Abstract
Over 4 billion years of evolution, multiple mutations, including nucleotide substitutions, gene and genome duplications and recombination, have established de novo genes that translate into proteins with novel properties essential for high-order cellular functions. However, molecular processes through which a protein evolutionarily acquires a novel function are mostly speculative. Recently, we have provided evidence for a potential evolutionary mechanism underlying how, in mammalian cells, phosphatidylinositol 5-phosphate 4-kinase β (PI5P4Kβ) evolved into a GTP sensor from ATP-utilizing kinase. Mechanistically, PI5P4Kβ has acquired the guanine efficient association (GEA) motif by mutating its nucleotide base recognition sequence, enabling the evolutionary transition from an ATP-dependent kinase to a distinct GTP/ATP dual kinase with its KM for GTP falling into physiological GTP concentrations—the genesis of GTP sensing activity. Importantly, the GTP sensing activity of PI5P4Kβ is critical for the manifestation of cellular metabolism and tumourigenic activity in the multicellular organism. The combination of structural, biochemical and biophysical analyses used in our study provides a novel framework for analysing how a protein can evolutionarily acquire a novel activity, which potentially introduces a critical function to the cell.
Keywords: energy metabolism, GTP, GTP sensing kinase, molecular evolution, PI5P4Kβ
Introduction
Kinases play critical roles in a wide variety of cellular processes, such as signal transduction, transcription and metabolism, and the human genome contains far over 500 kinases [1]. Even though there is diversity in their structure, substrate specificity and participating pathways, the vast majority of kinases, from bacteria to humans, use ATP as the physiological phosphate donor [2–5]. By contrast, a recent study challenges this dogma and suggests that, in higher animals, the specificity has been disrupted with important functional implications.
Applying a proteome-wide screening of GTP interacting proteins with a follow-up biochemical and the structure-based reverse genetics approach, we showed that the GTP sensing activity of phosphatidylinositol 5-phosphate 4-kinase β (PI5P4Kβ) is critical for metabolic adaptation and tumourigenesis [6,7]. Transformed cells that lack the GTP sensing activity of PI5P4Kβ cannot sustain their growth under xenograft conditions, demonstrating that the GTP sensing function endows a survival advantage. The critical GTP sensing activity role of PI5P4Kβ, especially in cancer cells, establishes this lipid kinase as a target of cancer therapeutics [7]. In line with the notion, increased GTP synthesis is reported in cancer cells [8] and the higher cellular GTP concentration can be a metabolic signature that the GTP sensor kinase can sense. Moreover, the finding that PI5P4Kβ can use GTP more efficiently than ATP reveals this to be the first GTP-utilizing kinase with a biologically relevant GTP-dependent activity.
Interestingly, a molecular evolutionary analysis of phosphoinositide sequences revealed that PI5P4Kβ was derived from an ATP-dependent kinase, PI4P5K [1,9], after the emergence of multicellular organisms [10]. This fact suggests that the GTP sensing activity of PI5P4Kβ can be related to a newly adopted cellular function, which modulates cellular processes of multicellular organisms, presumably through intracellular metabolic communications. In the recent study published in the journal, Structure, we uncovered the molecular process by which the PI5P4Kβ was evolved to demonstrate the GTP sensing activity [11]. An integrated structural and biochemical strategy we developed here to analyse the molecular evolution of PI5P4Kβ would provide a new framework to investigate the mechanism by which organisms develop a new protein function. Here, we will briefly summarize the strategy we adopted to analyse the molecular evolution of PI5P4Kβ and describe the importance of nondetrimental cryptic mutations, which pave the way for smooth evolutionary transition—leading to the acquisition of new protein activity. We will also discuss the principle of how the new protein activity renders a high-order function to cells.
The GTP sensing kinase, PI5P4Kβ: Its evolutional origin
PI5P4Kβ, also called the type II phosphatidylinositol 5-phosphate 4-kinase or the type II PIP kinase, is a kinase that phosphorylates PI(5)P to produce PI(4,5) P2 (4). While PI5P4Kβ regulates the amount of two lipid second messengers, PI(5)P and PI(4,5)P2, it has been suggested that a major role of PI5P4K is to regulate the levels of PI(5)P since the majority of PI(4,5)P2 is produced by another pathway by phosphatidylinositol 4-phosphate 5-kinase (PI4P5K) or the type I PIP kinase from PI(4)P [6,9]. The knock-out mice of the PI5P4Kβ display reduced body weight, resistance to obesity induced by high-fat diets and increased insulin sensitivity [12].
Since its identification in 1997 [13], PI5P4Kβ was undoubtedly considered to utilize ATP for its enzymatic reaction. However, our recent study uncovered that PI5P4Kβ is a unique GTP-utilizing kinase that preferably uses GTP over ATP [6,7]. Even in the presence of a physiological concentration of ATP (1–2 mM) [14], more than 50% of PI(4,5)P2 was produced by using GTP at 0.5 mM concentration [6]. PI5P4Kβ displayed a higher GTP-dependent kinase activity than ATP, regardless of PI(5)P levels and the types of divalent metal ions [6].
Molecular evolutionary analyses suggested that PI5P4K diverges from PI4P5K at the ancestral lineage of Choanoflagellates and Filasterea [10]. Interestingly, the ancestral PI4P5K is a canonical kinase that preferably utilizes ATP [6,9,15]. The accumulating evidence argues that PI5P4Kβ might have evolved from an ATP-type kinase and adopted a GTP-utilizing activity to function as a GTP sensor at a certain point during evolution. However, it remains elusive how PI5P4Kβ acquires the novel GTP-dependent kinase activity besides the original ATP-dependent activity. To address this central question, we first conducted a comprehensive structural analysis of structurally known kinases and G-proteins [11].
