Abstract
P-glycoprotein (P-gp) is known as the "multidrug resistance protein" because it contributes to tumor resistance to several different classes of anticancer drugs. The effectiveness of such polymers in treating cancer and delivering drugs has been shown in a wide range of in vitro and in vivo experiments. The primary objective of the present study was to investigate the inhibitory effects of several naturally occurring polymers on P-gp efflux, as it is known that P-gp inhibition can impede the elimination of medications. The objective of our study is to identify polymers that possess the potential to inhibit P-gp, a protein involved in drug resistance, with the aim of enhancing the effectiveness of anticancer drug formulations. The ADMET profile of all the selected polymers (Agarose, Alginate, Carrageenan, Cyclodextrin, Dextran, Hyaluronic acid, and Polysialic acid) has been studied, and binding affinities were investigated through a computational approach using the recently released crystal structure of P-gp with PDB ID: 7O9W. The advanced computational study was also done with the help of molecular dynamics simulation. The aim of the present study is to overcome MDR resulting from the activity of P-gp by using such polymers that can inhibit P-gp when used in formulations. The docking scores of native ligand, Agarose, Alginate, Carrageenan, Chitosan, Cyclodextrin, Dextran, Hyaluronic acid, and Polysialic acid were found to be −10.7, −8.5, −6.6, −8.7, −8.6, −24.5, −6.7, −8.3, and −7.9, respectively. It was observed that, Cyclodextrin possess multiple properties in drug delivery science and here also demonstrated excellent binding affinity. We propose that drug efflux-related MDR may be prevented by the use of Agarose, Carregeenan, Chitosan, Cyclodextrin, Hyaluronic acid, and/or Polysialic acid in the administration of anticancer drugs.
Keywords: Toxicity, Molecular docking, P-glycoprotein, In silico-ADMET, Natural polymers, MDR, Molecular dynamic simulation
Graphical abstract
1. Introduction
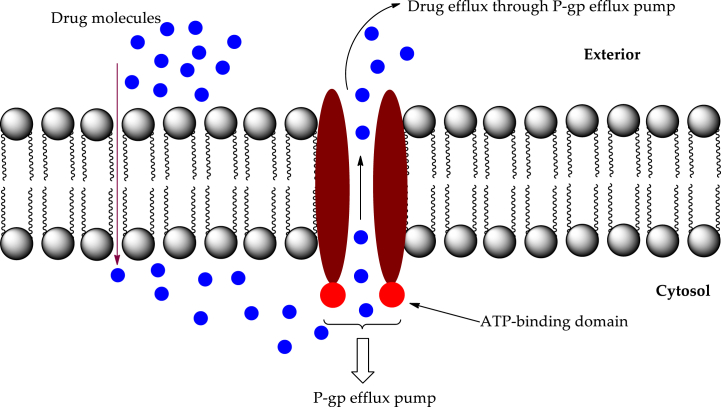
Not just in cancer but also in other illnesses, multidrug resistance (MDR) is the leading cause of treatment failure [1]. P-glycoprotein (P-gp) is an ATP-binding cassette (ABC) transporter that helps keep cells free of harmful substances, including toxins and xenobiotics. Among the many drug transporters, P-gp plays a role in the absorption and excretion of many different medicines. The concentrations of these compounds in the blood and different tissues, and consequently their effects, are controlled by this process [2,3]. P-gp operates as a transmembrane efflux pump, pushing its substrates from within the cell to outside the cell. In humans, P-gp is encoded by a small gene family that consists of two isoforms. Class I (MDR1/ABCB1) isoforms function as drug transporters, whereas class II (MDR2/3/ABCB4) isoforms are responsible for phosphatidylcholine export into the bile [4]. Drugs with molecular weights as low as 250 gm/mol (cimetidine) and as high as 1202 gm/mol (cyclosporin) may all be recognized and transported by a single P-gp molecule. P-gp was first discovered in various cancers [5,6]. The overexpression of P-gp in these cells impeded their ability to take up cytotoxic medicines. P-gp gained the name "multidrug resistance protein" because it increased tumour resistance to several types of anticancer medication [[7], [8], [9]]. The schematic presentation of drug efflux by P-gp is demonstrated in Fig. 1.
Fig. 1.
The mechanism of drug import and efflux through P-gp efflux pump.
To combat MDR in cancer cells, researchers are developing non-substrate medicinal compounds or formulations that allow the drug to sidestep the efflux pumps' trafficking [5,10,11]. Researchers have shown that co-administering efflux pump inhibitors with an efflux pump substrate improves bioavailability from oral dosing. By combining oral drug delivery with cancer therapy, the local treatment of gastro-intestinal carcinoma may be another potential application of efflux pump inhibitors. Eflux pump inhibitors may be broken down into two primary groups: small molecule inhibitors (SMIs) and polymeric inhibitors (PIs). First-generation SMIs are pharmacologically active drugs used for various therapeutic reasons and discovered to also block efflux pumps. Examples of such medications include quinine and verapamil. To prevent further pharmaceutical interactions, the second and third generations of SMIs, such as PSC833, GF120918, and KR30031, were created to specifically target and block efflux pumps [5,12,13]. Unfortunately, SMI-mediated toxicity, accumulation, or anti-targeting is a typical concern associated with the co-administration of SMIs.
The broad specificity of P-gp has been extensively investigated in various studies aimed at inhibiting the protein and thereby enhancing the effectiveness of anticancer drugs. The overarching approach employed thus far entails the development of compounds that possess the ability to either compete with anticancer drugs for transport or function as direct inhibitors of P-gp. Despite the significant progress achieved in vitro, the clinical setting lacks any existing compounds capable of effectively inhibiting P-gp-mediated resistance. The failure of the intervention can potentially be ascribed to the presence of toxicity, adverse drug interaction, and various pharmacokinetic challenges [14].
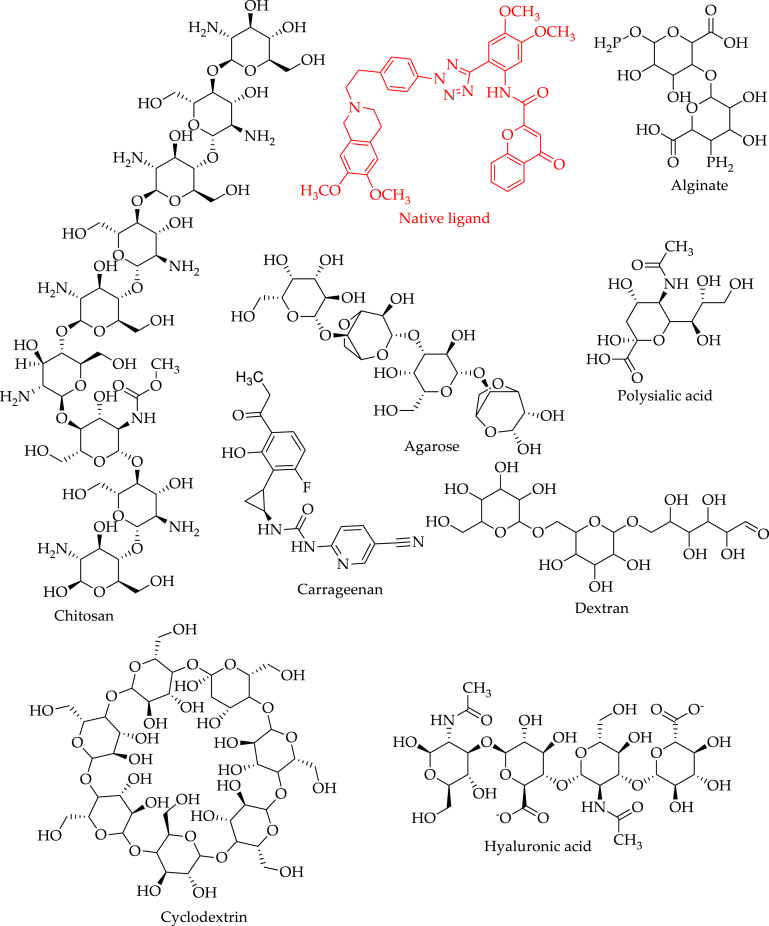
Pharmaceutical polymers are an integral part of drug delivery systems, whether it is an anticancer drug or any other medication. Some commonly used polymeric pharmaceutical excipients, such as Tween® 80 and pluronic® P85, have been shown to block efflux pumps in recent years, despite being thought to be completely safe and pharmacologically inactive [15]. Numerous in vitro and in vivo experiments have shown the usefulness of such polymers in cancer treatment and drug delivery [16,17]. In terms of the origin of the polymer, one may make a distinction between natural and synthetic polymers. The ADMET profile of all the selected natural polymers has been studied, and binding affinities were investigated through a computational approach using the recently released crystal structure of P-gp with PDB ID: 7O9W. In order to provide more reliability, the molecular dynamic simulation was also carried out. As far as we are aware, no article has reported the computational analysis of natural polymers on P-gp to date; this could be because the crystal structure of P-gp was not previously released until April 17th, 2021. Recently, Sameer Urgaonkar et al. deposited the crystal structure of P-gp, which we have used in the present study [18]. The structures of natural polymers and native ligand present in the crystal protein are illustrated in Fig. 2. In present investigation, we aimed to recommend natural polymers with P-gp inhibition potential that can improve the efficacy of anticancer drug formulations.
Fig. 2.
The structures of native ligand present in the crystal structure of P-gp enzyme and natural polymers.
