Abstract
Background
Evidence showed that N6-methyladenosine (m6A) is strongly associated with male germline development. However, the role of m6A methylation on circRNAs in amphibians remains unknown. In this study, we conducted m6A sequencing analysis to explore the m6A transcriptome-wide profile of circRNAs in testis tissues of Xenopus laevis (X. laevis) with and without treatment with 100 µg/L atrazine (AZ).
Results
The analysis showed that m6A modification of circRNAs enriched in sense overlapping in testes of X. laevis. We identified the differential m6A modification sites within circRNAs in testes of AZ-exposed X. laevis and compared that with animals from control group. The results showed that a total of 1507 methylated m6A sites was induced by AZ (760 up-methylated and 747 down-methylated). The cross-analysis exhibited a negative correlation of differentially methylated m6A peaks and circRNAs expression level. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that 20 key pathways may be involved in the mechanism of testis damage of AZ-exposed X. laevis.
Conclusions
These findings indicated that differentially m6A-methylated circRNAs may play important roles in abnormal testis development of AZ-exposed X. laevis. This study is the first report about a map of m6A modification of circRNAs in male X. laevis and provides a basis for further studying on the function and mechanism of m6A methylation of circRNAs in the testis development of amphibian.
Supplementary Information
The online version contains supplementary material available at 10.1186/s41021-023-00279-0.
Keywords: RNA methylation, M6A, CircRNA, Amphibious, Atrazine
Introduction
N6-methyladenosine (m6A), as the most widespread post-transcriptional RNA modification, has attracted extensive attention in the field of epigenetics [1]. Emerging evidence suggested that m6A modifications played an important role in RNA metabolism, and its dynamic regulation was significantly correlated with gene expression [2]. Additionally, a previous study reported that knockout of RNA m6A regulators in the testis led to abnormal metabolism of the RNAs, which eventually caused spermatogenetic disorders and infertility [3]. Recently, it is reported that the process of spermatogenesis was influenced by gene regulation at transcriptional, post-transcriptional and epigenetic levels [4]. RNA m6A modification played an important role in spermatogenesis [5].
Circular RNAs (circRNAs) are a class of non-coding RNA with the length more than 200 nucleotides [6]. Its closed-loop structures lacking a 5’-cap structure and a 3’-poly (a) tail, were generated by back-splicing of linear mRNAs [7]. They were abundant, stable and highly conserved in evolution [8]. CircRNAs have pivotally important role in regulating gene expression by sequestering specific miRNAs like a sponge or buffering inhibition of mRNA targets [9]. At present, m6A modification has been identified in circRNAs, and most m6A circRNAs are expressed in a cell type-specific manner suggesting that m6A circRNAs play different biological functions in specific cell types [10]. For example, Xu et al. found that METTL3/FTO/YTHDF1/2-mediated m6A modification can enhance the stability of circRNA-SORE and induce sorafenib resistance in hepatocellular carcinoma (HCC) [11]. METTL3-mediated m6A methylation regulates the expression of circMETTL3 and promotes breast cancer cell proliferation and migration [12]. M6A modification improves the expression of circCUX1 and makes hypopharyngeal squamous cell carcinoma radioresistant [13]. Mettl14-mediated m6A methylation could also export circGFRa1 to cytoplasm from nucleus and promote female germline stem cells (FGSCs) self-renewal [14]. Tang et al. found that m6A modification modulates the biogenesis of a subset of circRNAs in male germ cells and further regulates spermatogenesis [15]. To our knowledge, the m6A transcriptome-wide map of circRNAs in amphibians remains to be explored.
Studies have found that environmental pollution could negatively affect amphibians’ health status and fertility, ultimately leading to a significant decrease in their population [16–18]. Atrazine (2-chloro-4-ethylamino-6-isopropylamino-s-triazine, AZ), an endocrine disrupting chemical (EDCs), is a highly effective triazine herbicide widely used in China, the USA and other countries [19]. It is often detected in groundwater, surface and drinking water [20]. It has been reported that AZ could cause metamorphosis of tadpoles and gonadal dysplasia of in several frog species [21–23], such as demasculinization and complete feminization of X. laevis [21]. This may be one of the factors causing global amphibian declines [24, 25]. In our previous study, we examined the damage of X. laevis exposed to AZ (0.1, 10, or 100 µg/L) for 90 days in the water environment. The results showed that AZ caused the decrease of gonad weight and gonad somatic index and the histological damage of testis in X. laevis [26]. In addition, we reported that m6A transcriptome-wide map of an amphibian species X. laevis. These findings provided clues to reveal the role of m6A which may affect amphibian testis development [27]. However, the mechanism of reproductive toxicity in male X. laevis chronically exposed to AZ remains unclear. Therefore, it is necessary to investigate the potential changes of m6A modification of circRNAs.
Thus, this study aimed to address whether changes in m6A modification of circRNAs are associated with abnormal testicular development in X. laevis chronically exposed AZ. We comprehensively analyzed the m6A modification profiles of circRNAs in normal and 100 µg/L AZ-exposed X. laevis for 180 days, respectively. Then, we predicted the signal pathways in which dysregulated m6A methylation of circRNAs involved by KEGG pathway analysis. Meanwhile, our work will provide a basis for further studying on the function and mechanism of m6A methylation of circRNAs in the abnormal testis development of X. laevis and also provide novel insights into its function and biological significance in amphibians.
Methods
Ethic approval
All animal experiments were carried out according to relevant guidelines and care regulations. All procedures complied with the “Principles of Animal Care”. The protocol was assessed and approved by the Committee on the Ethics of Animal Experiments of Shandong Academy of Occupational Health and Occupational Medicine [28].
Animal treatment
Three pairs of adult male and female X. laevis were purchased from the Chinese Academy of Sciences (Beijing, China). The offspring were generated by natural mating. Laboratory freshwater produced by UV treatment and carbon filtration was used for the acclimatization of frogs in the laboratory and for all subsequent exposures. The X. laevis were kept at an average water temperature of 22 ± 2 °C at pH 7.5, under 12 h light and 12 h dark cycle. Tadpoles were fed fairy shrimp (Artemia nauplii) eggs in a young age daily and pork liver three times per week ad libitum when the tadpoles completed metamorphosis [26].
