Abstract
Newcastle disease virus {NDV (avian paramyxovirus type 1 [APMV1])} isolates were recovered from imported exotic birds confiscated following importation into the United States, from waterbirds in the United States, and from poultry. The exotic birds probably originated from Central and South America, Asia, and Africa. The NDV isolates were initially characterized as highly virulent because of a short mean death time in embryonated chicken eggs. The isolates were typed as neurotropic or viscerotropic velogenic by intracloacal inoculation of adult chickens. Intracerebral pathogenicity index values for the virulent NDV isolates ranged from 1.54 to 1.90, compared to a possible maximum value of 2.0. These isolates had a dibasic amino acid motif in the fusion protein cleavage site sequence required for host systemic replication. Sequence differences were detected surrounding the fusion protein cleavage site and the matrix protein nuclear localization signal, indicating evolution of highly virulent NDV. Phylogenetically, these isolates were categorized with other highly virulent NDV strains that caused outbreaks in southern California poultry during 1972 and in cormorants in the north central United States and southern Canada during 1990 and 1992. These isolates are related to NDV that may have the APMV1 strain chicken/Australia/AV/32 or a related virus as a possible progenitor. Recent virulent NDV isolates and those recovered during disease outbreaks since the 1970s are phylogenetically distinct from current vaccine viruses and standard challenge strains.
Outbreaks of Newcastle disease were first reported in poultry from Java, Indonesia, and Newcastle-upon-Tyne in 1926, and the disease is now recognized worldwide (3). It is caused by Newcastle disease virus (NDV), a member of the family Paramyxoviridae, designated avian paramyxovirus 1 (APMV1). The enveloped virus has a negative-sense single-stranded genome of approximately 15 kb which codes for an RNA-directed RNA polymerase, hemagglutinin-neuraminidase protein, fusion protein, matrix protein, phosphoprotein, and nucleoprotein (3). The virus has a wide host range, with 27 of the 50 orders of birds reported to have been infected by NDV (21). Infectious virus may be transmitted by ingestion or inhalation, which is the basis of mass application vaccination procedures for poultry (27). NDV isolates are characterized by pathogenesis in chicken and may be categorized into three main pathotypes, depending on severity of disease (2, 3). Lentogenic isolates are of low virulence and cause mild respiratory or enteric infections. Viruses of intermediate virulence that cause primarily respiratory disease are mesogenic, while virulent viruses that cause high mortality are velogenic. Velogenic forms of NDV are further classified as neurotropic or viscerotropic based on clinical manifestation (3) and are “list A” agents that require reporting to the Office of International Epizootes (29).
The primary molecular determinants for NDV pathogenicity are the fusion protein cleavage site amino acid sequence (16, 28) and the ability of specific cellular proteases to cleave the fusion proteins of different pathotypes (17, 30). Dibasic amino acids surrounding glutamine at position 114 are present in the fusion protein cleavage site of mesogenic or velogenic strains, while lentogenic NDV isolates lack this motif (16, 28). The presence of dibasic amino acids in the fusion protein sequence allows for systemic spread of velogenic NDV, whereas replication of lentogenic NDV is limited to mucosal surfaces of the host (30). This is also the major factor in differentiating velogenic and mesogenic NDV from lentogenic NDV isolates in cell culture. All NDV isolates will replicate in chicken embryo kidney cells (23), presumably because of the presence of a required protease (30). However, lentogens must have added proteases for replication in avian fibroblasts or mammalian cell types, whereas mesogenic and velogenic NDV isolates do not have this requirement (22, 23, 28).
Viscerotropic velogenic viruses have entered the United States via importation of exotic birds (9, 15, 31, 39), and a virus of psittacine origin caused the early 1970s outbreak in the southern California area (44, 45). During 1990 and 1992, cormorants in the north central United States and southern Canada developed widespread Newcastle disease attributed to neurotropic velogenic viruses (6, 18, 37, 46). Previously we used a set of degenerate oligonucleotide primers for consistent single-tube reverse transcription-PCR (RT-PCR), followed by direct automated nucleotide sequencing of the amplified product. Subsequent phylogenetic analysis allowed for reliable pathotype prediction and was used to determine the molecular epidemiology of Newcastle disease (37). These techniques were used to demonstrate that virulent cormorant isolates from the 1990 and 1992 outbreak were possibly related to isolates of psittacine origin (37, 38).
