Abstract
A 44-year-old woman developed Legionella pneumophila pneumonia after cerebral surgery. Initially, one colony from a clinical specimen and two colonies from water samples, all belonging to serogroup 12, did not match when their DNA restriction patterns were compared. When additional colonies from the water specimens were analyzed, a serogroup 12 strain complementary to that found in the clinical specimen was identified. Other colonies from the clinical specimen were identified as serogroup 12 strains complementary to those identified from the water. In addition, the same serogroup 1 strain was isolated from the patient and the water system.
Legionella pneumophila is recognized as an important pathogen causing nosocomial pneumonia. Patients with an impaired immune system or after surgery have a high risk of acquiring legionellosis (12, 19). Although different species, serogroups, and monoclonal subtypes of Legionella have been isolated from single environmental sources, infections by multiple strains have rarely been reported (1, 4, 5, 10, 16). One possible explanation for this is that only a single colony or a few colonies are subtyped from clinical samples. Nevertheless, the majority of clinical cases are likely to be caused by single strains. We report here on the usefulness of molecular subtyping techniques (2) for detecting Legionella strains harbored in a hospital water supply system and involved in a case of infection with multiple strains of L. pneumophila.
Case report.
A 44-year-old female patient with a history of breast carcinoma presented with tinnitus, dizziness, and nausea. Magnetic resonance tomography revealed a metastasis of the breast carcinoma in the left cerebellopontine angle, which was removed through a left suboccipital craniectomy. On the first postoperative day (POD), the patient could be extubated and showed unchanged neurological findings, except that a paralysis affecting swallowing had developed. Twelve hours later, the patient developed an obstructive hydrocephalus caused by local swelling at the operative site. Despite ventricular drainage and antiedematous therapy, the patient lost consciousness on the second POD and had to be reintubated and ventilated. On the seventh POD, the body temperature increased to 39.5°C and a chest X ray disclosed bilateral pulmonary infiltrates. Blood cultures, tracheal secretions, and urine and cerobrospinal fluid specimens were sterile by conventional bacteriological techniques. Treatment with ciprofloxacin and clindamycin was started but did not improve the patient’s condition. On the 13th POD, serology for L. pneumophila became positive. Despite changing the therapy to rifampin, erythromycin, and ciprofloxacin in standard dosage, the patient died from multiorgan failure on the 15th POD, before the culture for Legionella became positive.
Microbiological methods for Legionella.
Cultures of tracheal secretions were performed with selective buffered charcoal-yeast extract agar (BMPA-BCYE; Oxoid, Wesel, Germany) made in-house. Subsequent to the growth of Legionella from the clinical specimen, water samples were collected from taps and outlets in the patient’s room and from other locations in the ward. Showers are not in use in the intensive care unit. Additional water samples were cultured 3 months later, because the number of colonies kept was not sufficient for further typing. The hospital water system is approximately 40 years old and supplies four separate buildings. One-liter samples were collected after the water reached constant temperatures, which ranged from 35 to 45°C. The water samples were plated and unconcentrated, and after filtration of MWY-BCYE agar plates (Oxoid) revealed Legionella counts of between 10 and 20 CFU/ml. No attempt was made to culture specimens from the cold water system, and attempts to recover legionella-infected fluids from respiratory equipment were unsuccessful.
Serological typing of Legionella strains was performed with a genus-specific monoclonal antibody (MAb), 22-1 (9); a commercially available fluorescein isothiocyanate (FITC)-labeled, L. pneumophila-specific MAb (Fresenius, Oberursel, Germany); and rabbit antisera for all 15 serogroups of L. pneumophila (14). MAbs to react with serogroup 12 strains were prepared in our laboratory and used as described previously (8, 13). Serogroup 1 strains were subtyped with MAbs according to the method described by Joly et al. (11). Epidemiologically unrelated L. pneumophila strains were taken from our strain collection. For macrorestriction analysis (MRA), chromosomal DNAs were digested overnight with SfiI, AscI, and NotI (New England Biolabs, Schwahlbach, Germany) and separated with the CHEF III System (BioRad Laboratories, Munich, Germany) (14). Computer-assisted analysis of the restriction patterns was performed with the software package GelCompare (Applied Maths, Kortrijk, Belgium).
