Abstract
Citrobacter freundii is characterized by AmpC β-lactamases that develop resistance to β-lactam antibiotics. The production of extended-spectrum β-lactamase (ESBL) is substantially high in Escherichia coli, C. freundii, Enterobacter cloacae, and Serratia marcescens, but infrequently explored in C. freundii. The present investigation characterized the ESBL C. freundii and delineated the genes involved in decrease in antibiotics resistance. We used the VITEK-2 system and Analytical Profile Index (API) kit to characterize and identify the Citrobacter isolates. The mRNA level of AmpC and AmpR was determined by RT-qPCR, and gel-shift assay was performed to evaluate protein-DNA binding. Here, a total of 26 Citrobacter strains were isolated from COVID-19 patients that showed varying degrees of antibiotic resistance. We examined and characterized the multidrug resistant C. freundii that showed ESBL production. The RT-qPCR analysis revealed that the AmpC mRNA expression is significantly high followed by a high level of AmpR. We sequenced the AmpC and AmpR genes that revealed the AmpR has four novel mutations in comparison to the reference genome namely; Thr64Ile, Arg86Ser, Asp135Val, and Ile183Leu while AmpC remained intact. The ΔAmpR mutant analysis revealed that the AmpR positively regulates oxidative stress response and decreases β-lactam and aminoglycosides resistance. The AmpC and AmpR high expression was associated with resistance to tazobactam, ampicillin, gentamicin, nitrofurantoin, and cephalosporins whereas AmpR deletion reduced β-lactam and aminoglycosides resistance. We conclude that AmpR is a positive regulator of AmpC that stimulates β-lactamases which inactivate multiple antibiotics.
Keywords: C. freundii, β-lactamases, AmpR mutation, AmpC, Oxidative stress, Aminoglycosides resistance
1. Introduction
The emergence of multidrug resistant bacteria in healthcare sectors presents a serious challenge to public health and clinicians [1]. The β-lactam antibiotics are widely used in hospitalized patients and resistance develops among microbes because of the β-lactamase enzyme production [2]. Various clinical isolates have become a global health concern because of β-lactam resistant strains including C. freundii, Escherichia coli, Pseudomonas aeruginosa, Enterobacter cloacae, Klebsiella pneumonia, and Serratia marcescens which are equipped with β-lactamases [3,4]. The genus Citrobacter belongs to the family of Enterobacteriaceae and members are found in soil, water, and sewage. Citrobacter can colonize human and animal gastrointestinal tracts and is widely associated with nosocomial infections [5]. C. freundii develops resistance to antibiotics by several mechanisms including the production of β-lactamases that hydrolyze the β-lactam ring of antibiotics. The β-lactamases have been classified by Ambler and Bush-Jacobi-Medeiros based on their molecular structure and functional similarities. Ambler classified β-lactamases into four classes; A, C, and D classes which use a serine whereas class B uses the metal zinc as an enzyme active center [6]. In the updated Bush-Jacobi-Medeiros classification, β-lactamases are classified into three groups based on the functional similarities between substrate and inhibitor profiles [7]. Group 1 commonly known as cephalosporinases consist of Ambler class C β-lactamases (AmpC enzymes). This group involves chromosomal and plasmid encoded genes that code for ESBL enzymes. The enzymes are induced by β-lactam antibiotics and the bacteria showed resistance to penicillin, cephamycin, tazobactam, and cephalosporins but were sensitive to cefepime and carbapenems. The well-known example of group 1 β-lactamases genes include AmpC of E. coli , CMY-2 of E. cloacae , and blaCMY of C. freundii that reduce the effectiveness of β-lactam antibiotics by conferring resistance to the narrow and broad-spectrum antibiotics. Group 2 commonly known as serine β-lactamases, has serine in the active center and includes Amber classes A and D. Group 3 comprises metallo-β-lactamases that include class B of Amber classification [7,8]. Within the genome of C. freundii, the β-lactamase blaCMY (AmpC) genes are present that produce β-lactamases [9]. The AmpC β-lactamase is regulated by the AmpR which is located upstream of the AmpC [10,11]. The AmpR mutant showed low expression of AmpC whereas constitutive expression of AmpR resulted in high production of AmpC [12]. The inducible vector encoded AmpC β-lactamases with AmpR have been studied in E. cloacae, Morganella morganii, and C. freundii. For instance, ACT-1, DHA-1, and CMY-13 are plasmid-encoded β-lactamases and their inducible expression gave resistance to cephalosporins in E. cloacae [13], M. morganii [14], and C. freundii [15] respectively. Previous reports have shown that most of the Gram negative bacilli such as E. coli reconstitute and recycled their cell wall from cell wall degraded products such as anhydromuropeptides [16]. In C. freundii, the presence of β-lactam antibiotic increased the cytoplasmic anhydromuropeptides which bind to AmpR and activated AmpC while the absence of activating ligand like peptidoglycan recycled product repressed AmpC expression [17,18]. For example in E. coli, unhydrolyzed anhydromuropeptide accumulation in the cytoplasm activated AmpR which induced AmpC expression [19]. Previously, Ryuichi and coworkers determined the AmpC β-lactamase expression level with or without AmpR and their results suggested that the resistance of CFE-1 (plasmid-encoded AmpC β-lactamase) to cephalosporins is due to the substitution of Asp135Ala in AmpR of C. freundii [20]. Among the 13 Citrobacter species, C. freundii is considered an opportunistic pathogen that colonizes the respiratory and urinary tract of hospitalized patients [21]. To date, there is no comprehensive report of Citrobacter infection in COVID-19 patients, and the present report delineated an ESBL C. freundii that showed multidrug resistance profile from hospitalized patients. We hypothesized the involvement of AmpC and AmpR genes in the development of β-lactam resistance and speculated new mechanism of resistance. Initially, the strains were identified by the VITEK-2 system and characterized by an API test kit. We analyzed the relative expression levels of AmpC and AmpR and examined the effects of AmpR on the expression of AmpC. We aim to elucidate the mechanism by which C. freundii acquired β-lactam and aminoglycosides resistance and corroborated the role of AmpR in the production of ESBL.
2. Materials and methods
2.1. Samples processing and identification
We collected bacterial samples from COVID-19 patients between the years 2020 and 2021 and screened 3492 samples and detected bacterial coinfection and superinfection in hospitalized patients [[22], [23], [24]]. The sources of specimens were nasopharyngeal and endotracheal swabs. The bacterial isolates from COVID-19 patients that were resistant to antibiotics were refreshed from −20 °C and cultured on tryptic soy agar (TSA) and Luria Bertani (LB) agar. The VITEK-2 system and 16 S rRNA sequencing were used for the characterization and identification of the isolates. Briefly, the isolates were subcultured on TSA plates, and the bacterial culture turbidity was adjusted with a densitometer to the McFarland 0.5 standard in normal saline. The VITEK-2 identification Gram-negative bacterial cards were loaded into the VITEK-2 system and the test cards were automatically filled with a bacterial suspension and incubated for 3 h. The cards were read by kinetic fluorescence measurement every 15 min and the data was analyzed by the VITEK-2 system software and the results were generated automatically [25]. Afterward, the reference identification and characterization were done with the API 20E system (bioMérieux, Inc., France). Next, the MICs of C. freundii to tazobactam, ampicillin, gentamicin, ciprofloxacin, cefotaxime, and azithromycin were determined by broth microdilution method according to Clinical and Laboratory Standards Institute guidelines as previously described [26,27].
