Abstract
Plants cope with various recurring stress conditions that often induce DNA damage, ultimately affecting plant genome integrity, growth, and productivity. The CROWDED NUCLEI (CRWN) family comprises lamin-like proteins with multiple functions, such as regulating gene expression, genome organization, and DNA damage repair in Arabidopsis (Arabidopsis thaliana). However, the mechanisms and consequences of CRWNs in DNA damage repair are largely unknown. Here, we reveal that CRWNs maintain genome stability by forming repairing nuclear bodies at DNA double-strand breaks. We demonstrate that CRWN1 and CRWN2 physically associate with the DNA damage repair proteins RAD51D and SUPPRESSOR OF NPR1-1 Inducible 1 (SNI1) and act in the same genetic pathway to mediate this process. Moreover, CRWN1 and CRWN2 partially localize at γ-H2AX foci upon DNA damage. Notably, CRWN1 and CRWN2 undergo liquid–liquid phase separation to form highly dynamic droplet-like structures with RAD51D and SNI1 to promote the DNA damage response (DDR). Collectively, our data shed light on the function of plant lamin-like proteins in the DDR and maintenance of genome stability.
CROWDED NUCLEI proteins associate with γ-H2AX foci and interact with SUPPRESSOR OF NPR1-1 INDUCIBLE 1 and RAD51D to ensure effective DNA damage repair and maintenance of genome stability.
Introduction
In the complex growth environment, most organisms are constantly subjected to DNA damage caused by various endogenous factors and exogenous genotoxic stresses (Jackson and Bartek 2009; Mehta and Haber 2014), including reactive oxygen species (ROS), drought, UV light, and heavy metals. As the unrepaired DNA damage poses a significant threat to genome integrity and stability, it is essential for the organism to initiate DNA damage response (DDR). DNA double-strand breaks (DSBs) are particularly hazardous damage, which are repaired by 2 main mechanisms: nonhomologous end joining (NHEJ) and homologous recombination (HR) (Nisa et al. 2019). NHEJ is active throughout the cell cycle and directly joins the break ends which have no respect for the original sequence, resulting in mutations and loss of genetic information (Deriano and Roth 2013; Gentric et al. 2021). Conversely, HR takes place only in the S/G2 phases of the cell cycle and repairs the broken DNA with sister chromatid as a template for the restoration of the original sequence (Symington 2016; Hernandez Sanchez-Rebato et al. 2021).
Plants and multicellular animals have largely conserved DNA damage repair processes, involving the protein kinases ataxia telangiectasia mutated (ATM) and ATM and RAD3-related (ATR), which are mainly activated by DSBs and single-stranded DNA lesions, respectively (Amiard et al. 2013; Shiloh 2014; Pedroza-Garcia et al. 2022). Upon activation of DDR, both ATM and ATR are able to phosphorylate histone variant H2AX to produce γ-H2AX, which accumulates at the DSB sites and acts as a marker of DNA damage (Spampinato 2017; Gentric et al. 2021). The cells then initiate the DDR signaling cascades and ultimately result in transcriptional reprogramming of downstream genes, chromatin remodeling, cell cycle perturbation, recruitment of DNA repair machinery, and programmed cell death (PCD) (Yoshiyama 2016; Mahapatra and Roy 2020; Wang, La, et al. 2021). Compared to other multicellular organisms, sessile plants are ubiquitously more susceptible to genotoxic stress and DNA damage. SUPPRESSOR OF GAMMA RESPONSE1 (SOG1), a plant-specific transcription factor, is regarded as a functional analog of the tumor suppressor p53 (Yoshiyama 2016; Wei et al. 2021). As a master transcription factor in controlling DDR, SOG1 is phosphorylated by ATM and ATR kinases and mediates a variety of downstream mechanisms, including cell cycle processes, DDR, and PCD (Weimer et al. 2016; Ryu et al. 2019; Pedroza-Garcia et al. 2022).
