Abstract
Purpose:
We describe the clinical and genomic landscape of the non–small cell lung cancer (NSCLC) cohort of the American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) Biopharma Collaborative (BPC).
Experimental Design:
A total of 1,846 patients with NSCLC whose tumors were sequenced from 2014 to 2018 at four institutions participating in AACR GENIE were randomly chosen for curation using the PRISSMM data model. Progression-free survival (PFS) and overall survival (OS) were estimated for patients treated with standard therapies.
Results:
In this cohort, 44% of tumors harbored a targetable oncogenic alteration, with EGFR (20%), KRAS G12C (13%), and oncogenic fusions (ALK, RET, and ROS1; 5%) as the most frequent. Median OS (mOS) on first-line platinum-based therapy without immunotherapy was 17.4 months [95% confidence interval (CI), 14.9–19.5 months]. For second-line therapies, mOS was 9.2 months (95% CI, 7.5–11.3 months) for immune checkpoint inhibitors (ICI) and 6.4 months (95% CI, 5.1–8.1 months) for docetaxel ± ramucirumab. In a subset of patients treated with ICI in the second-line or later setting, median RECIST PFS (2.5 months; 95% CI, 2.2–2.8) and median real-world PFS based on imaging reports (2.2 months; 95% CI, 1.7–2.6) were similar. In exploratory analysis of the impact of tumor mutational burden (TMB) on survival on ICI treatment in the second-line or higher setting, TMB z-score harmonized across gene panels was associated with improved OS (univariable HR, 0.85; P = 0.03; n = 247 patients).
Conclusions:
The GENIE BPC cohort provides comprehensive clinicogenomic data for patients with NSCLC, which can improve understanding of real-world patient outcomes.
Translational Relevance.
While prospective clinical trials are essential for identifying therapies that improve outcomes for patients with cancer, trials cannot address all the gaps in our knowledge, particularly when it comes to questions about patient outcomes, mechanisms of disease, acquisition of treatment resistance, and response to therapy. Observational data from patients treated in routine clinical practice (“real-world data”) integrated with genomic sequencing information, allows for more detailed explorations and guides research. American Association for Cancer Research's Project Genomics Evidence Neoplasia Information Exchange (GENIE) Biopharma Collaborative provides high-quality real-world data that integrates rigorously annotated clinical data and corresponding tumor genomic data. Here, we describe the standardized clinical curation of 1,846 patients with non–small cell lung cancer whose tumors have undergone genomic profiling. The publicly available dataset enables real-world estimates of time to event outcomes and exploration of the associations between somatic alterations and response to therapies.
Introduction
Prospective clinical trials are the backbone of clinical cancer research, but many factors, including narrow eligibility criteria and lack of diversity among trial participants, limit the generalizability of prospective clinical trial results (1, 2). Real-world data, broadly defined as any data collected outside of a prospective clinical trial, can provide nuanced understanding of treatment effectiveness in a larger sample and address gaps in knowledge not addressed by clinical trials (3, 4). Collaborative repositories such as The Cancer Genome Atlas (5) have provided large-scale genomic data, which has led to transformational insights into the molecular landscape of various cancers (6, 7). Analyses of these data have resulted in the identification of mutational signature across cancer types (8) and improved prognostication by molecular characteristics (9). However, the addition of data describing clinical treatment and patient outcomes requires laborious curation of medical records and is beyond the scope of most publicly available genomic resources.
The American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) is a publicly available registry of next-generation sequencing (NGS) results from participating cancer institutions internationally (10). With the GENIE version 13.0-public release, data from 167,423 tumor samples and 148,268 patients were made available for analysis. In 2019, AACR launched the Biopharma Collaborative (GENIE BPC), a 5-year, multiphase research collaboration with a coalition of 10 biopharmaceutical companies to add deep clinical annotation to select cohorts of patients from a subset of institutions providing data in the main GENIE Registry. The project applies the PRISMM framework (Pathology; Radiology; Imaging; Signs and Symptoms; tumor Markers; Medical oncology assessments) for extraction of clinical data from electronic health records (EHR) by trained curators and formulation of a set of “real world” standards by which to categorize oncologic outcomes (11). Curated data undergo a multistep quality control and review process (12), which allows for compilation of clinical diagnosis, treatment, and outcome data, integrated with comprehensive tumor genomic characterization, for thousands of patients with cancer, a scale not typically available to the public. Here, we describe the clinical and genomic landscape of the non–small cell lung cancer (NSCLC) GENIE BPC cohort, the first of six cancer cohorts to be publicly released as part of phase I of Project GENIE BPC. The GENIE BPC NSCLC 2.0-public data release is available for download (https://www.synapse.org/#!Synapse:syn27056172/wiki/616601) and can be visualized and analyzed using the cBioPortal interface (https://genie.cbioportal.org/study/summary?id=nsclc_public_genie_bpc; de Bruijn I, et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. Forthcoming 2023).
Lung cancer is the second most common cancer in the United States for men and women (13), with over 200,000 new diagnoses in 2021. NSCLC comprises 80%–85% of lung cancer cases, the vast majority of which are driven by cigarette smoking (14). Deep molecular profiling has underscored the molecular heterogeneity of NSCLC. Complex tumor genomics and high tumor mutational burden (TMB) are often present in patients with heavy smoking histories while oncogene-driven tumors are frequently discovered in patients who never smoked (15). While the development of 10 effective matched targeted therapies for oncogene-driven NSCLC may have contributed to declines in lung cancer–related mortality in the last decade (16), nuanced understanding of how somatic alterations impact response to treatment outcomes is needed to further improve outcomes for all patients with lung cancers. With detailed clinicogenomic curation of 2,004 tumor samples from 1,846 patients, the GENIE BPC NSCLC cohort provides an opportunity to gain new insights into this clinically and molecularly heterogeneous disease.
Materials and Methods
Patient selection
The study was performed in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained for each subject or subject's guardian and human investigations were approved after approval by Institutional Review Boards (IRB) at respective institutions. Patients were selected for the GENIE BPC NSCLC cohort from the GENIE registry (10). Eligibility criteria were as follows: NGS reported at Dana-Farber Cancer Institute (DFCI), Memorial Sloan Kettering Cancer Center (MSKCC), or Vanderbilt-Ingram Cancer Center (VICC) between January 1, 2015, and December 31, 2017, or at Princess Margaret Cancer Centre-University Health Network (UHN) between January 1, 2014, and December 31, 2017; patient age ≥18 years at time of genomic sequencing; minimum of 2 years possible follow-up after sequencing; and eligible OncoTree Diagnosis (17) code. Eligible codes were NSCLC, ciliated muconodular papillary tumor, large cell lung carcinoma, lung adenocarcinoma, pleomorphic carcinoma of the lung, lung squamous cell carcinoma, poorly differentiated NSCLC, lung adenosquamous carcinoma, mucoepidermoid carcinoma of the lung, lymphoepithelioma-like carcinoma of the lung, clear cell carcinoma of the lung, large cell lung carcinoma with rhabdoid phenotype, giant cell carcinoma of the lung, or basaloid large cell carcinoma of the lung. Of 6,152 patients screened from GENIE v8.0-public, 4,982 met all eligibility criteria. Patients from each institution were randomly sampled for a total of 1,846 patients (DFCI, n = 696; MSKCC, n = 869; VICC, n = 226; UHN, n = 55) whose phenomic data were curated using the PRISSMM framework (11). For a subset of patients in the GENIE BPC NSCLC cohort that were treated with immune checkpoint inhibitors (ICI) at MSKCC, data from an MSKCC IRB/Privacy Board–approved separately maintained, retrospective database with RECIST assessments by a thoracic radiologist were available.
