Abstract
Introduction
The independent and causal cardiovascular disease risk factor lipoprotein(a) (Lp(a)) is elevated in >1.5 billion individuals worldwide, but studies have prioritised European populations.
Methods
Here, we examined how ancestrally diverse studies could clarify Lp(a)’s genetic architecture, inform efforts examining application of Lp(a) polygenic risk scores (PRS), enable causal inference and identify unexpected Lp(a) phenotypic effects using data from African (n=25 208), East Asian (n=2895), European (n=362 558), South Asian (n=8192) and Hispanic/Latino (n=8946) populations.
Results
Fourteen genome-wide significant loci with numerous population specific signals of large effect were identified that enabled construction of Lp(a) PRS of moderate (R2=15% in East Asians) to high (R2=50% in Europeans) accuracy. For all populations, PRS showed promise as a ‘rule out’ for elevated Lp(a) because certainty of assignment to the low-risk threshold was high (88.0%–99.9%) across PRS thresholds (80th–99th percentile). Causal effects of increased Lp(a) with increased glycated haemoglobin were estimated for Europeans (p value =1.4×10−6), although inverse effects in Africans and East Asians suggested the potential for heterogeneous causal effects. Finally, Hispanic/Latinos were the only population in which known associations with coronary atherosclerosis and ischaemic heart disease were identified in external testing of Lp(a) PRS phenotypic effects.
Conclusions
Our results emphasise the merits of prioritising ancestral diversity when addressing Lp(a) evidence gaps.
Keywords: biomarkers, epidemiology, genome-wide association study, genetic association studies
WHAT IS ALREADY KNOWN ON THIS TOPIC
Lipoprotein(a) (Lp(a)) is a highly heritable cardiovascular disease (CVD) risk factor for which pivotal clinical trials are underway.
Despite being one of the most variable CVD risk factors across populations, the majority of Lp(a) research has been performed in European ancestral populations, limiting the reach and generalisability of the evidence base that informs clinical and public health decision making to five-sixths of the global population.
WHAT THIS STUDY ADDS
By applying innovations in statistical genetics to five ancestrally diverse populations, we demonstrate how increasing ancestral diversity helps clarify the role of Lp(a) in disease pathogenesis, identify individuals with elevated Lp(a) and enable causal inference studies.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Important insights into Lp(a) can be enabled by inclusion of modestly sized populations of non-European ancestry. Increased awareness of the benefits of ancestral diversity will provide an essential foundation for future studies that aim to maximise the benefits and minimise the risks of therapeutic Lp(a) lowering in everyone.
Introduction
Lipoprotein(a) (Lp(a)), a highly atherogenic and prothrombotic lipoprotein, is an independent and causal cardiovascular disease (CVD) risk factor that is elevated in an estimated 1.5 billion individuals worldwide.1 2 Strong and consistent evidence linking Lp(a) with CVDs has motivated the development of Lp(a)-reducing therapies that are in pivotal clinical trials.3 Despite clinical, regulatory and public health interest in Lp(a), several major evidence gaps remain. First, the role of Lp(a) in the pathogenesis of non-CVD phenotypes remains largely unexplored despite the potential for broad phenotypic effects.4–8 Opportunities to anticipate adverse effects of therapeutic Lp(a) lowering, to illuminate mechanisms of action and to identify novel treatment indications through drug repurposing are therefore missed.9 10 Second, Lp(a) is not routinely measured in clinical practice, as few guidelines advocate universal measurement and the performance of commercially available assays varies.11 12 As a result, populations with very high lifetime CVD risk remain unidentified. Third, Lp(a) is distinguished from other CVD risk factors by pronounced ancestral differences.13 The causes and consequences of these ancestral differences are poorly understood, but may reflect heterogeneity in Lp(a)’s genetic architecture.14 Finally, Lp(s) is not found in commonly studied laboratory animals, limiting the reach of mechanistic studies.15 Causal inference studies could help bridge this gap, although few studies outside European populations or CVD outcomes have been published.4 16 As a result, opportunities to inform Lp(a) and maximise its utility in clinical medicine and public health remain incompletely realised.
Lp(a)’s high heritability (h2=70%–90%), oligogenic genetic architecture and relative stability across the lifespan offer several avenues to address these evidence gaps.17 18 These features have enabled estimation of highly accurate Lp(a) polygenic risk scores (PRS) that, by aggregating variants into a single score, explain as much as 60% of Lp(a) trait variance.16 19 20 Few other CVD risk factors are predicted with such high accuracy. However, highly predictive Lp(a) PRS are only available for European populations and these European-derived PRS are not portable to other populations. This lack of portability reflects heterogeneity in the genetic architecture of Lp(a) across ancestral populations21 as well as the broader challenge of limited ancestral diversity in genetics research.22 23
Here, we constructed ancestry specific Lp(a) PRS using seven different approaches to maximise accuracy for populations of African, East Asian, European and South Asian ancestry. These PRS are publicly available and, for non-European populations, outperformed existing Lp(a) PRS.24 We then showed how these PRS and the genome-wide association studies (GWAS) from which they were derived can inform efforts examining clinical application of Lp(a) PRS, clarify Lp(a)’s genetic architecture, enable causal inference studies and identify unexpected phenotypic effects, even when the size of non-European populations is modest.
Methods
Study populations
We included data from two sources: The Population Architecture through Genomics and Environment (PAGE) study and the UK Biobank (online supplemental table 1 and text). The PAGE study25 is a consortium funded since 2008 by the National Institutes of Health to examine the genetic underpinnings of common complex diseases and phenotypes in ancestrally diverse US populations. Five PAGE studies were included for PRS construction and testing: the Atherosclerosis Risk in Communities study (ARIC),26 the Coronary Artery Risk in Young Adults study (CARDIA),27 the Jackson Heart Study (JHS),28 the Multi-Ethnic Study of Atherosclerosis (MESA)29 and the Women’s Health Initiative (WHI).30 A sixth PAGE study, the Mount Sinai BioMe biobank (BioMe), was included to examine external application of PRS.
openhrt-2023-002382supp001.pdf (905.4KB, pdf)
The UK Biobank is a publicly available, longitudinal study of England, Wales and Scotland residents.31 For all studies except BioMe, eligible participants had genotypic data and Lp(a) measures and were categorised into one of four populations with large enough numbers to support ancestry specific Lp(a) GWAS: African, East Asian, European or South Asian. For BioMe, eligible participants had imputed GWAS data, inpatient ICD-9 and ICD-10 codes, and were categorised into three populations with large enough number to support self-reported race/ethnicity-specific analyses: European, African and Hispanic/Latino.