2.2. Growth conditions and culture media
C. freundii can grow on tryptic soy broth, LB broth, and Muller-Hinton (MH) agar (Sigma-Aldrich, Darmstadt, Germany). The antibiotics were purchased from the local market and stock solutions of antibiotics were preserved at −20 °C. The MIC was performed on cation-adjusted MH broth and the antibiotic disc assay was performed on TSA as previously described [27,28]. For the API 20E test kit, C. freundii 24 h culture suspension was made in normal saline and inoculated to each test tube in the kit strip. The strips were incubated at 37 °C for 24 h and seven digits code was generated according to the manufacturer's instructions.
2.3. API 20E test kit protocol
The API 20E test strip consists of twenty tests reaction that are widely used for the identification of Enterobacteriaceae. The C. freundii colonies were suspended in normal saline and inoculated into an API 20E test strip. The test tubes ADH, URE, ODC, H2S, and LDC tubes were overlaid with mineral oil to concede the anaerobic reactions whereas the CIT, GEL, and VP were filled with saline suspension. The strip was incubated at 37 °C for 24 h and the test reagent such as ferric chloride was added to TDA, Kovacs reagent to IND, and one drop of VP reagent 1(40% KOH) and VP Reagent 2 (α-naphthol) to VP were added. The color change was observed and the seven digits code was generated from the reactions and evaluated by the API catalog.
2.4. ESBL production assay
ESBL production was tested by double disc assay comprising cephalosporin (cefotaxime 30 μg ) alone and cephalosporin plus clavulanic acid (30 μg). The zone of inhibition was compared between the single antibiotic disk (cefotaxime) and combination disks (cefotaxime plus clavulanic acid) and the zone ≥ 5 mm for the combination disks relative to the single antibiotic disk was treated as ESBL positive [29]. The gentamicin (CN10μg), tobramycin (TOB10μg), tazobactam (TZP110μg), ampicillin (AMP30μg), nitrofurantoin (F110μg), ciprofloxacin (CIP2μg), cefotaxime (CTX30μg), and cefotaxime plus clavulanic acid (CTX-CVA30μg) discs were used to check the resistance pattern of C. freundii.
2.5. Genome extraction, PCR, and gene sequencing
C. freundii strain was cultured in LB broth at 37 °C and the genomic DNA was extracted from overnight culture by Thermo Scientific DNA Purification Kit. The genomic regions of AmpR and AmpC were amplified from the genomic DNA of C. freundii using the primers listed in Table 1. Primers were constructed from the available genome sequence of C. freundii (NCBI accession number NZ_CP033744.1). The PCR program consists of the following steps; step1-initial denaturation at 95 °C for 5 min; step2-denaturation at 95 °C for 30 s, primer annealing at 55 °C for 1 min or 50 s, extension at 72 °C for 1 min, 35 cycles; step3-final extension at 72 °C for 5 min.
Table 1.
All the primers and plasmids used in this study are listed in table.
| Strain | Relevant genotype | Source |
|---|---|---|
| C. freundii ESBL | Clinical isolate | Hospital |
| ΔAmpR | C. freundii AmpR mutant | This study |
| E. coli DH5α | Transformation | TransGen |
| E. coli BL21 | Protein expression | TransGen |
| Plasmids | ||
| pKD4 | Template for FLP recognition target site, Kanr, Ampr | [26] |
| pKD46 | Temperature-sensitive, λ Red recombinase plasmid, Kanr, Ampr | [26] |
| pCP20 | Temperature-sensitive, FLP recombinase plasmid, Ampr, Cmr | [26] |
| pET28a (+) |
Recombinant protein expression in E. coli BL21, Kanr |
Novagen |
|
Primer name |
Oligonucleotide (5′-3′) |
Application |
| AmpR- forward-1 | AGGCTTAATGATGACGCGTAGCTATATCCCTCTGTAGGCTGGAGCTGCTTC | AmpR mutant |
| AmpR-reversed-2 | GTTAATTATCAGGCCCGCCATTGGCGGGCCTTTTCATATGAATATCCTCCTTAG | AmpR mutant |
| AmpR-forward-3 | ATGACGCGTAGCTATATCCCTCTTAAC | PCR |
| AmpR-reversed-4 | GTGCAGCACCCCGGTCAACCAACGGGA | PCR |
| AmpR-forward-rt1 | GTGACGCATTCTGCCATCAGCCAGC | RT-qPCR |
| AmpR-reversed-rt2 | TTCCTGGGTCTGTTTAGTGGCAA | RT-qPCR |
| AmpC-forward-7 | TGATTTCATGATGAAAAAATCGATATGC | PCR |
| AmpC-reversed-8 | CAGTTATTGCAGTTTTTCAAGAATGCGC | PCR |
| AmpC-forward-rt3 | GAGCAGGCTATTCCGGGCATGGCCG | RT-qPCR |
| AmpC-reversed-rt4 | ATAGCGTCGCCGCCCAACACGCCGT | RT-qPCR |
2.6. RNA extraction and RT-qPCR
C. freundii overnight culture was harvested, washed with ddH2O, and dissolved in 1 ml of RNAiso Plus. The cell lysis was performed by 0.1-mm silica beads and the lysates were centrifuged at 12,000 rpm for 30 min. Further, the extract was treated with DNase I enzyme for residual DNA removal and the purity of RNA was determined from the ratio of absorbance at 260/280 nm. Reverse transcription and qPCR were performed by PrimeScript 1st strand cDNA synthesis kit and SYBR Premix Ex-Taq reagent kit respectively. The RT-qPCR analysis was performed by StepOne real-time PCR system (Applied Biosystems, USA). The gene signal was normalized with rpoD gene cDNA abundance. All primers are listed in Table 1.
2.7. Electrophoretic mobility shift assay (EMSA)
EMSA was performed to detect AmpR and AmpC proteins binding to the promoter region. Initially, AmpR and AmpC proteins were expressed in pET28a (+) vector in E. coli BL21, and purified through Ni+2-NTA histidine affinity column. The biotin-labeled probe (60bp) was incubated with AmpR and AmpC protein separately for 1 h at 37 °C. The 12 μl reaction mixture contains probe (3 fmol), protein (8 pmol), and lysis buffer. The reaction was stopped by heating at 95 °C for 5 min. The sample was run on 4% native polyacrylamide gel in 0.5 × Tris-borate-EDTA buffer at 400 mA for 30 min. A nylon membrane was used to transfer the DNA band and detected by a chemiluminescent detection kit (Thermo Fisher). Image Quant LAS 4000 mini was used to observe the membrane.
