Abstract
No clear conclusions have yet been reached regarding the accuracy of magnetic resonance imaging (MRI) radiomics in distinguishing high-grade glioma (HGG) from low-grade glioma (LGG). In the present study, a meta-analysis was conducted to determine the diagnostic value of MRI radiomics in differentiating between HGG and LGG, in order to guide their clinical diagnosis. PubMed, Embase and the Cochrane Library databases were searched up to November 2022. The search included studies in which true positive, false positive, true negative and false negative values for the differentiation of HGG from LGG were reported or could be calculated by retrograde extrapolation. Duplicate publications, research without full text, studies with incomplete information or unextractable data, animal studies, reviews and systematic reviews were excluded. STATA 15.1 was used to analyze the data. The meta-analysis included 15 studies, which comprised a total of 1,124 patients, of which 701 had HGG and 423 had LGG. The pooled sensitivity and specificity of the studies overall were 0.92 (95% CI: 0.89–0.95) and 0.89 (95% CI: 0.85–0.92), respectively. The positive and negative likelihood ratios of the studies overall were 7.89 (95% CI: 6.01–10.37) and 0.09 (95% CI: 0.07–0.12), respectively. The pooled diagnostic odds ratio of the studies was 85.20 (95% CI: 54.52–133.14). The area under the summary receiver operating characteristic curve was 0.91. These findings indicate that radiomics may be an accurate tool for the differentiation of glioma grades. However, further research is needed to verify the most appropriate of these technologies.
Keywords: magnetic resonance imaging radiomics, high-grade glioma, low-grade glioma, diagnostic value, meta-analysis
Introduction
Glioma is the most commonly occurring primary malignant brain tumor, with a frequency of 80% of all malignant brain cancers (1). The World Health Organization classifies gliomas into grades I–IV, of which grades I and II are defined as low-grade gliomas (LGGs) and grades III and IV as high-grade gliomas (HGGs) (1). LGGs have a long survival period, whereas HGGs are highly aggressive and have a poor prognosis (2). Most gliomas are treated with surgery, and HGGs often require adjuvant radiation therapy or chemotherapy after surgery to prevent short-term recurrence, whereas LGGs require close observation (3). Therefore, it is important to accurately grade tumors prior to surgery. Pathology is currently the gold standard for glioma grading; however, it is an invasive method and usually performed postoperatively. A growing body of research has focused on noninvasive methods for accurate tumor grading.
Magnetic resonance imaging (MRI) is an important method for the diagnosis of gliomas, and a variety of MRI techniques, including magnetic resonance spectroscopy (MRS), diffusion-weighted imaging (DWI) and pulse-weighted imaging, are used to grade gliomas (4,5). Diagnostic imaging relies heavily on the subjective experience the radiologist has with imaging data. By contrast, radiomics involves the transformation of medical images into quantitative, extractable and high-dimensional data, which include histograms, texture features and shape features (6). Radiomics has been primarily used in tumor research and has gradually become a tool for the extraction of diagnostic and prognostic information from conventional images. There has been a considerable amount of research on the association between MRI radiomics and lesion features, survival and perioperative outcomes in various malignancies (7). However, no clear conclusions have yet been reached regarding the accuracy of MRI radiomics in distinguishing HGG from LGG. Therefore, MRI radiomics has not been widely used in clinical practice. To the best of our knowledge, the present study is the first meta-analysis to examine the diagnostic value of MRI radiomics in the differentiation of HGG from LGG to guide the clinical diagnosis of glioma.
Materials and methods
Literature inclusion and exclusion criteria
The inclusion criteria were as follows: i) Retrospective or prospective studies evaluating the efficacy of radiomics in differentiating between LGG and HGG; ii) use of histopathology as the gold standard; and iii) the true-positive (TP), true-negative (TN), false-positive (FP) and false-negative (FN) values were either stated directly or could be indirectly extracted from the retrieved literature.
The exclusion criteria were as follows: i) small sample size (n<10), animal experiments, reviews, systematic reviews, case reports and conference papers; ii) studies for which no data were available; and iii) duplicate reports or studies based on the same data.
Search strategy
PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase (https://www.embase.com/) and the Cochrane Library (https://www.cochranelibrary.com/) were used in this meta-analysis. They were searched between their formation and November 2022. The search terms were: (‘glioma’ OR ‘gliomas’) AND (‘radiomics’ OR ‘texture features’ OR ‘texture analysis’ OR ‘histogram’) AND (‘magnetic resonance imaging’ OR ‘magnetic resonance image’ OR ‘MRI’).