Genomic data
As described previously (10), all centers in AACR Project GENIE provide mutation data in mutation annotation format or variant cell format as well as browser extensible data files for each assay panel. Full details of each center's clinical sequencing pipeline are available in the GENIE data guide (GENIE 10.1 public release; AACR; RRID: SCR_009197); summary of gene panels used is provided in Supplementary Table S1. NGS panels varied with respect to genes that were sequenced; patients with sequencing performed on a panel that did not include a particular gene were excluded from all analyses involving that gene (e.g., a patient sequenced on a panel that did not include STK11 is excluded from any analyses involving STK11). Details of gene coverage of each panel are publicly available (https://www.synapse.org/#!Synapse:syn26706786). Somatic oncogenic and likely oncogenic alterations according to OncoKB version 4.0 were used for analyses (17, 18).
Clinical data curation
Trained curators extracted clinical data from the EHR using the PRISSMM framework, as described previously (11, 12, 19). Curators reviewed the text of all pathology reports and recorded tumor characteristics (e.g., histology). All imaging reports (other than plain radiographs and ultrasounds) and one note from a medical oncologist (if available) or advanced practice provider per month were curated. Radiologist and medical oncologist assessments were reviewed for the presence of cancer and whether the cancer was improving/responding, stable, mixed, or progressing/worsening/enlarging; radiologist assessments and pathology reports were also reviewed for sites of disease. All cancer-directed drugs, including start and stop dates, were curated. Cancer-directed drugs include cytotoxic chemotherapies, immunotherapies, targeted therapies, and hormone therapies. A break in treatment of ≥8 weeks was used to indicate the end of a regimen; even if all drugs in the regimen were reinitiated 8+ weeks later, this was considered a new regimen. Additional details regarding curation and the clinical variables available are provided in the Analytic Data Guide that accompanies each data release (20). Data are stored in a Research Electronic Data Capture (REDCap; refs. 21, 22) database at each participating institution and uploaded to Sage Bionetworks for aggregation across institutions. A detailed description of the data processing and quality assurance pipeline has been published previously (12).
Statistical analyses
Descriptive statistics were used to summarize patient and cancer characteristics, including treatment regimens received. If a patient had multiple NSCLC diagnoses with associated genomic sequencing, the earliest was selected for analysis. Patient characteristics were descriptively compared with large phase III, practice-changing trials without molecular eligibility criteria for NSCLC [JMDB (23), KEYNOTE-189 (24), and KEYNOTE-042 (25)].
Survival analyses were performed to assess the association between genomic alterations with overall survival (OS) and progression-free survival (PFS) by treatment received: patients who received first-line platinum chemotherapy regimens (excluding regimens containing pembrolizumab, targeted therapies, or investigational agents), first-line EGFR tyrosine kinase inhibitors (TKI), or second-line and later single-agent ICI. For patients treated with second-line or later ICI therapy, TMB was quantified across gene panels. Unadjusted TMB values based on the number of nonsynonymous mutations per megabase (Mb) of genome covered were transformed to standardized z-scores within each sequencing panel using a power transformation to convert right-skewed TMB distributions to normal distributions (26). Samples that were sequenced on hotspot panels were excluded from analyses of TMB because the mutation rate is not reflective of the actual TMB; samples that were the sole sample sequenced on a panel were also excluded because it was not possible to normalize TMB across one sample.
Four definitions of real-world PFS from the start of a cancer-directed regimen are derived from the PRISSMM data model (19): (i) PFS-imaging (PFS-I; time to disease worsening documented in an imaging report or death), (ii) PFS-medical oncologist (PFS-M, time to first indication of disease worsening in medical oncologist note or death), (iii) PFS-I-or-M (time to first indication of disease worsening documented in an imaging report or medical oncologist assessment, whichever is earliest, or death), and (iv) PFS-I-and-M (time to the latter of disease worsening that was documented in both an imaging report and medical oncologist assessment or death). Patients without progression were censored at the start of a subsequent cancer-directed regimen, if applicable, or on the last known alive date in the absence of a subsequent regimen. On the basis of prior work examining the PRISSMM PFS endpoints as candidate surrogate outcomes for OS, PFS-I-and-M was used for the primary analyses because it demonstrated the strongest correlation with OS; analyses of PFS-I, PFS-M, and PFS-I-or-M are shown in the supplement (19). PFS endpoints were restricted to patients who did not progress prior to the NGS report date. PFS endpoints were reported among patients with advanced disease (i.e., de novo stage IV disease and patients diagnosed with stage I–III disease who later developed distant metastasis, where the anatomic site of distant metastasis was defined using ICD-O-3 codes from an imaging or pathology report).
Time to next treatment (TTNT) was defined as the time from the start of a drug regimen to the next treatment, last known alive date, or death. Receipt of a subsequent treatment (for any cancer, if applicable) and death were considered events; patients without an event were censored at the last known alive date.
Kaplan–Meier estimates and log-rank tests were used to explore univariable associations between genomic alterations and time-to-event endpoints. Multivariable Cox proportional hazards models were fit adjusted for stage at diagnosis (I–III vs. IV), age at diagnosis (years), smoking status (current, former, never smoker), and time from regimen start to NGS report date (months). Survival analyses account for the left truncated nature of the data, entering patients into the risk set at the time of their NGS report date (27). Note that given the left truncated nature of the data, the risk table may show an increasing number of patients at risk due to patients entering the risk set at the time of genomic sequencing report. In the case that a patient's first index cancer diagnosis was associated with multiple NGS reports, the earliest sequencing report was selected. Patients whose sequencing reports were returned postmortem or after the last known alive date were excluded from all time-to-event analyses. Median survival [95% confidence interval (CI)] estimates and HRs (95% CI) are reported.
The analyses presented in this article were facilitated by the cbioportalR and genieBPC R packages. cbioportalR utilizes the cBioPortal API to pull data into R from cBioPortal databases. The genieBPC R package, developed by the GENIE BPC Statistical Coordinating Center, establishes an infrastructure for downloading the GENIE BPC data from Synapse into R, processing the data to build an analytic cohort, and visualizing the drug regimen data using a sunburst plot. Both packages are available on CRAN (28). Analyses were performed in R version 4.1.0 (29).
Data availability
The GENIE BPC NSCLC 2.0-public data release is available for download at https://www.synapse.org/#!Synapse:syn27056172/wiki/616601. The data can be downloaded after creating a free online account.