At Nieuwkoop-Faber (NF) stage 47 (13 d post-hatch), mixed sex tadpoles from the same parents were discretionarily divided into two groups. One group was exposed to AZ at a dose of 100 µg/L (dissolved in solvent vehicle DMSO (0.01%)) for 180 days, and the other group was exposed to 0.01% DMSO only. Test solutions were renewed by 50% replacement every 48 h. The animals were monitored daily for health status and morphological changes. All of the females were excluded in this study. On day 180, the X. laevis in the control and AZ-exposed groups were sacrificed, respectively. The testes were isolated and weighed, and finally stored at − 80 °C for further analysis [8, 27].
CircRNAs preparation
Each group was recommended to include at least three biological replicates [29]. We randomly selected six testes (three from controls and three from 100 µg/L AZ-exposed groups) for circRNAs analysis. Total RNA from testes was extracted using Trizol reagent (Invitrogen Corporation, CA, USA) according to the manufacturer’s instruction. The concentration and purity of RNAs were measured using a NanoDrop® ND-2000 spectrophotometer (Thermo, Waltham, MA, USA). RNA integrity was examined by denaturing gel electrophoresis experiments [8]. RNA samples were further purified and converted to double-stranded cDNA for microarray analysis according to the Agilent ® Xenopus 4 × 44 k Gene Expression Microarray protocols.
CircRNAs m6A MeRIP sequencing
M6A of circRNAs were sequenced by MeRIP sequencing using Illumina HiSeq sequencer. In short, the fragmented RNA was incubated with anti-m6A polyclonal antibody (Synaptic Systems, Goettingen, Germany, 202,003) in IPP buffer for 2 h at 4 °C. Under the same conditions, the mixture was immune precipitated by incubation with protein-A beads (Thermo Fisher). Then, bound RNA was eluted and then extracted according to the manufacturer’s instructions. Purified RNA was used for RNA-seq library generation with NEBNext® Ultra™ RNA Library Prep Kit (NEB). Input samples without immune precipitation and m6A IP samples were subjected to 150 bp paired-end sequencing on Illumina HiSeq sequencer. Paired-end reads were obtained, and quality control was performed by Q30. More specific operations have been elucidated in our previous study [26, 27].
Data analysis
After sequencing, reads were trimmed with 3’ adaptor and low-quality readings were removed to produce clean reads with Cutadapt software (v1.9.3). Clean reads from all libraries were aligned to genome by bowtie 2 software [30]. Then circRNAs were detected and identified with find circ software [31], the circBase database [32] was also used to annotate the identified circRNAs, clean reads of all libraries were mapped to genome using hisat2 software (v2.04) [33]. Methylated sites on circRNAs were detected by MACS software [34], and differentially methylated sites of circRNAs were identified by diffReps [35]. In addition, the overlapping sites between the peaks, identified by both software based on the circRNAs exons were chosen for further analyzed. Pathway enrichment analysis of differentially methylated protein coding genes was performed through KEGG pathway database.
Results
AZ induced physiological changes of X. laevis
Compared with froglets in the control, the mortality of froglets significantly increased, the weight and gonadosomatic index (GSI) of the testis were significantly reduced, the metamorphosis time of tadpole was prolonged in AZ-exposed X. laevis. Histopathological results showed large empty spaces and irregular shape of seminiferous lobules in the testicular tissue. The number of germ cells in testicular tissue was significantly reduced. These results have been reported in our previous paper [26].
M6A sites within circRNAs in the testes of control and AZ-exposed X. laevis
The m6A transcriptome-wide profile of circRNAs were performed in testes in three biological replicates from the controls (n = 3) and AZ-exposed X. laevis (n = 3). We found that there were 1680 m6A peaks were shared in controls and AZ-exposed X. laevis, while 276 m6A peaks were identified in controls but absent in AZ-exposed X. laevis, and 1357 m6A peaks were identified in AZ-exposed X. laevis but absent in controls (Fig. 1).
Fig. 1.
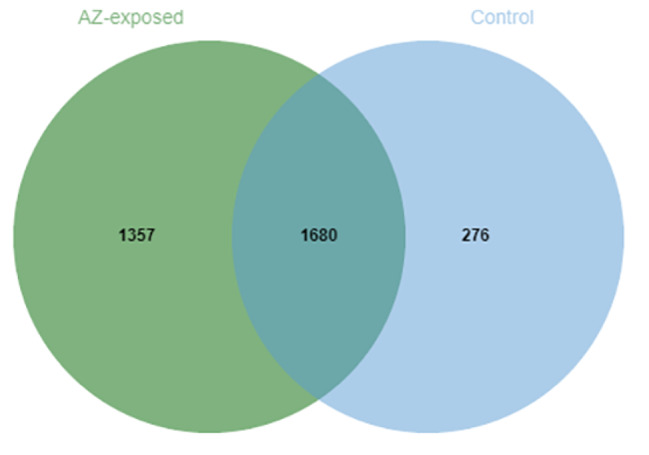
Venn diagram showing the overlap of m6A peaks within circRNAs in the testes of control and AZ-exposed X. laevis
Distribution of m6 A-methylated circRNAs in the testes of control and AZ-exposed X. laevis
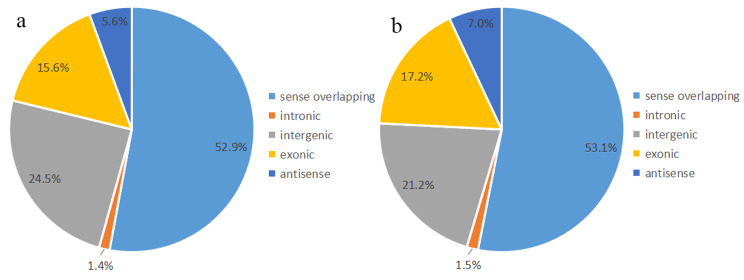
To further illustrate the distribution profiles of m6A-methylated circRNAs, we analyzed the genomic position of total m6A-methylated circRNAs in the testes of control and AZ-exposed X. laevis, respectively. We found that m6A-methylated circRNAs in both groups were divided into 5 groups: antisense, exonic, intergenic, intronic, sense overlapping. Particularly, we found that the most of m6A-methylated circRNAs were concentrated in sense overlapping (52.9% in control and 53.1% in AZ-exposed groups) (Fig. 2a and b).
