The authors show that murine Clec4A4 and human CLEC4A on conventional dendritic cells are negative immune checkpoint regulators that impair antitumor immune responses, suggesting hCLEC4A as a potential target for immune checkpoint blockade tumor immunotherapy.
Abstract
Clec4A4 is a C-type lectin receptor (CLR) exclusively expressed on murine conventional dendritic cells (cDC) to regulate their activation status. However, the functional role of murine Clec4A4 (mClec4A4) in antitumor immunity remains unclear. Here, we show that mClec4A4 serves as a negative immune checkpoint regulator to impair antitumor immune responses. Deficiency of mClec4A4 lead to a reduction in tumor development, accompanied by enhanced antitumor immune responses and amelioration of the immunosuppressive tumor microenvironment (TME) mediated through the enforced activation of cDCs in tumor-bearing mice. Furthermore, antagonistic mAb to human CLEC4A (hCLEC4A), which is the functional orthologue of mClec4A4, exerted protection against established tumors without any apparent signs of immune-related adverse events in hCLEC4A-transgenic mice. Thus, our findings highlight the critical role of mClec4A4 expressed on cDCs as a negative immune checkpoint molecule in the control of tumor progression and provide support for hCLEC4A as a potential target for immune checkpoint blockade in tumor immunotherapy.
Introduction
Dendritic cells (DC) are considered essential antigen (Ag)-presenting cells (APC) that play pivotal roles in organizing the immune system, linking innate information gathered by recognizing invading microbes through a variety of pattern recognition receptors (PRR) to tailored adaptive responses (1–3). DCs comprise two functionally distinguishable principal subsets, classical or conventional DCs (cDC) and plasmacytoid DCs (pDC; refs. 1–3). Furthermore, cDCs are subdivided into type 1 cDC (cDC1) and type 2 (cDC2) lineages, which differ in developmental pathway and function (1–3). For the initiation of primary T-cell responses against microbial infection, DCs recognize and process microbial Ag to present their antigenic peptides in the context of MHC in conjunction with costimulatory molecules and cytokines for the differentiation of naïve T cells to effector T (Teff) cells (1–3). On the other hand, DCs are implicated as critical for the maintenance of immune homeostasis under steady-state and certain environmental conditions by generating immune tolerance through mechanisms including the induction of clonal deletion and anergy of Ag-specific T cells as well as the generation of CD4+Foxp3+ regulatory T cells (Treg; ref. 3).
Clec4A4, also known as dendritic cell immunoreceptor 2 (DCIR2), is a member of the group II transmembrane C-type lectin receptors (CLR). It has an immunoreceptor tyrosine-based inhibition motif (ITIM) in its cytoplasmic portion and is exclusively expressed on murine cDC2 (4, 5). We have previously shown that self-interaction of murine Clec4A4 (mClec4A4) through binding of the glutamic acid–proline–serine (Glu–Pro–Ser; EPS) motif (6, 7) with oligosaccharide resides anchored in the N-glycosylation site within the carbohydrate-recognition domains (CRD), constitutively delivers inhibitory signaling via ITIM and is required for the suppressive effect of mClec4A4 on the activation of cDC2 (8). Furthermore, we have revealed that mClec4A4 negatively controls inflammation, Ag-specific T-cell responses, and autoimmune disorders as well as microbial infection through the regulation of the function of cDC2 (8). On the other hand, we have shown that human CLEC4A (hCLEC4A) exhibits a similar molecular basis to mClec4A4 and exerts an ITIM-mediated suppressive function that requires its homotypic interaction through the binding of the EPS motif with the N-glycosylation site within the CRD in human cDCs, suggesting that hCLEC4A is a functional orthologue of mClec4A4 (9). Furthermore, we have also shown that antagonistic mAb to hCLEC4A, which inhibits its self-interaction, enhances the activation of human cDCs (9).
As negative immune checkpoints have emerged as inhibitory pathways that play key roles in the regulation of the durability of immune responses, while maintaining self-tolerance to prevent autoimmunity, these pathways also have been found to enable tumors to evade immune destruction (10–12). Growing evidence supports the notion that cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and programmed cell death-1 (PD-1) expressed on T cells function as negative immune checkpoint regulators of T-cell function (10–12). Furthermore, blocking mAbs against CTLA-4 and the PD-1/programmed death ligand 1 (PD-L1) axis are currently used as immune checkpoint inhibitors (ICI) for tumor immunotherapy in the clinical settings (10–12). Although these ICIs can provide unprecedented durable curative effects on several types of tumors (10–12), there are still many patients that do not respond to these therapeutic approaches and some tumor types remain largely refractory to these therapies (13–15). Furthermore, the use of these ICIs reportedly has the potential to cause detrimental inflammatory side effects, which are often termed immune-related adverse events (irAE) triggered by the loss of T-cell self-tolerance (16–18).
In this study, we used Clec4a4−/− mice to show that mClec4A4 constitutes a unique regulatory CLR endowed with negative immune checkpoint function on cDC that impairs antitumor immune responses, leading to the promotion of tumor development. We further demonstrated that antagonistic mAb to hCLEC4A exerted a therapeutic effect against tumor development in hCLEC4A-transgenic (Tg) mice. Collectively, mClec4A4 mediated negative immune checkpoint function impacting antitumor immunity, and hCLEC4A may serve as a promising candidate for immune checkpoint blockade therapy in humans with cancer.
Materials and Methods
Mice
All mice used in this study were 8- to 12-week-old female mice. The following strains were used: C57BL/6 mice used as wild-type (WT) mice were purchased from Japan Clea. B6.CD45.1+OT-I T-cell receptor (TCR) Tg mice harboring ovalbumin (OVA)-specific CD8+ T cells (B6.CD45.1+OT-I mice) and B6.CD45.1+OT-II TCR Tg mice harboring OVA-specific CD4+ T cells (B6.CD45.1+OT-II mice) were bred in-house by crossing B6.OT-I mice and B6.OT-II mice, kindly provided by Dr. Takashi Saito (RIKEN Center for Integrative Medical Sciences, Japan), with CD45.1+ B6 mice (8, 19, 20). B6.Clec4a4−/− mice were generated previously (8) and have been deposited as B6.Cg-Clec4a4<tm1.1Ksat> in the RIKEN BioResource Center (accession number; RBRC09657). B6.hCLEC4A-Tg mice were generated as described below (Generation of hCLEC4A-Tg mice). All mice were bred and maintained under specific pathogen-free conditions in the animal facility at the University of Miyazaki, and all experiments were conducted in accordance with a protocol approved by the Animal Experiment Committee and Gene Recombination Experiment Committee in University of Miyazaki (Miyazaki, Japan).
Generation of hCLEC4A-tg mice
B6.hCLEC4A-Tg mice were generated using the pDOI-6 vector, a kind gift from Dr. Satoshi Ishido (Hyogo College of Medicine, Japan; ref. 21), which allows the expression of reporter cDNA at high levels in APCs under the control of the murine invariant chain (Ii) promoter. The cDNA of hCLEC4A, which was custom-made using GeneArt (Life Technologies) in a pMA-RQ vector backbone, and each of the 5′- and 3′-ends was tagged with ClaI. Following digestion of the cDNA of hCLEC4A with ClaI, the fragment was ligated into the ClaI site of the pDOI-6 vector. The linearized clone insert was released from the vector backbone by digestion with XhoI and PvuI, gel-purified, and microinjected into the pronuclei of fertilized C57BL/6 oocytes. The founder line with high transgene expression was chosen for further analysis. The mutant mice were genotyped by PCR using ii-promoter-F (5′-AGA CAC ACA GCA GCA GCA-3′) and clec4a-R (5′-TCA TCT CAC AAA CTG ACC TTT GA-3′) with the product being 4,426 base pairs (bp). The transgenic founders were mated with C57BL/6 mice, and offspring were screened by the same PCR genotyping to establish B6.hCLEC4A-Tg mice.
Cell lines
An OVA-transfected derivative of the B16 murine melanoma cell line (B16-OVA; ref. 22) and the murine colon adenocarcinoma cell line MC38 (23–25) were kindly provided by Dr. Shin-ichiro Fujii (RIKEN Center for Integrative Medical Sciences, Japan) in 2015 and 2020, respectively. B16-OVA and MC38 cells were cultured in RPMI1640 (FUJIFILM Wako Pure Chemical, 189–02025) supplemented with an antibiotic–antimycotic (FUJIFILM Wako Pure Chemical, 161–23181) and 10% heat-inactivated FCS (Sigma-Aldrich, 173012) at 37°C in a humidified atmosphere of 5% CO2 and air. Cell lines were not authenticated since provision and were cultured for fewer than 20 passages before use. Cell lines were tested for Mycoplasma using a Mycoplasma Detection Kit (InvivoGen).
Cell isolation
To prepare single-cell suspensions from spleen, inguinal lymph nodes (LN), which were used as tumor-draining LNs (tdLN) after tumor inoculation, axillary LNs (AxiLN), and mesenteric LNs (MesLN; refs. 8, 19, 20, 26), tissue samples were digested with collagenase type III (Worthington Biochemical, LS004182) at 37°C for 20 minutes, and were ground between glass slides. Splenocytes were treated with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich, R7757–100 mL) before suspension. Single-cell suspensions of leukocytes were obtained by forcing through a 100-μm cell strainer (BD Biosciences). CD11c+ DCs were purified by AutoMACS with CD11c MicroBeads UltraPure, mouse (Miltenyi Biotec, 130–125–835). In some experiments, CD11c+ DCs were sorted into MHC II+CD11c+CD8α– cDCs in spleen with high purity (each > 99%) using a FACSAriaII Cell Sorter (BD Biosciences) after staining with fluorescein-conjugated mAbs to MHC II (BD Biosciences, 557000), CD11c (BD Biosciences, 550261), and CD8α (BD Biosciences, 561966). CD4+ T cells or CD8+ T cells were purified from splenocytes of WT mice, B6.CD45.1+OT-II mice (CD45.1+Vα2+OT-II CD4+ T cells), and/or B6.CD45.1+OT-I mice (CD45.1+Vα2+OT-I CD8+ T cells) with mouse CD4 T lymphocyte Enrichment Set-DM and (BD Biosciences, 558131) or mouse CD8 T lymphocyte Enrichment Set-DM (BD Biosciences, 558471). Tumor tissues were ground between glass slides, and CD45+ leukocytes were purified by AutoMACS with mouse CD45 Microbeads (Miltenyi Biotec, 130–052–301).