Viscerotropic velogenic NDV isolates from imported exotic birds (9, 31) and poultry were provided by the Diagnostic Virology Laboratory, National Veterinary Services Laboratory, Animal Plant Health Inspection Service, U.S. Department of Agriculture, Ames, Iowa (DVL, NVSL, APHIS, USDA). Fourteen of these isolates obtained during 1986 through 1996 were used for further characterization (Table 1). Several NDV isolates used for comparative purposes included pathogenic (neurotropic velogenic) NDV isolates obtained from cormorants during outbreaks in 1992 (originally provided by the National Wildlife Health Research Center, Madison, Wis.) (6, 46). Subsequently, neurotropic velogenic NDV was isolated from turkeys (turkey/U.S./43084/92) in a commercial flock in North Dakota (37). Viscerotropic velogenic isolates from psittacines (cockatiel/U.S./FL/80 and mixed species/U.S./Largo/71) (8, 10, 14) and an NDV isolate made during the 1972–1974 outbreak of Newcastle disease in California (chicken/U.S./CA1083/72) (44) were also included for analysis. Strains chicken/U.S./B1/48 (19) and chicken/Australia/QV4/66 (40) were included as lentogenic vaccine-type viruses used by the poultry industry worldwide. Two mesogenic isolates of intermediate virulence included for sequence analysis were the vaccine strain chicken/U.S./Roakin/48 (7) and an anhinga isolate (anhinga/U.S./44083/93) from a captive population (DVL, NVSL, APHIS USDA). Velogenic viruses also included were previously characterized isolates, such as the viscerotropic velogenic chicken/U.K./Herts/33 (4), neurotropic velogenic chicken/Australia/AV/32 (1, 26), and chicken/U.S./GB/48 (35, 36) isolates. Initial characterization of all viral isolates was accomplished by hemagglutination inhibition (HI) with NDV-specific polyclonal antisera. Virulence of NDV isolates was evaluated by intracloacal inoculation of chickens and determination of an intracerebral pathogenicity index (ICPI) (2).
TABLE 1.
NDV isolates for which nucleotide sequence analysis was completed for molecular epidemiology
| APMV1 isolatea | Pathotypeb | ICPI |
|---|---|---|
| Chicken/Australia/AV/32 | N | 1.66 |
| Chicken/UK/Hertsfordshire/33 | V | 1.99 |
| Chicken/U.S./B1/48 | L | 0.13 |
| Chicken/U.S. (Tex.)/GB/48 | N | 1.74 |
| Chicken/U.S./Roakin/48 | M | 1.60 |
| Chicken/Australia/QV4/66 | L | 0.39 |
| Mixed species/U.S./Largo/71 | V | 1.86 |
| Chicken/U.S./CA1083 (Fontana)/72 | V | 1.80 |
| Cockatiel/U.S. (Fla.)/FL/80 | V | 1.88 |
| Amazon/U.S. (Conn.)/36501/89 | V | 1.75 |
| Cockatoo/Indonesia/14698/90 | V | 1.84 |
| African Gretimneh/Zimbabwe/28534/90 | V | 1.74 |
| Partridge/Singapore/37182/90 | V | NDc |
| Parakeet/Myanmar/11592/91 | V | 1.78 |
| Dy.Nap.parrot/U.S. (Ind.)/27492/91 | V | 1.78 |
| Y.Hd.parrot/U.S. (Mich.)/27993/91 | V | 1.90 |
| Parrot/U.S. (Ill.)/27994/91 | N | 1.89 |
| Cormorant/U.S. (Minn.)/40068/92 | N | 1.46 |
| Turkey/U.S. (N.Dak.)/43084/92 | N | 1.43 |
| R.F.parakeet/Tanzania, Belgium, China/28710/93 | V | 1.88 |
| Finch/Tanzania/6324/95 | N | 1.71 |
| Anhinga/U.S. (Fla.)/44083/93 | M | 1.54 |
| Mixed species/Mexico/37821-550-1/96 | V | 1.75 |
| Chicken/Mexico/37821-550-2/96 | V | 1.75 |
| Yellow cheek/U.S. (Tex.)/27345/96 | V | 1.80 |
| Amazon parrot/U.S. (Mo.)/31378/96 | V | 1.83 |
Isolate is given as bird type/country of origin/accession number/year of isolation as designated by NVSL, APHIS, USDA. Previously uncharacterized viruses included isolates 36501/89, 14698/90, 28534/90, 11592/91, 27492/91, 27993/91, 27994/91, 28710/93, 6324/95, 37821-550-1/96, 37821-550-2/96, 27345/96, and 31378/96.
