ABSTRACT
Environmental DNA analyses of fungal communities typically reveal a much larger diversity than can be ascribed to known species. Much of this hidden diversity lies within undescribed fungal lineages, especially the early diverging fungi (EDF). Although these EDF often represent new lineages even at the phylum level, they have never been cultured, making their morphology and ecology uncertain. One of the methods to characterize these uncultured fungi is a single-cell DNA sequencing approach. In this study, we established a large data set of single-cell sequences of EDF by manually isolating and photographing parasitic fungi on various hosts such as algae, protists, and micro-invertebrates, combined with subsequent long-read sequencing of the ribosomal DNA locus (rDNA). We successfully obtained rDNA sequences of 127 parasitic fungal cells, which clustered into 71 phylogenetic lineages belonging to seven phylum-level clades of EDF: Blastocladiomycota, Chytridiomycota, Aphelidiomycota, Rozellomycota, and three unknown phylum-level clades. Most of our single cells yielded novel sequences distinguished from both described taxa and existing metabarcoding data, indicating an expansive and hidden diversity of parasitic taxa of EDF. We also revealed an unexpected diversity of endobiotic Olpidium-like chytrids and hyper-parasitic lineages. Overall, by combining photographs of parasitic fungi with phylogenetic analyses, we were able to better understand the ecological function and morphology of many of the branches on the fungal tree of life known only from DNA sequences.
IMPORTANCE
Much of the diversity of microbes from natural habitats, such as soil and freshwater, comprise species and lineages that have never been isolated into pure culture. In part, this stems from a bias of culturing in favor of saprotrophic microbes over the myriad symbiotic ones that include parasitic and mutualistic relationships with other taxa. In the present study, we aimed to shed light on the ecological function and morphology of the many undescribed lineages of aquatic fungi by individually isolating and sequencing molecular barcodes from 127 cells of host-associated fungi using single-cell sequencing. By adding these sequences and their photographs into the fungal tree, we were able to understand the morphology of reproductive and vegetative structures of these novel fungi and to provide a hypothesized ecological function for them. These individual host-fungal cells revealed themselves to be complex environments despite their small size; numerous samples were hyper-parasitized with other zoosporic fungal lineages such as Rozellomycota.
KEYWORDS: early diverging fungi, parasite, phylogeny, single-cell analysis
INTRODUCTION
Estimates on the number and diversity of fungi have been radically altered by the widespread adoption of culture-independent methods, such as metabarcoding and metagenomics (1 - 3). These studies highlight the gap between the formally described fungal taxa and the estimated diversity, suggesting perhaps only 5%–10% of all fungal species have been described (4, 5). Moreover, they often identify major gaps in our knowledge of fungal phylogeny, such as entirely new lineages of fungi that were previously undetected (6 - 9). As novel as these sequence-based discoveries can be, one of the major hurdles to really understanding fungal diversity is a phenotypic characterization of the novel fungal lineages that comprise the so-called dark matter fungi found in metabarcoding studies (10). One approach to breaking through this barrier is the development of single-cell sequencing methods that rely on direct observations of cells through microscopy that can then be isolated and subjected to DNA sequencing and phylogenetic comparison to novel lineages from environmental DNA surveys (11 - 15). This way, information on both the habitat (e.g., host or substrate) and morphology can be obtained for these dark matter lineages.
Single-cell methods are particularly appropriate for studying the early diverging fungi (EDF), which are primarily microscopic and often unicellular. Metabarcoding studies show that many habitats are rich in novel EDF (8, 9, 16). Knowledge of the full diversity of EDF is growing, and new phyla are continuing to be described in this part of the tree (17 - 19). The fact that the undescribed EDF have never been cultured is likely because many of these fungi are parasitic (20 - 22). These fungi comprise a large portion of communities and are thus also ecologically relevant (16, 23 - 25). EDF are involved in ecosystem functions such as organic matter decomposition and nutrient cycling, making ecosystems more complex, and thus contribute to food web stability (26, 27). However, the morphology and ecological role of these EDF are speculative because they are recognized based only on environmental sequences. In this study, we endeavored to illuminate the morphology of uncultured EDF by isolating, photographing, and DNA sequencing parasitic fungi from several different types of freshwater habitats.
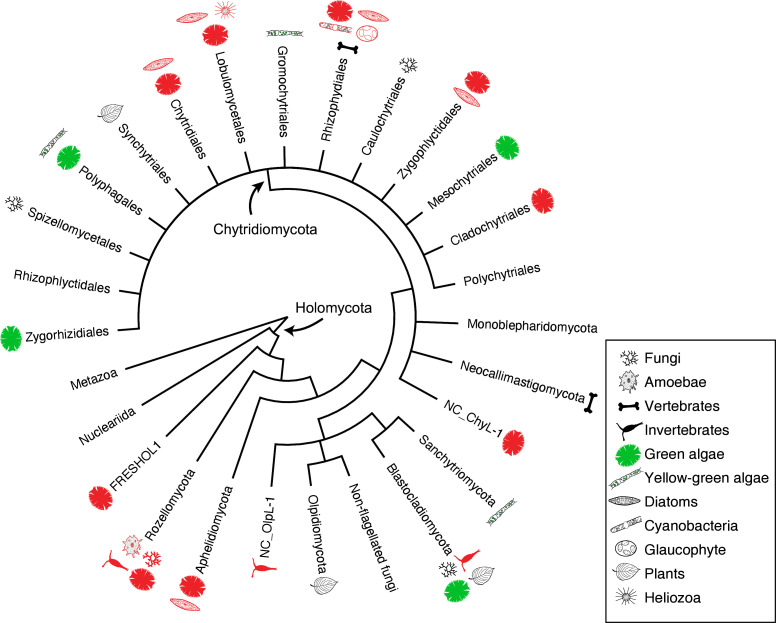
Our data fill in gaps in the constantly improving phylogenetic overview of EDF, which in the last two decades has been dramatically changed by extensive molecular phylogenetic analyses. Chytridiomycota (so-called chytrids) was divided into four independent phyla, Blastocladiomycota, Neocallimastigomycota, Monoblepharidomycota, and Chytridiomycota sensu stricto (28 - 30), plus the recent addition of phyla Olpidiomycota (18, 31) and Sanchytriomycota (17) (Fig. 1). In addition to chytrids sensu lato, Aphelidiomycota (=Aphelida, so-called aphelids, endoparasites of algae) and Rozellomycota (=Cryptomycota, so-called rozellids, endoparasites of fungi, animals, and protists) were recognized as the most basal lineages of fungi along with Microsporidia (5, 31). In some classifications, aphelids, rozellids, and Microsporidia have been treated as sister lineages of the true fungi because of the absence of a cell wall during the trophic phase and the presence of a phagotrophic nutrient strategy (although Microsporidia lack this feature) (22, 32, 33). The phylogeny and taxonomy of chytrids sensu lato have in the last two decades been biased toward culture-based observations and analyses mainly on saprotrophic chytrids (34). Parasitic chytrids as well as aphelids and rozellids can also be investigated by culture-based studies in which a parasite and its host are cultivated together and incorporated into phylogenetic analyses (20, 21, 35, 36), and these data have been vital for understanding host range across chytrid orders (Fig. 1). These culture-based studies have shown that algal parasites often represent new orders, families, or genera (20, 21, 37 - 41). Importantly, the orders Mesochytriales (20) and Zygophlyctidales (21) brought into formal definition novel clades that had previously only been known from environmental sequences (23, 42). Although further investigation of parasitic taxa is important to clarify the diversity of EDF, culture-based studies of parasitic taxa are difficult and time-consuming. Single-cell sequencing approaches can overcome some of these challenges and can be scaled up to higher throughput (11, 15).
Fig 1.
Schematic tree showing the phylogenetic relationships among the early diverging fungal phyla and orders in Chytridiomycota and their host range. Illustrations of each lineage indicate hosts of parasitic taxa. Red colored illustrations indicate hosts of single cells isolated in this study.
In this study, a single-cell isolation approach was employed along with long-read sequencing techniques (34, 43) to comprehensively isolate parasitic EDF on various hosts and determine their phylogenetic position based on ribosomal DNA (rDNA) sequences. Sequence data for 127 parasitic fungal cells were successfully obtained and revealed 71 lineages, many of which were phylogenetically distinguished from described taxa. Additionally, single-cell lineages were compared with long-read metabarcoding data from similar habitats (44) to assess the overlap between culture-independent methods.
RESULTS
Phylogenetic position of isolated single cells
Over 300 individual cells of chytrid-, aphelid-, and Microsporidia-like fungi associated with their various hosts such as green algae, diatoms, cyanobacteria, protists, and micro-invertebrates were isolated (Fig. 2 to 4). A single-cell pipeline was applied to 259 isolated cells (excluding some duplicated samples and putative non-fungal cells such as oomycetes and cercozoans, data not shown), and fungal rDNA sequences were successfully obtained for 129 cells by the Oxford Nanopore Technologies (ONT) or Sanger method (see Table S1 in the supplemental material). Excluding the two zygomycetous sequences (PSC016 and PSC279, Table S1), 127 sequences were used for subsequent analyses. Based on the phylogenetic analysis on the concatenated data set of 18S-5.8S-28S rDNA sequences (Fig. 5 to 10, full tree along with the photos of isolated cells is available as “pursuit_tree.html” at Deep Blue repository, https://dx.doi.org/10.7302/7000), the 127 cells were categorized into 71 lineages distributed among seven phylum-level clades of EDF: Blastocladiomycota, Chytridiomycota, Aphelidiomycota, Rozellomycota, and three clades of unknown phyla.
Fig 2.
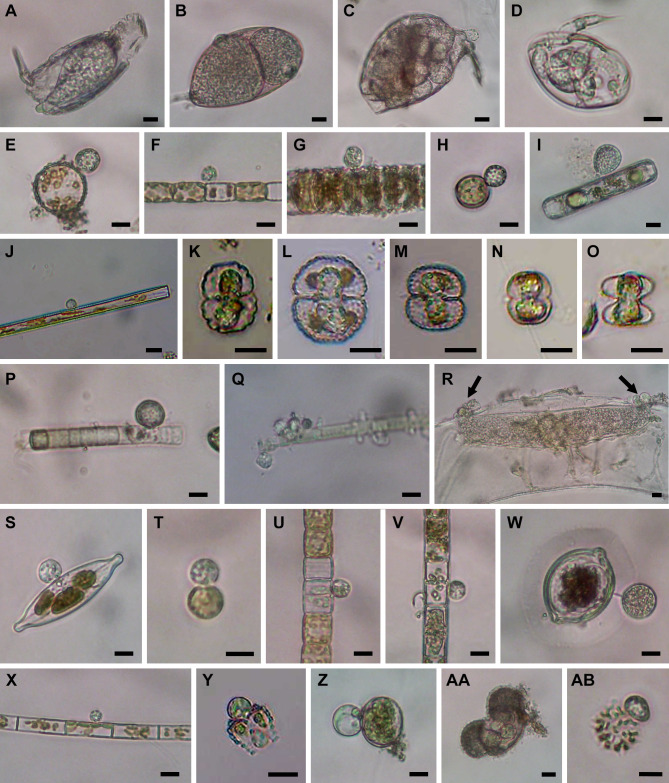
Microscopic images of isolated cells. (A) Olpidium-like chytrid PSC-L1 in rotifer. (B) Olpidium-like chytrid PSC-L2 in rotifer egg. (C) Olpidium-like chytrid PSC-L3 in rotifer. (D) Olpidium-like chytrid PSC-L4 in rotifer. (E–G) Chytrids PSC-L5 on Stephanodiscus spp. (E and G) and Stephanodiscus binderanus (F). (H) Chytrid PSC-L6 on Stephanodiscus sp. (I) Chytrid PSC-L7 on Pinnularia sp. (J) Chytrid PSC-L8 on Ulnaria sp. (K–O) Olpidium-like chytrids PSC-L9 in Cosmarium spp. (K–N) and Staurastrum sp. (O). (P and Q) Chytrid PSC-L10 on Oscillatoriales spp. (R) Hyper-parasitic chytrid PSC-L11 (arrows) attaching on elongated oomycete zoosporangium inside Spirogyra sp. (S) Chytrid PSC-L12 on Craticula sp. (T) Chytrid PSC-L13 on Conticribra sp. (U) Chytrid PSC-L14 on Stephanodiscus binderanus. (V) Chytrid on PSC-L15 on Aulacoseira sp. (W) Chytrid PSC-L16 on Desmidium sp. (X) Chytrid PSC-L17 on Aulacoseira sp. (Y) Chytrid PSC-L18 on Staurastrum sp. (Z) Chytrid PSC-L19 on Glaucocystis sp. (AA) Olpidium-like chytrid PSC-L20 in pine pollen. (AB) Chytrid PSC-L21 on Stauridium sp. All scale bars are 10 µm.
Fig 4.
