) GTGTTGGCCTCTCTGGTTTTC and (reverse) AAGAGCAGTTGTTCAGGGCT
human COL6A1: (forward) ATGTGCTCTTGCTGTGAATGC and (reverse) GAAGTTCTGCAGGCCAATGC
human HGSNAT: (forward) CACCTTCAGGGGGATTGCTC and (reverse) TACAAACCACGGGAACACGA
human RPS18: TGC GAG TAC TCA ACA CCA ACA (forward) and CTT CGG CCC ACA CCC TTA AT (reverse)
human GAPDH: CAA TGA CCC CTT CAT TGA CC (forward) and GAC AAG CTT CCC GTT CTC AG (reverse)
HCoV-OC43 M: ATG TTA GGC CGA TAA TTG AGG ACT AT (forward) and AAT GTA AAG ATG GCC GCG TATT (reverse)
HCoV-OC43 orf1ab: TGG ATT TTG GCG GGA TGG AA (forward) and GAG ACG GGC ATC TAC ACT CG (reverse)
SARS-CoV-2 E: ACAGGTACGTTAATAGTTAATAGCGT and ATATTGCAGCAGTACGCACACA
Melt curve analysis was used to assess whether single reaction products were produced. Expression was calculated relative to the housekeeping gene RPS18, with equivalent results also obtained relative to GAPDH.
siRNA
HCT-8 or Calu-3 cells were transfected with siRNA against IRE1α (D-004951-01-0005, Dharmacon), XBP1 (D-009552-02-005, D-009552-0005, Dharmacon), or no-target control siRNA (D-001210-03-05, Dharmacon) using Lipofectamine RNAiMax Transfection Reagent (Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence microscopy
After fixation in 2% paraformaldehyde, cells were permeabilized with 0.2% Triton-X100 in phosphate-buffered saline (PBS) and blocked with 3% bovine serum albumin + 0.2% Triton-X100 in PBS. Cells were labeled with anti-dsRNA mouse monoclonal antibody J2 (SCICONS, catalog no. 10010200) and Alexa Fluor 488 secondary antibody (Invitrogen, catalog no. A-21202). TO-PRO-3 (Invitrogen, catalog no. T3605) was used to label nuclei. For HCoV-OC43 infected cells, a Cytation 1 fluorescent cell imaging system (BioTek) was used for image acquisition (10× objective), and Gen5 software (BioTek) was used for image processing and subsequent analysis. For dsRNA staining, total fluorescence intensity was measured with four wells per condition using identical capture settings for the target of interest. The total fluorescence intensity from the uninfected DMSO-negative control was subtracted from all condition wells. For SARS-CoV-2-infected cells, images were collected with an EVOS FLoid cell imaging station (ThermoFisher). For quantification of dsRNA staining of SARS-CoV-2-infected cells, the total number of cells was determined by counting the number of nuclei, and the percentage of dsRNA-positive cells was calculated using this and the number of cells with positive dsRNA staining.
Coronavirus spike-mediated pseudovirus entry assay
Lentivirus pseudotyped with SARS-CoV-2 spike was generated in HEK293T cells, and pseudovirus entry assay was performed as previously described (28). Plasmids expressing the HIV-1 Gag and pol (pHDM-Hgpm2), HIV-1 Rev (pRC-CMV-rev1b), HIV-1 Tat (pHDM-tat1b), C-terminally truncated SARS CoV2 spike (pHDM-SARS-CoV-2delta21 Spike), and luciferase/GFP reporter (pHAGE-CMV-Luc2-IRES-ZsGreen-W) were co-transfected into HEK293T cells using Lipofectamine 3000 (ThermoFisher) according to the manufacturer’s instructions. Plasmids were a kind gift from Dr. Jesse D. Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). Pseudovirus was harvested 60 hours post-transfection and stored at −80°C. 293T-ACE2 cells were pre-treated with IRE1α inhibitors for 2.5 hours prior to pseudovirus infection. Luciferase reporter gene expression was assessed 72 hours post-infection by quantifying relative luminescence units using the ONE-GLO Luciferase Assay System (Promega).