Genotyping and imputation
After genotyping using one of several assays, imputation was performed across studies using the UK10K and 1000 Genomes Project (UK Biobank) or 1000 Genomes Project (PAGE) phase 3 reference panels (online supplemental table 2). In addition to study-specific protocols, we excluded variants meeting any of the following criteria on a population-specific and study-specific basis given the large differences in study size: minor allele frequency (MAF) <0.001; imputation quality score <0.4; or effective minor allele count Neff<30, where Neff =2f(1−f)Nq and f is minor allele frequency, N is the sample size and q is the imputation quality.
Statistical methods
Genome-wide association studies
We estimated Lp(a) ancestry-specific and study-specific genetic effects using generalised linear models of the form where was a vector of inverse-normalised Lp(a), was an identity link function, denoted confounders (age, sex, 15 ancestral principal components and study centre, when appropriate) and was variant dosage. Linear models were implemented in SUGEN (ARIC, CARDIA, WHI),32 EMMAX (JHS),33 SNPTEST (MESA)34 or SAIGE (UK Biobank),35 accounting for relatedness as appropriate. Quality control and visualisation of GWAS results were performed using the EasyQC36 and EasyStrata37 packages.
Ancestry-specific GWAS results were combined using inverse-variance weighted, genomic inflation-corrected meta-analysis.38 Variant effect heterogeneity was assessed with the Cochran’s Q test within ancestry. Variants with p values <5×10−9 (Bonferroni correction for 10M tests) were considered genome-wide significant.39 Our ancestry-specific Lp(a) GWAS summary statistics will be available on the NHGRI-EBI GWAS catalogue pending publication given submission requirements (https://www.ebi.ac.uk/gwas/).
Identification of independent/secondary signals
We used GCTA-COJO,40 the ‘cojo-slct’ method, to perform forward model selection to identify independently associated variants across each set of ancestry-specific GWAS meta-analyses. To account for linkage disequilibrium (LD) in each ancestry-specific analysis, we identified a subset of genetically unrelated UK Biobank participants and calculated LD between variant pairs in 10 Mb windows.
Variant-based heritability and percentage variance explained
Lp(a) narrow sense-heritability was estimated by ancestry using GCTA and unrelated UK Biobank participants.41 42 Briefly, a genetic relationship matrix was created by ancestry for each chromosome (1–22), including variants imputed with high quality (>0.7). After combining the matrices, we fit ancestry-specific linear models adjusting for age, sex, 15 ancestral principal components and study centre using restricted maximum likelihood to estimate the percentage variance of Lp(a) explained by genome-wide variants and LPA variants.
Lp(a) PRS estimation
We examined seven approaches to estimate Lp(a) ancestry specific PRS. These approaches were distinguished by the statistical method, the data used to construct variant weights, the genomic region examined (LPA locus or genome-wide) and the LD reference panel (online supplemental table 3). For approaches that used external PRS,43 Pruning and Thresholding44 and LDpred2,45 the discovery data were independent of the target data. For approaches that used GCTA-COJO40 to identify independent signals across ancestry-specific GWAS meta-analyses and Crosspred,46 cross-validation was used to address a lack of independence between the discovery and target data.
PRS performance at the study level was compared using the incremental R2 after accounting for age, age2, sex, 15 ancestral principal components and study centre for continuous Lp(a) and the area under the receiver operator curve (AUC) when Lp(a) was dichotomised. When estimating AUC using Lp(a) PRS as the predictor, measured Lp(a) was dichotomised at 125 nmol/L, the Lp(a) clinical risk enhancer threshold47; participants with Lp(a) levels at or above 125 nmol/L were classified as having high Lp(a). The 95% CI for R2 were obtained from 1000 non-parametric bootstrap replicates of the parameters reporting their quantile at 2.5% and 97.5%.
Certainty in PRS estimation
To inform the use of PRS for the identification of individuals who may benefit from Lp(a) testing, we quantified PRS certainty at the individual level for the best performing PRS within each ancestral population.48 Briefly, the variance of each participant’s assigned PRS was estimated across four PRS stratification thresholds (the 80th, 90th, 95th and 99th PRS percentiles). Selection of the 90th, 95th and 99th percentiles was data driven, whereas the 80th percentile was selected because it corresponded to a mean Lp(a) level of 125 nmol/L in African and European populations (online supplemental figure 1). A percentile threshold corresponding to 125 nmol/L was not examined in East Asian and South Asian populations because either the mean Lp(a) level for the highest percentiles did not exceed 100 nmol/L (East Asians) or there were too few participants in the percentiles with mean Lp(a) >125 nmol/L (South Asians), which could reflect modest PRS accuracy. We then computed a 95% credible interval for the individual PRS point estimates, which is interpreted as the interval in which the true PRS is expected to fall with 95% probability. Finally, we calculated the proportion of individuals whose 95% credible intervals were fully contained within their assigned threshold range as a measure of certainty.
BioMe phenome-wide association study
To characterise the degree to which Lp(a) association studies reported consistent results across populations, we conducted a phenome-wide association study (PheWAS) in BioMe participants of White, African American and Hispanic/Latino self-identified race/ethnicity using R.49 A genetically inferred Lp(a) value, which we substituted for measured Lp(a), was constructed for each BioMe participant with genotypic data. For White and Hispanic/Latino BioMe participants, we used the approach and weights from the external PRS developed in a European ancestral population, as an Lp(a) PRS for Hispanic/Latinos was unavailable. For African American BioMe participants, we used the independent signal (genome-wide) approach developed in PAGE and UK Biobank participants of African ancestry. Using these genetically inferred Lp(a) measures, we then estimated race/ethnic-specific associations with a maximum of 1004 phecodes with ≥20 cases derived from ICD-9 and ICD-10 inpatient codes, adjusting for age, sex and 15 ancestral principal components. Because statistical power to evaluate >1000 phecodes would be modest, we restricted our attention to phenotypes that were nominally significant (ie, p value <0.05) and were previously associated with Lp(a).
