Abstract
Adeno-associated virus (AAV)-mediated gene transfer has shown promise in rescuing mouse models of genetic hearing loss, but how viral capsid and promoter selection affects efficacy is poorly characterized. Here, we tested combinations of AAVs and promoters to deliver Tmprss3, mutations in which are associated with hearing loss in humans. Tmprss3tm1/tm1 mice display severe cochlear hair cell degeneration, loss of auditory brainstem responses, and delayed loss of spiral ganglion neurons. Under the ubiquitous CAG promoter and AAV-KP1 capsid, Tmprss3 overexpression caused striking cytotoxicity in vitro and in vivo and failed to rescue degeneration or dysfunction of the Tmprss3tm1/tm1 cochlea. Reducing the dosage or using AAV-DJ-CAG-Tmprss3 diminished cytotoxicity without rescue of the Tmprss3tm1/tm1 cochlea. Finally, the combination of AAV-KP1 capsid and the EF1α promoter prevented cytotoxicity and reduced hair cell degeneration, loss of spiral ganglion neurons, and improved hearing thresholds in Tmprss3tm1/tm1 mice. Together, our study illustrates toxicity of exogenous genes and factors governing rescue efficiency, and suggests that cochlear gene therapy likely requires precisely targeted transgene expression.
Keywords: Tmprss3, cochlea, hair cells, hearing loss, supporting cells, gene therapy
Graphical abstract

Cheng and colleagues show that selection of viral capsids and promoters can affect the toxicity of exogenous gene transfer of mouse Tmprss3 and its efficacy of rescue of hair cell loss and hearing loss caused by Tmprss3 deficiency. Tmprss3 deficiency causes hair cell loss and hearing loss in humans.
Introduction
Nearly 1 in 500 children in the United States is born with hearing loss, 65% of which are caused by genetic mutations.1 More than 70% of genetic hearing loss is attributed to autosomal recessive, nonsyndromic deafness (ARNSD) mutations,2 up to 10% of which are caused by transmembrane protease serine 3 (TMPRSS3) mutations. Missense mutations of TMPRSS3 cause variable onset and degrees of sensorineural hearing loss, leading to prelingual (DFNB10) or postlingual (DFNB8) ARNSD.2,3,4 As one of the most common causal genes of hearing loss among adult cochlear implant recipients,5 TMPRSS3 mutations currently lack a biological treatment that prevents or reverses the course of disease.
The mammalian cochlea is composed of distinct sensory and non-sensory cell types that are all essential for auditory function (Figure 1A). As mechanoreceptors, hair cells convert mechanical stimuli to electrical signals, are intercalated by supporting cells, and relay auditory input centrally via spiral ganglion neurons. Mice deficient in Tmprss3 (Y260X), where a nonsense mutation results in a truncated protease domain, exhibit normal cochlear development followed by rapid hair cell degeneration during the onset of hearing at postnatal day (P) 12.6 This leads to a complete loss of auditory brainstem responses (ABRs), implicating a requirement of Tmprss3 for hair cell survival and cochlear function.
Figure 1.
Tmprss3 deficiency causes cochlear hair cell degeneration and hearing loss
(A) In situ hybridization (RNAScope) of P5 wild-type (WT) cochlea (middle turn and counterstained with hematoxylin) revealed Tmprss3 mRNA expression in hair cells and supporting cells, interdental cells, inner phalangeal cells, lesser epithelial ridge, outer sulcus, and Rosenthal’s canal. Schematic depicting hair cell and supporting cell subtypes. (B) The TMPRSS3 protein consists of 453 amino acids, with a transmembrane (TM) domain, a low-density lipoprotein receptor class A (LDRA), a scavenger receptor cysteine-rich domain (SRCR), and a C-terminal serine protease. The mutation was generated by targeted mutation through homologous recombination in exon 1. (C) At P21, Tmprss3tm1/+ littermates had ABR thresholds that were indistinguishable from WT littermates, whereas Tmprss3tm1/tm1 mice demonstrated no ABR responses across all frequencies tested. (D–F) Immunostaining of P12 cochleae showed no loss or disorganization of hair cells and supporting cells among WT, Tmprss3tm1/+, and Tmprss3tm1/tm1 littermates prior to the onset of hearing. (G–L) Substantial inner and outer hair cell loss and disorganized supporting cells were observed in the P21 and P120 Tmprss3tm1/tm1 mice. No cell loss in WT or Tmprss3tm1/+ cochleae. (M–N) Quantification in the middle cochlear turn showing significant loss of hair cells in P21 and P120 Tmprss3tm1/tm1 cochleae and medial supporting cell loss at P120. (O–Q) Cross sections of Rosenthal’s canal at P21 showing no spiral ganglia neuron degeneration. (R–T) At P120, there was a noticeable loss of spiral ganglion neurons in the Tmprss3tm1/tm1 cochleae. (U–V) Quantitative analysis showing a significant loss of TuJ1+ spiral ganglion neurons, but not Sox2+ glial cells, in P120 Tmprss3tm1/tm1 cochleae. Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Two-way ANOVA with Tukey’s multiple comparison. n = 3–6. IHC, inner hair cell; OHC, outer hair cell; DC, Deiters’ cell; IPhC, inner phalangeal cell; IPC, inner pillar cell; OPC, outer pillar cell; SGN, spiral ganglion neuron.
Previous histological and single-cell RNA sequencing studies showed that Tmprss3 is broadly expressed in sensory hair cells (outer and inner), supporting cells (Deiters’, pillar, and inner phalangeal cells) and a subset of spiral ganglion neurons in the embryonic and postnatal cochleae.7,8,9,10,11,12 Tmprss3 encodes a serine protease and has been postulated to regulate ionic homeostasis of the cochlea.6,7,8,9,13 Both epithelial sodium channels (ENaCs) and calcium-activated potassium (BK) channels are candidate downstream targets of Tmprss3,9,10,11,12,13 yet loss of function of ENaC does not cause hearing loss in humans, while deletion of BK channels leads to hearing loss only after 8 weeks of age in mice, well after the onset of hearing.14,15 At present, the exact function of Tmprss3 and the pathogenesis of DFNB8/10 are unclear.
In the inner ear, adeno-associated virus (AAV) capsids have demonstrated low immunogenicity and variable degrees of transduction efficacy in multiple cell types.16 To rescue mouse models of hearing loss caused by hair cell mutations, independent studies have used AAV capsid types exhibiting high tropism for cochlear hair cells and employed ubiquitous promoters to overexpress genes of interest.17,18,19,20,21,22,23,24 However, whether this approach is effective for mutations affecting multiple cochlear cell types and whether selection of viral capsids and promoters affects the overall efficacy of rescue are not known.
Here, we used recombinant AAV (rAAV) vectors in an attempt to restore Tmprss3 expression and function in Tmprss3tm1 knockout mice (Tmprss3tm1/tm1). Our studies demonstrate that Tmprss3 is required for hair cell survival and auditory function. During rescue experiments, we discovered that the engineered KP1 capsid was capable of transducing cochlear cells with high efficiency.25 However, when administering the Tmprss3 transgene under control of the ubiquitous promoter (CAG)26 we observed cytotoxicity both in vitro and in vivo. Treatment with a selective viral capsid (AAV-DJ27) and decreasing vector dose reduced toxicity but failed to prevent hair cell loss or auditory dysfunction. In contrast, by using the EF1α core promoter,28 hair cell survival and auditory function were partially rescued in Tmprss3tm1 knockout mice. Together, we have established that the input capsid, promoter, and vector dose dictate cytotoxicity and efficacy in rescuing the Tmprss3-deficient cochlea.
Results
Tmprss3 is expressed in multiple cochlear cell types
To characterize Tmprss3 expression in the mouse cochlea, we performed RNAScope in situ hybridization. In the embryonic (E) day 18, postnatal 1- and 5-day-old (P1 and 5) wild-type cochlea, Tmprss3 mRNA was robustly expressed in the organ of Corti, including the inner hair cells, outer hair cells, and supporting cell subtypes (Figures 1A, S1A, and S1B). We did not observe a tonotopic gradient in expression. To a lesser extent, Tmprss3 transcripts were also detected in the greater epithelial ridge, lesser epithelial ridge, interdental cells, lateral cochlear wall, and select spiral ganglion neurons (Figure 1A). This is consistent with previous single-cell RNA sequencing and in situ hybridization data.11,12,25,29 We next assessed the effects of Tmprss3 deficiency by examining the Tmprss3tm1 mouse line (Figure 1B), which is a knockout model generated through targeted mutation by homologous recombination in exon 1.30 Using probes designed to detect the deleted sequences (BaseScope), we found that Tmprss3-exon1-2 transcripts were absent in the P1 Tmprss3tm1/tm1 cochlea, whereas Tmprss3 transcripts remained detectable in different cell types in the P1 wild-type cochlea (Figures S1E and S1F). These results indicate that Tmprss3 mRNA expression is effectively abolished in the Tmprss3tm1/tm1 cochlea.
Tmprss3 deficiency leads to hair cell degeneration and cochlear dysfunction
To determine whether Tmprss3 is required for cochlear maturation, we first examined cochleae in P5 Tmprss3tm1/tm1 mice and found no evidence of hair cell or supporting cell loss with cell counts comparable to those of Tmprss3tm1/+ and wild-type mice (Table S1). Similarly, each turn of the P12 and P13.5 Tmprss3tm1/tm1 cochleae showed comparable counts and organization of sensory hair cells and supporting cells to Tmprss3tm1/+ and wild-type cochleae (Figures 1D–1F, 1M, S1K, and S1L; Table S1), suggesting that Tmprss3 is not required for hair cell patterning or survival from P5 to 13.5.