2. Results
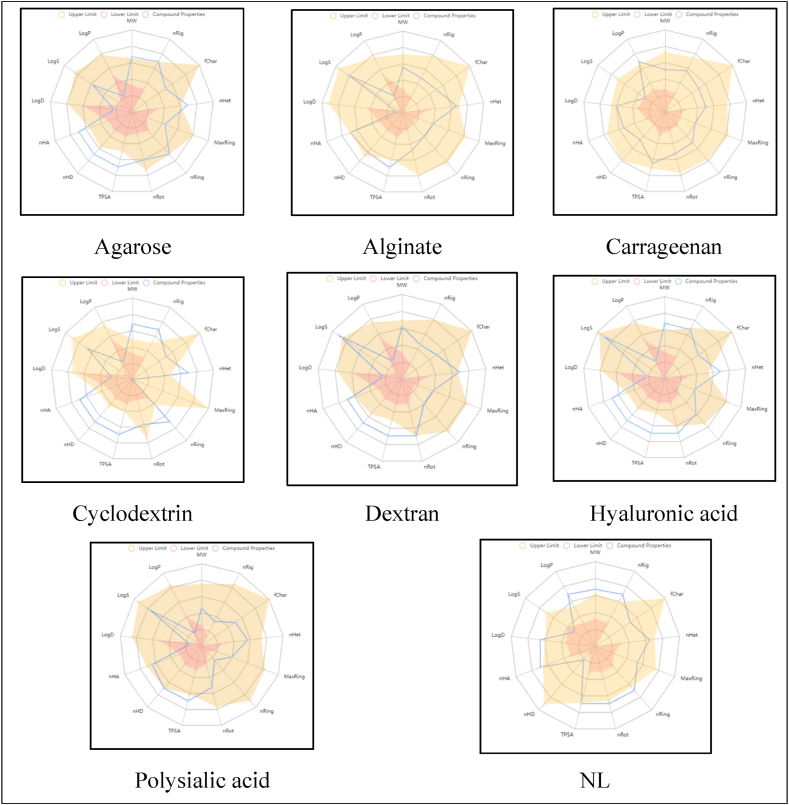
The physicochemical properties of molecules are tabulated in Table 1. Table 2 displays the results of several parameter calculations. They include the QED, NPscore, Lipinski Rule, Pfizer Rule, GSK Rule, Golden Triangle Rule, and Chelator Rule. The bioavailability of natural polymers and native ligand are shown in Table 3, along with absorption metrics including Caco-2 and MDCK permeability, P-gp-inhibitor and substrate ability, human intestinal absorption (HIA), and F20% and F30% bioavailability. The distribution and metabolism profiles are depicted in Table 4. Table 5 summarizes the excretion and toxicity profiles. The physicochemical radar obtained from the ADMETlab 2.0 online server is depicted in Fig. 3. An environmental toxicity profile (bioconcentration factors: IGC50, LC50FM, and LC50DM) is demonstrated in Table 6. The 3D-ribbon view of molecules superimposed in the allosteric site of P-gp is depicted in Fig. 4. The 2D-docking poses of molecules docked with P-gp are illustrated in Fig. 5. The RMSD results of the native ligand and carrageenan complex system with 7O9W are given in Fig. 6, Fig. 7.
Table 1.
Physicochemical properties of natural polymers and native ligand.
| Code | Physicochemical Property |
|||||||
|---|---|---|---|---|---|---|---|---|
| Molecular Weight | Volume | nHA | nHD | nRot | TPSA | logS | logP | |
| NL | 688.26 | 688.601 | 13 | 1 | 12 | 143.07 | −5.54 | 5.16 |
| Agarose | 630.2 | 539.336 | 19 | 10 | 8 | 285.37 | −1.238 | −2.806 |
| Alginate | 418.04 | 338.86 | 12 | 6 | 5 | 192.44 | −0.267 | −3.47 |
| Carrageenan | 368.13 | 361.571 | 7 | 3 | 6 | 118.34 | −4.193 | 3.294 |
| Chitosan | 1525.64 | 1339.277 | 48 | 37 | 28 | 808 | −0.614 | −7.142 |
| Cyclodextrin | 1134.37 | 974.194 | 35 | 21 | 7 | 554.05 | −0.777 | −4.572 |
| Dextran | 504.17 | 440.778 | 16 | 11 | 11 | 276.52 | 0.724 | −4.089 |
| Hyaluronic acid | 776.23 | 672.241 | 25 | 14 | 14 | 399.71 | 0.133 | −3.602 |
| Polysialic acid | 309.11 | 275.092 | 10 | 7 | 6 | 176.78 | −0.401 | −1.054 |
Table 2.
Drug-likeness properties of natural polymers and native ligand.
| Code | Medicinal Chemistry |
||||||
|---|---|---|---|---|---|---|---|
| QED | NPscore | Lipinski Rule | Pfizer Rule | GSK Rule | Golden Triangle | Chelator Rule | |
| NL | 0.189 | −0.942 | Rejected | Accepted | Rejected | Rejected | 0 |
| Agarose | 0.119 | 1.451 | Rejected | Accepted | Rejected | Rejected | 0 |
| Alginate | 0.242 | 1.075 | Rejected | Accepted | Rejected | Accepted | 0 |
| Carrageenan | 0.718 | −0.778 | Accepted | Accepted | Accepted | Accepted | 0 |
| Chitosan | 0.038 | 0.384 | Rejected | Accepted | Rejected | Rejected | 0 |
| Cyclodextrin | 0.113 | 0.65 | Rejected | Accepted | Rejected | Rejected | 0 |
| Dextran | 0.125 | 1.697 | Rejected | Accepted | Rejected | Rejected | 0 |
| Hyaluronic acid | 0.087 | 1.107 | Rejected | Accepted | Rejected | Rejected | 0 |
| Polysialic acid | 0.268 | 1.575 | Accepted | Accepted | Accepted | Accepted | 0 |
Table 3.
An absorption parameters of natural polymers and native ligand.
| Code | Absorption |
||||||
|---|---|---|---|---|---|---|---|
| Caco-2 Permeability | MDCK Permeability | Pgp-inhibitor | Pgp-substrate | HIA | F20% | F30% | |
| NL | −5.438 | 6.3e-05 | 1.0 | 0.019 | 0.053 | 0.031 | 0.046 |
| Agarose | −6.633 | 0.000301 | 0.001 | 0.985 | 1.0 | 0.252 | 1.0 |
| Alginate | −6.548 | 0.000498 | 0.0 | 0.817 | 0.993 | 0.655 | 0.999 |
| Carrageenan | −5.602 | 7e-06 | 0.005 | 0.029 | 0.007 | 0.001 | 0.009 |
| Chitosan | −6.417 | 0.005978 | 0 | 1 | 1 | 0.999 | 1 |
| Cyclodextrin | −6.641 | 0.006405 | 0.0 | 0.999 | 1.0 | 1.0 | 1.0 |
| Dextran | −6.202 | 0.000478 | 0.003 | 0.948 | 0.993 | 0.948 | 1.0 |
| Hyaluronic acid | −7.129 | 0.000548 | 0.0 | 0.998 | 1.0 | 0.996 | 1.0 |
| Polysialic acid | −6.581 | 0.000226 | 0.001 | 0.992 | 0.969 | 0.993 | 0.998 |
Table 4.
Distribution and metabolism profile of natural polymers and native ligand.
| Code | Distribution |
Metabolism |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPB | VD | BBB Penetration |
Fu (%) | CYP1A2 |
CYP2C19 |
CYP2C9 |
CYP2D6 |
CYP3A4 |
||||||
| inhibitor | substrate | inhibitor | substrate | inhibitor | substrate | inhibitor | substrate | inhibitor | substrate | |||||
| NL | 93.20 | 1.28 | 0.032 | 2.260 | 0.068 | 0.916 | 0.797 | 0.828 | 0.91 | 0.717 | 0.062 | 0.89 | 0.955 | 0.927 |
| Agarose | 14.59 | 0.044 | 0.355 | 53.72 | 0.0 | 0.007 | 0.011 | 0.05 | 0.0 | 0.005 | 0.005 | 0.064 | 0.004 | 0.001 |
| Alginate | 12.98 | 0.31 | 0.407 | 72.73 | 0.001 | 0.009 | 0.015 | 0.034 | 0.001 | 0.256 | 0.003 | 0.092 | 0.007 | 0.0 |
| Carrageenan | 94.64 | 0.357 | 0.374 | 4.151 | 0.234 | 0.543 | 0.443 | 0.073 | 0.76 | 0.914 | 0.417 | 0.736 | 0.42 | 0.294 |
| Chitosan | −7.84 | −0.872 | 0.813 | 67.23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.007 | 0.007 | 0 |
| Cyclodextrin | 7.042 | −1.086 | 0.792 | 40.21 | 0.0 | 0.0 | 0.001 | 0.034 | 0.0 | 0.0 | 0.0 | 0.015 | 0.001 | 0.0 |
| Dextran | 8.948 | 0.152 | 0.352 | 60.85 | 0.0 | 0.004 | 0.002 | 0.039 | 0.0 | 0.047 | 0.0 | 0.054 | 0.001 | 0.0 |
| Hyaluronic acid | 7.569 | 0.226 | 0.448 | 67.00 | 0.0 | 0.0 | 0.013 | 0.023 | 0.0 | 0.005 | 0.001 | 0.017 | 0.011 | 0.0 |
| Polysialic acid | 12.22 | 0.287 | 0.298 | 82.89 | 0.003 | 0.019 | 0.017 | 0.046 | 0.002 | 0.063 | 0.001 | 0.063 | 0.011 | 0.004 |
Table 5.
