Abstract
A nested PCR specific for the Mycoplasma pneumoniae P1 gene was used to diagnose mycoplasma infection in two cohort patients with severe pneumonia within 24 h of tissue receipt. A postmortem diagnosis of M. pneumoniae infection was obtained for the first patient, who died without the collection of appropriate paired samples for serodiagnosis. An open-lung biopsy obtained from the second patient allowed a quick, definitive diagnosis and proper selection of therapy.
Mycoplasma pneumoniae is a causative agent of primary atypical pneumonia and tracheobronchitis. Primarily a respiratory pathogen, it has been estimated to cause up to 20% of the approximately 500,000 cases of community-acquired pneumonia requiring hospitalization in the United States each year (11). These figures are in good agreement with estimates that up to 35% of outpatient pneumonia cases and up to 18% of pneumonia cases requiring hospitalization are caused by M. pneumoniae (10). In approximately 15% of patients with these infections, extrapulmonary complications develop, including neurologic, dermatologic, renal, musculoskeletal, cardiovascular, gastrointestinal, and immunologic manifestations (1, 2–5, 8, 9).
M. pneumoniae respiratory illness is underdiagnosed because culture procedures are time-consuming, expensive, and of limited clinical value since isolation has to be pursued for at least 6 weeks. Serologic diagnosis can be problematic because of coincidental mycoplasma titers and the frequent need for paired acute- and convalescent-phase sera. Newer detection methods targeting protein antigens or nucleic acids hold promise for more rapid diagnosis. Here we describe the use of a nested-PCR assay to detect M. pneumoniae DNA in tissue samples from two patients with severe pneumonia. With this approach, a definitive diagnosis was available within 24 h.
Both patients were part of a confirmed M. pneumoniae outbreak that involved five clinical cases in a community hospital in Middlesex County, N.J. The outbreak was confirmed by testing paired sera for a fourfold rise in titer to M. pneumoniae by culture, by PCR, and by the presence of clinical signs of disease consistent with an atypical mycoplasmal pneumonia. In the spring, a 28-year-old male (nonsmoker) experienced a sore throat, followed by a productive cough. Approximately 5 days after onset, he became severely dyspneic and was admitted to the hospital with acute respiratory failure and seizures. The patient was febrile at admission and throughout hospitalization, with a temperature reaching >40.5°C. A chest X ray at admission was normal; a second X ray revealed bilateral basilar interstitial infiltrates. The patient died 2 weeks after admission. No respiratory specimens were available for analysis. At autopsy, lung tissue was taken, placed on dry ice, and sent to the Respiratory Diseases Laboratory Section at the Centers for Disease Control and Prevention because a bacterial etiology was suspected.
One week later, a 42-year-old male (nonsmoker) noted a slight cough, sweating, and a temperature of 39.1 to 39.3°C. The cough became productive, and 1 week after onset he was admitted to the hospital with dyspnea and headaches. A chest X ray revealed bilateral basilar infiltrates and unilateral hilar adenopathy. This patient underwent an open-lung biopsy because of a suspected infectious etiology, and a small sample was sent on dry ice to the Centers for Disease Control and Prevention for evaluation. This sample was received 2 days after the autopsy sample.
Both the autopsy tissue from the first patient and the open-lung biopsy tissue from the second patient were thawed and minced and cultured in SP4 broth and diphasic media (14) by a 105-fold dilution before processing for PCR. Because the autopsy tissue appeared to be severely contaminated, antibiotics were added to the medium to suppress contamination. DNA was extracted from the minced tissue by using reagents supplied with the EnviroAmp Legionella Detection Kit (Perkin-Elmer, Norwalk, Conn.). Minced tissue was placed into 100 μl of room temperature EnviroAmp lysis buffer and boiled for 10 min. Samples were centrifuged at 12,000 rpm in a microcentrifuge for 15 min, and then supernatant fluids from this extraction were run in a nested-PCR assay. Total processing time from tissue mincing to obtaining a preparation suitable for PCR analysis was 1 h.
Primers P1-40, P1-178, P1-285, and P1-331 (Table 1) targeted to the P1 cytadhesin gene of M. pneumoniae were used to amplify species-specific DNA (6, 7). These primers, when previously tested in simulated throat swabs, detected levels of <40 genome copies. They have been tested with DNA from eight related Mycoplasma species, including M. genitalium, and have been found to be specific for M. pneumoniae DNA. There is also no reactivity with Ureaplasma urealyticum or with Acholeplasma laidlawii DNA (6).
TABLE 1.