Results
Clinical landscape
Baseline patient characteristics for the 1,846 patients are noted in Table 1. Baseline patient characteristics of the GENIE BPC cohort were compared with patients enrolled in three contemporaneous practice-changing, phase III NSCLC clinical trials without genomic marker eligibility criteria [JMDB (30), KEYNOTE-189 (24), and KEYNOTE-042 (25); Supplementary Table S2]. The GENIE BPC cohort features a higher proportion of women compared with all three clinical trials (58% vs. 29%–41%) and more Black patients than all three clinical trials combined (n = 95 vs. 73).
Table 1.
Patient and cancer characteristics.
| Characteristics | GENIE BPC n = 1,846, n (%) |
|---|---|
| Age at diagnosis | |
| Median (range) | 65 (18–88) |
| Sex | |
| Female | 1,064 (58) |
| Male | 782 (42) |
| Race | |
| White | 1,541 (83) |
| Asian | 120 (7) |
| Black | 95 (5) |
| Other | 31 (2) |
| Unknown | 59 (3) |
| Ethnicity | |
| Non-Spanish, non-Hispanic | 1,733 (94) |
| Spanish/Hispanic/Latino NOS | 12 (1) |
| Unknown | 101 (5) |
| Smoking status | |
| Current smoker | 258 (14) |
| Former smoker | 1,166 (63) |
| Never smoker | 419 (23) |
| Unknown | 3 (<1) |
| Histology | |
| Adenocarcinoma | 1,150 (62) |
| Squamous cell | 171 (9) |
| Other histology | 268 (15) |
| Unknown | 257 (14) |
| Stage at diagnosis | |
| I | 499 (27) |
| II | 176 (10) |
| III | 359 (19) |
| Stage I–III NOS | 12 (<1) |
| IV | 797 (43) |
| Unknown | 3 (<1) |
| Distant metastases postdiagnosis a | 583 (56) |
| Concurrent cancer diagnoses | |
| One NSCLC diagnosis | 1,307 (71) |
| Multiple cancer diagnoses | 539 (29) |
| Institution | |
| DFCI | 696 (38) |
| MSKCC | 869 (47) |
| UHN | 55 (3) |
| VICC | 226 (12) |
Abbreviation: NOS, not otherwise specified.
aPercentage reported among individuals diagnosed with stage I–III disease.
The most common systemic therapies received by patients in the cohort were platinum-based combination chemotherapies (e.g., 21% of patients treated with carboplatin with pemetrexed, 10% of patients treated with cisplatin and pemetrexed; Supplementary Table S3). The most frequent ICI monotherapies received were nivolumab (14%) and pembrolizumab (9%). Serial treatments patients with stage IV disease are visualized graphically (Fig. 1A). A median of nine medical oncology notes, 14 imaging reports, and four pathology reports were reviewed per patient, highlighting the broad scope of clinical characterization for this cohort of patients (Supplementary Table S4).
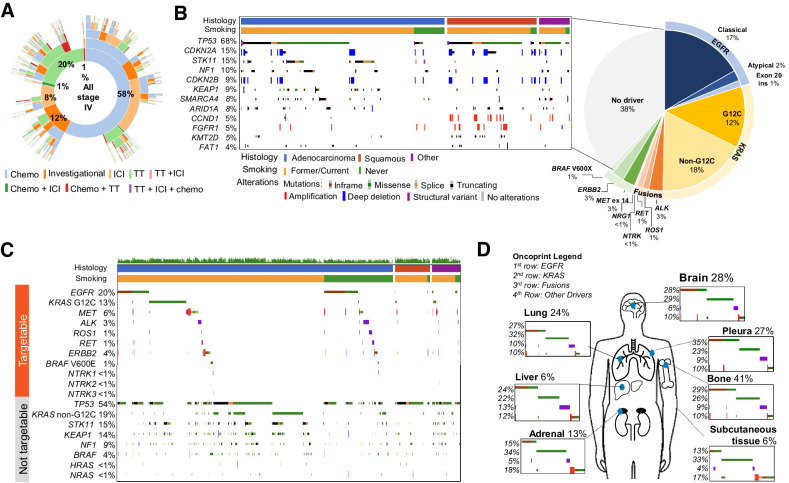
Figure 1.
Landscape of GENIE BPC NSCLC cohort. A, Treatments: Cancer-directed treatment regimens received are depicted using a sunburst plot, where the innermost ring represents the distribution of first treatment regimens received, the second ring represents the distribution of second treatment regimens received, and so on, such that the proportion of patients with a given treatment trajectory is easily visualized. Regimen line was defined as the order of the regimen given after the date of metastasis (i.e., a first-line regimen is the first regimen given following a de novo stage IV lung cancer diagnosis, or the first regimen given following evidence of distant metastasis for a patient diagnosed with stage I–III disease). B, Genomics of NSCLC. Distribution of oncogenic drivers in the cohort, with description to the left highlighting the most frequent alterations in samples without an identified driver. C, Graph of notable targetable and nontargetable alterations. Only oncogenic or likely oncogenic alterations are included. Patients are filtered according to histology and smoking history, shown at top of plot. D, Sites of disease; 797 samples out of 1,846 are annotated with sites of metastatic disease at diagnosis. Chemo, chemotherapy; TT, targeted therapy.
Genomic landscape
Nearly half (44%) of patients in the cohort harbored an oncogenic alteration for which there are either FDA approved or National Comprehensive Cancer Network (NCCN) (31) recommended therapies, with alterations in EGFR (20%) and KRAS G12C (13%) the most frequently observed (Fig. 1B). In addition to 18% of patients carrying KRAS non-G12C driver alterations (which have no approved targeted therapies), 38% of patients had no identifiable druggable target. Among patients without identifiable drug targets, alterations in TP53 (67%), STK11 (15%), and CDKN2A (12%) were frequent. By sorting NSCLC according to histology and smoking status, we visualize the enrichment of oncogenic alterations in KRAS, TP53, STK11, and KEAP1 in adenocarcinoma tumors from patients with former smoking histories compared with enrichment for oncogenic driver alterations in EGFR, ALK, ROS1, and RET in those without smoking history (Fig. 1C). A paucity of targetable oncogenic drivers is also observed in tumors with squamous histology.
The range of clinical characteristics and inclusion of curated sites of disease enables exploration of genomic alterations by a range of features. As an example, in Fig. 1D is shown the prevalence of common oncogenic driver alterations (EGFR, KRAS, fusion alterations, and others) in the most frequent sites of metastases, which are annotated for 797 patients diagnosed with de novo stage IV NSCLC. Oncogenic driver alterations in EGFR were found in 35% of pleural metastases, compared with 13% of subcutaneous metastases, where KRAS alterations were found in 33% of samples with subcutaneous metastases.