Flow cytometry
Cells obtained from untreated naïve mice or B16-OVA–bearing mice were stained with fluorescein-conjugated mAbs after Fc blocking with anti-mouse CD16/CD32 mAb (clone 2.4G2, BD Biosciences, 553140). The mAbs are listed in Supplementary Table S1. Fluorescence staining was analyzed with a FACSVerse flow cytometer (BD Biosciences) and FlowJo software (Tree Star, version 10.9.0). The flow cytometry gating strategies to identify cDC subsets and T-cell subsets in spleen, LNs, and tumor tissues are shown in Supplementary Fig. S1.
qRT-PCR
Total RNA from cells of tumor tissues obtained from B16-OVA–bearing mice was extracted by using RNeasy Plus Micro Kit (QIAGEN, 74034), and the first-strand cDNA was synthesized from 100 ng of total RNA with oligo(dT)20 primer using the PrimeScript RT Master Mix (Takara Bio, RR036A) according to the manufacturer's instructions. Transcriptional expression levels were analyzed, as described previously (26) by using SYBR Premix Ex Taq II (Takara Bio, RR820L) on Thermal Cycler Dice (Takara Bio) with specific primer pairs listed in Supplementary Table S2 after normalization for Gapdh expression by the 2–ΔΔCt method. Each sample contained three biological replicates.
RNA sequencing and analysis
RNA sequencing (RNA-seq) and analysis were performed at Rhelixa. In brief, total RNA from resident MHC IImedCD11chi cDCs in TdLNs in B16-OVA–bearing mice was extracted by using RNeasy Plus Micro Kit as described above, and full-length cDNA was prepared from total RNA (10 ng) by SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio, Z4889N). RNA-seq analysis was performed using the NovaSeq 6000 (Illumina) in the paired-end 2 × 100-bp cycle mode with NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, E7770L). The quality of the raw paired-end sequence reads was assessed with FastQC (Version 0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Low quality (<20) bases and adapter sequences were trimmed by Trimmomatic software (Version 0.38) with the following parameters: ILLUMINACLIP: path/to/adapter.fa:2:30:10 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:15 MINLEN:36. The trimmed reads were aligned to Mus musculus genome assembly GRCm38 (mm10) as the reference genome using RNA-seq aligner HISAT2 (Version 2.1.0). The HISAT2-resultant .sam files were converted into .bam files with samtools (4) and used to estimate the abundance of uniquely mapped reads with featureCounts (version 1.6.3). The raw counts were normalized with transcripts per million (TPM). On the basis of the normalized read counts, comparative analyses of all samples were conducted by hierarchical clustering, principal component analysis (PCA), correlation analysis, and heat maps. The tree diagram was made by hierarchical clustering with Wald method using each pair of Euclidean distances. Each sample was projected onto the 2D plane of the first and second PCA axes. Scatter plots were constructed by calculating Pearson correlation coefficients of each count between a pair of samples. Heat maps were created by calculating Z-scores of the count data using stats (Version 3.6.1) and gplots (Version 3.0.1.1) R packages. Differentially expressed genes (DEG) were detected using DESeq2 (Version 1.24.0) with the threshold of |log2 fold change (FC)| > 1 and Padj < 0.05 calculated by Benjamini and Hochberg (BH) method.
Tumor growth assay and treatment
C57BL/6 mice, B6.Clec4a4−/− mice, and B6.hCLEC4A-Tg mice were inoculated subcutaneously with B16-OVA (1 × 105) or MC38 (5 × 105) in the right flank. Tumor size was measured every day after the inoculation for 18 to 23 days using a digital caliper (CP-15CP; Mitsutoyo), and tumor volumes were approximated using the ellipsoidal formula: length × width × height × 0.52. For the depletion of CD4+ T cells or CD8+ T cells, tumor-bearing mice received anti-CD4 (clone GK1.5; 100 μg/mouse) or anti-CD8α (clone 3.155; 500 μg/mouse to start followed by 200 μg/mouse), which were a kind gift of Dr. Akihiko Yoshimura (Keio University School of Medicine, Japan), or control Ab (rat IgG2a, clone 54447, Wako, 552-61371; 200 μg/mouse), by intraperitoneal injection every 3 days starting from the day of tumor inoculation until the end of the experiment. For the blockade of hCLEC4A or PD-1, anti-hCLEC4A (ref. 9; clone A77–1, mouse IgG2c; 100 μg/mouse), neutralizing anti–PD-1 (clone: 29F.1A12, rat IgG2a, Biolegend, 135202; 100 μg/mouse for MC38 or 200 μg/mouse for B16-OVA), or control Ab (mouse IgG2a, clone 20102, Wako, 554-60471; 200 μg/mouse) was administered intraperitoneally to mice bearing tumors of approximately 100 mm3 every 3 days for a total of five times. Alternatively, spleen, inguinal LNs indicated as peripheral LNs (PLN), AxiLNs, MesLNs, skin, lung, liver, small intestine (SI), and tumor tissues were obtained from tumor-bearing mice on day 21 after tumor inoculation. In some experiments, body weight was measured every day after the inoculation for 21 days.
Adoptive transfer
For Ag-specific priming of CD4+ T cells or CD8+ T cells in vivo (8, 19, 20), CD45.1+OT-II CD4+ T cells or CD45.1+OT-I CD8+ T cells isolated by the protocol described above (see Cell isolation) were labeled with eFluor 670 (Thermo Fisher Scientific, 65–0840–85; 2.5 μmol/L) at 37°C for 10 minutes and then washed twice with cold PBS. Subsequently, eFluor 670–labeled CD45.1+OT-II CD4+ T cells or CD45.1+OT-I CD8+ T cells (each 5 × 106 cells/mouse) were intravenously injected into untreated naïve mice or B16-OVA–bearing mice on day 21 after tumor inoculation. After 3 days, the gated CD45.1+OT-II CD4+ T cells or CD45.1+OT-I CD8+ T cells in spleen and PLNs were analyzed for eFluor 670 dilution to detect dividing cells by flow cytometry analysis.
Culture of CD11c+ DCs
CD11c+ cDCs obtained by the protocol described above (see Cell isolation) from spleen and PLNs from untreated naïve mice or spleen and TdLNs from B16-OVA–bearing mice on day 21 after tumor inoculation were cultured with or without lipopolysaccharide (LPS, Sigma-Aldrich, L2637–5MG; 1 μg/mL) in RPMI1640 supplemented with an antibiotic–antimycotic and 10% heat-inactivated FCS as described above for 16 hours in 48-well culture plates. Subsequently, the culture supernatants were collected and stored at −80°C until assayed for cytokines.
Detection of cytokines by ELISA
Culture supernatants were assayed for mouse IL6 and mouse TNFα using IL6 Mouse Uncoated ELISA Kit with plates (Thermo Fisher Scientific, 88–7064–86) and TNFα Mouse Uncoated ELISA Kit with Plates (Thermo Fisher Scientific, 88–7324–76) according to the manufacturers’ instructions, and the absorbance was measured at 450 nm by microplate reader (iMark, Bio-Rad).
Enzyme-linked immunospot assay
OVA-specific responses of CD8+ T cells obtained from tumor tissues as described above (see Cell isolation) were quantified by the IFNγ enzyme-linked immunospot (ELISpot) assay with ELISpot Plus: mouse IFNγ [horseradish peroxidase (HRP); Mabtech, 3321–4HPW-2] and OVA257–264 peptide (SIINFEKL; 1 μg/mL, MBL, TS-5001-P) according to the manufacturers’ instructions. Pictures of each well were captured using AlphaImager Image Analysis System (Alpha Innotech), and individual spots were counted per well.
Ag-presentation assay
eFluor 670–labeled CD45.1+OT-II CD4+ T cells (105) or CD45.1+OT-I CD8+ T cells (105) were cultured with CD11c+ cDCs (104) obtained from PLNs in untreated naïve mice or TdLNs and tumor tissues in B16-OVA–bearing mice on day 21 after tumor inoculation for 3 days in 96-well round-bottomed plates (8). After 3 days, the gated CD45.1+OT-II CD4+ T cells or CD45.1+OT-I CD8+ T cells were analyzed by flow cytometry for eFluor 670 dilution to detect dividing cells.
Histopathologic assessment
Tissues from the skin, lung, liver, and SI were fixed with 4% paraformaldehyde (PFA) in PBS and embedded in paraffin (26). Tissue sections were stained with hematoxylin and eosin (H&E). In brief, paraffin-embedded tissues were cut into 5-μm-thick sections and placed onto silane-coated slide glasses. The sections were deparaffinized with toluene and rehydrated through graded ethanol series. Tissues were stained with Mayer Hematoxylin Solution (FUJIFILM Wako Pure Chemical, 131–09665) and Eosin Alcohol Solution (FUJIFILM Wako Pure Chemical, 050–06041), then dehydrated with graded ethanol series and cleared with xylene (FUJIFILM Wako Pure Chemical, Wako 242–00087; ref. 26). The stained slides were examined with a bright-field microscopy (BX53; Olympus).
Statistical analysis
All statistical analysis was performed using Prism 6 (GraphPad Software). Data are expressed as the mean ± SD from three to 10 individual samples in a single experiment, and we performed at least three independent experiments. The statistical significance of the differences between the values obtained was evaluated by unpaired two-tailed Student t test. A P < 0.01 was considered significant.
Data and materials availability
RNA-seq data reported in this study have been deposited in DNA Data Bank of Japan's BioProject under accession number DRA016613. All other data associated with this study are available within the article and its Supplementary Data files or on request from the corresponding author.