Fig. 2.
Distribution of m6A-methylated circRNAs in the testes of control and AZ-exposed X. laevis a: Pie charts showing the percentage of the distribution positions of m6A-methylated circRNAs in control group. b: Pie charts showing the percentage of the distribution positions of m6A-methylated circRNAs in AZ-exposed group
Differentially m6 A modification sites of circRNAs in X. laevis exposed to 100 µg/L AZ
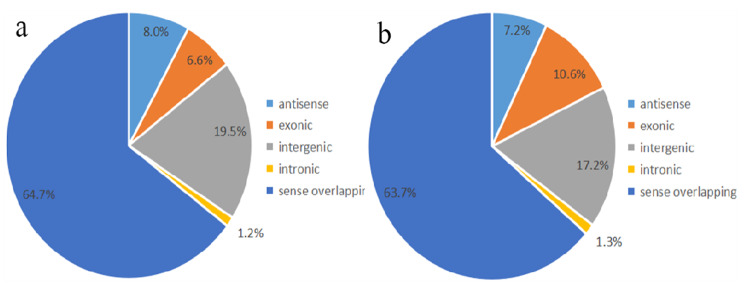
There were 1507 differentially methylated m6A sites were identified. Among them, 760 m6A methylated sites were significantly up-regulated, 747 m6A methylated sites were significantly down-regulated (Table S1 and Fig. 3). Tables 1 and 2 showed that top ten up-and down-methylated m6A sites of circRNAs with the highest fold change (FC) values. To further illustrate the distribution profiles of differentially methylated circRNAs, we found most significantly methylated circRNAs belonged to sense overlapping (Fig. 4a and b).
Fig. 3.
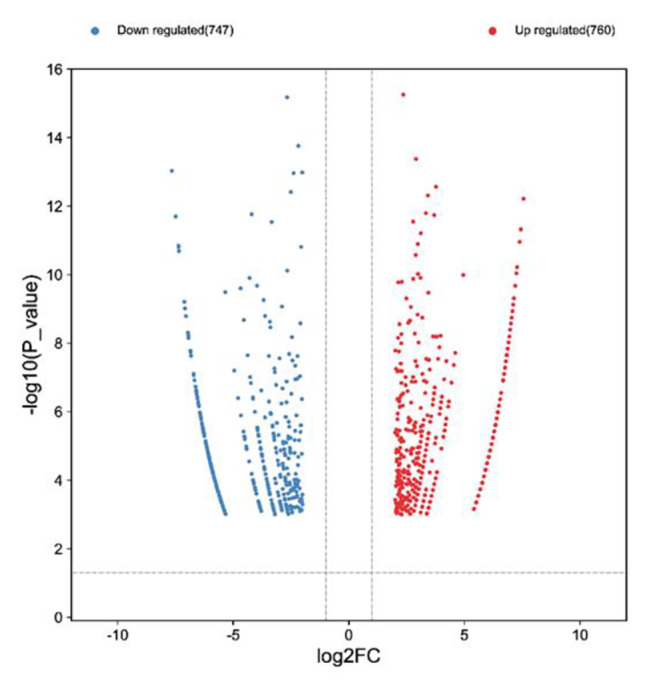
Volcano plots showing –log10 (P_value) versus log2FC in m6A methylated sites on circRNAs in the testes of control and AZ-exposed X. laevis. Red circles denote significantly up-regulated m6A methylated sites, whereas blue circles denote significantly down-regulated m6A methylated sites (p < 0.05 and fold change ≥ 4)
Table 1.
The top 10 up-methylated m6A sites of circRNAs
| chrom | txStart | txEnd | circRNA | Foldchange | Regulation |
|---|---|---|---|---|---|
| NC_030725.1 | 138,173,201 | 138,173,760 | NC_030725.1:138171762–138,186,798+ | 1047.5 | up |
| NC_030730.1 | 60,836,021 | 60,836,560 | NC_030730.1:60770414–60,848,459+ | 544.8 | up |
| NC_030741.1 | 38,394,381 | 38,394,940 | NC_030741.1:38391889–38,396,199+ | 428.8 | up |
| NC_030734.1 | 51,326,541 | 51,327,120 | NC_030734.1:51319661-51329727- | 188 | up |
| NC_030729.1 | 14,574,441 | 14,575,060 | NC_030729.1:14566445-14575976- | 187.9 | up |
| NC_030741.1 | 94,059,881 | 94,060,280 | NC_030741.1:94040702-94069184- | 173.4 | up |
| NC_030732.1 | 110,818,421 | 110,818,980 | NC_030732.1:110818083-110819953- | 167.4 | up |
| NC_030732.1 | 59,329,921 | 59,330,840 | NC_030732.1:59317677–59,337,960+ | 155.5 | up |
| NC_030738.1 | 116,486,141 | 116,487,120 | NC_030738.1:116446394-116502224- | 152.5 | up |
| NC_030741.1 | 20,961,581 | 20,962,140 | NC_030741.1:20954726–20,967,910+ | 146.6 | up |
txStart/txEnd: Start/end position of the differentially methylated RNA sites
Table 2.
