DELLA proteins coordinate with the chromatin-remodeling complex subunit BRAHMA and NF-YC transcription factors to regulate flowering time in Arabidopsis by forming a gibberellin-sensitive module.
Abstract
Gibberellin (GA) plays a key role in floral induction by activating the expression of floral integrator genes in plants, but the epigenetic regulatory mechanisms underlying this process remain unclear. Here, we show that BRAHMA (BRM), a core subunit of the chromatin-remodeling SWItch/sucrose nonfermentable (SWI/SNF) complex that functions in various biological processes by regulating gene expression, is involved in GA-signaling-mediated flowering via the formation of the DELLA–BRM–NF-YC module in Arabidopsis (Arabidopsis thaliana). DELLA, BRM, and NF-YC transcription factors interact with one another, and DELLA proteins promote the physical interaction between BRM and NF-YC proteins. This impairs the binding of NF-YCs to SOC1, a major floral integrator gene, to inhibit flowering. On the other hand, DELLA proteins also facilitate the binding of BRM to SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1). The GA-induced degradation of DELLA proteins disturbs the DELLA–BRM–NF-YC module, prevents BRM from inhibiting NF-YCs, and decreases the DNA-binding ability of BRM, which promote the deposition of H3K4me3 on SOC1 chromatin, leading to early flowering. Collectively, our findings show that BRM is a key epigenetic partner of DELLA proteins during the floral transition. Moreover, they provide molecular insights into how GA signaling coordinates an epigenetic factor with a transcription factor to regulate the expression of a flowering gene and flowering in plants.
IN A NUTSHELL.
Background: The timing of floral induction is tightly controlled by environmental cues and intrinsic signals. The critical role of gibberellin (GA) in this process has been extensively studied in past decades. DELLA proteins serve as the central regulatory hubs of GA signaling. The mechanism of GA-dependent transcription involves the recruitment of DELLA proteins to transcription factors. In general, epigenetic modifiers are believed to cooperate with transcription factors to regulate gene expression, but how epigenetic regulation participates in GA-dependent transcription of the floral integrator genes in plants remains unclear.
Question: Which epigenetic modifiers participate in GA-dependent transcription of the floral integrator genes? What is the detailed molecular mechanism involved in this process?
Findings: BRAHMA (BRM), a core catalytic subunit of the SWI/SNF-type chromatin-remodeling complex, is involved in GA-signaling-mediated flowering via the formation of the DELLA–BRM–NF-YC module in Arabidopsis. DELLA proteins promote the interaction of BRM with the transcription factor NUCLEAR FACTOR Y-C (NF-YC), impairing the binding of NF-YC to the floral integrator gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), resulting in late flowering. Meanwhile, DELLA proteins accelerate the binding of BRM to SOC1. In the presence of GA, GA-triggered DELLA degradation disturbs the DELLA–BRM–NF-YC module and the H3K4me3 level at SOC1 chromatin increases, resulting in higher gene expression and early flowering.
Next steps: We will investigate whether the BRM–NF-Y module is also responsive to other phytohormone signals and how BRM integrates different phytohormone signals during plant development.
Introduction
In flowering plants, the precise transition from vegetative growth to reproductive development is crucial for successful propagation. The timing of this transition is tightly controlled by environmental cues and intrinsic signals. Six flowering pathways have been identified in Arabidopsis (Arabidopsis thaliana) via various genetic and molecular biological studies: the photoperiod, vernalization, thermosensory, autonomous, gibberellin (GA), and age pathways (Michaels 2009; Amasino 2010; Li et al. 2016a; Bao et al. 2020). These pathways converge to regulate the expression of various floral integrator genes, including FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and LEAFY (LFY), which subsequently activate several downstream floral meristem identity genes, such as LFY, APETALA1 (AP1), and FRUITFULL (FUL), to initiate the formation of floral meristems (Kardailsky et al. 1999; Blazquez and Weigel 2000; Abe et al. 2005; Wigge et al. 2005; Lee and Lee 2010).
The crucial role of GA in floral induction in Arabidopsis has been extensively studied in the past 2 decades (Wilson et al. 1992; Griffiths et al. 2006; Porri et al. 2012; Wang et al. 2016). Prior to floral initiation, the levels of bioactive GAs increase, which promotes flowering by activating the expression of SOC1 and LFY in the shoot apex (Eriksson et al. 2006). Bioactive GAs can bind to their receptor GA-INSENSITIVE DWARF 1 (GID1) and, in turn, recruit DELLA proteins (DELLAs) for ubiquitination and degradation. This process, which is mediated by the F-box type E3 ubiquitin ligase SLEEPY 1 (SLY1), modulates plant responses to GAs (Dill et al. 2004; Ueguchi-Tanaka et al. 2005; Willige et al. 2007; Murase et al. 2008). DELLA proteins play pivotal negative roles in the GA signal transduction pathway. Arabidopsis contains 5 DELLAs: REPRESSOR OF ga1-3 (RGA), GA-INSENSITIVE (GAI), RGA-LIKE1 (RGL1), RGL2, and RGL3 (Sun and Gubler 2004). DELLAs can interact with epigenetic factors, such as SWI3C and PICKLE (Sarnowska et al. 2013; Zhang et al. 2014; Park et al. 2017), suggesting a connection between GA signaling and the epigenetic regulatory mechanisms. In addition, DELLAs can mediate transcriptional control to repress flowering by interacting with many transcription factors, such as CONSTANS (CO), MYC3, FLOWERING LOCUS C (FLC), SQUAMOSA PROMOTER BINDING-LIKEs (SPLs), WRKY75, bHLH48, bHLH60, and nuclear factor Y (NF-Y; Yu et al. 2012; Hou et al. 2014; Wang et al. 2016; Li et al. 2016b, 2017; Bao et al. 2019).
In eukaryotes, NF-Y complexes, which comprise 3 distinct subunits including NF-YA, NF-YB, and NF-YC, regulate the expression of their target genes by binding to DNA with the central pentamer CCAAT box (Nardini et al. 2013). To date, several NF-YA, NF-YB, and NF-YC transcription factors have been shown to modulate flowering in Arabidopsis (Ben-Naim et al. 2006; Wenkel et al. 2006; Cai et al. 2007; Kumimoto et al. 2008, 2010; Siriwardana et al. 2016). The best understood role of the NF-Y subunits (NF-Ys) during this process is their transcriptional activation of FT. Briefly, NF-Y subunits form the canonical NF-YB/NF-YC/NF-YA and non-canonical NF-YB/NF-YC/CO complexes, which bind to the distal enhancer bearing a CCAAT box and the proximal CO-responsive elements (COREs), respectively, in the FT promoter. The interaction of the 2 distally separated DNA-bound complexes is stabilized by the formation of a chromatin loop (Cao et al. 2014; Gnesutta et al. 2017a, b; Myers and Holt 2018). Additionally, NF-Ys can recruit different transcription factors and histone modifiers to regulate flowering (Liu et al. 2018; Luo et al. 2018; Myers and Holt 2018; Hwang et al. 2019; Li et al. 2021). We previously demonstrated that NF-Ys, acting as flowering activators, are sequestered from binding to SOC1 by interacting with DELLAs, which results in the downregulation of SOC1 expression and the repression of flowering (Hou et al. 2014). Although the molecular and genetic relationships between NF-Ys and DELLAs have been characterized in recent years (Hou et al. 2014; Liu et al. 2016; Hu et al. 2018), the detailed molecular mechanism underlying how DELLAs inhibit NF-Y activity during flowering remains unclear.
Chromatin-remodeling complexes (CRCs) play key roles in transcriptional regulation in eukaryotes. SWItch/sucrose non-fermentable (SWI/SNF) complexes are the best studied ATP-dependent CRCs, which utilize the energy derived from ATP hydrolysis to regulate the interaction between histones and DNA (Clapier and Cairns 2009; Clapier et al. 2017). BRAHMA (BRM) is a key catalytic subunit in the SWI/SNF complex that serves as an important regulator of the growth and development of Arabidopsis by modifying the expression of its target genes (Farrona et al. 2004, 2011; Hurtado et al. 2006; Kwon et al. 2006; Tang et al. 2008; Han et al. 2012; Wu et al. 2012; Vercruyssen et al. 2014; Li et al. 2015, 2022; Yang et al. 2015; Zhang et al. 2017). Several studies over the past 2 decades have revealed the role of BRM in regulating flowering. BRM negatively regulates flowering by repressing the expression of CO, FT, and SOC1 (Farrona et al. 2004, 2011; Hurtado et al. 2006). BRM also directly activates the expression of SHORT VEGETATIVE PHASE (SVP) and TARGET OF FLC AND SVP1 (TFS1) by altering chromatin modifications at these loci (Li et al. 2015; Richter et al. 2019). Although recent studies have shown that BRM associates with transcription factors to regulate flowering (Richter et al. 2019; Yang et al. 2022), the relationship between BRM and NF-Ys in this process remains unclear.