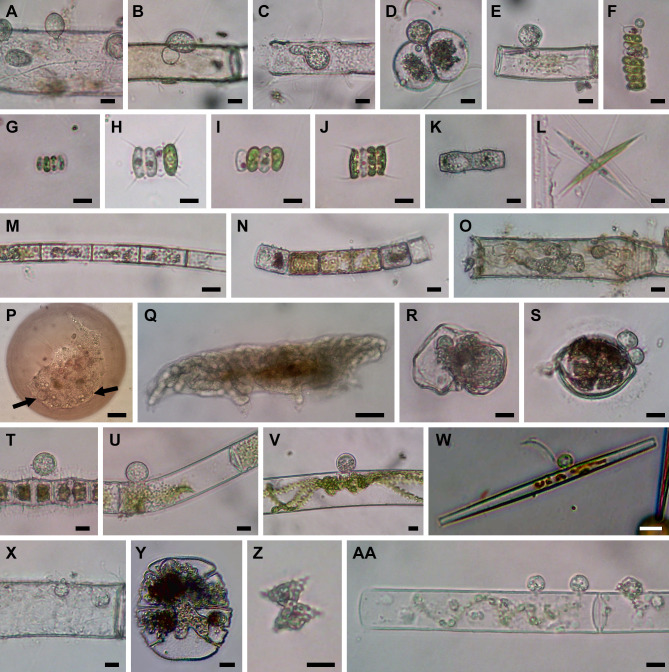
Microscopic images of isolated cells. (A) Chytrid PSC-L48 on Oedogonium sp. (B) Chytrid PSC-L49 on Oedogonium sp. (C) Chytrid PSC-L50 in Oedogonium sp. (D and E) Chytrid PSC-L51 on Cosmarium sp. (D) and Oedogonium sp. (E). (F) Chytrid PSC-L52 on Desmodesmus sp. (G and H) Aphelid PSC-L53 in Scenedesmus sp. (G) and Desmodesmus sp. (H). (I and J) Aphelid PSC-L54 in Scenedesmus sp. (I) and Desmodesmus sp. (J). (K) Two aphelids PSC-L55 and L59 in Bambusina sp. (L) Aphelid PSC-L58 in Ankistrodesmus sp. (M) Aphelid PSC-L56 in Aulacoseira sp. (N) Aphelid PSC-L57 in Melosira varians. (O) Isolated cell of rozellid PSC-L60 including Oedogonium sp. and endobiotic, tube-shaped zoosporangia. (P) Microsporidia-like rozellid PSC-L61 (indicated by arrows) in Arcella sp. (Q) Isolated cell of rozellids PSC-L62 including tardigrade and tube-shaped zoosporangia. (R) Isolated cell of rozellids PSC-L63 including putative broken rotifer body and endobiotic zoosporangium. (S−Y) Hyper-parasitic Rozella infecting parasitic chytrids: PSC-L64 in chytrids on Desmidium sp. (S) and Bambusina sp. (T), PSC-L65 in chytrid on Mougeotia sp. (U), PSC-L66 in chytrid on Spirogyra sp. (V), PSC-L67 in chytrid on Ulnaria sp. (W), PSC-L68 in chytrid in Oedogonium sp. (X), and PSC-L69 in Olpidium-like chytrid in Micrasterias truncata (Y). (Z) Staurastrum sp. harboring unknown fungus PSC-L70. (AA) Isolated cell of unknown fungus PSC-L71 including Spirogyra sp. and attaching chytrid-like sporangia.
Fig 5.
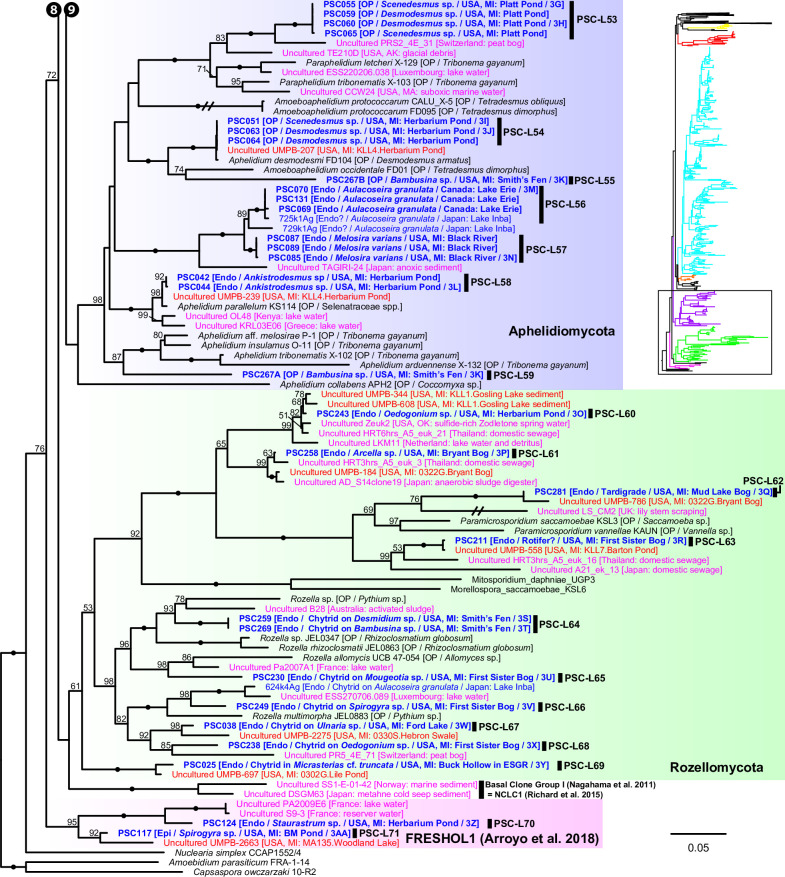
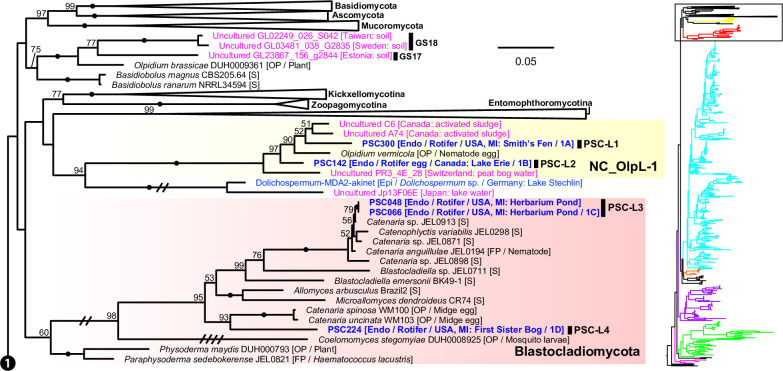
Maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set. Outer ring indicates the host/substrate of each culture or single cell. Brach color indicates sequence types (single cell, environmental DNA, PacBio OTU in this study, or culture/specimen). Blue circles on the tips indicate single-cell sequences obtained in this study. Red stars on the nodes indicate single-cell lineages reported in this study and the numbers correspond to the lineage numbers in the text (PSC-L1–71).
Fig 10.
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including Aphelidiomycota, Rozellomycota, the NCLC1 and FRESHOL1 clade, Nuclearia simplex, and outgroup taxa (two holozoan taxa).
We found five single-cell lineages that could not be placed into any phylum (i.e., phylum incertae sedis) (Fig. 5). Two lineages were Olpidium-like endoparasites of adult rotifers (PSC-L1; Fig. 2A) and rotifer eggs (PSC-L2; Fig. 2B) and formed a novel clade named NC_OlpL-1 (Novel Clade of Olpidium-like-1, Fig. 6). This clade also includes three environmental sequences and the unpublished sequence data of Olpidium vermicola. PSC-L52, an epibiotic chytrid on Desmodesmus sp. (Fig. 4F), was related to Rhizophydium scenedesmi strain EPG01 on Grasiella sp. (45). Along with some environmental sequences, these chytrids formed a distinct clade named NC_ChyL-1 (Novel Clade of Chytrid-like-1, Fig. 9), which is sister to Monoblepharidomycota, but statistical support for this relationship was not robust. Two lineages were placed in the clade FRESHOL1 reported previously (6) (Fig. 10). PSC-L70 was the cell of Staurastrum sp. filled by uncolored particles (Fig. 4Z). PSC-L71 was epibiotic chytrid-like cells on Spirogyra sp. (Fig. 4AA).
Fig 6.
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including Ascomycota, Basidiomycota, Mucoromycota, Entomopthoromycotina, Kickxellomycotina, Zoopagomycotina, Blastocladiomycota, and the NC_OlpL-1 clade. ML bootstrap values higher than 50% were shown on each branch. Black dots on branches indicate 100% bootstrap value. Double and quadruple slashes on branches indicate that length is reduced by half and quarter, respectively. Cultured fungi are labeled in black; saprotrophs are indicated as [S], and obligate [OP] and facultative [FP] parasites are indicated as [O(F)P / its host]. Single cells isolated in this study are labeled in bold blue and previously published sequences of single cells are labeled in blue; annotations are indicated as [Endo (endobiotic) or Epi (epibiotic) / host / isolation source / figure number if available]. Published environmental DNA sequences are labeled in pink and PacBio OTU sequences in this study are labeled in bold red; source of each sequence is described in parentheses.
Fig 9.
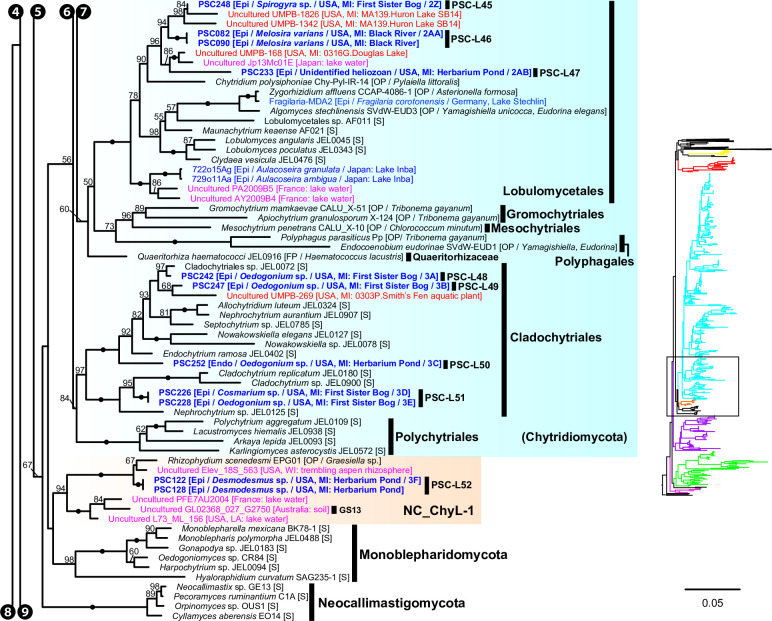
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including Monoblepharidomycota, Neocallimastigomycota, the NC_ChyL-1 clade, and orders Lobulomycetales, Gromochytriales, Mesochytriales, Polyphagales, Cladochytrilaes, and Polychytriales in Chytridiomycota.
Only two lineages, both endobiotic parasites of adult rotifers, were placed in the Blastocladiomycota (Fig. 5 and 6). PSC-L3 (Fig. 2C) was placed in the clade including Catenaria anguillulae and Catenophlyctis variabilis. PSC-L4 (Fig. 2D) was sister to two Catenaria spp. parasitic on midge eggs (46, 47).