The top 10 down-methylated m6A sites of circRNAs
| chrom | txStart | txEnd | circRNA | Foldchange | Regulation |
|---|---|---|---|---|---|
| NC_030741.1 | 26,736,741 | 26,737,300 | NC_030741.1:26724629–26,775,531+ | 3995.5 | down |
| NC_030726.1 | 38,143,141 | 38,143,700 | NC_030726.1:38131187-38150843- | 201.3 | down |
| NC_030737.1 | 3,714,221 | 3,714,760 | NC_030737.1:3707843-3738590- | 179.5 | down |
| NC_030728.1 | 135,118,701 | 135,119,340 | NC_030728.1:135115660–135,120,836+ | 165.5 | down |
| NC_030724.1 | 83,840,521 | 83,841,540 | NC_030724.1:83828634-83844198- | 165 | down |
| NC_030738.1 | 83,071,141 | 83,071,680 | NC_030738.1:83069676–83,126,138+ | 163.1 | down |
| NC_030738.1 | 11,481,761 | 11,482,340 | NC_030738.1:11462943–11,533,553+ | 150.7 | down |
| NC_030739.1 | 19,273,999 | 19,274,106 | NC_030739.1:19272571–19,278,583+ | 138.9 | down |
| NC_030724.1 | 152,774,701 | 152,776,080 | NC_030724.1:152754234–152,779,042+ | 135.9 | down |
| NC_030731.1 | 115,186,221 | 115,187,200 | NC_030731.1:115185766–115,230,884+ | 132.3 | down |
txStart/txEnd: Start/end position of the differentially methylated RNA sites
Fig. 4.
Overview the distribution of differentially m6A modification sites of circRNAs. a: Pie charts showing the percentage of up-methylated m6A in five segments. b: Pie charts showing the percentage of down-methylated m6A peaks in five segments
Conjoint analysis of m6 A-Seq and RNA-Seq Data of control and AZ-exposed X. laevis
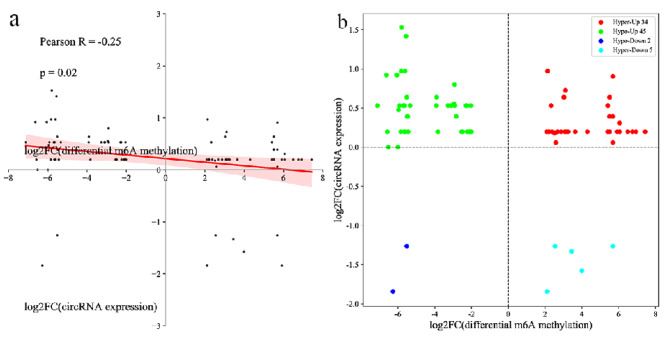
To elucidate the potential relationship between m6A methylation and gene expression, we performed a cross-analysis of m6A-seq and RNA-seq data. As shown in Table S2, in the case of combined analysis of differentially methylated m6A sites and differentially expressed circRNAs, circRNA“NC_030733.1:68560883-68632668-”,“NC_030737.1:31865134–31952348”, “NC_030737.1:83574596–83583489+”,“NC_030733.1:34483227-34511810-”,“NC_030733.1:68560883-68632668-”, “NC_030737.1:31865134-31952348-” were down-regulated in the testis of AZ-exposed X. laevis, but the m6A modification of which were up-regulated. However, circRNA “NC_030738.1:21126033–21131815+” was up-regulated in the testis of AZ-treated X. laevis, but the m6A modification of which was down-regulated. The result showed that differentially methylated m6A modification may negatively regulated the expression of circRNAs. Additionally, in the case of combined analysis of differentially methylated m6A sites and total circRNAs, we discovered a negative correlation of differentially methylated m6A peaks and circRNAs expression level (P = 0.02; Pearson R = -0.25) (Fig. 5a). The results of the four-quadrant diagram analysis showed that 760 hyper-methylated m6A sites were positive correlated with 34 up-regulated circRNAs transcripts, namely ‘hyper-up’, but 5 genes were found with down-regulated circRNAs transcripts, namely ‘hyper-down’. In the contrast, in 747 hypo-methylated m6A sites, we found 45 genes with up-regulated circRNAs transcripts and 2 genes with down-regulated circRNAs transcripts, named ‘hypo-up’ and ‘hypo-down’, respectively (Fig. 5b). Notably, we found that 57.0% (45/79) of the up-regulated circRNAs transcripts were associated with hypo-methylated m6A peaks. Meanwhile, the numbers of ‘hyper-down’ and ‘hypo-up’ genes were more than those of ‘hyper-up’ and ‘hypo-down’ genes. It indicated that m6A modifications may tend to be negatively correlated with circRNAs expression in the testes of X. laevis exposed to 100 µg/L AZ.
Fig. 5.
a: Dot plot of log2FC (circRNAs expression) against log2FC (differential m6A methylation) showing a negative correlation between overall m6A methylation and mRNA expression level (P = 0.02; Pearson R = -0.25). b: Four quadrant plots showing gene expression with differentially methylated m6A peaks
The enrichment pathways of genes related differentially m6A-methylated circRNAs by KEGG
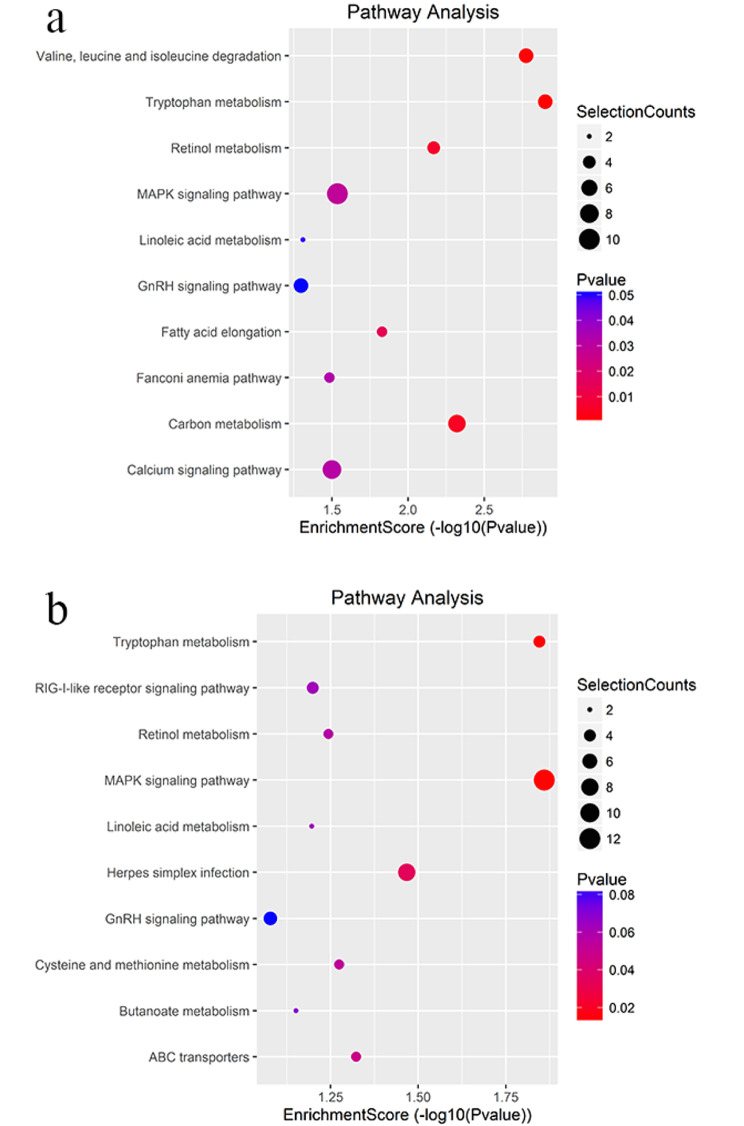
To explore the effect of differential m6A-methylated circRNAs in testis of AZ-exposed X. laevis. KEGG pathway analysis were performed for target genes related to the modified circRNAs. The result revealed that target genes related to up-methylated circRNAs were mainly enriched in ten pathways, such as Valine, Leucine and isoleucine degradation, Tryptophan metabolism, Retinol metabolism, MAPK signaling pathway, Linoleic acid metabolism, GnRH signaling pathway, Fatty acid elongation, Fanconi anemia pathway, Carbon metabolism and Calcium signaling pathway (Fig. 6a). Meanwhile, target genes related to down-methylated circRNAs were significantly involved in Tryptophan metabolism, RIG-I-Like receptor signaling pathway, Retinol metabolism, MAPK signaling pathway, Linoleic acid metabolism, Herpes simplex infection, GnRH signaling pathway, Cysteine and methionine metabolism, Butanoate metabolism and ABC transporters (Fig. 6b).