In this study, we show that BRM is involved in GA-signaling-mediated flowering by forming a GA-sensitive module with DELLAs and NF-YCs. Specifically, the interaction of BRM with NF-YCs inhibits its binding to SOC1, a major floral integrator gene, and the interaction between BRM with NF-YCs can be significantly promoted by DELLAs. Meanwhile, DELLAs also promote the binding of BRM to SOC1. Both mechanisms decrease the deposition of H3K4me3 at SOC1 chromatin, resulting in reduced SOC1 gene expression and late flowering. GA triggers the degradation of DELLAs to release the repression of BRM on NF-YC activity and SOC1 transcription, thus promoting early flowering. These findings provide important insights into the epigenetic regulatory mechanism by which GA signaling accelerates flowering via BRM in plants.
Results
DELLAs physically interact with BRM
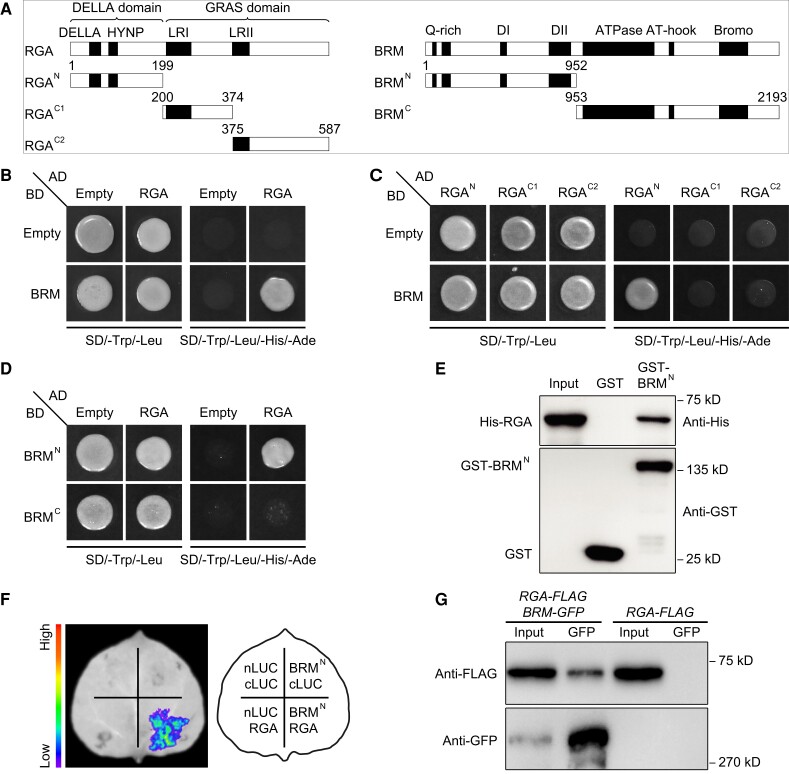
DELLAs are important repressors of the GA response that physically interact with other proteins to control flowering (Bao et al. 2020). However, few epigenetic factors associated with DELLAs that function in this process have been identified. To investigate potential epigenetic partners of DELLAs, we performed a series of yeast 2-hybrid assays. BRM, a core subunit of the SWI/SNF complex, strongly interacted with all 5 Arabidopsis DELLAs in yeast (Fig. 1, A and B; Supplemental Fig. S1). Region mapping assays showed that RGAN (amino acids 1 to 199) and BRMN (amino acids 1 to 952) were necessary and sufficient for the interaction of RGA with BRM (Fig. 1, A, C, and D). The N-terminal region of RGA (RGAN) contains a conserved DELLA domain (Zentella et al. 2016), and the N-terminal region of BRM (BRMN) normally serves as a docking site for the recruitment of transcription factors (Peirats-Llobet et al. 2016). We thus used BRMN instead of the full-length BRM protein in subsequent experiments. In vitro pull-down assays revealed that His-DELLAs were successfully pulled down by GST-BRMN but not by the GST control, suggesting that BRMN physically interacts with DELLA proteins in vitro (Fig. 1E; Supplemental Fig. S2).
Figure 1.
RGA physically interacts with BRM in vitro and in vivo. A) Schematic diagram of full-length RGA, BRM and their truncated derivatives used in the yeast 2-hybrid assay. The conserved domains are marked and the amino acid positions of these derivatives are numbered. B–D) Yeast 2-hybrid assays showing the interactions between RGA and BRM. Transformed yeast cells were grown on SD/-Trp/-Leu medium or SD/-Trp/-Leu/-His/-Ade medium. BD, GAL4 DNA-binding domain; AD, GAL4 DNA-activation domain. E) Pull-down assay showing direct interactions between His-RGA and GST-BRMN fusion proteins in vitro. His-RGA protein was incubated with immobilized GST-BRMN or GST protein. The pulled down proteins were detected by anti-His or anti-GST antibody. F) Split-LUC complementation imaging assay showing the interaction of RGA and BRMN in N. benthamiana cells. G) Co-IP assay showing the interaction of RGA and BRM in Arabidopsis. Plant extracts from 9-d-old 35S:RGA-FLAG pBRM:BRM-GFP (RGA-FLAG BRM-GFP) or RGA-FLAG seedlings under long-day conditions (LDs) were immunoprecipitated by using a GFP trap. The precipitated proteins were detected by anti-FLAG or anti-GFP antibody.
To further test the interaction between DELLAs and BRM in vivo, we used RGA as the representative DELLA homolog in subsequent analyses. We first performed split luciferase (split-LUC) complementation assays in Nicotiana benthamiana leaves. A strong interaction signal was observed when BRMN-nLUC and cLUC-RGA were co-expressed, whereas there was no signal in the negative controls (Fig. 1F). Next, we generated double transgenic Arabidopsis plants harboring 35S:RGA-FLAG pBRM:BRM-GFP (RGA-FLAG BRM-GFP) to perform a co-immunoprecipitation (co-IP) assay. RGA-FLAG co-immunoprecipitated with BRM-GFP (Fig. 1G), which confirms the interaction between RGA and BRM in Arabidopsis. These findings indicate that BRM interacts with DELLA proteins both in vitro and in vivo.
DELLAs regulate GA-mediated flowering via BRM
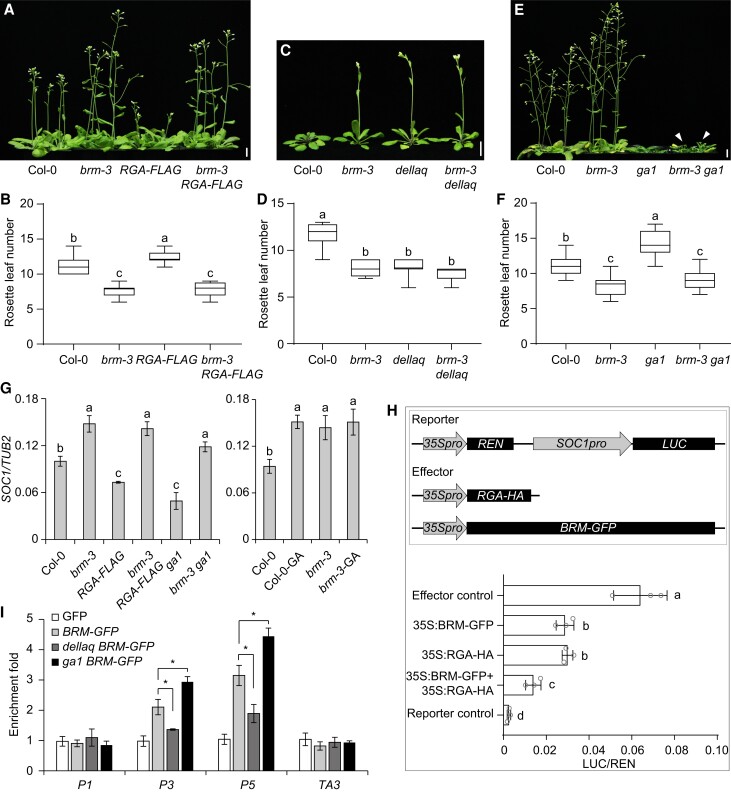
The role of BRM in flowering control was previously reported (Farrona et al. 2004; Li et al. 2015; Yang et al. 2022). In light of the DELLA–BRM interaction identified in this study, we speculated that these 2 proteins might function together to control flowering. Consistent with previous studies (Yan et al. 2020; Yang et al. 2022), we determined that both DELLAs and BRM negatively regulate flowering. Remarkably, the brm-3 mutant completely or partially restored the late-flowering phenotype of 35S:RGA-FLAG or pRGA:RGAΔ17 (GA-insensitive form of RGA; Yu et al. 2012) plants, respectively (Fig. 2, A and B; Supplemental Fig. S3, A and B). The quadruple mutant (dellaq) of RGA, GAI, RGL1, and RGL2 exhibited an early flowering phenotype, as expected, and the brm-3 dellaq mutant displayed a similar flowering phenotype to either the brm-3 or dellaq mutant (Fig. 2, C and D). Furthermore, the brm-3 mutant significantly rescued the late-flowering phenotype of ga1 (Fig. 2, E and F), a GA-deficient mutant in which DELLA proteins highly accumulate. These observations suggest that BRM may be epistatic to DELLA genes during flowering.