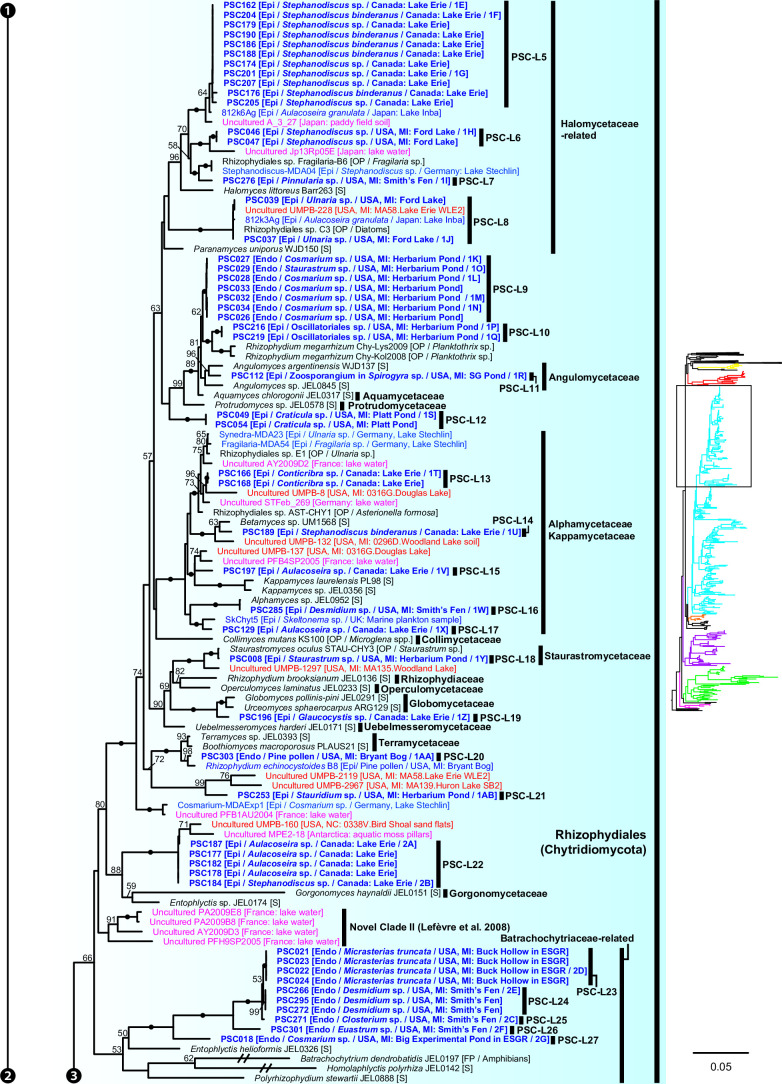
Most (n = 47) lineages were placed in Chytridiomycota, distributed among five orders: Rhizophydiales, Zygophlyctidales, Chytridiales, Lobulomycetales, and Cladochytriales (Fig. 5). Rhizophydiales was the most abundant in our isolates including 23 lineages (Fig. 7) on various hosts or substrates: 10 on diatoms (PSC-L5–8, 12–15, 17, 22; Fig. 2E through J, S through V, X, and 3A and B), 8 on zygnematophycean green algae (PSC-L9, 16, 18, 23–27; Fig. 2K through O, W, Y and 3C through G), and 1 each on chlorophycean green algae (PSC-L21; Fig. 2AB), glaucophyte algae (PSC-L19; Fig. 2Z), cyanobacteria (PSC-L10; Fig. 2P and Q), pine pollen (PSC-L20; Fig. 2AA), and oomycetes (PSC-L11; Fig. 2R). PSC-L11 was a putative hyper-parasitic chytrid attached to an endobiotic oomycete zoosporangium parasitizing Spirogyra sp. (Fig. 2R, arrows). Of the Rhizophydiales lineages, 16 exhibited typical epibiotic zoosporangium morphology, but the other seven were endobiotic zoosporangia in zygnematophycean green algae (PSC-L9, 23–27; Fig. 2K through O and 3C through G) or pine pollen (PSC-L20; Fig. 2AA). Most of our Rhizophydiales cells were distinguished from any cultivated chytrids, while three lineages (PSC-L8, 16, 18) were nearly identical to cultures of parasitic or saprotrophic chytrids.
Fig 7.
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including order Rhizophydiales in Chytridiomycota.
Fig 3.
Microscopic images of isolated cells. (A and B) Chytrid PSC-L22 on Aulacoseira sp. (A) and Stephanodiscus sp. (B). (C) Olpidium-like chytrid PSC-L25 in Closterium sp. (D) Olpidium-like chytrid PSC-L23 in Micrasterias truncata. (E) Olpidium-like chytrid PSC-L24 in Desmidium sp. (F) Olpidium-like chytrid PSC-L26 in Euastrum sp. (G) Olpidium-like chytrid PSC-L27 in Cosmarium sp. (H) Chytrid PSC-L29 on Fragilaria sp. (I) Chytrid PSC-L28 on Stephanodiscus binderanus. (J) Chytrid PSC-L30 on Aulacoseira ambigua. (K) Chytrid PSC-L31 on Sphaerocystis sp. (L) Chytrid PSC-L32 on Mougeotia sp. (M) Chytrid PSC-L33 on Mougeotia sp. (N) Two chytrids PSC-L32 and L33 on Mougeotia sp. (O) Chytrid PSC-L34 on Desmidium sp. (P) Chytrid PSC-L35 on Bambusina sp. (Q) Chytrid PSC-L36 on Cosmarium sp. (R) Chytrid on Desmidium sp. (S) Chytrid PSC-L38 on Desmidium sp. (T) Hyper-parasitic chytrid PSC-L39 attaching on another chytrid on Stephanodiscus binderanus. (U) Chytrid PSC-L40 on Mougeotia sp. (V) Chytrid PSC-L41 on Spirogyra sp. (W) Chytrid PSC-L42 on Spirogyra sp. (X) Chytrid PSC-L43 on Mougeotia sp. (Y) Chytrid PSC-L44 on Desmidium sp. (Z) Chytrid PSC-L45 on Spirogyra sp. (AA) Chytrid PSC-L46 on Melosira varians. (AB) Chytrid PSC-L47 on unidentified heliozoan. All scale bars are 10 µm.
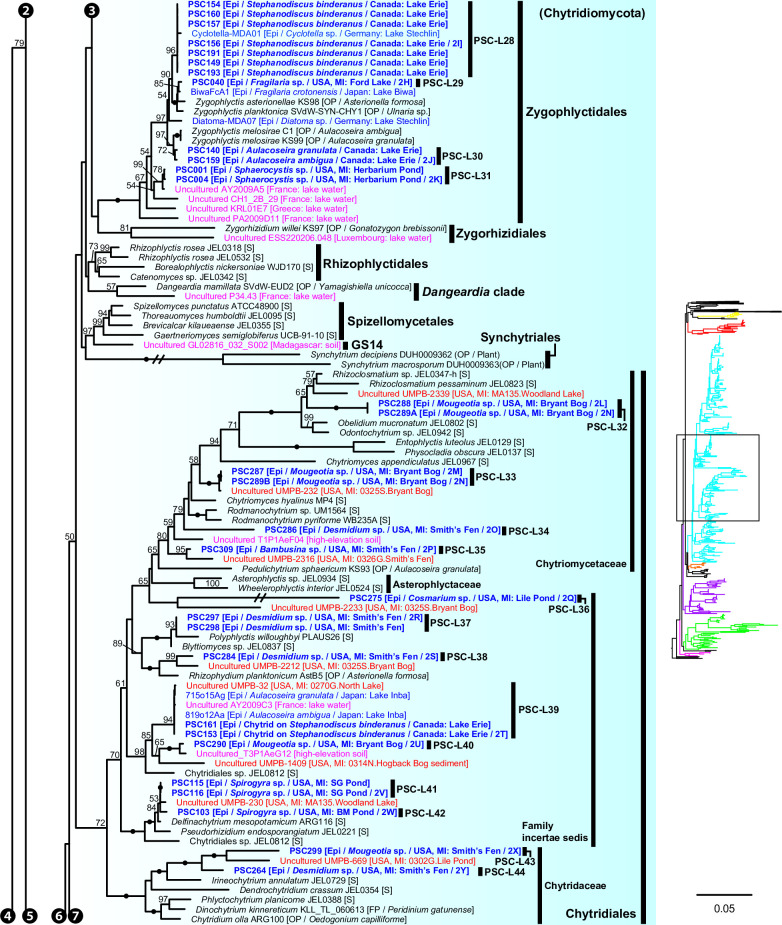
In Zygophlyctidales (Fig. 8), three lineages of diatom parasites (PSC-L28–30; Fig. 3H through J) formed a clade along with known diatom parasitic species, Zygophlyctis asterionellae, Z. planktonica, and Z. melosirae. An additional lineage, PSC-L31, was parasitic on the green alga Sphaerocystis sp. (Fig. 3K) and closely related to the environmental sequence AY2009A5 from a lake in France (48).
Fig 8.
Portion of maximum likelihood (ML) tree of 18S-5.8S-28S rDNA concatenated data set including orders Zygophlyctidales, Zygorhizidiales, Rhizophlyctidales, Spizellomycetales, Synchytriales, and Chytridiales in Chytridiomycota.
In Chytridiales (Fig. 8), 13 lineages were found, 12 of which were epibiotic chytrids on zygnematophycean green algae such as Bambusina (PSC-L35; Fig. 3P), Cosmarium (PSC-L36; Fig. 3Q), Desmidium (PSC-L34, 37, 38, and 44; Fig. 3O, R, S and Y), Mougeotia (PSC-L32, 33, 40, and 43; Fig. 3L through N, U, and X), and Spirogyra (PSC-L41 and 42; Fig. 3V and W). Regarding the cell PSC289 (Fig. 3N), sequences of two independent lineages (PSC-L32 and 33) were obtained by ONT sequencing, indicating that two morphologically similar chytrids infected a single host. An additional lineage, PSC-L39 (Fig. 3T), was a putative hyper-parasitic chytrid on the Zygophlyctidales chytrid PSC-L28 on S. binderanus (see Discussion). Six lineages belonged to the known families Chytriomycetaceae (n = 4) and Chytridiaceae (n = 2), but they were distinct from any described taxa. Outside of these families, four additional lineages (PSC-L36, 38, 39, and 40) related to environmental sequences were found. In contrast, three lineages were closely related to described taxa. PSC-L37 (Fig. 3R) could be morphologically identified as Polyphlyctis unispina which was originally found from the same location as our isolates (49). This lineage was sister to another species of the genus, P. willoughbyi. PSC-L41 and 42 (Fig. 3V and W) were sister to the saprotrophic chytrid Delfinachytrium mesopotamicum.
In Lobulomycetales (Fig. 9), three lineages were found: PSC-L45 on Spirogyra sp. (Fig. 3Z), PSC-L46 on Melosira varians (Fig. 3AA), and PSC-L47 on an unidentified heliozoan (Fig. 3AB). These lineages were related to environmental sequences and separated from the core Lobulomycetaceae clade including the type genus Lobulomyces.
Three lineages of putative saprotrophs on dead green algae were placed in Cladochytriales (Fig. 9). PSC-L48 (Fig. 4A) and PSC-L49 (Fig. 4B) on Oedogonium spp. were characterized by an epibiotic zoosporangium with a conspicuous endobiotic apophysis, resembling described taxa such as Chytridium lagenaria and C. schenkii (50, 51). PSC-L50 (Fig. 4C) could be an endobiotic zoosporangium with a discharge tube inhabiting the cell of dead Oedogonium sp. PSC-L51 on Cosmarium sp. (Fig. 4D) and Oedogonium sp. (Fig. 4E) was sister to Cladochytrium spp.
We found seven lineages in Aphelidiomycota (Fig. 10). Aphelid cells were recognizable based on host algal cells filled by a parasite cell with a conspicuous, red-colored residual body (Fig. 4G through N). Two lineages were parasitic on Desmodesmus and Scenedesmus and separated into independent clades: PSC-L53 (Fig. 4G and H) was placed in the clade including Paraphelidium spp. on Tribonema gayanum (52, 53) and PSC-L54 (Fig. 4I and J) was nearly identical to Aphelidium desmodesmi on Desmodesmus armatus (54). Similar to the cell PSC289 in Chytridiales, PSC267 on Bambusina sp. (Fig. 4K) included two distinct lineages: PSC-L55 sister to Amoeboaphelidium occidentale on Scenedesmus dimorphus (55, 56) and PSC-L59 sister to Aphelidium spp. parasitic on T. gayanum (57 - 60). Two lineages of diatom parasites, PSC-L55 on A. granulata (Fig. 4M) and PSC-L56 on M. varians (Fig. 4N), were distinct from described species. PSC-L58 on Ankistrodesmus sp. (Fig. 4L) was closely related to A. parallelum parasitic on selenastracean green alga (61).
In Rozellomycota, 10 single-cell lineages were found (Fig. 10). Many of them were recognized as epibiotic or endobiotic zoosporangia on algae (Fig. 4O, S through Y) or micro-invertebrates (Fig. 4Q and R). PSC-L61 was a cell of Arcella sp. harboring a sac-like structure including Microsporidia-like spores (arrows in Fig. 4P). This appearance is similar to endoparasites of amoebae such as Paramicrosporidium (62) and Morellospora (63), which produce Microsporidia-like spores but are phylogenetically placed in Rozellomycota and much shorter branched than canonical Microsporidia. PSC-L61 was distinguished from these previously reported Microsporidia-like taxa. Five lineages (PSC-L64–68) were isolated as epibiotic chytrids on green algae (Fig. 4S through V, X) and diatoms (Fig. 4W). However, they were positioned in the Rozella clade, which comprises endoparasites of chytrids and oomycetes, suggesting they were hyper-parasites of chytrids. PSC-L69 (Fig. 4Y), which showed the same morphology as PSC-L23 (Fig. 3D) in Rhizophydiales, was sister to all other Rozellomycota taxa. This lineage is also a putative Rozella-like hyper-parasite (see Discussion). PSC-L60 was a tube-shaped zoosporangium in Oedogonium sp. (Fig. 4O) and placed in the LKM11 clade (64). PSC-L62 in a tardigrade (Fig. 4Q) and PSC-L63 in a putative broken rotifer body (Fig. 4R) were related to Paramicrosporidium spp. These zoosporangium-like structures in PSC-L60, L62, and L63 may not correspond to rozellids, and the sequences could be derived from hyper-parasites of these zoosporangia or undetected contaminated cells.