Shortly after the onset of hearing around P14–14.5, Tmprss3tm1/tm1 cochleae showed rapid and extensive degeneration of both inner and outer hair cells (Figures S1M–S1P), corroborating previous results in Tmprss3 (Y260X) mice.6 By P21, all Myosin7a+ hair cells had significantly degenerated (p < 0.0001) in the Tmprss3tm1/tm1 cochleae, whereas wild-type and Tmprss3tm1/+ cochleae showed similar cell counts and organization (Figures 1G–1I and 1M; Table S1). As expected from the severe hair cell loss, P21 Tmprss3tm1/tm1 mice exhibited no detectable ABR at any frequency tested (Figure 1C), whereas both wild-type and Tmprss3tm1/+ mice displayed robust responses. These results demonstrate that Tmprss3 is required for hair cell survival and cochlear function after the second postnatal week.
Degeneration of supporting cells and spiral ganglion neurons in mature Tmprss3tm1/tm1 cochleae
In the juvenile and mature Tmprss3tm1/tm1 cochlea, sensory hair cell loss was the prominent feature. Sox2+ supporting cell subtypes, which expressed Tmprss3 mRNA between E18.5 and P5, appeared disorganized in the P21 Tmprss3tm1/tm1 cochlea, likely as a result of severe hair cell loss (Figures 1G–1I). However, we did not detect significant degeneration of supporting cells (Figure 1N; Table S1) or spiral ganglion neurons at this age (Figures 1O–1Q and 1U; Table S1). By P120, in addition to hair cell loss, there was a moderate and variable degree of supporting cell loss in the Tmprss3tm1/tm1 cochleae (Figures 1J–1L and 1N). Relative to P21 Tmprss3tm1/tm1 cochleae, there were significantly fewer supporting cells in the apical and middle turns of the P120 Tmprss3tm1/tm1 cochlea (Table S1).
In the Rosenthal canal, no degeneration of spiral ganglion neurons or glia was observed in P21 Tmprss3tm1/tm1 mice (Figures 1O–1Q, 1U, and 1V; Table S1). However, there were significantly fewer Tuj1+ spiral ganglion neurons, but not Sox2+ glia, in each turn of the P120 Tmprss3tm1/tm1 cochlea relative to age-matched Tmprss3tm1/+ and wild-type controls (Figures 1R–1V; Table S1). Together, these results indicate that Tmprss3 deficiency causes delayed loss of cochlear supporting cells and spiral ganglion neurons in the adult cochlea.
AAV-KP1 transduces multiple cochlear cell types with high efficacy
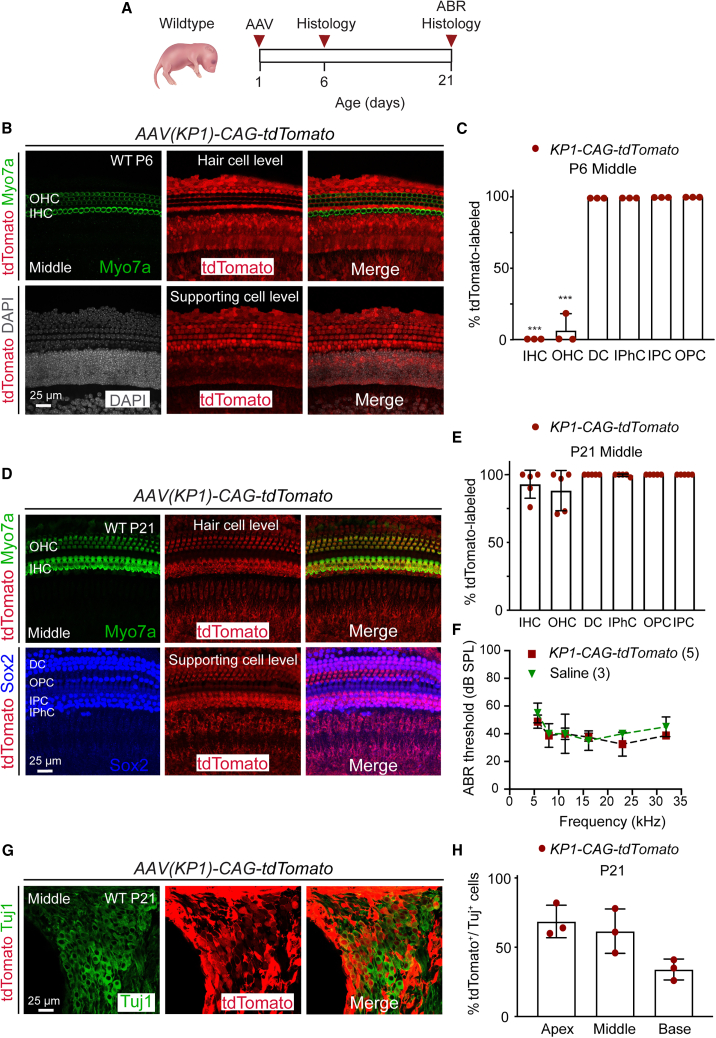
As Tmprss3 is expressed in multiple cochlear cell types, we postulated that a gene delivery approach that is ubiquitous and efficient would be needed to rescue the phenotype caused by its deficiency. The AAV-KP1 capsid was obtained from a screen of a shuffled AAV capsid library on primary human islet cells and shown to have transduction efficacy across multiple murine and human cell lines comparable with or higher than the AAV-DJ capsid.31,32 However, neither capsid had been systematically evaluated for its ability to transduce inner ear cell types. To characterize the tropism and efficacy of both chimeric capsids in the cochlea, we generated rAAV vectors carrying the tdTomato reporter expressed under a CAG promoter and packaged them using KP1 and DJ capsids (AAV-KP1-CAG-tdTomato and AAV-DJ-CAG-tdTomato). Viral capsids were injected via a posterior semicircular canal approach into P1 pups (1 μL injected over 3 min, 1.0 × 109 vector genomes [vg]) (Figure 2A).
Figure 2.
Tropism and efficacy of AAV-KP1-CAG-tdTomato in the cochlea
(A) Schematic showing AAV injection at P1 in WT pups and examination at P6 and P21. (B) Robust tdTomato expression in supporting cells (bottom) but not hair cells (top) in the P6 cochlea (middle turn shown). (C) Quantification of labeled hair cells and supporting cells. (D) Robust tdTomato expression in both hair cells and supporting cells (top and bottom) at P21. (E) Quantitative analysis of tdTomato-labeled hair cells and supporting cell subtypes. (F) Both saline- and AAV-KP1-CAG-tdTomato-injected animals showed normal ABR thresholds at P21. (G) Only a subset of Tuj1+ spiral ganglion neurons were tdTomato labeled at P21. (H) Spiral ganglion neurons were partially transduced in all three cochlear turns, with the apex showing the highest rate. Data shown as mean ± SD. ∗∗∗p < 0.001. Two-way ANOVA with Tukey’s multiple comparison. n = 3–5.
Five days after injection with AAV-KP1-CAG-tdTomato (P6), no or minimal tdTomato expression was detected in inner and outer hair cells (apex, 0.0% and 0.0%; mid, 0.0% and 6.6% ± 11.5%; base, 0.0% and 0.0%, respectively), whereas supporting cell subtypes showed robust expression (Figures 2B and 2C; Table S2). Twenty days post injection (P21), there was broad tdTomato expression in both inner and outer hair cells (>90.4% ± 8.3% and 84.0% ± 14.0%, respectively) across all three turns, as well as sustained labeling of supporting cells (>99.6% in all three turns) (Figures 2D and 2E; Table S2). Transduction efficacy of spiral ganglion neurons at P21 was found to be lower than those of both hair cells and supporting cells (68.7% ± 11.7% apical, 61.7% ± 16.0% middle, and 34.0% ± 7.5% basal turns; Figures 2G and 2H; Table S2). The P21 contralateral, control cochleae showed no tdTomato expression in inner and outer hair cells with only occasional transduction of supporting cells in the basal turn in three of four animals (Deiters’ cells, 26.6% ± 31.1%; outer pillar cells, 25.5% ± 24.3%; inner pillar cells, 15.8% ± 14.6%; and inner phalangeal cells, 36.2% ± 28.4%; Figures S2A–S2D; Table S2).
After injection with AAV-DJ-CAG-tdTomato capsid at P1, P21 wild-type mice demonstrated tdTomato-labeled hair cells and supporting cells, albeit at lower rates than with the KP1 capsid (Figures S2E and S2F). In the middle turn, the transduction rates were 65.6% ± 29.3% in inner hair cells, 30.0% ± 19.5% outer hair cells, 54.7% ± 5.0% Deiters’ cells, 48.7% ± 15.9% inner pillar cells, 52.3% ± 30.2% outer pillar cells, and 76.4% ± 25.0% inner phalangeal cells (Figure S2F; Table S2). Similar to saline-injected animals, those injected with rAAV packaged with either KP1 or DJ capsids exhibited no detectable ABR threshold shifts at P21 (Figure 2F and S2G). Together, these data indicate that both AAV-KP1 and AAV-DJ capsids transduce cochlear hair cells and supporting cells with no adverse effects on cell survival or cochlear function, with the former more efficiently transducing cochlear cells in vivo. Furthermore, these data show the temporal differences in tdTomato expression where there is a delay in the onset of KP1-CAG-tdTomato expression in hair cells compared to that of DJ-CAG-tdTomato expression.
Overexpression of Tmprss3 is cytotoxic in vitro and in vivo
To begin examining the effects of exogenous Tmprss3, we generated a KP1 capsid-packaged rAAV vector expressing the mouse Tmprss3 construct under the control of a CAG promoter. During rAAV production, detachment and death of producer cells (HEK293T/17) were noted, requiring a shortening of the incubation period prior to rAAV harvest (30 h instead of 60–72 h). Cell proliferation decreased with increasing multiplicity of infection (MOI) of AAV-KP1-CAG-Tmprss3 but not for a control rAAV-factor IX expression vector or for non-transduced cells (Figures 3A–3D). Similarly, dose-dependent cytotoxicity was found with rAAV-KP1-CAG-Tmprss3 using HeLa cells (Figures S3A–S3D).