Literature screening and data extraction
Two researchers conducted the literature search, screened the literature and extracted relevant materials. When questions or conflicts arose, a third individual was consulted prior to a decision being made. The data extraction content included the author, year of publication, sample size, sex, age and the values of TP, FP, TN and FN used in the differentiation of HGG from LGG and the definition of HGG as positive and LGG as negative. If no TP, FP, TN or FN results were reported, data such as sensitivity, specificity and positive and negative predictive values were subjected to retrograde extrapolation to calculate these results.
Literature quality assessment
The QUADAS-2 tool for evaluating the quality of published literature (8) was used separately by two academics, and RevMan 5.3 (Cochrane) was used to construct a quality evaluation map.
Data synthesis and statistical analysis
The Cochran Q test and I2 value were used to investigate heterogeneity among studies, and meta-analyses were performed using a random-effects model when significant heterogeneity was identified (I2>50%, P<0.05) and a fixed-effects model when no significant heterogeneity was detected. Sensitivity analysis was performed by eliminating each included study one by one, and performing a summary analysis of the remaining studies. The ROC curve is plotted with sensitivity as the vertical coordinate and (1-specificity) as the horizontal coordinate. The larger the area under the curve (AUC), the higher the diagnostic accuracy. AUC values were calculated and were considered to indicate the following: 0.5<AUC≤0.6, ineffective; 0.6<AUC≤0.7, poor; 0.7<AUC≤0.8, average; 0.8<AUC≤0.9, good; and 0.9<AUC≤1.0, excellent. A Deeks' funnel plot was generated using Stata 15.1 software (StataCorp LP) to assess publication bias.
Results
Literature search
The database yielded 1,349 studies on this topic which were reduced to 695 following the exclusion of duplicates. Of these, 442 studies were selected after the examination of titles and abstracts indicated studies that included animal experiments (n=45), or were meta-analyses, reviews, systemic reviews (n=116), case reports (n=53) or conference reports (n=39). After reading the full text, 359 studies that did not report the outcomes of interest and 68 studies for which it was not possible to construct 2×2 tables were excluded. Finally, 15 papers that were read in their entirety were subjected to meta-analysis (Fig. 1).
Figure 1.
Flow diagram for the selection of studies.
Baseline characteristics and quality assessment of the included studies
Baseline characteristics
The 15 publications evaluated in the present meta-analysis included a total of 1,124 patients, of which 701 had HGG and 423 had LGG. The age of the patients in the HGG group ranged from 43.0 to 66.0 years, while the age of the patients in the LGG group ranged from 34.6 to 63.2 years, which was considered comparable. Seven studies included patients from Asia, whereas the remaining studies included patients from Europe and the USA (Table I) (9–23).
