Abstract
Gene editing nucleases (GENs), represented by CRISPR/Cas9, have become major tools in biomedical research and offer potential cures for many human diseases. Gene editing therapy (GETx) studies in animal models targeting genes such as proprotein convertase subtilisin/kexin type 9 (PCSK9), apolipoprotein C3 (APOC3), angiopoietin Like 3 (ANGPTL3) and inducible degrader of the low-density lipoprotein receptor (IDOL) have demonstrated the benefits and advantages of GETx in managing atherosclerosis. Here we present our views on this brand new therapeutic option for cardiovascular diseases (CVD).
Keywords: cardiovascular diseases, CRISPR/Cas9, gene editing therapy
Gene editing nucleases (GENs), represented by Zinc Finger Nuclease (ZFN), Transcription Activator-Like Effector Nuclease (TALEN), and the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein-9 (CRISPR/Cas9) have become major tools in biomedical research [1]. ZFN, TALEN, and CRISPR/Cas9 are efficient in generating double strand breaks (DSB) in the specific genomic loci that can be repaired by error-prone non-homologous end joining (NHEJ) leading to a functional knockout of the targeted gene or used to integrate a DNA sequence at a specific locus through homologous recombination (HR).
GENs offer potential cures for many human diseases. Recently, gene editing therapy (GETx) studies in animal models targeting genes such as proprotein convertase subtilisin/kexin type 9 (PCSK9), apolipoprotein C3 (APOC3), angiopoietin Like 3 (ANGPTL3), and inducible degrader of the low-density lipoprotein receptor (IDOL) have demonstrated the benefits and advantages of GETx in managing atherosclerosis. Here we present our views on this brand-new therapeutic option for cardiovascular diseases (CVD).
Opportunities
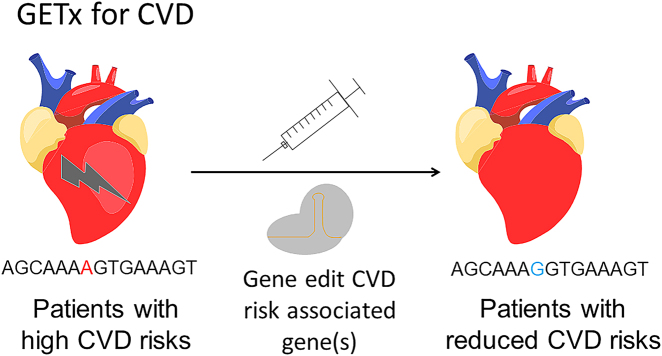
The 2020 Nobel Prize in Chemistry is awarded to Drs. Charpentier and Doudna for their contributions in the development of CRISPR/Cas9, the dominant GEN that is used nowadays [2]. GEN based therapy, referred to as GETx in this article, offers new options for treating, and maybe curing many diseases, including genetic diseases, infectious diseases, and cancer. The provocative question whether GETx can be employed for treating CVDs was asked by Gupta in 2014, 1 year after Cas9 debuted [3]. The idea is based on the consideration that CVD, likely many other diseases, is genetically predisposed; hence modifying the gene(s) “for the good” will bring therapeutic benefits [4]. The methodology is straightforward (Figure 1): (i) identify a gene whose mutation is cardioprotective. In most cases, such mutation leads to loss of function (LoF), and there are many known candidate genes, for example, PCSK9, cholesteryl ester transfer protein (CETP), ANGPTL3, and ApoC3 [4]; (ii) use Cas9 (or other gene editing tools) to produce these favorable mutation(s); (iii) the patient enjoys a lifelong reduction of CVD risks, without the need to receive repetitive treatments. The process analogs that of vaccination in which one shot render protection of infectious diseases. Indeed, some people refer to Cas9 therapy for CVD as “vaccination for CVD” [5].
Figure 1:
Illustration of GETx for CVD. CVD, cardiovascular diseases; GETx, gene editing therapy.
GETx for CVD, however, differs fundamentally from that of vaccination. GEN works at the DNA level [6]. For example, in CRISPR/Cas9, a guide RNA (gRNA) complementary to the target locus is used as the “GPS” to guide the nuclease (i.e., Cas9) to the right place for action, where a double stranded break (DSB) is generated followed by repair through either the error-prone NHEJ or the homology directed repair (HDR) pathway. The former one is exploited for gene knockout applications whereas the latter one is used to achieve knock-in by providing a donor template. Both knockout and knock-in efficiencies can reach double digits in vitro and in vivo, although in general knockout is easier and of higher success rate because NHEJ is the dominant DNA repair pathway. This could work in favor for GETx for CVD, as the goal in most cases is to generate a LoF mutation.