Initial virus pathotype as provided by NVSL, APHIS, USDA following intracloacal chicken inoculations. L, lentogen; M, mesogen; V, viscerotropic velogenic; N, neurotropic velogenic.
ND, not determined.
For nucleotide sequence analysis, NDV isolates were replicated in embryonated eggs (2), and RNA was extracted (11) directly from allantoic fluid as described previously (37). Oligonucleotide RT-PCR primers designed to amplify regions of the fusion protein gene, including the fusion protein cleavage site (43) and the matrix protein gene (38) region encoding the nuclear localization signal of the matrix protein (12), were reported previously (37). A single-tube RT-PCR for genomic NDV RNA was completed as described previously (25, 37), with Superscript (Life Technologies, Gaithersburg, Md.) (24) and Amplitaq (Perkin-Elmer) polymerase (32). Amplification products were purified with Microcon (AMICON) spin filters and spectrophotometrically quantified. Direct double-stranded nucleotide sequencing (34) was completed with Taq polymerase (Applied Biosystems, Inc.) with the oligonucleotide primers used for RT-PCR, fluorescence-labelled dideoxynucleotides, and an automated nucleic acid sequencer (41). Nucleotide sequence editing, analysis, prediction of amino acid sequences, and alignments were conducted with GeneWorks 2.5 software (IntelliGenetics, Mountain View, Calif.). The phylogenetic analysis presented was completed with the Phylogenetic Analysis Using Parsimony (PAUP) (42) software with a heuristic search and 1,000 bootstrap replicates.
Listed in Table 1 are NDV isolates from imported pet and exotic birds for which ICPI values and nucleotide and amino acid sequence information have been obtained. Several reference isolates along with viruses from poultry and waterbirds were included for comparative purposes. As best as could be determined, the recently recovered isolates were from birds having origins in Central and South America, Asia, and Africa. Many of these birds were quarantined prior to direct entry into the United States. However, several viruses were obtained from birds following importation into the United States. These viruses have the state of origin within the United States in which they were isolated designated in parentheses. A variety of birds, including parrots, parakeets, cockatiels, partridges, finches, and a timneh, harbored NDV. The virus isolates chosen for analysis were obtained prior to and following the 1990-to-1992 Newcastle disease outbreaks in cormorants and turkeys (37) and were chosen to represent the period from 1989 through 1996 to chronologically surround these outbreaks.
Initial biological characterization of the NDV isolates included mean death time determinations in embryonated chicken eggs and intracloacal inoculation of chickens. All virulent viruses produced a mean death time in embryonated chicken eggs of less than 60 h, indicating these viruses were probably highly pathogenic for chickens. Intracloacal inoculation of chickens revealed that the majority of velogenic NDV isolates from exotic birds produced viscerotropic lesions (Table 1). However, a velogenic virus, parrot/U.S./27994/91, was isolated from a psittacine bird, and another velogenic virus, finch/Tanzania/6324/95, was isolated that produced neurologic symptoms following intracloacal inoculation of chickens. Values following determination of an ICPI ranged from 1.71 to 1.90 for highly virulent NDV isolates (Table 1). These values conform to results obtained with standard challenge viruses such as the neurotropic isolate chicken/U.S. (Tex.)/GB/48 (ICPI of 1.74) and the viscerotropic isolate chicken/U.S./CA1083 (Fontana)/72 (ICPI of 1.80), which demonstrates that these viruses are highly virulent for chickens.