Results
Deficiency of mClec4A4 inhibits tumor progression
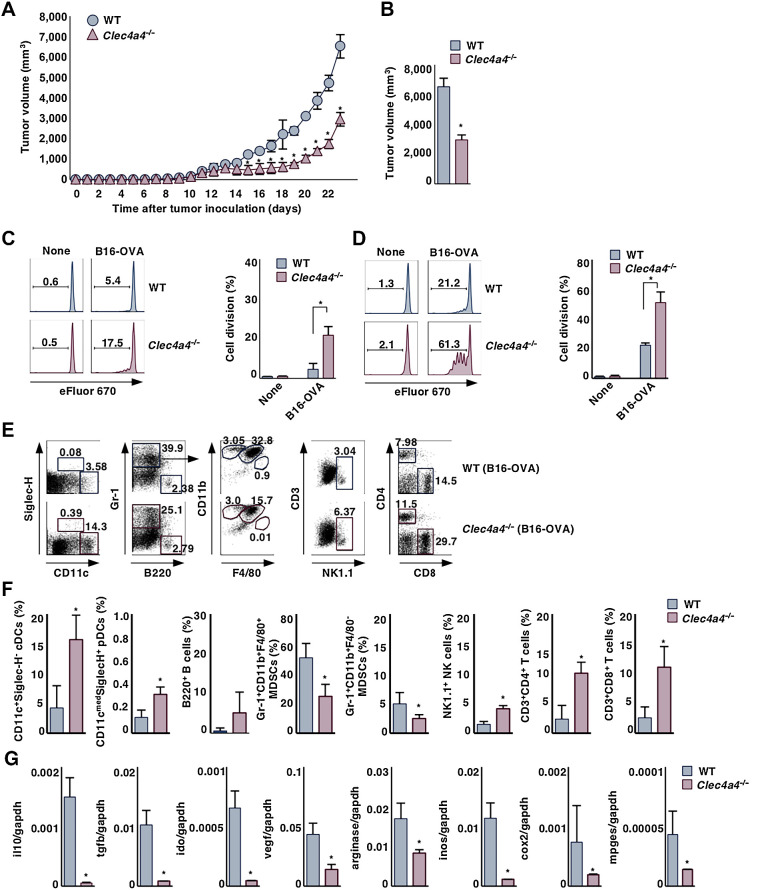
To begin investigating whether mClec4A4 affects tumor progression and antitumor immunity mediated through the control of cDC function, we utilized the B16-OVA experimental model in which the poorly immunogenic B16 murine melanoma tumor cell line has been engineered to express OVA (22) to assess the generation of Ag-specific T-cell responses. To evaluate the relevance of mClec4A4 in antitumor immunity in vivo, we compared tumor growth in WT and Clec4a4−/− mice. Clec4a4−/− mice displayed a significant reduction in tumor growth for the duration of the experiment (Fig. 1A and B). We also observed that Clec4a4−/− mice exhibited a potent suppression of the growth of the immunogenic murine colon adenocarcinoma cell line MC38 (23–25) compared with WT mice (Supplementary Fig. S2A and S2B). These results indicated that deficiency of mClec4A4 suppressed tumor development.
Figure 1.
Deficiency of mClec4A4 abrogates tumor progression. A and B, WT and Clec4a4−/− mice were inoculated with B16-OVA, and tumor growth was monitored. A, Tumor volume for 23 days. B, Tumor volume on day 23; n > 6 per group. P < 0.01 compared with WT mice. WT and Clec4a4−/− mice that had been inoculated with or without B16-OVA were adoptively transferred with eFluor 670–labeled CD45.1+OT-II CD4+ T cells (C) or CD45.1+OT-I CD8+ T cells (D) on day 21 after tumor inoculation. Cell division profile (left) and proportion (right) among CD45.1+OT-II CD4+ T cells (C) or CD45.1+OT-I CD8+ T cells (D) in PLNs in untreated naïve mice or tdLNs in tumor-bearing mice on day 3 after the adoptive transfer; n > 6 per group. Numbers in the histogram represent the proportion of the dividing cells. P < 0.01 compared with WT mice. E–G, WT and Clec4a4−/− mice were inoculated with B16-OVA. E and F, Cell surface expression profile (E) and proportion (F) of leukocytes in tumor tissues on day 21 after tumor inoculation; n > 6 per group. P < 0.01 compared with WT mice. Numbers in the dot plot represent the proportion of the indicated cell populations among leukocytes. G, Transcriptional expression indicated immunosuppressive molecules in tumor tissues on day 21 after tumor inoculation; n > 6 per group. P < 0.01 compared with WT mice. All data are representative of at least three independent experiments.
To address the contributions of CD4+ or CD8+ T cells to tumor regression in the context of mClec4A4 deficiency, we administered CD4- or CD8α-depleting mAb into B16-OVA–bearing mice (Supplementary Fig. S2C–S2F). Treatment of tumor-bearing WT mice or Clec4a4−/− mice with anti-CD4 led to a significant reduction of tumor growth, implying that this treatment not only depleted antitumor CD4+ T cells but also certain CD4+ immunosuppressive populations, such as CD4+ Foxp3+ Tregs, myeloid-derived suppressor cells (MDSC; refs. 27, 28), and pDCs, all of which can impair antitumor immune responses (29). On the other hand, the elimination of CD8+ T cells by anti-CD8α resulted in aggressive tumor growth in tumor-bearing Clec4a4−/− mice. Collectively, these results indicated that the elimination of CD8+ T cells enhanced tumor progression in the context of mClec4A4 deficiency.
To determine the role of mClec4A4 in the initiation of Ag-specific T-cell responses in tumor-bearing mice, we adoptively transferred eFluor670-labeled OT-II CD4+ T cells or OT-I CD8+ T cells, which express the OVA-specific IT-II and OT-1 TCRs, respectively (8, 19, 20), into untreated naïve or B16-OVA–bearing mice, and monitored their Ag-specific division in spleen, PLNs, and tdLNs. Whereas tumor-bearing WT mice exhibited a more potent Ag-specific division of OT-I CD8+ T cells than OT-II CD4+ T cells in TdLNs, their apparent responses were not detected in spleen (Fig. 1C and D; Supplementary Fig. S3). On the other hand, tumor-bearing Clec4a4−/− mice displayed more potent Ag-specific divisions in tdLNs than tumor-bearing WT mice (Fig. 1C and D). We did not observe Ag-specific divisions of OT-II CD4+ T cells and OT-I CD8+ T cells in spleen and PLNs in both untreated naïve mice (Fig. 1C and D; Supplementary Fig. S3). These results indicated that the deficiency of mClec4A4 promoted Ag-specific priming of T cells under tumor-bearing conditions.
Deficiency of mClec4A4 alters the immunosuppressive features of the tumor microenvironment
Tumors appear to operate multiple mechanisms to impair the functions of effector immune cells and generate immunosuppressive immune cell subsets in the tumor microenvironment (TME) and thereby circumvent antitumor immunity and facilitate their progression (30–32). To clarify how the deficiency of mClec4A4 can inhibit tumor development, we examined the leukocytes in lymphoid tissues in B16-OVA–bearing WT mice and Clec4a4−/− mice. In WT mice, tumor development was associated with increased frequencies of Gr-1+CD11b+F4/80+ MDSCs, Gr-1+CD11b+F4/80– MDSCs, and Gr-1+CD11blowF4/80+ macrophages in spleen, whereas it was associated with decreased frequencies in spleen of CD11c+Siglec-H– cDCs, B220+ B cells, CD3+CD4+ T cells, and CD3+CD8+ T cells (Supplementary Fig. S4). In contrast, the increased frequencies of MDSCs were attenuated in spleen in tumor-bearing Clec4a4−/− mice when compared with tumor-bearing WT mice, whereas the reduction in the frequencies of CD11c+Siglec-H– cDCs and B220+ B cells and the enhancement of the frequency of Gr-1+CD11blowF4/80+ macrophages were similarly observed (Supplementary Fig. S4). On the other hand, tumor progression was associated with reduced frequencies of CD11c+Siglec-H– cDCs, CD3+CD4+ T cells, and CD3+CD8+ T cells, enhanced frequency of B220+ B cells, and slightly increased frequency of MDSCs in tdLNs in WT mice (Supplementary Fig. S5). Similar changes in the frequencies of B220+ B cells, MDSCs, CD3+CD4+ T cells, and CD3+CD8+ T cells were observed in tdLNs in Clec4a4−/− mice following tumor transplantation, whereas the frequency of CD11 cmedSiglec-H+ pDCs was higher than tumor-bearing WT mice (Supplementary Fig. S5).
We also compared the tumor-infiltrating leukocytes in B16-OVA–bearing WT and Clec4a4−/− mice (Fig. 1E and F). When compared with tumor-bearing WT mice, tumor-bearing Clec4a4−/− mice exhibited marked infiltration of CD11c+Siglec-H– cDCs, CD11 cmedSiglec-H+ pDCs, NK1.1+ natural killer (NK) cells, CD3+CD4+ T cells, and CD3+CD8+ T cells in tumor tissues, whereas they displayed significant reduction in the accumulation of Gr-1+CD11b+F4/80+ MDSCs and Gr-1+CD11b+F4/80– MDSCs. Taken together, these results indicated that deficiency of mClec4A4 promoted or diminished the infiltration of inflammatory leukocytes or MDSCs, respectively, in lymphoid and tumor tissues in tumor-bearing mice.
Tumor cells and certain tumor-infiltrating immune cells secrete diverse soluble factors that facilitate the emergence of an immunosuppressive TME that attenuates antitumor immune responses, leading to the appearance of progressively growing tumors. We therefore compared the transcriptional expression of immunosuppressive molecules in tumor tissues in B16-OVA–bearing WT mice and Clec4a4−/− mice (Fig. 1G). Tumor tissues obtained from WT mice expressed transcripts encoding IL10, TGFβ, indoleamine 2,3-dioxygenase (IDO), VEGF, arginase, inducible nitric oxide synthase (iNOS), COX2, and membrane-associated prostaglandin E2 synthase (mPGES). Clec4a4−/− mice displayed lower transcriptional expression of these immunosuppressive molecules in tumor tissues than WT mice. Collectively, these results indicated that the deficiency of mClec4A4 affected the formation of the immune regulatory milieu of the TME.