Comprehensive analysis of ATP and GTP recognition leads to identifying the GEA motif
The ATP recognition by kinases was comprehensively analysed by all protein, PI and IP kinases structures deposited in the protein data bank (PDB) [11]. We identified 968 interactions in these 660 kinase structures and found that the N(1) and NH2(6) positions of ATP adenine base typically form two hydrogen bonds between the mainchain amide and the carbonyl groups from the (i + 2)th and ith residues, respectively (Fig. 1A).
Fig. 1.
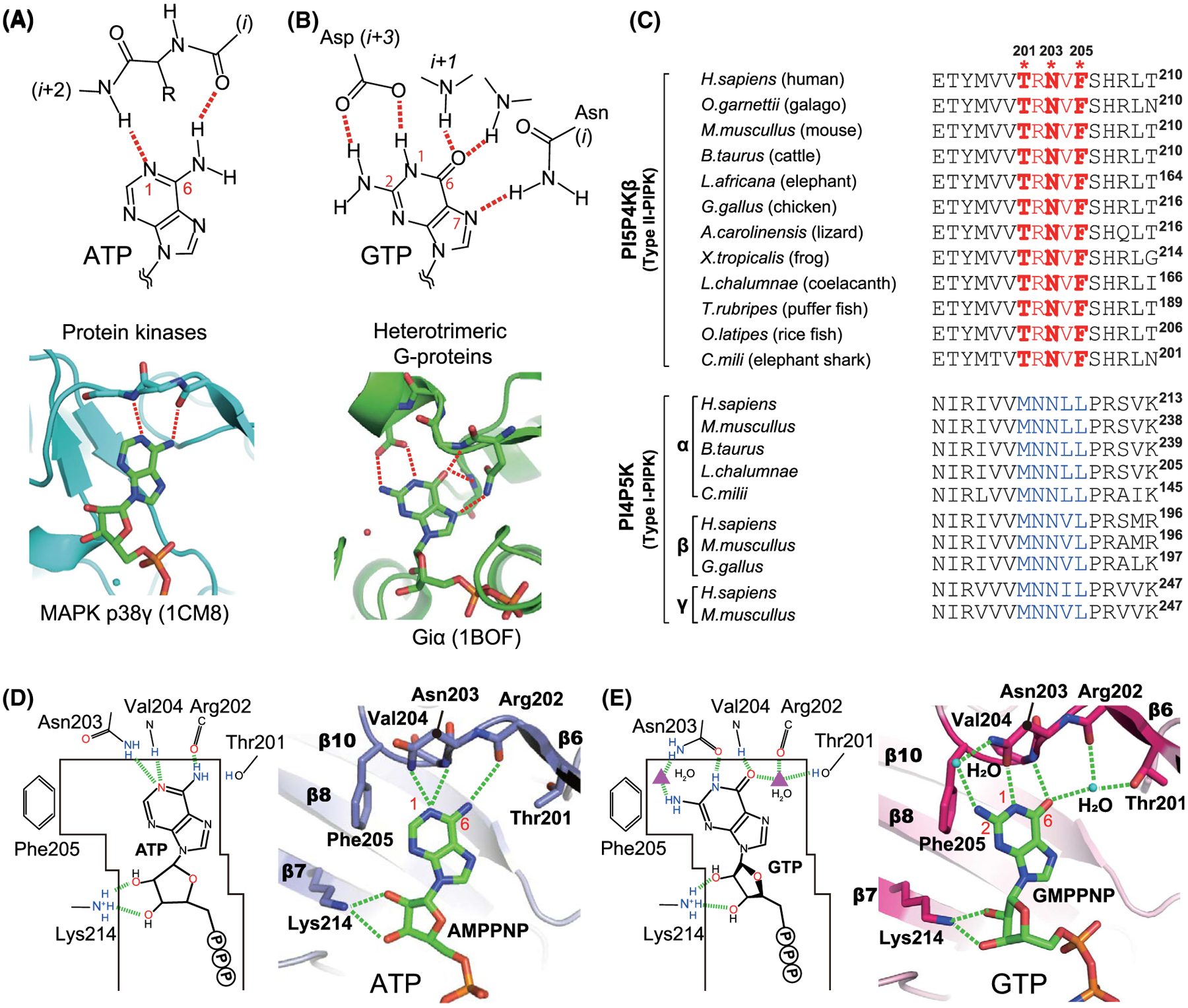
Comparison of nucleotide recognition modes of kinases, G-proteins and PI5P4Kβ. (A and B) Typical nucleotide recognition modes of (A) kinases and (B) G-proteins. (C) Sequence alignment of PI5P4Kβ and PI4P5K family proteins. The GTP-recognizing GEA motif is highlighted in red. For PI4P5K family proteins, the ATP-recognizing MNNψL sequences are coloured in blue. Asterisks indicate the positions Thr-201, Asn-203 and Phe-205 in human PI5P4Kβ. (D and E) Distinct ATP- and GTP-binding modes of human PI5P4Kβ. (A) ATP- and (B) GTP-binding modes of PI5P4Kβ determined in our previous study [1,11]. The hydrogen bonds between purine nucleotide triphosphates (PNTs) and PI5P4Kβ are shown in red dotted lines. PDB ID for the ATP- and GTP-bound structures are 6K4H and 6K4G, respectively. Figure is adapted from [11]. The structure graphics were generated by the PyMOL Molecular Graphics Software (version 2.3.0a0 Open-Source). The multiple sequence alignment was generated by the CLUSTAL.