Excretion and toxicity profile of native ligand and natural polymers.
| Code | Excretion |
Toxicity |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL | T1/2 | H-HT | DILI | AMES toxicity | Rat oral acute toxicity |
FDAMDD | Skin sensitization | Carcinogenicity | Eye corrosion | Eye irritation | Respiratory toxicity | |
| NL | 6.982 | 0.078 | 0.546 | 0.988 | 0.209 | 0.755 | 0.902 | 0.042 | 0.24 | 0.003 | 0.007 | 0.903 |
| Agarose | 0.622 | 0.872 | 0.065 | 0.077 | 0.086 | 0.137 | 0.0 | 0.461 | 0.041 | 0.003 | 0.028 | 0.028 |
| Alginate | 1.579 | 0.752 | 0.127 | 0.892 | 0.03 | 0.401 | 0.001 | 0.019 | 0.01 | 0.003 | 0.006 | 0.024 |
| Carrageenan | 6.66 | 0.277 | 0.37 | 0.954 | 0.438 | 0.907 | 0.933 | 0.089 | 0.078 | 0.003 | 0.009 | 0.402 |
| Chitosan | −2.095 | 0.982 | 0.122 | 0.044 | 0.027 | 0 | 0 | 0.04 | 0 | 0.004 | 0.004 | 0.007 |
| Cyclodextrin | −0.191 | 0.905 | 0.124 | 0.765 | 0.038 | 0.015 | 0.0 | 0.009 | 0.001 | 0.003 | 0.004 | 0.002 |
| Dextran | 0.607 | 0.746 | 0.116 | 0.863 | 0.084 | 0.039 | 0.0 | 0.007 | 0.002 | 0.003 | 0.007 | 0.004 |
| Hyaluronic acid | 0.671 | 0.897 | 0.252 | 0.935 | 0.056 | 0.0 | 0.0 | 0.016 | 0.001 | 0.003 | 0.01 | 0.0 |
| Polysialic acid | 1.461 | 0.724 | 0.142 | 0.183 | 0.018 | 0.0 | 0.003 | 0.054 | 0.002 | 0.003 | 0.039 | 0.009 |
Fig. 3.
Physicochemical radar of native ligand and natural polymers obtained from ADMETlab 2.0 web server. Where; MW, molecular weight; nRig, number of rigid bonds; fChar, formal charge; nHet, number of heteroatoms; MaxRing, number of atoms in the biggest ring; nRing, number of rings; nRot, number of rotatable bonds; TPSA, topological polar surface area; nHD, number of hydrgen bond donors; nHA, number of hydrogen bond accaptors; LogD, logP at physiological pH 7.4; LogS, log of the aqueous solubility; LogP, log of the octanal/water partition co-efficient.
Table 6.
Environmental toxicity profile of native ligand and natural polymers.
| Code | Environmental toxicity |
|||
|---|---|---|---|---|
| Bioconcentration factors | IGC50 | LC50FM | LC50DM | |
| NL | 1.14 | 4.778 | 5.866 | 5.654 |
| Agarose | 0.285 | 2.406 | 1.549 | 2.904 |
| Alginate | 0.068 | 3.01 | 3.394 | 4.128 |
| Carrageenan | 0.458 | 3.545 | 4.753 | 6.817 |
| Chitosan | 0.199 | 2.078 | −0.209 | 4.974 |
| Cyclodextrin | 0.667 | 1.327 | −0.235 | 1.939 |
| Dextran | 0.073 | 1.713 | −0.217 | 2.992 |
| Hyaluronic acid | 0.001 | 0.86 | 1053 | −0.175 |
| Polysialic acid | −0.108 | 0.843 | 0.81 | 1.067 |
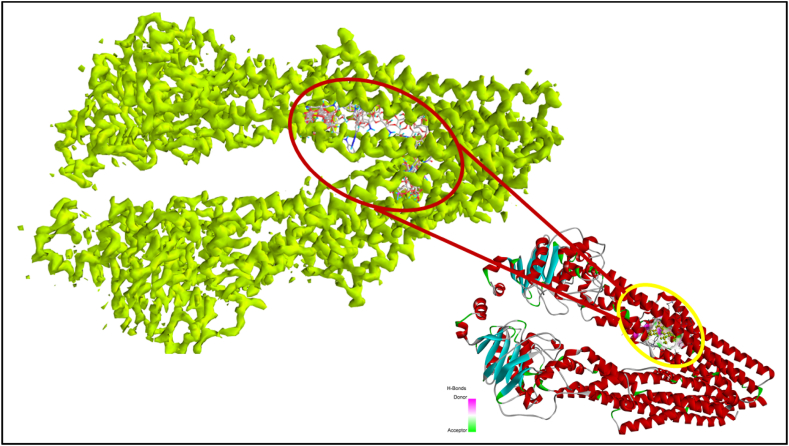
Fig. 4.
3D-ribbon view of molecules superimposed in the allosteric site of P-gp; Blue and red ribbon view displays the active cavity of native ligand whereas; light green view shows binding of natural polymers in the same cavity.
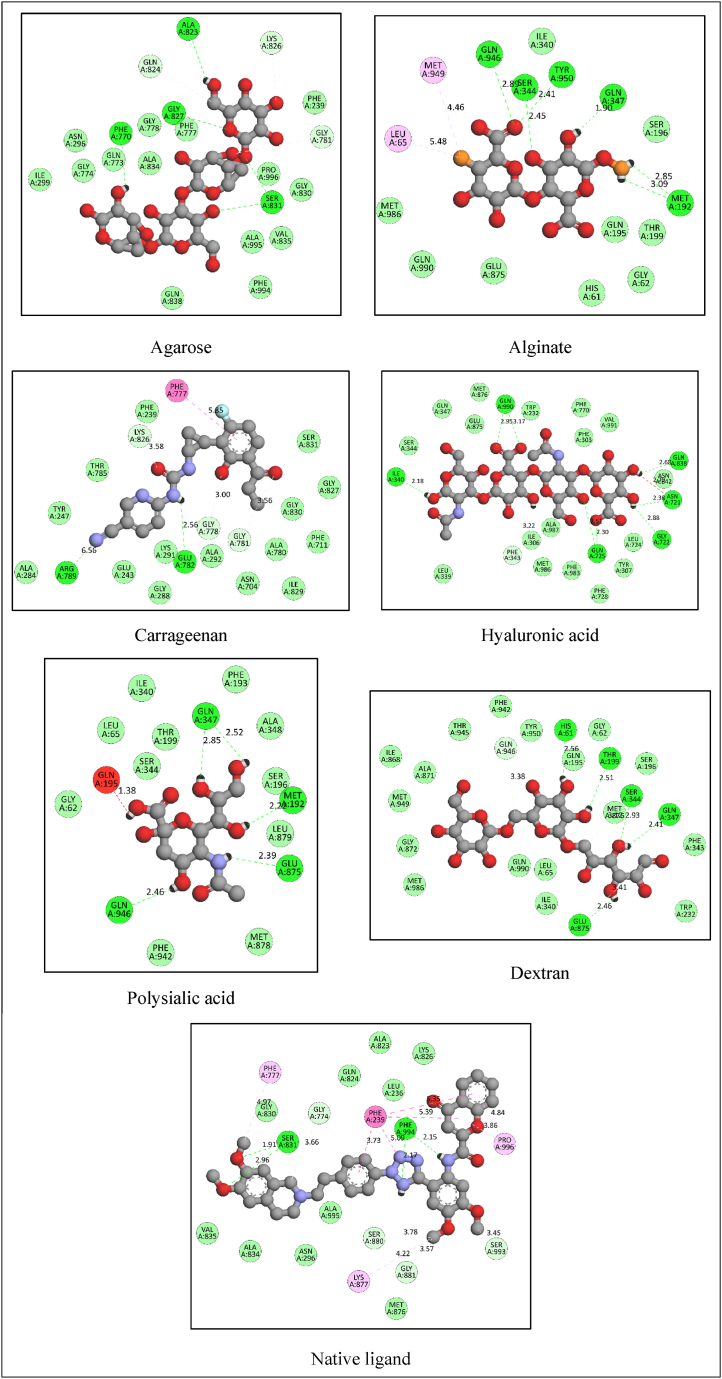
Fig. 5.
The 2D-docking poses of molecules docked with P-gp (2D-docking poses of Chitosan and Cyclodextrin were not generated by software, might be due to over bulkiness of the molecules).
Fig. 6.
Protein ligand interactions in RMSD of native ligand complex.
Fig. 7.
Protein ligand interactions in RMSD of Carageenen complex.
3. Discussion
3.1. In silico ADMET analysis
In addition to being used as active components in finished dosage forms, polymeric compounds also come into contact with pharmaceuticals in the form of processing aids and packaging. Natural polymers account for the vast majority of excipients in traditional dosage forms; several of them are on the Generally Recognized as Safe (GRAS) list due to their lengthy history of usage in pharmaceutical marketing [19]. Still, we calculated all the ADMET and drug-likeness properties to get more insight into their drug-like behavior. To accomplish goals like self-regulated drug delivery, long-term administration of protein medicines, and drug targeting to particular organs in the body, polymers with unique or numerous features will need to be created. All of these call for the creation of intelligent polymeric systems that can detect and react to normal and abnormal bodily functions. The use of polymers in the creation of novel pharmaceuticals will continue to be crucial. The physicochemical properties of molecules are tabulated in Table 1. Although these parameters are applicable to check the oral bioavailability of the compound, we calculated them to compare the properties of native ligand present in the crystal structure of enzyme with polymers. It was observed that the structure of native ligand was also bulky and resembled with the structures of polymers. The molecular weights of all the polymers, including the native ligand, were not ideal except for alginate, carrageenan, and polysialic acid. The measurement of the topological polar surface area (TPSA) is a valuable tool in assessing a drug's capability to traverse cell membranes. It provides a comprehensive evaluation of the influence exerted by polar atoms, including oxygen and nitrogen, along with their associated hydrogen atoms, on the overall molecular surface area. The concept of TPSA has demonstrated its utility in the optimization of various compound properties, such as cellular potency, intestinal permeability, and blood-brain barrier permeation. The phenomenon of compound permeation through membranes is a multifaceted and not fully comprehended process. It is important to acknowledge that TPSA calculations do not purport to account for scenarios involving active transport or efflux mechanisms. The maximum TPSA for transcellularly delivered orally active medicines should be about 120 Å2 [20]. Moreover, logS and logP values were significantly good. As these polymers are ideal for the delivery of drugs to the target organs and to make controlled-released formulations, so they can reach the site of action where they can impart some pharmacological action. Generally, film formation, thickening, gelling (controlled release), adhesion, pH-dependent solubility (controlled release), and solubility in organic solvents are desired polymer characteristics in formulation science [21,22]. Depending on their ability to dissolve in water, pharmaceutical polymers may be classified as either water-soluble or water-insoluble [23]. Agarose is soluble in hot water; alginate, carrageenan, and polysialic acid are insoluble in water; cyclodextrin is slightly soluble in water; and dextran, and hyaluronic acid are highly soluble in water.