Western blot
Protein extraction from cultured cells was performed with Protein Extraction Reagent Type 4 (MilliporeSigma) with added HALT protease Inhibitor Cocktail (ThermoFisher), phenylmethylsulfonyl fluoride (PMSF) protease inhibitor (ThermoFisher), and Benzonase nuclease (MilliporeSigma), mixed with loading buffer and heated at 95°C for 10 minutes under reducing conditions. Serum specimens from patients with RT-PCR-confirmed SARS-CoV-2 infection were collected from the clinical laboratories at the University of Washington and Harborview Medical Centers in Seattle, WA, USA, between March and May of 2020 (71). Specimens were collected under an institutional review board-approved waiver of consent from patients receiving care in the intensive care unit. Samples from healthy blood donors obtained prior to the COVID-19 pandemic were used as normal controls. Specimens were stored at −80°C before testing. Thawed specimens were diluted 1:30 with digestion buffer Protein Extraction Reagent Type 4 (MilliporeSigma) with added HALT protease Inhibitor Cocktail (ThermoFisher), PMSF protease inhibitor (ThermoFisher), and benzonase nuclease (MilliporeSigma), mixed with loading buffer and heated at 95°C for 10 minutes under reducing conditions. Proteins were separated by SDS-PAGE using Any kD TGX stain-free gels (Bio-Rad) and transferred to nitrocellulose membranes. Membranes were probed with rabbit polyclonal anti-XBP1 (Invitrogen, catalog no. PA5-27650) or rabbit monoclonal anti-XBP1 (Cell Signaling Technology, catalog no. D2C1F) and mouse monoclonal anti-vinculin (Santa Cruz Biotechnology, catalog no. sc-73614) antibodies, followed by incubation with secondary antibody, donkey anti-rabbit IRDye 800CW (LI-COR, catalog no. 926-32213), and goat anti-mouse IRDye 680RD (LI-COR, catalog no. 926-68073). The blots were imaged with an Odyssey Infrared Imaging System (LI-COR Biosciences) and relative density units were calculated with Image Studio Lite Version.
Statistics
The unpaired two-tailed Student’s t-test or the Mann–Whitney U test was used for comparisons between the two groups. P values of less than 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
S.L.F. was supported by the NIAID of the NIH under Award Nos R01AI162684 and R21AI153487.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We thank Dr. Jesse D. Bloom (Fred Hutchinson Cancer Research Center, Seattle, Washington) for kindly sharing cells and reagents. We thank Dr. Balaji Manicassamy (University of Iowa, Iowa City, Iowa, USA) for kindly providing stocks of single-cycle infectious SARS-CoV-2 virus replicon particles.
The following reagents were deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, and NIH: SARS-related coronavirus 2, Isolate USA-WA1/2020, NR-52281, human coronavirus, OC43, and NR-52725.
The authors declare that they have no conflict of interest.
Contributor Information
Susan L. Fink, Email: sfink@uw.edu.
Sara Cherry, University of Pennsylvania, Philadelphia, Pennsylvania, USA .
Matthew B. Frieman, University of Maryland School of Medicine, Baltimore, Maryland, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.00540-23.
Regulated IRE1α-dependent decay.
sXBP1 protein.
IRE1α is required for optimal HCoV-OC43.
IRE1α inhibitors reduce viral protein and are not cytotoxic.
IRE1α inhibitors decrease SARS-CoV-2.
Pretreatment with tert-butyl hydroperoxide.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. 2020. The molecular virology of coronaviruses. J Biol Chem 295:12910–12934. doi: 10.1074/jbc.REV120.013930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hetz C, Zhang K, Kaufman RJ. 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21:421–438. doi: 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529. doi: 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- 4. Schröder M, Kaufman RJ. 2005. The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed] [Google Scholar]
- 5. Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. doi: 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- 6. Sriburi R, Jackowski S, Mori K, Brewer JW. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167:35–41. doi: 10.1083/jcb.200406136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated IRE1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186:323–331. doi: 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belmadani S, Matrougui K. 2019. Broken heart: a matter of the endoplasmic reticulum stress bad management? WJC 11:159–170. doi: 10.4330/wjc.v11.i6.0000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eizirik DL, Pasquali L, Cnop M. 2020. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nat Rev Endocrinol 16:349–362. doi: 10.1038/s41574-020-0355-7 [DOI] [PubMed] [Google Scholar]