Figure 2.
BRM is epistatic to DELLAs in regulating flowering time. A and B) Flowering phenotypes of Col-0, brm-3, 35S:RGA-FLAG (RGA-FLAG), and brm-3 RGA-FLAG. C and D) Flowering phenotypes of Col-0, brm-3, dellaq, and brm-3 dellaq. E and F) Flowering phenotypes of Col-0, brm-3, ga1, and brm-3 ga1. White arrowheads show flowering. Scale bars in A), C), and E), 1 cm. Values in B), D), and F) are shown as boxplots, with the box representing the interquartile range, the central line indicating the median, and the whiskers showing the minimum or maximum value (n ≥ 20). Different lowercase letters indicate statistically significant differences (1-way ANOVA, P < 0.01). G) RT-qPCR analysis of SOC1 expression in 9-d-old Col-0, brm-3, RGA-FLAG, brm-3 RGA-FLAG, ga1, brm-3 ga1, GA-treated Col-0 (Col-0-GA), and GA-treated brm-3 (brm-3-GA) seedlings. For the Col-0-GA and brm-3-GA samples, 8-d-old Col-0 and brm-3 seedlings treated with 100 μM GA3 for 24 h were collected. TUB2 was used as an internal control. H) Transient expression assay indicating that the expression of SOC1 is co-regulated by BRM and RGA. I) ChIP analysis of BRM-GFP binding to the SOC1 locus in Col-0, dellaq, and ga1 mutant. Nine-day-old Col-0, p35S:GFP (GFP), BRM-GFP, dellaq BRM-GFP, and ga1 BRM-GFP seedlings were collected for ChIP assay. All plants used for analyses were grown at 22 °C under LDs. The enrichment of a TA3 genomic fragment was used as the negative control. Values in G), H), and I) represent means ± Sd of 3 independent experiments. Different lowercase letters indicate statistically significant differences (1-way ANOVA, P < 0.01). Asterisks indicate significant differences between the selected samples (*P < 0.01, Student's t-test).
The flowering genes SOC1 and FT contribute to the regulation of GA-mediated flowering (Bao et al. 2020). We thereby investigated whether DELLAs and BRM co-regulate the expression of these 2 flowering genes by reverse transcription quantitative PCR (RT-qPCR). Consistent with the flowering phenotypes, the transcript levels of SOC1 and FT were markedly altered in brm-3, 35S:RGA-FLAG, pRGA:RGAΔ17, and ga1 plants compared with those in Col-0, and the loss of BRM function resulted in the substantial de-repression of SOC1 and FT expression in 35S:RGA-FLAG, pRGA:RGAΔ17, and ga1 plants (Fig. 2G; Supplemental Figs. S3C and S4). The expression of SOC1 in the brm-3 dellaq mutant was not examined because no seeds from the homozygous brm-3 dellaq mutant plants could be obtained due to its abnormal fertility. We thus treated the brm-3 mutant with GA instead of the brm-3 dellaq mutant prior to RT-qPCR. GA treatment did not further promote the expression of SOC1 and FT in the brm-3 background, even though GA treatment significantly increased their expression in Col-0 plants (Fig. 2G; Supplemental Fig. S4), which is also consistent with the flowering phenotypes of the brm-3 and brm-3 dellaq mutants (Fig. 2, C and D).
Chromatin immunoprecipitation (ChIP) assays showed that BRM was enriched at the chromatin of SOC1 but not FT (Supplemental Fig. S5). We thus asked whether the DELLA–BRM module co-regulates flowering by altering the expression of SOC1. Transient expression assays in N. benthamiana leaves revealed that BRM or RGA decreased the activity of LUC driven by the SOC1 promoter, and the co-expression of BRM and RGA substantially increased the inhibition of LUC driven by the SOC1 promoter (Fig. 2H), suggesting that the DELLA–BRM module negatively regulates the expression of SOC1.
DELLAs can affect the binding of transcription factors to their target genes (Hou et al. 2014; Liu et al. 2016; Hu et al. 2018). To explore how the DELLA–BRM module regulates SOC1 expression, we performed ChIP assays to determine whether DELLAs affect the association of BRM to the SOC1 promoter. The enrichment of BRM at SOC1 chromatin was drastically reduced in the dellaq mutant but increased in the ga1 mutant background (Fig. 2I). However, the level of BRM or RGA protein was not altered in the dellaq/ga1 or brm-3 mutant compared with Col-0 plants, respectively (Supplemental Figs. S6 and S7), indicating that DELLAs enhance the binding of BRM to SOC1 rather than altering the protein level of BRM during flowering. Consistent with this observation, GA significantly reduced the binding of BRM-GFP to SOC1, whereas paclobutrazol (PAC), an inhibitor of GA biosynthesis, increased the binding of BRM-GFP to SOC1, even when the protein levels of BRM-GFP were not altered by these treatments (Supplemental Fig. S8). Overall, these findings suggest that DELLAs regulate the expression of SOC1 via BRM to control flowering.
BRM physically interacts with NF-YCs
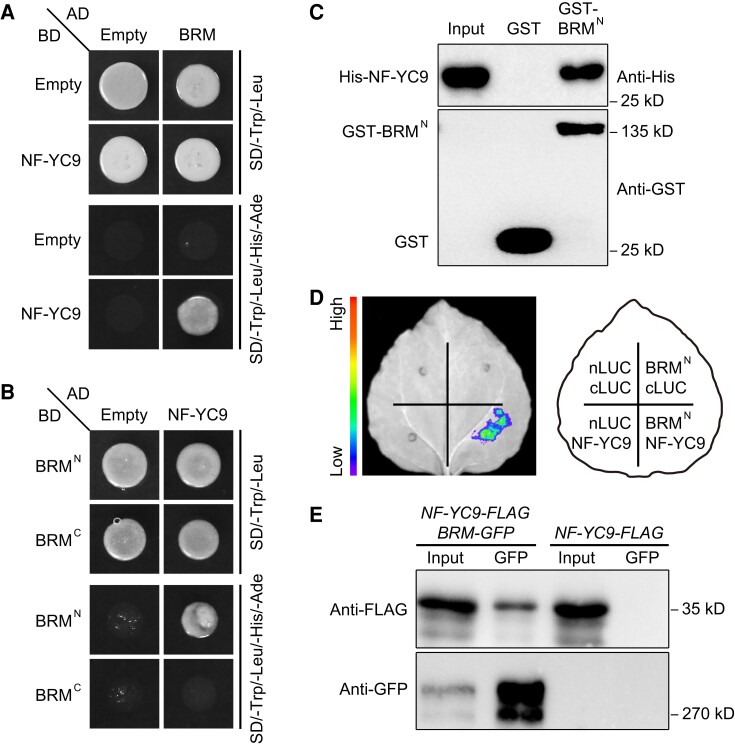
We previously showed that DELLAs function together with NF-Y subunits to control flowering in the GA pathway (Hou et al. 2014). Considering that DELLAs also interact with BRM, we investigated the potential biological relationships among these proteins. NF-YC3, NF-YC4, and NF-YC9 have redundant functions in regulating flowering (Kumimoto et al. 2010; Liu et al. 2018). All 3 of these proteins interacted with BRM in yeast (Fig. 3A; Supplemental Fig. S9A). Region mapping assays revealed that BRMN, but not BRMC, mediates interactions with NF-YCs (Fig. 3B; Supplemental Fig. S9B). In in vitro pull-down assays, His-NF-YC3, His-NF-YC4, and His-NF-YC9 could be pulled down by GST-BRMN but not by the GST control, suggesting that BRMN physically interacts with NF-YCs in vitro (Fig. 3C; Supplemental Fig. S10).
Figure 3.
BRM physically interacts with NF-YC9 in vitro and in vivo. A) Yeast 2-hybrid assay showing the interactions between BRM and NF-YC9. B) Yeast 2-hybrid assay showing that BRMN directly interacts with NF-YC9. Transformed yeast cells were grown on SD/-Trp/-Leu medium or SD/-Trp/-Leu/-His/-Ade medium. C) Pull-down assay showing direct interactions between GST- BRMN and His-NF-YC9 fusion proteins in vitro. His-NF-YC9 protein was incubated with immobilized GST- BRMN or GST protein. The pulled down proteins were detected by anti-His or anti-GST antibody. D) Split-LUC complementation imaging assay showing the interaction of BRMN with NF-YC9 in N. benthamiana cells. E) Co-IP assay showing the interaction of NF-YC9 and BRM in Arabidopsis. Plant extracts from 9-d-old nf-yc9-1 pNF-YC9:NF-YC9-FLAG pBRM:BRM-GFP (NF-YC9-FLAG BRM-GFP) or NF-YC9-FLAG seedlings under LDs were immunoprecipitated by GFP trap. The precipitated proteins were detected by anti-FLAG or anti-GFP antibody.