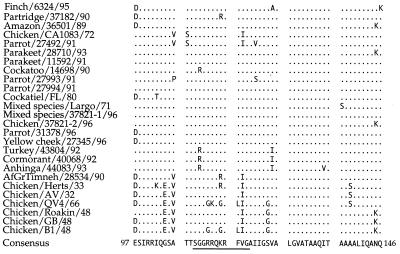
Following nucleotide sequencing of amplification products, predicted amino acid sequences were determined for the fusion protein cleavage site of all isolates listed in Table 1 (Fig. 1). Lentogenic vaccine isolates chicken/U.S./B1/48 and chicken/Australia/QV4/66 have the 109SGGGR(K)QGRLIG119 sequence at the fusion protein cleavage site, while the mesogenic and velogenic viruses have the sequence 109SGGRRQK(R)RFV[I]G119 containing the diagnostic pair of dibasic amino acids [RRQK(R)R] associated with the primary molecular determinant of virulence. Many of the recent exotic isolates share a V-for-I substitution at position 118 that is present in the mixed species/U.S./Largo/71 pet bird isolate, but not in the chicken/U.S./CA1083 (Fontana)/72 virus fusion protein cleavage site sequence. This amino acid substitution is also present in the sequence from the turkey/U.S./43804/92 and the cormorant/U.S./40068/92 viruses. The turkey/U.S./43804/92, cormorant/U.S./40068/92, and anhinga/U.S./44083/93 viruses also share an R-for-G substitution at position 110 with the cockatoo/Indonesia/14698/90 isolate. Among the U.S. NDV isolates obtained prior to 1950 and the chicken/Australia/QV4/66 virus, a G-for-S substitution occurs at position 124. Also, an E rather than G occurs at position 104 amino terminal to the fusion protein cleavage site among all isolates recovered prior to 1970.
FIG. 1.
Alignment of predicted amino acid sequences surrounding the fusion protein cleavage site. Amino acid sequences were derived by computer translation of primary nucleotide sequences of the resultant 254-bp amplification products. The fusion protein cleavage site sequence from position 109 to position 119 is underlined.
Alignments of predicted amino acid sequences encompassing the matrix protein nuclear localization signal were completed following computer translation of nucleotide sequences from the NDV matrix protein gene amplification products (Fig. 2). Several characteristic amino acid substitutions occur in the matrix protein indicative of NDV isolates from the United States recovered prior to 1950. These include an R-for-K substitution at positions 217 and 247, the T-for-S substitution at residue 243, and an S-for-R substitution at position 263. Residue 259 located within the nuclear localization signal appears to be highly variable with primarily charged amino acids located at that position. At position 229, a G-for-S substitution is shared by the cormorant/U.S./40068/92 and turkey/U.S./43804/92 isolates with the anhinga/U.S./44083/93 isolate.
FIG. 2.
Alignment of predicted amino acid sequences surrounding the matrix protein nuclear localization signal. Amino acid sequences were derived by computer translation of primary nucleotide sequences of the resultant 232-bp amplification products. The matrix protein nuclear localization signal from position 247 to position 263 is underlined.
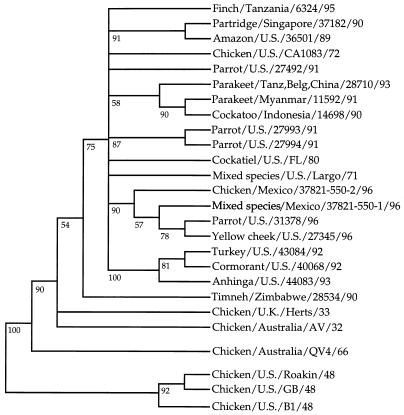
Following alignment, contiguous nucleotide sequence information from the fusion protein and matrix protein gene amplification products was used to determine phylogenetic relationships among the NDV isolates presented in Table 1. Using maximum parsimony analysis, two major phylogenetic clades of NDV isolates were delineated (Fig. 3). All of the virulent velogenic NDV isolates obtained from 1989 to 1996 were related to chicken/Australia/AV/32 and chicken/UK/Herts/33 as possible progenitor-type viruses. Recent NDV isolates highly virulent for chickens are all related to viruses considered exotic to North America. This includes chicken/U.S./CA1083 (Fontana)/72 and cormorant/U.S. (Minn.)/40068/92 isolated during previous outbreaks of Newcastle disease in the United States. This group of viruses contains both neurotropic and viscerotropic NDV as well as one mesogenic virus from an anhinga. However, representative virulent NDV isolates recovered in the United States prior to 1970 contain only neurotropic viruses and are not phylogenetically related to NDV obtained since the chicken/U.S./CA1083 (Fontana)/72 isolate was isolated. Phylogenetic relationships among NDV isolates correlate well with observed sequence differences surrounding the fusion protein cleavage site (Fig. 1) and matrix protein nuclear localization signal (Fig. 2).
FIG. 3.
