ABSTRACT
CRISPR/Cas9 genome editing technology has been implemented in almost all living organisms. Its editing precision appears to be very high and therefore could represent a big change from conventional genetic engineering approaches. However, guide RNA binding to nucleotides similar to the target site could result in undesired off-target mutations. Despite this, evaluating whether mutations occur is rarely performed in genome editing studies. In this study, we generated CRISPR/Cas9-derived filamentous fungal strains and analyzed them for the occurrence of mutations, and to which extent genome stability affects their occurrence. As a test case, we deleted the (hemi-)cellulolytic regulator-encoding gene xlnR in two Aspergillus niger strains: a wild type (WT) and a non-homologous end-joining (NHEJ)-deficient strain ΔkusA. Initial phenotypic analysis suggested a much higher prevalence of mutations in the WT compared to NHEJ-deficient strains, which was confirmed and quantified by whole-genome sequencing analysis. Our results clearly demonstrate that CRISPR/Cas9 applied to an NHEJ-deficient strain is an efficient strategy to avoid unwanted mutations.
IMPORTANCE
Filamentous fungi are commonly used biofactories for the production of industrially relevant proteins and metabolites. Often, fungal biofactories undergo genetic development (genetic engineering, genome editing, etc.) aimed at improving production yields. In this context, CRISPR/Cas9 has gained much attention as a genome editing strategy due to its simplicity, versatility, and precision. However, despite the high level of accuracy reported for CRISPR/Cas9, in some cases unintentional cleavages in non-targeted loci—known as off-target mutations—could arise. While biosafety should be a central feature of emerging biotechnologies to minimize unintended consequences, few studies quantitatively evaluate the risk of off-target mutations. This study demonstrates that the use of non-homologous end-joining-deficient fungal strains drastically reduces the number of unintended genomic mutations, ensuring that CRISPR/Cas9 can be safely applied for strain development.
KEYWORDS: Aspergillus niger, biosafety, CRISPR-Cas9, risk assessment, ΔkusA
INTRODUCTION
Filamentous fungi are widely used as cell factories for the production of industrially relevant compounds, such as organic acids, proteins, enzymes, and secondary metabolites (1, 2). One fundamental aspect for the future of fungal biotechnology is the improvement of production strains by precise genetic engineering. The simplicity of the CRISPR/Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) systems has been key to widespread implementation of this genome-editing approach in almost all living organisms, including filamentous fungi. The precision that is possible by CRISPR/Cas9 systems represents a big change from conventional genetic engineering approaches, which largely rely on random chemical- or radiation-induced mutagenesis, or untargeted insertions of genes into the genomes through genetic engineering.
CRISPR/Cas9 genome-editing technology is based on the ability of the Cas9 nuclease to cut a specific DNA sequence, generating double-strand breaks (DSBs). Its specificity relies on a 20-bp guide RNA (gRNA) sequence that is designed to target a genetic sequence through the interaction with the protospacer adjacent motive (PAM) “NGG.”
After the DNA break takes place, DSBs are detected as potential damage and have to be repaired for the survival of the species. In fungi, two DNA repair mechanisms exist: the error-prone non-homologous end-joining (NHEJ) and homology-directed repair (HDR). With NHEJ, DNA breaks are re-ligated by the Ku70 and Ku80 heterodimeric protein complex, frequently leading to imprecise DNA repair and loss of genetic functionality (3). In contrast, HDR occurs in the presence of a homologous duplex DNA template (referred to as donor DNA, dDNA) via homologous recombination (HR) (4), which can be used to precisely introduce or remove the DNA sequence of interest in the target organism.
Biosafety should be a central feature of emerging biotechnologies to minimize unintended consequences. Genetic mutations can randomly appear through normal cell division, especially when species are cultured for many generations in the laboratory (5). Additionally, random mutations may also be caused by the mutagenic potential of the transformation process. In fact, it has been reported that transformation enhances the appearance of genetic variations, including chromosomal rearrangements, in different organisms such as bacteria (6), plants (7), and fungi (8, 9). Therefore, ensuring a safe application of CRISPR technologies in the target organisms is a key factor to consider. Despite the high level of accuracy reported for CRISPR/Cas9, annealing of the gRNA to sequences with similarity to the target sequence leads to unspecific DNA recognition and could result in unintentional cleavages in non-targeted loci, known as off-target mutations, thus restricting its application (10). It has been reported that gRNA can tolerate sequence mismatches that still allow Cas9 DSBs generation (11). Basically, the “seed region” (the first 8–12 PAM proximal nucleotides) of the gRNA is vital for recognition, and the potential of off-targeting increases when the mismatches locate further away from the PAM sequence (11). There are some reports of unintended Cas9-induced off-target effects in different organisms, including plants (12, 13), animal, and human cell lines (10, 14), and to an even lesser extent in filamentous fungi (15, 16) and yeast (17). Searching for off-target mutations is far from routine during the experimental procedures in most studies, sometimes leading to unexpected phenotypes misattributed to the originally intended mutation. In filamentous fungi, the off-target analysis has only been partially exploited in Ustilago maydis (15), Aspergillus fumigatus (9), Magnaporthe oryzae (4), and Trichoderma reesei (16). However, in all these cases, the number of mutants analyzed is too small to draw robust conclusions, and the off-target effects were analyzed only in wild-type (WT) strains. In this study, we analyzed a large set of CRISPR/Cas9-derived fungal mutant strains to determine whether CRISPR/Cas9 itself induces off-target mutations, and to which extent genome stability plays a role in the accumulation of off-target or random mutations. As a test case, we deleted the (hemi-)cellulolytic regulator-encoding gene xlnR (18) in two strains of the industrial workhorse Aspergillus niger: a WT strain and an NHEJ-deficient strain in which the kusA gene was deleted. The A. niger kusA gene encodes the ortholog of the Ku70 protein in other eukaryotes, and its deletion has resulted in an improved homologous integration efficiency of more than 80% compared to the 7% reported in A. niger WT with no negative effect on growth and biological cycle of the fungus (19). XlnR is a transcription factor involved in the process of (hemi-)cellulose utilization in ascomycete fungi, and was first identified in A. niger (18), being one of the best characterized regulators in this species. Additionally, XlnR together with AraR controls the pentose catabolic pathway, which is required for the utilization of D-xylose and L-arabinose (20). Additionally, nine more regulator-encoding genes (araR, gaaR, rhaR, galX, amyR, inuR, clrB, and creA) (21) were stepwise deleted in the ΔkusA strain. Finally, the effect of the transformation process and the presence of Cas9 in the accumulation of genetic mutations, as well as the off-target mutations caused by CRISPR/Cas9 in both genetic backgrounds or in the ΔkusA strain upon consecutive application of CRISPR/Cas9, were assessed.
RESULTS AND DISCUSSION
Phenotypic characterization of CRISPR/Cas9-derived transformants points to an increase in the number of mutations in A. niger wild type
Key safety and technical considerations for CRISPR/Cas9 application include genomic specificity and editing efficiency (22). In this study, we aimed to evaluate whether the application of a transient CRISPR/Cas9 system induces off-target mutations in the filamentous ascomycete fungus A. niger, and how different genomic backgrounds could affect the accumulation of these and other random mutations.