2.8. Construction of AmpR mutant
The AmpR mutant was constructed by the method of Datsenko and Wanner [30]. Following the protocols, primers pair AmpR-forward-1 and AmpR-reverse-2 were designed containing homologous flanking sites of AmpR and kanamycin resistance gene (Table 1). The AmpR-forward-1 and AmpR-reverse-2 primers were used to amplify the kanamycin resistance gene flanked by FLP recognition sites from plasmid pKD4. The fragment was cloned into temperature-sensitive (above 30 °C) lambda red recombinase vector pKD46 and transferred to C. ferundii competent cells. Kanamycin resistant C. ferundii was cured of pKD46 at 37 °C and the kanamycin resistance gene was removed by temperature-sensitive FLP recombinase plasmid pCP20 at 43 °C. The in-frame deletion of AmpR was confirmed by DNA sequencing.
2.9. Hydrogen peroxide sensitivity assay
Hydrogen peroxide (H2O2) sensitivity was determined by disc diffusion assay in 5 mM H2O2. Briefly, the C. freundii wild type, and its ΔAmpR overnight culture were 100 times diluted and grown for 2.5 h in a TSB medium. The culture OD600 was measured (OD600 = 2) and a 200 μl culture was spread on TSA plates (2 × 109 CFU/ml). A blank disc was impregnated in 5 mM H2O2 and placed onto the culture plates and zones of inhibition were measured after 24 h.
2.10. Statistical analysis
All reference genes and protein sequences were downloaded from the NCBI database and analyzed by ESPript 3.0 software. Student's t-test (unpaired) was performed by Graph pad prism 8. The significance level was considered P < 0.05.
3. Results
3.1. Prevalence of Citrobacter species
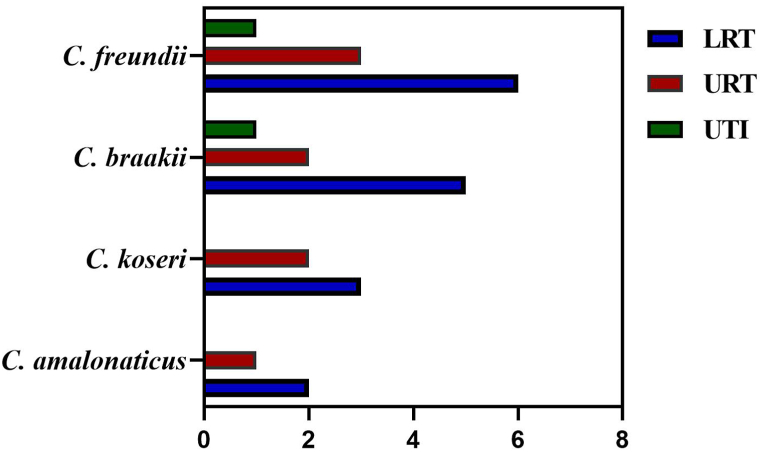
During the pandemic, a total of 3492 COVID-19 hospitalized patients were screened for bacterial coinfection and 26 isolates were identified as Citrobacter species by the VITEK-2 system. Among the 26 Citrobacter spp., 17 were from males and 9 from females. C. freundii was the most abundant species (10 isolates), followed by C. braakii (8 isolates), C. koseri (5 isolates), and C. amalonaticus (3 isolates). The majority of samples were isolated from the lower respiratory tract, upper respiratory tract, and urinary tract infections (Fig. 1). Out of the 26 strains, 11 were MDR of which one was ESBL producer. All 26 cases have comorbidities, with the most common type being pneumonia and urinary tract infection. Among the antibiotics, tazobactam, ampicillin, cefotaxime, gentamicin, tobramycin, and nitrofurantoin were not effective against ESBL strain while sensitive to imipenem and amikacin.
Fig. 1.
The prevalence of Citrobacter species in COVID-19 patients. The UTI (urinary tract infection), LRT (lower respiratory tract), and LRT (upper respiratory tract) sites were colonized by Citrobacter spp., and 26 species were isolated. The bars indicated the number of C. freundii, C. braakii, C. koseri, and C. amalonaticus isolated from LRT, URT, and UTI.
3.2. Characterization of C. freundii ESBL
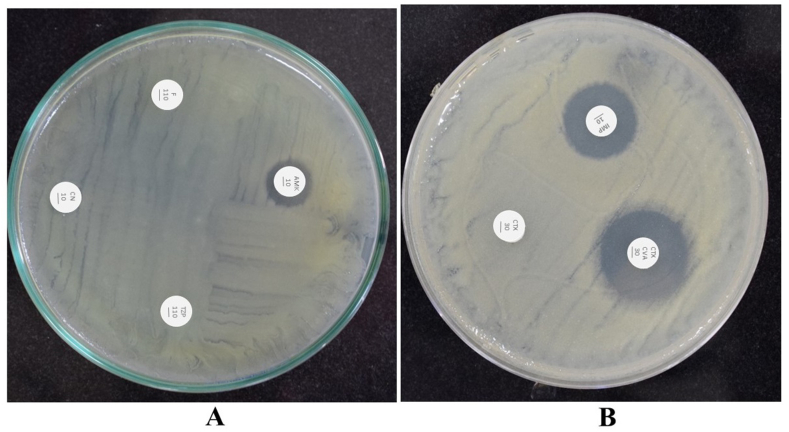
Initially, the clinical isolates were identified by the VITEK-2 system and followed by API. By API 20E test kit, positive results with ≥89% probability were confirmed as C. freundii. The results of C. freundii twenty test reactions are provided in Table 2. Further, the genome was extracted and 16 S rRNA sequencing was performed that validated the Citrobacter identification by VITEK-2 and API test kit. The C. freundii strain displayed resistance to antibiotics such as tazobactam (110 μg), cefotaxime (30 μg), ampicillin (30 μg), gentamicin (10 μg), and nitrofurantoin (110 μg) (Fig. 2A). We performed an ESBL phenotypic assay which revealed that the C. freundii strain displayed an 8 mm zone of inhibition on the combination disc (cefotaxime and clavulanic acid) relative to cefotaxime alone that confirmed the production of ESBL by C. freundii (Fig. 2B). In the ESBL assay, cefotaxime alone did not kill C. freundii while the addition of clavulanic acid inhibited β-lactamases and efficiently kill C. freundii. Furthermore, we determined the MIC of the ESBL strain by two-fold broth microdilution method in cation-adjusted MH broth that showed high MICs to tazobactam (256 μg/ml), ampicillin (128 μg/ml), cefotaxime (128 μg/ml), gentamicin (16 μg/ml), tobramycin (16 μg/ml), and ciprofloxacin (≤4 μg/ml). These findings conclude that C. freundii produced β-lactamases that inactivated β-lactam antibiotics.
Table 2.