We initially investigated one colony from the clinical specimen and two colonies from the water samples. They belonged to serogroup 12 and were indistinguishable by using MAbs (Table 1). When macrorestriction patterns were compared, the clinical isolate was unrelated to the two strains from water (Fig. 1). There were three possible reasons for this discrepancy. First, the patient could have acquired the infection in the rehabilitation center in which she stayed prior to the operation. However, since the incubation period for legionellosis ranges from 2 to 10 (16) days and the patient was in the hospital for 9 days before developing the infection, a nosocomial origin of the pneumonia was strongly suspected. Second, we could have overlooked the causative strain in the environmental samples if more than one strain was present. Third, the patient may have suffered from simultaneous infection with several different strains.
TABLE 1.
L. pneumophila serogroup 12 strains typed by using MAbs and MRA
| Type of isolate and strain | Origin of strain | Reactivity against the following MAba:
|
PFGE profileb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 33-1 | 33-2 | 33-3 | 33-4 | 18-2 | 40-1 | SfiI | AscI | NotI | ||
| Clinical | ||||||||||
| Köln P1 | This study | + | + | o | + | ++ | ++ | A | A | A |
| Köln P5 | This study | + | + | o | + | ++ | ++ | A | A | A |
| Köln P2 | This study | + | + | o | + | ++ | ++ | B | B | B |
| Köln P4 | This study | + | + | o | + | ++ | ++ | B | B | B |
| Environmental | ||||||||||
| Köln U1 | This study | + | + | o | + | ++ | ++ | B | B | B |
| Köln U2 | This study | + | + | o | + | ++ | ++ | B | B | B |
| Köln U3 | This study | + | + | o | + | ++ | ++ | A | A | A |
| Köln U5 | This study | + | + | o | + | ++ | ++ | A | A | A |
| Unrelated | ||||||||||
| ATCC 43130 | Patient, United States | ++ | ++ | ++ | ++ | o | o | C | C | C |
| Heidelberg U9b | Water, hospital H | ++ | ++ | ++ | + | ++ | ++ | D | D | D |
| Heidelberg U1a | Water, hospital H | ++ | ++ | ++ | + | ++ | ++ | D | D | D |
| Wien 46-2 | Water, city W | ++ | ++ | ++ | ++ | ++ | ++ | E | E | E |
| Dresden 2152 | Water, city D1 | ++ | ++ | ++ | ++ | ++ | o | F | F | F |
| Dresden 2149 | Water, city D1 | ++ | ++ | ++ | ++ | ++ | o | F | F | F |
| Schwerin 2 | Patient, city S | ++ | ++ | ++ | ++ | o | o | G | G | G |
| DK 4 | Water, city K | + | o | o | + | ++ | ++ | H | H1 | H |
| W 779-2 | Water, city D2 | ++ | ++ | ++ | ++ | + | o | I | H | I |
| W 356-7 | Water, city D3 | + | + | ++ | + | + | ++ | K | A | K |
| Schottl 20 | Water, city N2 | ++ | ++ | ++ | ++ | o | o | L | I | L |
| Schottl 21 | Water, city N2 | ++ | ++ | ++ | ++ | o | o | L | I | L |
| Schottl 22 | Water, city N1 | ++ | ++ | ++ | ++ | o | o | M | I | L |
| WS 27-3 | Water, hospital D | ++ | ++ | ++ | ++ | + | o | N | A | M |
| W 330-2 | Water, hospital D | ++ | ++ | ++ | ++ | + | o | N | A | M |
| W 29-3 | Water, hospital D | ++ | ++ | ++ | ++ | + | o | N | A | M |
| W 401-6 | Water, hospital D | ++ | + | + | + | o | o | O | H | E |
| W 406 | Water, hospital D | ++ | + | + | + | o | o | O | H | E |
| W 30-1 | Water, hospital D | ++ | + | + | + | o | o | O | H | E |
++, optical density of >0.6 in the enzyme-linked immunosorbent assay; +, optical density ranging from 0.2 to 0.6; o, optical density of <0.2.