Challenges
Many challenges remain to be addressed towards clinical applications of GETx [7]. At the forefront are GEN associated genotoxicity. GENs could cause high off-target insertions or deletions (indels). This is an intrinsic concern with Cas9 as well as other nucleases. In certain extreme examples, the off-target indel rates could be even higher than those at the on-target [8]. Intensive research is currently aimed to improve the specificity of the nucleases by using various approaches, such as specificity improving mutagenesis [9] and fusion of HDR-promoting motif [8], highlighting the pressing needs in this area.
Furthermore, GENs cause on-target genotoxicity. In precise gene editing (PGE) applications, after DSB generation, while a certain percentage of alleles are repaired by HDR, many are repaired by NHEJ leading to on-target indels. In most cases, the on-target indel rates are higher than the PGE rates. The potential genotoxicity caused by on-target indels, in our opinion, is even higher than those of off-target indels for at least two reasons: (i) the on-target indel rates are sometimes higher than off-target ones [8]; (ii) the on-target indels will likely cause consequences at this clinically significant locus.
Another major challenge is to realize efficient and safe in vivo delivery. In vivo delivery is a general challenge for nucleic acid therapeutics. In 2017, the U.S. Food and Drug Administration (FDA) approved LUXTURNA, an adeno-associated virus (AAV) based gene therapy for the treatment of biallelic RPE65 mutation – associated retinal dystrophy. This landmark approval positions AAV to the central stage of GETx as the choice of delivery vehicle [10]. The direct challenge using AAV for Cas9-based therapy is that the size of the most popular Cas9 variant spCas9 is 4.2 kb, approaching AAV’s packaging capacity (4.7 kb), leaving little room for other elements (e.g. promoter, etc.). The situation is aggravated if other variants of larger size, such as Cas9 base editor, are used. The use of AAV in the context of gene editing is comprehensively reviewed elsewhere [11].
There are also specific considerations related to GETx for CVD. For example, unlike genetic diseases which are mostly caused by monogenic mutations, CVDs are a complicated syndrome wherein many genes are involved. So ideally, the targeted mutations shall be the ones that exist in the human allele pool and are known for not only providing cardioprotection but also not associated with increased other health risks such as cancer. Secondly, both AAV and Cas9 are new tools. The long-term efficacy and safety profiles of the AAV-Cas9 therapy are yet to be determined, especially given that CVD are chronicle diseases. Thirdly, although several tissue specific AAV subtypes have been developed to minimize the germline contribution, the possibility that the DNA edits go into the germline still exists and poses an ethical concern. At the technical level, many questions remain to be answered: when to initiate such therapy (i.e. at infant age or later)? How many cells need to be edited to achieve therapeutic effects (i.e. 10%, 50%, 100%)? What cells are to be targeted (i.e. liver cells, cardiomyocytes, all cells), etc.?
Actions
In the United States, the National Institutes of Health (NIH) has launched a Somatic Cell Genome Editing (SCGE) consortium program [12]. The four major objectives of the NIH SCGE program are to: (i) expand the toolbox of GENs. (ii) develop effective in vivo delivery systems; (iii) design new assays for testing the safety and efficacy of GETx; and (iv) distribute the knowledge, methods, and tools to the scientific community. Other countries including China are expected to launch similar programs. These NIH SCGE objectives, although generally applicable for any GETx, are instructive for future work in our efforts to advance GETx for CVD.
Firstly, new GENs with improved efficacy and safety outcomes are needed. Base editors and prime editors, which do not generate DSB in their editing processes, represent safer choices than the conventional spCas9 [13]. For example, Chadwick, Wang, and Musunuru reported in vivo base editing of PCSK9 in mice, leading to the reduction of plasma cholesterol levels and importantly without evidence of off-target mutagenesis [14]. Other emerging Cas9 variants, such as Hifi-Cas9 [15], HypaCas9 [16], and miCas9 [8] could also contribute to the improvements of safety by dramatically reduce off-target and even on-target indel events.
Secondly, in light of the cargo size limitation of AAV and its potential immunogenicity and tumorigenicity concerns, other in vivo delivery methods, especially non-viral means, are being actively pursued. Lipid nanoparticles (LNP) are proven extremely successful in delivering mRNA-based vaccines in our ongoing battle against SARS-CoV-2. Will they be useful for in GETx for CVD? Another consideration is that how to improve the targeting specificity for non-liver organs/tissues? While liver is a relatively “easy to target” organ, other CVD organs such as the heart are much more challenging, for which incorporating tissue specific ligands or local delivery may provide some help.