We used CRISPR/Cas9 plasmids harboring both cas9 and gRNA to effectively target xlnR, and dDNA was also included to allow DNA repair by HR. Deletion mutants of xlnR gene have been shown to grow poorly on xylan as the sole carbon source (23, 24). Therefore, this makes xlnR a good candidate for its disruption using CRISPR/Cas9, as it facilitates transformant selection by phenotypic characterization. Additionally, for control experiments, A. niger strains resulting from transformations using empty plasmids (with no cas9), and plasmids with cas9 but no gRNA were included.
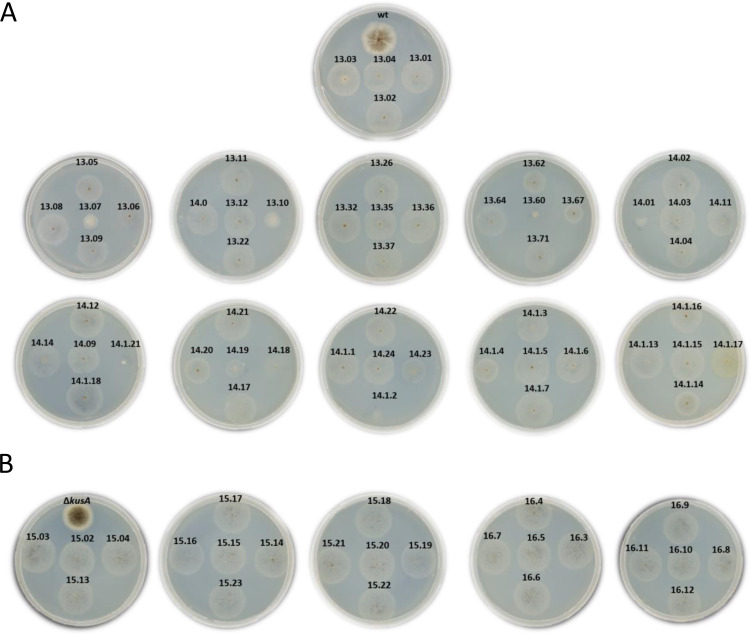
All the 78 mutants generated after transformation with CRISPR/Cas9 plasmids containing cas9 and gRNA were analyzed on xylan-containing plates (Fig. 1) and confirmed by PCR (Fig. S1). From these 78 mutants, 54 ΔxlnR mutants were obtained in A. niger WT genetic background (Fig. 1A), while 24 were obtained in the NHEJ-deficient strains ΔkusA (Fig. 1B). All of them showed reduced growth on xylan plates, confirming the disruption of the targeted gene and 100% on-target activity of the designed gRNA. However, 13 WT-derived A. niger ΔxlnR strains (ID 13.06, 13.07, 13.10, 13.60, 13.67, 14.01, 14.18, 14.19, 14.23, 14.1.2, 14.1.4, 14.1.14, and 14.1.21) showed unexpected growth phenotypes after 4 days of growth (Fig. 1A). In contrast, none of the ΔxlnR mutants generated in the NHEJ-deficient strains showed unexpected growth phenotypes under the same culture conditions (Fig. 1B). Taken together, these results may indicate that the aberrant phenotypes observed in the 24% of the WT-derived ΔxlnR strains obtained are as consequence of the application of CRISPR/Cas9. Also, these results preliminary suggested a higher genetic instability of the WT genome in terms of mutation accumulation, in which both NHEJ and HDR DNA repair mechanisms take place, compared to the NHEJ-deficient strains, in which only HDR is possible. To further confirm these hypotheses, a selection of control and transformant strains from both genetic backgrounds were selected for whole-genome sequencing (WGS) and analyzed for the presence of genetic variants (see Fig. S1 and sections below).
Fig 1.
Phenotypic characterization of A. niger ΔxlnR strains in wild-type (A) and ΔkusA (B) genetic backgrounds. Strains were grown on 1% xylan-containing minimal medium plates for 4 days at 30°C.
The genetic variations induced by the transformation process are mitigated in NHEJ-deficient A. niger strains
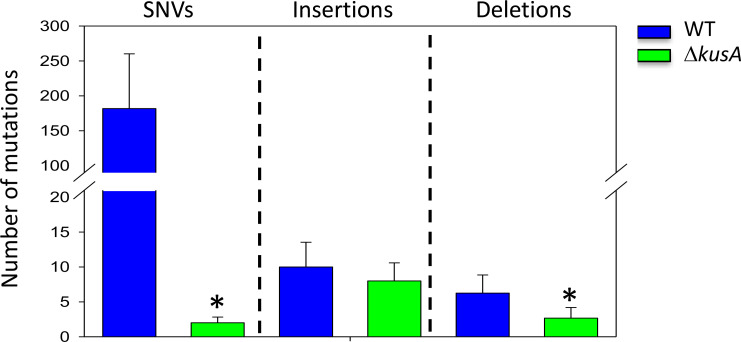
In this study, we transformed both A. niger WT and ΔkusA backgrounds with the empty ANEp8-pyrG plasmid, which did not contain cas9, and analyzed the resulting strains by WGS to determine the frequency of genetic mutations that may occur due to the transformation process. This also provides a baseline to compare the possible impact of CRISPR/Cas9 on off-target/random mutations. Comparative bioinformatics analyses revealed that multiple genomic mutations were present in both genetic backgrounds, accumulating single-nucleotide variants (SNVs), insertions, and deletions after the protoplast-mediated transformation process (Fig. 2). However, the average number of genetic variations detected in the ΔkusA strains (2 SNVs, 8 insertions, and 2 deletions) was generally lower compared to the WT (181 SNVs, 10 insertions, and 6 deletions) (Table S1). In this context, there was a statistically significant difference in the average number of detected SNVs and deletions between the WT and the ΔkusA SNVs (P = 0.001) and deletions (P = 0.034) between the WT and the ΔkusA strain after transformation (Table S3). These results confirm the occurrence of genetic mutations during the transformation process used in this study, which is a polyethylene glycol (PEG)-mediated transformation procedure commonly used by the scientific community nowadays not only for Aspergillus species (9, 25 - 27), but also for Penicillium (28 - 32), Talaromyces (33), and more phylogenetically distant fungal species such as Trichoderma (34), Neurospora (35), Stagonospora (36), and Dichomitus (37), for instance. This standard transformation protocol might induce cellular stress as it requires the enzymatic digestion of the cell wall for protoplast release, followed by recovery on an osmotically stabilized agar medium (9). Additionally, these results indicate that by disrupting the NHEJ repair mechanism, the genomic integration of mutations is prevented, severely reduced, and/or cannot survive in the resulting transformants. This supports a previous suggestion (38) that the ΔkusA genetic background is a more stable genotype, and therefore more suitable for further transformation procedures in terms of mutation accumulation.
Fig 2.