Fig. 6.
The annotated significant pathways targeted by the enrichment score of the differentially m6A-methylated circRNAs-related genes including up-methylated (a) and down-methylated (b) in testis of X. laevis exposed to 100 µg/L AZ. The horizontal axis is the -log10 (P-value) for the pathway and the vertical axis is the pathway category
Discussion
Studies have shown that AZ could cause histological damage of the testis, reduced sperm count and generate abnormal sperm [36]. In addition, X. laevis exposed to AZ also showed damaged germ cell and reduced testicular weight [37]. These results were consistent with the previous results of our study [26].
M6A methylation has been shown to be one of the most abundant internal modifications of RNA in higher eukaryotes [38, 39]. It was characterized by dynamic reversibility, wide spready and unique patterns [10]. Emerging evidence have suggested that m6A methylation played important biological functions in RNA modification, tumorigenesis, fat metabolism, reproductive development, and so on [39, 40]. And the role of m6A circRNAs has also been gradually elucidated. Hence, we explored m6A transcriptome-wide map of circRNAs identified in the testis of normal and AZ-treated X. laevis, to predict the role of m6A modification of circRNAs in abnormal testicular development. In this study, we investigated the changes of m6A modification of circRNAs in the testis of X. laevis by exogenous environmental stimulation. Our findings showed that the number of m6A sites within circRNAs increased in the testis of AZ-exposed X. laevis compared with that of controls. The majority of m6A methylated circRNAs and differential m6A methylated ones enriched in sense overlapping. Interestingly, studies have found that the back splicing tends to occur predominantly in m6A-enriched sites in male germ cells. The change allows the most circRNAs contain large open reading frames to ensure long-term and stable protein production for specific physiological processes in the absence of corresponding linear mRNA [15, 41]. And aberrant m6A methylation may affect the abnormal spermatogenesis [42]. Therefore, we speculated that m6A modification enriched in sense overlapping may play an important role in abnormal development of testis in amphibian species.
To further explore the potential role of m6A modification in the testes of AZ-exposed X. laevis, we performed combined analysis of m6A-Seq and RNA-Seq. Results revealed that a negative correlation between differentially methylated peaks and circRNAs expression levels. It is reported that circRNAs were characterized by covalently closed structure and resistance to exonucleases, but m6A modification could induce the degradation process of circRNAs by regulating their stability [29]. Park et al. found that m6A-methylated circRNAs were cleaved by endoribonucleases via a YTHDF2-HRSP12-RNase P/MRP axis [43]. This may affect the biological function of circRNAs [44], such as m6A modification of circNSUN2 could promote the liver metastasis of colorectal cancer by forming a circNSUN2/IGF2BP2/HMGA2 RNA-protein ternary complex [12]. Additionally, it has been reported that the specific role of m6A modification on gene expression largely depended on the type of m6A ‘readers’ [45]. Hence, we speculated that m6A modification in circRNAs may play a potential role in the abnormal testicular development of AZ-exposed X. laevis. In the present study, we only examined circRNAs expression, more functional experiments will be needed to further verify the regulation role of m6A modification on gene expression.
We further analyzed the role of differentially m6A-methylated circRNAs related target genes in AZ-treated X. laevis for which 20 key pathways were obtained by KEGG pathway annotation method. In this study, the most GeneRatio term was “MAPK signaling pathway”. Studies have shown that some exogenous stimulants could activate MAPK signaling pathway, affected Sertoli cells (SCs) proliferation and blood-testis barrier (BTB) structure [46, 47]. Interestingly, In the process of spermatogenesis, SCs not only provided mechanical and nutritional support [48, 49], but also protected germ cells by forming an immune protective environment through the BTB [50, 51]. Therefore, we assumed that MAPK signaling pathway regulated by circRNAs with m6A modifications might mediate the abnormal testis development of AZ-exposed X. laevis.