The API 20E test results of ESBL C. freundii strain.
| S. No. | Test (Enzyme/Substrate) | Negative reaction | Positive reaction | C. freundii results |
|---|---|---|---|---|
| 1. | O-nitrophenyl-beta-d-galactopyranoside (ONPG) | Colorless | Yellow | + |
| 2. | Arginine dihydrolase (ADH) | Yellow | Red or orange | + |
| 3. | Lysine decarboxylase (LDC) | Yellow | Red or orange | – |
| 4. | Ornithine decarboxylase (ODC) | Yellow | Red or orange | – |
| 5. | Citrate utilization (CIT) | Pale to green | Blue green | + |
| 6. | Hydrogen sulfide (H2S) | Colorless | Black deposits | + |
| 7. | Urease (URE) | Yellow | Red or orange | – |
| 8. | Tryptophan deaminase (TDA) | Pale yellow or brownish | Brown red | _ |
| 9. | Indole (IND) | Pale yellow or colorless | Red (within 2-mints) | _ |
| 10. | Voges–Proskauer (VP) | Pale yellow or colorless | Red or pink (within 10-mints) | _ |
| 11. | Gelatinase (GEL) | No black diffusion | Black diffusion | – |
| 12. | Glucose (GLU) | Blue or blue-green | Red or yellow | + |
| 13. | Mannitol (MAN) | Blue or blue-green | Red or yellow | + |
| 14. | Inositol (INO) | Blue | Yellow | – |
| 15. | Sorbitol (SOR) | Blue green | Yellow | + |
| 16. | Rhamnose (RHA) | Blue | Yellow | + |
| 17. | Sucrose fermentation (SAC) | Blue | Yellow | + |
| 18. | Melibiose (MEL) | Blue green | Yellow | + |
| 19. | Amygdalin (AMY) | Blue | Yellow | – |
| 20. | Arabinose (ARA) | Blue | Yellow | + |
Fig. 2.
(A) Antibiotic disc diffusion assay revealed that C. freundii is resistant to gentamicin (CN10), nitrofurantoin (F110), and tazobactam (TZP110) while sensitive to amikacin (AMK10) with a zone of inhibition size of 2 mm (B) The ESBL phenotypic assay showed ESBL production and C. freundii was resistant to cefotaxime (CFX30) while sensitive to cefotaxime plus clavulanic acid (CTX and CVA30) with a zone of inhibition size of 8 mm. C. freundii was sensitive to imipenem (IMP10) with a zone of inhibition size of 5 mm.
3.3. Identification of mutations in AmpR sequence in C. freundii ESBL strain
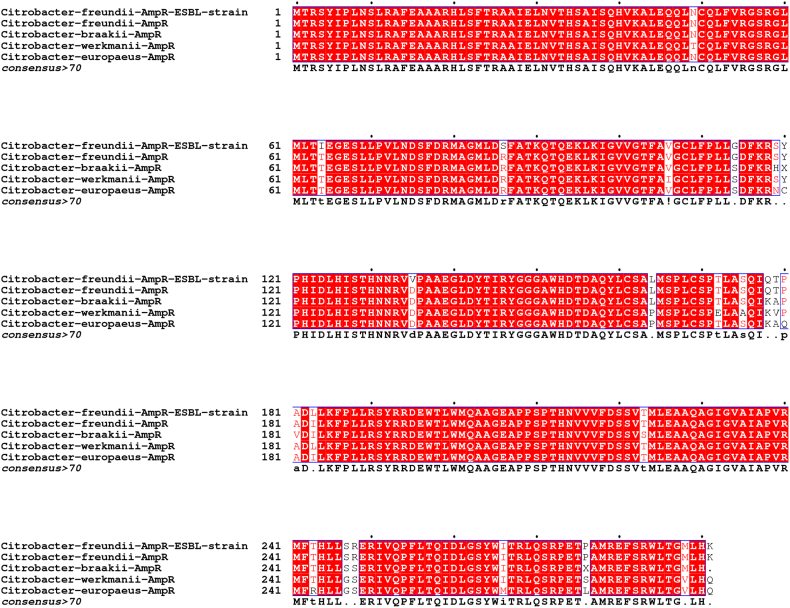
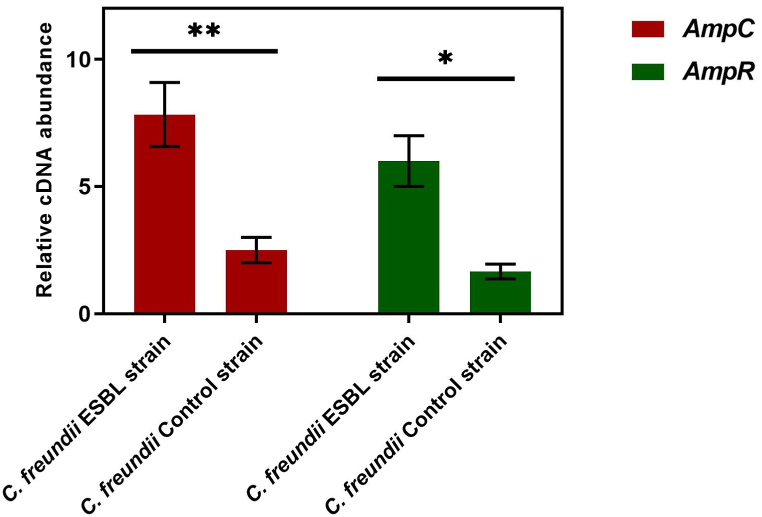
Six strains of Citrobacter were studied by DNA amplification using primers listed in Table 1, and AmpC and AmpR genes were detected in C. freundii strains. The AmpC sequence analysis of the C. freundii ESBL strain revealed that the AmpC gene was identical to the previously reported chromosome-encoded class C cephalosporinase of the C. freundii whereas the AmpR gene showed 99% similarity. The ESBL C. freundii AmpR gene sequence analysis revealed four point mutations compared to the C. freundii reference genome (NCBI accession number NZ_CP033744.1). From the translation of AmpR DNA sequences of the ESBL strain, we detected four mutations in the AmpR amino acids sequence namely; Thr64Ile, Arg86Ser, Asp135Val, and Ile183Leu (Fig. 3). Further, we determined the mRNA level of the ESBL strain by RT-qPCR which revealed that the AmpR mRNA level was high (P < 0.05) relative to the control strain (β-lactam sensitive). We hypothesized that a change in AmpR mRNA level might alter the AmpC transcript, so we compared the AmpC mRNA expression level of the ESBL strain versus the control strain. The results showed that the AmpC mRNA level was approximately three fold higher than the control strain and was highly significant (P < 0.01) (Fig. 4). We proposed that mutations in AmpR increased the expression level of its mRNA that ectopically regulated AmpC expression which resulted in the overproduction of AmpC β-lactamase that inactivated the antibiotics. Furthermore, we also analyzed the AmpD DNA sequence because previously the AmpD mutations were associated with the overproduction of AmpC in E. cloacae [31]. From the genomic sequence of ESBL strain, we did not detect any change in the DNA sequence of AmpD that further confirmed the AmpC β-lactamase high production by AmpR mutations. Thus, we conclude that the genetic changes in the transcriptional regulator AmpR affected AmpC expression that increased the production of AmpC which caused a reduction in the potency of β-lactam antibiotics. Collectively, we reported chromosomal AmpR mutations that increased AmpC expression and resulted in an escalation in β-lactam antibiotics resistance in the clinical isolate of C. freundii.
Fig. 3.
The AmpR amino acid sequence of C. freundii ESBL strain was aligned with C. freundii, C. braakii, C. werkmanii, and C. europaeus from the NCBI database and mutations of isoleucine, serine, valine, and leucine were detected in comparison with C. freundii reference genome (NCBI accession number NZ_CP033744.1).