Mendelian randomisation
Causal effects of Lp(a) on two phenotypes with inconsistent evidence of association with Lp(a) (estimated glomerular filtration rate (eGFR) and glycated haemoglobin (hbA1c))8 50 were estimated in UK Biobank participants without chronic kidney disease or diabetes, respectively, by population using Mendelian randomisation (MR). MR is a form of instrumental variable (IV) analysis based on the concept that if X (Lp(a)) affects outcome Y (eGFR or hbA1c), factors affecting X (ie, Lp(a) PRS, G) must also affect Y.51 G therefore serves as an IV to estimate causal effects of X on Y. Strengths of MR include G-Y associations that are assumed to be robust to confounding from variables other than ancestry, which can be addressed through adjustment. Random assignment of G at conception also enables assessment of temporality of the G-Y association and, by extension, the X-Y association. Causal effects were estimated in SAS (PROC SYSLIN) using two-stage least squares after examination of the pleiotropy and instrument strength MR assumptions and adjusting for age, sex, study centre and 15 ancestral principal components.51 52 To facilitate comparison, we also conducted standard association analysis using linear regression.
Results
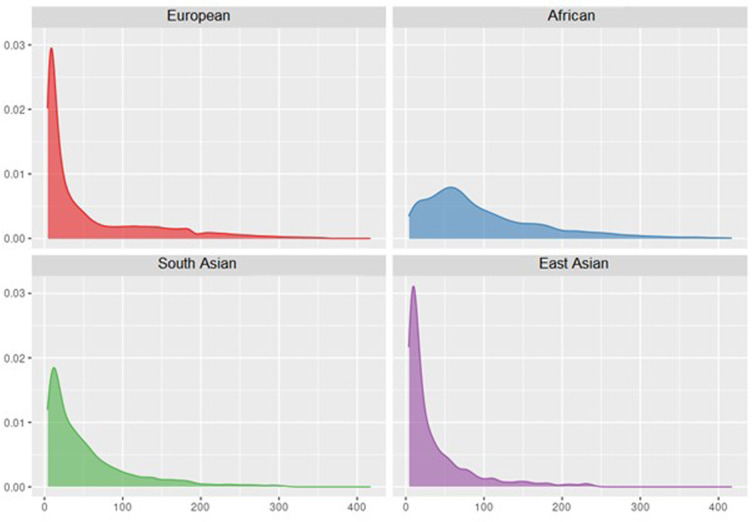
A total of 407 799 participants of African (n=25 208), East Asian (n=2895), European (n=3 62 558), South Asian (n=8192) and Hispanic/Latino (n=8946) race/ethnicity or ancestry were included (online supplemental table 1). Study participants spanned early to late adulthood (age range: 18 to >80 years) and 56% were female. As exemplified by UK Biobank participants, the distribution of Lp(a) varied by ancestry (figure 1, online supplemental table 4). An estimated 21%, 4%, 14% and 8% of African, East Asian, European and South Asian UK Biobank participants had Lp(a) values >125 nmol/L.
Figure 1.
Distributions of lipoprotein(a) by ancestry in n=3 57 096 UK Biobank participants at study baseline.
Lp(a) ancestry specific GWAS
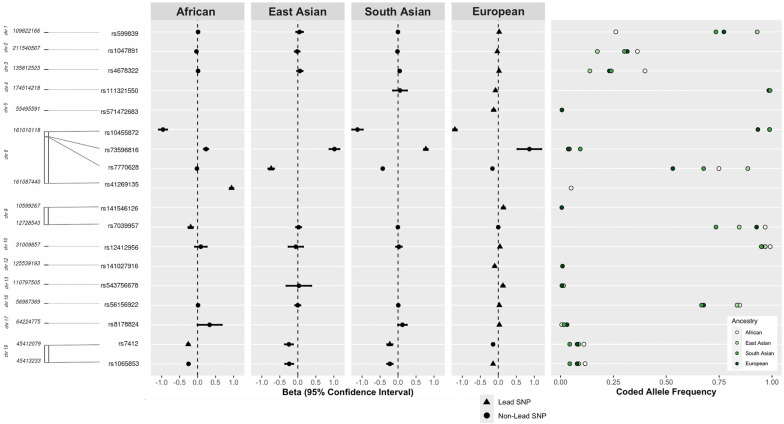
Fourteen genome-wide significant Lp(a) loci were identified, with little evidence of within-ancestry heterogeneity: 3 loci for African, 1 locus for East Asian, 13 loci for European and 2 loci for South Asian participants (figure 2, online supplemental table 5 and figure 2). The major chromosome 6 LPA locus was the most pronounced signal, harboured numerous independent secondary signals (online supplemental tables 6 and 7) and had lead variants with very strong (SD increment per effect allele copy: |0.74–1.21|) effects. LPA also demonstrated a unique genetic architecture for each ancestral population. The African LPA lead variant (rs41269135, A effect allele) was monomorphic (ie, only the non-effect allele G was observed) in East Asian, European and South Asian participants, and the European LPA lead variant (rs10455872, A effect allele) was monomorphic in East Asian participants. Other notable Lp(a) loci included APOE on chromosome 19, which was identified at genome-wide significance levels for African (lead variant rs7412, T effect allele), European (lead variant rs1065853, T effect allele) and South Asian participants (lead variant rs7412, T effect allele). APOE lead variants rs7412 and rs1065853 were in high LD (eg, R2=1 in 1000 Genomes EUR) (online supplemental table 5), suggesting that these lead variants tag the same locus.
Figure 2.
Genome wide-significant (p<5×10−9) loci discovered in ancestry-specific lipoprotein(a) genome-wide association studies of African (n=19 333), East Asian (n=2895), European (n=354 843) and South Asian (n=8192) Population Architecture through Genomics and Environment study and UK Biobank participants.
Variant-based heritability and percentage variance explained
Ancestry specific Lp(a) variant-based heritability estimates ranged from 13.1% in East Asian to 38.4% in African participants for the entire genome (online supplemental table 8). To reduce uncertainty and better capture the oligogenic architecture of Lp(a), we also estimated variant-based heritability restricting to the LPA locus. When restricting to the LPA locus, ancestry-specific variant-based heritability estimates approximately doubled in magnitude (LPA-specific h2 range: 22.5% in East Asian to 76.1% in European participants). However, smaller increases were observed for African participants (LPA-specific h2=45.7%; genome-wide h2=38.4%).