Different letters indicate different patterns (difference in one band was designated as a new type). PFGE, pulsed-field gel electrophoresis.
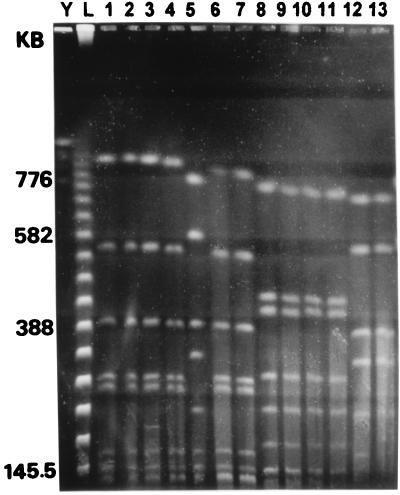
FIG. 1.
MRA of SfiI-cleaved DNAs of L. pneumophila strains isolated from the patient and from the hospital water supply. DNA sizes are indicated on the left. Lanes: Y, yeast chromosomal DNA standard; L, lambda concatemers; 1, Köln P2 (sg12); 2, Köln U1 (sg12); 3, Köln U2 (sg12); 4, Köln P4 (sg12); 5, Köln P1 (sg12); 6, Köln U6 (sg12); 7, Köln U8 (sg12); 8, Köln P3 (sg1); 9, Köln P6 (sg1); 10, Köln U4 (sg1); 11, Köln U7 (sg1); 12, Köln U3 (sg12); and 13, Köln U5 (sg12).
Therefore, we analyzed additional colonies. Of the eight colonies grown from the clinical sample, four belonged to serogroup 1, monoclonal subtype Bellingham, and four belonged to serogroup 12. Of the 47 environmental colonies serotyped, 5 were non-L. pneumophila species, since they reacted with a genus-specific MAb but not with the FITC-labeled, L. pneumophila-specific MAb. Of 42 environmental L. pneumophila strains, 19 belonged to serogroup 1, subtype Camperdown; 3 belonged to serogroup 1, subtype Bellingham; 6 belonged to serogroup 6; and 14 belonged to serogroup 12. All serogroup 12 strains were indistinguishable by MAb typing.
Altogether, 8 colonies from the clinical specimen and 12 colonies from the water samples were analyzed by MRA. Selected SfiI restriction patterns of these strains are given in Fig. 1. MRA with NotI and AscI confirmed the identities of the serogroup 12 strains (Table 1) and of the serogroup 1 strain (monoclonal subtype Bellingham) isolated here from the clinical specimen and from the hospital water system. The serogroup 12 strains belonged to two different genotypes, each of which showed quite different restriction patterns. According to the definition of Tenover et al. (18), these strains were unrelated. By computer-assisted analysis, the similarity coefficient of Jaccard revealed a large genetic distance between the SfiI patterns for these two strains (Fig. 2). Thus, we are sure that one strain was not derived from the other.
FIG. 2.
Dendrogram generated from the Jaccard similarity coefficient computed for 14 serogroup 12 and 33 serogroup 1 (monoclonal subtype Bellingham) strains after pulsed-field gel electrophoresis of SfiI-restricted chromosomal DNAs. Strains from our patient are in boldface and are underlined. For strains that were isolated from the same source and were indistinguishable, one representative strain is shown.
To ensure that the causative strains in our study could be differentiated from epidemiologically unrelated strains, we compared the macrorestriction patterns of 19 serogroup 12 isolates and 33 strains typed as serogroup 1, monoclonal subtype Bellingham. Unrelated strains displayed considerable DNA polymorphism, as summarized in Table 1 and Fig. 2. All three enzymes readily grouped identical strains isolated from the same source.
In summary, we are certain that the three strains were transferred from the hospital water supply to the patient. The identities of the clinical and environmental isolates and the DNA polymorphisms of unrelated Legionella strains exclude the rehabilitation center, which is located at a distance of several kilometers from the hospital, as a source of infection.