Deficiency of mClec4A4 inhibits the alteration in activation status of cDCs under tumor-bearing conditions
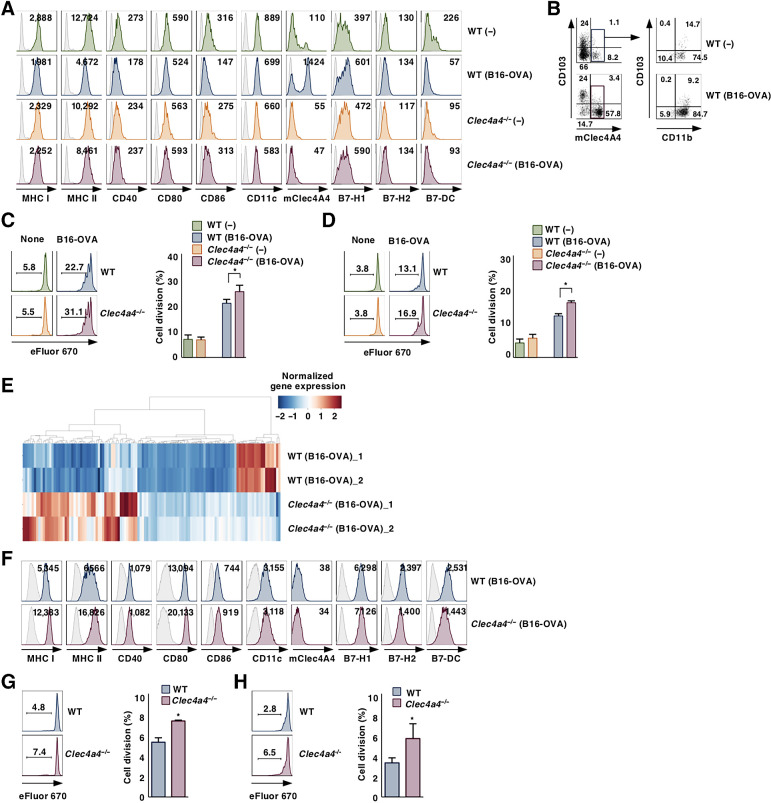
To determine how mClec4A4 controls the activation status of cDCs under tumor-bearing conditions, we characterized cDCs in lymphoid tissues in B16-OVA–bearing WT and Clec4a4−/− mice. Under tumor-bearing conditions, WT mice and Clec4a4−/− mice exhibited similar proportions of XCR1+ cDC1 and SIRPα+ cDC2 in spleen and tdLNs (Supplementary Figs. S6A, S6B, S7A, and S7B). Although tumor progression was associated with reduced expression of MHC class I (MHC I), CD40, CD80, B7-H1, and B7-H2 in splenic CD8α– cDC2 in WT mice, it was associated with enhanced expression of MHC I, CD40, CD80, CD86, B7-H1, and B7-H2 in Clec4a4−/− mice (Supplementary Fig. S6C and S6D). Furthermore, splenic CD8α– cDC2 obtained from Clec4a4−/− mice showed enhanced capacity to produce inflammatory cytokines upon stimulation with LPS as compared with those obtained from WT mice under normal and tumor-bearing conditions (Supplementary Fig. S6E). Following tumor transplantation in WT mice, resident MHC IImedCD11chi cDCs in tdLNs showed reduced expression of MHC I, MHC II, CD40, CD86, CD11c, and B7-DC (Fig. 2A; Supplementary Fig. S7C), while migratory MHC IIhiCD11 cmed cDCs in tdLNs exhibited reduced expression of MHC I, MHC II, CD40, and CD11c (Supplementary Fig. S8A and S8B). On the other hand, the increased expression of B7-H1 was observed in both cDC subsets in TdLNs in WT mice after tumor transplantation (Fig. 2A; Supplementary Figs. S7C, S8A, and S8B). Tumor progression was associated with high expression of mClec4A4 in resident MHC IImedCD11chiCD11b+CD103– cDC2 and resident MHC IImedCD11chiCD11b+CD103+ cDC2 in tdLNs in WT mice (Fig. 2A and B; Supplementary Fig. S7C), although migratory MHC IIhiCD11 cmed cDCs in tdLNs did not express it (Supplementary Fig. S8A and S8B). We also observed that tumor progression was associated with little or no effect on the expression of mClec4A4 on cDCs in other LNs in WT mice (Supplementary Fig. S8C and S8D). In contrast, tumor progression did not reduce the expression levels of MHC I, MHC II, CD40, CD80, CD86, B7-H2, and B7-DC in resident MHC IImedCD11chi LN cDCs, but not migratory MHC IIhiCD11 cmed LN cDCs, obtained from Clec4a4−/− mice, whereas it enhanced or reduced the expression of B7-H1 on resident MHC IImedCD11chi LN cDCs or migratory MHC IIhiCD11 cmed LN cDCs obtained from Clec4a4−/− mice (Fig. 2A; Supplementary Figs. S7C, S8A, and S8B). Furthermore, tumor progression was associated with reduced capacity of resident MHC IImedCD11chi cDCs in tdLNs from WT mice to produce inflammatory cytokines in response to LPS stimulation, whereas this was not seen for those cells from tdLNs in Clec4a4−/− mice (Supplementary Fig. S7D). We also observed that resident MHC IImedCD11chi cDCs obtained from tdLNs in B16-OVA–bearing Clec4a4−/− mice showed a higher capacity to present OVA for the activation of OT-II CD4+ T cells and OT-I CD8+ T cells than those from B16-OVA–bearing WT mice, while resident MHC IImedCD11chi cDCs obtained from PLNs in both untreated naïve mice failed to activate them (Fig. 2C and D). Gene expression profiling revealed that resident MHC IImedCD11chi cDCs obtained from tdLNs in tumor-bearing Clec4a4−/− mice had enhanced expression of about 60 genes related to inflammatory cytokines and chemokines as well as signaling components, whereas they had reduced expression of about 20 genes encoding inhibitory molecules as compared with those from tumor-bearing WT mice (Fig. 2E; Supplementary Tables S3–S6).
Figure 2.
Deficiency of mClec4A4 suppresses the change in activation status of cDCs under tumor-bearing conditions. WT and Clec4a4−/− mice were inoculated with or without B16-OVA. Cell surface expression profile of resident MHC IImedCD11chi cDCs (A) and mClec4A4+ cells among resident MHC IImedCD11chi cDC subsets (B) in PLNs in untreated naïve mice or tdLNs in tumor-bearing mice on day 21 after tumor inoculation; n > 6 per group. Numbers in the histogram represent mean fluorescence intensity (MFI; A). Numbers in the dot plot represent the proportion of the indicated cell populations (B). eFluor 670–labelled CD45.1+OT-II CD4+ T cells (C) or CD45.1+OT-I CD8+ T cells (D) were cultured with resident MHC IImedCD11chi cDCs from PLNs in untreated naïve mice or tdLNs in tumor-bearing mice on day 21 after tumor inoculation; n > 6 per group. Cell division profile (left) and proportion (right) among CD45.1+OT-II CD4+ T cells (C) or CD45.1+OT-I CD8+ T cells (D). Numbers in the histogram represent the proportion of dividing cells. P < 0.01 compared with WT mice. E, Gene expression signature of resident MHC IImedCD11chi cDCs in TdLNs in tumor-bearing mice on day 21 after tumor inoculation. Heat map shows DEGs (log2-FC > 1 or ≤1) determined using RNA-seq data from immune-related genes of resident MHC IImedCD11chi cDCs isolated from tumor-bearing Clec4a4−/− mice as compared with those from tumor-bearing WT mice. Normalized Z score values were calculated for each DEG. The distribution of the expression level of DEGs is shown on the color key legend (up, red; down, blue). F, Cell surface expression profile of cDCs in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the histogram represent mean MFI. eFluor 670–labeled CD45.1+OT-II CD4+ T cells (G) or CD45.1+OT-I CD8+ T cells (H) were cultured with cDCs from tumor tissues on day 21 after tumor inoculation; n > 6 per group. Cell division profile (left) and proportion (right) among CD45.1+OT-II CD4+ T cells (G) or CD45.1+OT-I CD8+ T cells (H). Numbers in the histogram represent the proportion of dividing cells. P < 0.01 compared with WT mice. All data are representative of at least three independent experiments.
Having demonstrated the infiltration of cDCs into tumor tissues in the context of mClec4A4 deficiency, we sought to determine differences in their activation status in tumor tissues between WT mice and Clec4a4−/− mice. WT and Clec4a4−/− mice had similar constituencies of XCR1+ cDC1 and SIRPα+ cDC2 in tumor tissues (Supplementary Fig. S9A and S9B). Tumor-infiltrating cDCs obtained from Clec4a4−/− mice exhibited higher expression of MHC I, MHC II, CD80, CD86, and B7-H1, and lower expression B7-H2 and B7-DC than those obtained from WT mice (Fig. 2F; Supplementary Fig. S9C). Furthermore, tumor-infiltrating cDCs obtained from B16-OVA–bearing Clec4a4−/− mice displayed a higher capacity to present OVA for the activation of OT-II CD4+ T cells and OT-I CD8+ T cells than those obtained from B16-OVA–bearing WT mice (Fig. 2G and H). Collectively, these results indicated that the deficiency of mClec4A4 inhibited changes in the activation status of cDCs under tumor-bearing conditions.
Deficiency of mClec4A4 improves the activation status of T cells
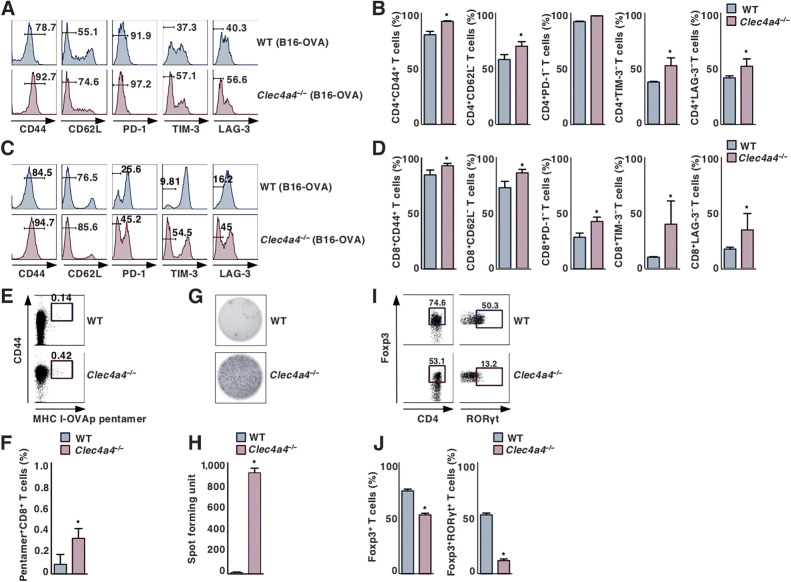
To evaluate how mClec4A4 controls antitumor T-cell responses, we characterized T cells in lymphoid tissues in B16-OVA–bearing WT and Clec4a4−/− mice. Similar expression patterns of CD44 and CD62 L in CD4+ T cells and CD8+ T cells in spleen, PLNs, and tdLNs were observed between normal and tumor-bearing conditions in WT mice, while they did not express any immune checkpoint molecules, including PD-1, T-cell Ig and mucin-domain containing-3 (TIM-3), and lymphocyte activation gene-3 (LAG-3) (Supplementary Figs. S10A, S10B, S11A, and S11B). In contrast, tumor progression was associated with an increase in the fractions of CD4+CD44+CD62L– T cells and CD8+CD44+CD62L– T cells, which are known as effector memory T (TEM) cells (33), in these lymphoid tissues in Clec4a4−/− mice (Supplementary Figs. S10A, S10B, S11A, and S11B). On the other hand, tumor-bearing Clec4a4−/− mice showed enhanced generation of MHC I-OVA pentamer+CD44highCD8+ T cells in TdLNs, but not spleen, as compared with tumor-bearing WT mice (Supplementary Figs. S10C, S10D, S11C, and S11D). Collectively, these results indicated that the deficiency of mClec4A4 enhanced the induction of TEM cells and cytotoxic T lymphocytes (CTL) in lymphoid tissues under tumor-bearing conditions.