Table I.
Baseline characteristics of the included studies.
| Sample size | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| First author/s, year | Country | HGG | LGG | Age, years | Sex, male/female | TP | FP | FN | TN | Sensitivity (%) | Specificity (%) | (Refs.) |
| Law et al, 2007 | USA | 61 | 31 | 43.0 (4.0–85.0) | 61/31 | 58 | 4 | 3 | 27 | 95.1 | 87.0 | (9) |
| Jakab et al, 2011 | Hungary | 14 | 26 | HGG, 47.3±15.4; LGG, 34.6±15.9 | HGG, 5/9; LGG, 13/13 | 12 | 3 | 2 | 23 | 85.7 | 88.5 | (10) |
| Kang et al, 2011 | Korea | 21 | 6 | - | - | 18 | 0 | 3 | 6 | 85.7 | 100.0 | (11) |
| Kim et al, 2013 | Korea | 54 | 9 | - | - | 46 | 0 | 8 | 9 | 85.2 | 100.0 | (12) |
| Santarosa et al, 2016 | Italy | 17 | 9 | 55.4 (22.0–79.0) | 15/11 | 17 | 0 | 0 | 9 | 100.0 | 100.0 | (13) |
| Skogen et al, 2016 | Norway | 68 | 27 | HGG, 44.0 | - | 63 | 5 | 5 | 22 | 93.0 | 81.0 | (14) |
| (21.0–70.0); LGG, 56.0 (24.0–81.0) | ||||||||||||
| Hsieh et al, 2017 | China | 34 | 73 | - | - | 33 | 6 | 1 | 67 | 97.0 | 92.0 | (15) |
| Cho and Park, 2017 | Korea | 54 | 54 | - | - | 48 | 5 | 6 | 49 | 89.0 | 91.0 | (16) |
| Bisdas et al, 2018 | UK | 18 | 19 | 63.2±7.6 | 21/16 | 14 | 4 | 4 | 15 | 77.0 | 79.0 | (17) |
| Ditmer et al, 2018 | USA | 80 | 14 | 52.0 (10.0–84.0) | 59/35 | 74 | 2 | 6 | 12 | 93.0 | 86.0 | (18) |
| Su et al, 2018 | China | 30 | 10 | - | - | 29 | 1 | 1 | 9 | 96.7 | 90.0 | (19) |
| Tian et al, 2018 | China | 111 | 42 | HGG, 51.2±13.3; LGG, 39.0±13.2 | HGG, 63/48; LGG, 18/24 | 108 | 5 | 3 | 37 | 97.3 | 88.1 | (20) |
| Alis et al, 2020 | Turkey | 32 | 28 | HGG, 66.0 | - | 28 | 3 | 4 | 25 | 87.5 | 89.2 | (21) |
| (33.0–78.0); LGG, 46.0 (27.0–68.0) | ||||||||||||
| Zhang et al, 2020 | China | 65 | 43 | HGG, 51.3 (21.0–78.0); LGG, 43.2 (24.0–66.0) | HGG, 36/29; LGG, 25/18 | 62 | 5 | 3 | 38 | 95.0 | 88.0 | (22) |
| Malik et al, 2021 | USA | 42 | 32 | HGG, 62.0 (46.0–79.0); LGG, 41.0 (22.0–71.0) | HGG, 28/14; LGG, 24/12 | 38 | 4 | 4 | 28 | 91.0 | 86.0 | (23) |
Values are presented as the mean ± SD or median (quartile). HGG, high-grade glioma; LGG, T low-grade glioma; P, true-positive; FP, false-positive; FN, false-negative; TN, true-negative.
Quality assessment
In the assessment of patient selection, only one study was found to have a high risk of bias and the remainder had an unclear or low risk of bias. In the remaining three QUADAS-2 domains, namely index test, reference standard, flow and timing, most studies were found to be of low risk. Patient selection, index test and reference standard were also evaluated for applicability concerns. With regard to the reference standard domain, one study was high-risk and the rest were unclear or low risk. As for applicability concern, only one study showed high risk in ‘patient selection’ and no high risk in ‘index test’. Overall, the quality of the studies included in this review was indicated to be acceptable (Figs. 2 and 3).
Figure 2.

Methodological quality summary graphs.
Figure 3.
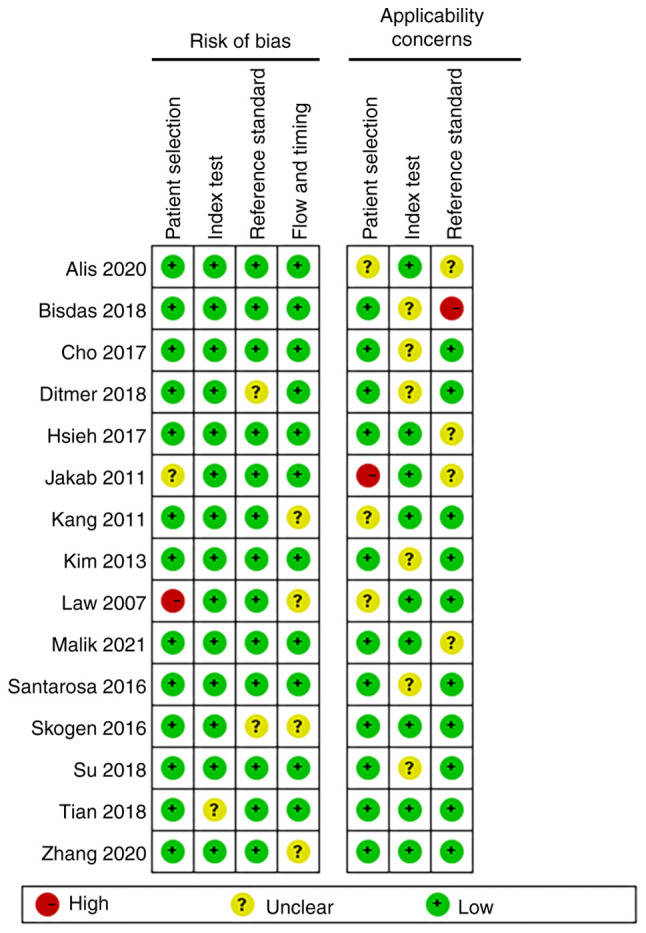
Methodological quality of individual studies.