“GnRH signaling pathway” was also included in the result of KEGG. GnRH, as a local bioregulator, controlled the secretion of gonadotropin of the pituitary gland, including follicle stimulating hormone and luteinizing hormone, which were critical components affecting sperm development [52]. In addition, it was reported that GnRH could also be synthesized in seminiferous tubules, and as a paracrine mediator of spermatogenesis [53]. In GnRH deficient males, there would be incomplete testicular growth and maturation, cryptorchidism, androgen deficiency and other symptoms [54]. Therefore, m6A-methylation of circRNAs involved in “GnRH signaling pathway” may play an important role in the growing development and functional activity of testis of AZ-exposed X. laevis.
In addition, it has been shown that Ca2+, which was produced by the Calcium Signaling Pathway, could directly control metabolism, secretion, fertilization, proliferation and other processes [55]. And it also participated in the cAMP-induced steroidogenesis in Leydig cells [56]. Therefore, we speculated that “Calcium Signaling Pathway” regulated by m6A-methylated circRNAs may involve testicular dysplasia of AZ-exposed X. laevis. Most of these pathways in the result were related to metabolism such as “Linoleic acid metabolism” and “Fatty acid elongation” signaling pathways. Fatty acid (FA) mainly included monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) [57], while Linoleic acid were essential PUFA [58]. Its synthesis was mediated by rate-limiting elongation and desaturation enzymes, which were regulated by fatty acid desaturase 2 (FADS2) and elongation of very-long-chain fatty acids-like 2 (ELOVL2) [58]. It has been found that arrested spermatogenesis and lacked mature spermatozoa in the testis of mice with FADS2 and ELOVL2 gene knockout [59, 60]. Additionally, it was reported that the FA metabolic disorders could occur in infertile men [57]. Hence, the role of m6A-methylated circRNAs involved in “Linoleic acid metabolism” and “Fatty acid elongation” signaling pathways deserve further study in male reproductive damage. Consequently, the results predicted that m6A methylation of circRNAs may regulate the differential expression of these target genes in abnormal testis development of X. laevis exposed to 100 µg/L AZ.
Conclusion
We detected the m6A transcriptome-wide profile of circRNAs in the testes of control and AZ-exposed X. laevis. The results showed that AZ could alter expression profile in 1507 m6A methylated peaks within circRNAs in which 760 were significantly up-methylated and 747 significantly down-methylated, and they mainly enriched in sense overlapping. Conjoint analysis indicated that a negative correlation of differentially methylated m6A peaks and circRNAs expression level, suggesting a regulatory role of m6A modification of circRNAs in amphibious gene expression. KEGG pathway analysis revealed that m6A related to “MAPK signaling pathway”, “GnRH signaling pathway”, “Calcium signaling pathway” and pathways related to metabolism such as “Linoleic acid metabolism” and “Fatty acid elongation” maybe play a pivotal role in the abnormal testis development of AZ-exposed X. laevis. Our study provided a basis for further studies on the function and mechanism of m6A methylation of circRNAs in the abnormal testis development of X. laevis. Meanwhile, this study presented the first m6A transcriptome-wide map of circRNAs in amphibian species X. laevis. This may help to further understand the role of m6A methylation in testis development and spermatogenesis in amphibians.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely appreciate Cloud-seq Biotech Ltd. Co. (Shanghai, China) and Yuxing Medtech Ltd. Co. (Ji’nan, China) who provided high throughput sequencing instrument.
Abbreviations
- M6A
N6-methyladenosine
- X. laevis
Xenopus laevis
- AZ
Atrazine
- circRNAs
Circular RNAs
- EDCs
Endocrine disrupting chemicals
- FC
Fold change
- HCC
Hepatocellular carcinoma
- FGSCs
Female germline stem cells
- GSI
Gonadosomatic index
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-activated protein kinases
- SCs
Sertoli cells
- BTB
Blood-testis barrier
- GnRH
Gonadotropin-releasing hormone
- FA
Fatty acid
- MUFA
Monounsaturated fatty acids
- PUFA
Polyunsaturated fatty acids
- FADS2
Fatty acid desaturase 2
- ELOVL2
Elongation of very-long-chain fatty acids-like 2
Authors’ Contributions
SL and HM conceived and supervised the study; SH and JQ designed experiments; YW and BC conducted the experiments; ZW and KX prepared all figures and tables; ZX wrote the main manuscript text; SL and PC analyzed the data and revised the manuscript. All authors reviewed the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by the Health Commission of Shandong Province (202012010629; 202112010222; M-20222055), Ji’nan Science and Technology Bureau (202019108; 202019109), National Natural Science Foundation of China (81573198; 30901214), the Innovation Project of Shandong Academy of Medical Sciences, Academic Promotion Programme of Shandong First Medical University (2019QL001), Ministry of Science and Technology of PRC (2018ZX09711001–011).
Data Availability
All data generated and analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
The protocol was approved by the Committee on the Ethics of Animal Experiments of Shandong Academy of Occupational Health and Occupational Medicine.
Consent for publication
All authors have approved the publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xin Zhang and Linlin Sai contributed equally to this work.
