CD36 promotes blast migration and extramedullary disease in acute myeloid leukemia and represents a critical target that can be exploited for clinical prognosis and patient treatment.
Abstract
Identifying mechanisms underlying relapse is a major clinical issue for effective cancer treatment. The emerging understanding of the importance of metastasis in hematologic malignancies suggests that it could also play a role in drug resistance and relapse in acute myeloid leukemia (AML). In a cohort of 1,273 AML patients, we uncovered that the multifunctional scavenger receptor CD36 was positively associated with extramedullary dissemination of leukemic blasts, increased risk of relapse after intensive chemotherapy, and reduced event-free and overall survival. CD36 was dispensable for lipid uptake but fostered blast migration through its binding with thrombospondin-1. CD36-expressing blasts, which were largely enriched after chemotherapy, exhibited a senescent-like phenotype while maintaining their migratory ability. In xenograft mouse models, CD36 inhibition reduced metastasis of blasts and prolonged survival of chemotherapy-treated mice. These results pave the way for the development of CD36 as an independent marker of poor prognosis in AML patients and a promising actionable target to improve the outcome of patients.
Significance:
CD36 promotes blast migration and extramedullary disease in acute myeloid leukemia and represents a critical target that can be exploited for clinical prognosis and patient treatment.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of diseases characterized by uncontrolled proliferation of clonal neoplastic hematopoietic precursor cells and impaired function of normal hematopoiesis, leading to neutropenia, anemia, and thrombocytopenia (1). If untreated, patients die of infection, bleeding, or organ failure, usually in a matter of weeks. Induction therapy, which combines 7 days of cytarabine (AraC) and 3 of anthracycline, is highly effective in killing leukemic cells, but despite a high rate of complete remission after these cytotoxic agents, most patients relapse (2). In AML patient-derived xenografts (PDX) in immunodeficient mice (nonobese diabetic scid gamma mouse, i.e., NSG) treated with AraC, we found CD36, a plasma membrane receptor and fatty acid transporter, among the most strongly upregulated genes in chemoresistant cells (3). In this study, we investigated the clinical relevance of this observation and the mechanisms by which CD36 expression may contribute to chemoresistance and relapse.
We and others have shown that AML blasts are highly dependent on fatty acid metabolism and oxidative phosphorylation (OXPHOS) to bypass chemotherapy (3–5). In a number of tumors (6–8), an increased CD36 expression has been found to support cancer cell metabolism through the transport of fatty acids. However, CD36 is a multiliganded receptor also binding molecules such as oxidized low-density lipoprotein (oxLDL; ref. 9) or thrombospondins 1 and 2 (TSP1 and TSP2; ref. 10), and CD36 is involved in a number of functions besides metabolism, including angiogenesis, tumor immunity, and metastasis (11–13). Metastatic cancers are largely resistant to treatments as compared with primary tumor cells and represent the most frequent cause of death in cancer patients. In solid cancers as in leukemia (8, 14), metastatic niches provide a microenvironment that fuels cell metabolism, protecting tumor cells from stresses and chemotherapies. Among the different mechanisms of relapse, the entry of leukemic blasts into a senescent-like state is an important driver of chemoresistance and AML relapse (15). Senescent cells can stimulate tumor progression through their senescence-associated secretory phenotype (SASP), which comprises proinflammatory and proinvasive molecules that can stimulate motility, invasion, and metastasis (16).
Until recently (17), because leukemic cells circulate in lymph and blood as their normal counterparts do, leukemias were generally not referred to as metastatic diseases. However, the presence at diagnosis of an extramedullary disease (EMD) in AML patients has been described for decades, with an incidence ranging from 2.5% (18) to 31% (19). The prognostic significance of EMD in AML was found as of no value in some studies (20), whereas others found EMD to convey a poor prognosis, independent of other established risk factors (18, 19, 21, 22). These discrepancies between studies partly result from the difficulty to accurately score EMD. As recently shown in a multicenter cohort of AML patients, EMD was significantly associated with a decreased median overall survival (OS) and was predictive of a reduced even-free survival (EFS) only when histology was added to the clinical examination (22). This highlights the need to better understand the molecular mechanisms driving EMD and to identify reliable biomarkers. The impact of EMD in AML relapse after chemotherapy also needs to be deciphered.
Here, to uncover the molecular mechanisms behind dissemination of leukemic blasts in extramedullary organs, we combined in vitro and in vivo experiments using AML cell lines and patient cells, including single-cell analysis, alongside a clinical study from a large cohort of 1,273 AML patients, some of whom have received an intensive chemotherapy regimen and have been followed for at least three years. Our findings highlight CD36 as an actionable driver of leukemic blast metastasis and unravel new mechanisms contributing to AML relapse.
Materials and Methods
Patients
Inclusion criteria were newly diagnosed AML or relapse according to the WHO 2008 classification and age 15 years or older. This study included 1,273 patients admitted at the Hematology Department of Toulouse University Hospital-IUCT-O and/or registered in the regional oncology network from January 1, 2016, to December 31, 2020 [Toulouse University Hospital (TUH) cohort]. Data were gathered in an electronic clinical research form. A written informed consent was obtained from all patients in accordance with the Declaration of Helsinki, allowing the collection of clinical and biological data in an anonymized database. Samples from AML patients were stored in the HIMIP collection (BB-0033-00060). In accordance with French law, the HIMIP collection was declared to the Ministry of Higher Education and Research (DC 2008-307 collection 1) and obtained a transfer agreement (AC 2008-129) after approval by the Comité de Protection des Personnes Sud-Ouest et Outremer II (ethics committee).
Cytogenetic and molecular risk classifications were in accordance with the Medical Research Council and ELN 2010 classifications, respectively (23, 24). Details on treatment with first-line chemotherapy regimen used over time have been reported elsewhere (25). The decision-making process with regard to intensive or nonintensive treatment was based on initial characteristics such as white blood cell count, cytogenetics, age, secondary AML, performance status, and comorbidities. EMD status was determined at AML diagnosis by clinical examination.
Endpoints, including response, EFS, LFS, cumulative incidence of relapse (CIR), and OS, were assessed according to standard criteria (1). Bone marrow assessment in patients treated with intensive chemotherapy was performed after blood recovery or in case of delayed recovery between days 35 and 45.
In vivo animal studies
NSG (NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ) mice were used for the transplantation of AML cell lines or primary AML samples. Mice were housed in sterile conditions using HEPA-filtered microisolators and fed with irradiated food and sterile water in the Animal core facility of the Cancer Research Center of Toulouse. All animals were used in accordance with a protocol reviewed and approved by the Institutional Animal Care and Use Committee of Région Midi-Pyrénées. Six- to 9-week-old male or female mice were randomly assigned to experimental groups before cell injection or drug treatments.
Primary patients sample phenotyping
Multiparameter flow cytometry was performed on whole bone marrow (BM) or blood specimens using a standard stain–lyse–wash procedure with ammonium chloride lysis. 1 × 105 cells were stained per analysis tube (see antibody list in Supplementary Table S1), and data were acquired on at least 1 × 104 blasts when specimen quality permitted. Data on standardized 8- to 10-color staining combinations were acquired on Navios instruments and analyzed using Kaluza (Beckman-Coulter). A blast gate including CD45 dim mononuclear cells was analyzed according to cytomorphologic data.
Xenograft mouse models
NSG mice were produced at the Genotoul Anexplo platform in Toulouse (France), using breeders obtained from Charles River Laboratories. To assess response to chemotherapy in PDX models, mice (6–9-week-old) were sublethally treated with busulfan (30 mg/kg) 24 hours before injection of leukemic cells. Leukemia samples were thawed at 37°C, washed in IMDM 20% FBS, and suspended in Hank's Balanced Salt Solution (HBSS) at a final concentration of 2–10 × 106 cells in 200 μL for tail-vein injection; 8 to 18 weeks after AML cell transplantation, once cell engraftment was confirmed by flow cytometry on peripheral blood or BM aspirates, NSG mice were treated with daily intraperitoneal injection of 60 mg/kg AraC or vehicle (PBS) for 5 days. AraC was kindly provided by the pharmacy of the TUH. Mice were sacrificed on day 8 after AraC treatment to harvest human leukemic cells from murine BM.
To establish CLDX, NSG mice were treated with busulfan (20 mg/kg) 24 hours before injection of AML cell lines. Cells were thawed and washed as previously described, suspended in HBSS at a final concentration of 0.2–2 × 106 in 200 μL before intravenous injection. Mice were treated with daily intraperitoneal injection of 30 mg/kg AraC for 5 days (starting day 10 for U937 and day 17 for OCIAML3 after injection) and sacrificed at day 8 or later as indicated. When indicated, mice received concomitantly with AraC until sacrifice three times weekly an intravenous injection of PBS (100 μL) containing either 5 μg of monoclonal CD36Ab FA6.152 (Stem Cell Technologies 60084) or 5 μg of mouse control IgG (Stem Cell Technologies 60070). Mice' survival time was also determined. All animals were used in accordance with a protocol reviewed and approved by the Institutional Animal Care and Use Committee of Région Midi-Pyrénées.
14C palmitate uptake and oxidation assay
Cells were preincubated for 3-hour with [1–14 C] palmitate (1 μCi/mL; PerkinElmer) and nonlabeled (cold) palmitate. Palmitate was coupled to a fatty acid-free BSA in a molar ratio of 5:1. Following incubation, 14CO2 and 14C-ASM were measured as previously described (26). Briefly, the assayed medium is transferred into a custom-made Teflon 48-well trapping plate. The plate was clamped and sealed, and perchloric acid was injected through the perforations in the lid into the medium, which drives CO2 through the tunnel into an adjacent well, where it was trapped in 1N NaOH. Following trapping, the media were spun twice and 14C-ASM was measured by scintillation counting. Aliquots of NaOH and medium were transferred into scintillation vials, and radioactivity was measured on a multipurpose scintillation counter (LS 6500; Beckman-Coulter). All assays were performed in triplicates, and data were normalized to viable cell numbers.
RNA extraction and real-time RT-qPCR.
Total cell RNA was isolated and extracted using the ZYMO RNA kit (Zymo). For mouse tissues, total RNA was isolated by Qiazol extraction and purified using RNeasy minicolumns (Qiagen). 300 to 1,000 ng total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystem 4368814), SYBR Green PCR Master Mix (Applied Biosystem 4309155), and 300 nmol/L primers on an Applied Biosystem (Massachusetts, United States) StepOne instrument. Relative gene expression was determined using the 2−ΔCT or 2−ΔΔCT method and normalized to m36B4 or hRPLP0 expression as described in figure legends. Primer sequences are provided in Supporting Information and Methods.
Migration assay
For migration assays, 24 mm diameter, 8 μm pore transwells (Corning 353097) were used. Cell suspension in 0% FBS αMEM medium was added in the upper chamber. The lower chamber was filled with 500 μL of medium with or without 10% FBS. As indicated, anti-CD36 blocking antibodies (1 μg/mL; FA6-152, Stem Cell Technologies 60084 or Cayman chemical JC63.1 10009893) and/or recombinant TSP1 (2 nmol/L; R&D Systems 3074-TH) were added in the upper chamber. Leukemic cells were allowed to migrate at 37°C for 24 hours. Inserts were removed and cell concentration was measured by cell counting with trypan blue coloration.
IHC
Subcutaneous adipose tissue samples were immediately immersed in 10% neutral buffered formalin fixative after sampling for 24 hours before storage in PBS at 4°C. After paraffin embedding, 3-μm thickness paraffin sections were dewaxed (successive toluene and descending alcohol baths) and stained with hematoxylin and eosin. Labeling of 3-μm sections of paraffin-embedded adipose tissues samples was performed after antigen retrieval (Ptlink low pH, reference K8005, Dako) for 30 minutes, using anti–human-specific Ku-80 antibody (2753S Cell Signaling Technology, rabbit monoclonal antibody, dilution 1:150) and incubated for 50 minutes at room temperature. Staining was carried out with Dakostainer automated system using a biotinylated anti-rabbit antibody (reference ABK125 Microm) for 25 minutes at room temperature, followed by HRP (1:150, 25 minutes room temperature) and DAB as a chromogen. The stained slides were imaged by light microscopy on a Nikon Eclipse Ci-L microscope with a DS138 Fi3 Camera and NIS Elements D software.
Publicly accessible transcriptomic databases of AML patients used in this study
The following AML cohorts were used as indicated: Gene-Expression Omnibus (GEO) accession no. GSE14468 (Verhaak cohort; ref. 27), BeatAML cohort (28), and The Cancer Genome Atlas (TCGA) cohort (29). FAB M3 patients and patients who did not receive intensive chemotherapy were removed for analysis. Patients with OS value inferior to 1 year but still alive were removed in Kaplan–Meier curves. Other publicly accessible transcriptomic databases of AML patients used in this study were GSE40871, Klco and colleagues (30); GSE97631, Farge and colleagues (3); GSE146544, Duy and colleagues (15).
Establishment of the CD36 gene signature
Global analysis of frequency versus genes on normalized Affymetrix data showed that CD36 expression followed a Pareto distribution in TCGA and BeatAML databases, whereas it followed a normal distribution in the Verhaak database. Only patients with nonzero CD36 expression were considered. In order to standardize the cutoff of differential expression among the three databases and establish experimental groups, based on a 95% confidence interval (i.e., P < 0.05) of CD36 expression, patients with a z-score > 1.64 were categorized as “high CD36” and < 1.64 as the “baseline group.” Welch t test of means for difference in variances was performed between the two groups to determine the differentially expressed genes. Then, data points were filtered to preserve overexpressed genes with a P < 0.05 in the high CD36 (positive “statistic”) group. Corresponding test statistic values were then listed in decreasing order. Gene symbols were assigned unique Ensembl IDs to mitigate redundancies from gene synonyms and awkward formatting. The top 1,000 genes in each database were used to select the common genes among the different databases.
Statistical analysis
For univariate survival analyses of OS and LFS in patients, Kaplan–Meier survival curves were drawn and differences in survival functions were tested using the log-rank test. Univariate survival analyses used cumulative incidence functions and Gray's test for relapse (CIR), because nonrelapse mortality was treated as competing events. Hazard ratios (HR) and 95% confidence intervals (CI) were assessed using a standard Cox model for OS and EFS and a proportional subdistribution hazard model, which is an extension of the Cox model for the situation of competing risks, for CIR. All reported P values were two-sided and the significance threshold was < 0.05. Statistical analyses were performed on STATA version 13 (STATA Corp).
We assessed the statistical analysis of the difference between two sets of data using a two-tailed (nondirectional) nonparametric Mann–Whitney test or unpaired test according to samples distribution, with or without Welch correction depending on samples variance. For graphs with control condition normalized to 1, we used a one-sample t test with a hypothetical value set to 1. For multiple comparisons, one-way ANOVA or two-way ANOVA were performed with multiple comparison tests. For paired tests, the Wilcoxon test was used. For contingency analysis of clinical data, we addressed Fisher exact test on raw data before normalization in percentages. For survival analyses, we used log-rank (Mantel–Cox) test. Analyses were performed using GraphPad Prism (v9). A P value of less than 0.05 indicates significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Data availability
All raw data generated in this study are available upon request from the corresponding author. The data analyzed in this study were obtained from GEO at GSE14468, GSE40871, GSE97631, and GSE146544. Data from the BeatAML study were obtained from http://www.vizome.org, and TCGA AML data set (29) was obtained from https://cbioportal.org.
Results
CD36 is an independent prognostic marker of AML progression
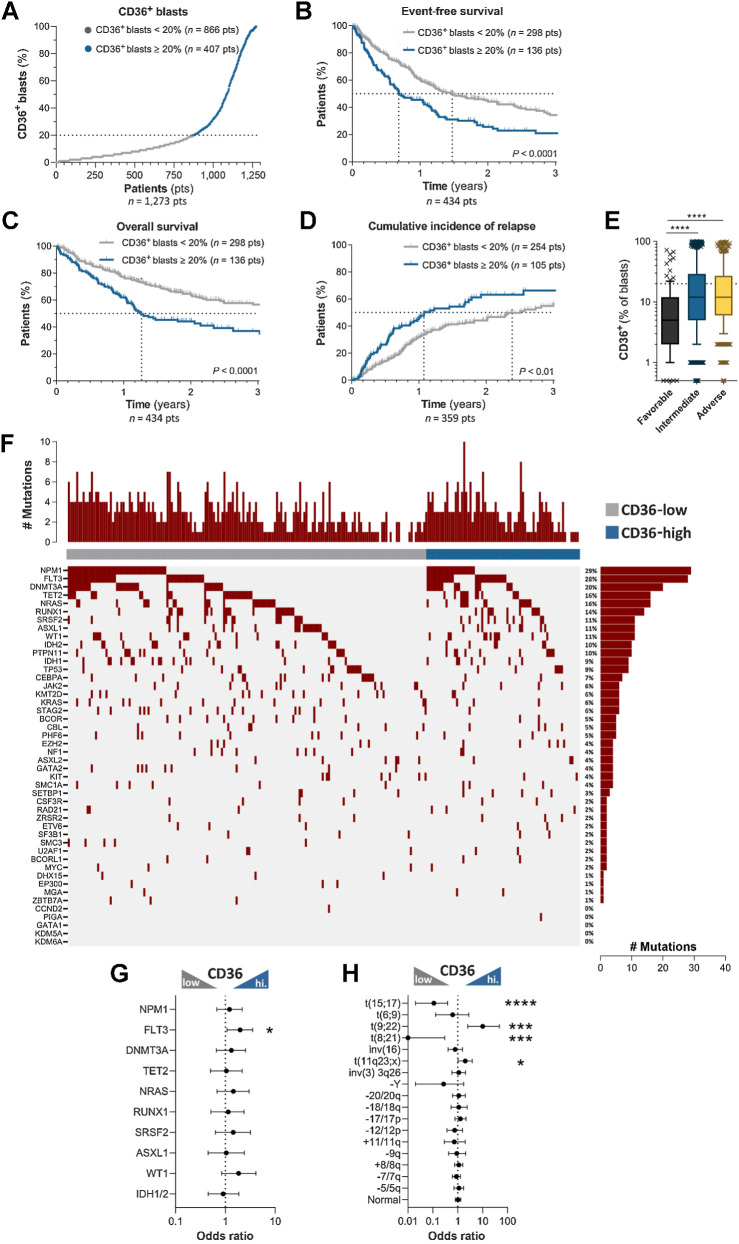
To study if CD36 has a clinical prognostic value, we measured CD36 protein expression by flow cytometry in AML blasts of 1,273 AML patients from the TUH. Based on the percentage of blasts expressing CD36 at diagnosis, AML patients were stratified as CD36-high (CD36-expressing blasts ≥ 20%, 407/1273, 32%) versus CD36-low (Fig. 1A). In this cohort, 435 AML patients were treated with an intensive chemotherapy regimen combining 3 days of daunorubicin or 5 days of idarubicin and 7 days of cytarabine. Although several features impact patients’ prognosis, as already well described (1), a multivariate analysis revealed that CD36 was significantly associated with a worse EFS (HR: 1.55; 95% CI: 1.17–2.05, P = 0.002), and a worse OS (HR: 1.69; 95% CI: 1.18–2.41, P = 0.005; Table 1). Median EFS of CD36-high patients was half that of CD36-low patients (252 days vs. 538 days, respectively; HR:1.65, P < 0.0001; Fig. 1B), whereas median OS, which was 462 days in CD36-high patients, was not reached after 3 years in CD36-low patients (HR: 1.88, P < 0.0001; Fig. 1C). Furthermore, a high CD36 protein expression at diagnosis was associated with an increased CIR after intensive chemotherapy (Fig. 1D), and a multivariate analysis showed that a high expression of CD36 was associated with a shorter CIR (SHR: 1.53; 95% CI: 1.07–2.19, P = 0.02; Table 1). We next characterized the molecular and cytogenetic profile of patients exhibiting a high percentage of CD36-expressing blasts. Consistent with our findings, intermediate and unfavorable karyotypes had a higher percentage of CD36-expressing blasts (Fig. 1E). To provide an overview of the genomic landscape of CD36 AML clusters, we plotted targeted NGS data from 224 AML patients to examine the co-occurrence of mutations. We also calculated odds ratios for recurrent cytogenetic abnormalities. A comparison of the genomic landscape in 224 AML did not reveal a pattern related to CD36 expression, except for FLT3 abnormalities (Fig. 1F and G). We next examined the set of recurrent karyotype abnormalities (n = 1,202) showing an enrichment of AML with KMT2A (11q23) or t(9;22) abnormalities in CD36-high group and an increase in t(15;17) and t(8;21) AML in CD36-low group (Fig. 1H).