Sensitivity and specificity
A meta-analysis was performed using a fixed-effects model due to the low heterogeneity in sensitivity (I2=42.08%) and specificity (I2=0.00%). The pooled sensitivity and specificity of the studies overall were 0.92 (95% CI: 0.89–0.95) and 0.89 (95% CI: 0.85–0.92), respectively (Fig. 4).
Figure 4.
Forest plot of the sensitivity and specificity of magnetic resonance imaging radiomics for the diagnosis of glioma. Q, Cochran's Q; df, degrees of freedom.
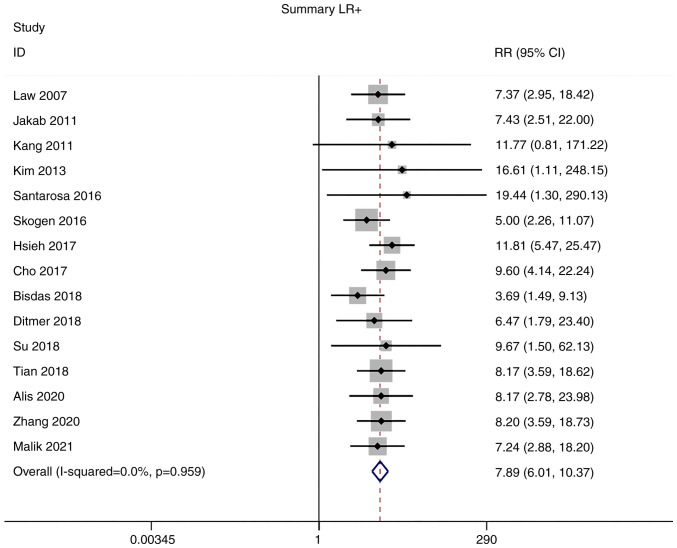
Positive and negative likelihood ratios (LRs)
Owing to the low heterogeneity in positive LR (I2=0.0%) and negative LR (I2=40.1%), meta-analyses were performed using a fixed-effects model. The pooled positive and negative LRs of the studies overall were 7.89 (95% CI: 6.01–10.37) and 0.09 (95% CI: 0.07–0.12), respectively (Figs. 5 and 6).
Figure 5.
Forest plot of the LR+ of magnetic resonance imaging radiomics in the diagnosis of glioma. LR+, positive likelihood ratio.
Figure 6.
Forest plot of the LR- of magnetic resonance imaging radiomics in the diagnosis of glioma. LR, negative likelihood ratio.
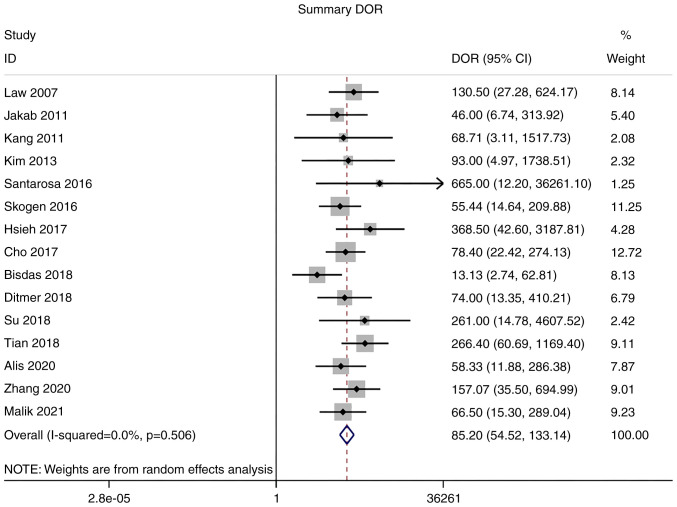
Diagnostic odds ratio (DOR)
As there was no significant heterogeneity in DOR (I2=0.0%), a meta-analysis of DOR was conducted using a fixed-effects model. The overall pooled DOR of the studies was 85.20 (95% CI: 54.52–133.14; Fig. 7).