Average number of random mutations caused by the transformation process. Average number of SNVs, insertions and deletions of the wild-type (WT, blue bars), and the ΔkusA strain (green bars) that were transformed with the ANEp8-pyrG plasmid in which cas9 is not present. Error bars represent the standard deviation (SD) of the replicate samples. Asterisks represent statistical significance for each of the different mutations (t test, P < 0.05). Note the break at Y-axis.
Finally, these results can provide an explanation for the percentage of WT ΔxlnR strains with unexpected morphologies detected after phenotypic analyses (see previous section), which can be attributed to the higher mutation accumulation rate in the WT.
The presence of Cas9 does not increase occurrence of mutations in A. niger
Apart from mutation that occurs due to the transformation process, we wanted to assess if the presence of Cas9 would increase the occurrence of genetic mutations, as increasing evidence has shown that high levels of Cas9 protein may have a toxic effect to host cells. In this sense, Cas9 endonuclease has been reported to exert toxicity against several organisms such as Saccharomyces cerevisiae (39), Cryptococcus neoformans (40), M. oryzae (4), and Fusarium venenatum (41). Additionally, some authors demonstrated that increasing Cas9 concentrations in the presence of functional gRNAs did not increase the occurrence of genomic mutations in A. fumigatus (9). However, there are no studies in which the genotoxicity of Cas9 without the presence of functional gRNAs that drive Cas9 endonuclease activity is reported to date.
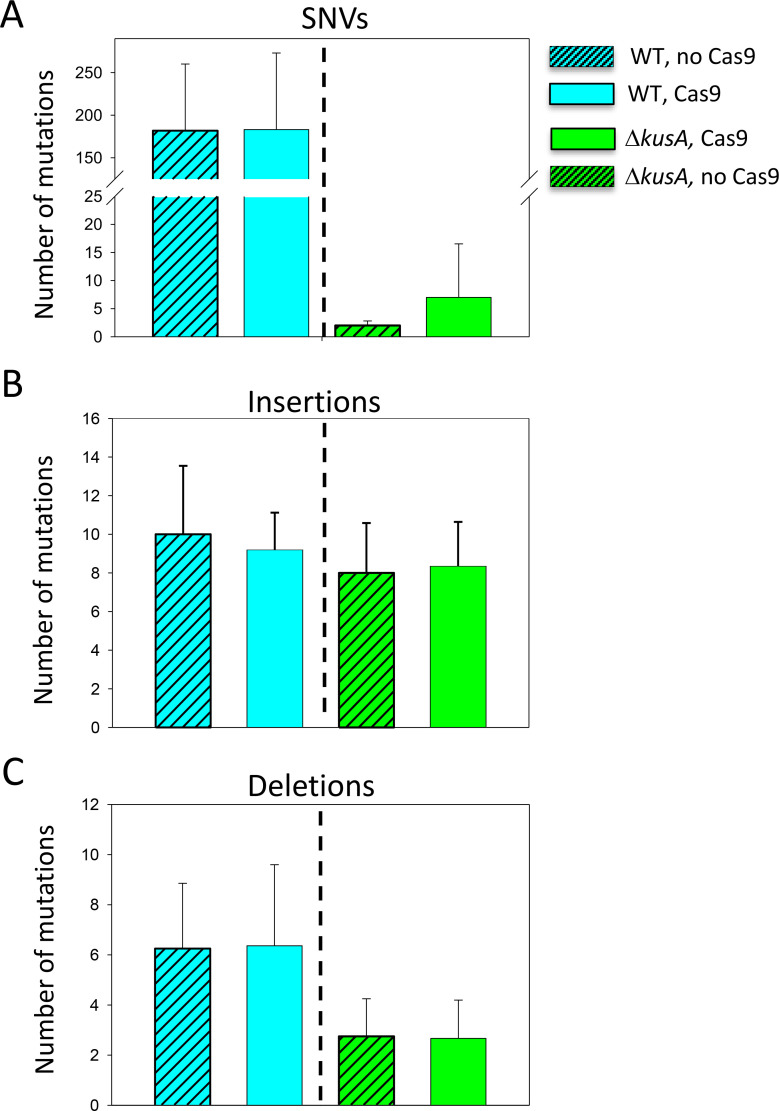
To determine if the presence of Cas9 (without any gRNA) in the cells increases the occurrence of genetic mutations, we compared and statistically analyzed the genetic variation identified after transformation in the presence and absence of Cas9 between the WT and ΔkusA strains. As shown in Fig. 3, there was no significant increase in the number of SNVs, insertions, and deletions when the ANEp8-Cas9-pyrG plasmid was transformed into the cells compared to the number of variations shown after transformation with an empty plasmid in any of the genetic backgrounds under study (P > 0.05) (Table S2). Thus, we show for the first time that the presence of Cas9 does not aggravate the occurrence of mutations in the absence of gRNAs. Additionally, the results confirmed that A. niger ΔkusA genetic background accumulates fewer genetic mutations, particularly SNVs (Fig. 3A, green bars) (Table S1), further demonstrating the higher genomic stability of the NHEJ-deficient strains compared to the WT in terms of mutation accumulation.
Fig 3.
Assessment of the number of random mutations caused by the presence of Cas9. Average number of SNVs (A), insertions (B), and deletions (C) in the wild-type (WT, blue bars) and ΔkusA strains (green bars) detected after transformation in the absence (patterned bars) or presence (solid bars) of Cas9 endonuclease. Error bars represent the standard deviation (SD) of the replicate samples. No difference in significance was found in any analysis (P < 0.05). Note the break in Y-axis of panel A.
NHEJ-deficient strains mitigate the impact of unintended mutations after the application of CRISPR/Cas9
To date, the putative occurrence of off-target mutations caused by CRISPR/Cas9 in the industrial workhorse A. niger had not been addressed. In addition, it has not been addressed so far whether the DNA repair mechanism used to repair DSBs or the use of NHEJ-deficient fungal strain would affect the occurrence of off-target mutations after CRISPR/Cas9 genome editing. In this study, we wanted to address these issues by comparing the number of genetic variants obtained in: (i) A. niger WT genetic background that repaired DSBs at the xlnR locus by NHEJ compared to HDR; (ii) the WT and ΔkusA genetic backgrounds each of them transformed with cas9-containing plasmid with no gRNA compared to that transformed with a functional CRISPR/Cas9 system; and (iii) CRISPR/Cas9-derived ΔxlnR strains in the WT compared to the ΔkusA genetic background.