Phylogenetic relationships of NDV isolates based on nucleotide sequences from a portion of the fusion protein and matrix protein genes. Amplification products with NDV RNA used as a template for RT-PCR were directly sequenced, and a phylogenetic tree was generated by parsimony analysis of the aligned contiguous nucleotide sequences. Numbers represent bootstrap confidence levels following 1,000 replications.
Previously we reported the use of degenerate oligonucleotide primers for amplification of nucleotide sequences from the genomes of divergent NDV isolates. The same primers were used to directly sequence the amplified products, and phylogenetic analysis was used to categorize NDV isolates (37). Two major phylogenetic branches of NDV isolates were identified (37) that conform to data obtained when simple distance matrix analysis is used (33, 43). Velogenic viruses obtained from exotic and other avian species since 1986 were primarily viscerotropic, with high ICPI values proving they are highly virulent for chickens. These viruses have entered the United States via importation of pet and exotic birds (9, 15, 31, 39), and a psittacine origin NDV isolate was epidemiologically linked to the major outbreak in southern California during the early 1970s (44, 45). These viruses group in a clade with chicken/Australia/AV/32 as the earliest reported isolate and include viruses isolated from birds in Mexico during 1996. Also, outbreaks of Newcastle disease in Taiwanese domestic poultry during 1995 were caused by viscerotropic velogenic NDV phylogenetically related to viruses in the chicken/Australia/AV/32 clade. It was suggested that both domestic and free-living birds were a source of virulent viruses in Taiwan (47).
Several amino acid sequence differences surrounding the fusion protein cleavage site and matrix protein nuclear localization signal were detected that correlate with the phylogenetic data. These included a V-for-I substitution at residue 118 within the fusion protein cleavage site sequence of velogenic NDV isolates obtained after 1972. As reported previously, the fusion protein cleavage site sequence from cormorants from the 1992 outbreak (18, 37) resembles the sequence from Republic of Ireland isolates causing disease in poultry during 1990 (5, 13). The matrix protein nuclear localization signal position 259 was again identified as a hypervariable site (37). This variability exists despite its location within a sequence required for matrix protein transport to the nucleus (12). Identification of several shared changes within the fusion protein and matrix proteins among virulent isolates is consistent with the quasispecies nature of RNA viruses (20). These differences occurred among virus isolates from a variety of birds with different geographic origins and indicate that multiple lineages of virulent NDV isolates are circulating among domestic, pet, and wild birds. This is particularly important, because some exotic species may harbor velogenic NDV isolates for extended periods of time (14). Consequently, highly virulent NDV isolates continue to circulate among birds other than chickens, and these isolates threaten commercial poultry worldwide.
Nucleotide sequence accession number.
Nucleotide sequences for portions of the fusion protein and matrix protein genes from the RT-PCRs were submitted to GenBank as a sense single-strand contiguous sequence for each NDV isolate. The accession numbers assigned to each new isolate (numbers only are given [for full designations, see Table 1]) are as follows: 11592/91, AF015507; 14698/90, AF015508; 27345/96, AF015509; 27492/91, AF015510; 27993/91, AF015511; 27994/91, AF015512; 28534/90, AF015513; 31378/96, AF015514; 28710/93, AF015515; 36501/89, AF015516; 37182/90, AF015517; 37821-550-1/96, AF015518; 37821-550-2/96, AF015520; and 6324/95, AF015519.
Acknowledgments
Gratitude is extended to the excellent technical assistance of Joyce Bennett for nucleotide sequence analysis and William Wilkes for assistance with ICPI tests. We thank the Molecular Genetics Instrumentation Facility at the University of Georgia for oligonucleotide synthesis. Appreciation is extended to Corrie Brown, Elizabeth Howerth, Mike Perdue, David Stallknecht and David Suarez for helpful critiques of the manuscript.
These investigations were supported by the U.S. Poultry & Egg Association grant no. 165 and USDA, ARS, CRIS project no. 6612-32000-015-00D-085.