Ancestry specific PRS performance
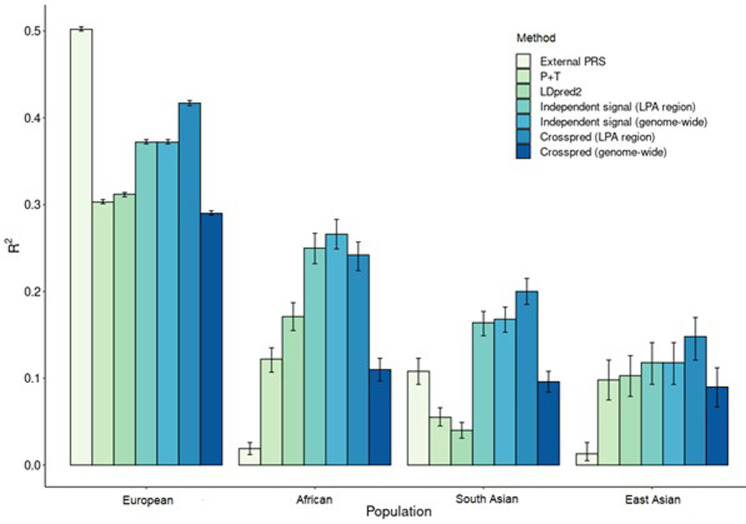
We evaluated seven approaches for estimating ancestry specific Lp(a) PRS (figure 3, online supplemental table 9). The external PRS that included 43 LPA variants43 was highly predictive in Europeans (R2=50%), but was not transferable across ancestries (R2 range: 2–11%). Lp(a) PRS prediction accuracy in non-European populations increased considerably when ancestry specific discovery data were used, with R2=12%–25% in African and R2=10%–12% in East Asian populations despite discovery populations that were modest in size (n=11 287 in Africans and n=706 in East Asians). For South Asians, increased prediction accuracy was achieved when using Crosspred, the approach that did not require an independent discovery population (R2=20%), which was unavailable. Prediction accuracy also was increased in East Asian populations (R2=15%) when Crosspred was used. When Lp(a) PRS were extended to include regions outside LPA, prediction accuracy was increased for African population (R2=27%), but was substantially decreased (R2 range: 9%–27%) for East Asian, European and South Asian populations. The most accurate PRS for each ancestral population showed good to excellent calibration (online supplemental figure 1) and varied in the ability to distinguish participants with high Lp(a) levels (AUC range: 0.66 (East Asians)–0.90 (Europeans)) (online supplemental table 9).
Figure 3.
Performance of lipoprotein(a) polygenic risk scores (PRS) by methods across study populations in n=385 263 participants of African (n=19 333), East Asian (n=2895), European (n=3 54 843) and South Asian (n=8192) ancestry from the Population Architecture through Genomics and Environment study and UK Biobank.
PRS certainty
We quantified certainty in PRS at the individual level across different PRS thresholds that might be used to identify high-risk populations (table 1). For the 90th–99th percentile thresholds, PRS assignment to the upper threshold was most certain for European participants (84.7%–94.4% of participants with 95% credible intervals fully contained in the upper threshold) and least certain for the East Asian participants (9.1%–19.6% of participants with 95% credible intervals fully contained in the upper threshold). In contrast, certainty in PRS assignment to the lower threshold was high for all populations (90th–99th percentile range: 88.6%–99.9%). We also examined an ancestry specific threshold corresponding to a mean Lp(a) ≥125 nmol/L, in African and European participants (online supplemental figure 1). This threshold occurred at the 80th percentile for both African and European participants. Approximately 94.2% of European and 65.5% of African participants with PRS assigned to the upper threshold had their 95% credible intervals fully contained in the upper threshold. Certainty in assignment to lower threshold remained high for both European (98.1%) and African (87.9%) participants.
Table 1.
PRS-based individual stratification uncertainty across four populations (European, African, South Asian and East Asian) and four thresholds in n=357 096 UK Biobank participants at study baseline
| Ancestry | PRS <percentile | PRS >percentile | ||
| Certain | Certain/N. certain+N. uncertain (SD) (%) | Certain | Certain/N. certain+N. uncertain (SD) (%) | |
| 99th percentile | ||||
| African | 7857 | 98.6 (0.1) | 31 | 38.3 (5.4) |
| East Asian | 2081 | 96.0 (0.4) | 2 | 9.1 (6.1) |
| European | 334 946 | 99.9 (0.01) | 2911 | 85.5 (0.6) |
| South Asian | 8016 | 98.8 (0.1) | 25 | 30.5 (5.1) |
| 95th percentile | ||||
| African | 7384 | 96.6 (0.2) | 239 | 59.3 (2.5) |
| East Asian | 1879 | 90.4 (0.7) | 14 | 12.7 (3.2) |
| European | 320 446 | 99.6 (0.01) | 15 986 | 94.4 (0.2) |
| South Asian | 7345 | 94.4 (0.3) | 185 | 45.1 (2.5) |
| 90th percentile | ||||
| African | 6736 | 93.0 (0.3) | 478 | 59.3 (1.7) |
| East Asian | 1746 | 88.6 (0.7) | 43 | 19.6 (2.7) |
| European | 298 579 | 98.0 (0.3) | 28 719 | 84.7 (0.2) |
| South Asian | 6683 | 90.7 (0.3) | 391 | 47.7 (1.7) |
| 80th percentile, corresponding to mean Lp(a) level of 125 nmol/L* | ||||
| African | 5660 | 87.9 (0.4) | 1055 | 65.5 (1.2) |
| European | 265 779 | 98.1 (0.03) | 63 844 | 94.2 (0.1) |
*Mean Lp(a) level for highest percentiles for East Asian ancestral population did not exceed 100 nmol/L. For South Asian populations, too few participants had PRS >125 nmol/L mean level.
Lp(a), lipoprotein(a); PRS, polygenic risk scores.
Application of PRS in external studies to identify potentially novel Lp(a) effects
A total of 23 circulatory system phenotypes were nominally associated with genetically inferred Lp(a) in at least one BioMe population (online supplemental table 10). Of note, established associations with ischaemic heart disease (IHD) and coronary atherosclerosis (CAD) only were observed for Hispanic/Latino participants (n=1400 IHD cases; n=1200 CAD cases). These observations may reflect increased statistical power in Hispanic/Latinos relative to African Americans (n=680 IHD cases; n=620 CAD cases) and whites (n=740 IHD cases; n=710 CAD cases).