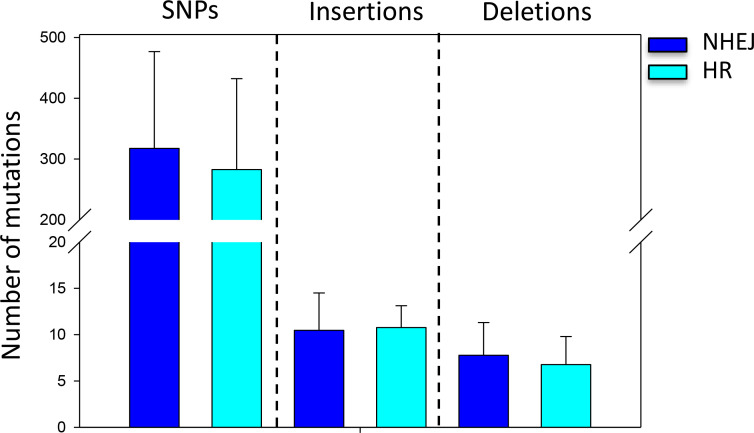
First, in order to analyze whether the DNA repair mechanisms influenced the number of genetic variants accumulated in the WT after a single CRISPR/Cas9 event, strains were transformed with the ANEp8-Cas9-pyrG plasmid that contained a xlnR-gRNA cassette together with a dDNA. In this situation, some strains repaired the DSBs via NHEJ, whereas others did this by HDR using the dDNA. NHEJ commonly resulted in short deletions and insertions, while the HDR resulted in full deletions (see example in Fig. S2). Results showed that there was no significant difference in the number of SNVs, insertions, or deletions (P > 0.05) in the WT strain regardless of the repair pathway (Fig. 4; Table S2). These results confirmed that neither the DNA repair mechanism, nor the presence of dDNA influenced the occurrence of mutations in the WT genetic background. Since there was no significant difference, these two categories were combined into a single category for subsequent analyses.
Fig 4.
Mutations arising in the wild type (WT) after xlnR disruption with CRISPR/Cas9 depending on the DNA repair mechanism. Average number of SNPs, insertions, and deletions in the WT ΔxlnR strains that underwent NHEJ (dark blue bars) or HR (light blue bars). Error bars represent the standard deviation (SD) of the replicate samples. No difference in significance was found in any analysis (P < 0.05). Note the break in Y-axis.
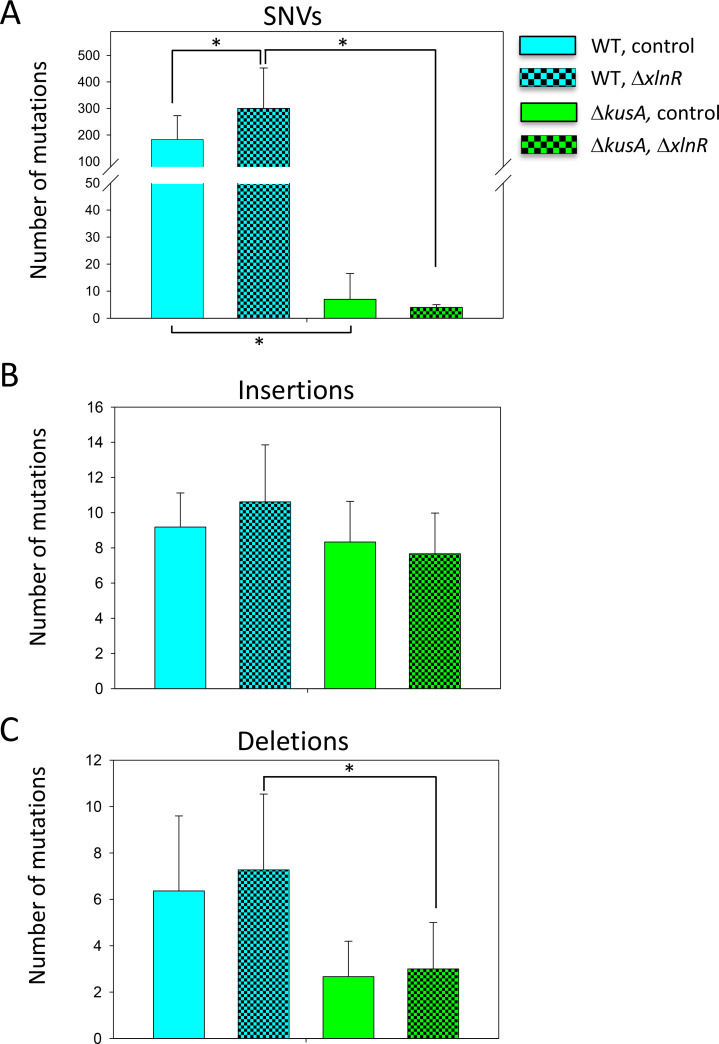
In parallel, we compared the two different background strains in which the xlnR gene was disrupted by CRISPR/Cas9 with their corresponding control strains that were transformed with cas9-containing plasmid but no gRNA. This way, we can determine if there is a significant increase in the number of mutations after the application of a functional CRISPR/Cas9 system within the cells. As shown in Fig. 5, there was a significant increase in the number of SNVs in the WT genetic background after the application of a functional CRISPR/Cas9 genome-editing event compared to the control (P = 0.023), with a 0.6-fold increase in this case (Table S1), while the number of insertions and deletions is more or less maintained and not significantly different in this genotype (Fig. 5, blue bars) (Table S2). In contrast, the number of SNVs, insertions, and deletions did not significantly change after the application of CRISPR/Cas9 in the NHEJ-deficient strains (Fig. 5, green bars) (Tables S1 and S2). Besides, there is statistical difference in the average number of SNVs and deletions between the WT ΔxlnR and ΔkusA ΔxlnR strains (P = 0.002 and P = 0.037, respectively), with a lower number of these mutations in the ΔkusA background strains (Fig. 5; Table S1). These results suggest that in the A. niger WT genetic background, there is a higher chance of genetic variant accumulation after the application of CRISPR/Cas9, which is mitigated in ΔkusA. Moreover, the number of mutations accumulated in ΔkusA is generally much lower than those in the WT, demonstrating that ΔkusA is a safer option when applying CRISPR/Cas9 in terms of mutation accumulation rate.
Fig 5.
Comparison of the number of mutations in xlnR deletion strains in the two different genetic backgrounds. Average number of SNVs (A), insertions (B), and deletions (C) in the wild-type strains (WT, blue bars) and in ΔkusA (green bars) which received the cas9-plasmid ANEp8-pyrG_cas9 without any functional guide (solid bars, control) and the strains that were transformed with the ANEp8-cas9-pyrG-xlnR plasmid (patterned bars). Error bars represent the standard deviation (SD) of the replicate samples. Difference in significance is represented by *P < 0.05. Note the break in Y-axis of panel A.