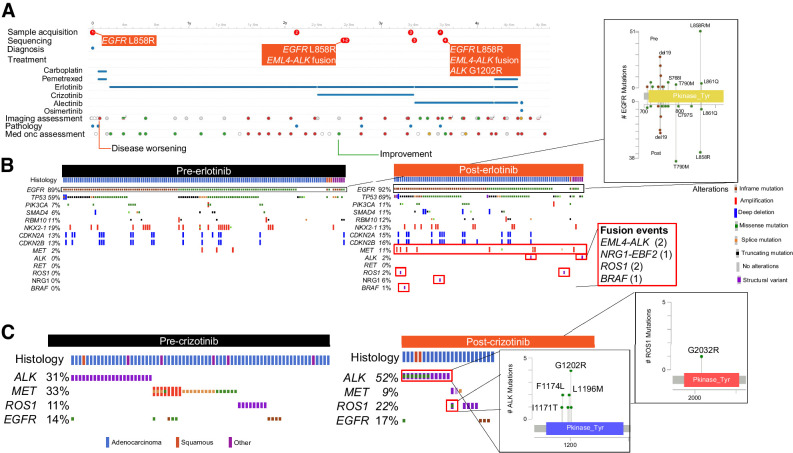
A subset of patients (n = 143, 8%) had multiple NGS tests, facilitating identification of genomic alterations acquired with the selective pressures of treatment. In the cBioPortal interface, allele frequencies of mutations over time can also be tracked. We highlight the longitudinal case of a patient to demonstrate how intrapatient genomic evolution can be traced using the single-patient view in cBioPortal (Fig. 2A). The patient (GENIE-DFCI-004022) was initially diagnosed with stage IV lung adenocarcinoma with EGFR L858R mutation and briefly treated with carboplatin and pemetrexed before starting erlotinib. The tumor acquired in stepwise fashion an EML4-ALK fusion on erlotinib followed by a second-site ALK G1202R resistance mutation during subsequent crizotinib therapy.
Figure 2.
Dynamic genomic changes over time. A, Patient snapshot. Timeline of all samples, treatments given, imaging, pathology and medical oncology assessments are provided for each patient, allowing for facile identification of genomic evolution with sequential treatment. B, Acquired alterations post-erlotinib: Most frequent genomic alterations observed for patients before and after treatment with erlotinib are shown; lollipop plots and inset boxes highlight acquisition of known mechanisms of resistance, including second-site EGFR alterations after erlotinib treatment, acquired gene fusions and MET amplification. C, Acquired alterations post-crizotinib. Plots identifying acquisition of the analogous gatekeeper mutations ROS1 G2032RR and ALK G1202R.
Alterations that are potentially acquired posttreatment can be identified by comparing genomic profiles for samples collected before and after specific treatments using cBioPortal. Among patients treated with erlotinib, 138 samples taken from 132 patients before receipt of erlotinib and 99 tumor samples from 90 patients obtained post-erlotinib are available; among these, 24 patients had paired NGS performed both pre- and post-erlotinib while the remainder are unpaired. Analysis of pre- and post-erlotinib samples demonstrates enrichment for second-site EGFR alterations (T790M, C797S), oncogenic fusions (ALK, NRG1, ROS1, and BRAF) and MET amplification after erlotinib treatment, which are known mechanisms of resistance to erlotinib (ref. 32; Fig. 2B). Similarly, NGS data are available on 80 samples taken before receipt of crizotinib from 67 patients and 25 samples taken after receipt of crizotinib from 22 patients, among which 7 patients had paired NGS performed both pre- and post-crizotinib. Comparison of the samples demonstrates enrichment for the analogous gatekeeper mutations in ROS1 (G2302R) and ALK (G1202R) after treatment (Fig. 2C). Such exploration can facilitate discoveries of new mechanisms of acquired resistance to available treatments.
Assessing the impact of genomic characteristics on survival
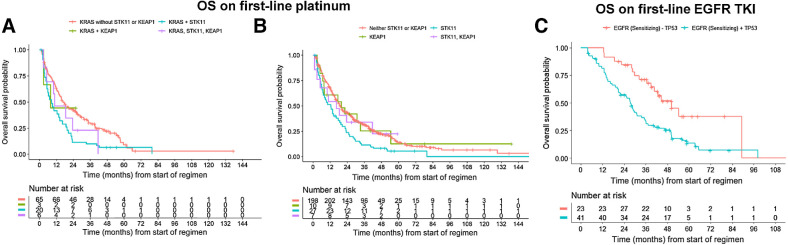
Somatic alterations can impact patient outcomes with systemic treatments that lack genomic selectivity, such as chemotherapy or immunotherapy. STK11 and KEAP1 comutations in KRAS-mutant NSCLC are known to influence survival during treatment with ICI (33, 34), but their impact on survival during platinum chemotherapy treatment is not well described. Patients who received first-line platinum chemotherapy harbored the following: KRAS alone (n = 113), KRAS and STK11 (n = 31), KRAS and KEAP1 (n = 4), KRAS and STK11 and KEAP1 (n = 8). Median OS from start of regimen was 16.6 months (95% CI, 13.3–26.5 months), 9.3 months (95% CI, 5.3–19.5 months), 7.9 months (95% CI, 3.0–not reached), and 10.8 months (95% CI, 5.1–not reached), respectively (Fig. 3A). However, neither OS nor PFS was statistically significantly different between the groups (Supplementary Fig. S1A–S1E). Similarly, across both KRAS-mutant and KRAS-wild-type patients treated with first-line platinum chemotherapy, STK11 and KEAP1 mutations were not significantly associated with prolonged OS or PFS (Fig. 3B, P = 0.07). Median OS was 19.1 months (95% CI, 16.7–24.9), 12.7 months (95% CI, 7.2–20.5), 20.0 months (95% CI, 7.8 months–not reached), and 18.5 months (95% CI, 10 months–not reached), respectively, for patients with STK11-wt/KEAP1-wt (n = 364), STK11 (n = 50), KEAP1 (n = 17), and STK11/KEAP1 (n = 12) mutations. There was also no association between STK11/KEAP1 mutation status with PFS (Supplementary Fig. S2A–S2E).
Figure 3.
Impact of genomics on survival. A, OS for patients with KRAS driver alterations treated with first-line platinum-containing chemotherapy, with or without co-occurring alterations in STK11 and KEAP1. HRs with 95% CI are shown in corresponding table. B, OS for all patients treated with first-line platinum chemotherapy, with or without STK11 and/or KEAP1 mutations. C, OS for patients with sensitizing EGFR alterations treated with first-line erlotinib, with or without co-occurring TP53 mutations. Note that due to the left truncated nature of the data, the risk table may show an increasing number of patients at risk due to patients entering the risk set at the time of genomic sequencing report.
To evaluate the prognostic implications of TP53 comutations in EGFR-mutant lung cancer, for which previously reported findings have been inconsistent (35–37), we examined the impact of TP53 comutations on patients with EGFR sensitizing alterations treated with first-line, first- or second-generation EGFR TKI monotherapy. There were 124 patients treated with erlotinib (n = 99), afatinib (n = 16), or gefitinib (n = 9). Of these, 71 had TP53/EGFR comutations, 37 only had EGFR sensitizing alterations, and 16 had unknown EGFR/TP53 mutation status. Median overall survival on first-line EGFR TKI was 50.3 mo (95% CI, 39.0–not reached) for patients with EGFR sensitizing alterations alone compared with 27.6 months (95% CI, 20.8–35.3) for patients with TP53 comutations (Fig. 3C). In multivariable analysis, TP53 mutation status was associated with worse OS (HR, 2.52; 95% CI, 1.39–4.57; P < 0.01) adjusted for stage at diagnosis, age, smoking history, and time between diagnosis and NGS.