Phylogenetic relationship between single cells and environmental sequences
The concatenated data set analysis showed that many of our single cells represented novel lineages distinguished from described taxa (Fig. 6 to 10). This result complements many environmental DNA studies that have reported unknown fungal lineages (23, 48). To examine overlap between our single-cell lineages and sequences only known from environmental DNA, we conducted a phylogenetic analysis on a comprehensive 18S rDNA data set including described taxa, environmental sequences available from NCBI database, with a focus on the phylum Chytridiomycota and phylum incertae sedis clades (see Fig. S1 in the supplemental material). In this analysis, new sequences of PacBio metabarcoding analyses primarily from Michigan, USA (44), were also used. Many of these PacBio sequences are derived from the same locations as the single cells isolated in this study, which gives a good opportunity to compare the two methods, metabarcoding and single-cell analysis, for exploring novel fungal diversity. The ML tree (see Fig. S1) showed that the vast majority of PacBio sequences represent entirely new lineages. Only a few of the single-cell lineages were closely related to PacBio environmental sequences. The PSC-L8 clade, in Rhizophydiales, included sequence UMPB-228 from Ford Lake, the same place where the two diatom parasites were isolated. In Chytridiales, PSC-L39 included UMPB-32 detected from multiple freshwater environments including Lake Erie (see Data S1 in the supplemental material) where some of the single cells were isolated. This lineage was also found in lakes in Japan (15) and France (48). Sequence UMPB-232 was the most abundant in Bryant Bog (see Data S1) and was related to PSC-L33 isolated from the same location. Although overlap of lineages in the PacBio and single-cell data sets was low in terms of species, multiple lineages of single cells had as their most closely related sequence an OTU from the PacBio data set, e.g., PSC-L13 and 21 in Rhizophydiales; PSC-L33, 35, 38, 43, and 44 in Chytridiales; PSC-L45 and 46 in Lobulomycetales; PSC-L49 in Cladochytriales; and PSC-L71 in the FRESHOL1 clade (see Fig. S1). On the other hand, both Mesochytriales and Polyphagales were represented by multiple OTUs in the PacBio data set but were absent in the single-cell data. Despite these exceptions, the general pattern was one of significant overlap of taxonomic genera and families in these two culture-independent approaches.
DISCUSSION
By utilizing single-cell techniques, 71 EDF lineages were sampled, many of which were newly recognized branches in the phylogeny of EDF, even at the phylum-level. The approach in the present study focused on targeting and sequencing individual EDF cells one at a time. Photographs of the isolated cells have implications for the ecology, morphology, and life cycle of these newly discovered EDF lineages. Using these data, we discuss the (i) ecological role of these uncultivated lineages, (ii) ecology and morphology of novel phylum-level clades, (iii) phylogenetic diversity of an enigmatic chytrid genus Olpidium, and (iv) unexpected recovery of hyper-parasitic lineages. Finally, technical advances and challenges of single-cell approaches used in this study are also discussed.
Shedding light on the ecological role of dark matter fungi
The cells isolated in this study were from diverse hosts, ranging from amoebae to invertebrates and especially algae. Some lineages are readily recognized as obligate algal parasites belonging to known parasite-specific groups such as Zygophlyctidales and Aphelidiomycota. Also found were many lineages of alga-associated cells in well studied orders such as Rhizophydiales, whose diversity has long been investigated based on numerous strains of saprotrophic taxa (65 - 67). Recently though several families of obligate algal parasites were described (37 - 68 - 68). Our data revealed further hidden diversity of putative parasitic lineages, representing new families or genera in the order. Although the isolated cells in this study were initially identified as “parasitic fungi,” some lineages we sampled in Chytridiales and Cladochytriales are putatively saprotrophic. When parasitic chytrids infect colonial algae, only dead cells are infected while living cells are uninfected (Fig. 2F, U, V and 3I, AA). In contrast, in some colonies, all algal cells are uncolored, or their chloroplasts are exhausted (Fig. 3O, P, R and Y), indicating the attaching chytrids grow on dead or moribund algae. Moreover, chytrids corresponding to PSC-L33 (Fig. 3M) in Chytridiales and PSC-L48 (Fig. 4A) in Cladochytriales were successfully isolated as pure cultures (data not shown).
These data also inform hypothetical ecological functions of lineages that were only known from metabarcoding approaches. For example, algal parasitic lineages PSC-L21 and 22 in Rhizophydiales formed independent clades along with some environmental sequences from aquatic environments, implicating a role for this clade as parasites of algae. Similarly, PSC-L45–47 in Lobulomycetales formed a novel clade including some environmental sequences from aquatic and soil environments, indicating that these lineages are parasites of algae and protists. Although Zygophlyctidales was previously thought to be composed only of diatom parasites (21), a lineage of a green algal parasite (PSC-L31) sister to an environmental sequence from a lake was found. This result indicates that other environmental sequence lineages in the order could exist as parasites of algae other than diatoms.
Generally, however, most of the single-cell sequences were poor matches at the species level to sequences from cultures or environmental DNA. This speaks to just how poorly we understand the true species level diversity of EDF, and how much work remains to be done in describing these fungi. In some cases, sequences from clades that were readily recovered with metabarcoding were not detected. The most striking case is Gromochytriales and Mesochytriales, together containing a total of three described species, all of which are obligate parasites (20, 69, 70). Despite limited described species, Mesochytriales is represented by numerous environmental DNA sequences (20). In this study, chytrids belonging to these orders were not identified with a single-cell approach. Instead, additional diversity was revealed based on phylogenetic analysis using PacBio metabarcoding data (see Fig. S1). Many of the sequences from lakes or ponds and some OTUs from Lake Erie, were related to Mesochytrium penetrans. This species is a parasite of a small green alga, Chlorococcum minutum (71), yet the collection strategy adopted for Lake Erie samples biased for larger colonial and filamentous forms. Further, most effort on Lake Erie was aligned with a winter science initiative (72), a season where diatoms are the dominant taxa associated with ice-cover in the lake (73). Thus, chytrids parasitic on smaller single cells or on taxa more prevalent during the summer may have been overlooked. More single-cell analyses on parasitic chytrids on various algae are necessary to reveal hidden taxa in the order.
While the pictures of the isolated cells can be informative in inferring their ecological role, sometimes they may be misleading. Specifically, each “cell” is actually a number of cells that include host, parasite, associated bacteria, and hyper-parasites. The latter were particularly common with some cells, such as PSC023, giving both an obvious chytrid pathogen as well as a likely Rozella hyper-parasite (74). Indeed, the majority of the Rozellomycota detected in this study were found as “by-catch” present in samples appearing as normal chytrids infecting algae (Fig. 4S through W). This result provides a slight cautionary tale that some of the sequences emerging from this approach may be most appropriately assigned not as parasites of the primary host but as hyper-parasites, an observation consistent with earlier results (15).
Discovery of novel clades of early diverging fungi
Our approach was successful in revealing novel diversity at many taxonomic levels: species, genera, families, and even phyla. The novelty at the phylum level is consistent with recent phylogenetic analyses that have revealed that some parasitic fungi represent novel lineages worthy of phylum-level distinction (17, 18). We found and characterized three phylum incertae sedis clades (Fig. 5), two of which are newly reported in the present study. The NC_OlpL-1 clade includes two single-cell lineages of Olpidium-like chytrids on rotifers and O. vermicola parasitic on nematode eggs (75), indicating that this clade represents animal-associated endobiotic chytrids. NC_OlpL-1 was sister to a previously reported undescribed phylum-level clade represented by a single-cell isolate of a Rhizosiphon-like chytrid on the cyanobacterium Dolichospermum from a lake in Germany (11). In the tree by Van den Wyngaert et al. (11), Dolichospermum parasites were sister to the Chytridiomycota + Monoblepharidomycota + Neocallimastigomycota clade without strong statistical support. Although these putative novel phylum clades are related to Kickellomycotina, Zoopagomycotina, and Entomophthoromycotina in our tree (Fig. 6), the exact phylogenetic position is uncertain. Phylogenomic analysis would clarify the evolutionary history and taxonomy of these enigmatic lineages.
The NC_ChyL-1 clade, which included isolates of epibiotic chytrids on Desmodesmus (PSC-L52) and Rhizophydium scenedesmi strain EPG01 on Graesiella sp (45), was sister to Monoblepharidomycota without strong statistical support. Previously, R. scenedesmi was shown to be sister to the genus Zygophlyctis in Chytridiomycota (45). However, another analysis showed that R. scenedesmi along with some environmental sequences were placed sister to Monoblepharidomycota (21) as with the present study. This clade could correspond to the clade GS13 defined by Tedersoo et al. (8) because one of their environmental sequences (GL02368_027_G2750 from Australian soil) was positioned within NC_ChyL-1.
The clade FRESHOL1 was originally defined by Arroyo et al. (6) in their metabarcoding analysis of the Paraná River in Argentina. This clade was sister to all other fungi including Aphelidiomycota and Rozellomycota as with our analysis. PSC-L70 was a cell of Staurastrum sp. filled with a putative endoparasite (Fig. 4Z). The isolate corresponding to PSC-L71 included chytrid zoosporangium-like cells on Spirogyra sp. (Fig. 4AAA). Although information on the life cycles of these two lineages is currently limited, there is the possibility that they are endoparasites of algae or chytrid-like organisms in these samples. Two deep-branching groups of fungi, Aphelidiomycota and Rozellomycota, are known as endoparasites of other organisms (22). The previously defined phylum-level clade NCLC1 is sister to Rozellomycota in our tree (Fig. 10) and is also comprised of putative endoparasites of marine diatoms (76). Given the phylogenetic position and host of the FRESHOL1 lineage, our findings strengthen the recently suggested hypothesis that the ancestor of Fungi sensu lato (including aphelids, rozellids, microsporidians, and canonical fungi) had a symbiotic relationship with cellulose-based cell-walled taxa (77). Further observations and phylogenetic analyses of the FRESHOL1 clade are pivotal to elucidate the early evolution of Holomycota lineages.
Phylogenetic diversity of Olpidium-like chytrids
We found Olpidium-like chytrids parasitic on various hosts such as adult rotifers (Fig. 2A, C, and D), rotifer eggs (Fig. 2B), desmid algae (Fig. 2K through O and 3C through G), and pine pollen (Fig. 2AA). The genus Olpidium is characterized by a holocarpic thallus, namely a simple thallus composed of only a zoosporangium without rhizoids (78). All species are endobiotic parasites of algae, plants, fungi, protists, and micro-invertebrates (78). Early molecular phylogenetic analyses (79, 80) revealed that plant root parasitic species are separated from core chytrid clades (e.g., Chytridiomycota and Blastocladiomycota) and related to zygomycetous fungi. A recent phylogenomic analysis showed that O. bornovanus parasitic on cucumber roots is sister to all terrestrial fungi (Dikarya + Mucoromycota + Zoopagomycota) (18). Olpidium-like chytrids obtained in the present study were not related to plant parasitic species (Olpidiomycota in Fig. 5) and were instead distributed among three other phylum-level clades.
Four lineages of rotifer parasites were placed in the NC_OlpL-1 clade (PSC-L1 and 2) and Blastocladiomycota (PSC-L3 and 4). The NC_OlpL-1 clade also included O. vermicola parasitic on nematode eggs. Apart from the plant parasitic lineage sister to Dikarya + zygomycetes (18), the NC_OlpL-1 clade is an additional putative independent phylum of Olpidium-like fungi. In Blastocladiomycota, PSC-L3 and 4 were related to taxa of the polyphyletic family Catenariaceae (81), which is characterized by polycentric thalli, consisting of catenulated zoosporangia connected by isthmuses (82). In both PSC-L3 and 4, multiple zoosporangia were seen in a single rotifer body but connections between zoosporangia were not visible. Some Olpidium species are known as rotifer parasites and often produce multiple zoosporangia in a single host, but early developmental stages have not been fully described (83 - 85). Some of these species could be related to Catenariaceae as with our rotifer parasites.