We next chose NF-YC9 as the representative of NF-YCs to investigate the interaction between BRM and NF-YCs in planta. Split-LUC complementation assays revealed a strong signal in N. benthamiana leaves when BRMN-nLUC and cLUC-NF-YC9 were co-expressed; however, no such signal was observed in the negative controls (Fig. 3D). Furthermore, a co-IP assay using nf-yc9-1 pNF-YC9:NF-YC9-FLAG pBRM:BRM-GFP (NF-YC9-FLAG BRM-GFP) Arabidopsis plants also showed that NF-YC9-FLAG protein could be co-immunoprecipitated with BRM-GFP (Fig. 3E). Taken together, these findings suggest that BRM interacts with NF-YCs both in vitro and in vivo.
BRM genetically interacts with NF-YCs during SOC1-mediated flowering
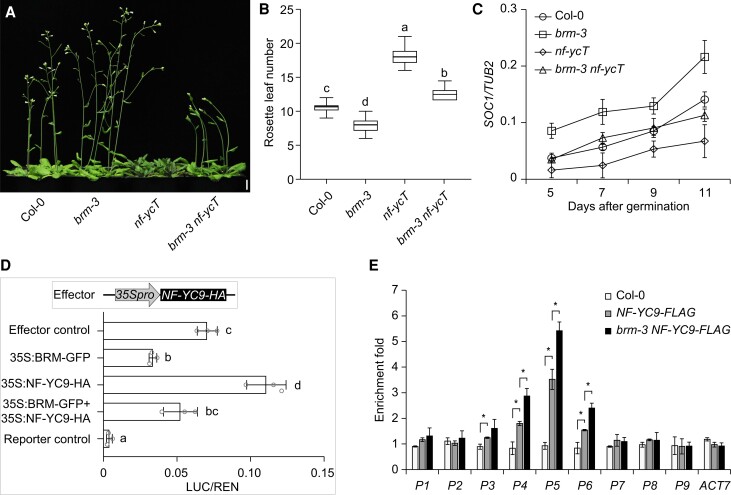
To clarify the biological function of the BRM–NF-YC interaction, we evaluated the genetic relationship between BRM and NF-YCs during flowering. Consistent with previous findings, brm-3 and nf-ycT (nf-yc3/4/9 triple mutant) plants showed early and late flowering, respectively (Liu et al. 2018; Yang et al. 2022). Notably, the quadruple brm-3 nf-ycT mutant flowered significantly earlier than the nf-ycT mutant and later than the brm-3 mutant (Fig. 4, A and B), suggesting that BRM and NF-YCs might antagonistically regulate flowering time. Consistently, the expression of SOC1 and FT was significantly higher in brm-3 nf-ycT than in nf-ycT but lower than in brm-3 (Fig. 4C; Supplemental Fig. S11). We also examined SOC1 promoter activity regulated by BRM and NF-YC9 using a dual-luciferase reporter assay in N. benthamiana leaves (Fig. 4D). The transient assay revealed that LUC activity was enhanced by NF-YC9; in contrast, LUC activity was inhibited by BRM (Fig. 4D). These findings suggest that BRM and NF-YCs antagonistically regulate the expression of SOC1.
Figure 4.
NF-YCs and BRM antagonistically regulate flowering by directly modulating SOC1 expression. A and B) Flowering phenotypes of Col-0, brm-3, nf-ycT, and brm-3 nf-ycT. Scale bar, 1 cm. Values in B) are shown as boxplots, with the box representing the interquartile range, the central line indicating the median, and the whiskers showing the minimum or maximum value (n ≥ 20). C) RT-qPCR analysis of SOC1 expression in developing Col-0, brm-3, nf-ycT, and brm-3 nf-ycT seedlings. TUB2 was used as an internal control. D) Transient expression assay indicating that the expression of SOC1 is regulated by NF-YC9 and BRM. Either the reporter or the relevant empty vector (Reporter control) was co-transformed with the effector or the relevant empty vector (Effector control) into N. benthamiana leaves. SOC1 promoter activity was calculated as the ratio of LUC to REN. E) ChIP analysis of NF-YC9-FLAG binding to the SOC1 locus. Nine-day-old Col-0, NF-YC9-FLAG, and brm-3 NF-YC9-FLAG seedlings were collected for ChIP assay. The enrichment of a ACT7 genomic fragment was used as the negative control. All plants used for analysis were grown at 22 °C under LDs. Values in C), D), and E) represent means ± Sd of 3 independent experiments. Different lowercase letters indicate statistically significant differences (1-way ANOVA, P < 0.01). Asterisks indicate significant differences between the selected samples (*P < 0.01, Student's t-test).
To characterize the opposite functions of BRM and NF-YCs in regulating the expression of SOC1, we performed ChIP assays using brm-3 nf-yc9-1 pNF-YC9:NF-YC9-FLAG (brm-3 NF-YC9-FLAG) plants. The enrichment of NF-YC9 on SOC1 rather than FT chromatin was higher in the brm-3 vs. the Col-0 background (Fig. 4E; Supplemental Fig. S12A). Given that the level of NF-YC9 protein was not affected by the loss of function of BRM (Supplemental Fig. S12B), the greater enrichment of NF-YC9 on SOC1 chromatin stems from the lack of BRM inhibition rather than alterations in the level of NF-YC9 protein.
Since both BRM and NF-YCs were shown to regulate the expression of genes via histone modifications (Hou et al. 2014; Yang et al. 2022), we examined the deposition of H3K4me3 and H3K27me3, key marks involved in transcriptional modulation at SOC1 chromatin. BRM and NF-YCs had opposite effects on the levels of H3K4me3 at the SOC1 locus, and these effects were abolished in the brm-3 nf-ycT mutant. In contrast, H3K27me3 deposition was only mediated by NF-YCs but not by BRM (Supplemental Fig. S13). Taken together, these results suggest that BRM regulates flowering time by antagonizing NF-YC activity via the deposition of H3K4me3 on SOC1 chromatin.
DELLA proteins enhance the interaction of BRM with NF-YCs
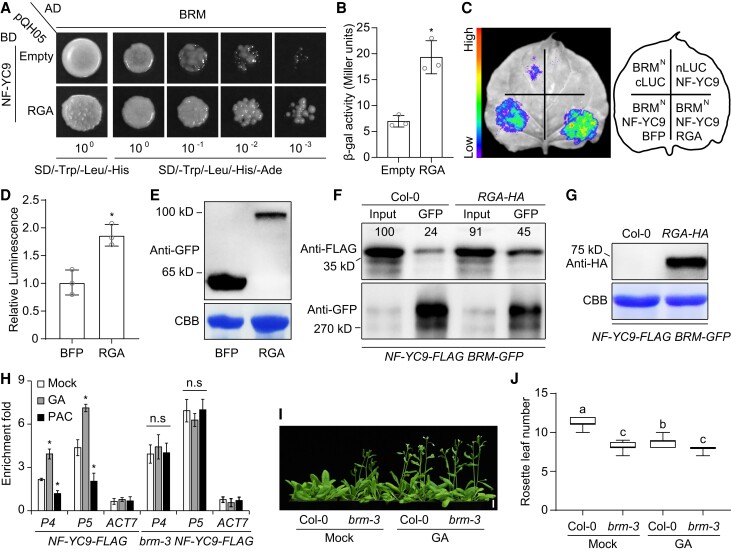
In light of the finding that DELLAs interact with both BRM and NF-YCs, we determined whether these proteins function together to regulate GA-mediated flowering. Among the 5 DELLAs in Arabidopsis, RGA and GAI play major roles in this process (Tyler et al. 2004); we thus selected these 2 DELLAs for further analyses. Yeast 3-hybrid assays revealed that the interaction between BRM and NF-YC9 was strengthened in the presence of RGA or GAI (Fig. 5, A and B; Supplemental Fig. S14, A and B). Split-LUC complementation assays also showed that the co-expression of RGA or GAI with BRM and NF-YC9 enhanced LUC activity compared with co-expression exclusively with BFP (Fig. 5, C and D; Supplemental Fig. S14, C and D), even when the protein level of BFP was markedly higher than that of RGA or GAI (Fig. 5E; Supplemental Fig. S14E). These results suggest that DELLAs increase the strength of the interaction between BRM and NF-YC9 in vivo. To confirm these findings, we verified the effect of RGA on the physical interaction between BRM and NF-YC9 via co-IP. We generated transgenic plants co-expressing NF-YC9-FLAG and BRM-GFP in the Col-0 and 35S:RGA-HA (RGA-HA) backgrounds and found that more NF-YC9-FLAG coimmunoprecipitated with BRM-GFP in the RGA-HA background than in the Col-0 background (Fig. 5, F and G; Supplemental Fig. S15).
Figure 5.