Table 2 displays the results of several parameter calculations. They include the QED, NPscore, Lipinski Rule, Pfizer Rule, GSK Rule, Golden Triangle Rule, and Chelator Rule. In 2012, the quantitative estimate of drug-likeness (QED) was established as an indicator of drug-likeness; it is an index of drug-likeness modelled using data on currently available drugs on the market. Computational methods and drug-like feature evaluation are common applications in today's small molecule drug development process. The QED of most of the compounds was rather appealing [24,25]. NPscores, were typically between −0.7 and 1.0. A higher score indicates a higher probability that the molecule in question is an NP [26,27]. In the context of drug discovery, it has been observed that the Lipinski rule of 5 (Rule of 5) offers valuable insights into the likelihood of encountering challenges related to absorption or permeation. Specifically, this rule posits that an increased number of hydrogen bond donors exceeding five, a count of hydrogen bond acceptors surpassing ten, a molecular weight exceeding 500, and a calculated logarithm of the partition coefficient (CLog P) greater than 5 are indicative of a higher probability of encountering poor absorption or permeation. According to Lipinski, it is important to note that the applicability of the Rule of 5 is limited to compounds that do not serve as substrates for active transporters [28]. In the present investigation, only carrageenan and polysialic acid accepted the Lipinski rule of 5.
Based on the established guidelines set forth by Pfizer, it has been determined that compounds exhibiting a high logarithm of the partition coefficient (log P) value, specifically exceeding 3, coupled with a low topological polar surface area (TPSA) value, specifically below 75, are indicative of a higher likelihood of toxicity [29]. All the polymer accepted the Pfizer rule. Based on the GSK rule, it has been observed that molecules with a molecular mass below 400 and a CLogP below 4 are more likely to possess an optimal ADME profile [30]. However, only carrageenan and polysialic acid accepted the GSK rule, despite the possibility that compounds fitting the GSK criteria would have a better ADMET profile. The Golden Triangle, a visualization tool, was developed to aid medicinal chemists in identifying drug candidates with desirable characteristics such as metabolic stability, permeability, and potency. This tool incorporates in vitro permeability, in vitro clearance, and computational data to guide the selection process. The criteria for inclusion in the Golden Triangle are a molecular weight (MW) ranging from 200 to 500 and a logarithmic distribution coefficient (logD) ranging from −2 to 5. By utilizing these parameters, researchers can focus their efforts on compounds that exhibit optimal properties for drug development [31]. All molecules except alginate, carrageenan, and polysialic acid rejected the Golden Triangle rule, despite the possibility that the compounds fulfilling the rule would have a better ADMET profile. It was observed that carrageenan and polysialic acid accepted all the rules and demonstrated drug-like properties, suggesting they can have a superior drug-likeness profile compared to other natural polymers selected for the study.
The bioavailability of natural polymers and native ligand are shown in Table 3, along with absorption metrics including Caco-2 and MDCK permeability, Pgp-inhibitor and substrate ability, human intestinal absorption (HIA), and F20% and F30% bioavailability. The Caco-2 human colon epithelial cancer cell line is used as a model for drug absorption in the digestive system. This model may be used to investigate drug efflux, forecast intestinal permeability, and establish whether or not a medication is safe for oral delivery. Fortunately, all of the molecules showed maximum Caco-2 permeability, which is reached when the value is greater than −5.15 log units [32]. Transfection of the MDR1 (ABCB1) gene into Madin-Darby canine kidney (MDCK) cells results in MDCK-MDR1 cells, which express the efflux protein P-gp. By measuring transport across the cell monolayer in both directions (from apical to basolateral and basolateral to apical), an efflux ratio can be calculated. This ratio serves as an indication of whether or not a substance moves via active efflux (which is mediated by P-gp). By using MDCK-MDR1, researchers may learn more about drug efflux and detect potential issues with drug permeability earlier on. Permeability of MDCK-MDR1 cells has been shown to be a reliable predictor of blood brain barrier permeability, complementing intestinal permeability as a predictor of BBB permeability [33]. All the molecules displayed medium to high MDCK permeability. In most cases, the evaluation of substrates is done using bidirectional transport assays, which indicate the apparent or net (but not the effective) transfer. A substance is considered to be a substrate if, when subjected to a transport test, it demonstrates a positive net efflux (that is, a greater active efflux by P-gp than passive inflow). Substrates are atoms or molecules that have at least one hydrogen bond acceptor pattern and a molecular mass that is greater than 450 gm/mol. In absorption properties, native ligand displayed P-gp inhibitory activity, whereas polymers showed P-gp substrate potential. Only native ligand and carrageenan displayed low human intestinal absorption (HIA), while other polymers demonstrated moderate-to-good HIA. The F20% and F30% bioavailability of all the molecules were within the range of low to high.
The distribution and metabolism profiles of polymers and native ligand are depicted in Table 4. Drugs with high protein binding (PPB, <90%) may have a low therapeutic index; almost all polymers had PPB less than 90%, whereas native ligands and carrageenan had very high PPB, indicating a lower therapeutic index. Volume distribution (VD; optimal 0.04–20L/kg) of all the molecules was within the range of acceptable limits. The Blood-Brain Barrier (BBB) is a highly specialized physiological mechanism comprised of Brain Microvascular Endothelial Cells (BMVEC). Its primary function is to safeguard the brain by preventing the entry of harmful substances from the bloodstream. Simultaneously, the BBB facilitates the transportation of vital nutrients to support the metabolic needs of brain tissues. Moreover, the BBB serves the crucial role of selectively preventing the entry of potentially harmful compounds into the brain, while facilitating the removal of such substances from the brain back into the bloodstream. The transportation of substances across the BBB is governed by a set of rigorous limitations imposed by various factors. These factors can be categorized into two main groups: physical factors, such as tight junctions, and metabolic barriers, which encompass enzymes and a wide range of transport systems. The BBB presents a notable obstacle in the delivery of therapeutic agents to the central nervous system (CNS) owing to its limited permeability [34]. In terms of BBB penetration, all of the polymers, including the natural ligand, fell into the mild to moderate range (0 indicates BBB−; >0 indicates BBB+). The overall drug effect is influenced by the introduction of substrate or the inhibition of cytochrome enzymes, as these enzymes play a vital role in drug metabolism. In the current study, native ligands inhibited Cytochrome enzyme, whereas polymers showed neither inhibition nor substrate against Cytochrome [35].
Table 5 summarizes the excretion and toxicity profiles of native ligand and natural polymers. Native ligand and carrageenan displayed moderate, whereas, the rest of the molecules showed low clearance rates (CL, high: >15 mL/min/kg; moderate: 5–15 mL/min/kg; low: <5 mL/min/kg). There was a uniformly short half-life among the molecules (T1/2, <3h). Several of the readings were within the allowable range, and the compounds' toxicity profiles hinted to beneficial qualities. None of the molecules showed human hepatotoxicity (H-HT). Native ligand, alginate, carrageenan, cyclodextrin, dextran, and hyaluronic acid showed slight drug-induced liver injury (DILI). All of the polymers and the native ligand tested negligible in terms of AMES toxicity, rat oral acute toxicity, maximum recommended daily dosage (FDAMDD), skin sensitization, carcinogencity, ocular corrosion/irritation, and respiratory toxicity. The physicochemical radar of the native ligand and polymers obtained from the ADMETlab 2.0 online server is depicted in Fig. 3. This radar diagram provides valuable insights into the optimal physicochemical characteristics of the molecules currently being investigated. The following information provides a concise overview of the drug-like properties associated with the molecules previously discussed in the preceding section [35].
An environmental toxicity profile (bioconcentration factors: IGC50, LC50FM, and LC50DM) of native ligand and natural polymers is demonstrated in Table 6. When considering the possibility for secondary poisoning and evaluating the hazards to human health posed by the food chain, bioconcentration factors are an important tool to apply. The bioconcentration factor is measured in ―log10[(mg/L)/(1000*MW)] [36,37]. Native ligand displayed a higher bioconcentration factor compared to natural polymers. Tetrahymena pyriformis served as the subject of a test that was devised in order to evaluate the toxicity of a number of different compounds. In this test, the Tetrahymena pyriformis 50% growth inhibition concentration was referred to as IGC50. A group contribution approach has been devised in order to correlate the acute toxicity (96-h LC50) to the fathead minnow (FM) for 397 different organic compounds, which is generally referred to as LC50FM [38]. Daphnia magna (D. magna) is an organism that is used as a model bioindicator in the field of ecotoxicology because of its extreme sensitivity to contaminants. The concentration at which 50% of the Daphnia in a test batch become immobilised throughout the course of a continuous period of exposure, which is typically 48 h, is referred to as the LC50DM [39]. The IGC50, LC50FM, and LC50DM values of native ligand and natural polymers were within acceptable limits.