Having demonstrated that deficiency of mClec4A4 promoted the accumulation of tumor-infiltrating lymphocytes (TIL), we next sought to determine differences in the activation status of TILs between WT and Clec4a4−/− mice. Clec4a4−/− mice exhibited higher proportions of CD4+CD44+CD62L– TILs and CD8+CD44+CD62L– TILs than WT mice (Fig. 3A–D). Moreover, CD4+ TILs and CD8+ TILs obtained from Clec4a4−/− mice displayed reduced expression of PD-1, TIM-3, and/or LAG-3 as compared with those obtained from WT mice (Fig. 3A-D). Furthermore, tumor-bearing Clec4a4−/− mice showed a marked accumulation of MHC I-OVA pentamer+CD44highCD8+ TILs and Ag-specific IFNγ-producing TILs in tumor tissues as compared with tumor-bearing WT mice (Fig. 3E–H). Taken together, these results indicated that deficiency of mClec4A4 promoted the infiltration of TILs with reduced expression of negative immune checkpoint molecules into tumor tissues.
Figure 3.
Deficiency of mClec4A4 promotes the activation status of TILs under tumor-bearing conditions. WT and Clec4a4−/− mice were inoculated with B16-OVA. Cell surface expression profile (A and C) and proportion (B and D) of CD4+ T cells (A and B) and CD8+ T cells (C and D) in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the histogram represent the proportion of the indicated cell populations. P < 0.01 compared with WT mice. Cell surface expression profile (E) and proportion (F) of MHC I-OVA pentamer+CD44highCD8+ T cells among CD8+ T cells in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the dot plot represent the proportion of the indicated cell populations. P < 0.01 compared with WT mice. OVA-specific IFNγ-producing T-cell response in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Representative picture (G) and spot counts (H) of IFNγ-producing cells. P < 0.01 compared with WT mice. Cell surface expression profile (I) and proportion (J) of CD4+Foxp3+ T cells and CD4+Foxp3+RORγt+ T-cells among CD4+ T cells in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the dot plot represent the proportion of the indicated cell populations. P < 0.01 compared with WT mice. All data are representative of at least three independent experiments.
To address the influence of mClec4A4 deficiency on the appearance of CD4+Foxp3+ Tregs under tumor-bearing conditions, we compared the frequency of these cells in lymphoid tissues in B16-OVA–bearing WT and Clec4a4−/− mice. In WT mice, tumor progression was associated with an enhanced proportion of CD4+Foxp3+ Tregs in TdLNs, but not spleen (Supplementary Figs. S10E, S10F, S11E, and S11F). On the other hand, tumor-bearing Clec4a4−/− mice showed a lower frequency of CD4+Foxp3+ Tregs in tdLNs than tumor-bearing WT mice (Supplementary Fig. S11E and S11F).
We also compared the accumulation of CD4+Foxp3+ Tregs in tumor tissues between B16-OVA–bearing WT and Clec4a4−/− mice. Although CD4+ T-cells infiltrated into tumor tissues in tumor-bearing WT mice (Fig. 1E and F), the majority of these cells were CD4+Foxp3+retinoic acid–related orphan receptor γt (RORγt)– Tregs and CD4+Foxp3+RORγt+ Tregs (refs. 34–36; Fig. 3I and J), which were different from those in lymphoid tissues (Supplementary Figs. S10G, S10H, S11G, and S11H). However, tumor-bearing Clec4a4−/− mice exhibited a lower frequency of tumor-infiltrating CD4+Foxp3+ Tregs than tumor-bearing WT mice (Fig. 3I and J). Collectively, these results indicated the deficiency of mClec4A4 reduced the accumulation of CD4+Foxp3+ Tregs in lymphoid and tumor tissues under tumor-bearing conditions.
Blockade of hCLEC4A with antagonistic anti-hCLEC4A has therapeutic effects in tumor-bearing hCLEC4A-tg mice
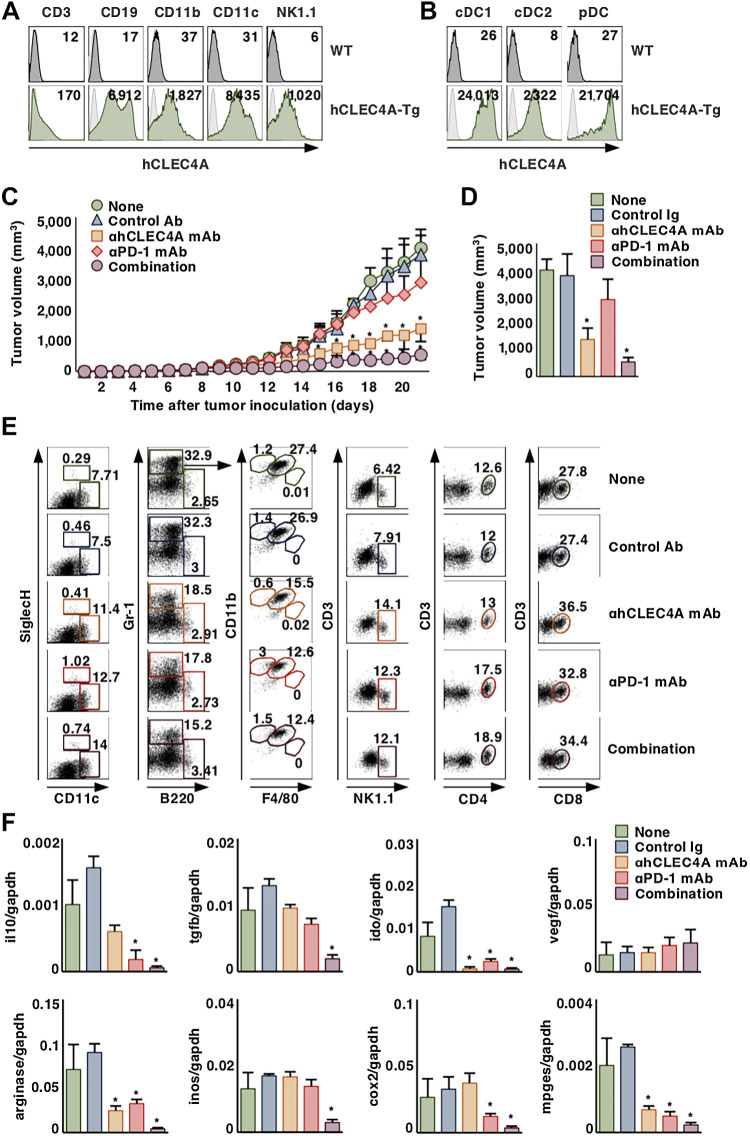
hCLEC4A can be considered a functional ortholog of mClec4A4 (8, 9), and we have recently generated an antagonistic mAb to hCLEC4A that enhances the function of cDCs, while it could not recognize mClec4A4 (9). Given that mClec4A4 acts as an immune checkpoint molecule expressed on cDCs in mice, hCLEC4A could also function as an immune checkpoint regulator for antitumor immunity in humans. To investigate the utility of antagonistic anti-hCLEC4A for protection against tumor development, we created hCLEC4A-Tg mice that express hCLEC4A under the control of the murine Ii promoter (21). Although hCLEC4A is mainly expressed on cDCs in human peripheral blood mononuclear cells (PBMC; ref. 9), CD11c+ cDCs displayed a higher expression of hCLEC4A than CD19+ B cells and CD11b+ macrophages, although NK1.1+ NK cells and CD3+ T cells had slight or no expression, in spleen, TdLNs, and tumor tissues in hCLEC4A-Tg mice (Fig. 4A; Supplementary Fig. S12A). Furthermore, CD11c+CD8α+ cDC1 exhibited a higher expression of hCLEC4A than CD11c+CD8α– cDC2 and CD11 cmedSiglec-H+ pDCs (Fig. 4B). On the other hand, splenic CD8α– cDC2 showed a normal expression of mClec4A4 in hCLEC4A-Tg mice (Supplementary Fig. S12B).
Figure 4.
Blockade of hCLEC4A with antagonistic anti-hCLEC4A suppresses tumor development in hCLEC4A-Tg mice. A, Cell surface expression profile of hCLEC4A on CD3+ T cells, CD19+ B cells, CD11b+ macrophages, CD11c+ DCs, and NK1.1+ NK cells in spleen in hCLEC4A-Tg mice. Numbers in the histogram represent MFI. B, Cell surface expression profile of hCLEC4A on CD11c+CD8α+ cDC1, CD11c+CD4+ cDC2 and CD11 cmedSiglec-H+ pDCs in spleen in hCLEC4A-Tg mice. Numbers in the histogram represent MFI. C–F, B16-OVA–bearing hCLEC4A-Tg mice (tumor volume; approximately 100 mm3) were treated with or without anti-hCLEC4A, anti–PD-1, or control Ab. Tumor growth was monitored. C, Tumor volume over 21 days. D, Tumor volume on day 21; n > 6 per group. P < 0.01 compared with untreated control mice. E, Cell surface expression profile of leukocytes in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the dot plot represent the proportion of the indicated cell populations among leukocytes. F, Transcriptional expression of the indicated immunosuppressive molecules in tumor tissues on day 21 after tumor inoculation; n > 6 per group. P < 0.01 compared with untreated control mice. All data are representative of at least three independent experiments.