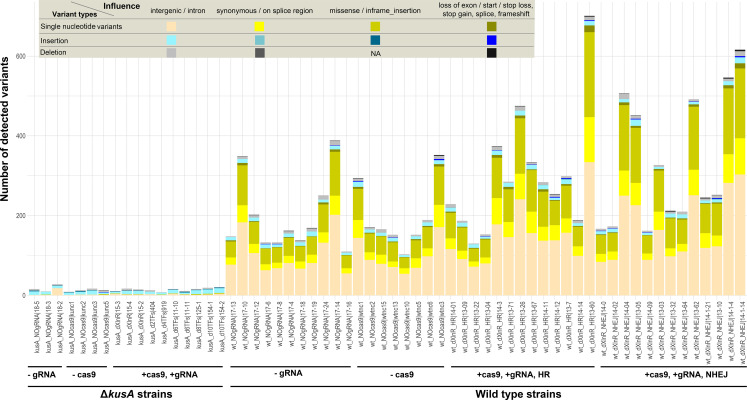
Having demonstrated the higher occurrence of mutation accumulation in the WT strain after the transformation process and the CRISPR/Cas9 application, we aimed to address the biological influence of the different variant types according to their location in the genome (Fig. 6). For the WT-derived strains, the majority of genetic variants detected (88–95%) are SNVs (Fig. 6, in green; Table S1). Whereas approximately half of the SNVs occurred in the intergenic or intron regions, the other major detected SNVs were related to splicing and protein-coding regions, and a small ratio of the detected SNVs was related to changes of start or stop codons or frameshift. Overall, many of these mutations may cause moderate effects on the function of the corresponding proteins, thus, resulting in unexpected morphologies that in the practice could be misattributed to the originally intended mutations. Noteworthy, we have detected a small number of insertion and deletion variants, which mainly occur in non-coding regions. The overall numbers and distribution of variants are consistent with a previous study performed in A. fumigatus (9). In contrast, we observed a much lower ratio of SNVs linked to non-coding regions in A. niger (~50%) than in A. fumigatus (~96%), which can be attributed to differences between species and/or analysis methods. In contrast, in the ΔkusA strains, only a limited number of genomic variants were detected. In these strains, SNVs only contributed to 10–30% of total variants (except for one sample with a ratio of 62%), with insertions being the predominant variants (Fig. 6, in blue; Table S1). Remarkably, most of the mutations took place in intergenic/intronic regions. Therefore, there is a small chance of affecting gene functionality.
Fig 6.
Distribution of the different types of genomic variants detected in each of the sequenced A. niger strains.
To further examine the genetic variants identified in both genetic backgrounds, we compared them with the predicted CRISPR/Cas off-targets using Cas-OFFinder. None of the variants were identified as putative Cas9 off-target by the software. However, we cannot discard the possibility that some of these mutations would have been caused by CRISPR/Cas9, especially in the WT, in which the mutation rate is much higher. In any case, using NHEJ-deficient strains mitigates the problem of mutation occurrence observed in the WT.
Editing of multiple loci is often required to either introduce multiple heterologous genes or fine-tune metabolic networks of microbial cell factories. The establishment of recyclable, marker-free CRISPR-Cas9 tools as the one used in this study (38) brought powerful genome engineering capabilities, considerably saving time and resources in strain construction programs (42). However, it has not been studied yet whether consecutive CRISPR/Cas9 transformations induced a higher accumulation of mutations compared to single transformations. For this, we stepwise obtained a 10-deletion mutant in the 10 studied transcription factors involved in plant biomass degradation (∆araR∆gaaR∆rhaR∆galX∆xlnR∆clrB∆amyR∆inuR∆clrA∆creA) (21) after five consecutive transformations using CRISPR/Cas9 in the NHEJ-deficient strain, in which we already saw a significant decrease in the accumulation of unintended mutations. Finally, the 10-deletion strain together with the intermediate 4-, 6-, and 8-deletion strains (see Materials and Methods) was analyzed by WGS (Fig. S3). Comparative analysis of the multi-deletion strains revealed again that only a small set of genetic variants accumulated in A. niger ΔkusA strains (Fig. S3A). Besides, all the 10 targeted transcription factors were precisely deleted, and only 23 extra genetic variants were accumulated after five rounds of consecutive CRISPR/Cas9 gene editing (Fig. S3B). All these data strongly support that NHEJ-deficient strains significantly mitigate the impact of the occurrence of unintended mutations during the application of CRISPR/Cas9.
Several studies have reported that numerous undesired mutations linked to CRISPR/Cas9 in different organisms (13, 43 - 45). Nevertheless, the assessment of CRISPR/Cas9-derived off-target mutations in filamentous fungi has been poorly exploited to date. In A. fumigatus WT, after WGS of six independent mutants, the CRISPR/Cas9 system was reported as a highly efficient and reliable method of gene targeting with no associated off-target mutations (9). In another study, after sequencing two independent CRISPR/Cas9-derived M. oryzae WT mutants, authors concluded that no off-target CRISPR-generated mutations occurred (4). In T. reesei, a constitutively active CRISPR/Cas9 system was reported to generate off-target mutations close to the sgRNA target site in the ura5 gene in seven independent mutants, although no WGS of these strains was performed (16). Finally, WGS of two independent bw2 knock-out strains revealed no obvious off-target effects in U. maydis (15). However, in all these studies, the number of mutants analyzed was very small to draw robust conclusions and only WT strains were taken into account. In this context, our study is the first one showing the occurrence of mutations in A. niger and the protective effect of the ΔkusA strains in the accumulation of such mutations in a large and representative set of CRISPR/Cas9-derived mutants.
Several research groups have put such an effort in developing technical solutions to address CRISPR/Cas9 safety concerns. These include reducing the levels of active Cas9 (46), reducing Cas9 lifetime (47, 48), shortening the gRNA sequence at the 5′-end region (49), or generating high fidelity Cas9 nucleases such as SpCas9-FokI, SpCas9-HF1, and eSp-Cas9, among others [reviewed in reference (50)]. In addition, optimum gRNA design is crucial to maintain editing specificity.
Bioinformatic tools are now available to search for potential Cas9 off-target sites, including Cas-Designer (51) and Cas-OFFinder (52), which we used in our study. Also, studies have been published on ways to minimize off-target mutations in CRISPR/Ca9 systems. For example, CRISPR Guide RNA Assisted Reduction of Damage (CRISPR GUARD) has been recently described as a method for protecting off-target sites by co-delivery of short gRNAs directed against off-target loci by competition with the on-target gRNA (53). However, based on our study, we propose that using NHEJ-deficient strains is one of the most efficient strategies to decrease the occurrence of unintended mutations after the application of CRISPR/Cas9 in filamentous fungi, especially for consecutive mutations.
In conclusion, as CRISPR/Cas9 genome-editing technology is widely used in many organisms, including filamentous fungi, the safety of genome-edited strains is a matter of discussion within the scientific community. In this study, we analyzed a large set of CRISPR/Cas9-derived fungal mutant strains to determine whether CRISPR/Cas9 induces off-target mutations in two different A. niger genetic backgrounds, the WT and NHEJ-deficient ΔkusA strains. Overall, we showed that the transformation process in itself is the most dominant cause of random mutations, but that this can be largely mitigated by using a NHEJ-deficient strain. CRISPR/Cas9 itself is a reliable and safe genome-editing technology when applied in a NHEJ-deficient strain. Thus, we strongly recommend the use of NHEJ-deficient strains during strain engineering due to their much higher genomic stability in terms of mutation accumulation. While we expect that the higher genome stability in NHEJ-deficient strains is a common phenomenon in fungi, similar evaluation of other species as presented here for A. niger should be performed to confirm this.