Primer sequences
| Primer | Sequence (5′-3′) |
|---|---|
| P1-40 | ATT CTC ATC CTC ACC GCC ACC |
| P1-178 | CAA TGC CAT CAA CCC GCG CTT AAC C |
| P1-285 | GTT GTC GCG CAC TAA GGC CCA CG |
| P1-331 | CGT GGT TTG TTG ACT GCC ACT GCC G |
In the first set of reactions, a final reaction volume of 100 μl consisted of 1× buffer II, 1.5 mM MgCl2 (both from Perkin-Elmer), deoxynucleoside triphosphates (dNTPs) at 125 μM per dNTP (Pharmacia Biotech, Piscataway, N.J.), 2 U of AmpliTaq polymerase (Perkin-Elmer), and 20 pmol (each) of the P1-40 and P1-331 primers. DNA in 20-, 10-, and 5-μl amounts was added. Samples were heated to 85°C before the addition of the polymerase. The initial run consisted of 25 cycles of 94°C for 30 s and 60°C for 30 s, followed by a 5-min extension at 72°C. Five microliters of this reaction mixture was used as a template in the second round of PCR with the same reaction mixture and with the inner primer pair P1-178 and P1-285. This run was identical to the first, with a 94°C, 45-s denaturation step. The expected sizes of PCR-generated bands were 285 bp for the P1-40–P1-331 primer pair and 107 bp for the P1-178–P1-285 primer pair. Samples were loaded onto 1% molecular biology-certified agarose (Bio-Rad, Richmond, Calif.), 2% NuSieve agarose gels (FMC Bioproducts, Rockland, Maine) and visualized with ethidium bromide.
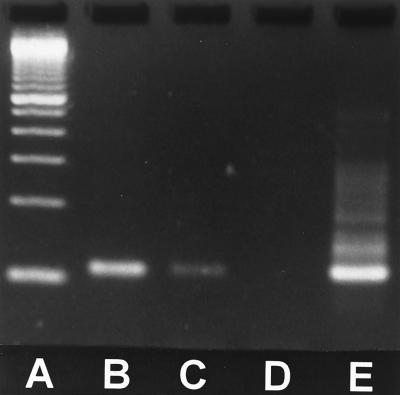
No bands were present for either sample following the first round of PCR (data not shown). Figure 1 shows the results of the nested reaction. Both the autopsy and the open-lung biopsy tissue generated a 107-bp band, consistent with the expected size of this portion of the P1 gene.
FIG. 1.
Nested M. pneumoniae PCR of lung tissue. Lanes: A, 100-bp molecular size markers; B, open-lung biopsy tissue from patient 2; C, autopsy lung tissue from patient 1; D, negative control; E, positive control showing 107-bp amplicon.
This is the first report of detection of M. pneumoniae in nonpreserved autopsy tissue or fresh tissue. Another PCR technique has been used previously to detect M. pneumoniae on formalin-fixed, paraffin-embedded lung tissue (15).
The extraction procedure from frozen tissue took approximately 1 h. The nested PCR consisted of two primer sets targeting the P1 gene of M. pneumoniae. Both PCR cycles have two steps (as opposed to the usual three steps), with annealing and extension occurring simultaneously, thus greatly decreasing the reaction time. One sample can be processed and run through both steps of the nested PCR in 1 day. A nonnested PCR with the P1-178 and P1-331 primer pairs, which we have used successfully with some other clinical samples, did not detect mycoplasmas in these specimens (data not shown). It may be that nested amplifications are able to detect smaller amounts of DNA, but it is more likely that the effects of inhibitors (human cells and cellular debris present in the tissue samples) were diminished by the initial amplification of specific target DNA followed by dilution into a second round of amplification (12, 13). In our experience, running multiple dilutions of target DNA can increase the chances for detection if initial attempts with undiluted samples fail.
M. pneumoniae was cultured from the open-lung biopsy tissue after 3 weeks of incubation. The autopsy tissue was grossly contaminated, and culture for M. pneumoniae was unsuccessful despite the addition of antibiotics to the medium and the use of appropriate dilution methods. Paired acute- and convalescent-phase sera, required for definitive serologic diagnosis of M. pneumoniae infections, were unavailable from this patient. A single serum sample was positive for M. pneumoniae by the rapid Remel enzyme immunoassay (a qualitative membrane-based enzyme immunoassay) (Remel, Lenexa, Kans.) and by the complement fixation test (both tests detect immunoglobulin G and M antibodies), but a single positive serum result does not provide a definitive diagnosis. Although serologic diagnosis was not possible, the positive PCR result from the lung tissue, together with the patient’s clinical signs and close association with other confirmed mycoplasma cases, allowed confirmative diagnosis. In the second case, rapid diagnosis from the lung biopsy specimen allowed the appropriate use of antibiotics for both the patient and symptomatic close contacts, thus limiting the scope of the outbreak. This information could be crucial, because mycoplasmas are unresponsive to some of the antibiotics commonly used for treatment of community-acquired pneumonia.
The P1 PCR target is an immunodominant cytadherence protein specific for M. pneumoniae. M. genitalium has a cytadherence protein, MgPa, which shares serologic cross-reactivity with P1, but the MgPa gene is not amplified by these primer pairs (data not shown). Subsequently, fresh and autopsy lung samples from other suspect pneumonia patients have been tested by nested PCR, with negative results, thus serving as negative controls for the assay.