In our case, the causative strains were present in the central and peripheral hot water supply system. They may have infected the patient through aerosol released from taps or following aspiration (4, 12). However, we are not certain how the legionellae were transmitted, since respiratory equipment was always filled with sterile water. The patient never used an incentive respirometer. Respiratory circuits were changed daily and treated in an automatic sterilizer. Therefore, in addition to the malignancies, paralysis of deglutition after surgery was probably the major risk factor applicable to our patient. Another possible mode of transmission was the aspiration of mouth rinsing fluid containing ethereal oils, albeit this fluid should have been diluted with sterile water rather than tap water. No further cases have been observed despite intensive surveillance by culture and serological tests.
Few cases of simultaneous infections with different Legionella species or L. pneumophila serogroups have been reported (1, 4, 5, 10, 16). The clinical course was not different from that for infections caused by a single strain, and our study probably has no relevance for antibiotic treatment or for pathogenicity, since all Legionella strains are sensitive to erythromycin, rifampin, and ciprofloxacin (6). It is safe to assume that any species or serogroup may produce infection if a sufficiently compromised patient is exposed to a large inoculum, although strains may vary in their degrees of virulence (3).
There are few recommendations from the literature as to the number of colonies which should be selected for subtyping. Harrison et al. (8) recommended that the number of colonies tested should equal the square root of the number of colonies grown. Horbach et al. (10) typed 10 to 20 colonies and found three of seven infections to be caused by multiple strains. During the last 3 years, we have observed eight cases of Legionella pneumonia proven by culture. In all of these cases, we serotyped 8 to 12 colonies from the clinical samples. Only the infection described above involved multiple strains.
The fact that we identified two genetically different serogroup 12 strains, from the patient and from the environment, suggests that in some cases serotyping, even with MAbs, is not sufficient to discriminate between individual strains. Thus, the clinical isolate did not match the environmental ones by genetic fingerprinting, and additional colonies had to be analyzed. The costs for MRA were calculated to be $22 per sample (17). Thus, even an intensive molecular-typing study is much cheaper than environmental sampling and/or eradication measures (7) carried out in the wrong location, which cost several thousand U.S. dollars. In addition, the true source of the infection would have remained unidentified, serving as a potential source for additional cases.
Acknowledgments
We are grateful to Eckhard Budde (Schwerin, Germany), Ron Fallon (Glasgow, United Kingdom), John Kurtz (Nottingham, United Kingdom), Matthias Maiwald (Heidelberg, Germany), Sören Uldum (Copenhagen, Denmark), and Günther Wewalka (Vienna, Austria) for providing unrelated L. pneumophila strains. We thank Jutta Möller, Sylvia Petsche, and Sigrid Gäbler for technical assistance and Volker Bellmann for preparing the photographs.
This study was supported by the Deutsche Forschungsgemeinschaft (Lu 485/1-2).