3.2. Computational analysis
Computational drug discovery is an effective strategy that has the potential to speed up and reduce the financial burden of the drug discovery and development process. Computational drug discovery's use has grown and expanded as a consequence of the sudden rise in the availability of data on biological macromolecules and small compounds, and it is now employed in almost every stage of the drug discovery and development process. This consists of preclinical testing, lead discovery and optimization, target validation, and lead identification. Some of the most important computational drug development tools that have advanced in recent decades are molecular docking, pharmacophore modelling and mapping, de novo design, molecular similarity calculation, and sequence-based virtual screening [[40], [41], [42], [43]]. In the present study, we applied a molecular docking tool to understand the binding potential of natural polymers with P-gp (PBD ID: 7O9W), and the interactions are tabulated in Table S1 in the supplementary file. The docking results are elaborated in the section given below:
The binding affinity of the native ligand was −10.7 kcal/mol, and it made eight hydrogen (conventional and carbon) bonds with Phe994, Ser831, Ser993, Ser880, Gly881, and Gly774. It also showed hydrophobic interactions (Pi-Pi stacked, Pi-Pi T-shaped, alkyl, and Pi-alkyl) with Phe239, Lys877, Pro996, and Phe777.
Agarose is a water-soluble linear polysaccharide that is isolated from seaweed. It has shown promising properties in a variety of sectors, spanning from environmental engineering to medicine, and is most famously used in targeted drug administration. It has reversible thermogelling behavior, remarkable mechanical characteristics, strong bioactivity, and switchable chemical reactivity for functionalization. As a consequence of this, agarose has garnered a lot of interest in the process of fabricating improved delivery systems, since it may act as a sophisticated carrier for pharmaceuticals. Agarose displayed a −8.5 kcal/mol binding affinity and formed five conventional hydrogen bonds with Ala823, Phe770, Gly827, Ser831, and three carbon-hydrogen bonds with Gln824, Gly781, and Lys826. It does not demonstrate any kind of hydrophobic interaction with P-gp.
It is well known that alginates are among the most adaptable biopolymers, since they are used in a broad variety of applications. The thickening, gel-forming, and stabilizing qualities of alginate are often what determine whether or not it will be used as an excipient in pharmaceutical formulations. The demand for custom-made polymers has expanded in recent years as a result of an increasing emphasis on maintaining and improving control over the delivery of drugs. In the development of a controlled-release product, the use of hydrocolloids like alginate might be an important factor to consider. Hydration of alginic acid results in the production of a very viscous "acid gel" when the pH of the solution is low. In the presence of a divalent cation such as calcium ion, alginate may also be readily gelatinized into a solid. Alginate exhibited a docking score of −6.6 kcal/mol and formed six conventional hydrogen bonds with Gln347, Met192, Ser344, Gln946, and Tyr950. It also formed two hydrophobic (alkyl) interactions with Leu65 and Met949.
In recent years, carrageenan-based biomaterials have garnered a lot of attention because of their multifunctional qualities. These qualities include biodegradability, biocompatibility, and non-toxicity. Furthermore, carrageenan-based biomaterials have bioactive properties, such as antiviral, antibacterial, antihyperlipidemic, anticoagulant, antioxidant, antitumor, and immunomodulating properties. Because of their bioactive and physicochemical features, they have been used in pharmaceutical formulations as ideal biomaterials for drug administration and, more recently, in the development of tissue engineering. This has led to their widespread use in both fields. Carrageenan displayed a −8.7 kcal/mol binding affinity and formed three conventional hydrogen bonds with Glu782 and Arg789. It has formed three carbon-hydrogen bonds with Gly778, Gly781, and Lys826. It also showed one pi-pi stacked interaction with Phe777.
Chitosan is a mucopolysaccharide that is quite similar to cellulose in its chemical structure. Deacetylation of chitin is the chemical reaction that results in the formation of this chitin as a byproduct. This substance can also be found in crustacean shells and fungi cellular walls. The presence of the main amine at the C-2 position of the glucosamine residues is a distinguishing characteristic of chitosan, making it one of a kind. Chitosan has a variety of useful functional characteristics as a direct result of the presence of such a high amine content. Chitosan displayed a docking score of −8.6 kcal/mol and formed 11 conventional hydrogen bonds with Tyr307, Gln347, Asn721, Val991, Gln195, Gln725, Gln773, and Gln946. It has formed two carbon-hydrogen bonds with Glu875 and Gln774.
There is a class of cyclic oligosaccharides known as cyclodextrins. These molecules have a hydrophilic surface on the outside and a lipophilic cavity on the inside. Cyclodextrin molecules are often not able to pass through lipophilic membranes because of their comparatively large size and the huge number of hydrogen donors and acceptors that they contain. In the pharmaceutical industry, cyclodextrins have been used primarily as complexing agents to improve the aqueous solubility of poorly soluble pharmaceuticals as well as boost the bioavailability and stability of these medications. Cyclodextrins have been found to enhance medication delivery in studies conducted on both people and animals, and this improvement may be achieved with almost any kind of drug formulation. Cyclodextrin showed −24.5 kcal/mol of binding free energy and formed only one conventional hydrogen bond with Ala819 and two carbon-hydrogen bonds with Ala819 and Glu243. It has formed many electrostatic (attractive charge) interactions with Glu782 and Glu243.
As a way to stop cancer cells from growing, Dextran is used as a drug carrier. This effectively reduces the toxic and side effects of the drug in the body. Targeting makes it so that there is more of the active substance near the target tissue. This makes less damage happen to other organs and normal tissues. In the future, Dextran could be used to carry antitumor drugs to where they need to go. This would allow for slow-release chemotherapy and targeted drug delivery. Dextran showed −6.7 kcal/mol binding affinity and formed 08 conventional hydrogen bonds with Asn721, Gly722, Gln838, ILE340, Gln990, Gln725, and Gln990. It has formed one Pi-donor hydrogen bond with Phe343.
The formation of hyaluronic acid-drug conjugates involves the formation of prodrugs by the formation of covalent bonds between small molecule anticancer drugs and hyaluronic acid. These covalent connections are difficult to break in the circulation, but once they have reached their destination, they are hydrolyzed or enzymatically dissolved, which results in the release of the medicine. Hyaluronic acid-drug conjugates have the potential to enhance the solubility of the drug, alter the distribution and half-life of the drug in vivo, raise the accumulation of tumour tissue by boosting the osmotic retention effect, and more effectively exercise their therapeutic impact. Hyaluronic acid exhibited −8.3 kcal/mol binding affinity and demonstrated similar kind of interactions as shown by Dextran.
A homopolymer of sialic acid with either α-2,8 or α-2,9 links, or a combination of α-2,8 and α-2,9 linkages, is what makes up polysialic acid. Polysialic acid, which is made up of a α-2,8 bond, is non-immunogenic and biodegradable. It also works to lower the immunogenicity of protein polypeptides. In a similar manner, polysialic acid demonstrates the ability to avoid being consumed by phagocytes and to have a prolonged circulation period in vivo. Polysialic acid formed 05 conventional hydrogen bonds with Met192, Gln347, Gln946, and Glu875 with binding free energy of −7.9 kcal/mol. The binding interaction poses of molecules are illustrated in Fig. 4, Fig. 5.
Drug toxicity, quick disintegration, low specificity, and limited targeting are only some of chemotherapy's many drawbacks. Nanomedicine, which employs a wide variety of individually optimized drug delivery systems, has emerged as an integral part of cancer therapy in recent decades [44]. Smaller than 100 nm in size, the materials created by nanomedicine can be employed as drug nanocarriers thanks to their unique characteristics. These include, but are not limited to, their high specificity, small size, high solubility, and hydrophilicity. Nanocarriers accumulate in cancer tissue, which has leaky vasculature, resulting in an enhanced permeability and retention effect [45]. Because of their biocompatibility and biodegradability, natural polymers, also known as biopolymers, are particularly well-suited for use in medical applications such as cell-based transplantation, tissue engineering, and gene therapy. Natural polymers include various classes of polysaccharides and proteins [46].
It is possible to create semi-synthetic polymers that mimic human tissue structure components by chemically modifying the functional groups of natural polymers to accept synthetic molecules [46]. Synthetic polymers are more likely to be used in controlled drug delivery systems than biopolymers because there are many ways to design their structure and change their physical and chemical properties [45,47,48]. However, few delivery systems for pharmaceuticals have been able to meet therapeutic demands so far, despite polymeric nanomedicine's promising track record of offering sustained release of medications with reduced cytotoxicity and altered tumour retention [49]. Therefore, here we tried to gather more property-based information about some natural polymers to be used in the delivery of anticancer medicines.