Causal inference using PRS as instrumental variables
For eGFR, estimates from MR uniformly indicated no causal effect of Lp(a) (table 2) (p value range: 0.15–0.76). These results contrasted with results obtained using standard methods evaluating measured Lp(a), which instead suggested that Lp(a) was inversely and significantly associated with eGFR in European (p value =1.7×10−8), South Asian (p value =0.039) and African (p value =0.014) populations. For hbA1c, both statistical methods suggested that increased Lp(a) was associated with increased hbA1c in European ancestry participants (p values <1.4×10−6). Causal effects of Lp(a) on hbA1c suggested inverse associations in African and East Asian populations.
Table 2.
Mendelian randomisation causal estimates and standard estimates for lipoprotein(a) with glycated haemoglobin and estimated glomerular filtration rate in UK Biobank participants at study baseline by ancestral population
| Statistical method | African (n=7529) | East Asian (n=2156) | European (n=326 028) | South Asian (n=8048) | ||||
| Β (SE) | P value | Β (SE) | P value | Β (SE) | P value | Β (SE) | P value | |
| Estimated glomerular filtration rate | ||||||||
| MR | 0.019 (0.47) | 0.31 | 0.064 (0.99) | 0.15 | 0.0013 (0.0021) | 0.53 | 0.0062 (0.020) | 0.76 |
| Standard | 0.025 (0.010) | 0.014 | 0.010 (0.017) | 0.58 | 0.0086 (0.0015) | 1.7×10–8 | 0.027 (0.010) | 0.039 |
| Glycated haemoglobin | ||||||||
| MR | 0.065 (0.13) | 0.60 | 0.34 (0.23) | 0.14 | 0.044 (0.0091) | 1.4×10–6 | 0.12 (0.12) | 0.31 |
| Standard | 0.23 (0.70) | 5.0×10–3 | 0.035 (0.087) | 0.68 | 0.13 (0.0067) | 5.4×10–87 | 0.048 (0.053) | 0.36 |
MR, Mendelian randomisation.
Discussion
In this study, we demonstrated how the development and application of Lp(a) PRS in ancestrally diverse populations could help address longstanding Lp(a) evidence gaps. Importantly, studies of ancestrally diverse populations contributed unique information even when the PRS were less accurate than PRS in European populations. As new therapies for Lp(a) emerge, expanding ancestral diversity even further will help strengthen genetically informed risk prediction, enable novel biological insight and ensure that the promises of precision medicine are relevant for all populations.
Despite great interest in Lp(a) PRS and repeated calls for expanding ancestral diversity when constructing and testing PRS more generally, studies continue to prioritise European populations.22 Lp(a) is no exception. Three of five published studies that constructed Lp(a) PRS in the UK Biobank were restricted to European populations, although the UK Biobank included almost 20 000 participants of diverse ancestries.8 53 54 Of two prior studies that included African, East Asian and South Asian UK Biobank participants, Lp(a) PRS accuracy was poor to moderate (eg, R2 ranged 0.3% in Africans to 16% in South Asians55). Such low accuracy likely represents the application of discovery data and analytic methods that perform well in European population, but poorly in other populations. We built on these efforts by prioritising ancestrally diverse populations and sharing our results publicly to enable further efforts. Our results demonstrated that increasing PRS accuracy in non-European populations is feasible and, unlike PRS for many other cardiovascular phenotypes,56 does not require prohibitively large sample sizes. Indeed, inclusion of modestly sized samples of ancestry matched discovery data outperformed efforts that substituted discovery data collected in several hundred thousand participants of European ancestry, but these PRS remained considerably less accurate than results in European populations.
Adding further insight to the clinical application of Lp(a) PRS were estimates of individual PRS certainty. Certainty is an important companion metric to commonly reported population level measures like R2 that do not quantify variability in individual PRS estimates. We demonstrated that Lp(a) PRS showed promise as a ‘rule out’ for elevated Lp(a) because certainty of assignment to the low-risk threshold was high for individuals in all populations and across the four thresholds we evaluated. In contrast, while Lp(a) PRS may be useful to identify individuals of European ancestry for follow-up Lp(a) testing (ie, to ‘rule in’ potentially high Lp(a)), certainty was low for African, East Asian and South Asian participants. Improving Lp(a) PRS certainty for non-European populations assigned to high Lp(a) PRS thresholds—the individuals at highest risk of Lp(a)-associated CVD—is important because Lp(a) remains a ‘hidden CVD risk factor’. In addition to not being part of a standard lipid panel, clinical guidelines do not routinely recommend Lp(a) measurement.11 12 Even as Lp(a) measurement becomes more routine, integration of Lp(a) PRS risk thresholds into medical records or even direct to consumer testing57 could help broaden Lp(a) testing uptake in individuals with high PRS estimates. Although clinical application remains rare, Lp(a) PRS assignment and risk stratification could be performed using summary statistics and workflows, as presented herein, assuming genotypic data are available. These efforts are enabled by growing interests in merging electronic health records and genotypic data,58 the potential for Lp(a) PRS to capture lifetime elevations in Lp(a), the growing popularity of direct to consumer genetic testing, and in the USA, decreasing primary care receipt among individuals with no evident chronic medical conditions.59 Integrating Lp(a) PRS into electronic health record also could inform identification of clinical trials participants.54 Although Lp(a) presents a good ‘test case’ for PRS-informed screening given the modest to high accuracy of PRS, without additional work increasing PRS accuracy for everyone, these efforts will continue to benefit European populations primarily.
One outstanding question is how the genetic architecture of Lp(a) varies by ancestry and the phenotypic consequences of this variation. One potential major driver of Lp(a) ancestral differences are LPA kringle IV type 2 repeats (KIV-2).18 19 KIV-2 repeats are not called in array data, although methods that capture KIV-2 repeats from sequencing data are emerging.60 Because array data are more commonly available, it is unclear the degree to which these innovations will become integrated into PRS estimation. However, causal inference, particularly the development of genetic instruments for extremely low Lp(a), is one area where KIV-2 enabled PRS accuracy gains could be prioritised. Studies in European populations have not identified a strong genetic instrument for extremely low Lp(a).61 The phenotypic consequences of extremely low Lp(a) are poorly understood, although emerging therapies may reduce Lp(a) by ~80%62 and associations with increased type 2 diabetes risk have been reported.50 Like European populations, large proportions of East Asian populations harbour very low Lp(a) levels. These features, combined with marked ancestral heterogeneity in Lp(a)’s genetic architecture, motivate studies in East Asian populations that construct genetic instruments for extremely low Lp(a) and, if feasible, apply these instruments for causal inference.