REFERENCES
- 1.Aubert G, Bornstein N, Rayet I, Pozzetto B, Lenorand P H. Nosocomial infection with Legionella pneumophila serogroup 1 and 8 in a neonate. Scand J Infect Dis. 1990;22:367–370. doi: 10.3109/00365549009027062. [DOI] [PubMed] [Google Scholar]
- 2.Barbaree J M. Selecting a subtyping technique for use in investigations of legionellosis epidemics. In: Barbaree J M, Breiman R F, Dufour A P, editors. Legionella—current status and emerging perspectives. Washington, D.C: American Society for Microbiology; 1993. pp. 169–172. [Google Scholar]
- 3.Bezanson G, Fernandez R, Haldane D, Burbridge S, Marrie T. Virulence of patients and water isolates of Legionella pneumophila in guinea pigs and mouse L929 cells varies with bacterial genotype. Can J Microbiol. 1994;40:426–431. doi: 10.1139/m94-070. [DOI] [PubMed] [Google Scholar]
- 4.Dournon E, Buré A, Desplaces N, Carette M F, Mayaud C. Legionnaires’ disease related to gastric lavage with tap water. Lancet. 1982;i:797–798. doi: 10.1016/s0140-6736(82)91837-2. [DOI] [PubMed] [Google Scholar]
- 5.Dowling J N, Kroboth F J, Karpf M, Lee R B, Pasculle A W. Pneumonia and multiple lung abscesses caused by dual infection with Legionella micdadei and Legionella pneumophila. Am Rev Respir Dis. 1983;127:121–125. doi: 10.1164/arrd.1983.127.1.121. [DOI] [PubMed] [Google Scholar]
- 6.Edelstein, P. H. 1995. Antimicrobial chemotherapy for Legionnaires’ disease: a review. Clin. Infect. Dis. 21(Suppl. 3):265–276. [DOI] [PubMed]
- 7.Freije R M. Legionella control in health care facilities: a guide for minimizing risk. Indianapolis, Ind: HC Information Resources, Inc.; 1996. [Google Scholar]
- 8.Harrison T G, Saunders N A, Haththotuwa A, Doshi N, Taylor A G. Phenotypic variation amongst genotypically homogeneous Legionella pneumophila serogroup 1 isolates: implications for the investigations of outbreaks of Legionnaires’ disease. Epidemiol Infect. 1990;104:171–180. doi: 10.1017/s0950268800059331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helbig J H, Ludwig B, Lück P C, Groh A, Witzleb W, Hacker J. Monoclonal antibodies to Legionella Mip proteins recognize genus- and species-specific epitopes. Clin Diagn Lab Immunol. 1995;2:160–165. doi: 10.1128/cdli.2.2.160-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horbach I, Naumann D, Fehrenbach F J. Simultaneous infections with different serogroups of Legionella pneumophila investigated by routine methods and Fourier transform infrared spectroscopy. J Clin Microbiol. 1988;26:1106–1110. doi: 10.1128/jcm.26.6.1106-1110.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joly J R, McKinney R M, Tobin J H, Bibb W F, Watkins I D, Ramsay D. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J Clin Microbiol. 1986;23:768–771. doi: 10.1128/jcm.23.4.768-771.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korvick J A, Yu V L. Legionnaires’ disease: an emerging surgical problem. Ann Thorac Surg. 1987;43:341–347. doi: 10.1016/s0003-4975(10)60633-9. [DOI] [PubMed] [Google Scholar]
- 13.Lück P C, Helbig J H, Günther U, Assmann M, Blau R, Koch H, Klepp M. Epidemiologic investigation by macrorestriction analysis and by using monoclonal antibodies of nosocomial pneumonia caused by Legionella pneumophila serogroup 10. J Clin Microbiol. 1994;32:2692–2697. doi: 10.1128/jcm.32.11.2692-2697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lück P C, Köhler J, Maiwald M, Helbig J H. DNA polymorphisms in strains of Legionella pneumophila serogroup 3 and 4 detected by macrorestriction analysis and their use in epidemiologic investigation in nosocomial legionellosis. Appl Environ Microbiol. 1995;61:2000–2003. doi: 10.1128/aem.61.5.2000-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer R D, Edelstein P H, Kirby B D, Louie M H, Mulligan M E, Morgenstern A A, Finegold S M. Legionnaires’ disease: unusual clinical and laboratory features. Ann Intern Med. 1980;93:240–243. doi: 10.7326/0003-4819-93-2-240. [DOI] [PubMed] [Google Scholar]
- 16.Muder R R, Yu V L, Vickers R M, Rihs J, Shonnard J. Simultaneous infection with Legionella pneumophila and Legionella micdadei. Am J Med. 1983;74:609–614. doi: 10.1016/0002-9343(83)91018-5. [DOI] [PubMed] [Google Scholar]
- 17.Swaminatan B, Matar G M. Molecular typing methods. In: Pershing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 26–50. [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn W C. Legionella. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R Y, editors. Manual of clinical microbiology. Washington, D.C: American Society for Microbiology; 1995. pp. 533–544. [Google Scholar]