The other seven lineages of Olpidium-like chytrids were positioned in Rhizophydiales in Chytridiomycota (Fig. 7). Six of them were endoparasites of desmid algae: PSC-L9 related to Angulomycetaceae and PSC-L23–27 related to Batrachochytriaceae. PSC-L26 and 27 resemble O. untricuriforme in producing a branched tube-like zoosporangium (51). PSC-L24 on Desmidium sp. is similar to O. hyalothecae on Hyalotheca dissiliens (51); both infect algae of a filamentous clade in Desmidiaceae (86). Another lineage, PSC-L20 was an endobiotic chytrid in pine pollen and was related to Terramycetaceae. Rhizophydiales chytrids typically produce monocentric and epibiotic thalli with endobiotic rhizoidal systems (65). Exceptionally, Batrachochytrium dendrobatidis and Entophlyctis helioformis produce endobiotic thalli in amphibian skin cells and moribund green algal cells, respectively (87, 88). In B. dendrobatidis, rhizoids are rarely seen on zoosporangia in host skin in comparison to culture conditions (88). Simplification of thalli could occur easily in the endobiotic lifestyle. PSC-L23–27 were sister to E. helioformis, and these alga-associated endobiotic chytrids could be pivotal in investigating the evolution of nutritional modes and thallus morphology in Batrachochytriaceae.
Our phylogenetic analysis clearly showed that the genus Olpidium is polyphyletic, and that host generally tracks phylogeny. Tedersoo et al. (31) suggested accommodating Olpidium in the phylum Olpidiomycota based on the phylogenetic position of plant parasitic species of Olpidium. However, this taxonomic treatment should be examined by investigating more taxa, especially the type species, O. endogenum, which is known as a parasite on green algae of the genus Closterium.
Unexpected findings of hyper-parasites
In the present study, we found putative hyper-parasites within Chytridiomycota and Rozellomycota. The two lineages in Chytridiomycota were clearly recognizable as a chytrid zoosporangium on top of another parasite. PSC-L11 in Rhizophydiales was parasitic on an elongated zoosporangium inside Spirogyra sp. (Fig. 2R). The host of this chytrid could be an endoparasitic oomycete in algae. Regarding similar described species, Rhizophydium carpophilum is known as a parasite of oogonia and oospores of Saprolegnia and Achlya (89) and also reported as a hyper-parasite of endoparasitic Olpidiopsis infecting Achlya (90). Another hyper-parasitic chytrid isolated in the present study is PSC-L39 in Chytridiales, a spherical zoosporangium on an epibiotic chytrid parasite on S. binderanus (Fig. 3T). Its host could be the chytrid of PSC-L28 (Fig. 3I) in Zygophlyctidales because the shape of the zoosporangium is similar, and they were found in the same sample collected at Lake Erie. Currently, some 15 species are known as epibiotic chytrid parasites of other chytrids (78, 91). One of them, Septosperma anomalum, was reported as a hyper-parasite infecting diatom parasite such as Chytriomyces tabellariae on Tabellaria flocculosa (92) and Zygophlyctis asterionellae on Asterionella formosa (93). Our isolates are distinguished from S. anomalum based on the shape of zoosporangium. Also, S. anomalum produces a unique resting spore with septation, which was not observed in our sample. Unfortunately, DNA sequence data are currently not available for any chytrid species parasitic on other chytrids, preventing comparison with our isolates. PSC-L39 corresponds to the clade CH_D including single-cell isolates from Lake Inba in Japan (15). These isolates were recorded as a chytrid parasite on Aulacoseira spp. but there is a possibility that its hyper-parasitic nature was overlooked.
The other single-cell lineages of hyper-parasites were found in Rozellomycota. These were isolated as epibiotic or endobiotic chytrid parasites of green algae or diatoms, but they were phylogenetically related to Rozella spp. (PSC-L64–69). The genus Rozella is well known as an endoparasite of chytrids or oomycetes (94). Rozella invades the host as an unwalled cell, consumes host cytoplasm by phagocytosis, and ultimately fills the entire host cell. Due to this endoparasitic nature, chytrid zoosporangia infected by Rozella might be difficult to detect, although some species cause hypertrophy or abnormal septation of the host cell (36, 95). Therefore, infections by Rozella were likely overlooked in our isolates. The putative hosts of our Rozella isolates were speculated: PSC-L64 (Fig. 4S) on Chytridiales chytrid PSC-L38 on Desmidium sp. (Fig. 3S); PSC-L66 (Fig. 4V) on Lobulomycetales chytrid PSC-L45 on Spirogyra sp. (Fig. 3Z); PSC-L67 (Fig. 4W) on Rhizophydiales chytrid PSC-L8 on Ulnaria sp. (Fig. 2J); and PSC-L69 (Fig. 4Y) on Olpidium-like chytrid PSC-L23 on Micrasterias truncata (Fig. 3D). Indeed, single-cell genomic analysis on the amplified genome of isolate PSC023 (lineage PSC-L23) revealed that the genome included both the host as well as the putative hyper-parasite corresponding to PSC-L69 (74). However, in our ONT sequencing, only a chytrid rDNA sequence was obtained in PSC023. We assume that biased PCR amplification occurred in this sample. Phylogenomic analysis showed that the Rozellomycota genome in PSC023 was sister to Rozella spp. although PSC-L69 separated from the Rozella clade in the present study. Our finding of hyper-parasitic Rozella indicates cryptic diversity of endoparasites infecting chytrids. These findings need to be taken into consideration when using single-cell approaches to infer nutritional mode from the recovered genomes.
Technical advances and challenges using single-cell technique
The approach outlined presents both advances over traditional methods of single-cell genomics that involve fluorescence-activated cell sorting (96) as well as challenges. The primary advantages are that the method allows images of the target species to be obtained and that the success rate of going from cell to sequence is higher. Among the 259 cells processed with multiple displacement amplification (MDA), DNA sequence data were successfully generated for 139 cells (54%) in total. Excluding putative contaminants (e.g., cercozoans) and fungus-like organisms (e.g., oomycetes and hyphochytrids), 129 cells (50%) were categorized as fungal sequences (Table S1). This rate is higher than previous single-cell sequencing studies (single-cell sorting + whole genome amplification + PCR and sequencing) on planktonic prokaryotes and protists, which had a 5%–38% success rate (97 - 101). Moreover, photos accompanied these cells. While these cells are no longer available for morphological analysis, their amplified DNA with high concentration (147–1,600 ng/µL, Table S1) is present, which is facilitating ongoing genome sequencing. We believe that the high success rate of amplification and sequencing of the target cells is likely due to the fact that these fungal cells are actually comprised of multiple nuclei, in many cases representing the near mature reproductive stages of the chytrid zoosporangia that may contain 5–50 or more nuclei. A final technical advance is the combination of single-cell approaches with long rDNA PCR. Amplification of the majority of the coding bases of the rRNA operon in addition to the highly variable internal transcribed spacer region allows for robust phylogenetic placement as well as discrimination at the species level (102). Amplification of both 18S and 28S regions allows the data to be compared to multiple data sets, given that there are disparate uses of the two regions in both environmental DNA and systematics studies (48, 103).
There are also some disadvantages of the method. First, it is hard to scale up to a large number of cells because this is a manual approach in which each cell requires as much as an hour to find, clean, and pipette into a sample tube. Second, this approach requires considerable taxonomy, microscopy, and microbial natural history skills. These skills are lacking in most microbiology and mycology training. Third, there are biases in the targeting of hosts. Most of the isolated cells in this study were parasites of algae, although a few protists and micro-invertebrates were isolated. More diverse taxa could host parasitic EDF, but they may have been undersampled due to our limited ability to find them and diagnose them as infected. These biased isolations potentially hinder clarifying the diversity of EDF; such a limitation is less applicable to metabarcoding and metagenomic approaches. Finally, our samples are far from single cells, and often contain host cells, bacterial cells, and in several cases, hyper-parasites. This is both an advantage and disadvantage because it identifies interesting symbioses, but it also makes ascribing ecological function more complicated. Presence of host and bacterial DNA could limit the ability to sequence fungal genomes from these samples. In some cases, we were able to amplify host DNA in order to confirm species identity (data not shown), but in other cases, host DNA could not be recovered as presumably the parasite had already consumed it. Despite these disadvantages, the target single-cell isolation is a powerful method to investigate uncultured parasitic fungi, and its use will expand our understanding of the ecology and phylogeny of EDF.
MATERIALS AND METHODS
Sample collection and single-cell isolation
We collected 50–250 mL of water samples with detritus and/or plant material from ponds or lakes in Michigan in 2019–2021 (see Table S2 in the supplemental material). For Lake Erie, seston was collected by boat with a plankton net (≥20 µm) deployed 1–3 m from the surface, after which the collected material was transferred to a 50 mL conical centrifuge tube maintained at in situ water temperature in the dark. The samples were transferred to University of Michigan and incubated for ~1 month, at 20°C, under LED lighting. Water samples were observed using a Nikon TMS Inverted microscope (Nikon, Tokyo, Japan) to detect fungi associated with algae, micro-invertebrates, and protists. Detected fungal cells were photographed using Moticam X Camera (Motic, Hong Kong, China) or Dino-Eye Edge S Eyepiece Camera (AnMo Electronic Corporation, Taipei, Taiwan) digital cameras. Representative images were edited and assembled into plates using Adobe Photoshop. The cells were isolated manually using a manually prepared drawn-out glass capillary pipette. The isolated cells were washed by serial transfer in small drops (more than five) of UV-sterilized water, transferred into 200 µL PCR tubes with 1–2 µL of water, and kept at −80°C until DNA extraction.
Whole genome amplification, PCR, and sequencing
To conduct DNA extraction and whole genome amplification of isolated cells by multiple displacement amplification (MDA), we used the Qiagen REPLI-g Single Cell Kit (Qiagen, Germantown, Maryland, USA) and processed the samples as described in Davis et al. (14). The DNA concentrations of some of the MDA products were measured with the Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). To obtain rDNA sequences, we utilized the Oxford Nanopore Technology sequencing pipeline for zoosporic eufungi (34). To amplify the 18S-ITS1-5.8S-ITS2-28S rDNA operon, we performed long-range PCR with fungal specific primers, NS1short and RCA95m (43). We used TaKaRa LA Taq DNA polymerase (Takara Bio USA, San Jose, CA, USA) with the protocol described in Simmons et al. (34). For some samples, we used KOD Xtreme Hot Start DNA Polymerase (Merck Millipore, Burlington, MA, USA). We prepared 12.5 µL amplifications composed of the following: (i) 0.25 µL KOD extreme, (ii) 6.25 µL 2X Xtreme Buffer, (iii) 2.5 µL dNTPs, (iv) 0.75 µL each 5 µM barcoded primer NS1short/RCA95m, and (v) 2 µL 1/50 or 1/100 diluted MDA products. We performed PCR on an Eppendorf Mastercycler Pro S with the following conditions: (i) 95°C for 2 minutes, (ii) 10 cycles of denaturation at 98°C for 10 minutes, annealing at 55–50°C (0.5°C decrease per cycle) for 30 seconds, and extension at 68°C for 5 minutes, (iii) 30 cycles of 98°C for 10 seconds, 50°C for 30 seconds, and 68°C for 5 minutes. The PCR products (4.5–6 kbp) were assessed by electrophoresis. We generated long-read sequences with an Oxford Nanopore Technologies MinION device and MinKNOW software (Oxford Nanopore Technologies, Oxford, United Kingdom). We prepared pooled barcoded amplicon libraries with the ONT Ligation Sequencing Kit (LSK-109), following the manufacturer’s protocol. We generated fast5 sequencing reads in MinKNOW that we base-called in Guppy (ONT). With the resulting fastq reads, we quality filtered (104) with NanoFilt (105) and converted them to fasta files with Seqtk (https://github.com/lh3/seqtk). We demultiplexed the pooled data with MiniBar (106), assembled sequences in Canu 1.9 (107) with defined cut-off criteria (34, 104), polished sequences with Medaka (https://github.com/nanoporetech/medaka), and removed barcodes to produce the final rDNA operon sequences in Geneious 9.1.7 (Biomatters, Auckland, New Zealand). For samples that failed the long-range PCR for ONT sequencing, we attempted short-range PCR using primers: SR1.5 (108)/AU4v2 (109) and CRYPTO2-2F (109)/AU4v2 for partial 18S, ITS5 (110)/RCA95m for ITS and partial 28S, and LR0R (111)/RCA95m for partial 28S. PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific, USA). Sequencing analyses were performed with Genewiz sequencing service (NJ, USA) using the following primers: SR1.5, CRYPTO2-2F, NS4 (110), and AU4v2 for 18S, ITS3, ITS4, ITS5 (110) for ITS, and LR0R and LR5 (112) for 28S.
Phylogenetic analysis
To clarify phylogenetic positions of single cell isolates, a phylogenetic analysis of a concatenated data set of 18S, 5.8S, and 28S rDNA sequences was performed (see Table S3 in the supplemental material). The 5.8S rDNA sequences were extracted from the data of ITS1-5.8S-ITS2 using ITSx (113). Sequences were aligned using MAFFT v7.487 (114) with the “L-INS-I” method, and the alignment was trimmed using trimAl (115) with the “gappyout” method. The maximum likelihood (ML) tree was inferred with IQ-TREE 2 (116). The best model of each alignment was examined using ModelFinder (117) implemented in the IQ-TREE 2. According to the corrected Akaike information criterion (AICc), GTR + F + R9, JC + R4, and TIM3 + F + R9 models were selected for 18S, 5.8S, and 28S, respectively. An ML analysis was run with a partitioned model (118). The branch supports were assessed with standard non-parametric bootstrap analysis (100 replicates). The tree was visualized with FigTree (https://github.com/rambaut/figtree) and edited with Adobe Illustrator.