Fig. 4.
The RT-qPCR analysis showed high expression of AmpC and AmpR mRNA levels in the ESBL C. freundii strain relative to the control C. freundii strain (β-lactam sensitive). The asterisks represent the significance level of mRNA. ✱ P < 0.05, ✱✱ P < 0.01.
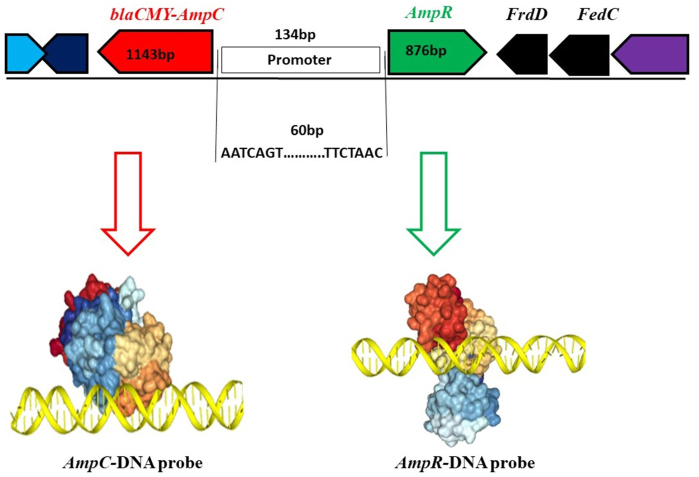
3.4. AmpR protein binds to the AmpC promoter
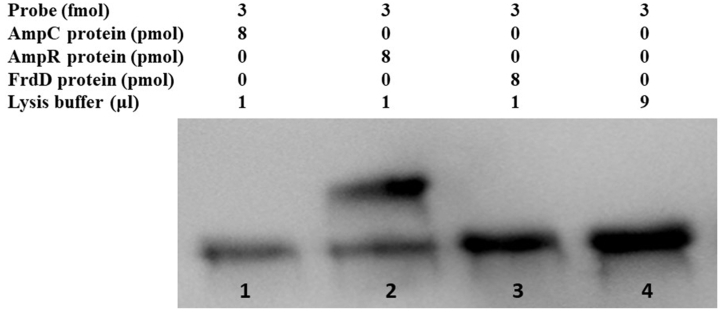
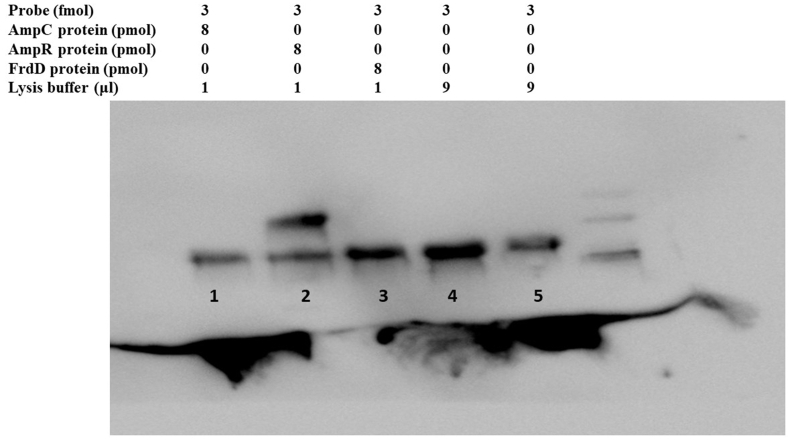
The AmpR is a transcriptional regulator of the LysR family and contains a helix-turn-helix domain. The promoter site of AmpC was predicted by Softberry BPROM that lay in the intergenic region between AmpR and AmpC. The intergenic region of AmpR and AmpC consists of 134 bp, and a 60bp DNA probe containing −10 and −35 regions was selected and protein-DNA docking was performed. The protein-DNA probe docking revealed that AmpR has a high docking score (−419.60) than AmpC (−191.13) which indicated a strong binding ability. By analyzing AmpR and AmpC sequence, a helix-turn-helix domain was detected in AmpR which showed binding affinity with the probe (Fig. 5). To validate the docking analysis, the 60bp probe was biotin labeled and protein binding affinity was tested by EMSA. The AmpR protein specifically binds to the 60 bp probe (lane 2) while the AmpC protein (lane 1) cannot bind to the probe which confirmed the docking results (Fig. 6, for uncropped image see Supplementary Fig. S1). The FrdD gene lies close to AmpR and was used as a control in EMSA which did not bind to the probe as well (lane 3). From these findings, we conclude that the AmpR binds to the AmpC promoter and regulates AmpC expression in C. freundii.
Fig. 5.
Docking of protein and DNA probe. The AmpR and AmpC proteins and probe (60bp) were evaluated for possible superimposition via HDOCK (http://hdock.phys.hust.edu.cn/). The AmpR helix-turn-helix domain showed a strong binding affinity with the probe while the AmpC protein did not show any specific binding to promoter.
Fig. 6.
EMSA results. The shift was observed in lane 2 between the AmpR protein and biotin labeled probe (60bp). Lane 1 represents AmpC protein plus probe; lane 2 AmpR protein plus probe; lane 3 FrdD protein plus probe; and lane 4 probe and lysis buffer. The uncropped and extended figure is provided as a Supplementary Material Fig. S1.
3.5. AmpR regulates C. freundii oxidative stress response
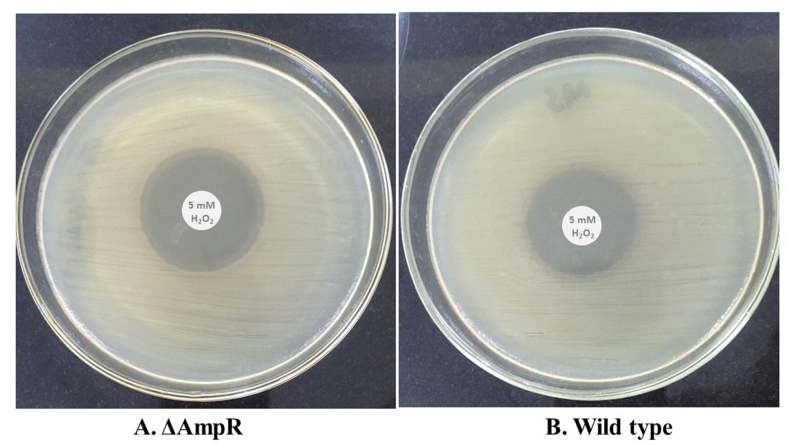
To discern the AmpR role in oxidative stress, we performed the H2O2 sensitivity assay between wild type and ΔAmpR. The C. freundii ΔAmpR was sensitive to H2O2 and showed a zone of 11.5 mm (Fig. 7A) whereas the wild type showed a zone of 10 mm (Fig. 7B). The ΔAmpR cell growth was more inhibited than the wild type by H2O2 which indicated reduced resistance to H2O2. These finding supported the fact that AmpR positively regulated oxidative stress response in C. freundii and protect the cells during a hostile environment.
Fig. 7.