RGA enhances the interaction of NF-YC9 with BRM. A) Yeast 3-hybrid assay showing the enhanced interaction between NF-YC9 and BRM in the presence of RGA. Transformed yeast cells were grown on SD/-Trp/-Leu/-His medium or SD/-Trp/-Leu/-His/-Ade medium. B) Quantitative yeast 3-hybrid assay defining the strength of the interaction in A). C and D) Split-LUC complementation imaging assay showing the enhanced interaction of NF-YC9 and BRMN by RGA in N. benthamiana cells C), and the quantification of luciferase activity D). E) The protein levels of BFP-GFP and RGA-GFP in N. benthamiana cells determined by probing with anti-GFP antibody. F) Co-IP assay showing that RGA promote the interaction of NF-YC9 and BRM in Arabidopsis. Plant extracts from 9-d-old NF-YC9-FLAG BRM-GFP 35S: RGA-HA (RGA-HA) or NF-YC9-FLAG BRM-GFP (Col-0) seedlings under LDs were immunoprecipitated by GFP trap. The precipitated proteins were detected by either anti-FLAG or anti-GFP antibody. The protein levels of NF-YC9-FLAG were analyzed using Image J and presented as numbers at the top. G) Protein expression analysis of RGA-HA in NF-YC9-FLAG BRM-GFP and NF-YC9-FLAG BRM-GFP RGA-HA lines. The immunoblot was probed with anti-HA antibody (Sigma, H3663). The bottom gels in E) and G) were stained with Coomassie Brilliant Blue as loading controls. H) ChIP analysis of NF-YC9-FLAG binding to the SOC1 locus in Col-0 and brm-3 backgrounds mock treated or treated with 100 μM GA3 or 10 μM PAC for 24 h. Nine-day-old NF-YC9-FLAG and brm-3 NF-YC9-FLAG seedlings under LDs were collected for ChIP assay. The enrichment of a TA3 genomic fragment was used as the negative control. Values in B), D), and H) represent means ± Sd of 3 independent experiments. n.s., no significance (Student's t-test). Asterisks in B), D), and H) indicate significant differences (*P < 0.01, Student's t-test). I and J) Flowering phenotypes of Col-0 and brm-3 grown at 22 °C under LDs treated with mock or 100 μM GA3 once every 2 d until bolting. Scale bar, 1 cm. Values in J) are shown as boxplots, with the box representing the interquartile range, the central line indicating the median, and the whiskers showing the minimum or maximum value (n ≥ 20). Different lowercase letters indicate statistically significant differences (1-way ANOVA, P < 0.01).
DELLA proteins, the key repressors in GA signaling, regulate the expression of SOC1 by inhibiting the binding of NF-Y to SOC1 (Hou et al. 2014). This observation, coupled with the results of the current study, suggests that DELLAs might recruit BRM to inhibit the binding of NF-Y to SOC1 by enhancing the physical interaction of BRM with NF-YCs. To test this hypothesis, we treated NF-YC9-FLAG and brm-3 NF-YC9-FLAG seedlings with GA or PAC and performed ChIP assays to evaluate the binding ability of NF-YC9 to SOC1 chromatin. As expected, GA or PAC treatment had a significant effect on the binding of NF-YC9 to the SOC1 promoter in the Col-0 background. However, the loss-of-function mutation of BRM abolished this effect (Fig. 5H), indicating that BRM is essential for the effect of GA signaling on the binding of NF-YC9 to SOC1. Consistent with this finding, the flowering of brm-3 plants was not responsive to GA treatment, even though GA treatment significantly accelerated the flowering of Col-0 plants (Fig. 5, I and J). Overall, our findings demonstrate that DELLA proteins facilitate the formation of the DELLA–BRM–NF-YC module, which is essential for GA-mediated flowering.
Discussion
GA signaling regulates flowering by inducing the degradation of DELLA proteins, which are the key repressors of the GA-signaling pathway (Sun 2008). DELLA proteins physically interact with a series of transcription factors and modulate their transcriptional activities on their target genes to regulate flowering (Bao et al. 2020). For instance, DELLAs prevent NF-Y from binding to the SOC1 locus via protein–protein interactions, which inhibits flowering (Hou et al. 2014). However, the molecular mechanisms underlying the ability of DELLAs to mediate the epigenetic regulatory effects of NF-Y on gene expression remain unclear.
Here, we show that DELLA proteins can recruit a catalytic subunit of the SWI/SNF complex, BRM, to inhibit the binding of NF-YCs to SOC1. In addition, DELLAs promote the binding of BRM to the SOC1 locus during GA-signaling-mediated flowering. In the presence of GA, DELLA proteins are degraded, which reduces the binding of BRM to chromatin and the strength of the interaction between BRM and NF-YCs, thus activating the expression of SOC1 and promoting flowering (Fig. 6). Notably, the expression of FT is also regulated by BRM, but it does not affect the binding strength of NF-YC9 to FT chromatin, suggesting that the DELLA–BRM–NF-YC module may not occur on the FT locus. BRM represses FT expression by directly altering the expression of its upstream repressor SVP (Lee et al. 2007), which is a direct target of BRM (Li et al. 2015). The differential regulation of SOC1 and FT suggests that the formation of the DELLA–BRM–NF-YC module may have spatial and temporal characteristics. Further studies on where and when the heterotrimer module functions in plants should help address this interesting issue.
Figure 6.
A proposed model illustrating how the DELLA–BRM–NF-YC module regulates GA-signaling-mediated flowering. In the absence of GA, DELLA proteins accumulate and promote the formation of the DELLA–BRM–NF-YC module, thereby inhibiting the binding of NF-YCs to the SOC1 locus by BRM. At the same time, DELLA proteins promote BRM binding to SOC1, thereby inhibiting the expression of SOC1 by decreasing the deposition of H3K4me3 at SOC1 chromatin, resulting in late flowering. In the presence of GA, DELLA proteins are ubiquitinated and degraded by the 26S proteasome. This releases the inhibition of NF-YCs by disturbing the DELLA–BRM–NF-YC module and reduces the binding of BRM to SOC1, which enhances the expression of SOC1 by increasing the deposition of H3K4me3 at SOC1 chromatin, resulting in early flowering. TF, transcription factor; YA, NF-YA; CO, CONSTANS. One (No GA) and 3 (GA) purple squares represent higher and lower H3K4me3 levels, respectively.
Several recent reports provide structural insights into how SWI/SNF complexes recognize and remodel nucleosomes in humans and yeast (Han et al. 2020; He et al. 2020; Mashtalir et al. 2020), illustrating the detailed role of SWI/SNF complexes in gene transcription. However, no evidence supports the function of the ATPases in these complexes (such as BRM in plants) in DNA recognition. Three DNA-binding factors are required for the binding of BRM to its targets (Li et al. 2016; Zhang et al. 2017; Yang et al. 2022). Among these, GNC is a GATA transcription factor that mediates the recruitment of BRM to SOC1 to regulate flowering. It is thus possible that GNC or other transcription factors recruit the DELLA–BRM module to SOC1 (Fig. 6). However, we cannot exclude the possibility that other NF-YCs besides those examined in our study mediate the recruitment of BRM to SOC1, since additional NF-YC family members in plants evolved to allow subtle adjustments to many different environmental conditions (Petroni et al. 2012). The crystal structure revealed that the NF-YC/NF-YB dimer coordinates with NF-YA or the CCT domain to specifically target DNA sequences (Gnesutta et al. 2017a, b; Shen et al. 2020). Therefore, NF-YC/NF-YB/NF-YA or a CCT domain-containing protein that represses flowering might also be involved in this process, but not CO, which functions as a positive regulator of flowering (Putterill et al. 1995).
BRM is a core SWI/SNF chromatin-remodeling ATPase that uses the energy from ATP hydrolysis to alter the position and occupancy of nucleosomes, leading to either the activation or repression of gene transcription (Clapier et al. 2017). BRM is involved in regulating several plant hormone pathways, such as the abscisic acid, auxin, cytokinin, and GA pathways (Han et al. 2012; Archacki et al. 2013; Efroni et al. 2013; Wu et al. 2015; Peirats-Llobet et al. 2016; Vain et al. 2019). In the GA pathway, BRM affects GA biosynthesis and signaling by regulating the expression of a large of GA-responsive genes (Archacki et al. 2013); however, the precise mechanism underlying how BRM regulates in GA signaling remains unclear. In the current study, we determined that BRM interacts with the GA-signaling hubs DELLA proteins and NF-YC transcription factors to form the DELLA–BRM–NF-YC module to inhibit the effects of NF-YCs. On the other hand, the binding of BRM to the chromatin of flowering-related genes can be accelerated by DELLA proteins. These mechanisms underlie the ability of BRM to regulate the expression of flowering genes and flowering via the GA-signaling pathway. These findings also demonstrate that BRM fine-tunes GA responses at multiple levels.