- 10. Morimoto RI. 2020. Cell-nonautonomous regulation of proteostasis in aging and disease. Cold Spring Harb Perspect Biol 12:a034074. doi: 10.1101/cshperspect.a034074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma Y, Wang C, Xue M, Fu F, Zhang X, Li L, Yin L, Xu W, Feng L, Liu P. 2018. The coronavirus transmissible gastroenteritis virus evades the type I interferon response through IRE1α-mediated manipulation of the microRNA miR-30a-5p/SOCS1/3 axis. J Virol 92:e00728-18. doi: 10.1128/JVI.00728-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fung TS, Liao Y, Liu DX. 2014. The endoplasmic reticulum stress sensor IRE1α protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. J Virol 88:12752–12764. doi: 10.1128/JVI.02138-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fink SL, Jayewickreme TR, Molony RD, Iwawaki T, Landis CS, Lindenbach BD, Iwasaki A. 2017. IRE1α promotes viral infection by conferring resistance to apoptosis. Sci Signal 10:eaai7814. doi: 10.1126/scisignal.aai7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bechill J, Chen Z, Brewer JW, Baker SC. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J Virol 82:4492–4501. doi: 10.1128/JVI.00017-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Versteeg GA, van de Nes PS, Bredenbeek PJ, Spaan WJM. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J Virol 81:10981–10990. doi: 10.1128/JVI.01033-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Echavarría-Consuegra L, Cook GM, Busnadiego I, Lefèvre C, Keep S, Brown K, Doyle N, Dowgier G, Franaszek K, Moore NA, Siddell SG, Bickerton E, Hale BG, Firth AE, Brierley I, Irigoyen N. 2021. Manipulation of the unfolded protein response: a pharmacological strategy against coronavirus infection. PLoS Pathog 17:e1009644. doi: 10.1371/journal.ppat.1009644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan C-P, Siu K-L, Chin K-T, Yuen K-Y, Zheng B, Jin D-Y. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol 80:9279–9287. doi: 10.1128/JVI.00659-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Regla-Nava JA, Alvarez E, Oliveros JC, Zhao J, Fett C, Perlman S, Enjuanes L. 2011. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog 7: e1002315. doi: 10.1371/journal.ppat.1002315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Favreau DJ, Desforges M, St-Jean JR, Talbot PJ. 2009. A human coronavirus OC43 variant harboring persistence-associated mutations in the S glycoprotein differentially induces the unfolded protein response in human neurons as compared to wild-type virus. Virology 395:255–267. doi: 10.1016/j.virol.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolpikova EP, Tronco AR, Hartigh A den, Jackson KJ, Iwawaki T, Fink SL. 2020. IRE1α promotes zika virus infection via XBP1. Viruses 12:278. doi: 10.3390/v12030278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bashir S, Banday M, Qadri O, Bashir A, Hilal N, Rader S, Fazili KM. 2021. The molecular mechanism and functional diversity of UPR signaling sensor IRE1. Life Sci 265:118740. doi: 10.1016/j.lfs.2020.118740 [DOI] [PubMed] [Google Scholar]
- 23. Moore K, Hollien J. 2015. IRE1-Mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol Biol Cell 26:2873–2884. doi: 10.1091/mbc.E15-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, Legge K, Monick MM. 2012. Influenza a viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem 287:4679–4689. doi: 10.1074/jbc.M111.284695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cross BCS, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A 109:E869–78. doi: 10.1073/pnas.1115623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, Koong AC. 2011. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 117:1311–1314. doi: 10.1182/blood-2010-08-303099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tufanli O, Telkoparan Akillilar P, Acosta-Alvear D, Kocaturk B, Onat UI, Hamid SM, Çimen I, Walter P, Weber C, Erbay E. 2017. Targeting IRE1 with small molecules counteracts progression of atherosclerosis. Proc Natl Acad Sci U S A 114:E1395–E1404. doi: 10.1073/pnas.1621188114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. doi: 10.3390/v12050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malicoat J, Manivasagam S, Zuñiga S, Sola I, McCabe D, Rong L, Perlman S, Enjuanes L, Manicassamy B. 2022. Development of a single-cycle infectious SARS-CoV-2 virus replicon particle system for use in biosafety level 2 laboratories. J Virol 96:e0183721. doi: 10.1128/JVI.01837-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bornstein SR, Dalan R, Hopkins D, Mingrone G, Boehm BO. 2020. Endocrine and metabolic link to coronavirus infection. Nat Rev Endocrinol 16:297–298. doi: 10.1038/s41574-020-0353-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jordan RE, Adab P, Cheng KK. 2020. Covid-19: risk factors for severe disease and death. BMJ 368:m1198. doi: 10.1136/bmj.m1198 [DOI] [PubMed] [Google Scholar]