3.3. Molecular dynamic simulation
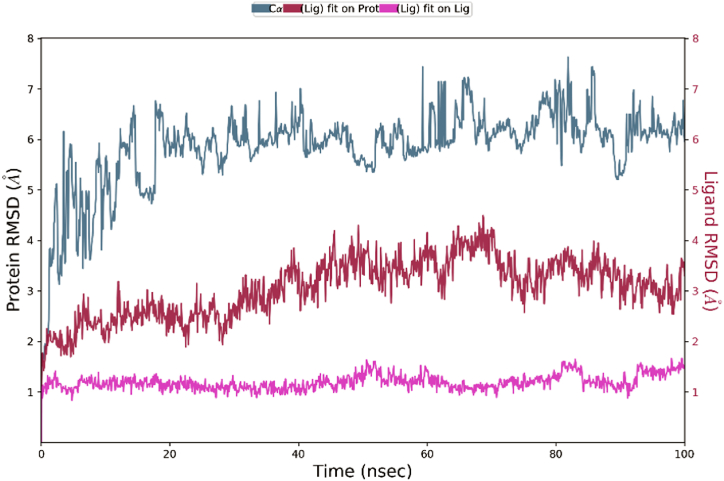
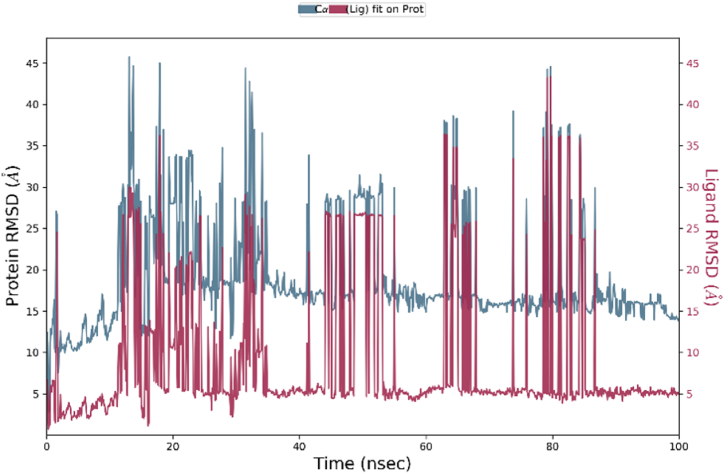
Molecular dynamic simulations (MDS) using the Desmond module confirmed the stability and dependability of molecules that successfully hit the P-glycoprotein. MDS were performed at 100 ns simulation intervals. Root mean square deviation (RMSD) and Root mean square fluctuation (RMSF) were calculated to determine a complex's stability and were used to validate docking score results. RMSD shows interactions of a compound with a target and was calculated accordingly by atom selection. RMSD was performed for ligands and proteins through structural confirmation. The plot of RMSD shows that the native ligand and carrageenan complex system was stable with P-glycoprotein (7O9W). The RMSD results of the native ligand and carrageenan complex system with 7O9W are given in Fig. 6, Fig. 7. The native ligand and carrageenan complex system -RMSD analysis was found to be stable but the carrageenan complex system shows minor deviation from the native ligand complex. RMSD of the native ligand complex was found between 1 Å to 4 Å which shows stable conformation with respect to simulation time. The RMSD of the carrageenan ligand complex was found between 5 Å to 15 Å which shows stable conformation with respect to simulation time.
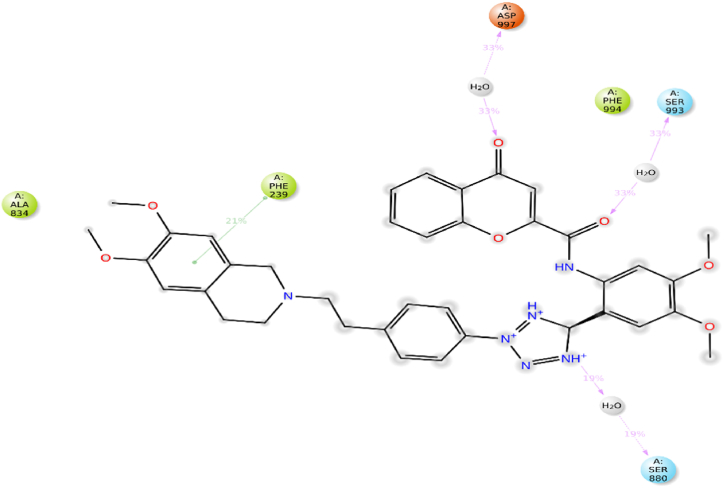
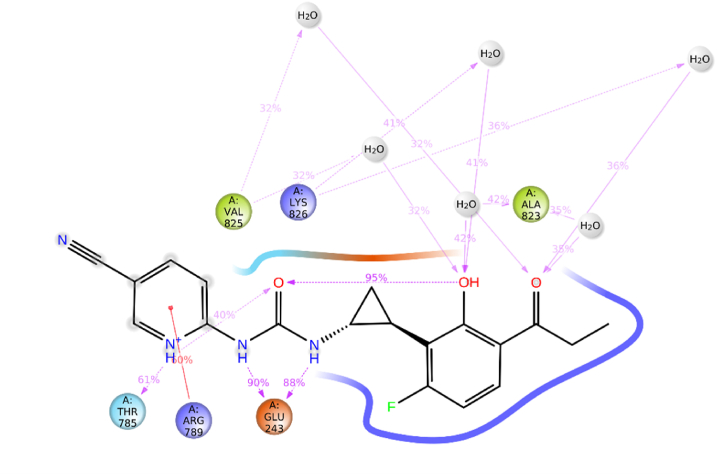
RMSF trajectory used for determination of mobility of molecules with ligand binding and local changes with protein chains. Protein RMSF was also used to analyze fluctuation and differences in the position of alpha-carbon atoms. RMSF of alpha carbon with native ligand and carrageenan complex system observed remarkable analysis. The carrageenan complex system shows minor fluctuations with respect to the native ligand complex system. The interactions with amino acids, if more than 30.0%, then interactions are strong between them with simulation time in the selected trajectory (0–100 ns) are shown. The native ligand with 7O9W had a strong interaction with SER_993 occurring at 33%, which showed the strongest interactions. Additionally, other amino acids also interacted, such as ASP_997 with 33%, less than 30% interactions are also observed which shows weak interactions with PHE_21% and SER_ 880 WITH 19% (Fig. 8). The carrageenan with 7O9W showed the strongest interactions percentage with respect to the native ligands. The Carrageenan shows a strong interaction with Glu_243 occurring at 90%, Additionally, other amino acids also interacted, such as Thr_785 with 61%, Arg_789 with 60%, Ala_823 with 42%, Val_825 with 32% and less than 30% interactions are not observed (Fig. 9).
Fig. 8.
2D plot of native ligand with protein.
Fig. 9.
2D plot of carrageenan with protein.
The properties of ligands also examine for the radius of gyration (rGyr), no. of internal H-bonds (IntraHB), the surface area of the molecule (MolSA), solvent accessible surface area (SASA), and polar surface area (PSA) analysed by Desmond simulation. The important property like the radius of gyration (rGyr) measures the flexibility and orientation of the ligand. Throughout the simulation time, the protein's average radius remains constant. Protein compression is illustrated by a decrease in gyration radius during the course of the simulation, while an increase in radius indicated the contrary. The overall results of native and Carrageenan complex system simulation results show that both complexes are stable in the pocket of the protein.
4. Methods and materials
4.1. In silico ADMET analysis
To aid medicinal chemists in the lead discovery, development, and optimization processes, a novel kind of tool known as an in silico ADMET evaluation model has been developed. Often used for estimating the pharmacokinetics and toxic properties of diverse substances, ADMETlab 2.0 is a completely redesigned version of the AMDETlab web server (https://admetmesh.scbdd.com/) [35].
4.2. Computational analysis
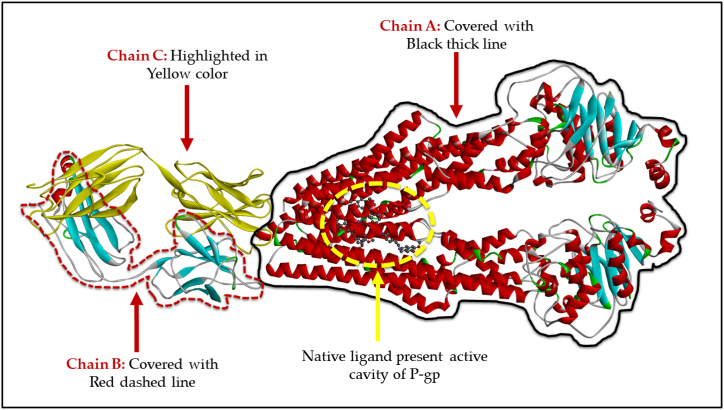
The native ligand and polymers were docked into the human P-glycoprotein crystal structure using Autodock vina in PyRx 0.8 version [50]. The structures of the natural polymers and the native ligand were downloaded from PubChem (sdf. File format) and then converted to mol file using ChemDraw Ultra 12.0 version. The use of the Universal Force Field (UFF) allowed for the optimization of each of the ligands in terms of lowering the amount of energy that was required by them [51]. The crystal structure of Encequidar-bound human P-glycoprotein in complex with UIC2-Fab was obtained from RCSB Protein Data Bank (PDB) with PDB ID: 7O9W (https://www.rcsb.org/structure/7O9W). First, the enzyme's structure was improved with the help of Discovery Studio Visualizer (version 19.1.0.18287); then, the enzyme was cleaned up and made ready for docking with the assistance of the same tool [52]. In order to facilitate molecular docking, a three-dimensional grid box with an exhaustiveness value of eight was designed. The dimensions of the box were as follows: size_x = 52.3306319041 Å; size_y = 36.7275092822 Å; size_z = 42.8046787888 Å [50]. In order to carry out the whole molecular docking procedure, as well as to find cavity and active amino acid residues, the strategy that was provided by Khan et al. was adopted [[53], [54], [55], [56], [57], [58], [59], [60], [61], [62]]. Fig. 10 illustrates the native ligand molecule in conjunction with the exposed cavity of the P-gp protein.
Fig. 10.
3D-ribbon view of P-gp with identified active cavity and native ligand present in it. Different chains are denoted using multiple colors.
4.3. Molecular dynamic simulation
The effectiveness of molecular docking analysis was evaluated by MDS. The Desmond module of the Schrödinger software was used to do the molecular dynamic simulation. The created complex's stability was evaluated using a molecular dynamics simulation that was run from 0 to 100 ns. We chose the orthorhombic box, 10 Å distant from the box's edges, for the complex's position using Desmond's system construction module. We fixed predefined (TIP3P) solvated water molecules. The native ligand with 7O9W was neutralized by adding 36 Na and 46 Cl ions with concentrations of 50 mM and 64 mM, respectively, and Carrageenan with 7O9W was neutralized by 23 Na ions (51 mM) and 35 Cl ions (77 mM). For simulation analysis, we employed the OPLS3e force field for protein, ligands, and ions. In the molecular dynamics module, we selected a load from a workspace in which the full system contained 45669 atoms of native ligands with 7O9W & 28624 atoms of carrageenan with 7O9W. The simulation time of a running project was fixed at 100 ns for the analysis of trajectories. The ensemble class NPT was selected in which the temperature was maintained at 300K and 1 atm of pressure was constant. The evaluation of molecular dynamics properties, like, RMSD, P-SSE graph, RMSF of protein and ligand, complex interactions protein-ligand contact, L-Torsions, and ligand properties such as PSA, SASA, molSA, intraHB, etc. were done [63,64].