East Asian populations are not the only population for Lp(a)’s ancestral heterogeneity could enable natural experiments that would be challenging to conduct in studies of European populations. For example, there is a limited understanding of how variants outside LPA regulate Lp(a).61 The genetic architecture of Lp(a) in African populations may be particularly well suited to examine this question because while the LPA locus explains almost all Lp(a) genetic variance in European populations, LPA accounts for ~50% of genetic variance in African populations.21 This finding is consistent with our observations that genome-wide Lp(a) heritability was highest in African populations and that the best performing PRS in African populations included variants outside LPA. However, African populations remain severely under-represented in Lp(a) GWAS.
Finally, we demonstrated how ancestrally diverse PRS could be applied in an external biobank to identify potentially novel associations. To date, available Lp(a) PheWAS have largely focused on cardiovascular traits in predominantly European populations.7 8 63 Continued prioritisation of European populations in Lp(a) PheWAS assumes no ancestral heterogeneity in the genetic architecture of Lp(a) or its phenotypic effects and that European populations can support well powered studies of all phenotypes of interest. Results from BioMe demonstrate how unrealistic these assumptions are. Although accuracy of the Lp(a) PRS in Hispanic/Latinos is likely not as high as accuracy in European populations due to reduced portability of European derived PRS, known associations with IHD and CAD only were identified in Hispanic/Latinos. It remains unknown how many other associations may be missed due to ongoing biases toward European populations.
Several limitations of the present study warrant consideration. First, our results may be limited by the lack of standardised methods used to measure Lp(a). However, little evidence of within-population effect heterogeneity was observed, suggesting modest influence. Second, KIV-2 repeats were not evaluated because sequencing data are not universally available. Studies have demonstrated the existence of common variants in LD with KIV-2 repeats, which captured some degree of KIV-2 repeat variability, thus reducing this concern.64 Third, while incorporating ancestry specific genetic information in developing PRS improved predictive values within each ancestral population substantially, there remained a significant imbalance in predictive performance. This limitation stresses that achieving equally informative PRS across ancestrally diverse populations will not be possible without more fundamental shifts in increasing diversity in genetic research.
In conclusion, the present study emphasises the need to expand scientific inquiry into Lp(a) through deliberate prioritisation of ancestrally diverse populations. Although we used PRS as an example of how such expansion could broaden understanding of Lp(a), we anticipate that there will be further gains from increased diversity that do not include genomics or PRS more specifically. At best, continued failure to expand Lp(a) research priorities to include ancestrally diverse populations will perpetuate Lp(a) evidence gaps. At worst, this continued failure will perpetuate health disparities by limiting the relevance of precision medicine in non-European populations.
Footnotes
Twitter: @jantonioglopez, @christy_avery
CLA and MG contributed equally.
Contributors: All authors reviewed this work and agree to publication. CLA acts as guarantor and accepts full responsibility for the work and the conduct of the study, had full access to the data, and controlled the decision to publish.
Funding: This work was supported by UK Biobank application 25953. The following grants supported this study: R01HL152828 (Avery, Ballou, Howard, North), R01HL151152 (Avery, Gignoux, Graff, North), R01HG010297, R01HG011345 (Avery, Gignoux, Graff, North), T32HL007055 (Lee) and F32HL149256 (Lee). Amgen Inc (Thousand Oaks, California) partially funded this study. The PAGE (Population Architecture Using Genomics and Epidemiology) programme is funded by the National Human Genome Research Institute with cofunding from the National Institute on Minority Health and Health Disparities and the National Heart, Lung, and Blood Institute. Assistance with data management, data integration, data dissemination, genotype imputation, ancestry deconvolution, population genetics, analysis pipelines and general study coordination was provided by the PAGE Coordinating Center (NI-HU01HG007419). Genotyping services were provided by the Center for Inherited Disease Research, which is fully funded through a federal contract from the National Institutes of Health (NIH) to The Johns Hopkins University, contract number HHSN268201200008I. Genotype data quality control and quality assurance services were provided by the Genetic Analysis Center in the Biostatistics Department of the University of Washington, through support provided by the Center for Inherited Disease Research contract. PAGE data and materials included in this report were funded through the following studies and organisations: (1) The Atherosclerosis Risk in Communities study (ARIC): The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Measurement of Lp(a) was supported by Denka Seiken. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. (2) The Coronary Artery Risk Development in Young Adults Study (CARDIA): The Coronary Artery Risk Development in Young Adults Study (CARDIA) is supported by contracts HHSN268201800003I, HHSN268201800004I, HHSN268201800005I, HHSN268201800006I and HHSN268201800007I from the National Heart, Lung, and Blood Institute (NHLBI). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). GWAS genotyping and data analyses were funded in part by grants U01-HG004729 and R01-HL093029 from the National Institutes of Health to Dr Myriam Fornage. (3) Women’s Health Initiative (WHI): The WHI programme is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts 75N92021D00001, 75N92021D00002, 75N92021D00003, 75N92021D00004, 75N92021D00005. (4) The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. (5) The MESA project is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420. Also supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute (NHLBI) grant R01HL105756. (6) BioMe: The Mount Sinai BioMe Biobank is supported by The Andrea and Charles Bronfman Philanthropies. We thank all participants and all our recruiters who have assisted and continue to assist in data collection and management. We are grateful for the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) programme was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for 'NHLBI TOPMed: Jackson Heart Study' (phs000964) was performed at the Northwest Genomics Center (HHSN268201100037C). Core support including centralised genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonisation, data management, sample-identity QC and general programme coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed. The project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR002490 (LMR). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the US Department of Health and Human Services.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data may be obtained from a third party and are not publicly available. All data are available through parent studies.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the institutional review board of the University of North Carolina at Chapel Hill (#19-2281) and complies with the Declaration of Helsinki. Participants gave informed consent to participate in the study before taking part.