Having demonstrated the predominant expression of hCLEC4A on murine DCs subsets in hCLEC4A-Tg mice, we assessed the antitumor effects of the blockade of hCLEC4A and/or PD-1 with mAb administered as monotherapy or combinational therapy to tumor-bearing hCLEC4A-Tg mice. Although the blockade of PD-1 had minimal suppression on the growth of B16-OVA, consistent with prior studies (37), the blockade of hCLEC4A markedly inhibited it (Fig. 4C and D). Furthermore, combination blockade of hCLEC4A and PD-1 showed more potent inhibition on the growth of B16-OVA than monotherapy blockade of hCLEC4A (Fig. 4C and D). In addition, tumor-bearing mice treated with blockade of hCLEC4A had no apparent signs of tissue damage of skin, lung, liver, and SI and no significant weight loss, and appeared similar to untreated tumor-bearing mice (Supplementary Fig. S13). Blockade of hCLEC4A and/or PD-1 exhibited a similar potent suppression of the growth of MC38 (Supplementary Fig. S14). Collectively, these results indicated that monotherapy with antagonistic anti-hCLEC4A was effective at inhibiting the tumor development, while the combination therapy with mAbs to hCLEC4A and PD-1 exerted enhanced antitumor effects.
We further addressed the impact of the blockade of hCLEC4A and/or PD-1 on the constitution of tumor-infiltrating leukocytes and the transcriptional expression of immunosuppressive molecules in tumor tissues in B16-OVA–bearing hCLEC4A-Tg mice. When compared with untreated tumor-bearing hCLEC4A-Tg mice, the blockade of hCLEC4A or PD-1 enhanced infiltrations of CD11c+Siglec-H– cDCs, CD11 cmedSiglec-H+ pDCs, NK1.1+ NK cells, CD3+CD4+ T cells, and/or CD3+CD8+ T cells in tumor tissues, whereas it reduced the accumulation of Gr-1+CD11b+F4/80+ MDSCs (Fig. 4E; Supplementary Fig. S15). On the other hand, the blockade of hCLEC4A or PD-1 reduced transcriptional expression of IL10, IDO, arginase, COX-2, and mPGES in tumor tissues (Fig. 4F). Furthermore, combination blockade of hCLEC4A and PD-1 not only yielded more profound reduction in the expression of these genes than each monotherapy but also suppressed the transcriptional expression of TGFβ and iNOS (Fig. 4F). Taken together, these results indicated that monotherapy with antagonistic anti-hCLEC4A was effective for improving the immunosuppressive TME, while the combination therapy with mAbs to hCLEC4A and PD-1 exerted additive effects.
Blockade of hCLEC4A enhances antitumor immune responses in hCLEC4A-tg mice
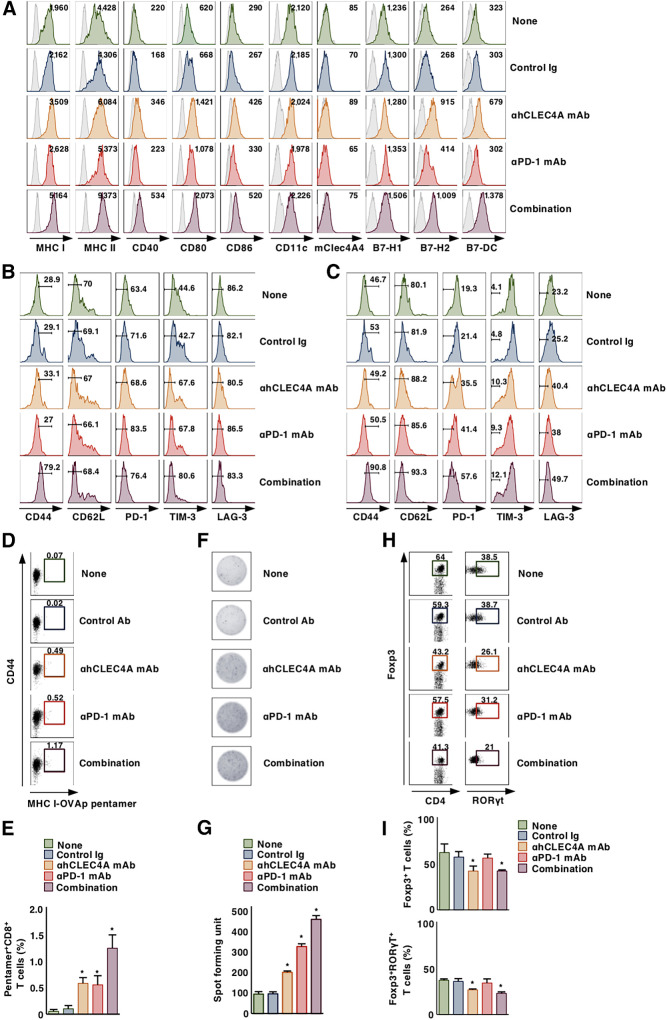
Having demonstrated the ability of immune checkpoint blockade through hCLEC4A to inhibit tumor progression, we addressed the effect of blockade of hCLEC4A with antagonistic anti-hCLEC4A on antitumor immune responses in tumor tissues in tumor-bearing hCLEC4A-Tg mice. Monotherapy with antagonistic anti-hCLEC4A as well as neutralizing anti–PD-1 had little or no effect on the proportions of XCR1+ cDC1 and SIRPα+ cDC2 in tumor tissues when compared with untreated hCLEC4A-Tg tumor-bearing mice (Supplementary Fig. S16A and S16B). On the other hand, monotherapy with antagonistic anti-hCLEC4A resulted in more potent enhancement of the expression of MHC I, MHC II, CD40, CD80, CD86, B7-H2, and B7-DC on tumor-infiltrating cDCs than monotherapy with neutralizing anti–PD-1 when compared with untreated tumor-bearing hCLEC4A-Tg mice. Moreover, their combination yielded further enhancement of the expression of MHC and costimulatory/coinhibitory molecules on tumor-infiltrating cDCs (Fig. 5A; Supplementary Fig. S16C). Collectively, these results indicated that the blockade of hCLEC4A with antagonistic anti-hCLEC4A enhanced the activation status of cDCs under tumor-bearing conditions.
Figure 5.
Blockade of hCLEC4A with antagonistic anti-hCLEC4A enhances antitumor immune responses in hCLEC4A-Tg mice. B16-OVA–bearing hCLEC4A-Tg mice (tumor volume; approximately 100 mm3) were treated with or without anti-hCLEC4A, anti–PD-1, or control Ab. A, Cell surface expression profile of cDCs in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the histogram represent MFI. Cell surface expression profile of CD4+ T cells (B) and CD8+ T cells (C) in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the histogram represent the proportion of the indicated cell populations. Cell surface expression profile (D) and proportion (E) of MHC I-OVA pentamer+CD44highCD8+ T-cells among CD8+ T cells in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the dot plot represent the proportion of the indicated cell populations. P < 0.01 compared with untreated control mice. OVA-specific IFNγ-producing T-cell response in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Representative picture (F) and spot counts (G) of IFNγ-producing cells. P < 0.01 compared with untreated control mice. Cell surface expression profile (H) and proportion (I) of CD4+Foxp3+ T cells and CD4+Foxp3+RORγt+ T cells among CD4+ T cells in tumor tissues on day 21 after tumor inoculation; n > 6 per group. Numbers in the dot plot represent the proportion of the indicated cell populations. P < 0.01 compared with untreated control mice. All data are representative of at least three independent experiments.
We also examined the activation status of TILs under conditions of blockade of hCLEC4A with antagonistic anti-hCLEC4A. Although monotherapy with antagonistic anti-hCLEC4A or neutralizing anti–PD-1 had little or no effect on the proportions of CD4+CD44+CD62L– TILs and CD8+CD44+CD62L– TILs, these treatments reduced the expression of PD-1, Tim-3, and/or LAG-3 on CD4+ TILs and CD8+ TILs, when compared with untreated hCLEC4A-Tg tumor-bearing mice (Fig. 5B and C; Supplementary Fig. S17). Combination therapy with mAbs specific for hCLEC4A and PD-1 also enhanced the proportions of CD4+CD44+CD62L– T cells and CD8+CD44+CD62L– T cells, and reduced expression of negative immune checkpoint molecules on CD4+ TILs and CD8+ TILs (Fig. 5B and C; Supplementary Fig. S17). In addition, blockade of hCLEC4A or PD-1 enhanced the accumulation of MHC I-OVA pentamer+CD44highCD8+ TILs and Ag-specific IFNγ-producing TILs as compared with untreated tumor-bearing hCLEC4A-Tg mice, and their combination further enhanced this (Fig. 5D–G). Taken together, these results indicated that blockade of hCLEC4A with antagonistic anti-hCLEC4A enhanced the effector functions of tumor-specific TILs.
We further studied the emergence of CD4+Foxp3+ Tregs in tumor tissues under the blockade of hCLEC4A with antagonistic anti-hCLEC4A in tumor-bearing hCLEC4A-Tg mice. Monotherapy with antagonistic anti-hCLEC4A, but not neutralizing anti–PD-1, suppressed the infiltration of CD4+Foxp3+RORγt– Tregs and CD4+ Foxp3+RORγ+ Tregs into tumor tissues as compared with untreated tumor-bearing hCLEC4A-Tg mice, while their combination therapy had little or no enhancement to the reduction in their accumulation (Fig. 5H and I). Collectively, these results indicated that blockade of hCLEC4A with antagonistic anti-hCLEC4A inhibited the accumulation of CD4+Foxp3+ Tregs in tumor tissues.
Discussion
Although much attention has been paid to several coinhibitory pathways as targets of immune checkpoint blockade to reinforce T-cell function for tumor immunotherapy (38–40), the intrinsic inhibitory immune checkpoint regulation of cDC function for the control of antitumor immunity remains to be determined. In this study, our findings suggest that mClec4A4 and hCLEC4A serve as unique CLRs function as immune checkpoint regulators of cDC subsets in mice and humans to impair antitumor immunity. Thus, hCLEC4A emerges as a potential target for a novel immune checkpoint blockade tumor immunotherapy.