MATERIALS AND METHODS
Strains, media, and growth conditions
The ascomycete A. niger N593 (pyrG−, referred to as WT) and A. niger CBS138852 (pyrG− ΔkusA) strains were used as parentals for transformation. Strains were grown at 30°C in Aspergillus minimal medium (MM) or complete medium (CM) (54) supplemented with 1% D-glucose and 1.22 g/mL uridine (Sigma-Aldrich, Zwijndrecht, the Netherlands). Conidia were subsequently harvested, dispersed in sterile Milli-Q H2O, and the concentration was determined using a hemocytometer. For phenotypic characterization of the transformants, 250 spores were inoculated on MM plates supplemented with 1% beechwood xylan and 1.22 g/mL uridine (Sigma-Aldrich).
Escherichia coli DH5α was used to propagate plasmids, and was grown in Luria-Bertani medium containing 50 µg/mL of ampicillin (Sigma-Aldrich).
DNA constructs, fungal transformation, and screening of transformants
Plasmid constructions for the generation of A. niger deletion strains in the WT and NHEJ-deficient strains were performed as previously described (23). The self-replicative CRISPR/Cas9 plasmid ANEp8-Cas9-pyrG was used (AddGene #117169) (38). As control for transformation, the non-cas9-containing version of the ANEp8 plasmid (ANEp8-pyrG, empty plasmid) was also used. The gRNA sequences to delete xlnR (ID: NRRL3_04034), gaaR (ID: NRRL3_08195), araR (ID: NRRL3_07564), rhaR (ID: NRRL3_01496), galX (ID: NRRL3_07290), clrA (ID: NRRL3_03544), clrB (ID: NRRL3_09050), amyR (ID: NRRL3_07701), inuR (ID: NRRL3_03593), and creA (ID: NRRL3_05946) were designed with no predicted off-targets and the highest on-target activity using Geneious 11.1.4 (https://www.geneious.com) based on the experimentally determined predictive model described by reference (55). The dDNA constructs were obtained by fusion PCR using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Bleiswijk, the Netherlands) and include 500–1,000 bp of the 5′ and 3′ flanking regions of the target gene for HR. All primers used were purchased from Integrated DNA Technologies and are listed in Table S3.
A. niger protoplast generation and transformation were performed as previously described (27). In brief, 1 µg of either ANEp8-Cas9-pyrG or ANEp8-pyrG plasmid was transformed into the cells, and 5 µg of the dDNA was co-transformed to allow HR. Different transformation experiments were carried out: (i) both genetic background strains (WT and ΔkusA) were transformed with a version of the plasmid that contained the cas9 gene (ANEp8-Cas9-pyrG) without gRNA (ii); both backgrounds were transformed with an empty plasmid that did not contain cas9 (ANEp8-pyrG); and (iii) both genetic backgrounds were transformed with a cas9-containing plasmid (ANEp8-Cas9-pyrG) and the gRNA cassette for targeting xlnR gene with dDNA. Purification of transformant strains was achieved by two consecutive single colony streaking, followed by cultivating the strains on uridine-containing plates to promote the loss of the self-replicating plasmids. Subsequently, transformants were grown on medium containing 5-fluoro-orotic acid in order to screen for colonies which lost the plasmid. For the construction of multi-deletion strains, the A. niger ΔkusA strain was repeatedly transformed with a cas9- and a gRNA cassette-containing plasmid (ANEp8-Cas9-pyrG) together with the corresponding dDNA, knocking out two genes in each transformation round, followed by the removal of the self-replicating plasmid. First, the gaaR and araR genes were simultaneously knocked out. The double deletion strain ΔgaaRΔaraR was used as background for the deletion of rhaR and galX, resulting in the quadruple deletion strain ΔgaaRΔaraRΔrhaRΔgalX, which was used for the generation of the ΔgaaRΔaraRΔrhaRΔgalXΔxlnRΔclrB sextuple deletion strain (56). Finally, the octuple ΔgaaRΔaraRΔrhaRΔgalXΔxlnRΔclrBΔamyRΔinuR deletion strain was generated by the deletion of amyR and inuR in the sextuple deletion strain, while the decuple deletion strain ΔgaaRΔaraRΔrhaRΔgalXΔxlnRΔclrBΔamyRΔinuRΔclrAΔcreA was generated by the double deletion of clrA and creA in the octuple deletion strain (R. S. Kun, S. Garriegues, R. P. de Vries, unpublished data). Genomic DNA from transformants was isolated using Wizard Genomic DNA Purification Kit (Promega, Leiden, the Netherlands), and PCR reactions were performed to analyze the transformant colonies using GoTaq Flexi DNA Polymerase (Promega) following the manufacturer’s instructions.
Genome sequencing and variant calling
To investigate the possible off-target effects, we sequenced all mutant and reference strains using the Illumina NovaSeq 6000 system at Centre d’expertise et de services Génome Québec (https://cesgq.com/home). The resulting data were 250 bp paired-end reads, which were cleaned up using Fastp v0.23.2 (57) with default parameters.
Variants were called using the Breseq tool with default settings (58, 59). The genome and gene annotations of A. niger NRRL3 were used as reference (https://mycocosm.jgi.doe.gov/Aspni_NRRL3_1/Aspni_NRRL3_1.home.html) (60). Variants with quality score lower than 30 were filtered out. Mutations were annotated using SnpEff v5.0 (61). Only the mutations detected that were supported with at least 10 reads and were not present in the initial parental strains (WT or ΔkusA strain) were considered for further analysis. To assess whether the detected variants were potentially caused by CRISPR-Cas9, off-targets were predicted using Cas-OFFinder (51) by allowing “three mismatches with one DNA/RNA bulge.” On/off-target mutations were additionally visually confirmed using the Integrative Genomics Viewer (62).
Statistical analysis
Differences between the samples were analyzed using Student two-tailed t test. Statistical analyses were performed using STATGRAPHICS 18 (https://www.statgraphics.com/). Statistical significance was regarded as P < 0.05.
ACKNOWLEDGMENTS
The authors acknowledge Prof. Adrian Tsang (Concordia University, Montreal, Canada) who kindly provided the ANEp8-pyrG and ANEp8-Cas9-pyrG plasmids.
R.S.K. and S.G. were supported by a grant of the Applied Science Division (TTW) of NWO and the Biotechnology and Safety Program of the Ministry of Infrastructure and Water Management 15807 to R.P.D.V.
S.G. and R.S.K. carried out the experiments. M.P. performed the bioinformatic analyses. S.G. and M.P. analyzed the data. S.G. conducted the statistical analyses and wrote the original manuscript. M.P. contributed to manuscript writing. R.P.D.V. and S.G. designed the experiments. R.P.D.V. supervised the research, reviewed and edited the manuscript, and obtained the funding. All authors read and approved the final manuscript.
Contributor Information
Ronald P. de Vries, Email: r.devries@wi.knaw.nl.
Marcio Rodrigues, Oswaldo Cruz Foundation, Curitiba, Brazil .
DATA AVAILABILITY
In total, we generated WGS data sets for 65 different strains, including both the reference and CRISPR/Cas9-edited strains. The related sequencing data have been deposited in NCBI Sequence Read Archive (SRA) database under the BioProject accession number PRJNA931827 and SRA accession numbers SRR23345953–SRR23346017.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.00668-23.