References
- 1.Tsimikas S, Fazio S, Ferdinand KC, et al. NHLBI working group recommendations to reduce Lipoprotein(A)-Mediated risk of cardiovascular disease and aortic stenosis. J Am Coll Cardiol 2018;71:177–92. 10.1016/j.jacc.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ference BA, Kastelein JJP, Catapano AL. Lipids and lipoproteins in 2020. JAMA 2020;324:595–6. 10.1001/jama.2020.5685 [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow DI, Rider DA, Yavari A, et al. Treatment and prevention of Lipoprotein(A)-Mediated cardiovascular disease: the emerging potential of RNA interference Therapeutics. Cardiovasc Res 2022;118:1218–31. 10.1093/cvr/cvab100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioannidou A, Watts EL, Perez-Cornago A, et al. The relationship between lipoprotein A and other lipids with prostate cancer risk: A multivariable Mendelian Randomisation study. PLoS Med 2022;19:e1003859. 10.1371/journal.pmed.1003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langsted A, Kamstrup PR, Nordestgaard BG. High Lipoprotein(A) and low risk of major bleeding in brain and Airways in the general population: a Mendelian randomization study. Clin Chem 2017;63:1714–23. 10.1373/clinchem.2017.276931 [DOI] [PubMed] [Google Scholar]
- 6.Fanshawe AE, Ibrahim M. The current status of lipoprotein (a) in pregnancy: a literature review. J Cardiol 2013;61:99–106. 10.1016/j.jjcc.2012.09.009 [DOI] [PubMed] [Google Scholar]
- 7.Satterfield BA, Dikilitas O, Safarova MS, et al. Associations of genetically predicted Lp(A) (lipoprotein [a]) levels with cardiovascular traits in individuals of European and African ancestry. Circ Genom Precis Med 2021;14:e003354. 10.1161/CIRCGEN.120.003354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emdin CA, Khera AV, Natarajan P, et al. Phenotypic characterization of genetically lowered human Lipoprotein(A) levels. J Am Coll Cardiol 2016;68:2761–72. 10.1016/j.jacc.2016.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diogo D, Tian C, Franklin CS, et al. Phenome-wide Association studies across large population cohorts support drug target validation. Nat Commun 2018;9:4285. 10.1038/s41467-018-06540-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rastegar-Mojarad M, Ye Z, Kolesar JM, et al. Opportunities for drug repositioning from Phenome-wide Association studies. Nat Biotechnol 2015;33:342–5. 10.1038/nbt.3183 [DOI] [PubMed] [Google Scholar]
- 11.Authors/Task Force M, Guidelines ESCCfP, Societies ESCNC . 2019 ESC/EAS guidelines for the management of Dyslipidaemias: lipid modification to reduce cardiovascular risk. Atherosclerosis 2019;290:140–205. [DOI] [PubMed] [Google Scholar]
- 12.Cegla J, France M, Marcovina SM, et al. Lp(A): when and how to measure it. Ann Clin Biochem 2021;58:16–21. 10.1177/0004563220968473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraft HG, Lingenhel A, Pang RW, et al. Frequency distributions of Apolipoprotein(A) Kringle IV repeat Alleles and their effects on Lipoprotein(A) levels in Caucasian, Asian, and African populations: the distribution of null Alleles is non-random. Eur J Hum Genet 1996;4:74–87. 10.1159/000472175 [DOI] [PubMed] [Google Scholar]
- 14.Sandholzer C, Hallman DM, Saha N, et al. Effects of the Apolipoprotein(A) size polymorphism on the Lipoprotein(A) concentration in 7 ethnic groups. Hum Genet 1991;86:607–14. 10.1007/BF00201550 [DOI] [PubMed] [Google Scholar]
- 15.Reyes-Soffer G, Ginsberg HN, Berglund L, et al. Lipoprotein(A): A genetically determined, causal, and prevalent risk factor for Atherosclerotic cardiovascular disease: A scientific statement from the American heart Association. ATVB 2022;42. 10.1161/ATV.0000000000000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S, Ference BA, Staley JR, et al. Association of LPA variants with risk of coronary disease and the implications for Lipoprotein(A)-Lowering therapies: A Mendelian randomization analysis. JAMA Cardiol 2018;3:619–27. 10.1001/jamacardio.2018.1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enkhmaa B, Anuurad E, Zhang W, et al. Heritability of apolipoprotein (a) traits in two-generational African-American and Caucasian families. J Lipid Res 2019;60:1603–9. 10.1194/jlr.P091249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zekavat SM, Ruotsalainen S, Handsaker RE, et al. Deep coverage whole genome sequences and plasma Lipoprotein(A) in individuals of European and African Ancestries. Nat Commun 2018;9:2606. 10.1038/s41467-018-04668-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kronenberg F, Utermann G. Lipoprotein(A): resurrected by Genetics. J Intern Med 2013;273:6–30. 10.1111/j.1365-2796.2012.02592.x [DOI] [PubMed] [Google Scholar]
- 20.Xie T, Wang B, Nolte IM, et al. Genetic risk scores for complex disease traits in youth. Circ Genom Precis Med 2020;13:e002775. 10.1161/CIRCGEN.119.002775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholz M, Kraft HG, Lingenhel A, et al. Genetic control of Lipoprotein(A) concentrations is different in Africans and Caucasians. Eur J Hum Genet 1999;7:169–78. 10.1038/sj.ejhg.5200290 [DOI] [PubMed] [Google Scholar]
- 22.Martin AR, Kanai M, Kamatani Y, et al. Clinical use of current Polygenic risk scores may exacerbate health disparities. Nat Genet 2019;51:584–91. 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538:161–4. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert SA, Gil L, Jupp S, et al. The Polygenic score catalog as an open database for reproducibility and systematic evaluation. Nat Genet 2021;53:420–5. 10.1038/s41588-021-00783-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matise TC, Ambite JL, Buyske S, et al. The next PAGE in understanding complex traits: design for the analysis of population architecture using Genetics and epidemiology (PAGE) study. Am J Epidemiol 2011;174:849–59. 10.1093/aje/kwr160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.THE ARIC INVESTIGATORS . The Atherosclerosis risk in communities (ARIC) study: design and objectives. The ARIC investigators. Am J Epidemiol 1989;129:687–702. 10.1093/oxfordjournals.aje.a115184 [DOI] [PubMed] [Google Scholar]
- 27.Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 1988;41:1105–16. 10.1016/0895-4356(88)90080-7 [DOI] [PubMed] [Google Scholar]
- 28.Taylor HA, Wilson JG, Jones DW, et al. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson heart study. Ethn Dis 2005;15:S6–4. [PubMed] [Google Scholar]
- 29.Bild DE, Bluemke DA, Burke GL, et al. Multi-ethnic study of Atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–81. 10.1093/aje/kwf113 [DOI] [PubMed] [Google Scholar]
- 30.Design of the women’s health initiative clinical trial and observational study. Controlled Clinical Trials 1998;19:61–109. 10.1016/S0197-2456(97)00078-0 [DOI] [PubMed] [Google Scholar]