Pembrolizumab has a tumor-agnostic FDA approval for patients with high TMB (≥10 mutations/Mb; ref. 38). High TMB has not been shown to be associated with improved OS in prospective clinical trials of ICI in patients with NSCLC (39), although meta- and retrospective analyses have suggested an association between TMB and survival on ICI treatment in NSCLC (40, 41). In univariable analysis, TMB z-score was associated with OS during second-line and later ICI treatment (HR, 0.85; 95% CI, 0.74–0.98; P = 0.03; n = 247) and with PFS-I- and-M (HR, 0.81; 95% CI, 0.70–0.94; P = 0.006). In multivariate analysis, adjusting for stage at diagnosis, age at diagnosis, smoking status, line of therapy, and time between diagnosis and NGS, the association between TMB z-score and PFS-I-and-M remained significant (HR, 0.84; 95% CI, 0.72–0.98; P = 0.03).
Real-world survival on standard therapies
As a large-scale dataset of patients from multiple institutions, the detailed curation available in the GENIE BPC dataset presents opportunities for real-world estimates of survival that may be used to answer questions ranging from patient-oriented (“how long do patients derive benefit from these treatments?”) to outcomes research assessing the efficacy-effectiveness gap for cancer therapeutics outside of trial settings (1). As proofs of principle, we provide here OS and PFS I-and-M estimates for several common treatments for NSCLC.
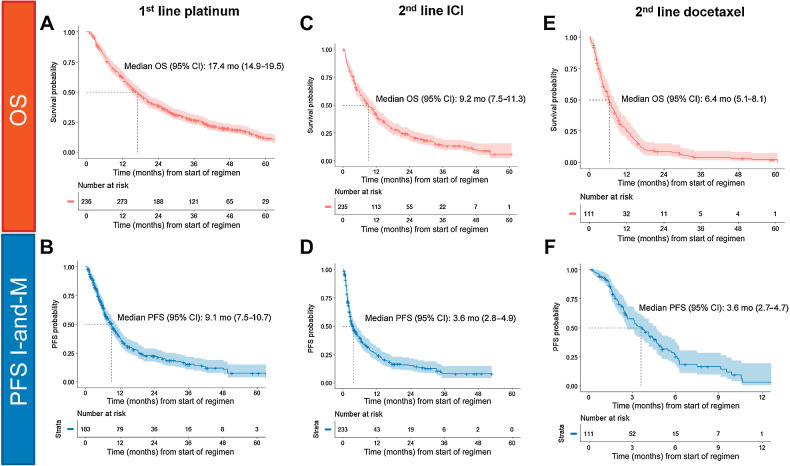
At the time when many patients in the GENIE BPC cohort received first-line treatment, platinum-doublet chemotherapy without immunotherapy was the preferred first-line treatment for patients with advanced NSCLC, with the exception of patients with oncogenic drivers who receive first-line matched targeted therapies (31). In the GENIE BPC dataset, 511 patients received a first-line regimen containing either cisplatin or carboplatin without concurrent pembrolizumab, targeted therapies, or investigational agents for the treatment of metastatic NSCLC. A total of 154 patients (30%) were initially stage I–III and later developed metastatic disease and 357 patients (70%) initially were diagnosed as stage IV. The most common treatments given were carboplatin and pemetrexed (n = 183, 36%), carboplatin, pemetrexed, and bevacizumab (n = 74, 14%), carboplatin and paclitaxel (n = 62, 12%), and cisplatin and pemetrexed (n = 61, 12%). Median OS from start of regimen was 17.4 months (95% CI, 14.9–19.5 months; Fig. 4A); median PFS I-and-M was 9.1 months (95% CI, 7.5–10.7 months; n = 340; Fig. 4B).
Figure 4.
Real-world survival estimates. A, OS for all patients treated with first-line platinum-containing chemotherapy regimens after metastatic index lung cancer diagnosis. B, PFS imaging-and-medical oncology (PFS I-and-M) for all patients treated with first-line platinum-containing chemotherapy. A total of 179 patients were excluded because of disease progression prior to sequencing test performed. C, OS for all patients treated with second-line or higher ICIs. D, PFS I-and-M for all patients treated with second-line or higher ICI E. OS for all patients treated with second-line or higher docetaxel-containing regimens. F, PFS I-and-M for all patients treated with second-line or higher docetaxel.
The most common second-line and later treatments received in the GENIE BPC cohort were ICI or docetaxel with or without ramucirumab. These were the preferred second-line treatments at the time following a series of landmark trials comparing single-agent anti-PD-(L)1 antibodies with docetaxel (31, 42–44). In total, 289 patients received ICI in the second-line or later setting and did not receive ICI in the first-line setting: nivolumab (n = 175), pembrolizumab (n = 73), atezolizumab (n = 39), durvalumab (n = 2). Median OS was 9.2 months (95% CI, 7.5–11.3 months; Fig. 4C) and median PFS I-and-M was 3.6 months (95% CI, 2.8–4.9 months; n = 263; Fig. 4D). A total of 141 patients received second-line docetaxel (n = 76 docetaxel, n = 65 docetaxel and ramucirumab). Median OS was 6.4 months (95% CI, 5.1–8.1; Fig. 4E) and median PFS-I-and-M was 3.6 months (95% CI, 2.7–4.7 months; n = 119; Fig. 4F).
Comparison of real-world outcome measures to RECIST
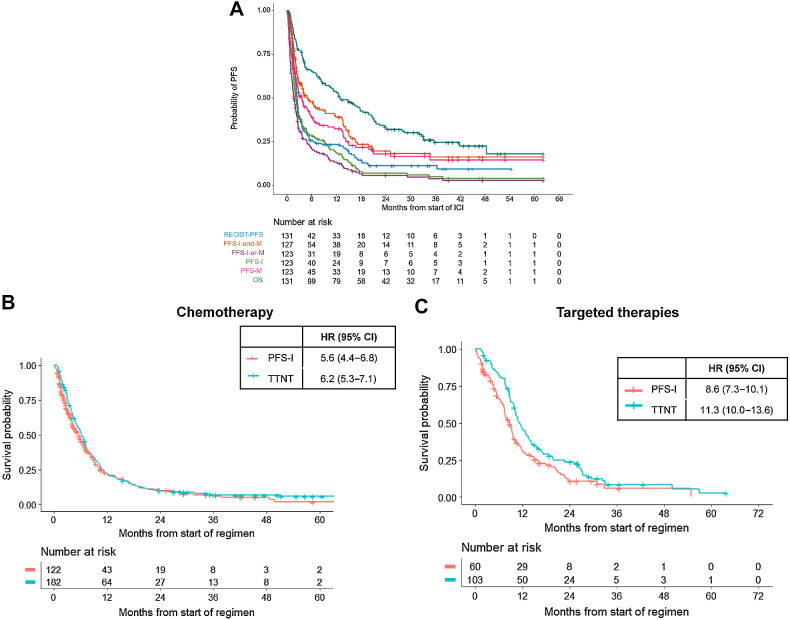
Real-world PFS estimated by PFS-I, PFS-I-and-M, PFS-M, and PFS-I-or-M may differ by months, with unclear implications for clinical practice (Supplementary Fig. S3A–S3C). For patients treated with first-line platinum chemotherapy, median PFS-I-or-M is 5.3 months (95% CI, 4.3–6.3) while the median PFS I-and-M is 9.1 months (95% CI, 7.5–10.7; Supplementary Fig. S3A). We sought to describe how real-world PFS (rwPFS) using the PRISSMM framework and PFS by RECIST compare. A total of 156 patients diagnosed with advanced disease and treated with ICI in the GENIE BPC NSCLC cohort also had RECIST measurements available. Median PFS-I, PFS-M, PFS-I-or-M, and PFS-I-and-M from initiation of ICI were 2.2 months (95% CI, 1.7–2.6), 3.6 months (95% CI, 2.4–5.7), 1.7 months (95% CI, 1.3–2.3), and 4.9 months (95% CI, 3.5–12), respectively, while median PFS-RECIST was 2.5 months (95% CI, 2.2–2.8; Fig. 5A).