Figures S1 to S3 and Tables S1 to S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Nielsen JC, Nielsen J. 2017. Development of fungal cell factories for the production of secondary metabolites: linking genomics and metabolism. Synth Syst Biotechnol 2:5–12. doi: 10.1016/j.synbio.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meyer V. 2021. Metabolic engineering of Filamentous fungi, p 765–801. In Nielsen J, Stephanopoulos G, Lee SY (ed), Metabolic engineering. doi: 10.1002/9783527823468 [DOI] [Google Scholar]
- 3. Davis AJ, Chen DJ. 2013. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foster AJ, Martin-Urdiroz M, Yan X, Wright HS, Soanes DM, Talbot NJ. 2018. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci Rep 8:14355. doi: 10.1038/s41598-018-32702-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Álvarez-Escribano I, Sasse C, Bok JW, Na H, Amirebrahimi M, Lipzen A, Schackwitz W, Martin J, Barry K, Gutiérrez G, Cea-Sánchez S, Marcos AT, Grigoriev IV, Keller NP, Braus GH, Cánovas D. 2019. Genome sequencing of evolved Aspergilli populations reveals robust genomes, transversions in A. flavus, and sexual aberrancy in non-homologous end-joining Mutants. BMC Biol 17:88. doi: 10.1186/s12915-019-0702-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bubendorfer S, Krebes J, Yang I, Hage E, Schulz TF, Bahlawane C, Didelot X, Suerbaum S. 2016. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter Pylori. Nat Commun 7:11995. doi: 10.1038/ncomms11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Márton L, Hrouda M, Pécsváradi A, Czakó M. 1994. T-DNA-insert-independent mutations induced in transformed plant cells during Agrobacterium co-cultivation. Transgenic Res 3:317–325. doi: 10.1007/BF01973592 [DOI] [PubMed] [Google Scholar]
- 8. Fierro F, Martín JF. 1999. Molecular mechanisms of chromosomal rearrangement in fungi. Crit Rev Microbiol 25:1–17. doi: 10.1080/10408419991299185 [DOI] [PubMed] [Google Scholar]
- 9. Al Abdallah Q, Souza ACO, Martin-Vicente A, Ge W, Fortwendel JR. 2018. Whole-genome sequencing reveals highly specific gene targeting by in vitro assembled Cas9-ribonucleoprotein complexes in Aspergillus fumigatus. Fungal Biol Biotechnol 5:11. doi: 10.1186/s40694-018-0057-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826. doi: 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJR, Boekema EJ, Dickman MJ, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 108:10092–10097. doi: 10.1073/pnas.1102716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawall K, Cotter J, Then C. 2020. Broadening the GMO risk assessment in the EU for genome editing technologies in agriculture. Environ Sci Eur 32:106. doi: 10.1186/s12302-020-00361-2 [DOI] [Google Scholar]
- 13. Sturme MHJ, van der Berg JP, Bouwman LMS, De Schrijver A, de Maagd RA, Kleter GA, Battaglia-de Wilde E. 2022. Occurrence and nature of off-target modifications by CRISPR-Cas genome editing in plants. ACS Agric Sci Technol 2:192–201. doi: 10.1021/acsagscitech.1c00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miao Y, Liu D, Li G, Li P, Xu Y, Shen Q, Zhang R. 2015. Genome-wide transcriptomic analysis of a superior biomass-degrading strain of A. fumigatus revealed active lignocellulose-degrading genes. BMC Genomics 16:459. doi: 10.1186/s12864-015-1658-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuster M, Schweizer G, Reissmann S, Kahmann R. 2016. Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet Biol 89:3–9. doi: 10.1016/j.fgb.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 16. Hao Z, Su X. 2019. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol 19:2. doi: 10.1186/s12896-018-0498-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakočiūnas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. 2015. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28:213–222. doi: 10.1016/j.ymben.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 18. van Peij NN, Visser J, de Graaff LH. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol 27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x [DOI] [PubMed] [Google Scholar]
- 19. Meyer V, Arentshorst M, El-Ghezal A, Drews A-C, Kooistra R, van den Hondel C, Ram AFJ. 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol 128:770–775. doi: 10.1016/j.jbiotec.2006.12.021 [DOI] [PubMed] [Google Scholar]
- 20. Battaglia E, Zhou M, de Vries RP. 2014. The transcriptional activators Arar and Xlnr from Aspergillus niger regulate expression of pentose catabolic and pentose phosphate pathway genes. Res Microbiol 165:531–540. doi: 10.1016/j.resmic.2014.07.013 [DOI] [PubMed] [Google Scholar]
- 21. Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, de Vries RP. 2017. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol Biofuels 10:152. doi: 10.1186/s13068-017-0841-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pineda M, Lear A, Collins JP, Kiani S. 2019. Safe CRISPR: challenges and possible solutions. Trends Biotechnol 37:389–401. doi: 10.1016/j.tibtech.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 23. Garrigues S, Kun RS, de Vries RP. 2021. Genetic barcodes allow traceability of CRISPR/Cas9-derived Aspergillus niger strains without affecting their fitness. Curr Genet 67:673–684. doi: 10.1007/s00294-021-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kun RS, Garrigues S, Di Falco M, Tsang A, de Vries RP. 2021. Blocking utilization of major plant biomass polysaccharides leads Aspergillus niger towards utilization of minor components. Microb Biotechnol 14:1683–1698. doi: 10.1111/1751-7915.13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyer V, Mueller D, Strowig T, Stahl U. 2003. Comparison of different transformation methods for Aspergillus giganteus. Curr Genet 43:371–377. doi: 10.1007/s00294-003-0406-3 [DOI] [PubMed] [Google Scholar]
- 26. Meng J, Németh Z, Peng M, Fekete E, Garrigues S, Lipzen A, Ng V, Savage E, Zhang Y, Grigoriev IV, Mäkelä MR, Karaffa L, de Vries RP. 2022. Galr, Galx and Arar co-regulate d-galactose and l-arabinose utilization in Aspergillus nidulans. Microb Biotechnol 15:1839–1851. doi: 10.1111/1751-7915.14025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kun RS, Meng J, Salazar-Cerezo S, Mäkelä MR, de Vries RP, Garrigues S. 2020. CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass. Enzyme Microb Technol 136:109508. doi: 10.1016/j.enzmictec.2020.109508 [DOI] [PubMed] [Google Scholar]
- 28. Garrigues S, Manzanares P, Marcos JF. 2022. Application of recyclable CRISPR/Cas9 tools for targeted genome editing in the Postharvest pathogenic fungi Penicillium digitatum and Penicillium expansum. Curr Genet 68:515–529. doi: 10.1007/s00294-022-01236-0 [DOI] [PubMed] [Google Scholar]
- 29. Ren Z, Su C, Yan J, Dai M, Zhao Y, Wang H, Zhang J. 2013. Establishment of a genetic transformation system for Penicillium brevicompactum to produce mycophenolic acid. Acta Microbiol Sin 53:1226–1232. [PubMed] [Google Scholar]
- 30. Salazar-Cerezo S, Kun RS, de Vries RP, Garrigues S. 2020. CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer. Enzyme Microb Technol 133:109463. doi: 10.1016/j.enzmictec.2019.109463 [DOI] [PubMed] [Google Scholar]