- 32. Huang S, Xing Y, Liu Y. 2019. Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease. J Biol Chem 294:18726–18741. doi: 10.1074/jbc.REV119.007036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martínez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C. 2017. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell 16:615–623. doi: 10.1111/acel.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu R, Zhang QH, Lu YJ, Ren K, Yi GH. 2015. Involvement of the IRE1α-XBP1 pathway and XBP1s-dependent transcriptional reprogramming in metabolic diseases. DNA Cell Biol 34:6–18. doi: 10.1089/dna.2014.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kučera O, Endlicher R, Roušar T, Lotková H, Garnol T, Drahota Z, Cervinková Z. 2014. The effect of tert-butyl hydroperoxide-induced oxidative stress on lean and steatotic rat hepatocytes in vitro. Oxid Med Cell Longev 2014:752506. doi: 10.1155/2014/752506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez-Carrasco R, Argüeso P, Fini ME. 2020. Dynasore protects ocular surface mucosal epithelia subjected to oxidative stress by maintaining UPR and calcium homeostasis. Free Radic Biol Med 160:57–66. doi: 10.1016/j.freeradbiomed.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rashid F, Dzakah EE, Wang H, Tang S. 2021. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res 296:198350. doi: 10.1016/j.virusres.2021.198350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siri M, Dastghaib S, Zamani M, Rahmani-Kukia N, Geraylow KR, Fakher S, Keshvarzi F, Mehrbod P, Ahmadi M, Mokarram P, Coombs KM, Ghavami S. 2021. Autophagy, unfolded protein response, and neuropilin-1 cross-talk in SARS-CoV-2 infection: what can be learned from other Coronaviruses. Int J Mol Sci 22:5992. doi: 10.3390/ijms22115992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen LC, Renner DM, Silva D, Yang D, Parenti NA, Medina KM, Nicolaescu V, Gula H, Drayman N, Valdespino A, Mohamed A, Dann C, Wannemo K, Robinson-Mailman L, Gonzalez A, Stock L, Cao M, Qiao Z, Moellering RE, Tay S, Randall G, Beers MF, Rosner MR, Oakes SA, Weiss SR. 2022. SARS-CoV-2 diverges from other betacoronaviruses in only partially activating the IRE1α/XBP1 endoplasmic reticulum stress pathway in human lung-derived cells. mBio 13:e0241522. doi: 10.1128/mbio.02415-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Schadewijk A, van’t Wout EFA, Stolk J, Hiemstra PS. 2012. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones 17:275–279. doi: 10.1007/s12192-011-0306-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su W-Q, Yu X-J, Zhou C-M. 2021. SARS-CoV-2 ORF3α induces incomplete autophagy via the unfolded protein response. Viruses 13:2467. doi: 10.3390/v13122467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xue M, Feng L. 2021. The role of unfolded protein response in coronavirus infection and its implications for drug design. Front Microbiol 12:808593. doi: 10.3389/fmicb.2021.808593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang X-P, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang H-Y, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459–468. doi: 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mori T, Hayashi T, Hayashi E, Su T-P. 2013. Sigma-1 receptor chaperone at the ER-Mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One 8:e76941. doi: 10.1371/journal.pone.0076941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grandjean JMD, Madhavan A, Cech L, Seguinot BO, Paxman RJ, Smith E, Scampavia L, Powers ET, Cooley CB, Plate L, Spicer TP, Kelly JW, Wiseman RL. 2020. Pharmacologic IRE1/xbp1s activation confers targeted ER proteostasis reprogramming. Nat Chem Biol 16:1052–1061. doi: 10.1038/s41589-020-0584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tam AB, Koong AC, Niwa M. 2014. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep 9:850–858. doi: 10.1016/j.celrep.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruan Q, Yang K, Wang W, Jiang L, Song J. 2020. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 46:1294–1297. doi: 10.1007/s00134-020-06028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Su L, Ma X, Yu H, Zhang Z, Bian P, Han Y, Sun J, Liu Y, Yang C, Geng J, Zhang Z, Gai Z. 2020. The different clinical characteristics of corona virus disease cases between children and their families in China – the character of children with COVID-19. Emerging Microbes & Infections 9:707–713. doi: 10.1080/22221751.2020.1744483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bryan A, Fink SL, Gattuso MA, Pepper G, Chaudhary A, Wener MH, Morishima C, Jerome KR, Mathias PC, Greninger AL. 2020. SARS-CoV-2 viral load on admission is associated with 30-day mortality. Open Forum Infect Dis 7:faa535. doi: 10.1093/ofid/ofaa535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult Inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi W-I, Kim IB, Park SJ, Ha E-H, Lee CW. 2021. Comparison of the clinical characteristics and mortality of adults infected with human coronaviruses 229E and OC43. Sci