Figure 7.
Forest plot of the DOR of magnetic resonance imaging radiomics for the diagnosis of glioma. Diagnostic odds ratio.
Summary receiver operating characteristic (SROC) curve analysis
An SROC curve analysis was performed, and the AUC of the SROC curve was calculated to be 0.91. This indicates the high diagnostic value of MRI radiomics for gliomas (Fig. 8).
Figure 8.

SROC curve of magnetic resonance imaging radiomics for the diagnosis of glioma. SROC, summary receiver operating characteristic; SENS, sensitivity; SPEC, specificity; AUC, area under the curve.
Publication bias
The Deeks' funnel plot for the studies of MRI radiomics in the diagnosis of glioma had a P-value of 0.36, indicating no significant publication bias in the studies included in the present meta-analysis (Fig. 9).
Figure 9.
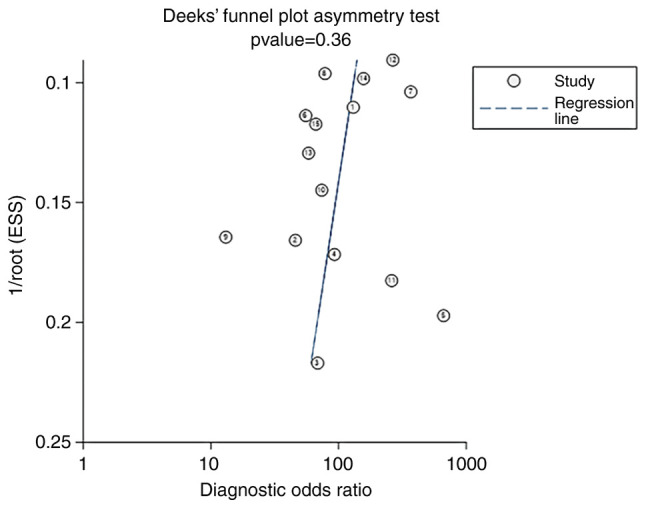
Deeks' funnel plot of magnetic resonance imaging radiomics for the diagnosis of glioma. ESS, effective sample size.
Sensitivity analysis
The 15 studies were subjected to a pooled analysis to determine whether any of the included studies had a disproportionate influence on the meta-analysis results. This was accomplished using sensitivity analyses that eliminated each study individually. According to the meta-analysis, no specific study substantially influenced the results, suggesting that the findings were consistent and credible (Fig. 10).
Figure 10.

Sensitivity of magnetic resonance imaging radiomics for the diagnosis of glioma.
Discussion
Histopathology is the gold standard for the diagnosis of gliomas; however, it is an invasive method in which tissue collection is necessary and can be harmful. However, MRI is also able to provide accurate information while avoiding unnecessary surgery. With the development of technology, an increasing number of metabolic and physiological MRI techniques, including diffusion tensor imaging, MRS, DWI and MRI are being used for glioma grading. These tests assess the malignancy of a tumor by identifying differences in the grayscale brightness and contrast of the pixels in an image (6). Radiomics uses tools such as MaZda, MATLAB, TexRAD, MISSTA, CAD and FireVoxel to identify new quantitative imaging markers without the need for additional acquisition equipment or tracers, thereby providing greater diagnostic capabilities than commonly used examination methods (24). In the present study, a meta-analysis was performed to systematically review the accuracy of radiomics in differentiating between LGG and HGG.