Figure 3.
Exogenous TMPRSS3 is cytotoxic in vitro and in vivo
(A–C) Transduction of 293T/17 (human embryonic kidney) cells with AAV-KP1-CAG-Tmprss3 resulted in cell death in a dose-dependent manner, whereas none was observed in no-virus controls. (D) Proliferation assay demonstrating that increasing the MOI with AAV-KP1-CAG-Tmprss3 significantly decreased 293T/17 cell counts. Controls using huF9-expressing rAAV and no virus showed higher viability. (E) Schematic showing the timeline of injection of viral vectors into WT or mutant (Tmprss3tm1/tm1) cochleae and subsequent examination at P6 and P21. (F) At high titers (1:1, 2.0 × 108 vg), AAV-KP1-CAG-Tmprss3 caused degeneration of OHCs and swelling of IHCs. (G) A lower titer (1:2, 1.0 × 108 vg) did not cause hair cell degeneration, although IHCs still appeared swollen (inset). (H) No degeneration was observed at a 1:10 dilution (2.0 × 107 vg). (I–K) Hair cell degeneration was not prevented in P21 Tmprss3tm1/tm1 cochleae with any titers of AAV-KP1-CAG-Tmprss3 tested. (L) Quantification at P21 showing significant hair cell loss in all three turns of the cochlea at 1:1, but not other dilutions, of AAV-KP1-CAG-Tmprss3. (M) Hair cells degenerated in each turn of P21 Tmprss3tm1/tm1 cochleae injected with any titers of AAV-KP1-CAG-Tmprss3, resulting in significantly fewer hair cells than saline-injected, WT controls. (N) ABR thresholds were significantly higher in the ears of P21 WT animals injected with the full 1:1 titer AAV-KP1-CAG-Tmprss3 relative to saline-injected WT controls. (O and P) Some elevation of ABR thresholds was observed at a 1:2 dilution, and no changes were observed at a 1:10 dilution. Tmprss3tm1/tm1 mice displayed no ABR responses across all three titers. (Q) No cell loss was detected in the P21 WT cochlea after AAV-DJ-CAG-Tmprss3 had been injected at P1 (2.0 × 108 vg), although IHC appeared swollen. (R) Sensory hair cell loss at P21 was not prevented by AAV-DJ-CAG-Tmprss3 in Tmprss3tm1/tm1 mice. (S) Hair cell counts in saline- and AAV-DJ-CAG-Tmprss3-injected WT cochleae and AAV-DJ-CAG-Tmprss3-injected Tmprss3tm1/tm1 cochleae. (T) WT ears injected with AAV-DJ-CAG-Tmprss3 showed elevated ABR thresholds at 8 and 16 kHz. Tmprss3tm1/tm1 ears treated with AAV-DJ-CAG-Tmprss3 had no ABR responses. Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Two-way ANOVA with Tukey’s multiple comparison. n = 3.
To determine whether AAV-related toxicity extends to cochlear tissues, we next administered AAV-KP1-CAG-Tmprss3 (2.0 × 108 vg) into the cochlea in P1 Tmprss3tm1/tm1 and wild-type mice (Figure 3E). In the wild-type cochlea, no degeneration or disorganization of hair cells was observed at P6 (Figures S3E and S3F), while striking disorganization and degeneration of hair cells was noted at P21 (Figure 3F). There were significantly fewer Myo7a+ hair cells in each cochlear turn (apex, 53.3 ± 2.1; middle, 51.0 ± 19.1; and base, 58.3 ± 21.1) than in saline-injected controls (apex, 102.7 ± 9.2; middle, 91.3 ± 7.0; and base, 95.0 ± 10.5) (p < 0.05; Figure 3L; Table S3). Sox2+ supporting cells also appeared disarrayed, but no degeneration was detected at P21 (Figure 3F; Table S3). As controls, no hair cell or supporting cell degeneration was detected in the contralateral ears at P21 (Figures S3G–S3I and S3M). Reducing the injected viral titers 1:2 and 1:10 (1.0 × 108 and 2.0 × 107 vg, respectively) prevented hair cell degeneration in the P21 wild-type cochleae, although inner hair cells appeared swollen after delivery of the former (1:2) titer rAAV (Figures 3G–3H and 3L). AAV-KP1-CAG-Tmprss3 at full or reduced titers failed to prevent degeneration of hair cells in P21 Tmprss3tm1/tm1 mice (Figures 3I–3K, 3M, and S3J–S3M).
Additionally, AAV-KP1-CAG-Tmprss3 injection resulted in elevated ABR thresholds that were significantly higher than those of saline-injected P21 wild-type animals (p < 0.01; Figure 3N) and non-injected ears (Figure S3N). Halving the titers lessened, and a 10-fold dilution prevented, ABR threshold shifts (Figures 3O–3P). However, AAV-KP1-CAG-Tmprss3 at full or reduced titers failed to rescue ABR thresholds in Tmprss3tm1/tm1 mice (Figures 3N–3P). Together, these results indicate that exogenous Tmprss3 causes cytotoxicity in vitro and in vivo and that decreasing transduction reduced toxicity but failed to prevent Tmprss3 deficiency-induced hair cell loss and auditory dysfunction.
Exogenous Tmprss3 toxicity is associated with multiple AAV capsid types
To verify that cytotoxicity can be reduced by decreasing transduction rates, we also used AAV-DJ to overexpress Tmprss3 in vivo. AAV-DJ-CAG-Tmprss3 (2.0 × 108 vg) was administered to P1 Tmprss3tm1/tm1 and wild-type mice. In the injected P21 wild-type cochlea, no hair cell loss was detected (Figures 3Q, 3S, S4, and S4B), while a small but significant ABR threshold shift across several frequencies was observed, suggesting some cytotoxicity similar to the lower titers of AAV-KP1-CAG-Tmprss3 (p < 0.05; Figure 3T). Moreover, AAV-DJ-CAG-Tmprss3 administration failed to prevent hair cell loss or ABR threshold shifts in Tmprss3tm1/tm1 mice (Figures 3R–3T, S4C, and S4D). Thus, Tmprss3-related cytotoxicity is likely dependent on transduction efficiency and can occur with both AAV-KP1 and AAV-DJ viral capsids. The presence of normal numbers of hair cells in DJ-CAG-Tmprss3-transduced wild-type cochleae as well as the presence of ABR thresholds, albeit significantly elevated, in these mice suggest that functional hair cells are present. Thus, viral transduction may be affecting supporting cells or cells outside of the organ of Corti. Additionally, using a capsid with lower transduction efficacy failed to prevent hair cell degeneration and auditory dysfunction caused by Tmprss3 deficiency.
AAV-KP1-EF1α-Tmprss3 gene vector is not cytotoxic in vitro or in vivo
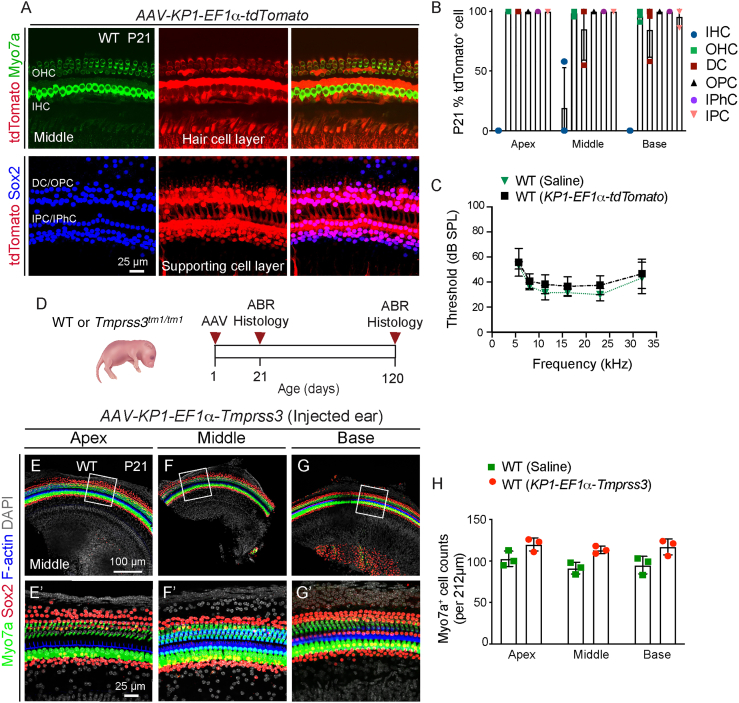
The promoter EF1α has been demonstrated to drive lower transgene expression relative to the CAG promoter in various tissue types.33,34,35 To determine whether this promoter can help abolish cytotoxicity, we generated an AAV-KP1 vector packaging Tmprss3 cDNA under the control of an EF1α core promoter. We next tested the transduction efficacy of AAV-KP1-EF1α-tdTomato in vivo. Five days post injection into wild-type cochleae, most inner and outer hair cells (apex, 91.5% ± 12.3% and 97.9% ± 3.4%; middle, 60.9% ± 36.8% and 75.4% ± 39.5%, base, 70.0% ± 19.6% and 79.3% ± 23.4%) and almost all supporting cell subtypes expressed tdTomato (Figures S4A and S4B). At 20 days post injection, both outer hair cells and supporting cells, but not inner hair cells, remained highly transduced (>95.8% in all three turns; Figures 4A and 4B; Table S3). Similar to AAV-KP1-CAG-tdTomato, no ABR threshold shifts were detected after AAV-KP1-EF1α-tdTomato administration (Figure 4C). In contralateral control cochleae, tdTomato expression was observed in some supporting cells of the basal turn (Deiters’ cells, 10.3% ± 17.9%; outer pillar cells, 10.7% ± 17.6%; inner pillar cells, 37.8% ± 41.6%; and inner phalangeal cells, 18.1% ± 16.8%; Table S2) but not in middle or apical turns.