PacBio metabarcoding analysis
Single-cell sequence data were compared to a large collection of environmental DNA sequences utilizing a recently developed PacBio metabarcoding data set of 18S amplicons (44). These data are complementary because many of the sampling localities are shared (i.e., are from aquatic habitats in Michigan). For this analysis, we extracted reference sequences of 339 PacBio OTUs that were putatively identified as Chytridiomycota using taxonomic assignment in Qiime v1.9.1 (119) with BLAST (120) and a curated version of the SILVA database (121), which was amended to include more EDF including Aphelidiomycota and Rozellomycota (available here as “Updated_Silva_Cryptos_Aphelids.txt”: https://github.com/Michigan-Mycology/Lab-Code-and-Hacks/tree/master/Cryptomycota_ecology/Data_files/). We performed further manual curation of the sequences identified as Chytridiomycota for putative chimeras by phylogenetic analyses. We prepared a reference data set of 18S rDNA of cultured Chytridiomycota species, single-cell sequences from our and previous studies, and outgroup taxa (see Table S4 in the supplemental material). The sequences were aligned and trimmed as described above. All OTU sequences were divided into two parts, the former and latter ~650 bp nucleotides. The original and divided PacBio sequences were added into the reference alignment using MAFFT with the “--add” and “--keeplength” options. Maximum likelihood trees were inferred using FastTree (122) with the “-gtr” option. The trees were visualized using FigTree. The OTUs of the following results were excluded in the subsequent analyses: (i) the phylogenetic positions of divided sequences were clearly different and (ii) the sequence was extremely long branched. After manual curation, 123 OTUs were excluded leaving 216 OTUs in the final data set (see Table S5 in the supplemental material). To examine the phylogenetic position of these OTUs, we performed an ML analysis. We prepared the 18S data set of almost the entire Chytridiomycota by adding environmental sequences available in GenBank (see Table S6 in the supplemental material) to the reference data set used above (see Table S4) and sequences were aligned and trimmed as above. Subsequently, the curated PacBio OTU sequences were added to this alignment as above. The ML tree was inferred using IQ-TREE 2 with the GTR + F + R6 model selected by ModelFinder. A standard non-parametric bootstrap analysis of 100 replicates was performed. The trees were visualized and edited as above.
ACKNOWLEDGMENTS
K.S. was supported by JSPS Overseas Research Fellowship (No. 201960485). This project was funded, in part, by the U.S. National Science Foundation grants DEB-1929738 and DEB-1354625 to T.Y.J. R.M.L.M. acknowledges the support from NSERC (RGPIN-2019–03943) and through the Great Lakes Center for Fresh Waters and Human Health supported by NIEHS (1P01ES02328939-01) and NSF (OCE-1840715). T.Y.J. is a fellow of CIFAR program Fungal Kingdom: Threats & Opportunities.
The authors are grateful to D. Peck, J. Anderson, A. Zastepa, and the Command and crew of USCGC NEAH BAY and CCGS LIMNOS who conducted sampling of Lake Erie in support of this study. Additionally, the authors thank Sally L. Glockling for providing the sequence data of Olpidium vermicola.
Contributor Information
Timothy Y. James, Email: tyjames@umich.edu.
Arturo Casadevall, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA .
DATA AVAILABILITY
The sequence data obtained in this study were deposited in GenBank under the accession numbers OQ687116–OQ687331 and OQ702805 https://www.ncbi.nlm.nih.gov/nuccore/OQ702805–OQ702950.
DIRECT CONTRIBUTION
This article is a direct contribution from Timothy James, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Luis Galindo, Oxford University, and Jeremy Wideman, Arizona State University.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01313-23.
OTU table showing the read abundance of PacBio OTUs in each sample.
Maximum likelihood (ML) tree of 18S rDNA data set.
List of single cell isolates with sequencing data.
List of sampling locations.
List of taxa and environmental sequences used for the phylogenetic analysis of the concatenated data set (18S, 5.8S, 28S rDNA).
List of sequences of cultures and single cell isolates used for the phylogenetic analysis of 18S rDNA data set.
List of the curated PacBio OTU sequences.
List of environmental sequences used for the phylogenetic analysis of 18S rDNA data set.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson K-H, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K. 2014. Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688 [DOI] [PubMed] [Google Scholar]
- 2. Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L. 2019. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 17:95–109. doi: 10.1038/s41579-018-0116-y [DOI] [PubMed] [Google Scholar]
- 3. Peay KG, Kennedy PG, Talbot JM. 2016. Dimensions of biodiversity in the earth mycobiome. Nat Rev Microbiol 14:434–447. doi: 10.1038/nrmicro.2016.59 [DOI] [PubMed] [Google Scholar]
- 4. Hawksworth DL, Lücking R. 2017. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr 5:5. doi: 10.1128/microbiolspec.FUNK-0052-2016 [DOI] [PubMed] [Google Scholar]
- 5. James TY, Stajich JE, Hittinger CT, Rokas A. 2020. Toward a fully resolved fungal tree of life. Annu Rev Microbiol 74:291–313. doi: 10.1146/annurev-micro-022020-051835 [DOI] [PubMed] [Google Scholar]
- 6. Arroyo AS, López-Escardó D, Kim E, Ruiz-Trillo I, Najle SR. 2018. Novel diversity of deeply branching holomycota and unicellular holozoans revealed by metabarcoding in middle Paraná river, Argentina. Front Ecol Evol 6:99. doi: 10.3389/fevo.2018.00099 [DOI] [Google Scholar]
- 7. Richards TA, Leonard G, Mahé F, Del Campo J, Romac S, Jones MDM, Maguire F, Dunthorn M, De Vargas C, Massana R, Chambouvet A. 2015. Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc Biol Sci 282:20152243. doi: 10.1098/rspb.2015.2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tedersoo L, Bahram M, Puusepp R, Nilsson RH, James TY. 2017. Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 5:42. doi: 10.1186/s40168-017-0259-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tedersoo L, Anslan S, Bahram M, Kõljalg U, Abarenkov K. 2020. Identifying the ‘unidentified’ fungi: a global-scale long-read third-generation sequencing approach. Fungal Divers 103:273–293. doi: 10.1007/s13225-020-00456-4 [DOI] [Google Scholar]
- 10. Grossart H-P, Wurzbacher C, James TY, Kagami M. 2016. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol 19:28–38. doi: 10.1016/j.funeco.2015.06.004 [DOI] [Google Scholar]
- 11. Van den Wyngaert S, Ganzert L, Seto K, Rojas-Jimenez K, Agha R, Berger SA, Woodhouse J, Padisak J, Wurzbacher C, Kagami M, Grossart H-P. 2022. Seasonality of parasitic and saprotrophic zoosporic fungi: linking sequence data to ecological traits. ISME J 16:2242–2254. doi: 10.1038/s41396-022-01267-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishida S, Nozaki D, Grossart H-P, Kagami M. 2015. Novel basal, fungal lineages from freshwater phytoplankton and Lake samples. Environ Microbiol Rep 7:435–441. doi: 10.1111/1758-2229.12268 [DOI] [PubMed] [Google Scholar]
- 13. Davis WJ, Amses KR, James ES, James TY. 2019. A new 18S rRNA phylogeny of uncultured predacious fungi (Zoopagales). Mycologia 111:291–298. doi: 10.1080/00275514.2018.1546066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis WJ, Jerônimo GH, Simmons DR, Quandt CA, James TY. 2019. Revisiting the aquatic phycomycete biota of the Douglas lake region since the time of Dogma and Sparrow. Philipp J Syst Biol 13:1–14. doi: 10.26757/pjsb2019b13001 [DOI] [Google Scholar]
- 15. Kagami M, Seto K, Nozaki D, Nakamura T, Wakana H, Wurzbacher C. 2021. Single dominant diatom can host diverse parasitic fungi with different degree of host specificity. Limnol Oceanogr 66:667–677. doi: 10.1002/lno.11631 [DOI] [Google Scholar]
- 16. Comeau AM, Vincent WF, Bernier L, Lovejoy C. 2016. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci Rep 6:30120. doi: 10.1038/srep30120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galindo LJ, López-García P, Torruella G, Karpov S, Moreira D. 2021. Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota. Nat Commun 12:4973. doi: 10.1038/s41467-021-25308-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang Y, Rochon D, Sekimoto S, Wang Y, Chovatia M, Sandor L, Salamov A, Grigoriev IV, Stajich JE, Spatafora JW. 2021. Genome-scale phylogenetic analyses confirm Olpidium as the closest living zoosporic fungus to the non-flagellated, terrestrial fungi. Sci Rep 11:3217. doi: 10.1038/s41598-021-82607-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voigt K, James TY, Kirk PM, de A Santiago ALCM, Waldman B, Griffith GW, Fu M, Radek R, Strassert JFH, Wurzbacher C, Jerônimo GH, Simmons DR, Seto K, Gentekaki E, Hurdeal VG, Hyde KD, Nguyen TTT, Lee HB. 2021. Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Divers 109:59–98. doi: 10.1007/s13225-021-00480-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karpov SA, Kobseva AA, Mamkaeva MA, Mamkaeva KA, Mikhailov KV, Mirzaeva GS, Aleoshin VV. 2014. Gromochytrium mamkaevae gen. & sp. nov. and two new orders: Gromochytriales and Mesochytriales (Chytridiomycetes). Persoonia 32:115–126. doi: 10.3767/003158514X680234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seto K, Wyngaert S, Degawa Y, Kagami M. 2020. Taxonomic revision of the genus Zygorhizidium: Zygorhizidiales and Zygophlyctidales ord. nov. (Chytridiomycetes, Chytridiomycota). Fungal Syst Evol 5:17–38. doi: 10.3114/fuse.2020.05.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karpov SA, Mamkaeva MA, Aleoshin VV, Nassonova E, Lilje O, Gleason FH. 2014. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front Microbiol 5:112. doi: 10.3389/fmicb.2014.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jobard M, Rasconi S, Solinhac L, Cauchie H-M, Sime-Ngando T. 2012. Molecular and morphological diversity of fungi and the associated functions in three European nearby lakes. Environ Microbiol 14:2480–2494. doi: 10.1111/j.1462-2920.2012.02771.x [DOI] [PubMed] [Google Scholar]
- 24. Picard KT. 2017. Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol 25:1–13. doi: 10.1016/j.funeco.2016.10.006 [DOI] [Google Scholar]
- 25. Hassett BT, Vonnahme TR, Peng X, Jones EBG, Heuzé C. 2020. Global diversity and geography of planktonic marine fungi. Botanica Marina 63:121–139. doi: 10.1515/bot-2018-0113 [DOI] [Google Scholar]
- 26. Lafferty KD, Allesina S, Arim M, Briggs CJ, De Leo G, Dobson AP, Dunne JA, Johnson PTJ, Kuris AM, Marcogliese DJ, Martinez ND, Memmott J, Marquet PA, McLaughlin JP, Mordecai EA, Pascual M, Poulin R, Thieltges DW. 2008. Parasites in food webs: the ultimate missing links. Ecol Lett 11:533–546. doi: 10.1111/j.1461-0248.2008.01174.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paseka RE, White LA, Van de Waal DB, Strauss AT, González AL, Everett RA, Peace A, Seabloom EW, Frenken T, Borer ET. 2020. Disease-mediated ecosystem services: pathogens, plants, and people. Trends Ecol Evol 35:731–743. doi: 10.1016/j.tree.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 28. James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R. 2006. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98:860–871. doi: 10.3852/mycologia.98.6.860 [DOI] [PubMed] [Google Scholar]