Hydrogen peroxide sensitivity assay. (A) The H2O2 assay revealed that the AmpR mutant has a greater zone of inhibition than the wild type (B). The ΔAmpR showed 11.5 mm whereas the wild type showed a 10 mm zone of inhibition.
3.6. AmpR mutant decreased antibiotics resistance
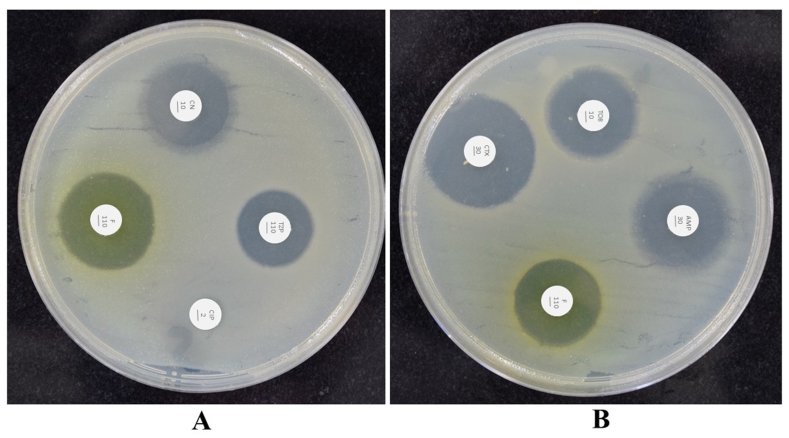
AmpR is required for β-lactam resistance through AmpC expression. The mutations in AmpR resulted in high expression of AmpC that governed β-lactams resistance. We constructed AmpR mutant strain that revealed a reduction in β-lactam resistance. The wild type C. freundii was resistant to tazobactam (110 μg), cefotaxime (30 μg), ampicillin (30 μg), penicillin (110 μg), gentamicin (10 μg), tobramycin (10 μg), and nitrofurantoin (110 μg) whereas AmpR mutant showed a decrease in resistance to tazobactam, gentamicin, tobramycin, nitrofurantoin, and cefotaxime (Fig. 8A and B). The AmpR mutant was highly sensitive to cefotaxime, nitrofurantoin, gentamicin, tobramycin, ampicillin, and tazobactam and showed a zone of inhibition size of 11.5 mm, 10 mm, 9 mm, 9 mm, 9.5 mm, and 8 mm respectively whilst no zone was observed in ciprofloxacin (Fig. 8A and B).
Fig. 8.
Antibiotic disc diffusion assay. (A and B) The ΔAmpR mutant was sensitive to gentamicin (CN10), tazobactam (TZP110), ampicillin (AMP30), nitrofurantoin (F110), cefotaxime (CTX10), and tobramycin (TOB10) whereas resistant to ciprofloxacin (CIP2).
4. Discussion
Citrobacter species are typically found in the intestinal and respiratory tracts of humans. Citrobacter infections may occur as sporadic or nosocomial outbreaks and are transmitted vertically as well as horizontally [32]. Nosocomial and community acquired Citrobacter infections are widely reported [33] and approximately 3–6% of all Enterobacteriaceae causing nosocomial infections accounted for Citrobacter [34,35]. Citrobacter has caused infections worldwide and various reports have concluded urinary and respiratory tracts as predominant sites of infections [36]. Our data reported Citrobacter coinfections during the COVID-19 pandemic and found the respiratory tract as the largely colonized site by Citrobacter species. So far, Citrobacter species have been isolated from surgical wounds, burns, skin pustules, sputum, urinary tract, lower respiratory tract, and perirectal abscesses [34,36,37]. In the present report, we detected C. freundii, C. braakii, C. koseri, and C. amalonaticus in the respiratory tract and urinary tracts, and found predominance in debilitated and elderly patients that suffered from COVID-19. In India, nosocomial, urinary tract, and neonatal septicemia were correlated with Citrobacter infections [38] which supported our results of Citrobacter coinfections in hospitalized patients. Earlier, Citrobacter bacteremia was detected in patients with malignancy, diabetes mellitus, hepatobiliary stones, and heart disease [39] and some studies reported C. koseri as the most prevalent species that caused gastrointestinal and urinary tract infections [36,39]. However, our data suggested C. freundii as the predominant species which caused infection in COVID-19 patients. Previously, C. freundii displayed resistance to tazobactam, gentamicin, cefazolin, ampicillin, norfloxacin, and other antibiotics in hospitalized patients [21,40], and our data also reported the prevalence of MDR in C. freundii. The reason might be the widespread misuse of antibiotics that resulted in the emergence of MDR strains. In countries such as China and India, MDR C. freundii was isolated that was sensitive to imipenem, amikacin, and third-generation cephalosporins [41,42]. These results are in accordance with our findings where C. freundii showed susceptibility to imipenem and amikacin. Importantly, we detected MDR C. freundii which was an ESBL producer that had mutations in the AmpR gene which resulted in AmpC high expression. Previously, AmpR mutations (Arg86Ser) increased the AmpC expression in E. cloacae [43] and plasmid-encoded AmpC (blaCFE-1) also showed AmpR-dependent constitutive expression of AmpC [44]. In addition, AmpC derepression was associated with defects in the AmpD gene in C. freundii [45], and decreased AmpD expression led to AmpC overproduction as well as increased β-lactam MICs in clinical isolates of E. coli, C. freundii, and P. aeruginosa [46]. Collectively, their results demonstrated that AmpD and AmpR modulated the expression of AmpC in several bacteria and conferred β-lactam resistance. Our findings conclude that the mutations in AmpR modulated the mRNA level of AmpC that leads to the production of β-lactamases and inactivation of β-lactam antibiotics. To date, neither C. freundii nor other bacteria have been reported with these AmpR mutations which are linked with high expression of AmpC, resistance to β-lactam, and aminoglycosides. Largely, the β-lactam resistance locus AmpR-AmpC is studied in P. aeruginosa and C. ferundii that consists of AmpR and AmpC genes that are divergently transcribed from promoters located in the intergenic region [47]. Generally, AmpC expression is controlled by AmpR in the presence or absence of inducers and is widely known as the transcriptional regulator of AmpC [17]. The AmpR-AmpC locus is highly conserved in Enterobacteriaceae, for instance, in E. coli and Shigella sonnei, β-lactams resistance is mediated by tandem duplications of the AmpC gene and promoter mutations located within the upstream of fumarate reductase operon [48,49] while in C. freundii, E. cloacae, and S. marcescens, AmpC expression is induced by β-lactam antibiotics that produce β-lactamases which inactivate β-lactam antibiotics [50,51]. Currently, we have shown that AmpR protein regulates AmpC expression by specifically binding to a 60bp fragment of AmpC promoter whilst AmpC could not bind. These results are in accordance with the previous findings where the AmpR helix turn helix motif was involved in DNA binding and autoregulation [18]. Similar results were also reported in P. aeruginosa, where in silico study revealed an AT-rich intergenic region of AmpR-AmpC as a putative AmpR binding site [52]. Besides, the AmpR deletion resulted in low expression of AmpC and a major decrease in β-lactam and aminoglycosides resistance in C. ferundii whereas similar results were only reported in P. aeruginosa [10,47]. In addition, the AmpR mutant showed increased sensitivity to H2O2 which confirmed AmpR role in protection against oxidative stress response. To date, there is no such report that delineated the involvement of AmpR in oxidative stress responses. Altogether, our results showed the isolation of ESBL C. freundii from the hospitalized patient and revealed AmpR mediated AmpC high expression and ΔAmpR changes in C. ferundii that are associated with a decrease in β-lactam and aminoglycosides resistance. The present study has several limitations. First, it did not characterize all Citrobacter species isolated and only focus on ESBL C. freundii. Secondly, all the Citrobacter species were isolated from COVID-19 patients and the rest of the hospitalized patients were not included. The current study highlighted the prevalence of Citrobacter coinfection in hospitalized patients but did not report the clinical data of patients. Lastly, it remains possible that certain Citrobacter strains were re-isolated via regular patient transfers or through the readmission of previously discharged patients during the study period.