Figure 4.
AAV-KP1-EF1α-Tmprss3 is not cytotoxic in the cochlea in vivo
(A) After injection of AAV-KP1-EF1α-tdTomato (1.0 × 109 vg/mL) at P1, there was robust tdTomato expression in OHCs (top) and supporting cell subtypes (lower) at P21 (middle turn shown). (B) Most OHCs and supporting cell subtypes and almost no IHCs were tdTomato labeled. (C) Relative to saline-injected ears, there were no detectable shifts in ABR thresholds in the AAV-KP1-EF1α-tdTomato-injected ears at P21. (D) Schematic showing the timeline of injection of AAV-KP1-EF1α-Tmprss3 into WT or mutant (Tmprss3tm1/tm1) cochleae and subsequent examination at P21 and P120. (E–G) AAV-KP1-EF1α-Tmprss3 (6.5 × 108 vg) injected into P1 WT mice did not cause any loss of sensory cells or supporting cells across all three turns at P21. (E′–G′) High-magnification images from (E–G). (H) Quantification of hair cells in saline- and AAV-KP1-EF1α-Tmprss3-injected WT cochleae. Data shown as mean ± SD. Two-way ANOVA with Tukey’s multiple comparison. n = 3.
Unlike AAV-KP1-CAG-Tmprss3, the AAV-KP1-EF1α-Tmprss3 construct did not diminish proliferation of 293T/17 cells at various MOIs, with rates similar to those of no-virus controls and those transduced with a control rAAV-factor IX prep (Figures S5C–S5F). Collectively, these results suggest that the use of the EF1α promoter abolished the cytotoxicity of exogenous Tmprss3 in vitro.
We next administered the AAV-KP1-EF1α-Tmprss3 (6.5 × 108 vg) vector into P1 wild-type mice (Figure 4D). At P21, we did not detect any degeneration of hair cells or supporting cells in any cochlear turns (Figures 4E–4H; Table S3). Collectively, these results suggest that the use of the EF1α promoter abolished the cytotoxicity of exogenous Tmprss3 in vitro and in vivo.
AAV-KP1-EF1α-Tmprss3 partially prevents cochlear degeneration and auditory dysfunction in Tmprss3tm1/tm1 mice
To assess its effects on hair cell degeneration and auditory dysfunction caused by Tmprss3 deficiency, we administered AAV-KP1-EF1α-Tmprss3 to P1 Tmprss3tm1/tm1 mice (Figure 4D). At P7, BaseScope in situ hybridization detected robust expression of Tmprss3 transgene in all supporting cell subtypes and, to a lesser degree, inner and outer hair cells in the organ of Corti of AAV-KP1-EF1α-Tmprss3-injected Tmprss3tm1/tm1 cochlea (Figures S6A–S6D″). We observed preservation of both inner and outer hair cells in all three turns of the P21 Tmprss3tm1/tm1 cochlea (Figures 5A–5C). Nearly complete hair cell survival in the middle and basal turns was observed, whereas, in the apical turn, survival was partial and variable, especially of the outer hair cells. Myo7a+ cell counts in all three turns of treated Tmprss3tm1/tm1 cochlea were significantly higher than those in contralateral, control cochlea (p < 0.001; Figure 5D; Table S3). Surprisingly, three of five Tmprss3tm1/tm1 animals showed survival of some hair cells in the contralateral, control cochlea, suggesting a low level of transport of virus between ears (Figures S7A–S7D).
Figure 5.
AAV-KP1-EF1α-Tmprss3 partially prevents degeneration and auditory dysfunction in Tmprss3tm1/tm1 mice
(A–C) After AAV-KP1-EF1α-Tmprss3 injection (6.5 × 108 vg) in P1 Tmprss3tm1/tm1 mice, most IHCs and OHCs were present in the middle and basal turns, while most IHCs and some OHCs remained in the apical turn at P21. (A′–C′) High-magnification images from (A)–(C). (D) Myo7a+ cell counts in each turn of treated Tmprss3tm1/tm1 cochlea were significantly higher than untreated Tmprss3tm1/tm1 cochlea and were similar to WT controls. (E–G) Hair cell survival persisted in each turn of P120-treated Tmprss3tm1/tm1 cochlea. (H) Each turn of P120-treated Tmprss3tm1/tm1 cochlea displayed significantly more hair cells than untreated Tmprss3tm1/tm1 cochlea and similar to WT controls. (I–K) Many SGNs were preserved in each turn of P120-treated Tmprss3tm1/tm1 cochlea, particularly the middle and basal turns. (L) Treated Tmprss3tm1/tm1 cochlea had higher SGN counts than untreated Tmprss3tm1/tm1 cochlea. (M) Raw ABR waveforms of P21 WT, untreated, and treated Tmprss3tm1/tm1 mice at 16kHz. (N and O). All treated Tmprss3tm1/tm1 ears demonstrated detectable ABR responses at P21 and P120, whereas untreated ears showed no responses. Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Two-way ANOVA with Tukey’s multiple comparison. n = 3–5.
In AAV-KP1-CAG-Tmprss3-injected Tmprss3tm1/tm1 cochlea, we detected robust expression of Tmprss3 transgene in both supporting cells and hair cells (Figures S6E–S6H″). Relative to AAV-KP1-EF1α-Tmprss3-injected Tmprss3tm1/tm1 cochlea, transgene appeared similarly expressed in supporting cells but higher among hair cells. How the level and pattern of transgene expression attributes to the lower cytotoxicity and ability of AAV-KP1-EF1α-Tmprss3 to prevent hair cell degeneration is currently unclear.
We further examined whether hair cell survival in P120-treated Tmprss3tm1/tm1 cochleae persisted and whether degeneration of supporting cells and spiral ganglion neurons was also prevented. Similar to P21, sensory hair cells were consistently present in the middle and basal turns of the treated Tmprss3tm1/tm1 cochlea, with survival being more variable in the apical turn. Treated Tmprss3tm1/tm1 displayed significantly more hair cells than the contralateral, control cochlea (p < 0.01; Figures 5E–5H; Table S3). As seen at P21, one out of the five P120, Tmprss3tm1/tm1 mice had a partial rescue of hair cells in the contralateral ear (Figures S7E–S7H). Both the counts and organization of Sox2+ supporting cells in the treated Tmprss3tm1/tm1 cochlea were similar to those in the littermate and wild-type controls, while some disorganization was observed in the apex (Figures 5E–5G; Table S3). Furthermore, spiral ganglion neuron survival in the apical and middle turns was significantly higher in the treated Tmprss3tm1/tm1 cochlea than in the contralateral cochlea (p < 0.01, Figures 5I–L; Table S3).
Finally, all five Tmprss3tm1/tm1 mice injected with AAV-KP1-EF1α-Tmprss3 showed detectable ABR responses across a range of frequencies at P21 and P120, when none of the non-injected Tmprss3tm1/tm1 mice or contralateral ears showed any responses at either age (Figures 5M–5O, S7M, S7N, and S7P; p < 0.0001). As expected, AAV-KP1-EF1α-Tmprss3 did not cause ABR threshold shifts in wild-type animals (Figure 5N). As a group, injected ears of Tmprss3tm1/tm1 mice showed significantly higher thresholds than wild-type animals at P21 and P120 (Figures 5N, 5O, S7M, and S7P). Individually, each injected ear of Tmprss3tm1/tm1 mice had ABR responses detected (Figures S7N and S7Q). However, only one of the three injected Tmprss3tm1/tm1 ears showed detectable distortion product otoacoustic emission (DPOAE) responses, whereas none of the non-injected, mutant ears showed responses at P21 (Figure S7O). This suggests that outer hair cell function was not well restored even though most injected mutant ears showed improved survival of hair cells. In summary, these data indicate that treatment with AAV-KP1-EF1α-Tmprss3 partially prevents hair cell degeneration and cochlear dysfunction.
Discussion
TMPRSS3 mutations cause DFNB8 and DFNB10 and are found in up to 11% of patients with sensorineural hearing loss.4,36 Previously, Guipponi and colleagues showed that Tmprss3 null mice exhibit severe degeneration of cochlear hair cells between P12 and 14.8 Here, we found that Tmprss3tm1 mutant mice also demonstrated extensive inner and outer hair cell loss around P14,32 confirming that Tmprss3 is required for hair cell survival after the onset of hearing. Strikingly, exogenous Tmprss3 delivered via an rAAV vector using the KP1 capsid and the CAG promoter was toxic to multiple cell lines, caused hair cell loss in wild-type cochlea in vivo, and failed to prevent hair cell degeneration in Tmprss3tm1/tm1 mice. Toxicity was diminished when using lower viral titers or by replacing the KP1 capsid with DJ, neither of which prevented hair cell degeneration in Tmprss3tm1/tm1 mice. Finally, by using the EF1α promoter, exogenous Tmprss3 was no longer cytotoxic in vitro and in vivo, prevented hair cell degeneration, and partially restored auditory function in Tmprss3tm1/tm1 mice. Recently, Du and colleagues reported similar findings of toxicity after overexpression of mouse Tmprss3 in the cochlea.37
Tmprss3 is broadly expressed in the embryonic and neonatal cochlea, with expression spanning sensory hair cells, a subset of spiral ganglion neurons, and non-sensory cells in the cochlea. This spatial pattern corroborates previous histological and single-cell RNA sequencing studies.7,8,9,10,11,12,25 Although TMPRSS3 is known to be a serine protease, its targets in the inner ear and exact function remain elusive.15 In this and previous studies, extensive loss of hair cells in Tmprss3-deficient mice indicates that Tmprss3 is critical for their survival.