Many proteins mediate the GA-signaling pathway by physically interacting with DELLA proteins. In general, interactions between DELLAs and transcription factors regulate the transcriptional activities or binding of transcription factors to their target genes (Yu et al. 2012; Fukazawa et al. 2014; Hou et al. 2014; Liu et al. 2016; Wang et al. 2016; Xu et al. 2016; Li et al. 2016b, 2017; Zhang et al. 2018; Bao et al. 2020). DELLAs can also disrupt the interaction of CO with NF-YB2 by the interactions among DELLAs, CO, and NF-YB2 (Xu et al. 2016) or compete with MYC2 to interact with JASMONATE-ZIM1 (JAZ1; Hou et al. 2010). These findings indicate that DELLA proteins modulate the functions of different protein partners. Indeed, our findings suggest that DELLAs enhance the association of BRM with NF-YCs to prevent NF-YCs from binding to the promoter of SOC1. Furthermore, DELLAs can promote BRM binding to the SOC1 promoter during flowering. These results also support the notion that the DELLA-mediated effects on GA signaling might partially depend on the interactions of DELLA proteins with CRCs (Sarnowska et al. 2013). Additional studies of the interactions between DELLAs and other proteins are needed to increase our understanding of the functions of DELLA proteins.
The epigenetic regulatory effects of the subunits of the NF-Y complex on plant growth and development have been a major focus of our research over the past decade (Hou et al. 2014; Tang et al. 2017; Liu et al. 2018; Zhang et al. 2021). Temporal changes in the interaction of NF-YCs with CURLY LEAF (CLF) occur and counteract the deposition of H3K27me3 at the FT locus to regulate flowering time (Liu et al. 2018). NF-Y can also modulate the level of H3K27me3 at the SOC1 locus and promote its expression by recruiting the H3K27 demethylase REF6 (Hou et al. 2014). BRM is a core catalytic subunit of the SWI/SNF-type CRC (Clapier et al. 2017). Recent studies have shown that BRM affects the level of H3K4me3 but not H3K27me3 at the SOC1 locus, suggesting that the effects of BRM on SOC1 expression are independent of the effects of the BRM-REF6 module (Li et al. 2015; Yang et al. 2022). In this study, we revealed that BRM and NF-YCs antagonistically regulate H3K4me3 occupancy at the SOC1 locus through physical interactions, thus altering flowering time. BRM can repress the binding of NF-YCs to SOC1 chromatin, which may be due to the competitive binding of BRM with NF-YCs to DNA or changes in the chromatin state induced by BRM. Given that NF-Ys are associated with multiple epigenetic factors, it will be interesting to investigate how NF-Ys orchestrate these factors and to uncover the fundamental roles of NF-Ys in epigenetic regulation during plant growth and development.
Materials and methods
Plant materials and growth conditions
All Arabidopsis (A. thaliana) plants used in this study are in the Col-0 background and were grown at 22 °C under long-day conditions (LDs, full-spectrum white fluorescent light intensity of 100 μmol m−2 s−1 with 16 h light/8 h dark photoperiod). All plant materials used in this study are listed in Supplemental Table S1. NF-YC-related plants, brm-3, p35S:GFP, pBRM:BRM-GFP, 35S:RGA-FLAG, pRGA:RGAΔ17, ga1, and dellaq were described previously (Yu et al. 2012; Yang et al. 2015; Hu et al. 2018; Yan et al. 2020; Zhang et al. 2021). To generate 35S:RGA-6HA, the coding region of RGA was inserted into the pGreen-35S:6HA vector (Hou et al. 2014). Primers used are listed in Supplemental Table S2. The floral dip method was used to generate the transgenic plants, and positive lines were selected by Basta treatment in soil. Seeds with a ga1 background were imbibed in 100 μM GA3 at 4 °C for 7 d and rinsed thoroughly with water before sowing.
Yeast 2-hybrid and 3-hybrid assays
The coding regions of genes for NF-YCs, BRM, and truncated versions of BRM were amplified and cloned into the EcoRI/PstI, NdeI/EcoRI, or NdeI/XmaI restriction site of pGBKT7 (Clontech). The coding regions of genes for NF-YCs, BRM, DELLAs, and truncated versions of RGA were amplified and cloned into the XmaI/BamHI, NdeI/XmaI, or NdeI/EcoRI restriction site of pGADT7 (Clontech). To analyze the effects of DELLAs on the interaction of BRM with NF-YC9, the RGA and GAI coding regions were amplified and cloned into the XhoI/XmaI or XmaI restriction site of pQH05 (Hou et al. 2014). Primers used are listed in Supplemental Table S2. Yeast 2-hybrid assays were performed using the Yeastmaker Yeast Transformation System 2 (Clontech). Yeast AH109 cells were co-transformed with specific bait and prey constructs. All yeast transformants were grown on SD/-Trp/-Leu medium for selection. To assess protein interactions, the transformed yeast cells were resuspended in liquid SD/-Leu/-Trp. Three microliters of suspended yeast cells were spotted onto SD/-Trp-Leu or SD/-Trp-Leu-His-Ade dropout plates to detect direct interactions between 2 proteins following incubation at 30 °C. Yeast 3-hybrid assays were performed as described previously (Hou et al. 2014). Measurement of β-galactosidase activity was performed according to the Yeast Protocols Handbook (Clontech) using chlorophenol red-β-D-galactopyranoside (Roche) as the substrate. Yeast 2-hybrid and 3-hybrid assays were repeated at least 3 times with similar results.
In vitro pull-down assay
The coding regions of genes for NF-YCs and DELLAs were cloned into the BamHI/HindIII, BamHI/SalI, SacI/XmaI, or KpnI/SalI restriction site of pQE30 (Qiagen) to produce His-NF-YCs and His-DELLAs constructs. The coding region of gene for BRMN was cloned into the BamHI/SalI restriction site of pGEX-4T-1 (Pharmacia) to produce GST-BRMN construct. Primers used are listed in Supplemental Table S2. These GST and His fusion recombinant proteins were induced and expressed in Escherichia coli Rosetta cells. The soluble His-NF-YC and His-DELLA proteins were extracted and immobilized on nickel-nitrilotriacetic acid agarose beads (Qiagen, 30210), while the soluble GST-BRMN and GST proteins were extracted and immobilized on Glutathione Sepharose Beads (GE Healthcare, 17-0756-01). For pull-down assays, the purified His-NF-YC or His-DELLA proteins were incubated with immobilized GST or GST-BRMN in binding buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 1 mM EDTA) at 4 °C overnight. After washing with binding buffer, proteins retained on the beads were subsequently resolved by SDS–PAGE and detected by immunoblotting with anti-HIS antibody (BGI, AbM59012-18-PU, 1: 10,000 dilution) or anti-GST antibody (TianGen, AB101-02, 1:10,000 dilution). Pull-down experiments were repeated 3 times with similar results.
Split-luciferase assay
The coding region of gene for BRMN was cloned into the KpnI/SalI restriction site of pCAMBIA1300-nLUC to produce BRMN-nLUC construct. The coding regions of genes for NF-YC9 and RGA were cloned into the KpnI/SalI restriction site of pCAMBIA1300-cLUC to produce cLUC-NF-YC9 and cLUC-RGA constructs (Chen et al. 2008). Primers used are listed in Supplemental Table S2. These constructs were transformed individually into Agrobacterium tumefaciens strain GV3101. GV3101 cells harboring the indicated constructs expressing nLUC or cLUC fused proteins were mixed at a 1:1 ratio and introduced into N. benthamiana leaves. To determine the effects of RGA and GAI on the interaction of NF-YC9 with BRMN, the coding regions of genes for RGA, GAI, and BFP (negative control) were cloned into the PstI restriction site of pGreen-35S:GFP vector (Qian et al. 2021) to produce 35S: RGA-GFP (RGA-GFP), 35S: GAI-GFP (GAI-GFP), and 35S: BFP-GFP (BFP-GFP) constructs, respectively. Primers used are listed in Supplemental Table S2. GV3101 cells harboring the constructs expressing BRMN-nLUC, cLUC-NF-YC9, and RGA-GFP, GAI-GFP, or BFP-GFP were mixed at a 1:1 ratio and infiltrated into N. benthamiana leaves. At 2 to 3 d after infiltration, the leaves were incubated with 1 mM d-luciferin sodium salt substrate (Abcam, ab145164) and kept in the dark for 10 min. The luminescence imaging workstation (Tanon 5200) was used to capture luciferase images. Split-luciferase experiments were repeated 3 times with similar results. LUC activity was measured using the Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer's instructions. The relative luminescence was presented with 3 independent experiments.
Co-IP assay
Seedlings with various genetic backgrounds were grown at 22 °C under LDs for 9 d and harvested for total protein extraction in co-IP buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 10 mM ZnSO4, 5 mM MgCl2, 1% Triton X-100, and 0.05% SDS). The total proteins were incubated with GFP trap beads (Chromotek, gtak-20) at 4 °C overnight. The beads were washed 3 times with co-IP buffer, and the precipitated proteins were eluted in 1×SDS loading buffer by boiling for 10 min. The immunoprecipitated proteins were separated on a 6% or 10% SDS–PAGE gel and detected by immunoblotting with anti-GFP (TransGen, HT801-01, 1:5,000 dilution) and anti-FLAG (Sigma, F3165, 1:10,000 dilution) antibodies. Co-IP experiments were repeated 3 times with similar results.