5. Conclusions
P-glycoprotein (P-gp) gained the name "multidrug resistance protein" because it increased tumor resistance to several types of anticancer medication. Pharmaceutical polymers are an integral part of drug delivery systems, whether it is an anticancer drug or any other medication. Numerous in vitro and in vivo experiments have shown the usefulness of such polymers in cancer treatment and drug delivery. To combat MDR in cancer cells, researchers are developing adjuvant compounds that can block efflux pumps, as well as novel therapeutic non-substrate agents or formulations that enable the medication to bypass efflux pump trafficking. Researchers have shown that co-administering efflux pump inhibitors with an efflux pump substrate improves bioavailability from oral dosing. Therefore, in the present investigation, we have selected some natural polymers to study their ability to inhibit P-gp, which can help to avoid drug efflux. Our aim was to suggest such polymers that can inhibit P-gp up to some extent and thus be a better option for the formulation of anticancer medications. The ADMET profile of all the selected polymers (agarose, alginate, carrageenan, cyclodextrin, dextran, hyaluronic acid, and polysialic acid) has been studied, and binding affinities were investigated through a computational approach using the recently released crystal structure of P-gp with PDB ID: 7O9W. The aim of the present study was to overcome MDR resulting from the activity of P-gp by using such polymers that can inhibit P-gp when used in formulations. The docking scores of native ligand, agarose, alginate, carrageenan, chitosan, cyclodextrin, dextran, hyaluronic acid, and polysialic acid were found to be −10.7, −8.5, −6.6, −8.7, −8.6, −24.5, −6.7, −8.3, and −7.9, respectively. The molecular dynamic study also reveals remarkable changes and showed the ligand complex is stable in a given protein complex. It was observed that cyclodextrin possesses multiple properties in drug delivery science and here also demonstrated excellent binding affinity. The results of the current investigation lead us to infer that if we can use agarose, carrageenan, chitosan, cyclodextrin, hyaluronic acid, and/or polysialic acid for the delivery of anticancer medication, it can help to avoid MDR resulting from drug efflux.
Author contribution statement
Kumaraswamy Gandla; Fahadul Islam; Mehrukh Zehravi; Anandakumar Karunakaran; Indu Sharma: Con-ceived and designed the experiments; Performed the experiments; Analysed and interpreted the data; Con-tributed reagents, materials, analysis tools or data; Wrote the paper.
Sanjay Kumar; Kumar Pratyush; Sachin A. Dhawale; Firzan Nainu; Sharuk L. Khan: Performed the experi-ments; Analysed and interpreted the data; Wrote the paper.
Md. Rezaul Islam; Kholoud Saad Al-Mugren; Mayeen Uddin Khandaker: Performed the experiments; Wrote the paper.
Falak A. Siddiqui; Talha Bin Emran: Conceived and designed the experiments; Contributed reagents, mate-rials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Funding
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R10), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R10), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Contributor Information
Kholoud Saad Al-Mugren, Email: ksalmogren@pnu.edu.sa.
Falak A. Siddiqui, Email: falakarjumand26@gmail.com.
Talha Bin Emran, Email: talhabmb@bgctub.ac.bd.
References
- 1.Wu C.-P., Ohnuma S., Ambudkar S.V. Discovering natural product modulators to overcome multidrug resistance in cancer chemotherapy. Curr. Pharm. Biotechnol. 2011;12:609–620. doi: 10.2174/138920111795163887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishna R., Mayer L.D. Modulation of P-glycoprotein (PGP) mediated multidrug resistance (MDR) using chemosensitizers: recent advances in the design of selective MDR modulators. Curr. Med. Chem. Anticancer. Agents. 2001;1:163–174. doi: 10.2174/1568011013354705. [DOI] [PubMed] [Google Scholar]
- 3.Aller S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P.M., Trinh Y.T., Zhang Q., Urbatsch I.L., Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323(80):1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques S.M., Šupolíková L., Molčanová L., Šmejkal K., Bednar D., Slaninová I. Screening of natural compounds as p-glycoprotein inhibitors against multidrug resistance. Biomedicines. 2021;9 doi: 10.3390/biomedicines9040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013:27–34. doi: 10.4137/DTI.S12519. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J.H., Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin. Pharmacokinet. 2003;42:59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Zhang H., Assaraf Y.G., Zhao K., Xu X., Xie J., Yang D.H., Chen Z.S. Overcoming ABC transporter-mediated multidrug resistance: molecular mechanisms and novel therapeutic drug strategies. Drug Resist. Updat. 2016;27:14–29. doi: 10.1016/j.drup.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Dallavalle S., Dobričić V., Lazzarato L., Gazzano E., Machuqueiro M., Pajeva I., Tsakovska I., Zidar N., Fruttero R. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist. Updat. 2020;50 doi: 10.1016/j.drup.2020.100682. [DOI] [PubMed] [Google Scholar]
- 9.Dai Y., Zhang F., Chen N., Wang G., Jia S., Zheng H. Analysis of P-glycoprotein structure and binding sites, in. 2nd Int. Conf. Inf. Sci. Eng. ICISE2010 - Proc. 2010:101–103. doi: 10.1109/ICISE.2010.5689524. [DOI] [Google Scholar]
- 10.Raghava K.M., Lakshmi P.K. Overview of P-glycoprotein inhibitors: a rational outlook. Brazilian J. Pharm. Sci. 2012;48:353–367. doi: 10.1590/S1984-82502012000300002. [DOI] [Google Scholar]
- 11.Melaine N., Liénard M.O., Dorval I., Le Goascogne C., Lejeune H., Jégou B. Multidrug resistance genes and P-glycoprotein in the testis of the rat, mouse, Guinea pig, and human. Biol. Reprod. 2002;67:1699–1707. doi: 10.1095/biolreprod.102.003558. [DOI] [PubMed] [Google Scholar]
- 12.Kuppens I.E.L.M., Witteveen E.O., Jewell R.C., Radema S.A., Paul E.M., Mangum S.G., Beijnen J.H., Voest E.E., Schellens J.H.M. A phase I, randomized, open-label, parallel-cohort, dose-finding study of elacridar (GF120918) and oral topotecan in cancer patients. Clin. Cancer Res. 2007;13:3276–3285. doi: 10.1158/1078-0432.CCR-06-2414. [DOI] [PubMed] [Google Scholar]
- 13.Lomovskaya O., Bostian K.A. Practical applications and feasibility of efflux pump inhibitors in the clinic - a vision for applied use. Biochem. Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Callaghan R., Luk F., Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab. Dispos. 2014;42:623–631. doi: 10.1124/dmd.113.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werle M. Natural and synthetic polymers as inhibitors of drug efflux pumps. Pharm. Res. (N. Y.) 2008;25:500–511. doi: 10.1007/s11095-007-9347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alakhov V.Y., Moskaleva E.Y., Batrakova E.V., Kabanov A.V. Hypersensitization of multidrug resistant human ovarian carcinoma cells by pluronic P85 block copolymer. Bioconjug. Chem. 1996;7:209–216. doi: 10.1021/bc950093n. [DOI] [PubMed] [Google Scholar]
- 17.Friche E., Jensen P.B., Sehested M., Demant E.J.F., Nissen N.N. The solvents Cremophor EL and Tween 80 modulate daunorubicin resistance in the multidrug resistant Ehrlich ascites tumor. Cancer Commun. 1990;2:297–303. [PubMed] [Google Scholar]
- 18.Urgaonkar S., Nosol K., Said A.M., Nasief N.N., Bu Y., Locher K.P., Lau J.Y.N., Smolinski M.P. Discovery and characterization of potent dual P-glycoprotein and CYP3A4 inhibitors: design, synthesis, cryo-EM analysis, and biological evaluations. J. Med. Chem. 2022;65:191–216. doi: 10.1021/acs.jmedchem.1c01272. [DOI] [PubMed] [Google Scholar]
- 19.Burdock G.A., Carabin I.G. Generally recognized as safe (GRAS): history and description. Toxicol. Lett. 2004;150:3–18. doi: 10.1016/j.toxlet.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Clark D.E. What has polar surface area ever done for drug discovery? Future Med. Chem. 2011;3:469–484. doi: 10.4155/fmc.11.1. [DOI] [PubMed] [Google Scholar]
- 21.Kumar N., Pahuja S., Sharma R. Pharmaceutical polymers - a review. Int. J. Drug Deliv. Technol. 2019;9:27–33. doi: 10.25258/ijddt.9.1.5. [DOI] [Google Scholar]
- 22.Pal N., Chauhan S. Encycl. Biomed. Polym. Polym. Biomater. 2015. Pharmaceutical polymers; pp. 5929–5942. [DOI] [Google Scholar]
- 23.Jones D. 2004. Pharmaceutical Applications of Polymers for Drug Delivery.http://books.google.co.uk/books?id=VQOZ0zrEB9IC [Google Scholar]
- 24.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosugi T., Ohue M. Quantitative estimate index for early-stage screening of compounds targeting protein-protein interactions. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ertl P., Roggo S., Schuffenhauer A. Natural product-likeness score and its applications in the drug discovery process. Chem. Cent. J. 2008;2 doi: 10.1186/1752-153x-2-s1-s2. [DOI] [PubMed] [Google Scholar]