While we cannot rule out dysfunction of other Tmprss3-expressing cell types (e.g., supporting cells) in Tmprss3tm1/tm1 mice, their survival is likely less dependent on Tmprss3, as we did not detect significant supporting cell degeneration. Moreover, spiral ganglion neuron degeneration observed at P120 is likely attributed to a loss of trophic support from hair cell loss rather than Tmprss3 deficiency.
In our study, high levels of exogenous Tmprss3 induced cytotoxicity in multiple cell lines in vitro and in the juvenile cochlea in vivo. Outside the inner ear, TMPRSS3 is highly expressed in several malignancies, including breast, ovarian, pancreatic, gastric, nasopharyngeal carcinoma, and brain gliomas.38,39,40,41,42,43 Several type II transmembrane serine proteases similar to TMPRSS3 are implicated in the development and progression of different types of cancer.44 Mechanistically, Tmprss3 overexpression can decrease E-cadherin levels and thereby disrupt cell-cell adhesion, leading to tumor invasiveness and metastasis.38,45 Based on these studies, we postulate that Tmprss3 overexpression similarly disrupts cell-cell adhesion, causing hair cells to be susceptible to degeneration after, but not prior to, the onset of hearing. Our model system has established a foundation to further investigate this potential mechanism.
Our study reveals that AAV-KP1-EF1α-Tmprss3 was effective in partially preventing hair cell loss and improving auditory function in Tmprss3tm1/tm1 mice, although outer hair cell function was not well restored. In situ hybridization showed that expression of Tmprss3 transgene was robust in supporting cells and remarkably lower in hair cells with the EF1α promoter. It is interesting that Tmprss3 transgene expression in hair cells appeared higher after treatment with AAV-KP1-CAG-Tmprss3 than AAV-KP1-EF1α-Tmprss3 with expression in supporting cells comparable between the two promoters. While AAV-KP1-EF1α-Tmprss3 partially rescued hearing function, AAV-KP1-CAG-Tmprss3 did not and moreover was cytotoxic to cochlear cells. One interpretation is that the Tmprss3 transgene is most critical for hair cell survival in the early postnatal cochlea, and the use of EF1α, and not CAG, promoter was able to restore Tmprss3 expression early. Alternatively, it is possible that hair cell survival does not directly depend on expression of Tmprss3 transgene within hair cells themselves but relies on an optimal level of expression in surrounding supporting cells, possibly via its effects on cell-cell junctions. Lastly, it is conceivable that a higher level of Tmprss3 transgene expression is needed to restore outer hair cell function. By increasing the titer or using a promoter (other than CAG) that drives Tmprss3 expression in outer hair cells, this may help to restore outer hair cell and overall cochlear function. Future work is necessary to delineate the function and downstream targets of Tmprss3 and the use of other promoters to drive Tmprss3 transgene expression in the cochlea.
Most early studies evaluating the efficacy of inner ear gene therapy have focused on mutations affecting sensory hair cells,17,46,47 with some recent studies examining mutations affecting non-sensory cells within the cochlea.48,49,50 The current study suggests that overexpression of Tmprss3 in supporting cells, rather than hair cells, may be more important for hair cell survival. However, this approach only partially prevents auditory function and fails to maintain outer hair cell function. While several studies have advocated the use of viral capsids with high transduction efficiency (e.g., AAV-ie, Anc80L65, and AAV2.7m8),51,52,53 it is unclear whether (1) broad transduction is necessary to rescue all cells expressing the gene of interest, (2) ectopic expression of a gene of interest can have deleterious effects in the inner ear, and (3) the level of transgene expression affects the efficacy of rescue. Even though AAV-KP1 robustly transduces multiple sensory and non-sensory cell types in the cochlea at rates comparable to several other viral capsids,51,52,53 AAV-KP1-CAG-Tmprss3 caused death of cochlear hair cells. Both decreasing viral titers and packaging with the less efficient AAV-DJ capsid reduced cell death and cochlear dysfunction. While these results suggest that cytotoxicity depends on the high transduction efficiency of cochlear cells, both approaches failed to rescue the function and cellular morphology of Tmprss3tm1 mutant cochlea. By using an EF1α core promoter, we eliminated cytotoxicity in cell lines and in the cochlea in vivo and partially rescued hair cell degeneration and cochlear dysfunction. The findings that rescue of hair cell survival and auditory function was partial and not complete are likely multifactorial and may include differences in spatiotemporal expression between exogenous and endogenous Tmprss3 as well as differences in the level of expression in cells of interest (e.g., hair cells). Nevertheless, since AAV-EF1α-Tmprss3 led to sustained hair cell survival at least to P120, this approach is promising and should guide future studies to further optimize rescue of hearing loss caused by Tmprss3 deficiency.
While some studies employed the CAG promoter within their viral vector constructs to successfully prevent cochlear dysfunction and degeneration in mouse models of hearing loss,22,46,54 this approach to drive Tmprss3 expression leads to cytotoxicity, especially when used with the broadly transducing viral capsid KP1. In the retina, the input dose, viral capsid, the encoded gene, the promoter driving transgene expression, and target cells have been demonstrated to govern cellular toxicity related to AAV administration.55 Moreover, broadly active promoters were found to be more toxic to the retinal pigment epithelium, and a weaker photoreceptor-specific promoter attenuated the toxicity.56 Here, there are several possible contributing factors to cytotoxicity of AAV-KP1-CAG-Tmprss3 in the cochlea, including ectopic transgene expression (e.g., stria vascularis, Reissner’s membrane), higher-than-native expression (hair cells and supporting cells), and differences in temporal expression. It is noteworthy that CAG-Tmprss3 contained the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), while EF1α-Tmprss3 did not. As WPRE has been reported to increase transgene expression,57 this sequence may further contribute to the toxicity observed. Lastly, Tmprss3 appears more highly expressed in hair cells after treatment with AAV-KP1-CAG-Tmprss3 than AAV-KP1-EF1α-Tmprss3, while that in supporting cells appears comparable between the two approaches. Thus, it is possible that the level of transgene expression in hair cells is more critical for hair cell function and in supporting cells for hair cell survival. The exact role of these factors warrants further investigation in future studies.
In summary, our results have demonstrated that the AAV-KP1 capsid has high transduction efficacy, but exogenous Tmprss3 under the CAG promoter led to cytotoxic effects in vitro and in vivo. Tmprss3-induced cytotoxicity was ameliorated by reducing transduction efficacy across cochlear cells using the AAV-DJ capsid and by using the EF1α core promoter. Collectively, our data indicate that precise spatial and temporal control of Tmprss3 expression is necessary for hair cell survival and cochlear function and further supports the need for a tailored approach to viral capsid and promoter selection to optimize gene therapy. These results may have important implications for the selection of viral capsids and promoters in human inner ear gene therapy.
Materials and methods
Mouse genotyping
The Tmprss3tm1/Lex mouse (MMRRC lab, stock # 032680, background C57/Bl6) strain was used. The Tmprss3tm1/tm1 mouse was described as having an absence of a startle response to 120 dB (prepulse inhibition assay).30 Mice of both genders were used. Genomic DNA was prepared from mouse tail tips. The genomic DNA template was produced by adding 180 μL of 50 mM NaOH to tissue biopsies and incubating at 98°C for 1 h and then 15°C for 2 min. Next, 20 μL of 1 M Tris-HCl was added, and the samples were vortexed. The following primers were used: Tmprss3 mutant forward (Fwd) (5′ GCA GCG CAT CGC CTT CTA TC), Tmprss3 mutant reverse (Rev) (5′ CAG AGC CTT AAC TCT CCA CG), and Tmprss3 wild-type Fwd (5′ TTC TAG GAC TTT GCT ATG ACC). All experiments were approved by the Institutional Animal Care and Use Committee (protocol #18606) at Stanford University.
In situ hybridization
Previously published protocols were followed.58,59 Briefly, temporal bone tissues harvested from P1–P6 mice were fixed in 4% paraformaldehyde (in PBS, pH 7.4, Electron Microscopy Services) for 22 h at 4°C. The tissues were then cryoprotected using a serial sucrose gradient over 2 days starting at 15%, 20%, 30% sucrose solution and then gradually increasing the optimal cutting temperature compound (OCT) (Tissue Tek) gradient of 30% sucrose: 50% OCT, to 30:70, and finally to 100% OCT. Next, tissues were stored at −80°C until further use. The sections were cut at 10 μm thickness and placed on Superfrost Plus slides (Fisher).
Tissue sections were hybridized with commercial probes from Advanced Cell Diagnostics (ACDbio) and counterstained with hematoxylin (Sigma-Aldrich) according to the manufacturer’s instructions for fixed frozen sections with colorimetric detection. Briefly, sections were washed in PBS (1×) for 5 min and then treated with H2O2 for 10 min. Next, sections were permeabilized using target retrieval reagent (ACDbio) and proteinase before hybridization. RNAScope Red v2.5 kit (catalog #323350) was used with the following probes: DapB (catalog #310043), Polr2a (catalog #312471), and Sox2 (catalog #401041). BaseScope v2 Red kit (catalog #323900) was used with the following probes: BaseScope Probe BA-Mm-Tmprss3-E1E2 (catalog #716911), BA-Mm-Ppib-1zz (catalog # 712351), BA-Dapb-1zz (catalog #701021) (ACDbio). The BaseScope Tmprss3 probe was diluted 1:20 and BaseScope step Amp 7 was performed for only 10 min to reduce signal intensity. RNA Polymerase II (Polr2) and Peptidylprolyl Isomerase B (PPib) are ubiquitously expressed in the cochlea and were selected as the positive controls. DapB gene is expressed by the Bacillus subtilis strain SMY, a soil bacterium, and not in mammalian tissues and was selected as a negative control (Figures S1C, S1D, S6G, and S6H). Wild-type and mutant cochleae were processed in parallel, with sections collected on the same slide and subjected to mRNA detection under identical conditions.