Quantitative RT-PCR
Growth conditions and treatment of seedlings were described in the text. Total RNA was extracted from the samples using a Plant RNA Kit (Promega, LS1040) and reverse transcribed to cDNA using MMLV-RTase (Promega, M1701) according to the manufacturer's protocols. Gene expression levels were determined by RT-qPCR on a Light Cycler 480 thermal cycler system (Roche) with KAPA SYBR Fast qPCR Kit Master Mix (Kapa Bio, KK4680). The relative expression level of each gene was quantified in triplicate and normalized to that of TUB2 (as an internal control). Primers used are listed in Supplemental Table S2.
Transient expression assay
To generate the pSOC1:LUC reporter construct, ∼2 kb SOC1 promoter was cloned into the HindIII/BamHI restriction site of the pGreenII 0800-LUC vector. The Renilla Luciferase (REN) gene under the control of the 35S promoter in the pGreenII 0800-LUC vector was used as the internal control. The coding region of BRM was cloned into the PstI restriction site of pGreen-35S:GFP to produce the 35S:BRM-GFP (BRM-GFP) construct and used as an effector. The coding region of NF-YC9 or RGA was cloned into the EcoRI/SpeI or HindIII/EcoRI restriction site of pGreen-35S:6HA to produce the 35:NF-YC9-HA or 35S:RGA-HA construct and used as another effector. Primers used are listed in Supplemental Table S2. These effector and reporter or control constructs were transformed individually into A. tumefaciens strain GV3101. GV3101 cells harboring the indicated constructs were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves. The LUC and REN activities were measured using the Dual-Luciferase Reporter Assay System according to the manufacturer's instructions. The LUC/REN ratio was presented with 3 independent experiments.
ChIP qPCR assay
Seedlings grown under LDs were used for the ChIP assays, which were performed as described previously (Hou et al. 2014). Briefly, seedlings at 9 d after germination were vacuum-infiltrated with 1% formaldehyde for cross-linking, which was stopped by adding 150 mM glycine. Chromatin was isolated from the samples and sonicated to generate DNA fragments with an average size of ∼500 bp. Subsequently, the chromatin complexes were immunoprecipitated by GFP trap beads or Protein G PLUS/Protein A agarose (Millipore, 16-201) plus anti-FLAG antibody at 4 °C overnight. The precipitated DNA fragments were recovered and quantified by qPCR with SYBR Premix ExTaq Mix using the primers shown in Supplemental Table S2. Relative enrichment fold was calculated by normalizing the amount of a target DNA fragment against that of a PP2A genomic fragment and then against the respective input DNA samples.
Observation of GFP fluorescence
GFP fluorescence in primary roots was observed under a confocal laser scanning microscope (Leica TCS SP5). A 488 nm laser was used to detect GFP excitation. All images were obtained with the same modifications and intensity parameters.
Statistical analysis
GraphPad Prism 8.0 and Microsoft Office Excel were used for statistical analysis of the numerical data. The statistically significant differences between 2 groups or multiple samples were determined by using a 2-tailed Student's t-test or 1-way ANOVA, respectively. Statistical data are provided in Supplemental Data Set 1.
Accession numbers
Sequence data from this article can be found in the TAIR website under the following accession numbers: NF-YC3 (AT1G54830), NF-YC4 (AT5G63470), NF-YC9 (AT1G08970), BRAHMA (AT2G46020), RGA (AT2G01570), GAI (AT1G14920), RGL1 (AT1G66350), RGL2 (AT3G03450), RGL3 (AT5G17490), SOC1 (AT2G45660), FT (AT1G65480), TUB2 (AT5G62690), ACT7 (AT5G09810), PP2A (AT1G69960), TA3 (AT1G37110), Cinful-like (AT4G03770).
Supplementary Material
Acknowledgments
The authors thank Dr Xiaoying Zhao from Hunan University for providing the 35S:RGA-FLAG seeds.
Contributor Information
Chunyu Zhang, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China.
Mingyang Jian, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Weijun Li, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Xiani Yao, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Cuirong Tan, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Qian Qian, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China.
Yilong Hu, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Xu Liu, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Xingliang Hou, Guangdong Provincial Key Laboratory of Applied Botany and State Key Laboratory of Plant Diversity and Prominent Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Author contributions
C.Z. and X.H. designed the research. C.Z., M.J., W.L., X.Y., and C.T. performed the research. C.Z., M.J., Q.Q., Y.H., X.L., and X.H. analyzed data; C.Z. and X.H. wrote the article.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. BRM interacts with DELLA proteins in yeast.
Supplemental Figure S2. BRMN interacts with DELLA proteins in vitro.
Supplemental Figure S3. The brm-3 mutant can rescue the late-flowering phenotype of pRGA:RGAΔ17 (RGAΔ17) plants.
Supplemental Figure S4. Expression analysis of FT.
Supplemental Figure S5. The binding of BRM on SOC1 and FT.
Supplemental Figure S6. The protein levels of BRM-GFP in BRM-GFP, dellaq BRM-GFP, and ga1 BRM-GFP.
Supplemental Figure S7. The protein levels of RGA in the brm-3 mutant.
Supplemental Figure S8. GA reduces BRM binding to SOC1.
Supplemental Figure S9. BRM interacts with NF-YC proteins in yeast.
Supplemental Figure S10. BRMN interacts with NF-YC proteins in vitro.
Supplemental Figure S11. Analysis of the regulation of FT by BRM and NF-YCs.
Supplemental Figure S12. BRM does not affect the binding of NF-YC9 to the FT locus.
Supplemental Figure S13. BRM and NF-YCs antagonistically regulate H3K4me3 level at SOC1.
Supplemental Figure S14. GAI enhances the interaction of NF-YC9 with BRM.
Supplemental Figure S15. Biological replicates for the Co-IP assays in Fig. 5F.
Supplemental Table S1. Sources of the plant materials.
Supplemental Table S2. List of primers used in this study.
Supplemental Data Set S1. Statistical analyses.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 32000416, 31970623), the Key-Area Research and Development Program of Guangdong Province (No. 2022B1111230001), the Guangdong Basic and Applied Basic Research Foundation (Nos. 2019A1515110885, 2020A1515110041, and 2021A1515012406), and the Guangzhou Municipal Science and Technology Project (No. 202002030057).