Analysis of Clec4a4−/− mice revealed that mClec4A4 deficiency led to reduced tumor progression, while the absence of CD8+ T-cells restored tumor progression in this context. Furthermore, the mClec4A4 deficiency enhanced the priming of Ag-specific CD8+ T cells. Therefore, these results suggest that mClec4A4 abrogates the capacity of cDC2 to mediate cross-presentation of tumor cell–associated Ag and prime naïve CD8+ T cells for the generation of tumor-specific CTLs, and that this leads to the progressive development of tumors.
In addition to the enhanced generation and accumulation of MDSCs in lymphoid tissues under tumor-bearing conditions, several inflammatory leukocytes as well as MDSCs and CD4+Foxp3+ Tregs infiltrated into tumor tissues. Furthermore, tumor tissues expressed the transcripts encoding certain immunosuppressive cytokines and metabolite-generating enzymes, possibly produced by MDSCs and CD4+Foxp3+ Tregs as well as other immunosuppressive populations. Therefore, these results suggest that tumor cells and stromal cells not only recruit inflammatory leukocytes, MDSCs, and CD4+Foxp3+ Tregs, but also convert the immunogenic leukocytes into a tolerogenic status through secretion of a variety of immunosuppressive mediators to establish an immunosuppressive TME, and that this inhibits antitumor immunity, subsequently promoting tumor progression. In contrast, in the context of mClec4A4 deficiency, there was not only reduced accumulation of MDSCs and CD4+Foxp3+ Tregs in lymphoid and tumor tissues, but also enhanced infiltration of cDCs, pDCs, NK cells, and T cells in tumor tissues, and this was accompanied by reduced transcriptional expression of several immunosuppressive mediators. Collectively, these results suggest that mClec4A4 promotes a tolerogenic TME mediated by promoting the induction and accumulation of MDSCs and CD4+Foxp3+ Tregs as well as their production of immunosuppressive mediators in lymphoid and tumor tissues, and inhibiting the infiltration of immunogenic leukocytes in tumor tissues, resulting in the abrogation of antitumor immunity and enhanced tumor development.
The activation status of DCs is linked with the expression patterns of MHC and costimulatory/coinhibitory molecules, and there was reduced expression of these molecules observed in splenic mClec4A4+ cDC2 as well as resident and migratory cDCs in tdLNs under tumor-bearing conditions. Furthermore, tumor progression was associated with an impaired ability of splenic mClec4A4+ cDC2 and resident cDCs in tdLNs to produce proinflammatory cytokines. These results suggest that the milieu of the TME-derived immunosuppressive mediators drives these cDC subsets toward tolerogenic DCs (3), favoring tumor progression. In contrast, splenic cDC2 or resident cDCs in tdLNs enhanced or retained expression of MHC molecules and several costimulatory/coinhibitory molecules under mClec4A4-deficient tumor-bearing conditions. In addition, mClec4A4 deficiency enhanced their ability to produce the proinflammatory cytokines even in the tumor-bearing states. Similarly, mClec4A4 deficiency enhanced the expression of MHC molecules and several costimulatory/coinhibitory molecules in tumor-infiltrating cDCs. It is conceivable that damage-associated molecular patterns (DAMP) released from the TME regulate the functions of various types of immune cells and elicit antitumor immune responses or generate chronic inflammation and immunosuppression, resulting in tumor regression or promotion (41, 42). Thus, these phenomena led us to hypothesize that mClec4A4 abrogates tumor-associated DAMP-mediated activation of splenic cDC2 and resident cDC2 in TdLNs, which promotes their tolerogenicity in the TME, possibly through the secretion of immunosuppressive cytokines and metabolites rather than inflammatory mediators, and that inhibits the activation of tumor-infiltrating cDCs, resulting in the attenuation of antitumor immune responses and promotion of tumor development.
Although mClec4A4 is predominantly expressed on cDC2 in spleen, resident cDCs in PLNs consisted of two subsets based on the expression of mClec4A4. Although the reason why there are two subsets of resident cDCs in PLNs with the differential expression of mClec4A4 remains unclear, these subsets might be generated from their common progenitor cells with distinct developmental pathways. Importantly, resident cDCs in tdLNs acquired high levels of expression of mClec4A4 during tumor progression. These phenomena imply that the immunosuppressive milieu of tumor-associated DAMPs and immunosuppressive cytokines drives the enhanced expression of mClec4A4 on resident cDCs in tdLNs, which contributes to the reinforcement of their tolerogenic function for the generation of tumor-associated immunosuppression, leading to tumor progression.
Different from T cells in lymphoid tissues under tumor-bearing conditions, tumor-infiltrating T cells expressed PD-1, TIM-3, and LAG-3 (43–46). These phenomena imply that expression of these immune checkpoint receptors is induced on T cells by complex components distinctively formed in the TME and that this differs from chronic viral infection and autoimmune disorders, where T cells receive persistent poor antigenic exposure in the context of the immunosuppressive cytokine milieu, leading to T-cell exhaustion. Collectively, these findings suggest that the milieu of the immunosuppressive TME drives tumor-infiltrating T cells to express the negative immune checkpoint molecules, leading to impairment of their antitumor effector functions.
Analysis of T cells under tumor-bearing conditions revealed that the deficiency of mClec4A4 promoted the induction of TEM cells and tumor-specific CTLs in lymphoid tissues. Thus, mClec4A4 could suppress the antigen-presenting functions of cDC2, limiting the generation of TEM cells and tumor-specific CTLs in lymphoid tissues. Furthermore, mClec4A4 deficiency not only enhanced the accumulation of tumor-specific TILs, but also reduced their expression of the negative immune checkpoint molecules, suggesting that tumor-infiltrating cDCs deliver antigenic and costimulatory/coinhibitory signals to tumor-specific TILs in the context of the mClec4A4-driven immunosuppressive TME to induce their alternative activation status with the expression of the negative immune checkpoint molecules. Taken together, these results suggest that mClec4A4 increases the threshold of responsiveness of cDC2 to tumor-associated DAMP in lymphoid and tumor tissues to promote tolerogenicity in TME, which impairs the quantity and quality of tumor-specific T-cell responses, leading to accelerated tumor development.
Although tumor progression enhanced the accumulation of CD4+Foxp3+RORγt– Tregs in tdLNs, CD4+ TILs mainly consisted of CD4+Foxp3+RORγt– Tregs and CD4+Foxp3+RORγt+ Tregs. It has been shown that CD4+Foxp3+RORγt+ Tregs, which were derived from CD4+Foxp3+RORγt– Treg cells, exhibited the suppressive activity against CD4+ Teff cell responses (35, 36), while their precursors and role in the control of antitumor immunity has been matter of debate. On the other hand, the analysis of the cytokine milieu in tumor tissues suggest that CD4+Foxp3+RORγt− Tregs convert into CD4+Foxp3+RORγt+ Tregs under the abundance of IL6 and IL1β in the suppressive TME, and CD4+Foxp3+RORγt+ Tregs as well as CD4+Foxp3+RORγt− Tregs impair antitumor responses of CD4+ Teff cells and CTLs. Thus, the TME could subvert antitumor immunity by promoting the expansion/induction, recruitment, and activation of CD4+Foxp3+ Tregs. In contrast, mClec4A4 deficiency reduced the accumulation of CD4+Foxp3+ Tregs in tdLNs and tumor tissues. Therefore, mClec4A4 could decrease the immunogenicity of the TME by enhancing the emergence of CD4+Foxp3+ Tregs through the control of cDC function to restrain antitumor immunity.
We have previously shown the critical roles of the EPS motif and the N-glycosylation site within the CRD in the homotypic interaction of hCLEC4A to transduce the ITIM-mediated downstream signals for the regulation of the function of human cDCs (9). Thus, our findings imply that hCLEC4A constitutively associates with itself through the binding of the EPS motif with oligosaccharide resides on glycans anchored at N-glycosylation site within the CRD on the cell surface of cDCs, and that this results in the ITIM-mediated suppression of their activation. Because the EPS motif within CRD in hCLEC4A potentially binds to several types of N-glycan (7), it is intriguing to hypothesize that other N-glycosylated specific ligand(s) for hCLEC4A with a higher affinity than itself may exist in normal and tumor cells of host as well as microbes, and that this could drive immune suppression under tumor-bearing conditions. Further studies will test these possibilities.
Having demonstrated the expression of hCLEC4A on murine DC subsets in hCLEC4A-Tg mice, this transgenic strain has provides the means to analyze the role of hCLEC4A in the control of DC function in vivo. We have also shown that splenic CD8α– cDC2 show normal expression of mClec4A4 in hCLEC4A-Tg mice. It is possible that the activation status of cDCs is regulated by various inhibitory molecules, including mClec4A4 and hCLEC4A, in hCLEC4A-Tg mice. Thus, these phenomena imply that the transgenic hCLEC4A and other existing inhibitory molecules, including mClec4A4, cooperatively constitute the immune equilibrium in hCLEC4A-Tg mice.
Although blockade of hCLEC4A with antagonistic anti-hCLEC4A removed the hCLEC4A-mediated functional suppression of cDCs (9), it might not completely cancel the functional inhibitory status of cDCs in hCLEC4A-Tg mice. Given that mClec4A4 and hCLEC4A could independently control the activation status of cDCs in human CLEC4A-Tg mice, further analysis of the activation status of cDCs under blockade of hCLEC4A with antagonistic anti-hCLEC4A in Clec4a4−/− hCLEC4A-Tg mice might provide the clues to elucidate their roles in generating the functional inhibitory status of cDCs in hCLEC4A-Tg mice.