The same strategy was applied to 128 G-protein structures in the PDB. In the G-protein structures, the N(1) and NH2(2) of the guanine base are simultaneously recognized by the sidechain carboxylate of Asp in the NKXD motif [16,17]. In most cases, the N (7) position forms a hydrogen bond with the Nδ of Asn in the NKXD motif. In addition, the O(6) creates a hydrogen bond(s) with a neighbouring i + 1th Lys and/or a remote mainchain amide group(s) (Fig. 1B).
Comparison of these ATP/GTP-binding modes with PI5P4Kβ revealed the dual specific nucleotide base recognition 201TRNVF205 sequence (residue number corresponds to that of human; Fig. 1C), which we designate as the GEA (Guanine Efficient Association) motif. The GEA motif binds to ATP similar to other kinases utilizing the mainchain amide group of Val-204 and carbonyl group of Arg-202 to recognize N(1) and NH2(6) positions of the adenine base, respectively (Fig. 1D). As for GTP-recognition mode, PI5P4Kβ uses amino acid sidechains to form hydrogen bonds to N(1), NH2(2) and O(6) of the guanine ring (Asn-203 for N(1) and NH2(2) and Thr-201 for O(6)). However, the interacting residues are distinct from those of the G-protein. Asn-203 in the 201TRNVF205 sequence is structurally located at the corresponding position of the conserved Asp residue of the G-protein, and its Oδ and Nδ atoms form direct and indirect hydrogen bonds with N(1) and NH2(2), respectively (Fig. 1E). In addition, Thr-201, Arg-202 and Val-204 in the TRNVF sequence form a hydrogen-bond network including O(6) of the guanine base with a water molecule (Fig. 1E). Such a GTP interaction is enabled by a 1.5 Å shift of the base moiety relative to the ATP. Although the presence of a water molecule in the hydrogen-bond network is unique in GTP recognition, a fragment molecular orbital (FMO) analysis supports its significant contribution to guanine base recognition. Taken together, PI5P4Kβ has a unique dual nucleotide recognition mode utilizing a short GEA motif, which could account for the ideal affinity of PI5P4Kβ to GTP to express GTP sensing function.
Promiscuous ITP and XTP recognition by the GEA motif
To further investigate the nucleotide base interactions via the GEA motif, we exploited the extensive structure and activity of human PI5P4Kβ in complex with 10 PNTs possessing different functional groups or protonation states at the 1st, 2nd, and 6th positions (Fig. 2A). For this purpose, the hydrolysis activity assay was assayed by NMR. NMR-based hydrolysis assay has been shown to faithfully recapitulate the characteristic GTP preference of the kinase [1]. Unlike radio-labelled PNT methods [6], this NMR-based strategy allows the activity of each nucleotide to be analysed in parallel in the same conditions without introducing any chemical modifications and additional reagents to detect the protein activity [11].
Fig. 2.
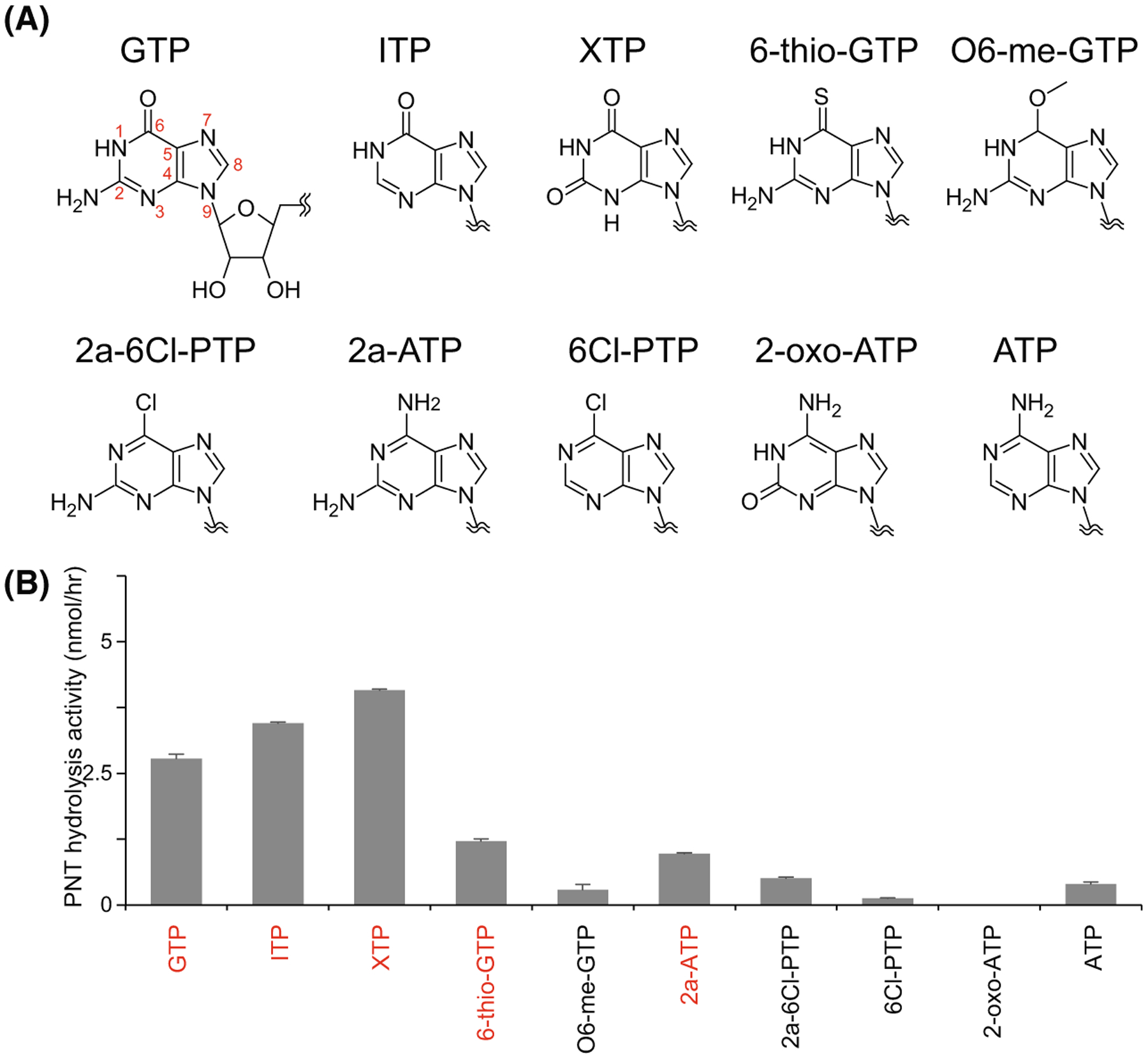
Promiscuous PNT hydrolysis activity of PI5P4Kβ. (A) Chemical structures of ATP and GTP, along with other PNTs used in this study. (B) PNT hydrolysis activity of PI5P4Kβ. 250 μM PNTs were mixed with 2 μM PI5P4Kβ. The average values from three experiments are shown with error bars (standard deviations; S.D.). Highly hydrolyzed nucleotides (>0.1 of the ratios) are indicated in red. Figure is adapted from [11].
The NMR activity assay revealed that PI5P4Kβ promiscuously hydrolyzed inosine triphosphate (ITP), xanthosine triphosphate (XTP), 6-thio-GTP and 2-amino-ATP (2a-ATP), in addition to GTP and ATP (Fig. 2B). The results indicate that NH2(2) is relatively dispensable, while NH(1) and O(6) are required for the binding to PI5P4Kβ. The crystal structures of PI5P4Kβ-ITP, PI5P4Kβ-XTP and PI5P4Kβ−2a-ATP complexes support the structural findings deduced from the NMR analysis [11]. While PI5P4Kβ can utilize at least four nucleotides present in human cells, the negligible cellular concentrations of XTP and ITP relative to ATP and GTP might not affect the ATP/GTP-dependent kinase activity of the WT PI5P4Kβ in living cells.
Dissecting the evolutionary path toward the GTP sensor
While enzymes are traditionally considered specific to a particular substrate, some can promiscuously utilize other substrates to catalyse different reactions [18,19]. The option of alternative substrates and/or interactions between proteins is in theory a wedge point for functional evolution [18,19]. Since PI5P4Kβ can react with GTP, ITP, XTP and ATP, we focussed on the evolution of the GEA motif of PI5P4Kβ and its substrate specificity.
Among the three amino acid residues that critically contribute to the nucleotide-binding in the GEA motif of PI5P4Kβ, Thr-201 and Phe-205, are substituted to Met and Leu, respectively, in PI4P5K, while Asn-203 is conserved in both PI5P4Kβ and PI4P5K (Fig. 1C). The comparison of genomic sequences and codon usage of 12 diversified vertebrates revealed that, although several nucleotide substitutions occurred at the codons corresponding to the GEA motif in PI5P4Kβ, amino acid sequences encoded by the codon were retained among these species, suggesting the region was under purifying selection [11]. Thus, in the evolutionary analyses, we focussed on the effect of the acquisition of Thr-201 and Phe-205 in the GEA motif in human PI5P4Kβ compared with original Met and Leu, respectively, in PI4P5K, for dissecting the evolution path toward the GTP sensor.
A trade-off between nucleotide specificity and the GTP-dependent activity in PI5P4Kβ
The PI5P4KβT201M_F205L double mutant, which has the same amino acids as the ancestral ATP-dependent kinase (i.e. PI4P5K), essentially abolished the hydrolysis activity for GTP, ITP and XTP and showed vigorous ATP-hydrolysis activity (Fig. 3A). Thus, the PI5P4KβT201M_F205L mutant is reverted from the GTP-dependent kinase to its ancestral ATP-dependent kinase. Intriguingly, the acquisition of Thr at the 201st position (see PI5P4KβF205L) makes the kinase more active to GTP, while largely retaining the ATP activity. However, the gain in the GTP-hydrolysis activity is associated with a much stronger activity to XTP. The acquisition of Phe at the 205th position (see PI5P4KβT201M) also did not affect its hydrolysis activity to ATP while slightly enhancing GTP-hydrolysis activity. Again, the hydrolysis activity to XTP and ITP was increased. The strongest GTP-hydrolysis activity was achieved when both Thr-201 and Phe-205 were acquired; however, the resultant WT PI5P4Kβ showed substantial activities to XTP and ITP. Thus, the GTP-hydrolysis activity coincides with extended activity to XTP and ITP, suggesting a trade-off between nucleotide specificity and the GTP-dependent kinase activity in the WT PI5P4Kβ. These results reveal that evolutional acquisition of the GEA motif results in the acquisition of GTP responsivity. It should be noted that we confirmed all mutant proteins retain their original protein folds, by determining their crystal structures [11].
Fig. 3.
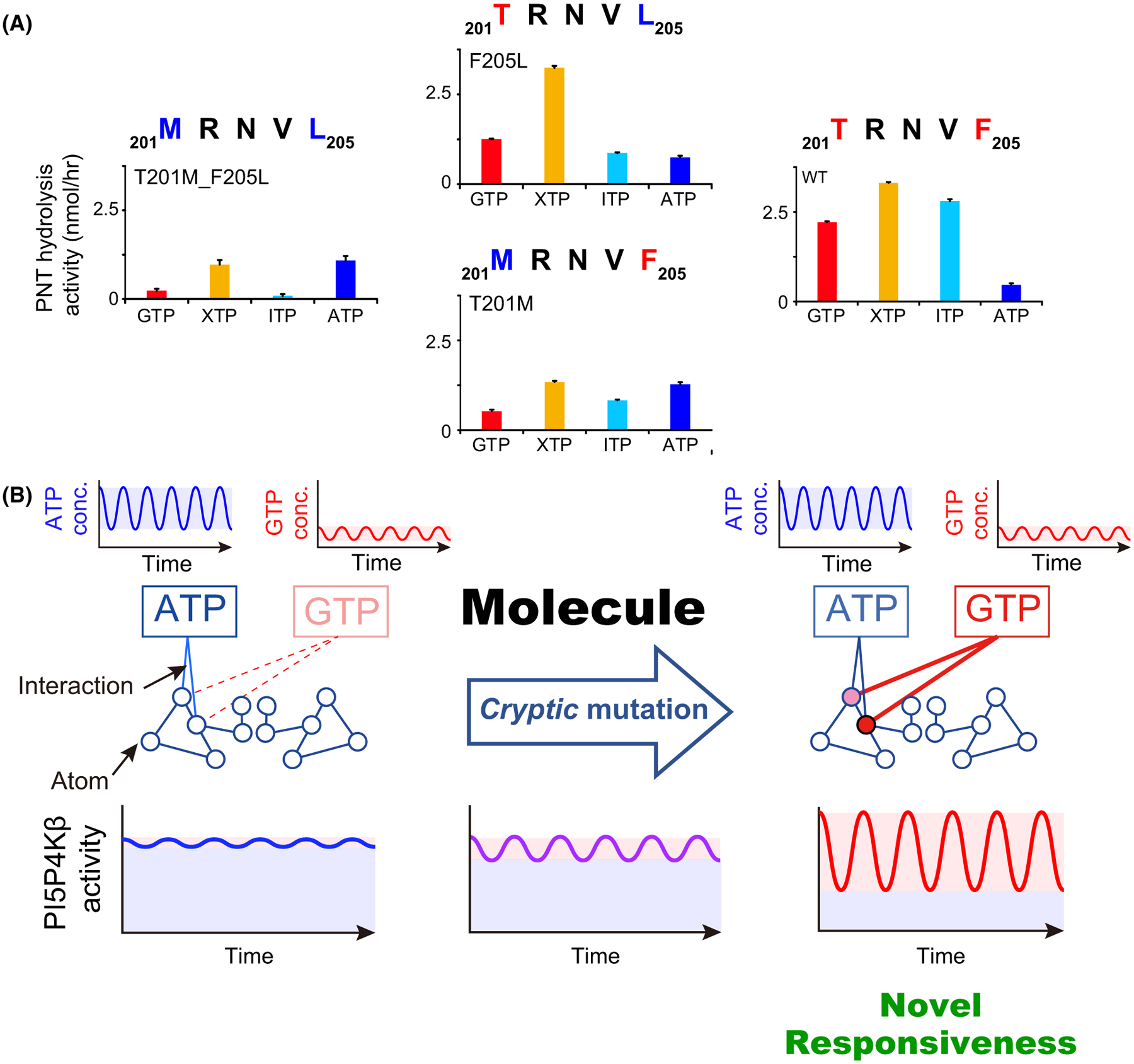
Molecular evolution of the GTP-dependent activity in PI5P4Kβ. (A) PNT hydrolysis activities of the WT PI5P4Kβ and T201M, N203D, N203A, F205L and T201M_F205L double mutants are shown along with the amino acid sequence of the GEA motif. The average values from the three experiments are shown with error bars (S.D.). Figure is modified from [11]. (B) Molecular evolution of the GTP-dependent activity in PI5P4Kβ. Two critical mutations induced the novel interaction with GTP, which endows the novel responsiveness to GTP while largely retaining basal ATP-dependent activity, in evolution pass via the cryptic mutations.
Role of cryptic mutations in the evolution of GTP sensing function
The evolutional analysis of the GEA motif unveiled that the gain in GTP-dependent kinase activity was achieved by loss of nucleotide specificity. This specificity change can be reconsidered in a cellular context where these nucleotides coexist in variable concentration ranges [14]. Substantial ATP-dependent activity is observed with PI5P4Kβ (Fig. 3A). Thus, considering the higher cellular concentrations of ATP than GTP [14], half of the PI5P4Kβ activity at most would originate from ATP at physiological nucleotide concentrations. By contrast, since the intracellular concentrations of ITP and XTP are negligibly low, the extended activity of XTP and ITP would not contribute to the biological function of PI5P4Kβ. In addition, since the concentration of ATP (1–5 mM) is much higher than the Km value of PI5P4Kβ for ATP (238 μM), the enzymatic activity originating from ATP is mostly insensitive to the physiological variations of ATP concentrations providing the ‘basal’ activity of PI5P4Kβ (Fig. 3B, blue shadow). However, the role of the ‘basal’ ATP-dependent activity of PI5P4Kβ is unclear as it can work as a damper that attenuates the small fluctuation in nucleotide concentrations, including those of ITP and XTP, and provides stability to the energy homeostasis system (Fig. 4, left).
Fig. 4.
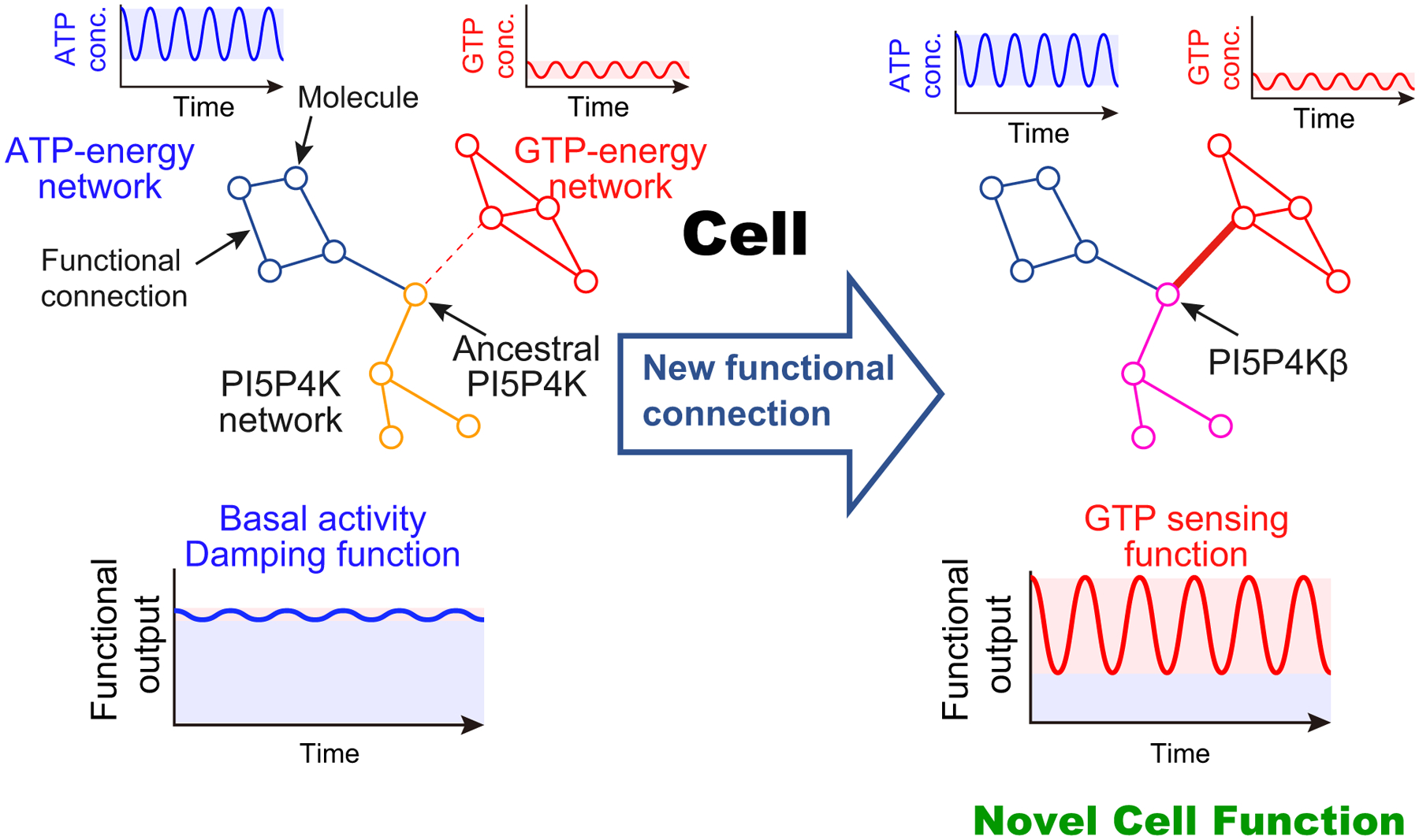
Evolutional acquisition of the GTP sensing function through rewiring of the functional cellular network. (Left) Ancestral PI5P4K only is connected to the ATP-energy network, and fluctuation in the ATP concentration was largely attenuated in the PI5P4K network to show the basal activity. (Right) Two critical mutations introduced the novel functional connection to the GTP-energy network, which allows the endowing of the novel GTP responsiveness to the PI5P4K network and a novel function to the cell.
On the other hand, the two mutations in PI5P4Kβ primarily change the responsiveness of the protein to the ambient cellular GTP concentration (Fig. 3B, red shadow). This leads to the establishment of an additional functional connection between GTP-energy homeostasis and the phosphoinositide signalling system in the cell (Fig. 4, right). Given the importance of the GTP concentration in controlling energy metabolism and proliferation under stress conditions [6], establishing a new mode of signalling that utilizes GTP likely introduces a high-order cellular function to the cell.
Toward this end, it would be interesting to examine in detail the fate of the original ATP-dependent activity in the context of evolutionary pressures. Neither PI5P4KβT201M nor PI5P4KβF205L loses its original ATP-dependent activity (Fig. 3A). Therefore, having the semi-optimal sequence (201TRNVL205 or 201MRNVF205), which might exist in the evolutional path toward the GTP-recognizing 201TRNVF205 sequence, is not detrimental to the protein as it would retain its ancestral basal ATP-dependent activity (Fig. 3B). Such functionally nondetrimental cryptic mutations might allow a smooth evolutional transition of an ATP-dependent kinase to a GTP sensing kinase and have less impact on the cellular functions. This notion would be supported by the fact that we have reported on establishing a cell line that only has PI5P4KβF205L [6]. Species with a single mutation in the GEA motif has not been identified to our knowledge. However, considering the probability of two mutations occurring simultaneously at different sites would be much rarer than having two mutations separately. We speculate that the period that the species with a single cryptic mutation in the GEA motif would exist as the bridge toward the fully functional GTP sensor.