Figure 5.
Comparison of RECIST and real-world endpoints. A, rwPFS versus PFS-RECIST. rwPFS metrics are compared with PFS determined by RECIST for patients treated with ICIs at MSKCC. B, Difference in TTNT and PFS-I for patients treated with first-line chemotherapy without oncogenic drivers. C, Difference in TTNT and PFS-I for patients with targetable alterations (EGFR, ALK, ROS1, MET) treated with targeted therapies.
We further hypothesized that PFS-I would closely align with TTNT, as medical oncologists would likely change treatment when imaging indicates disease growth. However, oncologists may tolerate disease progression for patients on targeted therapies longer than patients treated with chemotherapy, due to perceived tolerance and clinical benefit of oral targeted therapies. To examine this, we estimated PFS-I and TTNT for patients with EGFR, ALK, ROS1, RET, and MET oncogenic drivers treated with first-line targeted therapies to patients without these drivers treated with first-line chemotherapy. There were 438 patients without oncogenic drivers treated with first-line chemotherapy and 165 patients treated with erlotinib (n = 94), crizotinib (n = 25), osimertinib (n = 17), afatinib (n = 15), gefitinib (n = 7), alectinib (n = 7). Patients who progressed or began a new treatment regimen prior to undergoing NGS were excluded from these analyses (n = 204). Patients treated with chemotherapy had a median PFS-I of 5.6 months (95% CI, 4.4–6.8 months) and a TTNT of 6.2 months (95% CI, 5.3–7.1 months; Fig. 5B); patients with drivers treated with targeted therapies had a median PFS-I of 8.6 months (95% CI, 7.3–10.1 months) and a median TTNT of 11.3 months (95% CI, 10.0–13.6 months; Fig. 5C). In addition to prolonged PFS for first-line targeted therapies, these greater differences between TTNT and PFS-I suggest a potential pattern of clinicians maintaining patients on first-line targeted therapies beyond radiographic disease progression.
Discussion
In this work, we introduce the GENIE BPC NSCLC cohort, the first publicly released real-world dataset from the AACR GENIE BPC project integrating standardized clinical curation with high-quality genomic input. With data on over 2,000 sequenced samples from 1,846 patients, the dataset presents an opportunity to conduct translational and real-world analyses of patients with NSCLC that has the potential to yield novel insights and encourage new research directions.
We demonstrate that large-scale clinical curation combined with tumor genomic data in a multilayered approach is feasible and allows investigators to extend our understanding of the heterogeneous landscape of NSCLC. Genomic alterations enriched in specific patient subsets, such as by histology or disease site, can be easily identified, allowing for genomic characterization by site of metastasis. Detailed curation of disease progression and treatment response facilitates evaluation of how somatic alterations impact tangible patient outcomes such as survival. Integration of treatment histories with genomic features both at the intrapatient and interpatient level further enables identification of genomic evolution throughout treatment for a subset of patients with serial sequencing, presenting opportunities to identify mechanisms of acquired resistance.
Estimates of real-world survival on standard therapies may inform future research directions or establish survival benchmarks by which investigational therapies may be evaluated. In several analyses, OS on standard therapies estimated in the GENIE BPC cohort closely approximated OS from large-scale clinical trials. In the GENIE BPC dataset, median OS from start of platinum-based chemotherapy was 17.4 months. In the PARAMOUNT trial, which established maintenance pemetrexed as standard of care following initial platinum-pemetrexed doublet chemotherapy, median OS was 16.9 months (95% CI, 15.8–19.0 months; ref. 45). Similarly, median OS for patients treated with ICI in the second-line or later setting in the GENIE BPC cohort was 9.2 months, while median OS for patients with PD-L1 positive, previously treated NSCLC treated with pembrolizumab in the KEYNOTE-010 trial was 10.4 months (95% CI, 9.4–11.9 months; ref. 46). Survival estimates can be calculated for patients for whom an analogous clinical trial was not conducted and create a benchmark for future analyses.
Furthermore, the PRISSMM data curation model for the GENIE BPC standardizes the extraction of clinical data from the medical record, developing a set of criteria with which to evaluate response to treatments in a real-world setting. In a group of patients for whom GENIE BPC curation and independent RECIST evaluation of radiology outcomes were available, we demonstrate similarity between these measures of treatment response (47). PFS I-and-M and PFS-M, both of which require the medical oncology notes to indicate progressive disease, were notably longer than imaging-only assessments of disease progression (RECIST, PFS-I, and PFS-I-or-M), suggesting that medical oncologists may attribute early disease growth on ICI to pseudoprogression. Alternatively, radiology reports may note relatively minor increases in lesion size that are not considered clinically significant by medical oncologists, who continue therapy. While these data support the value of real-world endpoints, exploration is needed for other systemic treatments to further understand whether outcome measures typically used in prospective clinical trials and those used in observational datasets closely align.
There are limitations to our analyses and to how the data may be used. Primarily, the patients in this cohort were randomly selected from a group of patients who already had a targeted subset of their tumor genome sequenced. While targeted tumor DNA sequencing was part of the standard of care for patients with lung cancer during the period of this study at the institutions involved, it is likely that this inclusion criterion also introduces some bias in the group of patients available for analysis. The data are drawn from patients treated at four academic medical centers in North America, which is likely associated with a variety of referral biases. In addition, while each patient has a record of sequencing data, some oncogenic drivers, including EGFR and oncogenic fusions, may have been identified using other diagnostic techniques. In these cases, patients may have had other sequencing, IHC, or FISH testing not reported in the dataset that prompted initiation or change in therapy. Locoregional treatment, including radiation and surgery, and performance status measures are also not captured in the current dataset, potentially limiting the understanding of some patient's treatment courses and variations in outcomes. Further development of the PRISSMM data model to incorporate these treatment modalities is ongoing. In the statistical analyses shown here, several subgroups feature small sample sizes, limiting the ability to make formal comparisons adjusting for relevant confounding factors. TMB z-scores, which aim to standardize TMB derived across gene panels, do not correspond uniformly to equivalent TMB (i.e., TMB z-score of 0.17 corresponds to 11 mut/Mb using DFCI panel and 7 mut/Mb using MSK-IMPACT; ref. 26), limiting the interpretations of high TMB. Finally, analyses using any real-world data source should be rigorously evaluated for potential selection bias and confounding.