The pooled sensitivity and specificity of the studies overall were 0.92 (95% CI: 0.89–0.95) and 0.89 (95% CI: 0.85–0.92), respectively. Although the result of this study suggested that specificity was lower than sensitivity, a specificity of 0.89 indicated a low probability of missed diagnosis. In addition, positive and negative LRs of the studies were 7.89 (95% CI: 6.01–10.37) and 0.09 (95% CI: 0.07–0.12), respectively. The pooled DOR of the studies was 85.20 (95% CI: 54.52–133.14) and the area under the ROC curve was 0.91. AUC values were calculated an AUC of 0.9–1.0 was considered an ‘excellent’ diagnostic accuracy. Thus, an AUC of 0.91 suggests that MRI radiomics has high accuracy in distinguishing high-grade glioma from low-grade glioma. These results demonstrate that radiomics has high diagnostic value for glioma grading. A previous meta-analysis by Sohn and Bisdas (25) explored the diagnostic accuracy of machine learning-based radiomics in grading gliomas, and the pooled results also showed that sensitivity when diagnosing HGG was higher (96%; 95% CI: 0.93–0.98) than the specificity when diagnosing LGG (90%; 95% CI: 0.85-.93) (25). By contrast, the present study focuses on MRI-based imaging radiomics in grading gliomas. The results of this study further complement those of previous studies and highlight the diagnostic value of MRI-based imaging radiomics. A number of studies have shown the good prospects of radiomics for the resolution of clinical issues that cannot be addressed using conventional radiological diagnosis, and indicate that radiomics has stronger diagnostic capabilities than ordinary examination methods, suggesting that further consideration should be given to the standardization of its use in clinical practice (21,26,27). However, current studies rarely provide open access to the coding used for imaging data, feature extraction and model building, which makes it difficult to obtain open MRI radiomics data for secondary analysis and validation. In addition, if the sample size of the study is limited, it has been recommended that the number of feature parameters should be reduced to reduce the risk of overfitting (28).
The present review has certain limitations. First, most of the included studies were retrospective. Second, the majority of the studies were single-center studies involving patients who had undergone surgery, and there may have been admission and selection biases.
In conclusion, the present meta-analysis indicates that radiomics may be an accurate tool for the differentiation of glioma grades; however, further research is required to verify the most appropriate of these technologies.
Acknowledgements
Not applicable.
Funding Statement
Funding: Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JW conceived the study and wrote the manuscript. ZC and JC participated in searching and screening the studies, and performing data analysis. JW and JC confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Helseth R, Helseth E, Johannesen TB, Langberg CW, Lote K, Rønning P, Scheie D, Vik A, Meling TR. Overall survival, prognostic factors, and repeated surgery in a consecutive series of 516 patients with glioblastoma multiforme. Acta Neurol Scand. 2010;122:159–167. doi: 10.1111/j.1600-0404.2010.01350.x. [DOI] [PubMed] [Google Scholar]
- 3.Woodworth GF, McGirt MJ, Samdani A, Garonzik I, Olivi A, Weingart JD. Frameless image-guided stereotactic brain biopsy procedure: Diagnostic yield, surgical morbidity, and comparison with the frame-based technique. J Neurosurg. 2006;104:233–237. doi: 10.3171/jns.2006.104.2.233. [DOI] [PubMed] [Google Scholar]
- 4.Ryu YJ, Choi SH, Park SJ, Yun TJ, Kim JH, Sohn CH. Glioma: Application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS One. 2014;9:e108335. doi: 10.1371/journal.pone.0108335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson A, O'Connor JP, Parker GJ, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clin Cancer Res. 2007;13:3449–3459. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 6.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: Definitions, applications, biologic correlates, and challenges. Radiographics. 2017;37:1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Mulder F, Leeflang M, Wolff R, Whiting P, Bossuyt PM. QUAPAS: An adaptation of the QUADAS-2 tool to assess prognostic accuracy studies. Ann Intern Med. 2022;175:1010–1018. doi: 10.7326/M22-0276. [DOI] [PubMed] [Google Scholar]
- 9.Law M, Young R, Babb J, Pollack E, Johnson G. Histogram analysis versus region of interest analysis of dynamic susceptibility contrast perfusion MR imaging data in the grading of cerebral gliomas. AJNR Am J Neuroradiol. 2007;28:761–766. [PMC free article] [PubMed] [Google Scholar]