How would protein evolution be integrated into a new cellular function?
Taken together, the comprehensive biochemical and structural analysis of PI5P4Kβ uncovers the clear evolutionary path from ATP-dependent kinase to the ATP/GTP dual kinase with GTP sensing activity. The results illuminate how a new biological function could evolve by accumulating cryptic mutations, which are already present in genome sequences but without an immediate impact until other conditions come into play (Figs. 3B and 4). These ‘silent’ mutations without positive or negative impact and in that sense initially ‘cryptic’ only reveal themselves when an additional function for the cell evolves (Fig. 4, right). Since the functional cellular networks are already compartmentalized in a cell, a single new connection, as in the case of PI5P4Kβ, added to the cell would be readily functional leading to a substantial impact on the whole cellular system. Thus, small changes in substrate specificity could rewire the functional cellular network and could serve to efficiently introduce an entirely new set of high order functionally to the cell (Fig. 4). In this sense, the cryptic mutations that accumulate in a way that is functionally near neutral would represent a comparatively safe evolution path with extraordinary functional potential. Although in our example of PI5P4Kβ the signalling triggered by the change of GTP concentration is not fully elucidated, the notion of the functional connection between GTP sensing and phosphoinositide signalling strongly suggests this is a legitimate functional mechanism.
Our view of molecular evolution of the GTP sensing kinase is consistent with the observation that a protein with a new function is not necessarily constructed from de novo but is co-opted to existing ones by exaptation. Such an evolution is often associated with gene duplications, which provide additional room for yielding novel functions besides the original one [18,19]. This appears to be operative in the case of the evolution of PI5P4Kβ. In the ancestorial lineage, there was only one isotype of PI5P4K, until three isotypes (α, β, and γ) of PI5P4K appeared in vertebrates by two successive gene duplications. It should be noted that the other isotypes of PI5P4K, PI5P4Kα and PI5P4Kγ, have unqualified features as a GTP sensor, although they share high overall sequence similarity and can use GTP as phosphodonor [6,20]. It would also be interesting to determine the nucleotide specificity of the PI5P4Ks before vertebrates (e.g. insects and molluscs).
It should be noted that PI5P4Kβ is not only a promiscuous enzyme but also a ‘moonlighting’ protein [21,22], as it has been shown that PI5P4Kβ may act as scaffolds to recruit PI5P4Kα by heterodimerization [23] and the evolution of the moonlighting activity need to be clarified along with its high-order role in cellular functions.
A multidiscipline approach to unveil novel protein functions
The co-option of a new activity often occurs when the very weak corresponding activity already coexists with the original activity, as exemplified by PI5P4Kβ here. Other cases are reported in line with this notion [18,19]; however, such a promiscuous activity is often difficult to predict as those functions will not be performed by well-known protein motifs. Therefore, additional mechanistic and structural studies would be needed to improve our understanding and predictability of such a function. Although most of the studies of the protein activity, function and structure–activity relationships are conducted in a way that focusses on a known activity with a few particular substrates and a couple of complex structures, an exhaustive search for the possible substrates with a certain function of interest would be needed to discover a protein with a new function. Thus, a thorough study of its promiscuity with the physiological concentrations in mind would also be essential to unveil the way that the protein function in certain cellular conditions. In addition, extensive protein structural-activity relationship studies with those substrates should be conducted along with the quantum mechanic’s simulation for unveiling the origin of the promiscuity. The accumulation of the data describing such interaction in a quantitative manner might allow us to predict the possible range of substrates for a protein of interest, through machine learning processes. Furthermore, a strategic mutational study to track the evolution path would be needed to understand how the protein has evolved to acquire the novel function.
In this sense, our strategy in discovering a GTP sensing kinase, PI5P4Kβ and the follow-up studies on its promiscuity and evolution represents an integrated multidiscipline approach to unveil the way that a protein could evolve to express not only new biochemical but also novel biological functions to adapt environments. The difficulty lies in establishing a method to account for the whole range of different substrates and reactions. In the case of PI5P4Kβ, NMR has shown its ability to observe the reaction with multiple substrates in parallel. Although only 10 analogues were tested in our study, the capability of NMR to separately detect >100 metabolites and their reactions in a mixture without additional chemical modifications would provide a robust way to define the range of promiscuity of an enzyme. The cross-disciplinary approach used in this study offers a novel framework for analysing how proteins acquire a new function in evolution that renders a high-order cellular function, which might provide a survival advantage.