Immunohistochemistry
Cochleae were harvested and fixed in 4% paraformaldehyde in PBS for 30–40 min for processing. Cochleae from P12–120 mice were decalcified with 120 mM EDTA for 24–120 h at 4°C. Whole mounts were dissected into three turns with the removal of Reissner’s and tectorial membranes, and the stria vascularis was carefully removed.
Cryosections were prepared as described above. Tissues were washed in 0.1% Triton X-100 (in PBS) × 3 for 5 min (cryosections) or 15 min (whole mounts) and then blocked with 5% donkey serum, 0.1% Triton-100, 1% bovine serum albumin, and 0.02% sodium azide (NaN3) in PBS at pH 7.4 for 1 h at room temperature. Primary antibody inoculation was then performed in the same blocking solution overnight at 4°C. The following primary antibodies were used: rabbit anti-Myosin7a (1:500–1:1,000; Proteus Bioscience), goat anti-Sox2 (1:200–1:400; Santa Cruz Biotechnology), and mouse anti-Tuj1 (1:1,000; Neuromics). The following day, tissues were re-permeabilized with 0.1% Triton X-100 in PBS and incubated with secondary antibodies diluted in PBS containing 0.1% Triton X-100, 1% bovine serum albumin, and 0.02% NaN3 for 2 h at room temperature. Fluorescent-conjugated phalloidin (1:1,000, Invitrogen), DAPI (1:10,000, Invitrogen), and Alexa Fluor secondary antibodies (488, 546 or 647, 1:250–1:500; Life Technologies) were then used. After washing with PBS for 3 × 10 min (cryosection) or 30 min (whole mount), tissues were mounted in either anti-fade fluorescent mounting medium (DAKO) or ProlongGold (Thermo Fisher, catalog #P10144) and coverslipped for imaging.
Imaging and cell quantification
Whole mounts and cryosections were imaged as z stacks on Zeiss LSM700 and LSM880 (10× NA (numerical aperature) 0.3, 20× NA 0.8, and 40× NA 1.3 [oil]) confocal microscope. These images were captured at 1,024 × 1,024 (12-bit). Zen Black 2.3 (Carl Zeiss, Germany) was used.
For cell counting of whole-mount preparations, confocal images were analyzed using ImageJ software (NIH). Representative z stack images were taken on individual turns, and cells were counted from stacks and analyzed with ImageJ and Photoshop CS6 (Adobe Systems). For spiral ganglion neuron counting in sections, three sections per cochlea were used for quantification and averaged out (spiral ganglion neurons per 15,000 μm2).
Assessment of hearing function
ABRs and DPOAEs were measured as previously described.60 Briefly, mice were anesthetized (100 mg/kg ketamine and 10 mg/kg xylazine) and injected intraperitoneally. Three needle electrodes were placed as follows: one inferior to the tympanic bulla, referenced to an electrode on the vertex of the head, with a ground electrode placed in the hindlimb. Tone pip stimuli were delivered at frequencies ranging from 4 to 46 kHz (4.0, 5.7, 8.0, 11.3, 16.0, 22.3, 32, 46.1 kHz) up to an 80-dB sound pressure level (SPL) in 10-dB steps. In all, 512 trials at each frequency and intensity were conducted and averaged. Distortion product otoacoustic emissions were measured by a probe tip microphone placed in the auditory canal. The sound stimuli used to elicit the DPOAE were two 1-s sine wave tones of differing frequencies (F2/F1 ratio = 1.22). Frequencies ranged from 5.7 to 32 kHz and the two tones were stepped up from 20 to 80 dB SPL in 10-dB increments. The amplitude of the cubic distortion product was 2xF1-F2. The threshold was calculated as a DPOAE of two standard deviations above the noise floor for each frequency. For analysis of ABR and DPOAE, thresholds were manually scored in a blinded fashion by two individuals (K.A.A. and P.J.A.). A lack of a response was designated at the highest sound level, 80 dB SPL.
Vector constructs
The long isoform of Tmprss3 cDNA is 2,874 bp and is therefore within the packing capacity (∼4.8 kbp) of rAAV vectors. The mouse Tmprss3 gene was obtained from Origene (#MC216545) and cloned into the rAAV CAG-FLuc vector32 (plasmid #83281, Addgene) in place of the luciferase coding sequence using standard molecular biology techniques. The resulting construct (CAG-Tmprss3) contained AAV2 ITRs flanking the Tmprss3 sequence driven by a CAG promoter. A WPRE sequence between the coding sequence and the S40 late poly(A) signal was included to enhance expression.
Vector pAAV-EF1α-Tmprss3 was generated by replacing the respiratory syncytial virus (RSV) promoter region in plasmid pAAV-RHB61 with the first 212 nucleotides of the EF1α promoter from plasmid pAAV-EF1α-FLuc-WPRE-HGHpA (Addgene catalog #87951) using primers EF-core-NcoF (TATCTACCATGGGGCAGAGCGCACATCGCC) and EF-core-bluntR (CTGTGTTCTGGCGGCAAACCCG). Nco I and Ale I were used to ligate the promoter into the vector. The resulting vector, pAAV-EF1α-hAAT, was then digested with Ale I and Sal I to release the hAAT fragment. The Tmprss3 gene was amplified from pAAV-CAG-Tmprss3 with primers Tmp-bluntF (TGGTGTGCACCTCCAAGCGCCACCATGGCCGCTTCAGA) and Tmp-SalR (TACTAGTCGACCCAGCTCAACCTCAAGTCTTCAGATCTCTCTC), digested with Sal I, and ligated into the vector backbone. For cloning of plasmid pAAV-EF1α-TdRed, the hAAT transgene in vector pAAV-EF1α-hAAT was replaced in a similar manner as described above with the TdRed sequence obtained by amplification from plasmid pAAV-CAG-tdTomato (Addgene catalog #59462) using TdRed-bluntF (ATCCGGTACCGCCACCATGGTG) and TdRed-SalR (AGCTGTCGACTTACTTATACAGCTCATCCATG). All plasmids were sequence verified using Sanger sequencing. The EF1α-Tmprss3 construct did not contain a WPRE sequence, and the SV40 poly(A) signal was replaced with the bovine growth hormone (bGH) poly(A) sequence. Expression was confirmed by western blotting with an antibody specific for mouse Tmprss3 (Proteintech, #1793-1-AP).
rAAV production
293T/17 cells (ATCC #CRL-11268) were transfected with rep2-capKP1,32 the respective ITR-containing rAAV vector plasmid and pAd5 using the CaPO4 transfection method,32 or Transporter-5 transfection reagent (Polysciences, #26008). Virus was obtained from cell lysates 2 or 3 days post transfection and either purified using two rounds of CsCl centrifugation as previously described32 or using the AAV Pro All Serotype purification kit (Takara, #6666) according to the manufacturer’s instructions. Some rAAV preparations were further concentrated using Ultracel-100 spin columns (Millipore-Sigma, #UFC510008). Virus preparations were stored in aliquots at −80°C until use.
Viral genomes were isolated using the MinElute Virus Spin kit (Qiagen, #57704), and vector genome titers were determined using qPCR. For the Tmprss3-expressing rAAV preps, primers Tmp-qPCR-F (CACAGCAAGTACAAGCCAAAG) and Tmp-qPCR-R (GCTGGATGGT CTCGTCAAA) were used, while primer sets Td-qPCR-F (ATTACCTGGTGGAGTTCAAGAC) and Td-qPCR-R (GTCCTCGTTGTGTGAAGTGATA) were used to titer tdTomato-expressing rAAV preps. Copy number standards consisting of linearized and serially diluted (108–10/μL) plasmids were included on each qPCR plate. All AAVs used in this study were made in the same facility (Kay lab), with batches with higher titers diluted down to match those with lower titers.
In vitro transduction and transfection experiments
293T/17 cells or HeLa cells (ATCC #CCL-2) were seeded in 24-well plates, and, when they had reached a confluency of approximately 60%–70%, cells were transduced with rAAV at the MOI as indicated. Images were taken 3 days post transduction using a microscope with a built-in camera (Evos M5000, Invitrogen).
Cell proliferation assays
293T/17 cells or HeLa cells were seeded in 96-well plates at a density of 104 cells/well in 100 μL of medium and allowed to attach for 5 h. Cells were then transduced with rAAV diluted in 100 μL of medium in triplicate and assayed for proliferation at days 1, 2, 3, and 4 post transduction using the CellTiter 96 Aqueous One Solution assay according to the manufacturer’s instructions (Promega, #G3581). Standard curves were obtained using nontransduced cells seeded at various densities. Cells that had been transduced with a huFIX-expressing rAAV packaged with the KP1 capsid as well as nontransduced cells were included as controls.
In vivo gene transfer
To perform gene transfer experiments in vivo, P1 pups were anesthetized and injected via the posterior semicircular canal (PSCC) technique as previously described.62 Briefly, injection was performed using beveled glass microinjection pipettes, which were pulled from capillary glass on a P-2000 pipette puller (Sutter Instruments). Pups were anesthetized by rapid induction of hypothermia for 3–4 min on ice until loss of consciousness, and this state was maintained on a cooling platform for 10–15 min during the surgery. The surgical site was disinfected by scrubbing with Betadine and wiping with 70% ethanol. A postauricular incision was made to expose the PSCC and penetrate the tip of the micropipette. A total volume of 1 μL of either virus was unilaterally introduced at a rate of 300 nL/min into the left ear. The skin incision was closed using superglue. Body temperature was maintained on a 37°C warming pad for 30 min after surgery and before reintroduction into the parental cage.