- 31. Randhawa A, Pasari N, Sinha T, Gupta M, Nair AM, Ogunyewo OA, Verma S, Verma PK, Yazdani SS. 2021. Blocking drug efflux mechanisms facilitate genome engineering process in Hypercellulolytic fungus, Penicillium funiculosum Ncim1228. Biotechnol Biofuels 14:31. doi: 10.1186/s13068-021-01883-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seekles SJ, Teunisse PPP, Punt M, van den Brule T, Dijksterhuis J, Houbraken J, Wösten HAB, Ram AFJ. 2021. Preservation stress resistance of Melanin deficient conidia from Paecilomyces variotii and Penicillium roqueforti mutants generated via CRISPR/Cas9 genome editing. Fungal Biol Biotechnol 8:4. doi: 10.1186/s40694-021-00111-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang X, Hu X, Jan S, Rasheed SM, Zhang Y, Du M, Yang E. 2021. Development of CRISPR-Cas9 genome editing system in Talaromyces marneffei. Microb Pathog 154:104822. doi: 10.1016/j.micpath.2021.104822 [DOI] [PubMed] [Google Scholar]
- 34. Cardoza RE, Vizcaino JA, Hermosa MR, Monte E, Gutiérrez S. 2006. A comparison of the phenotypic and genetic stability of recombinant Trichoderma spp. generated by protoplast- and Agrobacterium-mediated transformation. J Microbiol 44:383–395. [PubMed] [Google Scholar]
- 35. Dhawale SS, Paietta JV, Marzluf GA. 1984. A new, rapid and efficient transformation procedure for Neurospora. Curr Genet 8:77–79. doi: 10.1007/BF00405435 [DOI] [PubMed] [Google Scholar]
- 36. Liu Z, Friesen TL. 2012. Polyethylene glycol (PEG)-mediated transformation in filamentous fungal pathogens, p 365–375. In Bolton MD, Thomma BPHJ (ed), Plant fungal pathogens: methods and protocols. Humana Press, Totowa, NJ. doi: 10.1007/978-1-61779-501-5 [DOI] [PubMed] [Google Scholar]
- 37. Daly P, Slaghek GG, Casado López S, Wiebenga A, Hilden KS, de Vries RP, Mäkelä MR. 2017. Genetic transformation of the white-rot fungus Dichomitus squalens using a new commercial protoplasting cocktail. J Microbiol Methods 143:38–43. doi: 10.1016/j.mimet.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 38. Song L, Ouedraogo J-P, Kolbusz M, Nguyen TTM, Tsang A. 2018. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS One 13:e0202868. doi: 10.1371/journal.pone.0202868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, Cate JHD. 2014. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife 3:e03703. doi: 10.7554/eLife.03703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Wei D, Zhu X, Pan J, Zhang P, Huo L, Zhu X. 2016. A ’suicide' CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci Rep 6:31145. doi: 10.1038/srep31145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong S, An K, Chen W, Zhou W, Sun Y, Wang Q, Li D. 2022. Evasion of Cas9 toxicity to develop an efficient genome editing system and its application to increase ethanol yield in Fusarium venenatum Tb01. Appl Microbiol Biotechnol 106:6583–6593. doi: 10.1007/s00253-022-12178-5 [DOI] [PubMed] [Google Scholar]
- 42. Adiego-Pérez B, Randazzo P, Daran JM, Verwaal R, Roubos JA, Daran-Lapujade P, van der Oost J. 2019. Multiplex genome editing of microorganisms using CRISPR-Cas. FEMS Microbiol Lett 366:fnz086. doi: 10.1093/femsle/fnz086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L. 2017. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun 8:15464. doi: 10.1038/ncomms15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Höijer I, Emmanouilidou A, Östlund R, van Schendel R, Bozorgpana S, Tijsterman M, Feuk L, Gyllensten U, den Hoed M, Ameur A. 2022. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat Commun 13:627. doi: 10.1038/s41467-022-28244-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kosicki M, Tomberg K, Bradley A. 2018. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771. doi: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. 2015. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11:316–318. doi: 10.1038/nchembio.1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim S, Kim D, Cho SW, Kim J, Kim J-S. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019. doi: 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramakrishna S, Kwaku Dad A-B, Beloor J, Gopalappa R, Lee S-K, Kim H. 2014. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24:1020–1027. doi: 10.1101/gr.171264.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284. doi: 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aquino-Jarquin G. 2021. Current advances in overcoming obstacles of CRISPR/Cas9 off-target genome editing. Mol Genet Metab 134:77–86. doi: 10.1016/j.ymgme.2021.08.002 [DOI] [PubMed] [Google Scholar]
- 51. Bae S, Park J, Kim J-S. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30:1473–1475. doi: 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park J, Bae S, Kim J-S. 2015. Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31:4014–4016. doi: 10.1093/bioinformatics/btv537 [DOI] [PubMed] [Google Scholar]
- 53. Coelho MA, De Braekeleer E, Firth M, Bista M, Lukasiak S, Cuomo ME, Taylor BJM. 2020. CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs. Nat Commun 11:4132. doi: 10.1038/s41467-020-17952-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Vries RP, Burgers K, van de Vondervoort PJI, Frisvad JC, Samson RA, Visser J. 2004. A new black Aspergillus species, A. Vadensis, is a promising host for homologous and heterologous protein production. Appl Environ Microbiol 70:3954–3959. doi: 10.1128/AEM.70.7.3954-3959.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol 32:1262–1267. doi: 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garrigues S, Kun RS, Peng M, Bauer D, Keymanesh K, Lipzen A, Ng V, Grigoriev IV, de Vries RP. 2022. Unraveling the regulation of sugar beet pulp utilization in the industrially relevant fungus Aspergillus niger. iScience 25:104065. doi: 10.1016/j.isci.2022.104065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sun L, Shou W. 2014. Identification of mutations in laboratory-evolved Microbes from next-generation sequencing data using Breseq. Methods Mol Biol:0554–6_12. doi: 10.1007/978-1-4939-0554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. doi: 10.1186/1471-2164-15-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aguilar-Pontes MV, Brandl J, McDonnell E, Strasser K, Nguyen TTM, Riley R, Mondo S, Salamov A, Nybo JL, Vesth TC, Grigoriev IV, Andersen MR, Tsang A, de Vries RP. 2018. The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi. Stud Mycol 91:61–78. doi: 10.1016/j.simyco.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cingolani P, Platts A, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, p iso–2. In Snpeff: SNPs in the genome of Drosophila melanogaster strain W1118. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S3 and Tables S1 to S3.
Data Availability Statement
In total, we generated WGS data sets for 65 different strains, including both the reference and CRISPR/Cas9-edited strains. The related sequencing data have been deposited in NCBI Sequence Read Archive (SRA) database under the BioProject accession number PRJNA931827 and SRA accession numbers SRR23345953–SRR23346017.