Acknowledgments
We thank Dr. Eric P. Smith for their excellent proofreading. This work was supported in part by Project for Promotion of Cancer Research and Therapeutic Evolution (P-PROMOTE; 22ama221112h0001 to KT, TS, and ATS) and Project for Cancer Research and Therapeutic Evolution (P-CREATE; 20cm0106173 to KT, TS, and ATS) from Japan Agency for Medical Research and Development (AMED), Creation of fundamental technologies by interdisciplinary research to coexist with infectious diseases including COVID-19 from Japan Science and Technology Agency (JST), CREST programme (JP20356709 to KT, SN, ATS, and TS), Generating research infrastructure and novel technologies for anti-infective drug and vaccine discovery from AMED (21gm1610003h0201 to KT), the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Numbers 22ama121001 (TS) and JP22ama121030 (KF) (support numbers 4582 and 2809), KAKENHI (grant numbers 22K18374, 21H02436 and 20H03378 to KT and 20H03165 to ATS) from Japan Society for the Promotion of Science (JSPS), UC College of Medicine Research Innovation grant, a Marlene Harris Ride Cincinnati grant and NIH grants (R01NS089815, R01CA255331, and R01GM144426 to ATS) Swiss Light Source (proposal numbers 20191094 and 20191134) and the public accession beam time of Photon Factory, Institute of Materials Structure Science and High Energy Accelerator Research Organization (Accession numbers: 2017G147, 2019G063, and 2021G054).
Abbreviations
- 2a-ATP
2-amino-ATP
- FMO
fragment molecular orbital
- GEA
guanine efficient association
- ITP
inosine triphosphate
- PDB
protein data bank
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PI(5)P
phosphatidylinositol 5-phosphate
- PI4P5K
phosphatidylinositol 4-phosphate 5-kinase
- PI5P4Kβ
phosphatidylinositol 5-phosphate 4-kinase β
- PNTs
purine nucleotide triphosphates
- XTP
xanthosine triphosphate
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Data availability statement
The structural data that support the findings of this study are openly available in Protein Data Bank: WT-GMPPNP complex, 6K4G; WT-AMPPNP complex, 6K4H. Other data are available from the corresponding authors upon reasonable request (kohtakeuchi@mol.f.u-tokyo.ac.jp).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T & Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 2.Clarke JH & Irvine RF (2011) The activity, evolution and association of phosphatidylinositol 5-phosphate 4-kinases. Adv Enzyme Regul 52, 40–45. [DOI] [PubMed] [Google Scholar]
- 3.Loijens JC, Boronenkov IV, Parker GJ & Anderson RA (1996) The phosphatidylinositol 4-phosphate 5-kinase family. Adv Enzyme Regul 36, 115–140. [DOI] [PubMed] [Google Scholar]
- 4.Shears SB & Wang H (2019) Inositol phosphate kinases: expanding the biological significance of the universal core of the protein kinase fold. Adv Biol Regul 71, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vadas O, Burke JE, Zhang X, Berndt A & Williams RL (2011) Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal 4, re2. [DOI] [PubMed] [Google Scholar]
- 6.Sumita K, Lo YH, Takeuchi K, Senda M, Kofuji S, Ikeda Y, Terakawa J, Sasaki M, Yoshino H, Majd N et al. (2016) The lipid kinase PI5P4K β; is an intracellular GTP sensor for metabolism and tumorigenesis. Mol Cell 61, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi K, Senda M, Lo YH, Kofuji S, Ikeda Y, Sasaki AT & Senda T (2016) Structural reverse genetics study of the PI5P4Kβ-nucleotide complexes reveals the presence of the GTP bioenergetic system in mammalian cells. FEBS J 283, 3556–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kofuji S, Hirayama A, Eberhardt AO, Kawaguchi R, Sugiura Y, Sampetrean O, Ikeda Y, Warren M, Sakamoto N, Kitahara S et al. (2019) IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol 21, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins GH, Fisette PL & Anderson RA (1994) Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem 269, 11547–11554. [PubMed] [Google Scholar]
- 10.Khadka B & Gupta RS (2019) Novel molecular signatures in the PIP4K/PIP5K family of proteins specific for different isozymes and subfamilies provide important insights into the evolutionary divergence of this protein family. Genes (Basel) 10, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi K, Ikeda Y, Senda M, Harada A, Okuwaki K, Fukuzawa K, Nakagawa S, Yu HY, Nagase L, Imai M et al. (2022) The GTP responsiveness of PI5P4Kβ evolved from a compromised trade-off between activity and specificity. Structure 30, 886–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB & Cantley LC (2004) Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol 24, 5080–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rameh LE, Tolias KF, Duckworth BC & Cantley LC (1997) A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390, 192–196. [DOI] [PubMed] [Google Scholar]
- 14.Traut T (1994) Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 140, 1–22. [DOI] [PubMed] [Google Scholar]
- 15.Loijens JC & Anderson RA (1996) Type I Phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem 271, 32937–32943. [DOI] [PubMed] [Google Scholar]
- 16.Wittinghofer A & Vetter IR (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem 80, 943–971. [DOI] [PubMed] [Google Scholar]
- 17.Vetter IR & Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304. [DOI] [PubMed] [Google Scholar]
- 18.Copley SD (2003) Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol 7, 265–272. [DOI] [PubMed] [Google Scholar]
- 19.Khersonsky O & Tawfik DS (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79, 471–505. [DOI] [PubMed] [Google Scholar]
- 20.Bazenet CE, Ruano AR, Brockman JL & Anderson RA (1990) The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J Biol Chem 265, 18012–18022. [PubMed] [Google Scholar]
- 21.Huberts DH & van der Klei IJ (2010) Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta 1803, 520–525. [DOI] [PubMed] [Google Scholar]
- 22.Jeffery CJ (2017) Moonlighting proteins – Nature’s Swiss Army knives. Sci Prog 100, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bultsma Y, Keune WJ & Divecha N (2010) PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J 430, 223–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structural data that support the findings of this study are openly available in Protein Data Bank: WT-GMPPNP complex, 6K4G; WT-AMPPNP complex, 6K4H. Other data are available from the corresponding authors upon reasonable request (kohtakeuchi@mol.f.u-tokyo.ac.jp).