Statistical analyses
Data were analyzed using Microsoft Excel (Microsoft) and GraphPad Prism (GraphPad). Two-tailed Student’s t tests or analysis of variance with post hoc tests were used to calculate statistical significance. p < 0.05 was considered statistically significant. Data are shown as mean ± SD. For all experiments, n values represent the number of animals examined.
Acknowledgments
We thank our laboratory for insightful comments on the manuscript and E. Huarcaya-Najarro for excellent technical support. This project was also supported by an NIH Shared Instrumentation Grant (S10-OD010580) from the National Center for Research Resources (NCRR) with contribution from Stanford's Beckman Center. This work was supported by American Neurotological Society research grant, American Society of Pediatric Otolaryngology research grant, and NIH Loan Repayment Program (K.A.A.); Stanford Maternal and Child Health Research Institute grant and Natural Sciences and Engineering Research Council grant (J.M.A.); NIH K08DC016034, RO1DC020574, and Triological Society and American College of Surgeon Clinician Scientist Development Award (R.F.N.); NIH R01AI116698 (M.K.); RO1DC01910, RO1DC021110, California Initiative in Regenerative Medicine, and the Yu and Oberndorf families (A.G.C.).
Author contributions
K.A.A., K.P., M.A.K., and A.G.C. designed experiments. K.A.A., K.P., I.A.L., Y.E., P.J.A., S.E.B., Y.S.C., W.D., R.F.N., and J.M.A. performed experiments. K.A.A., K.P., I.A.L., P.J.A., S.E.B., R.F.N., J.M.A., M.A.K., and A.G.C. analyzed data. K.A.A., P.J.A., J.M.A., M.A.K., and A.G.C. wrote the paper.
Declaration of interests
K.P. and M.A.K. are inventors of filed patents held by Stanford University.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.08.004.
Contributor Information
Mark A. Kay, Email: markay@stanford.edu.
Alan G. Cheng, Email: aglcheng@stanford.edu.
Supplemental information
Data and code availability
Original data generated in this research are included in the main figures or supplementary items. Additional original data are available upon request.
References
- 1.Morton C.C., Nance W.E. Newborn hearing screening--a silent revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Duman D., Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front. Biosci. 2012;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott H.S., Kudoh J., Wattenhofer M., Shibuya K., Berry A., Chrast R., Guipponi M., Wang J., Kawasaki K., Asakawa S., et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 4.Wattenhofer M., Sahin-Calapoglu N., Andreasen D., Kalay E., Caylan R., Braillard B., Fowler-Jaeger N., Reymond A., Rossier B.C., Karaguzel A., Antonarakis S.E. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum. Genet. 2005;117:528–535. doi: 10.1007/s00439-005-1332-x. [DOI] [PubMed] [Google Scholar]
- 5.Seligman K.L., Shearer A.E., Frees K., Nishimura C., Kolbe D., Dunn C., Hansen M.R., Gantz B.J., Smith R.J.H. Genetic Causes of Hearing Loss in a Large Cohort of Cochlear Implant Recipients. Otolaryngol. Head Neck Surg. 2022;166:734–737. doi: 10.1177/01945998211021308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fasquelle L., Scott H.S., Lenoir M., Wang J., Rebillard G., Gaboyard S., Venteo S., François F., Mausset-Bonnefont A.L., Antonarakis S.E., et al. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J. Biol. Chem. 2011;286:17383–17397. doi: 10.1074/jbc.M110.190652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guipponi M., Tan J., Cannon P.Z.F., Donley L., Crewther P., Clarke M., Wu Q., Shepherd R.K., Scott H.S. Mice deficient for the type II transmembrane serine protease, TMPRSS1/hepsin, exhibit profound hearing loss. Am. J. Pathol. 2007;171:608–616. doi: 10.2353/ajpath.2007.070068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guipponi M., Toh M.Y., Tan J., Park D., Hanson K., Ballana E., Kwong D., Cannon P.Z.F., Wu Q., Gout A., et al. An integrated genetic and functional analysis of the role of type II transmembrane serine proteases (TMPRSSs) in hearing loss. Hum. Mutat. 2008;29:130–141. doi: 10.1002/humu.20617. [DOI] [PubMed] [Google Scholar]
- 9.Guipponi M., Vuagniaux G., Wattenhofer M., Shibuya K., Vazquez M., Dougherty L., Scamuffa N., Guida E., Okui M., Rossier C., et al. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum. Mol. Genet. 2002;11:2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- 10.Gu S., Olszewski R., Taukulis I., Wei Z., Martin D., Morell R.J., Hoa M. Characterization of rare spindle and root cell transcriptional profiles in the stria vascularis of the adult mouse cochlea. Sci. Rep. 2020;10:18100. doi: 10.1038/s41598-020-75238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota M., Scheibinger M., Jan T.A., Heller S. Greater epithelial ridge cells are the principal organoid-forming progenitors of the mouse cochlea. Cell Rep. 2021;34:108646. doi: 10.1016/j.celrep.2020.108646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolla L., Kelly M.C., Mann Z.F., Anaya-Rocha A., Ellis K., Lemons A., Palermo A.T., So K.S., Mays J.C., Orvis J., et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 2020;11:2389. doi: 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina L., Fasquelle L., Nouvian R., Salvetat N., Scott H.S., Guipponi M., Molina F., Puel J.L., Delprat B. Tmprss3 loss of function impairs cochlear inner hair cell Kcnma1 channel membrane expression. Hum. Mol. Genet. 2013;22:1289–1299. doi: 10.1093/hmg/dds532. [DOI] [PubMed] [Google Scholar]
- 14.Peters T.A., Levtchenko E., Cremers C.W.R.J., Curfs J.H.A.J., Monnens L.A.H. No evidence of hearing loss in pseudohypoaldosteronism type 1 patients. Acta Otolaryngol. 2006;126:237–239. doi: 10.1080/00016480500388893. [DOI] [PubMed] [Google Scholar]
- 15.Pyott S.J., Meredith A.L., Fodor A.A., Vázquez A.E., Yamoah E.N., Aldrich R.W. Cochlear function in mice lacking the BK channel alpha, beta1, or beta4 subunits. J. Biol. Chem. 2007;282:3312–3324. doi: 10.1074/jbc.M608726200. [DOI] [PubMed] [Google Scholar]
- 16.Bankoti K., Generotti C., Hwa T., Wang L., O'Malley B.W., Jr., Li D. Advances and challenges in adeno-associated viral inner-ear gene therapy for sensorineural hearing loss. Mol. Ther. Methods Clin. Dev. 2021;21:209–236. doi: 10.1016/j.omtm.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akil O., Seal R.P., Burke K., Wang C., Alemi A., During M., Edwards R.H., Lustig L.R. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75:283–293. doi: 10.1016/j.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akil O., Dyka F., Calvet C., Emptoz A., Lahlou G., Nouaille S., Boutet de Monvel J., Hardelin J.P., Hauswirth W.W., Avan P., et al. Dual AAV-mediated gene therapy restores hearing in a DFNB9 mouse model. Proc. Natl. Acad. Sci. USA. 2019;116:4496–4501. doi: 10.1073/pnas.1817537116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isgrig K., Shteamer J.W., Belyantseva I.A., Drummond M.C., Fitzgerald T.S., Vijayakumar S., Jones S.M., Griffith A.J., Friedman T.B., Cunningham L.L., Chien W.W. Gene Therapy Restores Balance and Auditory Functions in a Mouse Model of Usher Syndrome. Mol. Ther. 2017;25:780–791. doi: 10.1016/j.ymthe.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimura H., Shibata S.B., Ranum P.T., Moteki H., Smith R.J.H. Targeted Allele Suppression Prevents Progressive Hearing Loss in the Mature Murine Model of Human TMC1 Deafness. Mol. Ther. 2019;27:681–690. doi: 10.1016/j.ymthe.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.György B., Meijer E.J., Ivanchenko M.V., Tenneson K., Emond F., Hanlon K.S., Indzhykulian A.A., Volak A., Karavitaki K.D., Tamvakologos P.I., et al. Gene Transfer with AAV9-PHP.B Rescues Hearing in a Mouse Model of Usher Syndrome 3A and Transduces Hair Cells in a Non-human Primate. Mol. Ther. Methods Clin. Dev. 2019;13:1–13. doi: 10.1016/j.omtm.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulon D., Papal S., Patni P., Cortese M., Vincent P.F., Tertrais M., Emptoz A., Tlili A., Bouleau Y., Michel V., et al. Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J. Clin. Invest. 2018;128:3382–3401. doi: 10.1172/JCI94351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nist-Lund C.A., Pan B., Patterson A., Asai Y., Chen T., Zhou W., Zhu H., Romero S., Resnik J., Polley D.B., et al. Improved TMC1 gene therapy restores hearing and balance in mice with genetic inner ear disorders. Nat. Commun. 2019;10:236. doi: 10.1038/s41467-018-08264-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan B., Askew C., Galvin A., Heman-Ackah S., Asai Y., Indzhykulian A.A., Jodelka F.M., Hastings M.L., Lentz J.J., Vandenberghe L.H., et al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat. Biotechnol. 2017;35:264–272. doi: 10.1038/nbt.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y.S., Cabrera E., Tucker B.J., Shin T.J., Moawad J.V., Totten D.J., Booth K.T., Nelson R.F. TMPRSS3 expression is limited in spiral ganglion neurons: implication for successful cochlear implantation. J. Med. Genet. 2022;59:1219–1226. doi: 10.1136/jmg-2022-108654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexopoulou A.N., Couchman J.R., Whiteford J.R. The CMV early enhancer/chicken beta actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol. 2008;9:2. doi: 10.1186/1471-2121-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 28.Kim D.W., Uetsuki T., Kaziro Y., Yamaguchi N., Sugano S. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. Gene. 1990;91:217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 29.Shrestha B.R., Chia C., Wu L., Kujawa S.G., Liberman M.C., Goodrich L.V. Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell. 2018;174:1229–1246.e17. doi: 10.1016/j.cell.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 31.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pekrun K., De Alencastro G., Luo Q.J., Liu J., Kim Y., Nygaard S., Galivo F., Zhang F., Song R., Tiffany M.R., et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 2019;4:e131610. doi: 10.1172/jci.insight.131610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Okada T., Nomoto T., Ke X., Kume A., Ozawa K., Xiao S. Promoter effects of adeno-associated viral vector for transgene expression in the cochlea in vivo. Exp. Mol. Med. 2007;39:170–175. doi: 10.1038/emm.2007.19. [DOI] [PubMed] [Google Scholar]