We have shown that monotherapy with antagonistic anti-hCLEC4A exhibited more potent inhibition on the development of established poorly immunogenic tumors than monotherapy with neutralizing anti–PD-1 in hCLEC4A-Tg mice, whereas their combination displayed a higher antitumor effect than either monotherapy. Analysis of tumor tissues in hCLEC4A-Tg mice revealed that monotherapy with antagonistic anti-hCLEC4A exhibited more potent enhancement of the expression of MHC and costimulatory molecules on tumor-infiltrating cDCs than monotherapy with neutralizing anti–PD-1. Because the administration of antagonistic anti-hCLEC4A did not deplete hCLEC4A-expressing murine leukocytes in hCLEC4A-Tg mice, our results suggest that the antagonistic anti-hCLEC4A enhances the activation status of hCLEC4A+ cDCs, possibly due to blockade of the inhibitory function of hCLEC4A in vivo. Furthermore, the protective effects of the monotherapy and the combination therapy were associated with enhanced infiltration of tumor-specific Teff cells with reduced expression of negative immune checkpoint molecules, and with reduced accumulation of MDSCs and CD4+Foxp3+ Tregs as well as reduced transcriptional expression of several immunosuppressive molecules in tumor tissues. On the other hand, there were no apparent signs of irAEs in the tumor-bearing mice receiving monotherapy with antagonistic anti-hCLEC4A, implying that blockade of hCLEC4A does not cause the loss of T-cell self-tolerance. Taken together, these results suggest that blockade of hCLEC4A with antagonistic anti-hCLEC4A, which potentiates the antigen-presenting function of hCLEC4A+ cDCs, could effectively suppress tumor development, even tumors unresponsive to current immune checkpoint blockade therapy, including PD-1/PD-L1 axis blockade therapy.
In conclusion, our findings suggest that mClec4A4 and hCLEC4A suppress TME-derived DAMP-mediated activation of cDCs in lymphoid and tumor tissues, and that this provides a tolerogenic environment in the TME that is mediated by promoting the induction and accumulation of MDSCs and CD4+Foxp3+ Tregs, and by inhibiting the infiltration and the function of cDCs and TILs in tumor tissues, leading to the abrogation of antitumor immunity. Thus, mClec4A4 and hCLEC4A act as negative immune checkpoint molecules predominantly expressed on cDC subsets that abolish antitumor immunity. Having demonstrated the therapeutic effect of blockade of hCLEC4A with antagonistic anti-hCLEC4A in hCLEC4A-Tg mice, antagonistic anti-hCLEC4A could be a potential candidate for the treatment of malignant tumors in humans. Furthermore, combination treatment strategies based on blockade of hCLEC4A and other ICIs as well as conventional tumor therapies may also be promising for generating potential improved treatment of patients with cancer.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (B; to K. Sato; 22H02924) and for Scientific Research (C; to T. Uto; 20K07703) from the Ministry of Education, Science and Culture of Japan, the Project for Cancer Research And Therapeutic Evolution (P-CREATE; to K. Sato; 16cm0106307h0001, 17cm0106307h0002, 18cm0106307h0003, 19cm0106307h0004, 20cm0106307h0005, and 21cm0106307h0006) and Project for Promotion of Cancer Research and Therapeutic Evolution (P-PROMOTE; to K. Sato; 22ama221311h0001) from Japan Agency for Medical Research and Development (AMED), Takeda Science Foundation (to T. Uto and T. Fukaya), the Naito Foundation (to K. Sato), Bristol Myers Squibb Foundation Grants (to K. Sato), GSK Japan Research Grant 2016 (to T. Fukaya), GSK Japan Research Grant 2018 (to T. Uto), Daiichi Sankyo Foundation of Life Science (to K. Sato), the Shin-Nihon Foundation of Advanced Medical Research (to T. Uto), and Kobayashi Foundation for Cancer Research (to T. Uto). We thank all members of the animal facility at University of Miyazaki, Yumiko Sato and Rumi Sunachi for secretarial assistance, and Yukari Kawagoe for technical help in cell sorting.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
T. Uto: Formal analysis and investigation. T. Fukaya: Investigation. S. Mitoma: Investigation. Y. Nishikawa: Investigation. M. Tominaga: Investigation. N. Choijookhuu: Investigation. Y. Hishikawa: Resources. K. Sato: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol 2014;14:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy TL, Grajales-Reyes GE, Wu X, Tussiwand R, Briseño CG, Iwata A, et al. Transcriptional control of dendritic cell development. Annu Rev Immunol 2016;34:93–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sato K, Uto T, Fukaya T, Takagi H. Regulatory dendritic cells. Curr Top Microbiol Immunol 2017;410:47–71. [DOI] [PubMed] [Google Scholar]
- 4. Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, et al. Differential antigen processing by dendritic cell subsets in vivo. Science 2007;315:107–11. [DOI] [PubMed] [Google Scholar]
- 5. Neubert K, Lehmann CH, Heger L, Baranska A, Staedtler AM, Buchholz VR, et al. Antigen delivery to CD11c+CD8− dendritic cells induces protective immune responses against experimental melanoma in mice in vivo. J Immunol 2014;192:5830–8. [DOI] [PubMed] [Google Scholar]
- 6. Bates EE, Fournier N, Garcia E, Valladeau J, Durand I, Pin JJ, et al. APCs express DCIR, a novel C-type lectin surface receptor containing an immunoreceptor tyrosine-based inhibitory motif. J Immunol 1999;163:1973–83. [PubMed] [Google Scholar]
- 7. Bloem K, Vuist IM, van der Plas AJ, Knippels LM, Garssen J, García-Vallejo JJ, et al. Ligand binding and signaling of dendritic cell immunoreceptor (DCIR) is modulated by the glycosylation of the carbohydrate recognition domain. PLoS One 2013;8:e66266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uto T, Fukaya T, Takagi H, Arimura K, Nakamura T, Kojima N, et al. Clec4A4 is a regulatory receptor for dendritic cells that impairs inflammation and T-cell immunity. Nat Commun 2016;7:11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nasu J, Uto T, Fukaya T, Takagi H, Fukui T, Miyanaga N, et al. Pivotal role of the carbohydrate recognition domain in self-interaction of CLEC4A to elicit the ITIM-mediated inhibitory function in murine conventional dendritic cells in vitro. Int Immunol 2020;32:673–82. [DOI] [PubMed] [Google Scholar]
- 10. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shin DS, Ribas A. The evolution of checkpoint blockade as a cancer therapy: what's here, what's next? Curr Opin Immunol 2015;33:23–35. [DOI] [PubMed] [Google Scholar]
- 12. Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. [DOI] [PubMed] [Google Scholar]
- 13. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: Mechanisms and strategies. Immunity 2018;48:417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kong YC, Flynn JC. Opportunistic autoimmune disorders potentiated by immune-checkpoint inhibitors anti-CTLA-4 and anti-PD-1. Front Immunol 2014;5:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdel-Wahab N, Shah M, Suarez-Almazor ME. Adverse events associated with immune checkpoint blockade in patients with cancer: A systematic review of case reports. PLoS One 2016;11:e0160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med 2018;378:158–68. [DOI] [PubMed] [Google Scholar]
- 19. Takagi H, Fukaya T, Eizumi K, Sato Y, Sato K, Shibazaki A, et al. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity 2011;35:958–71. [DOI] [PubMed] [Google Scholar]
- 20. Fukaya T, Murakami R, Takagi H, Sato K, Sato Y, Otsuka H, et al. Conditional ablation of CD205+ conventional dendritic cells impacts the regulation of T cell immunity and homeostasis in vivo. Proc Natl Acad Sci U S A 2012;109:11288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Santen H, Benoist C, Mathis D. A cassette vector for high-level reporter expression driven by a hybrid invariant chain promoter in transgenic mice. J Immunol Methods 2000;245:133–7. [DOI] [PubMed] [Google Scholar]
- 22. Brown DM, Fisher TL, Wei C, Frelinger JG, Lord EM. Tumours can act as adjuvants for humoral immunity. Immunology 2001;102:486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corbett TH, Griswold DP Jr, Roberts BJ, Peckham JC, Schabel FM Jr.. Tumor induction relationships in development of transplantable cancers of the colon in mice for chemotherapy assays, with a note on carcinogen structure. Cancer Res 1975;35:2434–9. [PubMed] [Google Scholar]
- 24. Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun 2018;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 2017;214:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uto T, Takagi H, Fukaya T, Nasu J, Fukui T, Miyanaga N, et al. Critical role of plasmacytoid dendritic cells in induction of oral tolerance. J Allergy Clin Immunol 2018;141:2156–67. [DOI] [PubMed] [Google Scholar]
- 27. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells 2020;9:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ueha S, Yokochi S, Ishiwata Y, Ogiwara H, Chand K, Nakajima T, et al. Robust antitumor effects of combined anti-CD4-depleting antibody and anti-PD-1/PD-L1 immune checkpoint antibody treatment in mice. Cancer Immunol Res 2015;3:631–40. [DOI] [PubMed] [Google Scholar]
- 30. Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer 2005;5:263–74. [DOI] [PubMed] [Google Scholar]
- 31. Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol 2006;6:715–27. [DOI] [PubMed] [Google Scholar]
- 32. Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol 2007;25:267–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 2013;31:137–61. [DOI] [PubMed] [Google Scholar]
- 34. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol 2015;6:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, et al. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol 2016;9:444–57. [DOI] [PubMed] [Google Scholar]
- 36. Kim BS, Lu H, Ichiyama K, Chen X, Zhang YB, Mistry NA, et al. Generation of RORγt+ antigen-specific T regulatory 17 cells from Foxp3+ precursors in autoimmunity. Cell Rep 2017;21:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharabi AB, Nirschl CJ, Kochel CM, Nirschl TR, Francica BJ, Velarde E, et al. Stereotactic radiation therapy augments antigen-specific PD-1-mediated antitumor immune responses via cross-presentation of tumor antigen. Cancer Immunol Res 2015;3:345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Le Mercier I, Lines JL, Noelle RJ. Beyond CTLA-4 and PD-1, the generation Z of negative checkpoint regulators. Front Immunol 2015;6:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity 2016;44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ElTanbouly MA, Zhao Y, Nowak E, Li J, Schaafsma E, Le Mercier I, et al. VISTA is a checkpoint regulator for naïve T cell quiescence and peripheral tolerance. Science 2020;367:eaay0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hernandez C, Huebener P, Schwabe RF. Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene 2016;35:5931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patidar A, Selvaraj S, Sarode A, Chauhan P, Chattopadhyay D, Saha B. DAMP-TLR-cytokine axis dictates the fate of tumor. Cytokine 2018;104:114–23. [DOI] [PubMed] [Google Scholar]
- 43. Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014;124:2246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis 2015;6:e1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74–80. [DOI] [PubMed] [Google Scholar]
- 46. Jie HB, Srivastava RM, Argiris A, Bauman JE, Kane LP, Ferris RL. Increased PD-1+ and TIM-3+ TILs during cetuximab therapy inversely correlate with response in head and neck cancer patients. Cancer Immunol Res 2017;5:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables