Highlights
-
•
Obesity and lipids cannot genetically increase the risk of allergic rhinitis.
-
•
Interleukin-6 might contribute to the development of this disease.
-
•
Adipocyte fatty acid-binding protein perhaps promoted the onset of this disease.
-
•
Bonferroni correction had the ability to eliminate these two correlations.
-
•
The above findings can be instructive for future research on allergic rhinitis.
Keywords: Adipocyte fatty acid-binding protein, Allergic rhinitis, Interleukin-6, Mendelian randomization, Obesity
Abstract
Objectives
Observational studies suggested that obesity may promote the development of allergic rhinitis. The aim of this study was to explore the association of obesity, lipids and adipokines with this allergic disease at the genetic level using Mendelian randomization strategies.
Methods
Summary data for three obesity indicators (such as body mass index), eight lipid indicators (such as triglycerides) and six adipokines (such as interleukin-6 and adipocyte fatty acid-binding protein) were collected, and suitable instrumental variables were extracted from these summary data according to the three main assumptions of Mendelian randomization. Three Mendelian randomization methods (such as inverse variance weighted) were used to detect the casual effect of the above indicators on allergic rhinitis risk. Sensitivity analyses were performed to assess heterogeneity and horizontal pleiotropy.
Results
After Bonferroni correction, the inverse variance weighted reported that elevated levels of interleukin-6 and adipocyte fatty acid-binding protein were nominally associated with the decreased risk of allergic rhinitis (OR = 0.870, 95% CI 0.765–0.990, p = 0.035; OR = 0.732, 95% CI 0.551–0.973, p = 0.032). The other Mendelian randomization methods supported these results. Obesity, lipids and other adipokines were not related to this allergic disease. Sensitivity analyses found no heterogeneity and horizontal pleiotropy in the study.
Conclusion
The study provided some interesting, but not sufficient, evidence to suggest that interleukin-6 and adipocyte fatty acid-binding protein might play a protective role in the development of allergic rhinitis at the genetic level. These findings should be validated by more research.
Level of evidence
This was a Mendelian randomized study with a level of evidence second only to clinical randomized trials, and higher than cohort and case-control studies.
Introduction
Allergic rhinitis is a common allergic and inflammatory disease in the upper respiratory tract.1 More than 500 million people are affected by this disease worldwide, and the prevalence’s of allergic rhinitis in China and several western countries are even higher, reaching between 12 and 30%.2, 3 Although the disease is not fatal, it can still cause a range of adverse effects if not effectively controlled. For example, severe allergic rhinitis may disrupt sleep, cause headaches and memory loss, trigger sinusitis and aggravate asthma.4, 5 Symptoms such as the runny nose induced by the disease can also affect the patient's image when socializing, causing social fear and depression.6 However, there is no cure for allergic rhinitis, and the available medications can only relieve the symptoms.7 Therefore, in-depth research into the pathogenesis and risk factors of the disease is necessary.
It is well known that obesity is a growing global health challenge. Several studies have confirmed the correlation between obesity and the development of asthma.8, 9 Because allergic rhinitis and asthma are very similar in their pathogenesis and there is a concept of “one airway, one disease”, the effect of obesity on allergic rhinitis has therefore attracted a great deal of attention globally.10 Then, an increasing number of studies have been carried out, suggesting that obesity and overweight may increase the risk of developing allergic rhinitis, and the correlation is implicated in inflammation and air pollution.11, 12, 13 However, these results are all obtained by observational studies and are susceptible to confounding factors and causal inversions. Therefore, a final conclusion still cannot be drawn.
Mendelian randomization (MR) studies are a form of genetics-based epidemiological research that allows for causal inference at the genetic level by replacing the traits under study with a series of genetic variants.14, 15 Such studies can effectively make up for the deficiencies of observational studies. However, there are still no published MR studies exploring the association between obesity and allergic rhinitis at present.
Taken together, this study proposed to adopt the MR approach to explore the impact of obesity on the risk of developing allergic rhinitis at the genetic level. Considering the close association of numerous peripheral lipids, lipoproteins, apolipoproteins and adipokines with obesity, this study also used this approach to explore the association of a range of lipids and adipokines with this allergic disease at the genetic level. The findings obtained from this study were expected to increase our knowledge and understanding of the pathogenesis of allergic rhinitis.
Methods
Summary data for exposures and outcome
Several obesity indicators, lipid indicators and adipokines were identified as the exposures in the study. The obesity indicators were Body Mass Index (BMI), Body Fat Percentage (BFP) and Waist-to-Hip Ratio (WHR). The lipid indicators were Triglycerides (TG), Total Cholesterol (TC), Low Density Lipoprotein (LDL) cholesterol, High Density Lipoprotein (HDL) cholesterol, non-HDL cholesterol, Lipoprotein A [Lp(a)], Apolipoprotein A-I (Apo A-I) and Apolipoprotein B (ApoB). The adipokines were Interleukin-6 (IL-6), Interleukin-8 (IL-8), Agouti-Related Protein (AGRP), Adipocyte Fatty Acid-binding Protein (A-FABP), leptin and resistin. In addition, allergic rhinitis was defined as the outcome of this study.
The summary data for BMI was obtained from a meta-analysis of Genome-Wide Association Studies (GWASs) for height and BMI in more than 700,000 individuals of European ancestry.16 The summary data for BFP was collected from a GWAS of the MRC Integrative Epidemiology Unit at the university of bristol (IEU) involving more than 450,000 individuals of European ancestry. The summary data for WHR was collected from a GWAS of the within family consortium including 85,978 individuals of European ancestry. The above summary data for BFP and WHR were downloaded from the IEU open GWAS project (https://gwas.mrcieu.ac.uk/), and the GWAS IDs were ukb-b-8909 and ieu-b-4830.
The summary data for TG, TC, LDL cholesterol, HDL cholesterol, non-HDL cholesterol were obtained from a series of lipid GWASs involving more than 1,300,000 individuals of European ancestry.17 The summary data for Lp(a) was collected from a GWAS of Neale lab using 273,896 individuals of European ancestry, and was also downloaded from the IEU open GWAS project with its GWAS ID of ukb-d-30790_raw. The summary data for Apo A-I and ApoB were obtained from a multivariable MR analysis evaluating the relationship between circulating apolipoproteins with risk of coronary heart disease using 393,193–439,214 individuals of European ancestry.18 The summary data for IL-6, IL-8, AGRP, A-FABP, leptin and resistin were collected from a GWAS for genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 European individuals.19
The summary data for allergic rhinitis was obtained from a FinnGen study including 340,880 Finnish individuals.20 These subjects were identified by reviewing the ICD-10 codes in the hospitalization records, and the ICD-10 codes for allergic rhinitis were J30.10, J30.19, J30.2, J30.3 and J30.4.
The detailed characteristics of these traits were listed in Table 1.
Table 1.
Characteristics of the summary data in the study.
| Traits (outcome/exposures) | Sources | Years | Population | Sex | Sample size | SNPsa | F statistic |
|---|---|---|---|---|---|---|---|
| Outcome | |||||||
| Allergic rhinitis | FinnGen | 2022 | European | Male and female | 340,880 | ‒ | ‒ |
| Obesity indicators | |||||||
| Body mass index | PMID: 30124842 | 2018 | European | Male and female | 681,275 | 464 | 31.076 |
| Body fat percentage | MRC-IEU | 2018 | European | Male and female | 454,633 | 337 | 25.671 |
| Waist-hip ratio | Within family consortium | 2022 | European | Male and female | 85,978 | 16 | 17.360 |
| Lipid indicators | |||||||
| Triglycerides | PMID: 34887591 | 2021 | European | Male and female | 1,320,016 | 363 | 55.714 |
| Total cholesterol | PMID: 34887591 | 2021 | European | Male and female | 1,320,016 | 333 | 77.232 |
| LDL cholesterol | PMID: 34887591 | 2021 | European | Male and female | 1,320,016 | 297 | 66.435 |
| HDL cholesterol | PMID: 34887591 | 2021 | European | Male and female | 1,320,016 | 412 | 60.039 |
| non-HDL cholesterol | PMID: 34887591 | 2021 | European | Male and female | 1,320,016 | 274 | 62.811 |
| Lipoprotein A | Neale lab | 2018 | European | Male and female | 273,896 | 16 | 2350.190 |
| Apolipoprotein A-I | PMID: 32203549 | 2020 | European | Male and female | 393,193 | 251 | 57.110 |
| Apolipoprotein B | PMID: 32203549 | 2020 | European | Male and female | 439,214 | 149 | 58.843 |
| Adipokines | |||||||
| Interleukin-6 | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 15 | 12.016 |
| Interleukin-8 | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 5 | 10.001 |
| Agouti-related protein | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 9 | 10.360 |
| Fatty acid-binding protein | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 10 | 10.158 |
| Leptin | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 14 | 10.017 |
| Resistin | PMID: 33067605 | 2020 | European | Male and female | 21,758 | 41 | 10.003 |
MRC-IEU, MRC Integrative Epidemiology Unit at the University of Bristol (IEU); LDL, Low-Density Lipoprotein; HDL, High-Density Lipoprotein.
The number of instrumental variables that satisfied the three assumptions of Mendelian randomization.
Instrumental variables
Single Nucleotide Polymorphism (SNP) is a DNA sequence polymorphism caused by a variation in a single nucleotide at the genomic level.21 It is the most common type of human heritable variation, accounting for over 90% of all polymorphisms. In the present study, suitable SNPs were used as instrumental variables, which was extracted from the above summary data according to the three main assumptions of MR.22
To meet the correlation assumption, all included SNPs must have a genome-wide correlation p-value of less than 5 × 10−8 (for obesity and lipid indicators) or 5 × 10−6 (for adipokines), and their F-statistics must be greater than 10. To satisfy the independence assumption, all SNPs with Linkage Disequilibrium (LD) were directly excluded using a clumping window of 10 MB and an r2 cutoff of 0.001. To satisfy the exclusivity assumption, all SNPs that were significantly associated with confounders or outcomes were directly excluded by phenoscanner (http://www.phenoscanner.medschl.cam.ac.uk/), and the potential confounders included nasal polyp, bronchial asthma, otitis media, sinusitis, allergic strep throat and epistaxis.1
Mendelian randomization analyses
This was a two-sample MR that adopted three main methods for causal inference, namely random-effect Inverse Variance Weighted (IVW), weighted median and MR-Egger.23, 24 Briefly, the IVW is by far the most accurate method for causal inference, when all SNPs are unaffected by horizontal pleiotropy. The other two methods are not as accurate but have wider applicability conditions. Of these, weighted median accepts that about 50% of SNPs are horizontally pleiotropic, while MR-Egger can provide results when all SNPs are affected by horizontal pleiotropy. The results of these two methods can be used as a complement to the IVW results. In the present study, all three methods reported Odds Ratios (ORs), 95% Confidence Intervals (95% CIs) and p-values. And correlations can be considered nominally significant when the p values for the IVW were less than 0.05 and the direction of the results obtained by the other two methods were the same as the direction of the IVW results. Due to the multiple comparison design (17 exposures and 1 outcome), Bonferroni-corrected p-values less than 0.003 (0.05/17) indicated statistical significance. In addition, scatter plots and forest plots were made to visualize the above results.
A series of sensitivity analyses were also performed.25 Heterogeneity among SNPs was assessed using Cochran’s Q test. Horizontal pleiotropy among SNPs was measured by MR-Egger intercept test and MR-PRESSO test. All above sensitivity analyses reported p-values, and the p-values less than 0.05 indicated heterogeneity or horizontal pleiotropy. Horizontal pleiotropy was also determined by funnel plot and leave-one-out test. If the funnel plot was relatively symmetrical, it suggested that there was no significant horizontal pleiotropy. And the leave-one-out test can eliminate each SNP in turn and observe the effect of each SNP on the pooled results.
All MR analyses were done using TwoSampleMR in R software (version 4.2.3).
Results
Sources of summary data in the study
In Table 1, there were seventeen exposures and one outcome in the study. Their summary data were all from European populations, containing subjects from both genders. The sample sizes were between 21,758 and 1,320,016. And a total of 5–464 suitable SNPs were selected for this study from the summary data of the seventeen exposures according to the three assumptions of MR described above, and they had F-statistics between 10.001 and 2350.190.
Effect of obesity and lipids on the risk of allergic rhinitis
In Table 2, the IVW reported that BMI, BFP and WHR were not associated with the risk of allergic rhinitis (OR = 1.042, 95% CI 0.935–1.162, p = 0.452; OR = 1.053, 95% CI 0.901–1.230, p = 0.517; OR = 0.061, 95% CI 0.001–3.781, p = 0.184). The weighted median and MR-Egger supported these results obtained from the IVW (p > 0.05).
Table 2.
Mendelian randomization estimates for obesity affecting on the risk of allergic rhinitis.
| Exposures | MR methods | Nº of SNPsa | p for MR | OR (95% CI) | p for Cochran’s Q | p for MR-Egger intercept | p for MR-PRESSO |
|---|---|---|---|---|---|---|---|
| Body Mass Index | |||||||
| IVW | 412 | 0.452 | 1.042 (0.935–1.162) | 0.082 | 0.937 | 0.086 | |
| Weighted median | 412 | 0.660 | 1.036 (0.885–1.213) | ||||
| MR-Egger | 412 | 0.874 | 1.029 (0.726–1.457) | ||||
| Body fat percentage | |||||||
| IVW | 286 | 0.517 | 1.053 (0.901–1.230) | 0.069 | 0.755 | 0.073 | |
| Weighted median | 286 | 0.628 | 1.056 (0.848–1.314) | ||||
| MR-Egger | 286 | 0.635 | 1.150 (0.646–2.046) | ||||
| Waist-hip ratio | |||||||
| IVW | 14 | 0.184 | 0.061 (0.001–3.781) | 0.273 | 0.640 | 0.297 | |
| Weighted median | 14 | 0.585 | 0.216 (0.001–53.070) | ||||
| MR-Egger | 14 | 0.856 | 0.426 (0.001–3605.636) | ||||
SNP, Single Nucleotide Polymorphisms; MR, Mendelian randomization; OR, Odds Ratio; CI, Confidence Interval; IVW, Inverse-Variance Weighted.
Number of SNPs used for causal inference in Mendelian randomization.
In Table 3, the IVW also reported that TG, TC, LDL cholesterol, HDL cholesterol, non-HDL cholesterol, Lp(a), Apo A-I and ApoB were not related to the risk of the disease (OR = 1.045, 95% CI 0.935–1.167, p = 0.436; OR = 1.002, 95% CI = 0.906–1.107, p = 0.976; OR = 0.961, 95% CI 0.869–1.063, p = 0.441; OR = 0.955, 95% CI 0.873–1.046, p = 0.322; OR = 0.980, 95% CI 0.886–1.084, p = 0.696; OR = 1.000, 95% CI 0.999–1.001, p = 0.587; OR = 0.923, 95% CI 0.831–1.025, p = 0.136; OR = 1.016, 95% CI 0.922–1.120, p = 0.751). And the weighted median and MR-Egger also provided some similar results (p > 0.05).
Table 3.
Mendelian randomization estimates for blood lipids affecting on the risk of allergic rhinitis.
| Exposures | MR methods | Nº of SNPsa | p for MR | OR (95% CI) | p for Cochran’s Q | p for MR-Egger intercept | p for MR-PRESSO |
|---|---|---|---|---|---|---|---|
| Triglycerides | |||||||
| IVW | 306 | 0.436 | 1.045 (0.935–1.167) | 0.084 | 0.568 | 0.093 | |
| Weighted median | 306 | 0.773 | 1.027 (0.855–1.235) | ||||
| MR-Egger | 306 | 0.979 | 0.997 (0.822–1.211) | ||||
| Total cholesterol | |||||||
| IVW | 277 | 0.976 | 1.002 (0.906–1.107) | 0.081 | 0.109 | 0.090 | |
| Weighted median | 277 | 0.846 | 0.985 (0.847–1.145) | ||||
| MR-Egger | 277 | 0.206 | 0.898 (0.760–1.061) | ||||
| LDL cholesterol | |||||||
| IVW | 253 | 0.441 | 0.961 (0.869–1.063) | 0.088 | 0.539 | 0.097 | |
| Weighted median | 253 | 0.327 | 0.926 (0.795–1.079) | ||||
| MR-Egger | 253 | 0.336 | 0.925 (0.790–1.084) | ||||
| HDL cholesterol | |||||||
| IVW | 355 | 0.322 | 0.955 (0.873–1.046) | 0.061 | 0.137 | 0.061 | |
| Weighted median | 355 | 0.249 | 0.922 (0.803–1.059) | ||||
| MR-Egger | 355 | 0.077 | 0.883 (0.769–1.013) | ||||
| non-HDL cholesterol | |||||||
| IVW | 228 | 0.696 | 0.980 (0.886–1.084) | 0.073 | 0.956 | 0.085 | |
| Weighted median | 228 | 0.654 | 0.968 (0.840–1.115) | ||||
| MR-Egger | 228 | 0.851 | 0.984 (0.831–1.165) | ||||
| Lipoprotein A | |||||||
| IVW | 13 | 0.587 | 1.000 (0.999–1.001) | 0.914 | 0.892 | 0.947 | |
| Weighted median | 13 | 0.554 | 1.000 (0.999–1.001) | ||||
| MR-Egger | 13 | 0.617 | 1.000 (0.999–1.001) | ||||
| Apolipoprotein A-I | |||||||
| IVW | 189 | 0.136 | 0.923 (0.831–1.025) | 0.156 | 0.652 | 0.163 | |
| Weighted median | 189 | 0.452 | 0.940 (0.799–1.105) | ||||
| MR-Egger | 189 | 0.233 | 0.890 (0.735–1.077) | ||||
| Apolipoprotein B | |||||||
| IVW | 123 | 0.751 | 1.016 (0.922–1.120) | 0.071 | 0.928 | 0.079 | |
| Weighted median | 123 | 0.607 | 1.037 (0.904–1.188) | ||||
| MR-Egger | 123 | 0.882 | 1.011 (0.876–1.167) | ||||
SNP, Single Nucleotide Polymorphisms; MR, Mendelian randomization; OR, Odds Ratio; CI, Confidence Interval; IVW, Inverse-Variance Weighted; LDL, Low-Density Lipoprotein; HDL, High-Density Lipoprotein.
Number of SNPs used for causal inference in Mendelian randomization.
Effect of adipokines on the risk of allergic rhinitis
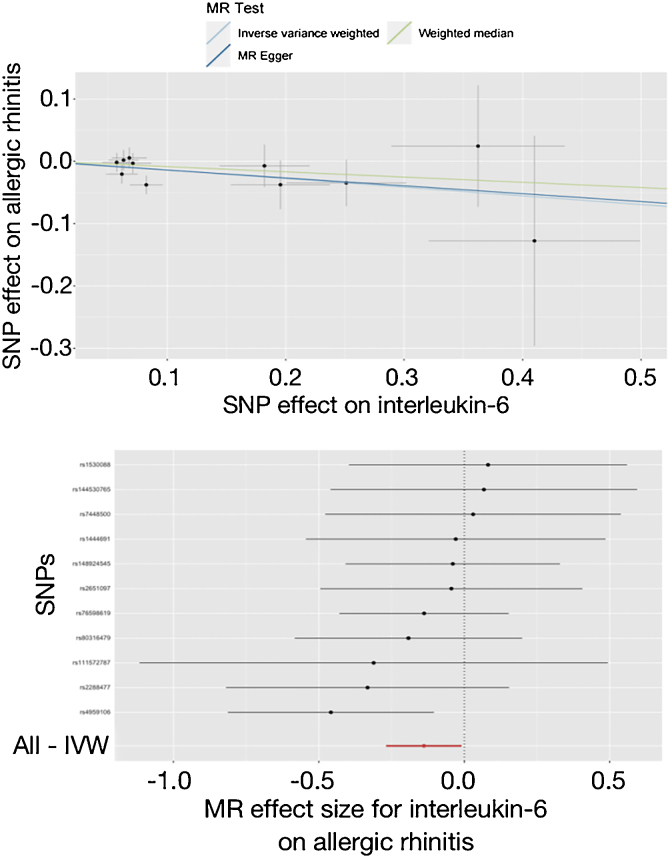
In Table 4, the IVW reported that elevated level of IL-6 was nominally associated with the decreased risk of allergic rhinitis (OR = 0.870, 95% CI 0.765–0.990, p = 0.035). Though the results of the weighted median and MR-Egger were not statistically significant (OR = 0.919, 95% CI 0.770–1.097, p = 0.351; OR = 0.881, 95% CI 0.679–1.143, p = 0.365), they were in the same direction as the IVW results. In Fig. 1, the scatter and forest plots visualized the above results. In Table 4, the Cochran’s Q did not find any heterogeneity (p = 0.777), and the MR-Egger intercept and MR-PRESSO did not report any horizontal pleiotropy or outliers (p = 0.918, p = 0.784). In Supplemental Fig. 12, the leave-one-out test also did not detect any significant outliers.
Table 4.
Mendelian randomization estimates for adipokines affecting on the risk of allergic rhinitis.
| Exposures | MR methods | Nº of SNPsa | p for MR | OR (95% CI) | p for Cochran’s Q | p for MR-Egger intercept | p for MR-PRESSO |
|---|---|---|---|---|---|---|---|
| Interleukin-6 | |||||||
| IVW | 11 | 0.035 | 0.870 (0.765–0.990) | 0.777 | 0.918 | 0.784 | |
| Weighted median | 11 | 0.351 | 0.919 (0.770–1.097) | ||||
| MR-Egger | 11 | 0.365 | 0.881 (0.679–1.143) | ||||
| Interleukin-8 | |||||||
| IVW | 4 | 0.280 | 1.146 (0.895–1.467) | 0.954 | 0.931 | 0.951 | |
| Weighted median | 4 | 0.416 | 1.124 (0.848–1.489) | ||||
| MR-Egger | 4 | 0.928 | 1.073 (0.278–4.144) | ||||
| Agouti-related protein | |||||||
| IVW | 7 | 0.172 | 0.842 (0.658–1.078) | 0.183 | 0.597 | 0.211 | |
| Weighted median | 7 | 0.646 | 0.935 (0.704–1.244) | ||||
| MR-Egger | 7 | 0.657 | 2.442 (0.060–100.176) | ||||
| Fatty acid-binding protein | |||||||
| IVW | 4 | 0.032 | 0.732 (0.551–0.973) | 0.398 | 0.581 | 0.466 | |
| Weighted median | 4 | 0.093 | 0.737 (0.517–1.052) | ||||
| MR-Egger | 4 | 0.427 | 0.411 (0.070–2.402) | ||||
| Leptin | |||||||
| IVW | 34 | 0.773 | 1.014 (0.923–1.113) | 0.150 | 0.857 | 0.143 | |
| Weighted median | 34 | 0.472 | 1.049 (0.921–1.194) | ||||
| MR-Egger | 34 | 0.987 | 0.998 (0.824–1.209) | ||||
| Resistin | |||||||
| IVW | 12 | 0.283 | 1.122 (0.909–1.384) | 0.064 | 0.353 | 0.068 | |
| Weighted median | 12 | 0.792 | 1.032 (0.814–1.309) | ||||
| MR-Egger | 12 | 0.295 | 2.495 (0.493–12.625) | ||||
SNP, Single Nucleotide Polymorphisms; MR, Mendelian randomization; OR, Odds Ratio; CI, Confidence Interval; IVW, Inverse-Variance Weighted.
Number of SNPs used for causal inference in Mendelian randomization.
Figure 1.
Effect of interleukin-6 on allergic rhinitis in the study. SNP, Single Nucleotide Polymorphisms; MR, Mendelian randomization; IVW, Inverse-Variance Weighted.
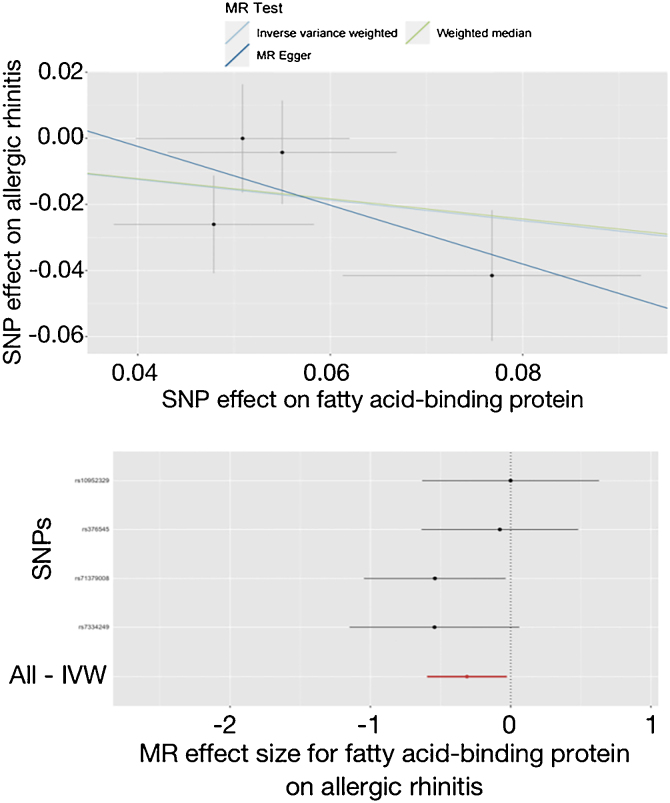
In Table 4, the IVW reported that higher level of A-FABP was nominally associated with the lower risk of allergic rhinitis (OR = 0.732, 95% CI 0.551–0.973, p = 0.032). The weighted median and MR-Egger results were in the same direction as the IVW direction (OR = 0.737, 95% CI 0.517–1.052, p = 0.093; OR = 0.411, 95% CI 0.070–2.402, p = 0.427). These results were visualized in Fig. 2. The Cochran’s Q, MR-Egger intercept and MR-PRESSO did not report any heterogeneity or horizontal pleiotropy in Table 4 (p = 0.398, p = 0.581, p = 0.466). The leave-one-out test did not find any outliers in Supplemental Fig. 15.
Figure 2.
Effect of adipocyte fatty acid-binding protein on allergic rhinitis in the study. SNP, Single Nucleotide Polymorphisms; MR, Mendelian randomization; IVW, Inverse-Variance Weighted.
In addition, the IVW did not detect any association of the other adipokines with the risk of allergic rhinitis (p > 0.05).
Discussion
As an allergic and inflammatory disease, the etiology and pathogenesis of allergic rhinitis are still unknown. However, genetic background is supposed to be one of the main pathogenic mechanisms. Thus, we investigated the genetic effects of obesity, lipids and adipokines on the risk of the disease using MR methods in the study, which contributed to our further understanding of the risk factors and pathogenesis of allergic rhinitis, and helped us to find suitable therapeutic targets and improve therapeutic strategies so as to reduce the incidence of this disease and improve its prognosis.
According to the IVW results in this study, although all obesity and lipid indicators cannot influence the risk of developing allergic rhinitis, the levels of two adipokines were potentially correlated with the risk of the disease. Briefly, for each SD increase in peripheral levels of IL-6 and A-FABP, the risk of developing allergic rhinitis decreased by approximately 10%–30%. Because the sensitivity analyses did not detect horizontal pleiotropy, the IVW results mentioned above were considered to be the most reliable. Moreover, the results obtained by the other two methods were directionally consistent with the IVW results, although none of them were statistically significant. Thus, MR can preliminarily confirmed the effect of IL-6 and A-FABP on the risk of allergic rhinitis. However, it should be noted that the IVW p-values for these two adipokines were only slightly lower than 0.05. More importantly, there was a design of multiple comparisons in this study. Therefore, we believed that these analyses had the potential for type I error, and therefore the Bonferroni correction was adopted. After the correction, the results showed that these two adipokines were only nominally associated with the risk of allergic rhinitis. These results were not sufficient to draw a conclusion, but they were certainly helpful for future research.
As mentioned above, several observational studies had reported that obesity contributed to the development of allergic rhinitis.11, 12, 13 But, the results of the present study at the genetic level did not support these findings. Considering that the pathogenesis of this allergic disease was complex, it certainly included multiple genetic and environmental factors. So, it was still possible that obesity may have an impact on the risk of developing allergic rhinitis through phenotypic-level mechanisms.
IL-6 was a classical inflammatory factor that can be secreted by several types of cells. In the peripheral circulation, approximately 1/3 of this factor was derived from adipose tissue and therefore IL-6 was also considered as an adipokine with inflammatory regulatory functions. Two previous observational studies found that there were rs1800795 (G/C at −174) and rs1800796 (G/A at −597) polymorphisms in IL-6 and that rs1800795 was associated with allergic rhinitis risk.26, 27 Of them, the study from China reported that individuals expressing the C allele of rs1800795 were at higher risk of developing this allergic disease, whereas the other study from Middle Asia concluded that individuals expressing the G allele were more susceptible to allergic rhinitis.26, 27 These contradictory results may be explained by the different study populations. Meanwhile, the present study included a European population and reported that peripheral IL-6 levels may nominally reduce the risk of this allergic disease, which did not consider genetic polymorphisms due to insufficient data. So, both the present MR study and the two observational studies mentioned above suggested that IL-6 had the potential to affect the development of allergic rhinitis, and the mechanism might be influenced by race and genetic polymorphism.
FABP were fatty acid carriers in a wide range of cells (including adipocytes) and played an important role in the physiological use of fatty acids in these cells. A previous animal study using mice explained the role of this proteins in allergic lung inflammation.28 Briefly, knockdown of FABP in the lungs can cause severe allergic lung inflammation, and up-regulation of this proteins suppressed allergy-associated immune cell activity. In addition, a high-fat diet may result in down-regulation of FABP expression in the lungs. The present study investigated the correlation between peripheral A-FABP levels and allergic rhinitis, and the obtained results appeared to be partially consistent with this animal study.
The main advantage of this study is the MR design. Compared to observational studies, this study allowed causal inferences to be made at the genetic level between the exposures and the outcome, avoiding many confounding factors and the appearance of causal inversions. So, the results obtained from this study were more reliable. Meanwhile, there were still some limitations in this study. For example, IL-6 was polymorphic, and it was likely that this polymorphism had a significant impact on allergic rhinitis. Also, closely related to this issue was the speculation that the effect of IL-6 on allergic rhinitis may be ethnospecific. In addition, the number of instrumental variables for several exposures (i.e., adipokines) might be somewhat inadequate due to the lack of sufficient summary data. All of these questions remained to be explored in future studies.
Conclusion
The present study provided some interesting, but not sufficient, evidence to suggest that IL-6 and A-FABP might play a protective role in the development of allergic rhinitis at the genetic level. However, it was important to emphasize that this was not a final conclusion, and the above findings should be validated by more research. In addition, the study did not find a genetic association of obesity or peripheral lipid levels with this allergic disease. These studies helped us to objectively validate previous observational studies and deepen our understanding of the risk factors and pathogenesis of allergic rhinitis.
Funding
Not applicable.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
We want to acknowledge the participants and investigators of the FinnGen study.
Footnotes
Peer Review under the responsibility of Associação Brasileira de Otorrinolaringologia e Cirurgia Cérvico-Facial.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.bjorl.2023.101306.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Bousquet J., Anto J.M., Bachert C., Baiardini I., Bosnic-Anticevich S., Walter Canonica G., et al. Allergic rhinitis. Nat Rev Dis Primers. 2020;6:95. doi: 10.1038/s41572-020-00227-0. [DOI] [PubMed] [Google Scholar]
- 2.Pang K., Li G., Li M., Zhang L., Fu Q., Liu K., et al. Prevalence and risk factors for allergic rhinitis in China: a systematic review and meta-analysis. Evid Based Complement Alternat Med. 2022;2022 doi: 10.1155/2022/7165627. 7165627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinart M., Keller T., Reich A., Fröhlich M., Cabieses B., Hohmann C., et al. Sex-related allergic rhinitis prevalence switch from childhood to adulthood: a systematic review and meta-analysis. Int Arch Allergy Immunol. 2017;172:224–235. doi: 10.1159/000464324. [DOI] [PubMed] [Google Scholar]
- 4.Ozoh O.B., Aderibigbe S.A., Ayuk A.C., Dede S.K., Egbagbe E., Babashani M. Health-related quality of life in asthma measured by the World Health Organization brief questionnaire (WHO-BREF) and the effect of concomitant allergic rhinitis—a population-based study. Clin Respir J. 2023 doi: 10.1111/crj.13608. [in press] [DOI] [PubMed] [Google Scholar]
- 5.Speth M.M., Hoehle L.P., Phillips K.M., Caradonna D.S., Gray S.T., Sedaghat A.R. Treatment history and association between allergic rhinitis symptoms and quality of life. Ir J Med Sci. 2019;188:703–710. doi: 10.1007/s11845-018-1866-2. [DOI] [PubMed] [Google Scholar]
- 6.Amritwar A.U., Lowry C.A., Brenner L.A., Hoisington A.J., Hamilton R., Stiller J.W., et al. Mental health in allergic rhinitis: depression and suicidal behavior. Curr Treat Options Allergy. 2017;4:71–97. doi: 10.1007/s40521-017-0110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiqui Z.A., Walker A., Pirwani M.M., Tahiri M., Syed I. Allergic rhinitis: diagnosis and management. Br J Hosp Med (Lond) 2022;83:1–9. doi: 10.12968/hmed.2021.0570. [DOI] [PubMed] [Google Scholar]
- 8.Reyes-Angel J., Kaviany P., Rastogi D., Forno E. Obesity-related asthma in children and adolescents. Lancet Child Adolesc Health. 2022;6:713–724. doi: 10.1016/S2352-4642(22)00185-7. [DOI] [PubMed] [Google Scholar]
- 9.Dixon A.E., Que L.G. Obesity and asthma. Semin Respir Crit Care Med. 2022;43:662–674. doi: 10.1055/s-0042-1742384. [DOI] [PubMed] [Google Scholar]
- 10.Chen M., Ge Y., Lin W., Ying H., Zhang W., Yu X., et al. Clinical features and nasal inflammation in asthma and allergic rhinitis. Clin Exp Immunol. 2022;208:25–32. doi: 10.1093/cei/uxac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Luo F., Han Y., Lou H., Tang X., Zhang L. Obesity/overweight and risk of allergic rhinitis: a meta-analysis of observational studies. Allergy. 2020;75:1272–1275. doi: 10.1111/all.14143. [DOI] [PubMed] [Google Scholar]
- 12.Han M.W., Kim S.H., Oh I., Kim Y.H., Lee J. Obesity can contribute to severe persistent allergic rhinitis in children through leptin and interleukin-1β. Int Arch Allergy Immunol. 2021;182:546–552. doi: 10.1159/000512920. [DOI] [PubMed] [Google Scholar]
- 13.Li R.L., Wu C.T., Chen S.M., Lue K.H., Lee S.S., Tsao C.Y., et al. Allergic rhinitis children with obesity are more vulnerable to air pollution: a cross sectional study. Sci Rep. 2023;13:3658. doi: 10.1038/s41598-023-30388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emdin C.A., Khera A.V., Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 15.Thanassoulis G., O’Donnell C.J. Mendelian randomization: nature’s randomized trial in the post-genome era. JAMA. 2009;301:2386–2388. doi: 10.1001/jama.2009.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham S.E., Clarke S.L., Wu K.H., Kanoni S., Zajac G.J.M., Ramdas S., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson T.G., Sanderson E., Palmer T.M., Ala-Korpela M., Ference B.A., Davey Smith G., et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 2020;17:e1003062. doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folkersen L., Gustafsson S., Wang Q., Hansen D.H., Hedman ÅK., Schork A., et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2:1135–1148. doi: 10.1038/s42255-020-00287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurki M.I., Karjalainen J., Palta P., Sipilä T.P., Kristiansson K., Donner K., et al. FinnGen: unique genetic insights from combining isolated population and national health register data. medRxiv. 2023 doi: 10.1101/2022.03.03.22271360. [Published online 3 March 2022] [DOI] [Google Scholar]
- 21.Tang M., Wang T., Zhang X. A review of SNP heritability estimation methods. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbac067. bbac067. [DOI] [PubMed] [Google Scholar]
- 22.Burgess S., Small D.S., Thompson S.G. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu S., Wang P., Fung W.K., Liu Z. A novel penalized inverse-variance weighted estimator for Mendelian randomization with applications to COVID-19 outcomes. Biometrics. 2022 doi: 10.1111/biom.13732. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yavorska O.O., Burgess S. Mendelian randomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S., Bowden J., Fall T., Ingelsson E., Thompson S.G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao N., Liu H.J., Sun Y.Y., Li Y.Z. Role of interleukin-6 polymorphisms in the development of allergic rhinitis. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15016987. [DOI] [PubMed] [Google Scholar]
- 27.Nasiri R., Movahedi M., Amirzargar A.A., Hirbod-Mobarakeh A., Farhadi E., Ansaripour B., et al. Association of interleukin 6 single nucleotide polymorphisms with allergic rhinitis. Int J Pediatr Otorhinolaryngol. 2014;78:1426–1429. doi: 10.1016/j.ijporl.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi S., Tayama S., Phung H.T., Kagawa Y., Miyazaki H., Takahashi Y., et al. Fatty acid-binding protein 5 limits ILC2-mediated allergic lung inflammation in a murine asthma model. Sci Rep. 2020;10:16617. doi: 10.1038/s41598-020-73935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.