Thirdly, the unique working mechanism (e.g. editing at the nucleotide level) of GETx requires new assay methods to quantify both the efficacy and safety outcomes at the DNA level. Guide-seq [17] is currently the gold standard in evaluating off-target editing events using cultured cells (i.e. not in vivo). An in vivo version, called VIVO [18], analyzes off-target editing events in animals (but not human cells). Ideally a combination of these two, i.e. analyzing off-target editing events in human cells in vivo (e.g. transplanting human cells to an immunodeficient host animal followed by gene editing), would provide more accurate information for this parameter. More powerful sequencing technologies, such as those that can have mega base read length, are certainly of use. Assay methods shall also be developed in evaluating the potential immunogenicity (caused by Cas9 elements or the delivery system), tumorigenicity (caused by undesirable gene editing or by viral vectors), and germline mutagenesis (caused by gene editing in germ cells) risks. Another consideration is that while most preclinical animal works are conducted in mice, this model system cannot predict long-term (i.e. >2 year) adverse effects due to their short lifespan. In this regard, longer lived models such as non-human primates, pigs, and rabbits are expected to make their contributions.
Lastly, how to engage public is of eminent significance for GETx for CVD. As pointed out by Li et al. [19], there is a gap between the application of the gene editing technologies which is driven by the researchers and the policies at the institutional, the national, and the international levels that govern these applications. So is there a gap between the minds of GETx researchers and the public. As the number one killer in both China and USA as well as many other countries, CVD affects billions of people. How will the public view GETx? How to explain the pros and cons? Whether and when will the society GET(x) ready for CVD? Perhaps as one of the first steps, efforts shall also be taken to inform clinicians of this emerging therapeutic option, and to encourage collaborations between researchers and doctors to conduct “bench to bed” research.
Perspectives
Human medicine has been driven by technology development. Will GENs represented by Cas9 prove to be another game changer for CV medicine development remains an open question? In 2015, about 18 million people died from CVD, of which three-quarters occurred in low- and middle-income countries, who cannot afford the cost and complexity of antibody-based or other biologics based “conventional” therapies which need multiple lifetime long administration [5]. A simpler vaccination-like treatment such as the proposed GETx therapy may greatly reduce the burden of medical care. At a smaller scale, before it is accepted by the entire society, GETx therapy could provide benefits to people who are at extremely high risks of CV events, by “resetting their odds” [5]. But the concerns are equally heavy on issues such as on-target efficiency, off-target mutagenesis, the possibility of germline editing, among others. Ethical debates aside, it is imperative for the scientific community to provide evidence to demonstrate the efficacy and to determine the safety of GETx for CVD.
Footnotes
Research funding: None declared.
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Ethical approval: The local Institutional Review Board deemed the study exempt from review.
References
- 1.Jacinto FV, Link W, Ferreira BI. CRISPR/Cas9-mediated genome editing: from basic research to translational medicine. J Cell Mol Med. 2020;24:3766–78. doi: 10.1111/jcmm.14916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crispr Developers Win 2020 Nobel Prize for Chemistry. Am J Med Genet A. 2021;185:8–9. doi: 10.1002/ajmg.a.61645. [DOI] [PubMed] [Google Scholar]
- 3.Gupta RM. One-shot, one cure with genome editing for dyslipidemia. Circ Cardiovasc Genet. 2014;7:967–8. doi: 10.1161/circgenetics.114.000958. [DOI] [PubMed] [Google Scholar]
- 4.Nishiga M, Qi LS, Wu JC. Therapeutic genome editing in cardiovascular diseases. Adv Drug Deliv Rev. 2021;168:147–157. doi: 10.1016/j.addr.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King A. A CRISPR edit for heart disease. Nature. 2018;555:S23–5. doi: 10.1038/d41586-018-02482-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–64. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 7.Alagoz M, Kherad N. Advance genome editing technologies in the treatment of human diseases: CRISPR therapy (Review) Int J Mol Med. 2020;46:521–34. doi: 10.3892/ijmm.2020.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, Ruan J, Song J, Wen L, Yang D, Zhao J, et al. MiCas9 increases large size gene knock-in rates and reduces undesirable on-target and off-target indel edits. Nat Commun. 2020;11:6082. doi: 10.1038/s41467-020-19842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid-Burgk JL, Gao L, Li D, Gardner Z, Strecker J, Lash B, et al. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020;78:794–800. doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeler AM, Flotte TR. Recombinant adeno-associated virus gene therapy in light of Luxturna (and Zolgensma and Glybera): where are we, and how did we get here? Annu Rev Virol. 2019;6:601–21. doi: 10.1146/annurev-virology-092818-015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–50. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha K, Sontheimer EJ, Brooks PJ, Dwinell MR, Gersbach CA, Liu DR, et al. The NIH somatic cell genome editing program. Nature. 2021;592:195–204. doi: 10.1038/s41586-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–44. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 14.Chadwick AC, Wang X, Musunuru K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler Thromb Vasc Biol. 2017;37:1741–7. doi: 10.1161/atvbaha.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–24. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–10. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–19. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Liu YG, Chen Y. Genome-editing technologies: the gap between application and policy. Sci China Life Sci. 2019;62:1534–8. doi: 10.1007/s11427-019-1566-1. [DOI] [PubMed] [Google Scholar]