References
- 1.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteom Bioinf. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Z, Niu Y, Li H. RNA N6-methyladenosine modification, spermatogenesis, and human male infertility. Mol Hum Reprod. 2021;27(6):gaab020. doi: 10.1093/molehr/gaab020. [DOI] [PubMed] [Google Scholar]
- 4.Yu Z, Guo R, Ge Y, Ma J, Guan J, Li S, et al. Gene expression profiles in different stages of mouse spermatogenic cells during spermatogenesis. Biol Reprod. 2003;69(1):37–47. doi: 10.1095/biolreprod.102.012609. [DOI] [PubMed] [Google Scholar]
- 5.Zhao TX, Wang JK, Shen LJ, Long CL, Liu B, Wei Y, et al. Increased m6A RNA modification is related to the inhibition of the Nrf2-mediated antioxidant response in di-(2-ethylhexyl) phthalate-induced prepubertal testicular injury. Environ Pollut. 2020;259:113911. doi: 10.1016/j.envpol.2020.113911. [DOI] [PubMed] [Google Scholar]
- 6.Sun A, Wang R, Yang S, Zhu X, Liu Y, Teng M, et al. Comprehensive profiling analysis of the N6-methyladenosine-modified circular RNA transcriptome in cultured cells infected with Marek’s disease virus. Sci Rep. 2021;11(1):11084. doi: 10.1038/s41598-021-90548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Tang J, Su Q, Chou WC, Zheng F, Guo Z, et al. Paraquat-induced oxidative stress regulates N6-methyladenosine (m6A) modification of circular RNAs. Environ Pollut. 2021;290:117816. doi: 10.1016/j.envpol.2021.117816. [DOI] [PubMed] [Google Scholar]
- 8.Sai L, Li L, Hu C, Qu B, Guo Q, Jia Q, et al. Identification of circular RNAs and their alterations involved in developing male Xenopus laevis chronically exposed to atrazine. Chemosphere. 2018;200:295–301. doi: 10.1016/j.chemosphere.2018.02.140. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Wu M, Fan Y, Li S, Lai Z, Huang Y, et al. Identification and characterization of circular RNAs in Qinchuan cattle testis. R Soc Open Sci. 2018;5(7):180413. doi: 10.1098/rsos.180413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu T, He B, Sun H, Xiong M, Nie J, Wang S, et al. Novel insights into the interaction between N6-methyladenosine modification and circular RNA. Mol Ther Nucleic Acids. 2022;27:824–37. doi: 10.1016/j.omtn.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19(1):163. doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Yang HY, Dai XY, Zhang X, Huang YZ, Shi L, et al. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int J Biol Sci. 2021;17(5):1178–90. doi: 10.7150/ijbs.57783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12(4):298. doi: 10.1038/s41419-021-03558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Tian G, Wu J. Novel circGFRα1 promotes Self-Renewal of female germline stem cells mediated by m6A writer METTL14. Front Cell Dev Biol. 2021;9:640402. doi: 10.3389/fcell.2021.640402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30(3):211–28. doi: 10.1038/s41422-020-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu NC, Seebacher F. Effect of the plastic pollutant bisphenol A on the biology of aquatic organisms: a meta-analysis. Glob Chang Biol. 2020;26(7):3821–33. doi: 10.1111/gcb.15127. [DOI] [PubMed] [Google Scholar]
- 17.Orton F, Säfholm M, Jansson E, Carlsson Y, Eriksson A, Fick J, et al. Exposure to an anti-androgenic herbicide negatively impacts reproductive physiology and fertility in Xenopus tropicalis. Sci Rep. 2018;8(1):9124. doi: 10.1038/s41598-018-27161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svanholm S, Säfholm M, Brande-Lavridsen N, Larsson E, Berg C. Developmental reproductive toxicity and endocrine activity of propiconazole in the Xenopus tropicalis model. Sci Total Environ. 2021;753:141940. doi: 10.1016/j.scitotenv.2020.141940. [DOI] [PubMed] [Google Scholar]
- 19.de Albuquerque FP, de Oliveira JL, Moschini-Carlos V, Fraceto LF. An overview of the potential impacts of atrazine in aquatic environments: perspectives for tailored solutions based on nanotechnology. Sci Total Environ. 2020;700:134868. doi: 10.1016/j.scitotenv.2019.134868. [DOI] [PubMed] [Google Scholar]
- 20.Solomon KR, Giesy JP, LaPoint TW, Giddings JM, Richards RP. Ecological risk assessment of atrazine in north american surface waters. Environ Toxicol Chem. 2013;32(1):10–1. doi: 10.1002/etc.2050. [DOI] [PubMed] [Google Scholar]
- 21.Hayes TB, Khoury V, Narayan A, Nazir M, Park A, Brown T, et al. Atrazine induces complete feminization and chemical castration in male african clawed frogs (Xenopus laevis) Proc Natl Acad Sci U S A. 2010;107(10):4612–7. doi: 10.1073/pnas.0909519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodeur JC, Sassone A, Hermida GN, Codugnello N. Environmentally-relevant concentrations of atrazine induce non-monotonic acceleration of developmental rate and increased size at metamorphosis in Rhinella arenarum tadpoles. Ecotoxicol Environ Saf. 2013;92:10–7. doi: 10.1016/j.ecoenv.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 23.LaFiandra EM, Babbitt KJ, Sower SA. Effects of atrazine on anuran development are altered by the presence of a nonlethal predator. J Toxicol Environ Health A. 2008;71(8):505–11. doi: 10.1080/15287390801907442. [DOI] [PubMed] [Google Scholar]
- 24.Hayes TB, Collins A, Lee M, Mendoza M, Noriega N, Stuart AA, et al. Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Proc Natl Acad Sci U S A. 2002;99(8):5476–80. doi: 10.1073/pnas.082121499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes T, Haston K, Tsui M, Hoang A, Haeffele C, Vonk A. Atrazine-induced hermaphroditism at 0.1 ppb in american leopard frogs (Rana pipiens): laboratory and field evidence. Environ Health Perspect. 2003;111(4):568–75. doi: 10.1289/ehp.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sai L, Dong Z, Li L, Guo Q, Jia Q, Xie L, et al. Gene expression profiles in testis of developing male Xenopus laevis damaged by chronic exposure of atrazine. Chemosphere. 2016;159:145–52. doi: 10.1016/j.chemosphere.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Sai L, Li Y, Zhang Y, Zhang J, Qu B, Guo Q, et al. Distinct m6A methylome profiles in poly(A) RNA from Xenopus laevis testis and that treated with atrazine. Chemosphere. 2020;245:125631. doi: 10.1016/j.chemosphere.2019.125631. [DOI] [PubMed] [Google Scholar]