Rep 11:4499. doi: 10.1038/s41598-021-83987-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Osada N, Kohara A, Yamaji T, Hirayama N, Kasai F, Sekizuka T, Kuroda M, Hanada K. 2014. The genome landscape of the African green monkey kidney-derived Vero cell line. DNA Res 21:673–683. doi: 10.1093/dnares/dsu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Marshall D, Lacy JM, Williams T, Yarid N. 2020. Histopathology and ultrastructural findings of fatal COVID-19 infections. Pathology. doi: 10.1101/2020.04.17.20058545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brahim Belhaouari D, Fontanini A, Baudoin J-P, Haddad G, Le Bideau M, Bou Khalil JY, Raoult D, La Scola B. 2020. The strengths of scanning electron microscopy in deciphering SARS-CoV-2 infectious cycle. Front Microbiol 11:2014. doi: 10.3389/fmicb.2020.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Caldas LA, Carneiro FA, Monteiro FL, Augusto I, Higa LM, Miranda K, Tanuri A, de Souza W. 2021. Intracellular host cell membrane remodelling induced by SARS-CoV-2 infection in vitro. Biol Cell 113:281–293. doi: 10.1111/boc.202000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380. doi: 10.1038/sj.emboj.7600903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81–93. doi: 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 59. Martinon F, Chen X, Lee A-H, Glimcher LH. 2010. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11:411–418. doi: 10.1038/ni.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. 2020. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev 54:62–75. doi: 10.1016/j.cytogfr.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herold T, Jurinovic V, Arnreich C, Lipworth BJ, Hellmuth JC, von Bergwelt-Baildon M, Klein M, Weinberger T. 2020. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol 146:128–136. doi: 10.1016/j.jaci.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smetana K, Brábek J. 2020. Role of interleukin-6 in lung complications in patients with COVID-19: therapeutic implications. In Vivo 34:1589–1592. doi: 10.21873/invivo.11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, Zhang Q, Wu J. 2020. Coronavirus infections and immune responses. J Med Virol 92:424–432. doi: 10.1002/jmv.25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burman A, Tanjore H, Blackwell TS. 2018. Endoplasmic reticulum stress in pulmonary fibrosis. Matrix Biol 68–69:355–365. doi: 10.1016/j.matbio.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thamsen M, Ghosh R, Auyeung VC, Brumwell A, Chapman HA, Backes BJ, Perara G, Maly DJ, Sheppard D, Papa FR. 2019. Small molecule inhibition of IRE1α kinase/RNase has anti-fibrotic effects in the lung. PLoS One 14:e0209824. doi: 10.1371/journal.pone.0209824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nasserie T, Hittle M, Goodman SN. 2021. Assessment of the frequency and variety of persistent symptoms among patients with COVID-19: A systematic review. JAMA Netw Open 4:e2111417. doi: 10.1001/jamanetworkopen.2021.11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Salamanna F, Veronesi F, Martini L, Landini MP, Fini M. 2021. Post-COVID-19 syndrome: the persistent symptoms at the post-viral stage of the disease. A systematic review of the current data. Front Med (Lausanne) 8:653516. doi: 10.3389/fmed.2021.653516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ghosh R, Wang L, Wang ES, Perera BGK, Igbaria A, Morita S, Prado K, Thamsen M, Caswell D, Macias H, Weiberth KF, Gliedt MJ, Alavi MV, Hari SB, Mitra AK, Bhhatarai B, Schürer SC, Snapp EL, Gould DB, German MS, Backes BJ, Maly DJ, Oakes SA, Papa FR. 2014. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158:534–548. doi: 10.1016/j.cell.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, Kiziltepe T, Ikeda H, Kawano Y, French M, Blumenthal M, Tam V, Kertesz NL, Malyankar UM, Hokenson M, Pham T, Zeng Q, Patterson JB, Richardson PG, Munshi NC, Anderson KC. 2012. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood 119:5772–5781. doi: 10.1182/blood-2011-07-366633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gabrail NY, Hamilton EP, Elias AD, Rimawi MF, Li C, Corvez MM, Li W, Feng Y, Wei J, Greene S, Patterson J, Zeng Q, Hui A-M. 2021. A phase 1/2 trial of ORIN1001, a first-in-class IRE1 inhibitor, in patients with advanced solid tumors. JCO 39:3080–3080. doi: 10.1200/JCO.2021.39.15_suppl.3080 [DOI] [Google Scholar]
- 71. Lerma LA, Chaudhary A, Bryan A, Morishima C, Wener MH, Fink SL. 2020. Prevalence of autoantibody responses in acute coronavirus disease 2019 (COVID-19). J Transl Autoimmun 3:100073. doi: 10.1016/j.jtauto.2020.100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regulated IRE1α-dependent decay.
sXBP1 protein.
IRE1α is required for optimal HCoV-OC43.
IRE1α inhibitors reduce viral protein and are not cytotoxic.
IRE1α inhibitors decrease SARS-CoV-2.
Pretreatment with tert-butyl hydroperoxide.