Data availability
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005:309(5737):1052–1056. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010:61(6):1001–1013. 10.1111/j.1365-313X.2010.04148.x [DOI] [PubMed] [Google Scholar]
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka-Nowicka R, et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS One. 2013:8(3):e58588. 10.1371/journal.pone.0058588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Hua C, Huang G, Cheng P, Gong X, Shen L, Yu H. Molecular basis of natural variation in photoperiodic flowering responses. Dev Cell. 2019:50(1):90–101.e3. 10.1016/j.devcel.2019.05.018 [DOI] [PubMed] [Google Scholar]
- Bao S, Hua C, Shen L, Yu H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J Integr Plant Biol. 2020:62(1):118–131. 10.1111/jipb.12892 [DOI] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006:46(3):462–476. 10.1111/j.1365-313X.2006.02706.x [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000:404(6780):889–892. 10.1038/35009125 [DOI] [PubMed] [Google Scholar]
- Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007:145(1):98–105. 10.1104/pp.107.102079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF III. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014:26(3):1009–1017. 10.1105/tpc.113.120352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009:78(1):273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017:18(7):407–422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004:16(6):1392–1405. 10.1105/tpc.020958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell. 2013:24(4):438–445. 10.1016/j.devcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell. 2006:18(9):2172–2181. 10.1105/tpc.106.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004:131(20):4965–4975. 10.1242/dev.01363 [DOI] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, March-Díaz R, Schmitz RJ, Florencio FJ, Turck F, Amasino RM, Reyes JC. Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS One. 2011:6(3):e17997. 10.1371/journal.pone.0017997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell. 2014:26(7):2920–2938. 10.1105/tpc.114.125690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N, Kumimoto RW, Swain S, Chiara M, Siriwardana C, Horner DS, Holt BF III, Mantovani R. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell. 2017a:29(6):1516–1532. 10.1105/tpc.16.00864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N, Saad D, Chaves-Sanjuan A, Mantovani R, Nardini M. Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol Plant. 2017b:10(4):645–648. 10.1016/j.molp.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006:18(12):3399–3414. 10.1105/tpc.106.047415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Sang Y, Rodrigues A, Biol F, Wu MF, Rodriguez PL, Wagner D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell. 2012:24(12):4892–4906. 10.1105/tpc.112.105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Reyes AA, Malik S, He Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature. 2020:579(7799):452–455. 10.1038/s41586-020-2087-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y. Structure of nucleosome-bound human BAF complex. Science. 2020:367(6480):875–881. 10.1126/science.aaz9761 [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010:19(6):884–894. 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun. 2014:5(1):4601. 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhou L, Huang M, He X, Yang Y, Liu X, Li Y, Hou X. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat Plants. 2018:4(5):289–298. 10.1038/s41477-018-0143-8 [DOI] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006:62(1–2):291–304. 10.1007/s11103-006-9021-2 [DOI] [PubMed] [Google Scholar]
- Hwang K, Susila H, Nasim Z, Jung JY, Ahn JH. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol Plant. 2019:12(4):489–505. 10.1016/j.molp.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999:286(5446):1962–1965. 10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008:228(5):709–723. 10.1007/s00425-008-0773-6 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF III. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010:63(3):379–391. 10.1111/j.1365-313X.2010.04247.x [DOI] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006:133(16):3223–3230. 10.1242/dev.02508 [DOI] [PubMed] [Google Scholar]
- Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot. 2010:61(9):2247–2254. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007:21(4):397–402. 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen C, Gao L, Yang S, Nguyen V, Shi X, Siminovitch K, Kohalmi SE, Huang S, Wu K, et al. The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 2015:11(1):e1004944. 10.1371/journal.pgen.1004944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet. 2016:48(6):687–693. 10.1038/ng.3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li X, Liu Y, Liu H. Flowering responses to light and temperature. Sci China Life Sci. 2016a:59(4):403–408. 10.1007/s11427-015-4910-8 [DOI] [PubMed] [Google Scholar]
- Li M, An F, Li W, Ma M, Feng Y, Zhang X, Guo H. DELLA proteins interact with FLC to repress flowering transition. J Integr Plant Biol. 2016b:58(7):642–655. 10.1111/jipb.12451 [DOI] [PubMed] [Google Scholar]
- Li T, Zhang R, Satheesh V, Wang P, Ma G, Guo J, An GY, Lei M. The chromatin remodeler BRAHMA recruits HISTONE DEACETYLASE6 to regulate root growth inhibition in response to phosphate starvation in Arabidopsis. J Integr Plant Biol. 2022:64(12):2314–2326. 10.1111/jipb.13345 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang G, Liang Y, Hu L, Zhu B, Qi D, Cui S, Zhao H. TCP7 interacts with Nuclear Factor-Ys to promote flowering by directly regulating SOC1 in Arabidopsis. Plant J. 2021:108(5):1493–1506. 10.1111/tpj.15524 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang H, Li X, Liang G, Yu D. Two DELLA-interacting proteins bHLH48 and bHLH60 regulate flowering under long-day conditions in Arabidopsis thaliana. J Exp Bot. 2017:68(11):2757–2767. 10.1093/jxb/erx143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun. 2016:7(1):12768. 10.1038/ncomms12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang Y, Hu Y, Zhou L, Li Y, Hou X. Temporal-specific interaction of NF-YC and CURLY LEAF during the floral transition regulates flowering. Plant Physiol. 2018:177(1):105–114. 10.1104/pp.18.00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Gao Z, Wang Y, Chen Z, Zhang W, Huang J, Yu H, He Y. The NUCLEAR FACTOR-CONSTANS complex antagonizes polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 2018:95(1):17–29. 10.1111/tpj.13926 [DOI] [PubMed] [Google Scholar]
- Mashtalir N, Suzuki H, Farrell DP, Sankar A, Luo J, Filipovski M, D’Avino AR, St Pierre R, Valencia AM, Onikubo T, et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell. 2020:183(3):802–817.e24. 10.1016/j.cell.2020.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. Flowering time regulation produces much fruit. Curr Opin Plant Biol. 2009:12(1):75–80. 10.1016/j.pbi.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008:456(7221):459–463. 10.1038/nature07519 [DOI] [PubMed] [Google Scholar]
- Myers ZA, Holt BF III. NUCLEAR FACTOR-Y: still complex after all these years? Curr Opin Plant Biol. 2018:45(Pt A):96–102. 10.1016/j.pbi.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013:152(1–2):132–143. 10.1016/j.cell.2012.11.047 [DOI] [PubMed] [Google Scholar]
- Park J, Oh DH, Dassanayake M, Nguyen KT, Ogas J, Choi G, Sun TP. Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions. Plant Physiol. 2017:173(2):1463–1474. 10.1104/pp.16.01471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats-Llobet M, Han SK, Gonzalez-Guzman M, Jeong CW, Rodriguez L, Belda-Palazon B, Wagner D, Rodriguez PL. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol Plant. 2016:9(1):136–147. 10.1016/j.molp.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012:24(12):4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012:139(12):2198–2209. 10.1242/dev.077164 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995:80(6):847–857. 10.1016/0092-8674(95)90288-0 [DOI] [PubMed] [Google Scholar]
- Qian Q, Yang Y, Zhang W, Hu Y, Li Y, Yu H, Hou X. A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 2021:231(1):137–151. 10.1111/nph.17314 [DOI] [PubMed] [Google Scholar]
- Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G. Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet. 2019:15(4):e1008065. 10.1371/journal.pgen.1008065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, et al. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 2013:163(1):305–317. 10.1104/pp.113.223933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Liu H, Guan Z, Yan J, Zheng T, Yan W, Wu C, Zhang Q, Yin P, Xing Y. Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell. 2020:32(11):3469–3484. 10.1105/tpc.20.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardana CL, Gnesutta N, Kumimoto RW, Jones DS, Myers ZA, Mantovani R, Holt BF III. NUCLEAR FACTOR Y, subunit A (NF-YA) proteins positively regulate flowering and act through FLOWERING LOCUS T. PLoS Genet. 2016:12(12):e1006496. 10.1371/journal.pgen.1006496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book. 2008:6:e0103. 10.1199/tab.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004:55(1):197–223. 10.1146/annurev.arplant.55.031903.141753 [DOI] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, Reyes JC, Wang A, Keller WA, Harada JJ, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008:147(3):1143–1157. 10.1104/pp.108.121996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Liu X, Liu X, Li Y, Wu K, Hou X. Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol Plant. 2017:10(2):260–273. 10.1016/j.molp.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004:135(2):1008–1019. 10.1104/pp.104.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005:437(7059):693–698. 10.1038/nature04028 [DOI] [PubMed] [Google Scholar]
- Vain T, Raggi S, Ferro N, Barange DK, Kieffer M, Ma Q, Doyle SM, Thelander M, Parizkova B, Novak O, et al. Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc Natl Acad Sci U S A. 2019:116(13):6463–6472. 10.1073/pnas.1809037116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jegu T, Archacki R, Van Leene J, Andriankaja M, et al. ANGUSTIFOLIA3 Binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014:26(1):210–229. 10.1105/tpc.113.115907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pan J, Li Y, Lou D, Hu Y, Yu D. The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 2016:172(1):479–488. 10.1104/pp.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006:18(11):2971–2984. 10.1105/tpc.106.043299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005:309(5737):1056–1059. 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007:19(4):1209–1220. 10.1105/tpc.107.051441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992:100(1):403–408. 10.1104/pp.100.1.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci U S A. 2012:109(9):3576–3581. 10.1073/pnas.1113409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife. 2015:4:e09269. 10.7554/eLife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 2016:590(4):541–549. 10.1002/1873-3468.12076 [DOI] [PubMed] [Google Scholar]
- Yan J, Li X, Zeng B, Zhong M, Yang J, Yang P, Li X, He C, Lin J, Liu X, et al. FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J Integr Plant Biol. 2020:62(11):1717–1740. 10.1111/jipb.12971 [DOI] [PubMed] [Google Scholar]
- Yang J, Xu Y, Wang J, Gao S, Huang Y, Hung FY, Li T, Li Q, Yue L, Wu K, et al. The chromatin remodelling ATPase BRAHMA interacts with GATA-family transcription factor GNC to regulate flowering time in Arabidopsis. J Exp Bot. 2022:73(3):835–847. 10.1093/jxb/erab430 [DOI] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M, Chen CY, Liu X, Luo M, Cui Y, et al. The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell. 2015:27(6):1670–1680. 10.1105/tpc.15.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvao VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 2012:24(8):3320–3332. 10.1105/tpc.112.101014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016:30(2):164–176. 10.1101/gad.270587.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Qian Q, Huang X, Zhang W, Liu X, Hou X. NF-YCs modulate histone variant H2A.Z deposition to regulate photomorphogenic growth in Arabidopsis. J Integr Plant Biol. 2021:63(6):1120–1132. 10.1111/jipb.13109 [DOI] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell. 2014:26(6):2472–2485. 10.1105/tpc.113.121848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li Y, Zhang X, Zha P, Lin R. The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol Plant. 2017:10(1):155–167. 10.1016/j.molp.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018:176(1):790–803. 10.1104/pp.17.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information.