- 31.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D-Y, Tao R, Kalsbeek WD, et al. Genetic Association analysis under complex survey sampling: the Hispanic community health study/study of Latinos. Am J Hum Genet 2014;95:675–88. 10.1016/j.ajhg.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide Association studies. Nat Genet 2010;42:348–54. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchini J, Howie B, Myers S, et al. A new Multipoint method for genome-wide Association studies by imputation of Genotypes. Nat Genet 2007;39:906–13. 10.1038/ng2088 [DOI] [PubMed] [Google Scholar]
- 35.Zhou W, Nielsen JB, Fritsche LG, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic Association studies. Nat Genet 2018;50:1335–41. 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide Association meta-analyses. Nat Protoc 2014;9:1192–212. 10.1038/nprot.2014.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler TW, Kutalik Z, Gorski M, et al. Easystrata: evaluation and visualization of stratified genome-wide Association meta-analysis data. Bioinformatics 2015;31:259–61. 10.1093/bioinformatics/btu621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of Genomewide Association scans. Bioinformatics 2010;26:2190–1. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fadista J, Manning AK, Florez JC, et al. The (In)Famous GWAS P-value threshold Revisited and updated for low-frequency variants. Eur J Hum Genet 2016;24:1202–5. 10.1038/ejhg.2015.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary Statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–75, 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the Heritability for human height. Nat Genet 2010;42:565–9. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trinder M, Uddin MM, Finneran P, et al. Clinical utility of Lipoprotein(A) and LPA genetic risk score in risk prediction of incident Atherosclerotic cardiovascular disease. JAMA Cardiol 2020;6:1–9. 10.1001/jamacardio.2020.5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Euesden J, Lewis CM, O’Reilly PF. Prsice: Polygenic risk score software. Bioinformatics 2015;31:1466–8. 10.1093/bioinformatics/btu848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Privé F, Arbel J, Vilhjálmsson BJ. Ldpred2: better, faster, stronger. Genetics 2020. 10.1101/2020.04.28.066720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mak TSH, Porsch RM, Choi SW. n.d. Polygenic scores for UK Biobank scale data. Genomics 10.1101/252270 [DOI] [Google Scholar]
- 47.Grundy SM, Stone NJ, Bailey AL, et al. AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/Apha/ASPC/NLA/PCNA guideline on the management of blood cholesterol: A report of the American college of cardiology/American heart Association task force on clinical practice guidelines. Circulation 2019;139:e1082–143. 10.1161/CIR.0000000000000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding Y, Hou K, Burch KS, et al. Large uncertainty in individual Polygenic risk score estimation impacts PRS-based risk stratification. Nat Genet 2022;54:30–9. 10.1038/s41588-021-00961-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Core Team . A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2021. [Google Scholar]
- 50.Gudbjartsson DF, Thorgeirsson G, Sulem P, et al. Lipoprotein(A) concentration and risks of cardiovascular disease and diabetes. J Am Coll Cardiol 2019;74:2982–94. 10.1016/j.jacc.2019.10.019 [DOI] [PubMed] [Google Scholar]
- 51.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian Randomisation studies: a guide, glossary, and checklist for Clinicians. BMJ 2018;362:k601. 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawlor DA, Harbord RM, Sterne JAC, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 53.Wu H, Luan J, Forgetta V, et al. Utility of genetically predicted Lp(A) (lipoprotein [a]) and Apob levels for cardiovascular risk assessment. Circ Genom Precis MED 2021:Circgen121003312. Circ Genom Precis Med 2021;14:e003312. 10.1161/CIRCGEN.121.003312 [DOI] [PubMed] [Google Scholar]
- 54.Dron JS, Wang M, Patel AP, et al. Genetic Predictor to identify individuals with high Lipoprotein(A) concentrations. Circ Genom Precis Med 2021;14:e003182. 10.1161/CIRCGEN.120.003182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinnott-Armstrong N, Tanigawa Y, Amar D, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet 2021;53:185–94.:1622. 10.1038/s41588-021-00956-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Sullivan JW, Raghavan S, Marquez-Luna C, et al. Polygenic risk scores for cardiovascular disease: A scientific statement from the American heart Association. Circulation 2022;146:e93–118. 10.1161/CIR.0000000000001077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elson SL, Furlotte NA, Hromatka BS, et al. Direct-to-consumer genetic testing for factor V Leiden and prothrombin 20210G>A: the consumer experience. Mol Genet Genomic Med 2020;8:e1468. 10.1002/mgg3.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall JL, Ryan JJ, Bray BE, et al. Merging electronic health record data and Genomics for cardiovascular research: A science advisory from the American heart Association. Circ Cardiovasc Genet 2016;9:193–202. 10.1161/HCG.0000000000000029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levine DM, Linder JA, Landon BE. Characteristics of Americans with primary care and changes over time, 2002-2015. JAMA Intern Med 2020;180:463–6. 10.1001/jamainternmed.2019.6282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukamel RE, Handsaker RE, Sherman MA, et al. Protein-coding repeat Polymorphisms strongly shape diverse human phenotypes. Science 2021;373:1499–505. 10.1126/science.abg8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coassin S, Kronenberg F. Lipoprotein(A) beyond the Kringle IV repeat polymorphism: the complexity of genetic variation in the LPA gene. Atherosclerosis 2022;349:17–35. 10.1016/j.atherosclerosis.2022.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsimikas S, Karwatowska-Prokopczuk E, Gouni-Berthold I, et al. Lipoprotein(A) reduction in persons with cardiovascular disease. N Engl J Med 2020;382:244–55. 10.1056/NEJMoa1905239 [DOI] [PubMed] [Google Scholar]
- 63.Kettunen J, Demirkan A, Würtz P, et al. Genome-wide study for circulating metabolites identifies 62 Loci and reveals novel systemic effects of LPA. Nat Commun 2016;7:11122. 10.1038/ncomms11122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanktree MB, Anand SS, Yusuf S, et al. Comprehensive analysis of Genomic variation in the LPA locus and its relationship to plasma Lipoprotein(A) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet 2010;3:39–46. 10.1161/CIRCGENETICS.109.907642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
openhrt-2023-002382supp001.pdf (905.4KB, pdf)
Data Availability Statement
Data may be obtained from a third party and are not publicly available. All data are available through parent studies.