- 29. Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao Y-J, Zhang N. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547. doi: 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 30. Powell MJ, Letcher PM. 2014. 6 Chytridiomycota, Monoblepharidomycota and Neocallimastigomycota, p 141–175. In McLaughlin DJ, Spatafora JW (ed), Systematics and evolution: Part A. Springer, Berlin, Heidelberg. doi: 10.1007/978-3-642-55318-9 [DOI] [Google Scholar]
- 31. Tedersoo L, Sánchez-Ramírez S, Kõljalg U, Bahram M, Döring M, Schigel D, May T, Ryberg M, Abarenkov K. 2018. High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Divers 90:135–159. doi: 10.1007/s13225-018-0401-0 [DOI] [Google Scholar]
- 32. Mikhailov KV, Karpov SA, Letcher PM, Lee PA, Logacheva MD, Penin AA, Nesterenko MA, Pozdnyakov IR, Potapenko EV, Sherbakov DY, Panchin YV, Aleoshin VV. 2022. Genomic analysis reveals cryptic diversity in aphelids and sheds light on the emergence of fungi. Curr Biol 32:4607–4619. doi: 10.1016/j.cub.2022.08.071 [DOI] [PubMed] [Google Scholar]
- 33. Torruella G, Grau-Bové X, Moreira D, Karpov SA, Burns JA, Sebé-Pedrós A, Völcker E, López-García P. 2018. Global transcriptome analysis of the aphelid Paraphelidium tribonemae supports the phagotrophic origin of fungi. Commun Biol 1:231. doi: 10.1038/s42003-018-0235-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simmons DR, Bonds AE, Castillo BT, Clemons RA, Glasco AD, Myers JM, Thapa N, Letcher PM, Powell MJ, Longcore JE, James TY. 2020. The Collection of Zoosporic Eufungi at the University of Michigan (CZEUM): introducing a new repository of barcoded Chytridiomyceta and Blastocladiomycota cultures. IMA Fungus 11:20. doi: 10.1186/s43008-020-00041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karpov SA, Mikhailov KV, Mirzaeva GS, Mirabdullaev IM, Mamkaeva KA, Titova NN, Aleoshin VV. 2013. Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist 164:195–205. doi: 10.1016/j.protis.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 36. Letcher PM, Longcore JE, Quandt CA, Leite D da S, James TY, Powell MJ. 2017. Morphological, molecular, and ultrastructural characterization of Rozella rhizoclosmatii, a new species in Cryptomycota. Fungal Biol 121:1–10. doi: 10.1016/j.funbio.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 37. Lepelletier F, Karpov SA, Alacid E, Le Panse S, Bigeard E, Garcés E, Jeanthon C, Guillou L. 2014. Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a chytrid infecting marine dinoflagellates. Protist 165:230–244. doi: 10.1016/j.protis.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 38. Van den Wyngaert S, Seto K, Rojas-Jimenez K, Kagami M, Grossart H-P. 2017. A new parasitic chytrid, Staurastromyces oculus (Rhizophydiales, Staurastromycetaceae fam. nov.), infecting the freshwater desmid Staurastrum sp. Protist 168:392–407. doi: 10.1016/j.protis.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 39. Seto K, Degawa Y. 2018. Collimyces mutans gen. et sp. nov. (Rhizophydiales, Collimycetaceae fam. nov.), a new chytrid parasite of Microglena (Volvocales, clade Monadinia). Protist 169:507–520. doi: 10.1016/j.protis.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 40. Seto K, Degawa Y. 2018. Pendulichytrium sphaericum gen. et sp. nov. (Chytridiales, Chytriomycetaceae), a new chytrid parasitic on the diatom, Aulacoseira granulata. Mycoscience 59:59–66. doi: 10.1016/j.myc.2017.08.004 [DOI] [Google Scholar]
- 41. Karpov SA, López-García P, Mamkaeva MA, Vishnyakov AE, Moreira D. 2016. Chytridiomycete Polyphagus parasiticus: molecular phylogeny supports the erection of a new chytridiomycete order. Mikol Fitopatol 50:362–366. [Google Scholar]
- 42. Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. 2008. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLOS ONE 3:e2324. doi: 10.1371/journal.pone.0002324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wurzbacher C, Larsson E, Bengtsson-Palme J, Van den Wyngaert S, Svantesson S, Kristiansson E, Kagami M, Nilsson RH. 2019. Introducing ribosomal tandem repeat barcoding for fungi. Mol Ecol Resour 19:118–127. doi: 10.1111/1755-0998.12944 [DOI] [PubMed] [Google Scholar]
- 44. Quandt CA, Marino JA, Simmons DR, Davis WJ, Hassett BT, Picard KT, James TY. 2023. Evaluating the diversity of the enigmatic fungal phylum Cryptomycota across habitats using 18S rRNA metabarcoding. Fungal Ecol 64:101248. doi: 10.1016/j.funeco.2023.101248 [DOI] [Google Scholar]
- 45. Ding Y, Peng X, Wang Z, Wen X, Geng Y, Zhang D, Li Y. 2018. Occurrence and characterization of an epibiotic parasite in cultures of oleaginous microalga Graesiella sp. J Appl Phycol 30:819–830. doi: 10.1007/s10811-017-1302-4 [DOI] [Google Scholar]
- 46. Martin WW. 1975. A new species of Catenaria parasitic in midge eggs. Mycologia 67:264–272. doi: 10.1080/00275514.1975.12019748 [DOI] [PubMed] [Google Scholar]
- 47. Martin WW. 1978. Two additional species of Catenaria (Chytridiomycetes, Blastocladiales) parasitic in midge eggs. Mycologia 70:461–467. doi: 10.1080/00275514.1978.12020247 [DOI] [PubMed] [Google Scholar]
- 48. Monchy S, Sanciu G, Jobard M, Rasconi S, Gerphagnon M, Chabé M, Cian A, Meloni D, Niquil N, Christaki U, Viscogliosi E, Sime-Ngando T. 2011. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing: fungal diversity in freshwater lakes. Environ Microbiol 13:1433–1453. doi: 10.1111/j.1462-2920.2011.02444.x [DOI] [PubMed] [Google Scholar]
- 49. Paterson RA. 1956. Additions to the phycomycete flora of the Douglas lake region. II. New chytridiaceous fungi. Mycologia 48:270–277. doi: 10.1080/00275514.1956.12024531 [DOI] [Google Scholar]
- 50. Karling JS. 1936. The endo-exogenous method of growth and development of Chytridium lagenaria. Am J Bot 23:619–627. doi: 10.1002/j.1537-2197.1936.tb09036.x [DOI] [Google Scholar]
- 51. Scherffel A. 1926. Einiges uber neue oder ungenügend bekannte chytridineen. Arch Protistenkd 54:167–260. [Google Scholar]
- 52. Karpov SA, Tcvetkova VS, Mamkaeva MA, Torruella G, Timpano H, Moreira D, Mamanazarova KS, López-García P. 2017. Morphological and genetic diversity of Opisthosporidia: new aphelid Paraphelidium tribonemae gen. et sp. nov.. J Eukaryot Microbiol 64:204–212. doi: 10.1111/jeu.12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karpov SA, Torruella G, Moreira D, Mamkaeva MA, López-García P. 2017. Molecular phylogeny of Paraphelidium letcheri sp. nov. (Aphelida, Opisthosporidia). J Eukaryot Microbiol 64:573–578. doi: 10.1111/jeu.12389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Letcher PM, Powell MJ, Lee PA, Lopez S, Burnett M. 2017. Molecular phylogeny and ultrastructure of Aphelidium desmodesmi, a new species in Aphelida (Opisthosporidia). J Eukaryot Microbiol 64:655–667. doi: 10.1111/jeu.12401 [DOI] [PubMed] [Google Scholar]
- 55. Letcher PM, Lopez S, Schmieder R, Lee PA, Behnke C, Powell MJ, McBride RC. 2013. Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the Cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS One 8:e56232. doi: 10.1371/journal.pone.0056232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Letcher PM, Powell MJ, Lopez S, Lee PA, McBride RC. 2015. A new isolate of Amoeboaphelidium protococcarum, and Amoeboaphelidium occidentale, a new species in phylum Aphelida (Opisthosporidia). Mycologia 107:522–531. doi: 10.3852/14-064 [DOI] [PubMed] [Google Scholar]
- 57. Karpov SA, Mamkaeva MA, Benzerara K, Moreira D, López-García P. 2014. Molecular phylogeny and ultrastructure of Aphelidium aff. melosirae (Aphelida, Opisthosporidia). Protist 165:512–526. doi: 10.1016/j.protis.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Karpov SA, Mamkaeva MA, Moreira D, López-García P. 2016. Molecular phylogeny of Aphelidium tribonemae reveals its sister relationship with A. aff. melosirae (Aphelida, Opisthosporidia). Protistology 10:97–103. doi: 10.21685/1680-0826-2016-10-3-4 [DOI] [Google Scholar]
- 59. Tcvetkova VS, Zorina NA, Mamkaeva MA, Karpov SA, , . 2019. Molecular phylogeny of Aphelidium arduennense sp. nov. – new representative of Aphelida (Opisthosporidia). Protistology 13:192–198. doi: 10.21685/1680-0826-2019-13-4-2 [DOI] [Google Scholar]
- 60. Karpov SA, Vishnyakov AE, López-García P, Zorina NA, Ciobanu M, Tcvetkova VS, Moreira D, , , . 2020. Morphology and molecular phylogeny of Aphelidium insulamus sp. nov. (Aphelida, Opisthosporidia). Protistology 14:191–203. doi: 10.21685/1680-0826-2020-14-4-3 [DOI] [Google Scholar]
- 61. Seto K, Nakada T, Tanabe Y, Yoshida M, Kagami M. 2022. Aphelidium parallelum, sp. nov., a new aphelid parasitic on selenastracean green algae. Mycologia 114:544–555. doi: 10.1080/00275514.2022.2039487 [DOI] [PubMed] [Google Scholar]
- 62. Corsaro D, Walochnik J, Venditti D, Steinmann J, Müller K-D, Michel R. 2014. Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res 113:1909–1918. doi: 10.1007/s00436-014-3838-4 [DOI] [PubMed] [Google Scholar]
- 63. Corsaro D, Walochnik J, Venditti D, Hauröder B, Michel R. 2020. Solving an old enigma: Morellospora saccamoebae gen nov., sp. nov. (Rozellomycota), a Sphaerita-like parasite of free-living amoebae. Parasitol Res 119:925–934. doi: 10.1007/s00436-020-06623-5 [DOI] [PubMed] [Google Scholar]
- 64. Corsaro D, Michel R, Walochnik J, Venditti D, Müller K-D, Hauröder B, Wylezich C. 2016. Molecular identification of Nucleophaga terricolae sp. nov. (Rozellomycota), and new insights on the origin of the Microsporidia. Parasitol Res 115:3003–3011. doi: 10.1007/s00436-016-5055-9 [DOI] [PubMed] [Google Scholar]
- 65. Letcher PM, Vélez CG, Barrantes ME, Powell MJ, Churchill PF, Wakefield WS. 2008. Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina. Mycol Res 112:759–782. doi: 10.1016/j.mycres.2008.01.025 [DOI] [PubMed] [Google Scholar]
- 66. Letcher PM, Powell MJ, Churchill PF, Chambers JG. 2006. Ultrastructural and molecular phylogenetic delineation of a new order, the Rhizophydiales (Chytridiomycota). Mycol Res 110:898–915. doi: 10.1016/j.mycres.2006.06.011 [DOI] [PubMed] [Google Scholar]
- 67. Letcher PM, Powell MJ, Davis WJ. 2015. A new family and four new genera in Rhizophydiales (Chytridiomycota). Mycologia 107:808–830. doi: 10.3852/14-280 [DOI] [PubMed] [Google Scholar]
- 68. Karpov SA, Reñé A, Vishnyakov AE, Seto K, Alacid E, Paloheimo A, Kagami M, Kremp A, Garcés E. 2021. Parasitoid chytridiomycete Ericiomyces syringoforeus gen. et sp. nov. has unique cellular structures to infect the host. Mycol Progress 20:95–109. doi: 10.1007/s11557-020-01652-x [DOI] [Google Scholar]
- 69. Karpov SA, Moreira D, Mamkaeva MA, Popova OV, Aleoshin VV, López-García P. 2019. New member of Gromochytriales (Chytridiomycetes)—Apiochytrium granulosporum nov. gen. et sp. J Eukaryot Microbiol 66:582–591. doi: 10.1111/jeu.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Karpov SA, Letcher PM, Mamkaeva MA, Mamkaeva KA. 2010. Phylogenetic position of the genus Mesochytrium (Chytridiomycota) based on zoospore ultrastructure and sequences from the 18S and 28S rRNA gene. Nova Hedwigia 90:81–94. doi: 10.1127/0029-5035/2010/0090-0081 [DOI] [Google Scholar]
- 71. Gromov BV, Mamkaeva KA, Pljusch AV. 2000. Mesochytrium penetrans gen. et sp. nov. (Chytridiales) - a parasite of the green alga Chlorococcum minutum (Chlorococcales), with an unusual behaviour of the sporangia. Nova hedwigia 71:151–160. doi: 10.1127/nova/71/2000/151 [DOI] [Google Scholar]