- 28.Qi X, Geng X, Zhang J, Qu B, Zhang X, Jia Q, et al. Comprehensive analysis of differences of N6-methyladenosine of lncRNAs between atrazine-induced and normal Xenopus laevis testis. Genes Environ. 2021;43(1):49. doi: 10.1186/s41021-021-00223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Transcriptome-wide mapping of N6-Methyladenosine by m6A-Seq. Methods Enzymol. 2015;560:131–47. doi: 10.1016/bs.mie.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 32.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8(6):e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu S, Zhang T, Wang Y, Zhou X, Wang S, Wang Z. Meta-analysis and experimental validation identified atrazine as a toxicant in the male reproductive system. Environ Sci Pollut Res Int. 2021;28(28):37482–97. doi: 10.1007/s11356-021-13396-6. [DOI] [PubMed] [Google Scholar]
- 37.Orton F, Carr JA, Handy RD. Effects of nitrate and atrazine on larval development and sexual differentiation in the northern leopard frog Rana pipiens. Environ Toxicol Chem. 2006;25(1):65–71. doi: 10.1897/05-136r.1. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Wang J, Geng X, Tu J, Gao H, Li L, et al. Circular RNA expression profile and m6A modification analysis in poorly differentiated adenocarcinoma of the stomach. Epigenomics. 2020;12(12):1027–40. doi: 10.2217/epi-2019-0153. [DOI] [PubMed] [Google Scholar]
- 40.Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16(11):1929–40. doi: 10.7150/ijbs.45231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Yuan XH, Jiang W, Lu YC, Huang QL, Yang Y, et al. Genome-wide map of N6-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis. World J Gastroenterol. 2021;27(43):7530–45. doi: 10.3748/wjg.v27.i43.7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong D, Zhang L, Huang K, Chen M, Chen Y, Liu Q et al. circRNA-miRNA-mRNA network analysis to explore the pathogenesis of abnormal spermatogenesis due to aberrant m6A methylation. Cell Tissue Res. 2023. [DOI] [PubMed]
- 43.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m6A-Containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74(3):494–507e498. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan L, Chen YG. Circular RNAs in Immune Response and viral infection. Trends Biochem Sci. 2020;45(12):1022–34. doi: 10.1016/j.tibs.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu T, Ling C, Hu M, Meng X, Deng Y, An H, et al. Effect of Nano-Titanium Dioxide on blood-testis barrier and MAPK signaling pathway in male mice. Biol Trace Elem Res. 2021;199(8):2961–71. doi: 10.1007/s12011-020-02404-4. [DOI] [PubMed] [Google Scholar]
- 47.Ni FD, Hao SL, Yang WX. Multiple signaling pathways in sertoli cells: recent findings in spermatogenesis. Cell Death Dis. 2019;10(8):541. doi: 10.1038/s41419-019-1782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crisostomo L, Alves MG, Gorga A, Sousa M, Riera MF, Galardo MN, et al. Molecular mechanisms and signaling pathways involved in the Nutritional support of spermatogenesis by sertoli cells. Methods Mol Biol. 2018;1748:129–55. doi: 10.1007/978-1-4939-7698-0_11. [DOI] [PubMed] [Google Scholar]
- 49.Yin J, Ni B, Tian ZQ, Yang F, Liao WG, Gao YQ. Regulatory effects of autophagy on spermatogenesis. Biol Reprod. 2017;96(3):525–30. doi: 10.1095/biolreprod.116.144063. [DOI] [PubMed] [Google Scholar]
- 50.Wang XX, Ying P, Diao F, Wang Q, Ye D, Jiang C, et al. Altered protein prenylation in sertoli cells is associated with adult infertility resulting from childhood mumps infection. J Exp Med. 2013;210(8):1559–74. doi: 10.1084/jem.20121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur G, Thompson LA, Dufour JM. Sertoli cells–immunological sentinels of spermatogenesis. Semin Cell Dev Biol. 2014;30:36–44. doi: 10.1016/j.semcdb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao F, Zou Y, Li H, Zhang Y, Liu X, Zhao X, et al. Decreased angiotensin receptor 1 expression in +/- AT1 knockout mice testis results in male infertility and GnRH reduction. Reprod Biol Endocrinol. 2021;19(1):120. doi: 10.1186/s12958-021-00805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chianese R, Ciaramella V, Scarpa D, Fasano S, Pierantoni R, Meccariello R. Anandamide regulates the expression of GnRH1, GnRH2, and GnRH-Rs in frog testis. Am J Physiol Endocrinol Metab. 2012;303(4):E475–487. doi: 10.1152/ajpendo.00086.2012. [DOI] [PubMed] [Google Scholar]
- 54.Adams TE. Using gonadotropin-releasing hormone (GnRH) and GnRH analogs to modulate testis function and enhance the productivity of domestic animals. Anim Reprod Sci. 2005;88(1–2):127–39. doi: 10.1016/j.anireprosci.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Berridge MJ. The Inositol Trisphosphate/Calcium Signaling Pathway in Health and Disease. Physiol Rev. 2016;96(4):1261–96. doi: 10.1152/physrev.00006.2016. [DOI] [PubMed] [Google Scholar]
- 56.Abdou HS, Villeneuve G, Tremblay JJ. The calcium signaling pathway regulates leydig cell steroidogenesis through a transcriptional cascade involving the nuclear receptor NR4A1 and the steroidogenic acute regulatory protein. Endocrinology. 2013;154(1):511–20. doi: 10.1210/en.2012-1767. [DOI] [PubMed] [Google Scholar]
- 57.Collodel G, Moretti E, Noto D, Iacoponi F, Signorini C. Fatty Acid Profile and Metabolism Are Related to Human Sperm Parameters and Are Relevant in Idiopathic Infertility and Varicocele. Mediators Inflamm. 2020; 2020:3640450. [DOI] [PMC free article] [PubMed]
- 58.Sun C, Zou M, Wang X, Xia W, Ma Y, Liang S, et al. FADS1-FADS2 and ELOVL2 gene polymorphisms in susceptibility to autism spectrum disorders in chinese children. BMC Psychiatry. 2018;18(1):283. doi: 10.1186/s12888-018-1868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zadravec D, Tvrdik P, Guillou H, Haslam R, Kobayashi T, Napier JA, et al. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J Lipid Res. 2011;52(2):245–55. doi: 10.1194/jlr.M011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroud CK, Nara TY, Roqueta-Rivera M, Radlowski EC, Lawrence P, Zhang Y, et al. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J Lipid Res. 2009;50(9):1870–80. doi: 10.1194/jlr.M900039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this published article and its supplementary information files.