REFERENCES
- 1.Albiston H E, Gorrie C J R. Newcastle disease in Victoria. Aust Vet J. 1942;18:75–79. [Google Scholar]
- 2.Alexander D J. Newcastle disease. In: Purchase H G, Arp L H, Domermuth C H, Pearson J E, editors. A laboratory manual for the isolation and identification of avian pathogens. 3rd ed. Kennett Square, Pa: American Association of Avian Pathologists, Inc.; 1989. pp. 114–120. [Google Scholar]
- 3.Alexander D J. Newcastle disease and other avian paramyxovirus infections. In: Calnek B W, Barnes H J, Beard C W, Reid W M, Yoder H W Jr, editors. Diseases of poultry. 9th ed. Ames: Iowa State University Press; 1991. pp. 496–519. [Google Scholar]
- 4.Alexander D J, Allan W H. Newcastle disease virus pathotypes. Avian Pathol. 1974;4:269–278. doi: 10.1080/03079457409353840. [DOI] [PubMed] [Google Scholar]
- 5.Alexander D J, Campbell G, Manvell R J, Collins M S, Parsons G, McNulty M S. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet Rec. 1992;130:65–68. doi: 10.1136/vr.130.4.65. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee M, Reed W M, Fitzgerald S D, Panigrahy B. Neurotropic velogenic Newcastle disease in cormorants in Michigan: pathology and virus characterization. Avian Dis. 1994;38:873–878. [PubMed] [Google Scholar]
- 7.Beaudette F R, Bivins J A, Miller B R. Newcastle disease immunization with live virus. Cornell Vet. 1949;39:302–334. [Google Scholar]
- 8.Brugh M, Beard C W. Atypical disease produced in chickens by Newcastle disease virus isolated from exotic birds. Avian Dis. 1984;28:482–487. [PubMed] [Google Scholar]
- 9.Bruning-Fann C, Kaneene J, Heamon J. Investigation of an outbreak of velogenic viscerotropic Newcastle disease in pet birds in Michigan, Indiana, Illinois, and Texas. J Am Vet Med Assoc. 1992;201:1709–1714. [PubMed] [Google Scholar]
- 10.Cheville N F, Stone H, Riley J, Ritchie A E. Pathogenesis of virulent Newcastle disease in chickens. J Am Vet Med Assoc. 1972;161:169–179. [PubMed] [Google Scholar]
- 11.Chomzcynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Coleman N A, Peeples M A. The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology. 1993;195:596–607. doi: 10.1006/viro.1993.1411. [DOI] [PubMed] [Google Scholar]
- 13.Collins M S, Bashiruddin J B, Alexander D J. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch Virol. 1993;128:363–370. doi: 10.1007/BF01309446. [DOI] [PubMed] [Google Scholar]
- 14.Erickson G A, Mare C J, Gustafson G A, Proctor S J, Miller L D, Carbey E A. Interactions between viscerotropic velogenic Newcastle disease virus and pet birds of six species. I. Clinical and serological responses, and viral excretion. Avian Dis. 1977;21:642–654. [PubMed] [Google Scholar]
- 15.Francis D W. Newcastle and psittacines, 1970–71. Poult Dig. 1973;32:16–19. [Google Scholar]
- 16.Glickman R L, Syddall R J, Iorio R M, Sheehan J P, Bratt M A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J Virol. 1988;62:354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoh B, Ohnishi Y, Inocencio N M, Esaki E, Nakayama K, Barr P J, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66:6391–6397. doi: 10.1128/jvi.66.11.6391-6397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heckert R A, Collins M S, Manvell R J, Strong I, Pearson J E, Alexander D J. Comparison of Newcastle disease viruses isolated from cormorants in Canada and the USA in 1975, 1990 and 1992. Can J Vet Res. 1996;60:50–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Hitchner S B, Johnson E P. A virus of low virulence for immunizing fowls against Newcastle disease (avian pneumoencephalitis) Vet Med. 1948;43:525–530. [PubMed] [Google Scholar]
- 20.Holland J J, de la Torre J C, Steinhauer D. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 21.Kaleta E F, Baldauf C. Newcastle disease in free-living and pet birds. In: Alexander D J, editor. Newcastle Disease. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 197–256. [Google Scholar]
- 22.Kaleta E F, Siegmann O, Jank Ladwig R, Glunder G. Isolation and biological properties of virulent subpopulations from lentogenic Newcastle disease virus strains. Comp Immunol Microbiol Infect Dis. 1980;2:485–496. doi: 10.1016/0147-9571(79)90090-0. [DOI] [PubMed] [Google Scholar]
- 23.King D J. Newcastle disease virus passage in MDBK cells as an aid in detection of a virulent subpopulation. Avian Dis. 1993;37:961–969. [PubMed] [Google Scholar]