- 10.Jakab A, Molnár P, Emri M, Berényi E. Glioma grade assessment by using histogram analysis of diffusion tensor imaging-derived maps. Neuroradiology. 2011;53:483–491. doi: 10.1007/s00234-010-0769-3. [DOI] [PubMed] [Google Scholar]
- 11.Kang Y, Choi SH, Kim YJ, Kim KG, Sohn CH, Kim JH, Yun TJ, Chang KH. Gliomas: Histogram analysis of apparent diffusion coefficient maps with standard-or high-b-value diffusion-weighted MR imaging-correlation with tumor grade. Radiology. 2011;261:882–890. doi: 10.1148/radiol.11110686. [DOI] [PubMed] [Google Scholar]
- 12.Kim H, Choi SH, Kim JH, Ryoo I, Kim SC, Yeom JA, Shin H, Jung SC, Lee AL, Yun TJ, et al. Gliomas: Application of cumulative histogram analysis of normalized cerebral blood volume on 3 T MRI to tumor grading. PLoS One. 2013;8:e63462. doi: 10.1371/journal.pone.0063462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santarosa C, Castellano A, Conte GM, Cadioli M, Iadanza A, Terreni MR, Franzin A, Bello L, Caulo M, Falini A, Anzalone N. Dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging for glioma grading: Preliminary comparison of vessel compartment and permeability parameters using hotspot and histogram analysis. Eur J Radiol. 2016;85:1147–1156. doi: 10.1016/j.ejrad.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol. 2016;85:824–829. doi: 10.1016/j.ejrad.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh KLC, Chen CY, Lo CM. Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med. 2017;83:102–108. doi: 10.1016/j.compbiomed.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Cho HH, Park H. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:3081–3084. doi: 10.1109/EMBC.2017.8037508. [DOI] [PubMed] [Google Scholar]
- 17.Bisdas S, Shen H, Thust S, Katsaros V, Stranjalis G, Boskos C, Brandner S, Zhang J. Texture analysis-and support vector machine-assisted diffusional kurtosis imaging may allow in vivo gliomas grading and IDH-mutation status prediction: A preliminary study. Sci Rep. 2018;8:6108. doi: 10.1038/s41598-018-24438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ditmer A, Zhang B, Shujaat T, Pavlina A, Luibrand N, Gaskill-Shipley M, Vagal A. Diagnostic accuracy of MRI texture analysis for grading gliomas. J Neurooncol. 2018;140:583–589. doi: 10.1007/s11060-018-2984-4. [DOI] [PubMed] [Google Scholar]
- 19.Su CQ, Lu SS, Han QY, Zhou MD, Hong XN. Intergrating conventional MRI, texture analysis of dynamic contrast-enhanced MRI, and susceptibility weighted imaging for glioma grading. Acta Radiol. 2019 Jun;60((6)):777–787. doi: 10.1177/0284185118801127. [DOI] [PubMed] [Google Scholar]
- 20.Tian Q, Yan LF, Zhang X, Zhang X, Hu YC, Han Y, Liu ZC, Nan HY, Sun Q, Sun YZ, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. 2018;48:1518–1528. doi: 10.1002/jmri.26010. [DOI] [PubMed] [Google Scholar]
- 21.Alis D, Bagcilar O, Senli YD, Isler C, Yergin M, Kocer N, Islak C, Kizilkilic O. The diagnostic value of quantitative texture analysis of conventional MRI sequences using artificial neural networks in grading gliomas. Clin Radiol. 2020;75:351–357. doi: 10.1016/j.crad.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Xiao J, Wu S, Lv F, Gong J, Jiang L, Yu R, Luo T. Deep convolutional radiomic features on diffusion tensor images for classification of glioma grades. J Digit Imaging. 2020;33:826–837. doi: 10.1007/s10278-020-00322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik N, Geraghty B, Dasgupta A, Maralani PJ, Sandhu M, Detsky J, Tseng CL, Soliman H, Myrehaug S, Husain Z, et al. MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J Neurooncol. 2021;155:181–191. doi: 10.1007/s11060-021-03866-9. [DOI] [PubMed] [Google Scholar]
- 24.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn CK, Bisdas S. Diagnostic accuracy of machine learning-based radiomics in grading gliomas: systematic review and meta-analysis. Contrast Media Mol Imaging. 2020;2020:2127062. doi: 10.1155/2020/2127062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Zhao W, Xie J, Lin H, Hu X, Li C, Shang Y, Wang Y, Jiang Y, Ding M, et al. Development and validation of a radiomics-based nomogram for predicting a major pathological response to neoadjuvant immunochemotherapy for patients with potentially resectable non-small cell lung cancer. Front Immunol. 2023;14:1115291. doi: 10.3389/fimmu.2023.1115291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Xia F, Wang X, Jin Y, Yan J, Wei X, Zhao Q. Multiclassifier radiomics analysis of ultrasound for prediction of extrathyroidal extension in papillary thyroid carcinoma in children. Int J Med Sci. 2023;20:278–286. doi: 10.7150/ijms.79758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, Sabater S. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.