- 34.Buck T.M., Wijnholds J. Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays. Int. J. Mol. Sci. 2020;21:4197. doi: 10.3390/ijms21124197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seita Y., Tsukiyama T., Azami T., Kobayashi K., Iwatani C., Tsuchiya H., Nakaya M., Tanabe H., Hitoshi S., Miyoshi H., et al. Comprehensive evaluation of ubiquitous promoters suitable for the generation of transgenic cynomolgus monkeysdagger. Biol. Reprod. 2019;100:1440–1452. doi: 10.1093/biolre/ioz040. [DOI] [PubMed] [Google Scholar]
- 36.Chung J., Park S.M., Chang S.O., Chung T., Lee K.Y., Kim A.R., Park J.H., Kim V., Park W.Y., Oh S.H., et al. A novel mutation of TMPRSS3 related to milder auditory phenotype in Korean postlingual deafness: a possible future implication for a personalized auditory rehabilitation. J. Mol. Med. 2014;92:651–663. doi: 10.1007/s00109-014-1128-3. [DOI] [PubMed] [Google Scholar]
- 37.Du W., Ergin V., Loeb C., Huang M., Silver S., Armstrong A.M., Huang Z., Gurumurthy C.B., Staecker H., Liu X., Chen Z.Y. Rescue of auditory function by a single administration of AAV-TMPRSS3 gene therapy in aged mice of human recessive deafness DFNB8. Mol. Ther. 2023 doi: 10.1016/j.ymthe.2023.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D., Qiu S., Wang Q., Zheng J. TMPRSS3 modulates ovarian cancer cell proliferation, invasion and metastasis. Oncol. Rep. 2016;35:81–88. doi: 10.3892/or.2015.4356. [DOI] [PubMed] [Google Scholar]
- 39.Wang J.Y., Jin X., Li X.F. Knockdown of TMPRSS3, a Transmembrane Serine Protease, Inhibits the Proliferation, Migration, and Invasion in Human Nasopharyngeal Carcinoma Cells. Oncol. Res. 2018;26:95–101. doi: 10.3727/096504017X14920318811695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S.L., Chen X., Wu T., Zhang X.W., Li H., Zhang Y., Ji Z.Z. Knockdown of TMPRSS3 inhibits gastric cancer cell proliferation, invasion and EMT via regulation of the ERK1/2 and PI3K/Akt pathways. Biomed. Pharmacother. 2018;107:841–848. doi: 10.1016/j.biopha.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 41.Wallrapp C., Hähnel S., Müller-Pillasch F., Burghardt B., Iwamura T., Ruthenbürger M., Lerch M.M., Adler G., Gress T.M. A novel transmembrane serine protease (TMPRSS3) overexpressed in pancreatic cancer. Cancer Res. 2000;60:2602–2606. [PubMed] [Google Scholar]
- 42.Huo J.F., Chen X.B. Knockdown of TMPRSS3 inhibits cell proliferation, migration/invasion and induces apoptosis of glioma cells. J. Cell. Biochem. 2019;120:7794–7801. doi: 10.1002/jcb.28054. [DOI] [PubMed] [Google Scholar]
- 43.Rui X., Li Y., Jin F., Li F. TMPRSS3 is a novel poor prognostic factor for breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:5435–5442. [PMC free article] [PubMed] [Google Scholar]
- 44.Netzel-Arnett S., Hooper J.D., Szabo R., Madison E.L., Quigley J.P., Bugge T.H., Antalis T.M. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 45.Tanabe L.M., List K. The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS J. 2017;284:1421–1436. doi: 10.1111/febs.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng R., Omar A., Gopal S.R., Chen D.H.C., Stepanyan R., Basch M.L., Dinculescu A., Furness D.N., Saperstein D., Hauswirth W., et al. Modeling and Preventing Progressive Hearing Loss in Usher Syndrome III. Sci. Rep. 2017;7:13480. doi: 10.1038/s41598-017-13620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chien W.W., Isgrig K., Roy S., Belyantseva I.A., Drummond M.C., May L.A., Fitzgerald T.S., Friedman T.B., Cunningham L.L. Gene Therapy Restores Hair Cell Stereocilia Morphology in Inner Ears of Deaf Whirler Mice. Mol. Ther. 2016;24:17–25. doi: 10.1038/mt.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Hou Z., Wang X., Jiang H., Neng L., Zhang Y., Yu Q., Burwood G., Song J., Auer M., et al. VEGFA165 gene therapy ameliorates blood-labyrinth barrier breakdown and hearing loss. JCI Insight. 2021;6:e143285. doi: 10.1172/jci.insight.143285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang Q., Wang J., Li Q., Kim Y., Zhou B., Wang Y., Li H., Lin X. Virally mediated Kcnq1 gene replacement therapy in the immature scala media restores hearing in a mouse model of human Jervell and Lange-Nielsen deafness syndrome. EMBO Mol. Med. 2015;7:1077–1086. doi: 10.15252/emmm.201404929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iizuka T., Kamiya K., Gotoh S., Sugitani Y., Suzuki M., Noda T., Minowa O., Ikeda K. Perinatal Gjb2 gene transfer rescues hearing in a mouse model of hereditary deafness. Hum. Mol. Genet. 2015;24:3651–3661. doi: 10.1093/hmg/ddv109. [DOI] [PubMed] [Google Scholar]
- 51.Tan F., Chu C., Qi J., Li W., You D., Li K., Chen X., Zhao W., Cheng C., Liu X., et al. AAV-ie enables safe and efficient gene transfer to inner ear cells. Nat. Commun. 2019;10:3733. doi: 10.1038/s41467-019-11687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isgrig K., McDougald D.S., Zhu J., Wang H.J., Bennett J., Chien W.W. AAV2.7m8 is a powerful viral vector for inner ear gene therapy. Nat. Commun. 2019;10:427. doi: 10.1038/s41467-018-08243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R., Vandenberghe L.H. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017;35:280–284. doi: 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emptoz A., Michel V., Lelli A., Akil O., Boutet de Monvel J., Lahlou G., Meyer A., Dupont T., Nouaille S., Ey E., et al. Local gene therapy durably restores vestibular function in a mouse model of Usher syndrome type 1G. Proc. Natl. Acad. Sci. USA. 2017;114:9695–9700. doi: 10.1073/pnas.1708894114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khabou H., Cordeau C., Pacot L., Fisson S., Dalkara D. Dosage Thresholds and Influence of Transgene Cassette in Adeno-Associated Virus-Related Toxicity. Hum. Gene Ther. 2018;29:1235–1241. doi: 10.1089/hum.2018.144. [DOI] [PubMed] [Google Scholar]
- 56.Xiong W., Wu D.M., Xue Y., Wang S.K., Chung M.J., Ji X., Rana P., Zhao S.R., Mai S., Cepko C.L. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl. Acad. Sci. USA. 2019;116:5785–5794. doi: 10.1073/pnas.1821000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patrício M.I., Barnard A.R., Orlans H.O., McClements M.E., MacLaren R.E. Inclusion of the Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances AAV2-Driven Transduction of Mouse and Human Retina. Mol. Ther. Nucleic Acids. 2017;6:198–208. doi: 10.1016/j.omtn.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansson L., Ebeid M., Shen J.W., Mokhtari T.E., Quiruz L.A., Ornitz D.M., Huh S.H., Cheng A.G. beta-Catenin is required for radial cell patterning and identity in the developing mouse cochlea. Proc. Natl. Acad. Sci. USA. 2019;116:21054–21060. doi: 10.1073/pnas.1910223116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Najarro E.H., Huang J., Jacobo A., Quiruz L.A., Grillet N., Cheng A.G. Dual regulation of planar polarization by secreted Wnts and Vangl2 in the developing mouse cochlea. Development. 2020;147 doi: 10.1242/dev.191981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huth M.E., Han K.H., Sotoudeh K., Hsieh Y.J., Effertz T., Vu A.A., Verhoeven S., Hsieh M.H., Greenhouse R., Cheng A.G., Ricci A.J. Designer aminoglycosides prevent cochlear hair cell loss and hearing loss. J. Clin. Invest. 2015;125:583–592. doi: 10.1172/JCI77424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu J., Williams J.A., Luke J., Zhang F., Chu K., Kay M.A. A 5' Noncoding Exon Containing Engineered Intron Enhances Transgene Expression from Recombinant AAV Vectors in vivo. Hum. Gene Ther. 2017;28:125–134. doi: 10.1089/hum.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talaei S., Schnee M.E., Aaron K.A., Ricci A.J. Dye Tracking Following Posterior Semicircular Canal or Round Window Membrane Injections Suggests a Role for the Cochlea Aqueduct in Modulating Distribution. Front. Cell. Neurosci. 2019;13:471. doi: 10.3389/fncel.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data generated in this research are included in the main figures or supplementary items. Additional original data are available upon request.