In summary, the publicly released GENIE BPC NSCLC data present a novel opportunity for clinicians and researchers to explore the diverse landscape of NSCLC. The first of its kind, our dataset includes larger numbers of patients with specific genomic alterations than are available in most traditional randomized clinical trials, allowing it to serve as an engine for hypothesis generation and to generate estimates of treatment effectiveness in the real world. Such analyses will ultimately improve outcomes for patients with NSCLC.
Supplementary Material
Supplemental Figure 1: OS and PFS for KRAS-mutated NSCLC treated with first-line platinum-based chemotherapy.
Supplemental Table 1
Supplemental Table 2
Supplemental Table 3
Supplemental Table 4
Supplemental Figure 2: OS and PFS for patients with NSCLC treated with first-line platinum-based chemotherapy.
Supplemental Figure 3: PFS estimates for patients with NSCLC treated with standard first- and second-line therapies.
Acknowledgments
The authors would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry as well as members of the AACR Project GENIE consortium for their commitment to data sharing; GENIE Coordinating Center, Sage Bionetworks, and/or cBioPortal staff who contributed substantively to the development and analysis of data, as well as writing the article. The authors would also like to acknowledge grant support from UL1 TR000445 from NCATS/NIH for use of REDcap database and the NCI at the NIH P30 CA00874 grant. Interpretations are the responsibility of study authors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
N.J. Choudhury reports personal fees from G1 Therapeutics, Sanofi, and OncLive; and other support from Wolters Kluwer, Amgen, Abbvie, Monte Rosa Therapeutics, Harpoon Therapeutics, and Merck outside the submitted work. J.A. Lavery reports other support from AACR Project GENIE BPC during the conduct of the study. S. Brown reports other support from AACR during the conduct of the study. J. Jee reports a patent license, MDSeq Inc. H. Rizvi reports personal fees from AstraZeneca outside the submitted work. K.C. Arbour reports personal fees from Sanofi-Genzyme; and other support from Revolution Medicines, Genentech, and Mirati outside the submitted work. M. Hellmann reports other support from AstraZeneca outside the submitted work; has a patent filed by Memorial Sloan Kettering related to the use of tumor mutational burden to predict response to immunotherapy (PCT/US2015/062208), pending and licensed by PGDx; and Equity/options from Factorial, Immunai, Shattuck Labs, Arcus, and Avail Bio. P.L. Bedard reports grants from AACR during the conduct of the study, as well as from AstraZeneca, Amgen, Bristol Myers Squibb, Bicara, GlaxoSmithKline, Genentech/Roche, Novartis, Merck, Lilly, SeaGen, Medicenna, Pfizer, and Zymeworks outside the submitted work; and as uncompensated advisory for Amgen, Merck, Zymeworks, SeaGen, Repare, Lilly, and Gilead. C. Yu reports other support from AACR during the conduct of the study. M. LeNoue-Newton reports other support from AACR during the conduct of the study, as well as from GE Healthcare outside the submitted work; and has a patent for 607172-WO-1 pending. C. Micheel reports grants from AACR during the conduct of the study. J.L. Warner reports grants from AACR during the conduct of the study, from Brown Physicians Incorporated outside the submitted work, and from NIH; personal fees from Westat, ASCO, Melax Tech, Flatiron, and Roche; and other support from HemOnc.org LLC. S.M. Sweeney reports grants from Amgen, AstraZeneca, Bayer Healthcare Pharmaceuticals, Boehringer-Ingelheim, Genentech, Merck, Bristol Myers Squibb, Janssen, Novartis, and Pfizer during the conduct of the study. K.L. Kehl reports grants from AACR during the conduct of the study. K.S. Panageas reports other support from AACR Project GENIE Biopharmaceutical Consortium during the conduct of the study. N. Schultz reports grants from AACR during the conduct of the study. D. Schrag reports grants from AACR during the conduct of the study and from Grail outside the submitted work; and personal fees from JAMA. G.J. Riely reports grants and nonfinancial support from AACR during the conduct of the study, as well as grants from Pfizer, Novartis, Takeda, Roche, Mirati, Lilly, Merck, and Rain Therapeutics outside the submitted work. No disclosures were reported by the other authors
Authors' Contributions
N.J. Choudhury: Conceptualization, investigation, methodology, writing–original draft, writing–review and editing. J.A. Lavery: Data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. S. Brown: Data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. I. de Bruijn: Resources, formal analysis, visualization, methodology. J. Jee: Data curation, formal analysis, methodology, writing–review and editing. T.N. Tran: Data curation, formal analysis, visualization, methodology, writing–review and editing. H. Rizvi: Resources, data curation, project administration, writing–review and editing. K.C. Arbour: Resources, writing–review and editing. K. Whiting: Formal analysis, writing–review and editing. R. Shen: Formal analysis, supervision, methodology, writing–review and editing. M. Hellmann: Resources, formal analysis, funding acquisition, writing–review and editing. P.L. Bedard: Resources, writing–review and editing. C. Yu: Resources, writing–review and editing. N. Leighl: Resources, writing–review and editing. M. LeNoue-Newton: Resources, writing–review and editing. C. Micheel: Resources, writing–review and editing. J.L. Warner: Resources, writing–review and editing. M.S. Ginsberg: Resources, formal analysis, writing–review and editing. A. Plodkowski: Resources, formal analysis, writing–review and editing. J. Girshman: Resources, formal analysis, writing–review and editing. P. Sawan: Resources, formal analysis, writing–review and editing. S. Pillai: Resources, project administration, writing–review and editing. S.M. Sweeney: Resources, funding acquisition, project administration, writing–review and editing. K.L. Kehl: Resources, investigation, writing–review and editing. K.S. Panageas: Resources, data curation, formal analysis, supervision, investigation, methodology, writing–review and editing. N. Schultz: Resources, supervision, project administration, writing–review and editing. D. Schrag: Resources, supervision, funding acquisition, project administration, writing–review and editing. G.J. Riely: Conceptualization, resources, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Phillips CM, Parmar A, Guo H, Schwartz D, Isaranuwatchai W, Beca J, et al. Assessing the efficacy-effectiveness gap for cancer therapies: a comparison of overall survival and toxicity between clinical trial and population-based, real-world data for contemporary parenteral cancer therapeutics. Cancer 2020;126:1717–26. [DOI] [PubMed] [Google Scholar]
- 2. Choudhury NJ, Riely GJ, Sabbatini PJ, Hellmann MD. Translating inspiration from COVID-19 vaccine trials to innovations in clinical cancer research. Cancer Cell 2021;39:897–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khozin S, Blumenthal GM, Pazdur R. Real-world data for clinical evidence generation in oncology. J Natl Cancer Inst 2017;109. [DOI] [PubMed] [Google Scholar]
- 4. Booth CM, Karim S, Mackillop WJ. Real-world data: towards achieving the achievable in cancer care. Nat Rev Clin Oncol 2019;16:312–25. [DOI] [PubMed] [Google Scholar]