5. Conclusion and future prospective
In summary, we have detected mutations in AmpR (Thr64Ile, Arg86Ser, Asp135Val, and Ile183Leu) that conferred resistance to β-lactam antibiotics in C. freundii. The ΔAmpR decreased β-lactam and aminoglycosides resistance and suppressed AmpC expression. The tetra mutant strain was an ESBL producer that reflected a changing pattern of drug resistance in C. freundii which altered the local susceptibility profile of the Citrobacter species in Pakistan. The data presented in this report illustrated the spectrum of Citrobacter infection in COVID-19 patients during the pandemic and revealed C. freundii is one of the commonest causes of coinfection in hospitalized patients. Knowingly, β-lactamase inhibitors have made a great impact in hospitals as adjunct therapies, however, the emergence of ESBL made clinical outcomes more complicated and the emergence of MDR strains. Antibiotic resistance breakers such as ESBL inhibitors represent a promising avenue of research for ESBL-producing Citrobacter species. Researchers need to explore the genetic determinants that bacteria adopt during resistance development, such as mutation and acquiring resistance genes. Besides, the control of antibiotic resistance is associated with proper antibiotic prescription, physician vigilance, and more proactive strategies to maximize the lifespans of current antibiotics through antimicrobial stewardship and public awareness.
Author contribution statement
Falak Naz Tariq: Mehreen Shafiq: Nadeem Khawar: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. GUL HABIB: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper. Haji Gul: Performed the experiments; Wrote the paper. Azam Hayat: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data. Mujaddad Ur Rehman: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data. Ihab Mohamed Moussa: Eman A. Mahmoud: Hosam O. Elansary: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Data availability statement
No data was used for the research described in the article.
Funding
The study was funded by the Office of Research Innovation and Commercialization, Abbottabad University of Science and Technology (Grant no AUST/ORIC/2021/375) and the Researchers Supporting Project number (RSPD2023R741), King Saud University.
Ethics approval
The study was approved by the Department of Microbiology, Abbottabad University of Science and Technology (AUST/ORIC/2021/375).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Researchers Supporting Project number (RSPD2023R741), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19486.
Contributor Information
Gul Habib, Email: gulhabib101@gmail.com.
Ihab Mohamed Moussa, Email: imoussa1@ksu.edu.sa.
Hosam O. Elansary, Email: helansary@ksu.edu.sa.
Appendix A. Supplementary data
The following is the supplementary data to this article:
figs1.
References
- 1.Vivas R., Barbosa A.A.T., Dolabela S.S., Jain S. Multidrug-resistant bacteria and alternative methods to control them: an overview. Microb. Drug Resist. 2019;25(6):890–908. doi: 10.1089/mdr.2018.0319. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit. Care. 2010;14(3):1–8. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Angelis G., Del Giacomo P., Posteraro B., Sanguinetti M., Tumbarello M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020;21(14):5090. doi: 10.3390/ijms21145090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gul H., Habib G., Khan I.M., Rahman S.U., Khan N.M., Liu Y. Genetic resilience in chickens against bacterial, viral and Protozoal pathogens. Front. Vet. Sci. 2022:1776. doi: 10.3389/fvets.2022.1032983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith A., Saiman L., Zhou J., Della-Latta P., Jia H., Graham P.L., III Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with Gram-negative bacilli in very low birthweight infants in the neonatal intensive care unit. Pediatr. Infect. Dis. J. 2010;29(9):831. doi: 10.1097/INF.0b013e3181e7884f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambler R.P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 7.Bush K., Jacoby G.A., Medeiros A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995;39(6):1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush K., Jacoby G.A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlow M., Hall B.G. Origin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 2002;46(5):1190–1198. doi: 10.1128/AAC.46.5.1190-1198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong K.-F., Jayawardena S.R., Indulkar S.D., Del Puerto A., Koh C.-L., Høiby N., Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005;49(11):4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartowsky E., Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC β‐lactamase. Mol. Microbiol. 1991;5(7):1715–1725. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 12.Nakano R., Okamoto R., Nagano N., Inoue M. Resistance to gram-negative organisms due to high-level expression of plasmid-encoded ampC β-lactamase blaCMY-4 promoted by insertion sequence ISEcp1. J. Infect. Chemother. 2007;13(1):18–23. doi: 10.1007/s10156-006-0483-6. [DOI] [PubMed] [Google Scholar]
- 13.Reisbig M.D., Hanson N.D. The ACT-1 plasmid-encoded AmpC β-lactamase is inducible: detection in a complex β-lactamase background. J. Antimicrob. Chemother. 2002;49(3):557–560. doi: 10.1093/jac/49.3.557. [DOI] [PubMed] [Google Scholar]
- 14.Giakkoupi P., Tambic-Andrasevic A., Vourli S., Skrlin J., Sestan-Crnek S., Tzouvelekis L., Vatopoulos A. Transferable DHA-1 cephalosporinase in Escherichia coli. Int. J. Antimicrob. Agents. 2006;27(1):77–80. doi: 10.1016/j.ijantimicag.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Miriagou V., Tzouvelekis L., Villa L., Lebessi E., Vatopoulos A., Carattoli A., Tzelepi E. CMY-13, a novel inducible cephalosporinase encoded by an Escherichia coli plasmid. Antimicrob. Agents Chemother. 2004;48(8):3172–3174. doi: 10.1128/AAC.48.8.3172-3174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 1995;17(3):421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg F., Westman L., Normark S. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. USA. 1985;82(14):4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindquist S., Lindberg F., Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J. Bacteriol. 1989;171(7):3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs C., Huang L., Bartowsky E., Normark S., Park J. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta‐lactamase induction. EMBO J. 1994;13(19):4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano R., Nakano A., Yano H., Okamoto R. Role of AmpR in the high expression of the plasmid-encoded AmpC β-lactamase CFE-1. mSphere. 2017;2:4. doi: 10.1128/msphere.00192-00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L., Chen D., Liu L., Lan R., Hao S., Jin W., Sun H., Wang Y., Liang Y., Xu J. Genetic diversity, multidrug resistance, and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front. Cell. Infect. Microbiol. 2018;8:233. doi: 10.3389/fcimb.2018.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habib G., Mahmood K., Gul H., Tariq M., Ain Q.U., Hayat A., Rehman M.U. Pathophysiology of methicillin-resistant Staphylococcus aureus superinfection in COVID-19 patients. Pathophysiology. 2022;29(3):405–413. doi: 10.3390/pathophysiology29030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habib G., Khan M.S.Z., Gul H., Hayat A., Rehman M.U. Elsevier; City: 2022. A Persistent High Ambient Temperature Waned the Community Spread of Severe Acute Respiratory Syndrome Coronavirus-2 in Pakistan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib G., Mahmood K., Ahmad L., Gul H., Hayat A., Rehman M.U. Clinical manifestations of active tuberculosis patients coinfected with severe acute respiratory syndrome coronavirus-2. Journal of Clinical Tuberculosis and Other Mycobacterial Diseases. 2023;31 doi: 10.1016/j.jctube.2023.100359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling T.K., Tam P., Liu Z., Cheng A.F. Evaluation of VITEK 2 rapid identification and susceptibility testing system against gram-negative clinical isolates. J. Clin. Microbiol. 2001;39(8):2964–2966. doi: 10.1128/JCM.39.8.2964-2966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wikler M. tenth ed. Clinical and laboratory standards institute.; 2009. Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard. [Google Scholar]
- 27.Habib G., Zhu J., Sun B. A novel type I toxin-antitoxin system modulates persister cell formation in Staphylococcus aureus. International Journal of Medical Microbiology. 2020;310(2) doi: 10.1016/j.ijmm.2020.151400. [DOI] [PubMed] [Google Scholar]
- 28.Habib G., Zhu Q., Sun B. Bioinformatics and functional assessment of toxin-antitoxin systems in Staphylococcus aureus. Toxins. 2018;10(11):473. doi: 10.3390/toxins10110473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarlier V., Nicolas M.-H., Fournier G., Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Clin. Infect. Dis. 1988;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 30.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko K., Okamoto R., Nakano R., Kawakami S., Inoue M. Gene mutations responsible for overexpression of AmpC β-lactamase in some clinical isolates of Enterobacter cloacae. J. Clin. Microbiol. 2005;43(6):2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doran T.I. The role of Citrobacter in clinical disease of children. Clin. Infect. Dis. 1999;28(2):384–394. doi: 10.1086/515106. [DOI] [PubMed] [Google Scholar]
- 33.Jones R.N., Jenkins S.G., Hoban D.J., Pfaller M.A., Ramphal R. In vitro efficacy of six cephalosporins tested against Enterobacteriaceae isolated at 38 North American medical centres participating in the SENTRY Antimicrobial Surveillance Program, 1997–1998. Int. J. Antimicrob. Agents. 2000;15(2):111–118. doi: 10.1016/s0924-8579(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 34.Lipsky B.A., Hook E.W., III, Smith A.A., Plorde J.J. Citrobacter infections in humans: experience at the Seattle Veterans Administration Medical Center and a review of the literature. Rev. Infect. Dis. 1980;2(5):746–760. doi: 10.1093/clinids/2.5.746. [DOI] [PubMed] [Google Scholar]
- 35.Lavigne J.-P., Defez C., Bouziges N., Mahamat A., Sotto A. Clinical and molecular epidemiology of multidrug-resistant Citrobacter spp. infections in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26(6):439–441. doi: 10.1007/s10096-007-0315-3. [DOI] [PubMed] [Google Scholar]
- 36.Ranjan K., Ranjan N. Citrobacter. An emerging health care associated urinary pathogen. Urol. Ann. 2013;5(4):313. [PMC free article] [PubMed] [Google Scholar]
- 37.Arens S., Verhaegen J., Verbist L. Differentiation and susceptibility of Citrobacter isolates from patients in a university hospital. Clin. Microbiol. Infection. 1997;3(1):53–57. doi: 10.1111/j.1469-0691.1997.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 38.Panigrahi P., Chandel D.S., Hansen N.I., Sharma N., Kandefer S., Parida S., Satpathy R., Pradhan L., Mohapatra A., Mohapatra S.S. Neonatal sepsis in rural India: timing, microbiology and antibiotic resistance in a population-based prospective study in the community setting. J. Perinatol. 2017;37(8):911–921. doi: 10.1038/jp.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih C.-C., Chen Y.-C., Chang S.-C., Luh K.-T., Hsieh W.-C. Bacteremia due to Citrobacter species: significance of primary intraabdominal infection. Clin. Infect. Dis. 1996;23(3):543–549. doi: 10.1093/clinids/23.3.543. [DOI] [PubMed] [Google Scholar]
- 40.Liu L., Qin L., Hao S., Lan R., Xu B., Guo Y., Jiang R., Sun H., Chen X., Lv X. Lineage, antimicrobial resistance and virulence of Citrobacter spp. Pathogens. 2020;9(3):195. doi: 10.3390/pathogens9030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sami H., Sultan A., Rizvi M., Khan F., Ahmad S., Shukla I., Khan H.M. Citrobacter as a uropathogen, its prevalence and antibiotics susceptibility pattern. CHRISMED J. Health Res. 2017;4(1):23. [Google Scholar]
- 42.Liu L., Lan R., Liu L., Wang Y., Zhang Y., Wang Y., Xu J. Antimicrobial resistance and cytotoxicity of Citrobacter spp. in maanshan anhui province, China. Front. Microbiol. 2017;8:1357. doi: 10.3389/fmicb.2017.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuga A., Okamoto R., Inoue M. ampR gene mutations that greatly increase class C β-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 2000;44(3):561–567. doi: 10.1128/aac.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakano R., Okamoto R., Nakano Y., Kaneko K., Okitsu N., Hosaka Y., Inoue M. CFE-1, a novel plasmid-encoded AmpC β-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob. Agents Chemother. 2004;48(4):1151–1158. doi: 10.1128/AAC.48.4.1151-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stapleton P., Shannon K., Phillips I. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob. Agents Chemother. 1995;39(11):2494–2498. doi: 10.1128/aac.39.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidtke A.J., Hanson N.D. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob. Agents Chemother. 2006;50(6):2030–2037. doi: 10.1128/AAC.01458-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balasubramanian D., Kumari H., Mathee K. Pseudomonas aeruginosa AmpR: an acute–chronic switch regulator. Pathogens disease. 2015;73(2):1. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole S.T., Guest J.R. Production of a soluble form of fumarate reductase by multiple gene duplication in Escherichia coli K12. Eur. J. Biochem. 1979;102(1):65–71. doi: 10.1111/j.1432-1033.1979.tb06263.x. [DOI] [PubMed] [Google Scholar]
- 49.Jaurin B., Grundström T., Normark S. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1982;1(7):875–881. doi: 10.1002/j.1460-2075.1982.tb01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole S., Nicolas M. Beta-lactam resistance mechanisms in gram-negative bacteria. Microbiol. Sci. 1986;3(11):334–339. [PubMed] [Google Scholar]
- 51.Lindberg F., Normark S. Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J. Bacteriol. 1987;169(2):758–763. doi: 10.1128/jb.169.2.758-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balasubramanian D., Schneper L., Merighi M., Smith R., Narasimhan G., Lory S., Mathee K. The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.