- 24.Kotewicz M L, Sampson C M, D’Alesio D E, Gerard G F. Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988;16:265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis J G, Chang G-J, Lanciotti R S, Trent D W. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Methods. 1992;38:11–24. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 26.McGinnes L W, Morrison T. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 1986;5:343–356. doi: 10.1016/0168-1702(86)90028-6. [DOI] [PubMed] [Google Scholar]
- 27.Meulmanns G. Control by vaccination. In: Alexander D J, editor. Newcastle disease. Boston, Mass: Kluwer Academic Publishers; 1988. pp. 318–332. [Google Scholar]
- 28.Nagai Y, Klenk H D, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72:494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 29.Office International des Epizootes. Manual for animal disease reporting to the OIE. Paris, France: World Organization for Animal Health; 1996. [Google Scholar]
- 30.Ogasawara T, Gotoh B, Suzuki H, Asaka J, Shimokata K, Rott R, Nagai Y. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 1992;11:467–472. doi: 10.1002/j.1460-2075.1992.tb05076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panigrahy B, Senne D A, Pearson J E, Mixson M A, Cassidy D R. Occurrence of velogenic viscerotropic Newcastle disease in pet and exotic birds in 1991. Avian Dis. 1993;37:254–258. [PubMed] [Google Scholar]
- 32.Saiki R K, Scharf F, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnhem N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi T, Toyoda T, Gotoh B, Inocencio N M, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution I. Multiple lineages defined by sequence variability of the hemagglutinin-neuraminidase gene. Virology. 1989;169:260–272. doi: 10.1016/0042-6822(89)90151-7. [DOI] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaper U M, Fuller F, Ward M D, Mehrotra Y, Stone H O, Stripp B R, De-Buysscher E V. Nucleotide sequence of the envelope protein genes of a highly virulent neurotropic strain of Newcastle disease virus. Virology. 1988;165:291–295. doi: 10.1016/0042-6822(88)90686-1. [DOI] [PubMed] [Google Scholar]
- 36.Schloer G M, Hanson R P. Relationship of plaque size and virulence for chickens of 14 representative Newcastle disease virus strains. J Virol. 1968;2:40–47. doi: 10.1128/jvi.2.1.40-47.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seal B S, King D J, Bennett J D. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J Clin Microbiol. 1995;33:2624–2630. doi: 10.1128/jcm.33.10.2624-2630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seal B S. Analysis of matrix protein gene nucleotide sequence diversity among Newcastle disease virus isolates demonstrates that recent disease outbreaks are caused by viruses of psittacine origin. Virus Genes. 1996;11:217–224. doi: 10.1007/BF01728661. [DOI] [PubMed] [Google Scholar]
- 39.Senne D A, Pearson J E, Miller L D, Gustafson G A. Virus isolations from pet birds submitted for importation into the United States. Avian Dis. 1983;27:731–744. [PubMed] [Google Scholar]
- 40.Simmons G C. The isolation of Newcastle disease virus in Queensland. Aust Vet J. 1967;43:29–30. doi: 10.1111/j.1751-0813.1967.tb04764.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith L M, Sanders J Z, Kaiser R J, Hughs P, Dodd C, Connell C R, Heines C, Kent S B H, Hood L E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:673–681. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 42.Swafford D. PAUP: phylogenetic analysis using parsimony. Version 3. Champaign, Ill: Illinois Natural History Survey; 1989. [Google Scholar]
- 43.Toyoda T, Sakaguchi T, Hirota H, Gotoh B, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology. 1989;169:273–282. doi: 10.1016/0042-6822(89)90152-9. [DOI] [PubMed] [Google Scholar]
- 44.Utterback W W, Schwartz J H. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971–1973. J Am Vet Med Assoc. 1973;163:1080–1088. [PubMed] [Google Scholar]
- 45.Walker J W, Heron B R, Mixson M A. Exotic Newcastle disease eradication program in the United States of America. Avian Dis. 1973;17:486–503. [PubMed] [Google Scholar]
- 46.Wobeser G, Leighton F A, Norman R, Myers D J, Onderka D, Pybus M J, Neufeld J L, Fox G A, Alexander D J. Newcastle disease in wild water birds in western Canada, 1990. Canadian Vet J. 1993;34:353–359. [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C-Y, Chang P-C, Hwang J-M, Shieh H K. Nucleotide sequence and phylogenetic analysis of Newcastle disease virus isolates from recent outbreaks in Taiwan. Avian Dis. 1997;41:365–373. [PubMed] [Google Scholar]