- 5. Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang JT, Lee YM, Huang RS. The impact of the Cancer Genome Atlas on lung cancer. Transl Res 2015;166:568–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hutter C, Zenklusen JC. The Cancer Genome Atlas: creating lasting value beyond its data. Cell 2018;173:283–5. [DOI] [PubMed] [Google Scholar]
- 8. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011;306:1557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. AACR Project GENIE Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schrag D. GENIE: real-world application. ASCO Annual Meeting; Chicago; June 4, 2018. [Google Scholar]
- 12. Lavery JA, Lepisto EM, Brown S, Rizvi H, McCarthy C, LeNoue-Newton M, et al. A scalable quality assurance process for curating oncology electronic health records: the project GENIE Biopharma collaborative approach. JCO Clin Cancer Inform 2022;6:e2100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 14. Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin 2018;68:31–54. [DOI] [PubMed] [Google Scholar]
- 15. Jordan EJ, Kim HR, Arcila ME, Barron DA, Chakravarty D, Gao J, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 2017;7:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med 2020;383:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kundra R, Zhang H, Sheridan R, Sirintrapun SJ, Wang A, Ochoa A, et al. OncoTree: a cancer classification system for precision oncology. JCO Clin Cancer Inform 2021;5:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kehl KL, Riely GJ, Lepisto EM, Lavery JA, Warner JL, LeNoue-Newton ML, et al. Correlation between surrogate end points and overall survival in a multi-institutional clinicogenomic cohort of patients with non-small cell lung or colorectal cancer. JAMA Netw Open 2021;4:e2117547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Statistical Coordinating Center at MSKCC. GENIE BPC analytic data guide NSCLC v2.0 public. American Association for Cancer Research; 2022.
- 21. Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O'Neal L, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non–small-cell lung cancer. J Clin Oncol 2008;26:3543–51. [DOI] [PubMed] [Google Scholar]
- 24. Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 25. Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819–30. [DOI] [PubMed] [Google Scholar]
- 26. Vokes NI, Liu D, Ricciuti B, Jimenez-Aguilar E, Rizvi H, Dietlein F, et al. Harmonization of tumor mutational burden quantification and association with response to immune checkpoint blockade in non–small-cell lung cancer. JCO Precis Oncol 2019;3:PO.19.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown S, Lavery JA, Shen R, Martin AS, Kehl KL, Sweeney SM, et al. Implications of selection bias due to delayed study entry in clinical genomic studies. JAMA Oncol 2022;8:287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lavery JA, Brown S, Curry MA, Martin A, Sjoberg DD, Whiting K. A data processing pipeline for the AACR project GENIE biopharma collaborative data with the {genieBPC} R package. Bioinformatics 2023;39;btac796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. R Core Team. A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria; 2021. [Google Scholar]
- 30. Scagliotti G, Novello S, von Pawel J, Reck M, Pereira JR, Thomas M, et al. Phase III study of carboplatin and paclitaxel alone or with sorafenib in advanced non-small-cell lung cancer. J Clin Oncol 2010;28:1835–42. [DOI] [PubMed] [Google Scholar]
- 31. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: non-small cell lung cancer in NCCN guidelines. v2; 2023. Available from:https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
- 32. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 2018;8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ricciuti B, Arbour KC, Lin JJ, Vajdi A, Vokes N, Hong L, et al. Diminished efficacy of programmed death-(Ligand)1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J Thorac Oncol 2022;17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 mutation on survival in patients with advanced EGFR-mutant non–small-cell lung cancer. JCO Precis Oncol 2018;2018:PO.18.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Labbé C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017;111:23–9. [DOI] [PubMed] [Google Scholar]
- 37. Shepherd FA, Lacas B, Le Teuff G, Hainaut P, Jänne PA, Pignon JP, et al. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2017;35:2018–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. FDA. KEYTRUDA (pembrolizumab) prescribing information; 2019.
- 39. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim S-W, Carcereny Costa E, et al. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. N Engl J Med 2019;381:2020–31. [DOI] [PubMed] [Google Scholar]
- 40. Rousseau B, Foote MB, Maron SB, Diplas BH, Lu S, Argilés G, et al. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N Engl J Med 2021;384:1168–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galvano A, Gristina V, Malapelle U, Pisapia P, Pepe F, Barraco N, et al. The prognostic impact of tumor mutational burden (TMB) in the first-line management of advanced non-oncogene addicted non-small-cell lung cancer (NSCLC): a systematic review and meta-analysis of randomized controlled trials. ESMO Open 2021;6:100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. [DOI] [PubMed] [Google Scholar]
- 43. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mazieres J, Rittmeyer A, Gadgeel S, Hida T, Gandara DR, Cortinovis DL, et al. Atezolizumab versus docetaxel in pretreated patients with NSCLC: final results from the randomized phase 2 POPLAR and phase 3 OAK clinical trials. J Thorac Oncol 2021;16:140–50. [DOI] [PubMed] [Google Scholar]
- 45. Paz-Ares LG, de Marinis F, Dediu M, Thomas M, Pujol J-L, Bidoli P, et al. PARAMOUNT: final overall survival results of the phase III study of maintenance pemetrexed versus placebo immediately after induction treatment with pemetrexed plus cisplatin for advanced nonsquamous non–small-cell lung cancer. J Clin Oncol 2013;31:2895–902. [DOI] [PubMed] [Google Scholar]
- 46. Herbst RS, Garon EB, Kim D-W, Cho BC, Gervais R, Perez-Gracia JL, et al. Five year survival update from KEYNOTE-010: pembrolizumab versus docetaxel for previously treated, programmed death-ligand 1–positive advanced NSCLC. J Thorac Oncol 2021;16:1718–32. [DOI] [PubMed] [Google Scholar]
- 47. Izano MA, Tran N, Fu A, Toland L, Idryo D, Hilbelink R, et al. Implementing real-world RECIST-based tumor response assessment in patients with metastatic non-small cell lung cancer. Clin Lung Cancer 2022;23:191–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: OS and PFS for KRAS-mutated NSCLC treated with first-line platinum-based chemotherapy.
Supplemental Table 1
Supplemental Table 2
Supplemental Table 3
Supplemental Table 4
Supplemental Figure 2: OS and PFS for patients with NSCLC treated with first-line platinum-based chemotherapy.
Supplemental Figure 3: PFS estimates for patients with NSCLC treated with standard first- and second-line therapies.
Data Availability Statement
The GENIE BPC NSCLC 2.0-public data release is available for download at https://www.synapse.org/#!Synapse:syn27056172/wiki/616601. The data can be downloaded after creating a free online account.