- 27.Menke J., Massa J., Koch O. Natural product scores and fingerprints extracted from artificial neural networks. Comput. Struct. Biotechnol. J. 2021;19:4593–4602. doi: 10.1016/j.csbj.2021.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benet L.Z., Hosey C.M., Ursu O., Oprea T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016 Jun 1. 2016:89–98. doi: 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ursu O., Rayan A., Goldblum A., Oprea T.I. Understanding drug-likeness. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011;1:760–781. doi: 10.1002/wcms.52. [DOI] [Google Scholar]
- 30.Walters W.P. Going further than Lipinski's rule in drug design. Expert Opin. Drug Discov. 2012;7:99–107. doi: 10.1517/17460441.2012.648612. [DOI] [PubMed] [Google Scholar]
- 31.Johnson T.W., Dress K.R., Edwards M. Using the Golden Triangle to optimize clearance and oral absorption. Bioorganic Med. Chem. Lett. 2009;19:5560–5564. doi: 10.1016/j.bmcl.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 32.Lee J.B., Zgair A., Taha D.A., Zang X., Kagan L., Kim T.H., Kim M.G., yeol Yun H., Fischer P.M., Gershkovich P. Quantitative analysis of lab-to-lab variability in Caco-2 permeability assays. Eur. J. Pharm. Biopharm. 2017;114:38–42. doi: 10.1016/j.ejpb.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Feng B., West M., Patel N.C., Wager T., Hou X., Johnson J., Tremaine L., Liras J. Validation of human MDR1-MDCK and BCRP-MDCK cell lines to improve the prediction of brain penetration. J. Pharm. Sci. 2019;108:2476–2483. doi: 10.1016/j.xphs.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Persidsky Y., Ramirez S.H., Haorah J., Kanmogne G.D. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J. Neuroimmune Pharmacol. 2006;1:223–236. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 35.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., Chen X., Hou T., Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkab255. W5–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garg R., Smith C.J. Predicting the bioconcentration factor of highly hydrophobic organic chemicals. Food Chem. Toxicol. 2014;69:252–259. doi: 10.1016/j.fct.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Arnot J.A., Gobas F.A.P.C. A review of bioconcentration factor (BCF) and bioaccumulation factor (BAF) assessments for organic chemicals in aquatic organisms. Environ. Rev. 2006;14:257–297. doi: 10.1139/A06-005. [DOI] [Google Scholar]
- 38.Martin T.M., Young D.M. Prediction of the acute toxicity (96-h LC50) of organic compounds to the fathead minnow (Pimephales promelas) using a group contribution method. Chem. Res. Toxicol. 2001;14:1378–1385. doi: 10.1021/tx0155045. [DOI] [PubMed] [Google Scholar]
- 39.Tkaczyk A., Bownik A., Dudka J., Kowal K., Ślaska B. Daphnia magna model in the toxicity assessment of pharmaceuticals: a review. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.143038. [DOI] [PubMed] [Google Scholar]
- 40.Leelananda S.P., Lindert S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016;12:2694–2718. doi: 10.3762/bjoc.12.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker J.M. 2011. Drug Designing and Discovery. [Google Scholar]
- 42.Macalino S.J.Y., Gosu V., Hong S., Choi S. Role of computer-aided drug design in modern drug discovery. Arch Pharm. Res. (Seoul) 2015;38:1686–1701. doi: 10.1007/s12272-015-0640-5. [DOI] [PubMed] [Google Scholar]
- 43.Walker J.M. 2011. Drug Design and Discovery: Methods and Protocols. [Google Scholar]
- 44.Hossen S., Hossain M.K., Basher M.K., Mia M.N.H., Rahman M.T., Uddin M.J. Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: a review. J. Adv. Res. 2019;15:1–18. doi: 10.1016/j.jare.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X., Wang L., Wei X., Zhou S. Polymer-based drug delivery systems for cancer treatment. J. Polym. Sci. Part A Polym. Chem. 2016;54:3525–3550. doi: 10.1002/pola.28252. [DOI] [Google Scholar]
- 46.Fathi M., Barar J. Perspective highlights on biodegradable polymeric nanosystems for targeted therapy of solid tumors. Bioimpacts. 2017;7:49–57. doi: 10.15171/bi.2017.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alsehli M. Polymeric nanocarriers as stimuli-responsive systems for targeted tumor (cancer) therapy: recent advances in drug delivery. Saudi Pharm. J. 2020;28:255–265. doi: 10.1016/j.jsps.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avramović N., Mandić B., Savić-Radojević A., Simić T. Polymeric nanocarriers of drug delivery systems in cancer therapy. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao X., Teng F., Shi C., Chen J., Wu S., Wang B., Meng X., Essiet Imeh A., Li W. Polymeric nanoparticles—promising carriers for cancer therapy. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.1024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 51.Rappé A.K., Casewit C.J., Colwell K.S., Goddard W.A., Skiff W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114:10024–10035. doi: 10.1021/ja00051a040. [DOI] [Google Scholar]
- 52.San Diego . Vol. 3.5. Accelrys Softw. Inc.; Release: 2012. https://www.3dsbiovia.com/about/citations-references/ (Accelrys Software Inc., Discovery Studio Modeling Environment). [Google Scholar]
- 53.Khan F.A., Sharuk L., Siddiui Beta-sitosterol: as immunostimulant, antioxidant and inhibitor of SARS-CoV-2 spike glycoprotein. Arch. Pharmacol. Ther. 2020;2 doi: 10.33696/pharmacol.2.014. [DOI] [Google Scholar]
- 54.Khan S.L., Sonwane G.M., Siddiqui F.A., Jain S.P., Kale M.A., Borkar V.S. Discovery of naturally occurring flavonoids as human cytochrome P450 (CYP3A4) inhibitors with the aid of computational chemistry. Indo Glob. J. Pharm. Sci. 2020;10:58–69. doi: 10.35652/igjps.2020.10409. [DOI] [Google Scholar]
- 55.Khan A., Unnisa A., Sohel M., Date M., Panpaliya N., Saboo S.G., Siddiqui F., Khan S. Investigation of phytoconstituents of Enicostemma littorale as potential glucokinase activators through molecular docking for the treatment of type 2 diabetes mellitus. Silico Pharmacol. 2021;10 doi: 10.1007/s40203-021-00116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shntaif A.H., Khan S., Tapadiya G., Chettupalli A., Saboo S., Shaikh M.S., Siddiqui F., Amara R.R. Rational drug design, synthesis, and biological evaluation of novel N-(2-arylaminophenyl)-2,3-diphenylquinoxaline-6-sulfonamides as potential antimalarial, antifungal, and antibacterial agents. Digit. Chinese Med. 2021;4:290–304. doi: 10.1016/j.dcmed.2021.12.004. [DOI] [Google Scholar]
- 57.Chaudhari R.N., Khan S.L., Chaudhary R.S., Jain S.P., Siddiqui F.A. Β-sitosterol: isolation from Muntingia calabura linn bark extract, structural elucidation and molecular docking studies as potential inhibitor of SARS-CoV-2 mpro (COVID-19) Asian J. Pharm. Clin. Res. 2020;13:204–209. doi: 10.22159/ajpcr.2020.v13i5.37909. [DOI] [Google Scholar]
- 58.Khan S., Kale M., Siddiqui F., Nema N. Novel pyrimidine-benzimidazole hybrids with antibacterial and antifungal properties and potential inhibition of SARS-CoV-2 main protease and spike glycoprotein. Digit. Chinese Med. 2021;4:102–119. doi: 10.1016/j.dcmed.2021.06.004. [DOI] [Google Scholar]
- 59.Unnisa A., Khan S.L., Sheikh F.A.H., Mahefooz S., Kazi A.A., Siddiqui F.A., Gawai N., Saboo S.G. In-silico inhibitory potential of triphala constituents against cytochrome P450 2E1 for the prevention of thioacetamide-induced hepatotoxicity. J. Pharm. Res. Int. 2021:367–375. doi: 10.9734/jpri/2021/v33i43a32499. [DOI] [Google Scholar]
- 60.Khan S.L., Siddiqui F.A., Shaikh M.S., Nema N.V., Shaikh A.A. Discovery of potential inhibitors of the receptor-binding domain (RBD) of pandemic disease-causing SARS-CoV-2 Spike Glycoprotein from Triphala through molecular docking. Curr. Chinese Chem. 2021;1 doi: 10.2174/2666001601666210322121802. [DOI] [Google Scholar]
- 61.Khan S.L., Siddiqui F.A., Jain S.P., Sonwane G.M. Discovery of potential inhibitors of SARS-CoV-2 (COVID-19) main protease (mpro) from nigella sativa (black seed) by molecular docking study. Coronaviruses. 2020;2:384–402. doi: 10.2174/2666796701999200921094103. [DOI] [Google Scholar]
- 62.Siddiqui F.A., Khan S.L., Marathe R.P., Nema N.V. Design, synthesis, and in silico studies of novel N-(2-Aminophenyl)-2,3- diphenylquinoxaline-6-sulfonamide derivatives targeting receptor- binding domain (RBD) of SARS-CoV-2 spike glycoprotein and their evaluation as antimicrobial and antimalarial agents. Lett. Drug Des. Discov. 2021;18:915–931. doi: 10.2174/1570180818666210427095203. [DOI] [Google Scholar]
- 63.Shaw D.E. Schrödinger Release; New York: 2021. Schrödinger: Desmond Molecular Dynamics System; p. 4. [Google Scholar]
- 64.Dhawale S., Pandit M., Thete K., Ighe D., Gawale S., Bhosle P., Lokwani D.K. In silico approach towards polyphenols as targeting glucosamine-6-phosphate synthase for Candida albicans. J. Biomol. Struct. Dyn. 2023 doi: 10.1080/07391102.2022.2164797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.