When interpreting results from antigen detection assays such as PCR, the results must be interpreted by taking into consideration the patient’s clinical signs. A positive PCR result from a secretion may indicate infection without disease. And because very little is known about the carriage of M. pneumoniae in the human respiratory tract, a positive PCR result from lung tissue must be interpreted carefully, all the while considering the patient’s clinical signs (this would be true with any antigen detection assay). Ideally, paired sera (acute- and convalescent-phase samples) should be tested whenever possible so that a patient can be evaluated for the characteristic rise in titer that is most strongly suggestive of current M. pneumoniae infection. In this report, the patients clearly had clinical signs consistent with mycoplasmal pneumonia, and the PCR results were very useful in arriving at a diagnosis in the absence of serology results from paired sera.
REFERENCES
- 1.Anikster Y, Glustein J Z, Weill M, Isacsohn M. Extrapulmonary manifestations of Mycoplasma pneumoniae infections. Isr J Med Sci. 1994;30:71–85. [PubMed] [Google Scholar]
- 2.Augstin E T, Gill V, Cunha B A. Mycoplasma pneumoniae meningoencephalitis complicated by diplopia. Heart Lung. 1994;23:436–437. [PubMed] [Google Scholar]
- 3.Broughton R A. Infections due to Mycoplasma pneumoniae in childhood. Pediatr Infect Dis J. 1986;5:71–85. doi: 10.1097/00006454-198601000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Cassell G H, Cole B C. Mycoplasmas as agents of human disease. N Engl J Med. 1981;304:80–89. doi: 10.1056/NEJM198101083040204. [DOI] [PubMed] [Google Scholar]
- 5.Davis C P, Cochran S, Lisse J, Buck G, DiNuzzo A R, Weber T, Reinarz J A. Isolation of Mycoplasma pneumoniae from synovial fluid samples in a patient with pneumonia and polyarthritis. Arch Intern Med. 1988;148:969–970. [PubMed] [Google Scholar]
- 6.Jensen J S, Søndergård-Anderson J, Uldum S A, Lind K. Detection of Mycoplasma pneumoniae in simulated clinical samples by polymerase chain reaction. APMIS. 1989;97:1046–1048. doi: 10.1111/j.1699-0463.1989.tb00516.x. [DOI] [PubMed] [Google Scholar]
- 7.Jensen J S, Uldum S A, Lind K. International Organization of Mycoplasmology Letters, vol. 2, poster session 1, no. 65. International Organization of Mycoplasmology; 1992. Clinical evaluation of a polymerase chain reaction assay for detection of Mycoplasma pneumoniae; p. 198. [Google Scholar]
- 8.Johnston C L W, Webster A D B, Taylor-Robinson D, Rapaport G A, Hughes G R V. Primary late-onset hypogammaglobulinaemia associated with inflammatory polyarthritis and septic arthritis due to Mycoplasma pneumoniae. Ann Rheum Dis. 1983;42:108–110. doi: 10.1136/ard.42.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koletsky R J, Weinstein A J. Fulminant Mycoplasma pneumoniae infection. Report of a fatal case, and a review of the literature. Am Rev Respir Dis. 1980;122:491–496. doi: 10.1164/arrd.1980.122.3.491. [DOI] [PubMed] [Google Scholar]
- 10.Luby J P. Pneumonia caused by Mycoplasma pneumoniae infection. Clin Chest Med. 1991;12:237–244. [PubMed] [Google Scholar]
- 11.Marston B J, Plouffe J F, File T M, Jr, Hackman B A, Salstrom S J, Lipman H P, Kolczak M S, Breiman R F. Incidence of community-acquired pneumonia requiring hospitalization. Arch Intern Med. 1997;157:1709–1718. [PubMed] [Google Scholar]
- 12.Miserez R, Pilloud T, Cheng X, Nicolet J, Griot C, Frey J. Development of a sensitive nested PCR method for the specific detection of Mycoplasma mycoides subsp. mycoides SC. Mol Cell Probes. 1997;11:103–111. doi: 10.1006/mcpr.1996.0088. [DOI] [PubMed] [Google Scholar]
- 13.Schoeb T R, Dybvig K, Keisling K F, Davidson M K, Davis J K. Detection of Mycoplasma pulmonis in cilia-associated respiratory bacillus isolates and in respiratory tracts of rats by nested PCR. J Clin Microbiol. 1997;35:1667–1670. doi: 10.1128/jcm.35.7.1667-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tully J G, Rose D L, Whitcomb R F, Wenzel R P. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly modified culture medium. J Infect Dis. 1979;139:478–482. doi: 10.1093/infdis/139.4.478. [DOI] [PubMed] [Google Scholar]
- 15.Williamson J, Marmion B P, Kok T, Antic R, Harris R J. Confirmation of fatal Mycoplasma pneumoniae infection by polymerase chain reaction detection of the adhesion gene in fixed lung tissue. J Infect Dis. 1994;170:1052–1053. doi: 10.1093/infdis/170.4.1052. . (Letter.) [DOI] [PubMed] [Google Scholar]