- 72. Zepernick BN, Denison ER, Chaffin JD, Bullerjahn GS, Pennacchio CP, Frenken T, Peck DH, Anderson JT, Niles D, Zastepa A, McKay RML, Wilhelm SW. 2022. Metatranscriptomic sequencing of winter and spring planktonic communities from lake erie, a laurentian great Lake. Microbiol Resour Announc 11:e0035122. doi: 10.1128/mra.00351-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beall BFN, Twiss MR, Smith DE, Oyserman BO, Rozmarynowycz MJ, Binding CE, Bourbonniere RA, Bullerjahn GS, Palmer ME, Reavie ED, Waters LMK, Woityra LWC, McKay RML. 2016. Ice cover extent drives phytoplankton and bacterial community structure in a large North-temperate lake: implications for a warming climate. Environ Microbiol 18:1704–1719. doi: 10.1111/1462-2920.12819 [DOI] [PubMed] [Google Scholar]
- 74. Amses KR, Simmons DR, Longcore JE, Mondo SJ, Seto K, Jerônimo GH, Bonds AE, Quandt CA, Davis WJ, Chang Y, Federici BA, Kuo A, LaButti K, Pangilinan J, Andreopoulos W, Tritt A, Riley R, Hundley H, Johnson J, Lipzen A, Barry K, Lang BF, Cuomo CA, Buchler NE, Grigoriev IV, Spatafora JW, Stajich JE, James TY. 2022. Diploid-dominant life cycles characterize the early evolution of fungi. Proc Natl Acad Sci U S A 119:e2116841119. doi: 10.1073/pnas.2116841119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barron GL, Szijarto E. 1986. A new species of Olpidium parasitic in nematode eggs. Mycologia 78:972–975. doi: 10.1080/00275514.1986.12025361 [DOI] [Google Scholar]
- 76. Chambouvet A, Monier A, Maguire F, Itoïz S, Del Campo J, Elies P, Edvardsen B, Eikreim W, Richards TA. 2019. Intracellular infection of diverse diatoms by an evolutionary distinct relative of the fungi. Curr Biol 29:4093–4101. doi: 10.1016/j.cub.2019.09.074 [DOI] [PubMed] [Google Scholar]
- 77. Galindo LJ, Torruella G, López-García P, Ciobanu M, Gutiérrez-Preciado A, Karpov SA, Moreira D, Folk R. 2023. Phylogenomics supports the monophyly of aphelids and fungi and identifies new molecular synapomorphies. Syst Biol 72:505–515. doi: 10.1093/sysbio/syac054 [DOI] [PubMed] [Google Scholar]
- 78. Sparrow FK. 1960. Aquatic Phycomycetes, . In Aquatic Phycomycetes. Ann Arbor, University of Michigan Press, Ann Arbor. doi: 10.5962/bhl.title.5685 [DOI] [Google Scholar]
- 79. James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822. doi: 10.1038/nature05110 [DOI] [PubMed] [Google Scholar]
- 80. Sekimoto S, Rochon D, Long JE, Dee JM, Berbee ML. 2011. A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evol Biol 11:331. doi: 10.1186/1471-2148-11-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Porter TM, Martin W, James TY, Longcore JE, Gleason FH, Adler PH, Letcher PM, Vilgalys R. 2011. Molecular phylogeny of the Blastocladiomycota (Fungi) based on nuclear ribosomal DNA. Fungal Biol 115:381–392. doi: 10.1016/j.funbio.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 82. Karling JS. 1977. Chytridiomycetarum Iconographia. J. Cramer. [Google Scholar]
- 83. Nowakowski L. 1876. Beitrag Zur Kenntniss der Chytridiaceen, p 73–100. In Beiträge Zur Biologie der Pflanzen [Google Scholar]
- 84. Karling JS. 1946. Brazilian chytrids. VIII. additional parasites of rotifers and nematodes. Lloydia 9:1–12. [Google Scholar]
- 85. Glockling SL. 1998. Isolation of a new species of rotifer-attacking Olpidium. Mycol Res 102:206–208. doi: 10.1017/S0953756297004693 [DOI] [Google Scholar]
- 86. Hall JD, Karol KG, McCourt RM, Delwiche CF. 2008. Phylogeny of the conjugating green algae based on chloroplast and mitochondrial nucleotide sequence data. J Phycol 44:467–477. doi: 10.1111/j.1529-8817.2008.00485.x [DOI] [PubMed] [Google Scholar]
- 87. Karling JS. 1928. Studies in the Chytridiales I. the life history and occurrence of Entophlyctis heliomorpha (Dang.) Fischer. Am J Bot 15:32–42. doi: 10.1002/j.1537-2197.1928.tb04876.x [DOI] [Google Scholar]
- 88. Berger L, Hyatt AD, Speare R, Longcore JE. 2005. Life cycle stages of the amphibian chytrid Batrachochytrium dendrobatidis. Dis Aquat Organ 68:51–63. doi: 10.3354/dao068051 [DOI] [PubMed] [Google Scholar]
- 89. Zopf W. 1884. Zur kenntniss der phycomyceten. I. Zur morphologie und biologie der ancylisteen und chytridiaceen. Nova Acta Acad Leop-Craol 47:143–236. doi: 10.5962/bhl.title.4084 [DOI] [Google Scholar]
- 90. Sparrow FK. 1932. Observations on the aquatic fungi of cold spring harbor. Mycologia 24:268–303. doi: 10.1080/00275514.1932.12020619 [DOI] [Google Scholar]
- 91. Karling JS. 1960. Parasitism among the chytrids. II Chytriomyces verrucosus sp. nov. and Phlyctochytrium synchytrii. Bull Torrey Bot Club 87:326–336. doi: 10.2307/2482628 [DOI] [Google Scholar]
- 92. Canter HM. 1949. Studies on british chytrids: IV. Chytriomyces tabellariae (Schröter) n.comb. parasitized by Septosperma anomalum (Couch) Whiffen. Trans Br Mycol 32:16–21. doi: 10.1016/S0007-1536(49)80031-3 [DOI] [Google Scholar]
- 93. Canter HM, Jaworski GHM. 1986. A study on the chytrid Zygorhizidium planktonicum Canter, a parasite of the diatoms Asterionella and Synedra. Nova Hedwigia 43:269–298. [Google Scholar]
- 94. Letcher PM, Powell MJ. 2018. A Taxonomic summary and revision of Rozella (Cryptomycota). IMA Fungus 9:383–399. doi: 10.5598/imafungus.2018.09.02.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Letcher PM, Longcore JE, James TY, Leite DS, Simmons DR, Powell MJ. 2018. Morphology, ultrastructure, and molecular phylogeny of Rozella multimorpha, a new species in Cryptomycota. J Eukaryot Microbiol 65:180–190. doi: 10.1111/jeu.12452 [DOI] [PubMed] [Google Scholar]
- 96. Stepanauskas R, Fergusson EA, Brown J, Poulton NJ, Tupper B, Labonté JM, Becraft ED, Brown JM, Pachiadaki MG, Povilaitis T, Thompson BP, Mascena CJ, Bellows WK, Lubys A. 2017. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat Commun 8:2134. doi: 10.1038/s41467-017-02128-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. 2011. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J 5:674–684. doi: 10.1038/ismej.2010.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sieracki ME, Poulton NJ, Jaillon O, Wincker P, de Vargas C, Rubinat-Ripoll L, Stepanauskas R, Logares R, Massana R. 2019. Single cell genomics yields a wide diversity of small planktonic protists across major ocean ecosystems. Sci Rep 9:6025. doi: 10.1038/s41598-019-42487-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li W, Podar M, Morgan-Kiss RM, Kelly RM. 2016. Ultrastructural and single-cell-level characterization reveals metabolic versatility in a microbial eukaryote community from an ice-covered Antarctic Lake. Appl Environ Microbiol 82:3659–3670. doi: 10.1128/AEM.00478-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, Stepanauskas R. 2012. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. 1. ISME J 6:113–123. doi: 10.1038/ismej.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PSG, Reitenga KG, Xie G, Poulton NJ, Gomez ML, Masland DED, Thompson B, Bellows WK, Ziervogel K, Lo C-C, Ahmed S, Gleasner CD, Detter CJ, Stepanauskas R, Ravel J. 2012. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS ONE 7:e35314. doi: 10.1371/journal.pone.0035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, Wingfield MJ, Aime MC, An K-D, Bai F-Y, Barreto RW, Begerow D, Bergeron M-J, Blackwell M, Boekhout T, Bogale M, Boonyuen N, Burgaz AR, Buyck B, Cai L, Cai Q, Cardinali G, Chaverri P, Coppins BJ, Crespo A, Cubas P, Cummings C, Damm U, de Beer ZW, de Hoog GS, Del-Prado R, Dentinger B, Diéguez-Uribeondo J, Divakar PK, Douglas B, Dueñas M, Duong TA, Eberhardt U, Edwards JE, Elshahed MS, Fliegerova K, Furtado M, García MA, Ge Z-W, Griffith GW, Griffiths K, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Guo L-D, Hagen F, Hambleton S, Hamelin RC, Hansen K, Harrold P, Heller G, Herrera C, Hirayama K, Hirooka Y, Ho H-M, Hoffmann K, Hofstetter V, Högnabba F, Hollingsworth PM, Hong S-B, Hosaka K, Houbraken J, Hughes K, Huhtinen S, Hyde KD, James T, Johnson EM, Johnson JE, Johnston PR, Jones EBG, Kelly LJ, Kirk PM, Knapp DG, Kõljalg U, Kovács GM, Kurtzman CP, Landvik S, Leavitt SD, Liggenstoffer AS, Liimatainen K, Lombard L, Luangsa-ard JJ, Lumbsch HT, Maganti H, Maharachchikumbura SSN, Martin MP, May TW, McTaggart AR, Methven AS, Meyer W, Moncalvo J-M, Mongkolsamrit S, Nagy LG, Nilsson RH, Niskanen T, Nyilasi I, Okada G, Okane I, Olariaga I, Otte J, Papp T, Park D, Petkovits T, Pino-Bodas R, Quaedvlieg W, Raja HA, Redecker D, Rintoul TL, Ruibal C, Sarmiento-Ramírez JM, Schmitt I, Schüßler A, Shearer C, Sotome K, Stefani FOP, Stenroos S, Stielow B, Stockinger H, Suetrong S, Suh S-O, Sung G-H, Suzuki M, Tanaka K, Tedersoo L, Telleria MT, Tretter E, Untereiner WA, Urbina H, Vágvölgyi C, Vialle A, Vu TD, Walther G, Wang Q-M, Wang Y, Weir BS, Weiß M, White MM, Xu J, Yahr R, Yang ZL, Yurkov A, Zamora J-C, Zhang N, Zhuang W-Y, Schindel D, Fungal Barcoding Consortium, Fungal Barcoding Consortium Author List . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lefèvre E, Letcher PM, Powell MJ. 2012. Temporal variation of the small eukaryotic community in two freshwater lakes: emphasis on zoosporic fungi. Aquat Microb Ecol 67:91–105. doi: 10.3354/ame01592 [DOI] [Google Scholar]
- 104. Simmons DR, Longcore JE, James TY. 2021. Polyrhizophydium stewartii, the first known rhizomycelial genus and species in the Rhizophydiales, is closely related to Batrachochytrium. Mycologia 113:684–690. doi: 10.1080/00275514.2021.1885206 [DOI] [PubMed] [Google Scholar]
- 105. De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. Nanopack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG, Prost S. 2019. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. Gigascience 8:giz006. doi: 10.1093/gigascience/giz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive K-MER weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. James TY, Porter D, Leander CA, Vilgalys R, Longcore JE. 2000. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can J Bot 78:336–350. doi: 10.1139/cjb-78-3-336 [DOI] [Google Scholar]
- 109. Lazarus KL, James TY. 2015. Surveying the biodiversity of the Cryptomycota using a targeted PCR approach. Fungal Ecol 14:62–70. doi: 10.1016/j.funeco.2014.11.004 [DOI] [Google Scholar]
- 110. White TJ, Bruns T, Lee S, Taylor JL.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322. In PCR protocols: a guide to methods and applications. Academic Press, New York. [Google Scholar]
- 111. Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycolo Res 98:625–634. doi: 10.1016/S0953-7562(09)80409-7 [DOI] [Google Scholar]
- 112. Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH, Bunce M. 2013. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. doi: 10.1111/2041-210X.12073 [DOI] [Google Scholar]
- 114. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, Teeling E. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol 65:997–1008. doi: 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 121. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OTU table showing the read abundance of PacBio OTUs in each sample.
Maximum likelihood (ML) tree of 18S rDNA data set.
List of single cell isolates with sequencing data.
List of sampling locations.
List of taxa and environmental sequences used for the phylogenetic analysis of the concatenated data set (18S, 5.8S, 28S rDNA).
List of sequences of cultures and single cell isolates used for the phylogenetic analysis of 18S rDNA data set.
List of the curated PacBio OTU sequences.
List of environmental sequences used for the phylogenetic analysis of 18S rDNA data set.
Data Availability Statement
The sequence data obtained in this study were deposited in GenBank under the accession numbers OQ687116–OQ687331 and OQ702805 https://www.ncbi.nlm.nih.gov/nuccore/OQ702805–OQ702950.