Figure 1.
CD36 expression in blasts at diagnosis is associated with human AML progression and relapse. A, AML patients from the TUH cohort (1,273 patients) were classified as low and high CD36 expressers (CD36-expressing blasts <20%, low, n = 866 and CD36-expressing blasts ≥20%, high, n = 407). B, Kaplan–Meier curve for event-free survival according to CD36 expression (CD36 low, n = 298; CD36 high, n = 136). C, Kaplan–Meier curve for overall survival according to CD36 expression (CD36 low, n = 298; CD36 high, n = 136). D, Cumulative incidence of relapse according to CD36 expression (CD36 low, n = 254; CD36 high, n = 105). B–D, log-rank test. E, Box plot of CD36 expression on blasts according to cytogenetic risk (n = 1,191). Box plot shows the 10th percentile, first quartile, median, third quartile, and 90th percentile. Mann–Whitney test was performed. F, Landscape of somatic mutations detected in diagnostic samples (n = 224) by sequencing with a panel of 52 genes. The number of mutations for each patient is shown at the top, whereas the frequencies of each mutation are located at the right. G, Forrest plot showing mutation enrichment at AML diagnosis based on blast CD36 level by logarithmic odds ratio. Fisher exact test was performed (P = 0.029 for FLT3). H, Forrest plot showing enrichment of recurrent cytogenetic anomalies at AML diagnosis according to CD36 expression by logarithmic odds ratio. Fisher exact test was performed (P < 0.0001 for t(15;17); P = 0.0008 for t(9;22); P = 0.0002 for t(8;21); P = 0.044 for t(11q23;x)). The circles (in the middle of the error bars) represent the odds ratios. The error bars represent 95% confidence interval of the odds ratio. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Table 1.
Multivariate analysis of the TUH cohort.
| HR/robust SHR (CIR) (95% CI) | Std. Err. | z | P>|z| | Statistical analysis | ||
|---|---|---|---|---|---|---|
| Event-free survival | CD36 | 1.55 (1.174–2.051) | 0.22 | 3.09 | 0.002 | ** |
| Age > 60 years | 1.64 (1.222–2.192) | 0.24 | 3.3 | 0.001 | ** | |
| WBC > 50 G/L | 1.85 (1.314–2.591) | 0.32 | 3.54 | 0.000 | *** | |
| Allo-SCT | 0.52 (0.375–0.709) | 0.08 | −4.09 | 0.000 | *** | |
| Favorable cytogenetics | 0.61 (0.361–1.03) | 0.16 | −1.85 | 0.064 | ns | |
| Adverse cytogenetics | 1.34 (0.955–1.892) | 0.23 | 1.7 | 0.090 | ns | |
| FLT3-ITD mutation | 1.31 (0.938–1.832) | 0.22 | 1.59 | 0.113 | ns | |
| NPM1 mutation | 0.55 (0.394–0.775) | 0.10 | −3.43 | 0.001 | ** | |
| AML secondary status | 1.53 (1.134–2.06) | 0.23 | 2.78 | 0.005 | ** | |
| Overall survival | CD36 | 1.69 (1.176–2.418) | 0.31 | 2.84 | 0.005 | ** |
| Age > 60 years | 1.39 (0.947–2.036) | 0.27 | 1.68 | 0.093 | ns | |
| WBC > 50 G/L | 1.84 (1.192–2.833) | 0.41 | 2.76 | 0.006 | ** | |
| Allo-SCT | 0.30 (0.187–0.47) | 0.07 | −5.17 | 0.000 | *** | |
| Favorable cytogenetics | 0.45 (0.203–1.003) | 0.18 | −1.95 | 0.051 | ns | |
| Adverse cytogenetics | 1.41 (0.903–2.204) | 0.32 | 1.51 | 0.131 | ns | |
| FLT3-ITD mutation | 1.46 (0.964–2.216) | 0.31 | 1.79 | 0.074 | ns | |
| NPM1 mutation | 0.61 (0.394–0.941) | 0.14 | −2.23 | 0.026 | * | |
| AML secondary status | 1.85 (1.272–2.697) | 0.36 | 3.22 | 0.001 | *** | |
| Cumulative incidence of relapse | CD36 | 1.53 (1.07–2.193) | 0.28 | 2.33 | 0.020 | * |
| Age > 60 years | 1.54 (1.088–2.192) | 0.28 | 2.43 | 0.015 | * | |
| WBC> 50 G/L | 1.57 (1.008–2.458) | 0.36 | 1.99 | 0.046 | * | |
| Allo-SCT | 0.77 (0.519–1.128) | 0.15 | −1.35 | 0.177 | ns | |
| Favorable cytogenetics | 0.76 (0.451–1.29) | 0.20 | −1.01 | 0.312 | ns | |
| Adverse cytogenetics | 0.86 (0.524–1.424) | 0.22 | −0.57 | 0.566 | ns | |
| FLT3-ITD mutation | 1.19 (0.763–1.869) | 0.27 | 0.78 | 0.437 | ns | |
| NPM1 mutation | 0.67 (0.437–1.014) | 0.14 | −1.9 | 0.058 | ns | |
| AML secondary status | 1.37 (0.921–2.05) | 0.28 | 1.56 | 0.120 | ns |
Note: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: Allo-SCT, allogeneic stem-cell transplantation; WBC, white blood cells.
Altogether, these clinical findings obtained on a large patient cohort reveal that a high percentage of blasts expressing CD36 at diagnosis is an independent prognostic marker of disease progression, thus warranting the study of CD36's contribution to AML progression and relapse.
CD36 inhibition delays AML relapse after chemotherapy
Using additional PDX, we confirmed our previous observation (3) that AraC treatment in vivo induced a sustained increase in CD36 expression at the plasma membrane of residual AML blasts (Supplementary Fig. S1A and S1B). To decipher whether CD36 is a promoter or a consequence of AML progression and relapse, we performed experiments using two human AML cell lines strongly expressing CD36 protein i.e., U937 and OCIAML3, U937 exhibiting two distinct subpopulations expressing low and high levels of CD36 at baseline, whereas OCIAML3 cells do not exhibit such phenotypical heterogeneity (Supplementary Fig. S1C). In both models, treating cells with AraC in vitro increased CD36 protein expression (Supplementary Fig. S1D). Using shRNA, we knocked-down CD36 and then engrafted these cells or their control counterparts into NSG mice. After engraftment, mice were treated with vehicle or AraC for 5 days, and survival experiments were performed. In both models, knocking down CD36 did not affect the OS of untreated mice (data not shown) but significantly increased the survival of AraC-treated mice (Fig. 2A and B). We then asked if CD36 inhibition might have a therapeutic potential once the disease was established. To this aim, we engrafted mice with U937 cells, and 10 days later randomized mice into four groups receiving vehicle only, AraC only, FA6-152 anti-CD36 blocking antibody only, or a combination of AraC and CD36 antibody. Whereas inhibition of CD36 did not increase the OS in untreated mice, AraC increased mice survival, and importantly, adding a CD36 antibody to AraC further significantly prolonged mice survival (Fig. 2C). Of note, neither CD36 knockdown with shRNA nor its inhibition with the FA6-152 blocking antibody affected the number of viable blasts (Supplementary Fig. S1E–S1H). Together, these data unveil a new therapeutic modality to potentialize chemotherapeutic effects, delaying the time to relapse.
Figure 2.
CD36 inhibition delays AML relapse after chemotherapy. A, Percentage of mice engrafted with U937 cells transduced with shCtrl (n = 11) or shCD36 (n = 11) and treated with vehicle or AraC, surviving over time after treatment. B, Same as A, with OCIAML3 [shCtrl (n = 18) and shCD36 (n = 22)]. C, Percentage of mice engrafted with U937 cells surviving over time after treatment with the indicated combinations of PBS or AraC administered for the first 5 days, with either control IgG or FA6-152 anti-CD36 antibody injected three times/week (n = 7 PBS-IGG; n = 7 PBS-anti-CD36; n = 7 AraC-IGG; n = 6 AraC-anti-CD36). D, Venn diagram of significantly cooverexpressed genes along with CD36 in TCGA, Verhaak, and BeatAML cohorts. E, Gene set enrichment analysis of CD36 gene signature in AraC versus untreated AML blasts from three different transcriptomic analyses. A–C, log-rank (Mantel–Cox) test. E, One sample t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We then questioned the mechanisms behind CD36-dependent relapse after chemotherapy. We first hypothesized that CD36 mediates lipid transfer in leukemic blasts. Surprisingly, CD36 silencing by shRNA, which reduced CD36 protein expression in basal as well as in AraC-treated cells (Supplementary Fig. S1I–S1K), did not affect cell neutral lipid content, measured by Bodipy staining (Supplementary Fig. S1L–S1M). Treatment with JC63.1 CD36 antibody, which blocks fatty acid and oxLDL uptake (31, 32), or with FA6-152 CD36 antibody, which in addition inhibits CD36 binding to TSP1/2 (33–36), both failed to affect lipid content (Supplementary Fig. S1N–S1O). Invalidation of CD36 using siRNA or FA6-152 CD36 blocking antibody did not affect 14C-palmitate uptake and oxidation (Supplementary Fig. S1P–S1AA), suggesting that CD36 is dispensable for lipid uptake in AML blasts.
To further investigate the role of CD36 in AML blast biology, we generated a CD36 gene signature comprising 249 genes that were all highly expressed along with CD36 in transcriptomic data from three AML patient databases (Fig. 2D; Supplementary Table S2). Interestingly, this signature was enriched in primary AML cells treated with AraC compared with control cells in two different databases (GSE40871, ref. 30; GSE146544, ref. 15), consistent with our previous observations (GSE97631, ref. 3; Fig. 2E).
CD36 triggers blast migration
A Gene Ontology (GO) analysis of the above-defined CD36 signature highlighted an enrichment in migration and adhesion processes (Fig. 3A), including the GO term “positive regulation of mononuclear cell migration” (Supplementary Fig. S2A), whereas no enrichment for GO terms related to lipid transport and metabolism was found (data not shown). Considering that CD36 involvement in metastasis is described in several tumors (6, 12, 37), we hypothesized that CD36 could drive leukemic blast migration. Knocking down CD36 with shRNA or siRNA decreased AML cell migration (Fig. 3B; Supplementary Fig. S2B and S2C). FA6-152 CD36 blocking antibody, but not JC63.1 antibody, significantly decreased U937 and OCIAML3 migration (Fig. 3C; Supplementary Fig. S2D). In fresh patient samples, FA6-152 antibody decreased migration (Fig. 3D). FA6-152 antibody blocks the binding of CD36 to its ligand TSP1, which is well known to promote cell migration in various tumor models (38–40). Invalidation of TSP1 using shRNA (Supplementary Fig. S2E and S2F) decreased migration of U937 (Fig. 3E) as well as of OCIAML3 cells (Supplementary Fig. S2G). Adding A6.1 anti-TSP1 antibody also decreased U937 and OCIAML3 cell migration, and noteworthy, CD36 knockdown with shRNA abolished this effect (Fig. 3F; Supplementary Fig. S2H). Conversely, recombinant TSP1 increased the migration of both U937 and OCIAML3 cells. This effect was hampered by the addition of CD36 blocking antibody (Fig. 3G; and Supplementary Fig. S2I), indicating that TSP1 promotes cell migration through its binding to CD36. As observed with FA6-152 anti-CD36 antibody, A6.1 TSP1 blocking antibody inhibited migration of fresh patient cells (Fig. 3H). Together, these data highlight that CD36 fosters AML blast migration, at least in part through its binding to TSP1.
Figure 3.
CD36 triggers blast migration. A, GO analysis of migration-related biological processes significantly enriched in the CD36 gene signature. B, Number of migrating shCtrl or shCD36 U937 cells in the lower chamber of the transwell assay (normalized to control; n = 5). C, As in B, but with CD36-blocking antibodies (FA6-152 or JC53.1; n = 4). D, Number of migrating cells in primary AML samples treated or not with CD36 blocking antibody (FA6-152; n = 7). E, Number of migrating shCtrl or shTSP1 U937 cells (normalized to control; n = 3). F, Number of migrating shCtrl or shCD36 U937 cells treated or not with TSP1-blocking antibody (A6.1; normalized to shCtrl; n = 5). G, Number of migrating U937 cells treated or not with recombinant TSP1 in the presence or not of CD36-blocking antibody (FA6-152; normalized to control; n = 3). H, As in D, but with TSP1-blocking antibody (A6.1; n = 5). I, Bioluminescence imaging of mice injected with the indicated AML cell line stably expressing a luciferase reporter from day 1 to day 64 after injection. J, Representative histologic section showing anti-human Ku-80 labeling of U937 AML blasts in the subcutaneous adipose tissue 17 days after xenograft. Positive brown nuclei highlight the infiltration of human blasts in murine adipose tissue either as dense infiltrate (top) or scattered isolated cells (bottom). Top scale bar, 50 μm; bottom, 10 μm. K, Percentage of viable OCIAML3 cells in the indicated tissues 14 days after orthotopic injection in the femur. L, Expression of CD36 [mean fluorescence intensity (MFI)] in viable OCIAML3 cells measured in the injected and contralateral femur and different organs of mice 14 days after orthotopic injection. Values are represented as mean ± SEM. B, C, and E, One sample t test. D and H, Paired t test. F and G, Ordinary one-way ANOVA with Tukey multiple comparisons test. L, Matched one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Because migration is usually associated with metastasis in solid tumors, we asked whether blasts could migrate and localize into extramedullary organs. To visualize AML cell distribution in vivo, we injected a large panel of bioluminescent AML cell lines into NSG mice. In all models, AML cells localized throughout the body as shown by the acquisition of bioluminescence in mice during the time course of AML progression, and on isolated organs at sacrifice (Fig. 3I; Supplementary Fig. S2J and S2K). Analyses of human-specific CD45 expression by RT-qPCR in NSG mice grafted with U937 cells confirmed the presence of human blasts in the BM, but also in different organs including the spleen, perigonadal adipose tissues, and liver (Supplementary Fig. S2L). IHC using a human-specific anti–Ku-80 antibody showed that AML blasts present in subcutaneous adipose tissues were scattered within the stroma, closely surrounding adipocytes (Fig. 3J), ruling out that the PCR results corresponded to blasts present in blood vessels. To further rule out that these extramedullary localizations might result from an experimental bias linked to the intravenous modality of blast injection, we injected OCIAML3 cells orthotopically into the right femur of NSG mice. Flow cytometry analyses revealed the presence of human CD45+ blasts in the contralateral femur, but also in the spleen, subcutaneous adipose tissue, perigonadal adipose tissue, and lung (Fig. 3K). This tissue distribution was comparable with that observed after intravenous injection of OCIAML3 cells (Supplementary Fig. S2M), establishing that leukemic cells are able to migrate out of the BM and engraft into distant locations. Interestingly, CD36 expression was increased in cells that had migrated from the site of injection, as shown comparing blasts from the BM of injected femurs to contralateral femurs and extramedullary organs excepting lungs (Fig. 3L). In addition, we pretreated OCIAML3 cells with IGG or FA6-152 anti-CD36 antibody, injected these cells intravenously into NSG mice, which were then treated with either IGG or FA6-152 antibody. Ten days later, whereas untreated control cells started engrafting in the BM, perigonadal adipose tissue (PGAT), liver and lungs, minimal to no engraftment was detected in mice injected with FA6-152 pretreated cells (Supplementary Fig. S2N), suggesting that even in the absence of chemotherapy, CD36 is required for AML cell extravasation and migration. Altogether, these results establish that AML blasts can migrate from the BM into different organs in xenograft models.
Residual blasts maintained their migratory properties after chemotherapy
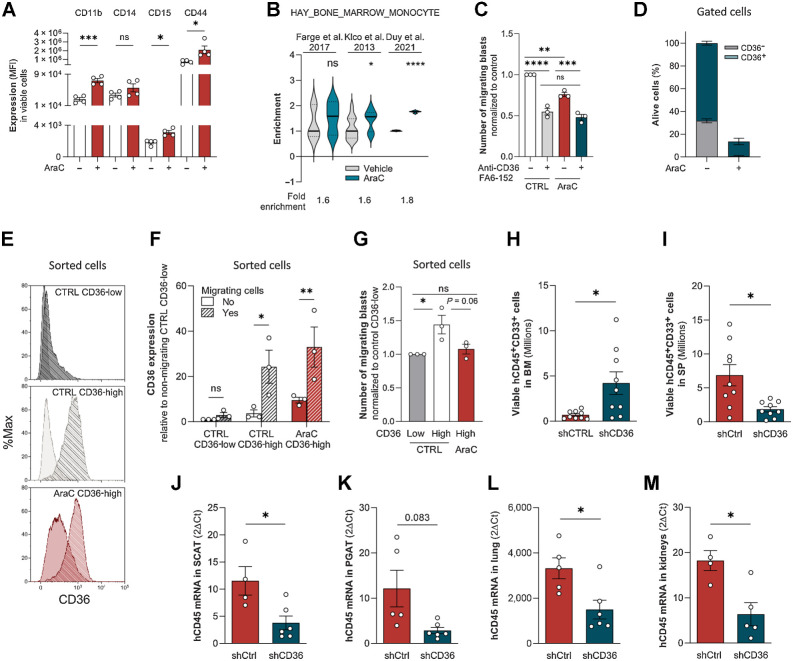
We observed that treating cells with AraC increased CD11b, CD14, CD15, and CD44 protein expression levels (Fig. 4A; Supplementary Fig. S3A), consistent with an AraC-induced partial differentiation of blasts toward a monocytic phenotype. Monocytes are particularly endowed with migratory capacities, and interestingly, a “bone marrow monocyte” gene signature was enriched in primary AML cells treated with AraC as compared with control cells in three databases (GSE40871, ref. 30; GSE97631, ref. 3; GSE146544, ref. 15; Fig. 4B). We thus questioned the migration ability of blasts bypassing chemotherapy. Although to a lesser extent than in control conditions, U937 cells that survived 4 days of AraC treatment were still able to migrate, and, noteworthy, in both control- and AraC-treated cells, CD36 blocking antibody FA6-152 decreased migration (Fig. 4C). We then studied the ability to migrate of viable CD36-high cells sorted after 4 days of treatment with AraC. Although about 83% of U937 cells died, the percentage of living CD36-positive cells increased from 70% in untreated cells up to 90% in resistant ones and very few CD36-low cells survived, precluding their analysis (Fig. 4D). CD36 expression was strongly increased in migrating compared with nonmigrating cells in both control and AraC-treated CD36-positive U937 cells (Fig. 4E and F). CD36-expressing blasts migrated more efficiently than CD36-negative cells in control condition, and, even though AraC partially decreased their migration, CD36-high cells were still efficiently migrating, similarly to untreated CD36-low cells (Fig. 4G).
Figure 4.
Residual blasts maintained their migratory properties after chemotherapy. A, CD11b, CD14, CD15, and CD44 cell-surface expression in viable control or AraC-treated (2 μmol/L) U937 cells for 96 hours (n = 4). B, Gene set enrichment analysis of “HAY_BONE_MARROW_MONOCYTE” gene signature in AraC versus untreated AML samples from three different transcriptomic analyses. C, Number of migrating cells in the presence or absence of CD36-blocking antibody (FA6-152) sorted from control or 2 μmol/L AraC-treated cells for 4 days (normalized to control; n = 3). D, Percentage of viable CD36-low and CD36-high cells in PBS or 2 μmol/L AraC-treated U937 cells for 4 days (n = 4). E, Representative histograms of CD36 expression in nonmigrating versus migrating CD36-low and CD36-high subpopulations sorted from control or AraC-treated U937 cells (2 μmol/L) for 4 days. Plain histogram, nonmigrating cells; striped histogram, migrating cells. F, Quantification of CD36 expression in nonmigrating versus migrating cells in the same conditions as in E (n = 3). G, Number of migrating cells from CD36-low and CD36-high subpopulations sorted from control or 2 μmol/L AraC-treated U937 cells for 4 days (normalized to Ctrl CD36-low cells; n = 3). H and I, Quantification in the BM (H) and spleen (I) of viable U937 cells transduced with shCtrl (n = 9) or shCD36 (n = 9) in mice relapsing after AraC treatment. J–M, Quantification of U937 cells by RT-qPCR (hCD45 mRNA normalized to m36B4) in SCAT (J), PGAT (K), lung (L), and kidneys (M) in mice relapsing after AraC treatment. Data expressed as 2−ΔCT (values multiplied by 1,000; n = 4–5 shCtrl, n = 5–7 shCD36). Values are represented as mean ± SEM. A, Mann–Whitney or unpaired t test depending on sample distribution. B, Unpaired t test. C and G, Ordinary one-way ANOVA with Tukey multiple comparisons test. F, Ordinary two-way ANOVA with multiple comparisons correction. H–M, Unpaired t test with or without Welch correction depending on sample variance. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We next engrafted mice with control or CD36 knocked-down U937 cells, treated mice with AraC for 5 days, and once disease relapse had translated into clinical signs, the presence of blasts was evaluated in different organs. At relapse, CD36 knockdown had increased the tumor burden in the BM (Fig. 4H), whereas it had reduced it in the spleen (Fig. 4I) and in several extramedullary tissues (Fig. 4J–M), albeit not in the liver (data not shown). Thus, the increased time to relapse in mice cotreated with chemotherapy and CD36 inhibition (Fig. 2A–C) likely does not result from a blast reduction in the BM but from a decreased blast dissemination into extramedullary organs.
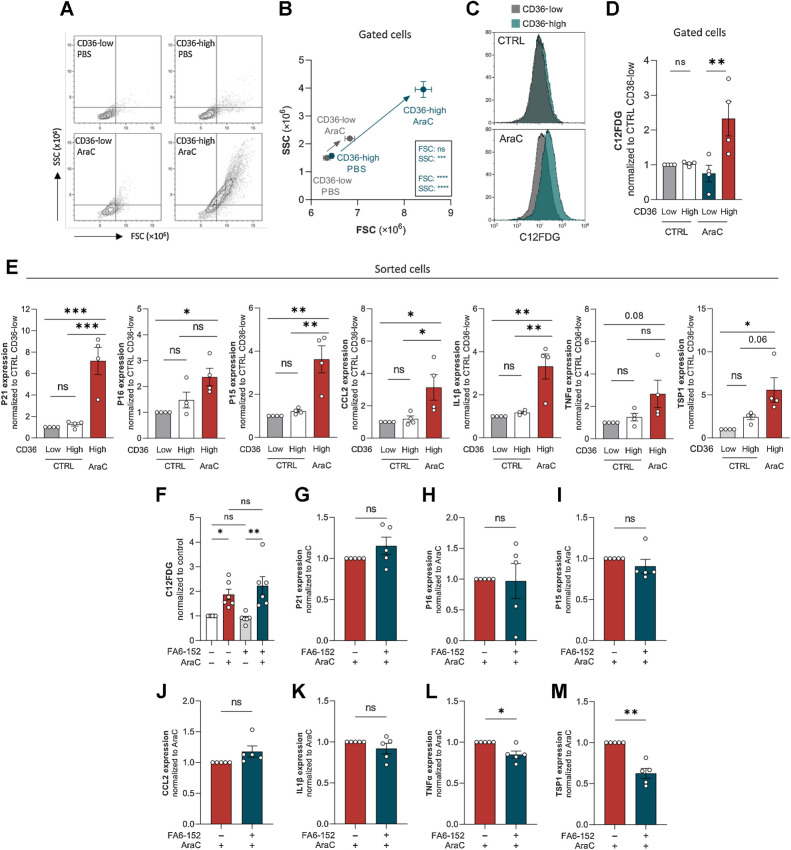
Senescent-like state induced by cytarabine chemotherapy occurs specifically in CD36-high AML blasts
We observed that AraC treatment increased both cell size and granularity, two common features of senescent cells, and noteworthily, this increase almost exclusively occurred within the CD36-high viable cell population (Fig. 5A and B). In addition, senescence in AraC-treated cells translated into an increased β-galactosidase activity, which also occurred selectively in CD36-high cells, as measured by a 2.5-fold increase in C12FDG fluorescence (Fig. 5C and D). To confirm these data, we treated U937 cells for 4 days with PBS or AraC, sorted viable CD36-high and -low cells, and analyzed the expressions of several cyclin-dependent kinase inhibitors (P21, P16, and P15), as well as that of cytokines of the SASP (CCL2, IL1b, and TNFa). As previously (Fig. 4D), the very small number of CD36-low cells surviving after AraC treatment hindered these analyses. Expressions of all these well-established senescence markers were markedly increased by AraC treatment in CD36-high cells (Fig. 5E). Similarly, size, granularity, β-galactosidase activity, and expressions of senescent markers, were all increased in chemoresistant OCIAML3 cells after 4 days of treatment with AraC (Supplementary Fig. S3B–S3D), specifically in CD36-high cells (Supplementary Fig. S3B and S3C). Interestingly, we found that TSP1 mRNA, which was found to be part of the SASP (41), was strongly increased by AraC (Fig. 5E; Supplementary Fig. S3D), especially within the CD36-high population (Fig. 5E). Of note, blocking CD36 with FA6-152 antibody did not alter AraC-induced increase in β-galactosidase activity of U937 or OCIAML3 cells (Fig. 5F; Supplementary Fig. S3E). However, a significant decrease in SASP-related genes TNFα and TSP1 mRNA level was observed in U937 cells, although no significant effect was observed regarding the expressions of other senescent markers (Fig. 5G–M).
Figure 5.
Senescent-like state induced by cytarabine chemotherapy occurs specifically in CD36-high AML blasts. A and B, FSC/SCC analysis on gated CD36-low and CD36-high cells from U937 treated or not with 2 μmol/L AraC for 4 days (n = 4). C, Representative histogram of C12FDG staining on gated CD36-low and CD36-high cells from U937 treated or not with 2 μmol/L AraC for 4 days. D, Quantification of C12FDG staining performed in C (normalized to control CD36-low cells; n = 4). E, Expression of senescence-associated genes measured by RT-qPCR in CD36-low and CD36-high subpopulations sorted from control or 2 μmol/L AraC-treated U937 cells for 4 days (normalized to Ctrl CD36-low cells). mRNA of the gene of interest normalized to the housekeeping gene; data expressed as 2ΔΔCt (n = 4). F, C12FDG staining on U937 cells treated or not with 2 μmol/L AraC and CD36 blocking antibody (FA6-152) for 4 days. G–M, Expression of senescence-associated genes measured by RT-qPCR in 2 μmol/L AraC-treated U937 cells with or without CD36 blocking antibody (FA6-152) for 4 days (normalized to AraC-treated cells). mRNA of the gene of interest normalized to the housekeeping gene; data expressed as 2ΔΔCt (n = 5). Values are expressed as mean ± SEM. B, Unpaired t test. D, E, and F, Ordinary one-way ANOVA with Tukey multiple comparisons test. G–M, One sample Wilcoxon or t test depending on sample distribution. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
To decipher if senescence-like state and migration are functionally related, we compared the senescent profile of AraC-treated migrating and nonmigrating cells. After 4 days of AraC or vehicle treatment, viable U937 cells were FACS-sorted and submitted to migration assay. In AraC-treated condition, β-galactosidase activity was increased in cells that had migrated compared with nonmigrating cells (Supplementary Fig. S3F–S3G), whereas no difference was found regarding expressions of CDKi- and SASP-related genes (data not shown). These data suggest that chemotherapy-resistant cells that migrate preserve their senescent-like profile and can even increase senescence-associated β-galactosidase activity. Together, these results show that chemotherapy increases the expressions of CD36 and its ligand TSP1 in blasts that retain their migration ability and triggers a senescent-like phenotype specifically in CD36-high blasts, suggesting that senescent-like state and migration properties coexist within CD36-high blasts bypassing chemotherapy.
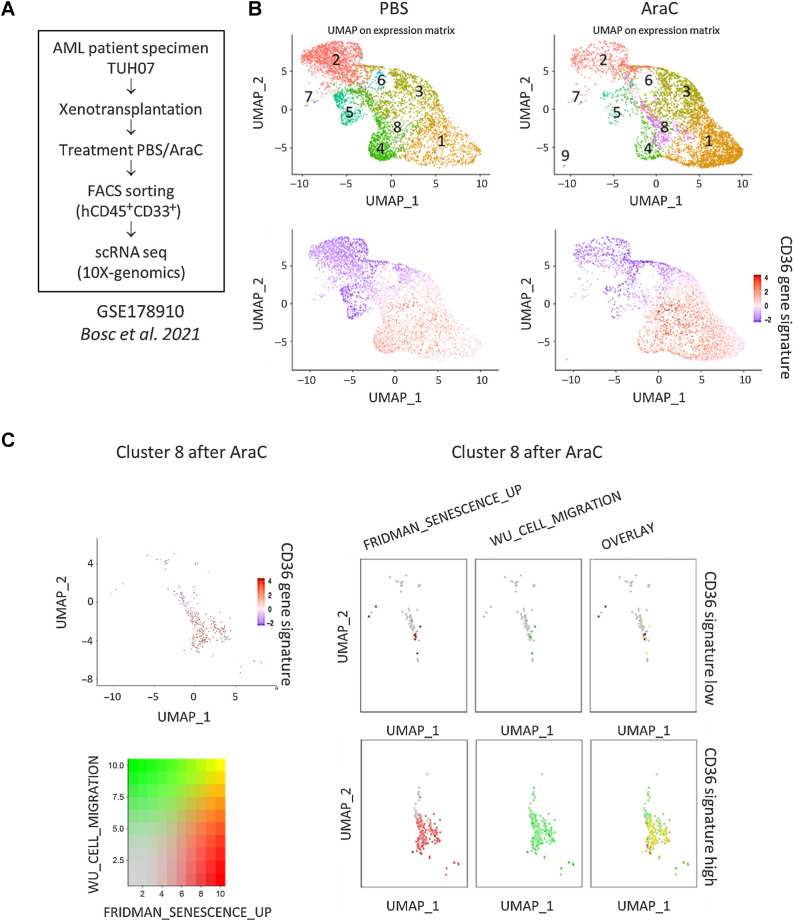
Single-cell transcriptomic analysis reveals enrichment of CD36-high, migration and senescence-like gene signatures in a cluster emerging after standard chemotherapy
We next asked whether primary AML patient cells persisting after chemotherapy also exhibit a senescent-like state associated with migration in vivo. To this aim, we analyzed the transcriptomes of more than 30,000 single viable AML cells from a PDX model (TUH07, ref. 42; Fig. 6A). The Seurat-based unsupervised hierarchical clustering analysis uncovered 8 distinct clusters before (PBS) and after AraC treatment (Fig. 6B). Although the cell count within each cluster varied before and after AraC treatment, cluster 8 exhibited the highest increase in cell count after AraC treatment (Supplementary Table S3). A detailed analysis of cluster 8 revealed heterogeneity within this cluster, which consisted of two subclusters exhibiting either high or low expression of the CD36 gene signature (Fig. 6C). Strikingly, the subcluster highly expressing the CD36 gene signature was significantly enriched in gene sets related to both senescence and migration processes, with a large proportion of cells exhibiting a strong overlay between the two signatures (Fig. 6C). Correlation plots emphasized the global colinearity among CD36, migration, and senescence across all cells, whereas the cluster 8 displayed the highest expression across all of the compared phenotypes (Supplementary Fig. S3H–S3J). These findings further strengthened that migration and senescence-like states coexist within high CD36-expressing blasts enriched after chemotherapy in vivo.
Figure 6.
Single-cell transcriptomic analysis reveals enrichment of CD36-high, migration, and senescence-like gene signatures in a cluster emerging after standard chemotherapy. A, Schematic representation of the single-cell RNA-seq experiment performed on the TUH07 PDX previously published in Bosc et al. 2021 (GEO accession GSE178910; ref. 42). B, Uniform Manifold Approximation and Projection (UMAP) plot of 31,604 single cells from PDX TUH07 using Seurat. Colors indicate k-means clusters (k = 9). C, Cluster 8 was isolated on the Seurat object and bifurcated into two groups “CD36-high” and “CD36-low” according to the enrichment level for the CD36 gene signature. Expression levels of gene sets related to senescence (FRIDMAN_SENESCENCE_UP) and migration (WU_CELL_MIGRATION) are shown.
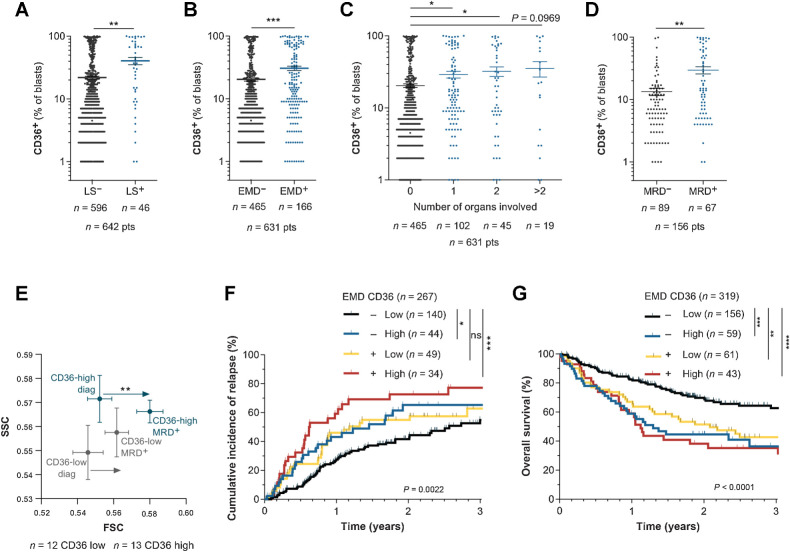
CD36 is positively associated with extramedullary disease in AML patients
We assessed the clinical relevance of these findings in patients from the TUH cohort. Interestingly, patients with hyperleukocytosis had a higher percentage of CD36+ blasts (Fig. 7A). The presence of EMD at diagnosis was documented in 631 patients and among them, 166 (26.3%) exhibited EMD with 1, 2, or more than 2 organs involved, in agreement with other reports (19, 20). In the TUH cohort, both EMD and a high CD36-positive blast percentage were independent prognostic factors of relapse and reduced OS (Supplementary Table S4). We found that the presence of an EMD was significantly correlated with a higher percentage of CD36+ blasts at diagnosis (Fig. 7B), regardless of the number of organs involved (Fig. 7C). Analysis of CD36 expression in the BM of patients from the TUH cohort showed a higher percentage of CD36-positive blasts in patients exhibiting minimal residual disease (MRD) after chemotherapy compared with patients without MRD (Fig. 7D). Using a CD36-gating strategy in diagnosis/MRD paired patient samples, we found a significant increase in cell size at MRD compared with diagnosis, a feature of senescent cells, specifically in CD36-high patients (Fig. 7E). Very interestingly, among patients classified as EMD-negative, a high CD36 expression identified a group at higher risk for both CIR and reduced OS (Fig. 7F and G). Together, these clinical data highlight that CD36 expression in blasts is positively associated with EMD in patients.
Figure 7.
CD36 is positively associated with extramedullary disease in AML patients. A, Percentage of CD36+ blasts in the BM of patients with (LS+) or without (LS−) leukocytosis at diagnosis (n = 596 LS−; n = 46 LS+). B, Percentage of CD36+ blasts in the BM of patients presenting (EMD+, n = 166) or not (EMD−, n = 465) clinical signs of EMD at diagnosis. C, Percentage of CD36+ blasts in the BM of patients depending on the number of organs involved in EMD (n = 465 for 0 organs; n = 102 for 1 organ; n = 45 for 2 organs; n = 19 for more than 2 organs). D, Percentage of CD36+ blasts in the BM of patients at diagnosis, exhibiting MRD or not after chemotherapy (n = 89 MRD−; n = 67 MRD+). E, FSC/SCC analysis on blasts from CD36 low (n = 12) and CD36 high (n = 13) patients followed up from diagnosis to MRD. F, Cumulative incidence of relapse in TUH patients according to CD36 expression and presence of EMD at diagnosis (n = 267). G, Overall survival in TUH patients according to CD36 expression and presence of EMD at diagnosis (n = 319). Values are represented as mean ± SEM. A, B, and D, Mann–Whitney test. C, Ordinary one-way ANOVA with Tukey multiple comparisons test. E, Wilcoxon matched-pairs signed rank test. F and G, Log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Discussion
Despite the availability in recent years of new treatments targeting key survival and metabolic vulnerabilities in leukemic blasts, AML remains an aggressive cancer with a poor prognosis, due to frequent relapses after chemotherapy. Here, our study highlights CD36 as a prognostic marker and therapeutic target of metastasis to thwart relapse in AML.
In patient- and cell line–derived xenografts in NSG mice, we have previously found and here confirmed that CD36 is overexpressed in chemoresistant blasts (3). Although CD36 has been shown to support fatty acid oxidation in chronic myeloid leukemia (8), our data suggest that, unlike FABP4 (43), CD36 is dispensable for lipid metabolism in AML blasts. Although inhibiting the lipid transport function of CD36 might require to knockout instead of knockdown its expression, as recently shown in AML (44), the absence of significant enrichment in GO terms related to lipid metabolism in the CD36 gene signature constitutes additional evidence of the uncoupling between CD36 and lipid metabolism in leukemic blasts.
Strikingly, our work demonstrated that CD36 fosters AML blast migration. In xenografted mice, a wide range of AML cell lines developed EMD in a variety of organs. This phenotype was also observed in the immuno-competent syngeneic AML model MLL/AF9 (45), showing that EMD in NSG mice does not result from an impaired immune response. Interestingly, AraC treatment, which increases CD36 expression, also triggers the differentiation of leukemic cells toward a monocytic phenotype. Monocytic cells migrate out of the marrow to perform their functions in tissues. Although chemotherapies were used in lymphomas to enhance CD34+ hematopoietic stem cell mobilization in order to collect them before autologous transplantation (46), their impact on leukemic blast mobilization has not been investigated. Importantly, while having no effect in untreated cells in contrast to findings obtained with MOLM13 cells (47), we demonstrated that CD36 inhibition delayed time to relapse after chemotherapy, as observed in an AML mouse model (44), this being associated with a decreased EMD.
Using a set of senescence markers, we demonstrated that chemotherapy-induced senescence occurs mostly, if not only, in CD36-high cells. Although CD36 inhibition did not impair β-galactosidase activity, it decreased the expression of some senescence-related secreted factors including TNFα and TSP1. These results are in accordance with a recently published paper (48) showing that CD36 inhibition in muscle senescent cells, although not impacting β-galactosidase activity nor the number of senescent cells, decreased the expression of some SASP-related genes. This confers to CD36 a role in the secretory phenotype of senescent cells, as previously described (49). Together, these data suggest that CD36, rather than contributing to leukemic blasts entry into senescence, could play a role in the secretory phenotype of senescent blasts treated with chemotherapy AraC. Importantly, we demonstrated that AraC-surviving blasts were still able to migrate. The preservation of migratory abilities of CD36-high cells bypassing chemotherapy and exhibiting a senescence-like state was further strengthened by single-cell analysis of a PDX treated with AraC (42). A cell cluster emerging after chemotherapy was not only overexpressing the CD36 gene signature but also enriched in both senescence and migration gene sets. Among the components of the SASP that could support leukemic blast migration, TSP1 was strongly increased by chemotherapeutic treatment, especially in the CD36-high cell population. The sustained induction of both CD36 and TSP1 by AraC treatment likely contributes to maintain the capacity of CD36-high cells to migrate, even after chemotherapeutic stress.
The clinical relevance of these results was analyzed in a large cohort of 1,273 AML patients from the TUH. CD36 was found by multivariate analysis to be an independent marker of AML progression, with a higher hazard ratio than major parameters that are characteristic of aggressive AML (FLT3-ITD, adverse cytogenetics, etc.). A previous study performed on a smaller cohort (266 patients) also reported the poor outcome of AML patients with a high CD36 expression (50). In the TUH cohort, the pejorative role of CD36 is further emphasized by the fact that patients with MRD after induction chemotherapy had a higher percentage of CD36-positive blasts at diagnosis than patients without MRD. Interestingly, both leukocytosis and EMD were significantly associated with a high percentage of CD36-positive blasts at diagnosis, and EMD was found to convey a poor prognosis, as recently shown (22). Importantly, among patients classified as EMD-negative, CD36 expression, whose evaluation is easily performed in routine practice, discriminated a group of patients with a higher CIR and a reduced OS. The impact of CD36 on patients' classification is reinforced by the fact that its expression is not clearly associated with mutational profiles commonly utilized in clinical practice for AML.
The present study has some limitations. Inhibition of CD36 by shRNA or blocking antibodies did not affect lipid uptake in leukemic blasts. These methods, however, do not result in complete loss of CD36 activity, and it is thus possible that the amount of protein remaining active is sufficient to allow normal lipid metabolism in blasts. CD36 knockout by genetic approaches, although not clinically easy to duplicate, could reveal a role of CD36 in lipid metabolism in leukemic blasts. We showed that CD36 fosters leukemic blasts migration, at least in part through its binding with TSP1, which was strongly released by blasts surviving chemotherapy. However, one cannot rule out the possibility that TSP1 may also be secreted by the microenvironment of different organs in response to chemotherapy, which could affect blast metastasis.
In conclusion, whereas most AML studies are focused on leukemic cells present in the BM, our study unravels the importance of blast dissemination and EMD triggered by AraC. It highlights the noncanonical role of CD36 in this process, opening a new therapeutic strategy through CD36 inhibition in combination with chemotherapy to impair this metastatic process.
Supplementary Material
Supplemental Text
Human antibodies for flow cytometry analyses (TUH cohort)
CD36 gene signature
Single cell analysis
Multivariate analysis of the TUH cohort
Figure S1
Figure S2
Figure S3
Acknowledgments
This work was supported by the Programme “Investissement d'Avenir” PSPC (IMODI; J.E. Sarry), the Laboratoire d'Excellence Toulouse Cancer (TOUCAN and TOUCAN2.0; contract ANR11-LABEX; J.E. Sarry), INCA (PLBIO 2020-010, DIALAML; J.E. Sarry), the Fondation Toulouse Cancer Santé (BADIPAML; J.E. Sarry), the Fondation ARC, the Cancéropôle Grand Sud-Ouest, the Ligue Nationale de Lutte Contre le Cancer, the association Prolific and the association GAEL. A. Sahal is a fellow from the European Regional Development Fund through the Interreg V-A Spain–France–Andorra (POCTEFA) program, project PROTEOblood (EFA360/19; J.E. Sarry). T. Farge has a fellowship from the Fondation Toulouse Cancer Santé and Fondation ARC. This work was supported by the National Research Agency (ANR: Agence Nationale de la Recherche) for the “Investissement d'avenir” (ANR-11-PHUC-001, CAPTOR research program; J.E. Sarry), and in part by ANR-21-CE14-0057-01 (C. Moro). We thank all members of mice core facilities (UMS006, ANEXPLO, Inserm, Toulouse) in particular Marie Lulka, Christine Campi, Sarah Gandarillas, and Cédric Baudelin for their support and technical assistance, and Eric Delabesse for the management of the Biobank BRC-HIMIP that is supported by CAPTOR. This work was granted access to the HPC resources of the CALMIP supercomputing center under the allocation 2019-T19001. Team J.E. Sarry is a member of OPALE Carnot Institute, The Organization for Partnerships in Leukemia. We thank Anne-Marie Benot, Muriel Serthelon, and Stéphanie Nevouet for their daily help with the administrative and financial management of our team. We are grateful to the CERT platform from RESTORE, in particular Marie-Laure Renoud and Jessica Fontaine, for flow cytometry facilities as well as Sophie Bonnel, Lucas Bourdens, Claire Maslo, and Anais Checkroun for their help in experiments. We thank Mrs. Zakaroff-Girard and Riant (Cytometry Core Facility, Inserm U1048, part of TRI Imaging Platform, Genotoul) and Mrs. Farce (The Technology Cluster's Cytometry and Cell-Sorting Platform, CRCT) for cell-sorting technical assistance. We thank Catherine Muller and Camille Attané from the Institut de Pharmacologie et de Biologie Structurale (IPBS), Beatrice Cousin, Vincent Cuminetti, and Louis Casteilla from STROMALab/RESTORE for very helpful discussions, and Armelle Yart from RESTORE for manuscript reviewing.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
T. Farge: Conceptualization, investigation, methodology, writing–original draft, writing–review and editing. J. Nakhle: Conceptualization, investigation, methodology, writing–review and editing. D. Lagarde: Investigation, writing–review and editing. G. Cognet: Investigation, writing–review and editing. N. Polley: Investigation, writing–review and editing. R. Castellano: Investigation, methodology, writing–review and editing. M.-L. Nicolau: Investigation. C. Bosc: Investigation. M. Sabatier: Investigation. A. Sahal: Investigation. E. Saland: Investigation. Y. Jeanson: Investigation. N. Guiraud: Investigation. E. Boet: Investigation. C. Bergoglio: Investigation. M. Gotanègre: Investigation. P.-L. Mouchel: Investigation. L. Stuani: Investigation. C. Larrue: Investigation. M. Sallese: Investigation. V. De Mas: Investigation. C. Moro: Methodology, writing–review and editing. C. Dray: Writing–review and editing. Y. Collette: Methodology, writing–review and editing. I. Raymond-Letron: Investigation. I. Ader: Conceptualization, funding acquisition, methodology, writing–review and editing. C. Récher: Conceptualization, funding acquisition, writing–review and editing. J.-E. Sarry: Conceptualization, funding acquisition, methodology, writing–original draft. F. Cabon: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing. F. Vergez: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing. A. Carrière: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov 2020;10:506–25. [DOI] [PubMed] [Google Scholar]
- 3. Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov 2017;7:716–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B, Porras DP, et al. Identification of chemotherapy-induced leukemic-regenerating cells reveals a transient vulnerability of human AML recurrence. Cancer Cell 2018;34:483–98. [DOI] [PubMed] [Google Scholar]
- 5. van Gastel N, Spinelli JB, Sharda A, Schajnovitz A, Baryawno N, Rhee C, et al. Induction of a timed metabolic collapse to overcome cancer chemoresistance. Cell Metab 2020;32:391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ladanyi A, Mukherjee A, Kenny HA, Johnson A, Mitra AK, Sundaresan S, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018;37:2285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng WW, Wilkins O, Bang S, Ung M, Li J, An J, et al. CD36-mediated metabolic rewiring of breast cancer cells promotes resistance to HER2-targeted therapies. Cell Rep 2019;29:3405–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell 2016;19:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Endemann G, Stanton LW, Madden KS, Bryant CM, White RT, Protter AA. CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem 1993;268:11811–6. [PubMed] [Google Scholar]
- 10. Asch AS, Barnwell J, Silverstein RL, Nachman RL. Isolation of the thrombospondin membrane receptor. J Clin Invest 1987;79:1054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng M, Cai X, Long L, Xie L, Ma H, Zhou Y, et al. CD36 promotes the epithelial-mesenchymal transition and metastasis in cervical cancer by interacting with TGF-beta. J Transl Med 2019;17:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017;541:41–5. [DOI] [PubMed] [Google Scholar]
- 13. Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal 2009;2:re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye H, Minhajuddin M, Krug A, Pei S, Chou CH, Culp-Hill R, et al. The hepatic microenvironment uniquely protects leukemia cells through induction of growth and survival pathways mediated by LIPG. Cancer Discov 2021;11:500–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duy C, Li M, Teater M, Meydan C, Garrett-Bakelman FE, Lee TC, et al. Chemotherapy induces senescence-like resilient cells capable of initiating AML recurrence. Cancer Discov 2021;11:1542–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kyjacova L, Saup R, Ronsch K, Wallbaum S, Dukowic-Schulze S, Foss A, et al. IER2-induced senescence drives melanoma invasion through osteopontin. Oncogene 2021;40:6494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whiteley AE, Price TT, Cantelli G, Sipkins DA. Leukaemia: a model metastatic disease. Nat Rev Cancer 2021;21:461–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang H, Brandwein J, Yi QL, Chun K, Patterson B, Brien B. Extramedullary infiltrates of AML are associated with CD56 expression, 11q23 abnormalities and inferior clinical outcome. Leuk Res 2004;28:1007–11. [DOI] [PubMed] [Google Scholar]
- 19. Cribe AS, Steenhof M, Marcher CW, Petersen H, Frederiksen H, Friis LS. Extramedullary disease in patients with acute myeloid leukemia assessed by 18F-FDG PET. Eur J Haematol 2013;90:273–8. [DOI] [PubMed] [Google Scholar]
- 20. Ganzel C, Manola J, Douer D, Rowe JM, Fernandez HF, Paietta EM, et al. Extramedullary disease in adult acute myeloid leukemia is common but lacks independent significance: analysis of patients in ECOG-ACRIN cancer research group trials, 1980–2008. J Clin Oncol 2016;34:3544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Byrd JC, Weiss RB, Arthur DC, Lawrence D, Baer MR, Davey F, et al. Extramedullary leukemia adversely affects hematologic complete remission rate and overall survival in patients with t(8;21)(q22;q22): results from cancer and leukemia group B 8461. J Clin Oncol 1997;15:466–75. [DOI] [PubMed] [Google Scholar]
- 22. Eckardt JN, Stolzel F, Kunadt D, Rollig C, Stasik S, Wagenfuhr L, et al. Molecular profiling and clinical implications of patients with acute myeloid leukemia and extramedullary manifestations. J Hematol Oncol 2022;15:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010;116:354–65. [DOI] [PubMed] [Google Scholar]
- 24. Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010;115:453–74. [DOI] [PubMed] [Google Scholar]
- 25. Bertoli S, Tavitian S, Huynh A, Borel C, Guenounou S, Luquet I, et al. Improved outcome for AML patients over the years 2000–2014. Blood Cancer J 2017;7:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laurens C, Bourlier V, Mairal A, Louche K, Badin PM, Mouisel E, et al. Perilipin 5 fine-tunes lipid oxidation to metabolic demand and protects against lipotoxicity in skeletal muscle. Sci Rep 2016;6:38310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009;94:131–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klco JM, Spencer DH, Lamprecht TL, Sarkaria SM, Wylie T, Magrini V, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood 2013;121:1633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mwaikambo BR, Sennlaub F, Ong H, Chemtob S, Hardy P. Activation of CD36 inhibits and induces regression of inflammatory corneal neovascularization. Invest Ophthalmol Vis Sci 2006;47:4356–64. [DOI] [PubMed] [Google Scholar]
- 32. Klenotic PA, Page RC, Li W, Amick J, Misra S, Silverstein RL. Molecular basis of antiangiogenic thrombospondin-1 type 1 repeat domain interactions with CD36. Arterioscler Thromb Vasc Biol 2013;33:1655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corbet C, Bastien E, Santiago de Jesus JP, Dierge E, Martherus R, Vander Linden C, et al. TGFbeta2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat Commun 2020;11:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nergiz-Unal R, Lamers MM, Van Kruchten R, Luiken JJ, Cosemans JM, Glatz JF, et al. Signaling role of CD36 in platelet activation and thrombus formation on immobilized thrombospondin or oxidized low-density lipoprotein. J Thromb Haemost 2011;9:1835–46. [DOI] [PubMed] [Google Scholar]
- 35. Ohgami N, Nagai R, Ikemoto M, Arai H, Kuniyasu A, Horiuchi S, et al. CD36, a member of class B scavenger receptor family, is a receptor for advanced glycation end products. Ann N Y Acad Sci 2001;947:350–5. [DOI] [PubMed] [Google Scholar]
- 36. Aburima A, Berger M, Spurgeon BEJ, Webb BA, Wraith KS, Febbraio M, et al. Thrombospondin-1 promotes hemostasis through modulation of cAMP signaling in blood platelets. Blood 2021;137:678–89. [DOI] [PubMed] [Google Scholar]
- 37. Gong J, Lin Y, Zhang H, Liu C, Cheng Z, Yang X, et al. Reprogramming of lipid metabolism in cancer-associated fibroblasts potentiates migration of colorectal cancer cells. Cell Death Dis 2020;11:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sargiannidou I, Qiu C, Tuszynski GP. Mechanisms of thrombospondin-1-mediated metastasis and angiogenesis. Semin Thromb Hemost 2004;30:127–36. [DOI] [PubMed] [Google Scholar]
- 39. Firlej V, Mathieu JR, Gilbert C, Lemonnier L, Nakhle J, Gallou-Kabani C, et al. Thrombospondin-1 triggers cell migration and development of advanced prostate tumors. Cancer Res 2011;71:7649–58. [DOI] [PubMed] [Google Scholar]
- 40. Hu C, Wen J, Gong L, Chen X, Wang J, Hu F, et al. Thrombospondin-1 promotes cell migration, invasion and lung metastasis of osteosarcoma through FAK dependent pathway. Oncotarget 2017;8:75881–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mikula-Pietrasik J, Sosinska P, Janus J, Rubis B, Brewinska-Olchowik M, Piwocka K, et al. Bystander senescence in human peritoneal mesothelium and fibroblasts is related to thrombospondin-1-dependent activation of transforming growth factor-beta1. Int J Biochem Cell Biol 2013;45:2087–96. [DOI] [PubMed] [Google Scholar]
- 42. Bosc C, Saland E, Bousard A, Gadaud N, Sabatier M, Cognet G, et al. Mitochondrial inhibitors circumvent adaptive resistance to venetoclax and cytarabine combination therapy in acute myeloid leukemia. Nat Cancer 2021;2:1204–23. [DOI] [PubMed] [Google Scholar]
- 43. Shafat MS, Oellerich T, Mohr S, Robinson SD, Edwards DR, Marlein CR, et al. Leukemic blasts program bone marrow adipocytes to generate a protumoral microenvironment. Blood 2017;129:1320–32. [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y, Guo H, Zhang Z, Lu W, Zhu J, Shi J. IL-6 promotes chemoresistance via upregulating CD36 mediated fatty acids uptake in acute myeloid leukemia. Exp Cell Res 2022;415:113112. [DOI] [PubMed] [Google Scholar]
- 45. Stavropoulou V, Kaspar S, Brault L, Sanders MA, Juge S, Morettini S, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell 2016;30:43–58. [DOI] [PubMed] [Google Scholar]
- 46. Montillo M, Tedeschi A, Rossi V, Cairoli R, Pungolino E, Intropido L, et al. Successful CD34+ cell mobilization by intermediate-dose Ara-C in chronic lymphocytic leukemia patients treated with sequential fludarabine and Campath-1H. Leukemia 2004;18:57–62. [DOI] [PubMed] [Google Scholar]
- 47. Zhang T, Yang J, Vaikari VP, Beckford JS, Wu S, Akhtari M, et al. Apolipoprotein C2-CD36 promotes leukemia growth and presents a targetable axis in acute myeloid leukemia. Blood Cancer Discov 2020;1:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moiseeva V, Cisneros A, Sica V, Deryagin O, Lai Y, Jung S, et al. Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration. Nature 2023;613:169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chong M, Yin T, Chen R, Xiang H, Yuan L, Ding Y, et al. CD36 initiates the secretory phenotype during the establishment of cellular senescence. EMBO Rep 2018;19:e45274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perea G, Domingo A, Villamor N, Palacios C, Junca J, Torres P, et al. Adverse prognostic impact of CD36 and CD2 expression in adult de novo acute myeloid leukemia patients. Leuk Res 2005;29:1109–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Text
Human antibodies for flow cytometry analyses (TUH cohort)
CD36 gene signature
Single cell analysis
Multivariate analysis of the TUH cohort
Figure S1
Figure S2
Figure S3
Data Availability Statement
All raw data generated in this study are available upon request from the corresponding author. The data analyzed in this study were obtained from GEO at GSE14468, GSE40871, GSE97631, and GSE146544. Data from the BeatAML study were obtained from http://www.vizome.org, and TCGA AML data set (29) was obtained from https://cbioportal.org.



![Figure 2. CD36 inhibition delays AML relapse after chemotherapy. A, Percentage of mice engrafted with U937 cells transduced with shCtrl (n = 11) or shCD36 (n = 11) and treated with vehicle or AraC, surviving over time after treatment. B, Same as A, with OCIAML3 [shCtrl (n = 18) and shCD36 (n = 22)]. C, Percentage of mice engrafted with U937 cells surviving over time after treatment with the indicated combinations of PBS or AraC administered for the first 5 days, with either control IgG or FA6-152 anti-CD36 antibody injected three times/week (n = 7 PBS-IGG; n = 7 PBS-anti-CD36; n = 7 AraC-IGG; n = 6 AraC-anti-CD36). D, Venn diagram of significantly cooverexpressed genes along with CD36 in TCGA, Verhaak, and BeatAML cohorts. E, Gene set enrichment analysis of CD36 gene signature in AraC versus untreated AML blasts from three different transcriptomic analyses. A–C, log-rank (Mantel–Cox) test. E, One sample t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/045d/10472106/e4459769aa47/2824fig2.jpg)
![Figure 3. CD36 triggers blast migration. A, GO analysis of migration-related biological processes significantly enriched in the CD36 gene signature. B, Number of migrating shCtrl or shCD36 U937 cells in the lower chamber of the transwell assay (normalized to control; n = 5). C, As in B, but with CD36-blocking antibodies (FA6-152 or JC53.1; n = 4). D, Number of migrating cells in primary AML samples treated or not with CD36 blocking antibody (FA6-152; n = 7). E, Number of migrating shCtrl or shTSP1 U937 cells (normalized to control; n = 3). F, Number of migrating shCtrl or shCD36 U937 cells treated or not with TSP1-blocking antibody (A6.1; normalized to shCtrl; n = 5). G, Number of migrating U937 cells treated or not with recombinant TSP1 in the presence or not of CD36-blocking antibody (FA6-152; normalized to control; n = 3). H, As in D, but with TSP1-blocking antibody (A6.1; n = 5). I, Bioluminescence imaging of mice injected with the indicated AML cell line stably expressing a luciferase reporter from day 1 to day 64 after injection. J, Representative histologic section showing anti-human Ku-80 labeling of U937 AML blasts in the subcutaneous adipose tissue 17 days after xenograft. Positive brown nuclei highlight the infiltration of human blasts in murine adipose tissue either as dense infiltrate (top) or scattered isolated cells (bottom). Top scale bar, 50 μm; bottom, 10 μm. K, Percentage of viable OCIAML3 cells in the indicated tissues 14 days after orthotopic injection in the femur. L, Expression of CD36 [mean fluorescence intensity (MFI)] in viable OCIAML3 cells measured in the injected and contralateral femur and different organs of mice 14 days after orthotopic injection. Values are represented as mean ± SEM. B, C, and E, One sample t test. D and H, Paired t test. F and G, Ordinary one-way ANOVA with Tukey multiple comparisons test. L, Matched one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/045d/10472106/94de82caa0e3/2824fig3.jpg)