In addition, the structural maintenance of chromosome 5/6 (SMC5/6), an evolutionarily conserved chromosomal ATPase protein complex, plays multiple roles in DSB repair (Murray and Carr 2008; Diaz and Pecinka 2018; Palecek 2018). It helps rescue the stalled replication forks and prevents the formation of certain recombinant intermediates. In Arabidopsis (Arabidopsis thaliana), SUPPRESSOR OF NPR1-1 INDUCIBLE 1 (SNI1), the homolog of Non-SMC Element 6 (NSE6), which is a subunit in yeast (Saccharomyces cerevisiae) SMC5/6 complex (Yan et al. 2013; Wang, Chen, et al. 2018; Zhu et al. 2021), performs important roles in DDR by promoting the DNA damage repair and activating cell cycle checkpoint (Wang, Chen, et al. 2018). Subsequent screening studies of SNI1 inhibitors (SSNs) identified DNA damage repair-related factors including Radiation sensitive 51 paralog (RAD51D/SSN1), SWIM domain-containing and Srs2-interacting protein 1 (SWS1/SSN2), Breast Cancer 2A (BRCA2A/SSN3), Radiation sensitive 17 (RAD17/SSN4), and Radiation sensitive 51 (RAD51) (Durrant et al. 2007; Song et al. 2011; Yan et al. 2013).
Despite these identified complexes and factors involving in DDR, how these molecules are dynamically assembled at DSB sites remains unknown. Liquid–liquid phase separation (LLPS) is a process for proteins and other nucleotides (for example, RNAs) to form membrane-free structures when their concentrations reach a critical threshold to allow them to assemble in the surrounding solution (Boeynaems et al. 2018). Recent studies have shown that human p53-binding protein 1 (53BP1) undergoes LLPS at DSB sites and promotes DSB repair (Kilic et al. 2019; Pessina et al. 2019; Zhang, Geng, et al. 2022). In S. cerevisiae, DNA-driven phase separation of Meiotic recombination protein 114 (REC114), Meiosis-specific protein 4 (Mei4), and Meiotic recombination protein 2 (Mer2) proteins plays a vital role in the initiation of recombination during meiosis (Claeys Bouuaert et al. 2021). The plant-specific histone methyltransferase SU(VAR)3-9-Related Protein 2 (MtSUVR2) promotes DDR through chromatin remodeling and LLPS in Medicago truncatula (Liu et al. 2022).
The nuclear lamina, a critical architectural feature under the inner nuclear membrane, performs important roles in the nucleus, including mechanical support for nuclear architecture, signal transduction, chromatin organization, gene regulation, and cell differentiation and metabolism (Stuurman et al. 1998; Dittmer and Misteli 2011; van Steensel and Belmont 2017; de Leeuw et al. 2018). Loss of function of genes encoding nuclear lamina components is associated with dysfunction of the nucleus such as the alteration of nuclear morphology and chromatin organization, which lead to various laminopathies in human including tumorigenesis, premature aging, and degenerative diseases (Redwood et al. 2011; Gibbs-Seymour et al. 2015; de Leeuw et al. 2018; Choi and Richards 2020; Graziano et al. 2021; Chang et al. 2022).
Although putative lamin orthologs were not found in plants, different classes of nuclear coiled-coil proteins have been considered putative nuclear lamin components (Masuda et al. 1993; Dittmer et al. 2007). The nuclear matrix constituent proteins (NMCPs) were identified as candidates to fulfill the roles of lamins across land plants (Masuda et al. 1993, 1997; Ciska et al. 2013; Guo et al. 2014, 2017). CROWDED NUCLEI 1-4 (CRWN1-4) proteins, the putative NMCP orthologs in Arabidopsis, are broadly expressed without obvious tissue specificity (Dittmer et al. 2007; Kimura et al. 2010; Hu et al. 2019). In Arabidopsis, a growing number of studies have demonstrated that functions of CRWNs in nuclear events, including regulation of chromatin organization, gene expression, and nuclear body formation apart from being structural proteins involved in the maintenance of the nuclear morphology (Meier et al. 2017; Groves et al. 2018; Sakamoto 2020).
Loss of function of CRWNs in Arabidopsis results in altered growth traits as well as changed responses to environmental and endogenous factors such as abscisic acid (ABA) (Zhao et al. 2016), copper (Sakamoto et al. 2020), salicylic acid (SA) (Choi et al. 2019), pathogens (Guo et al. 2017), ROS, and DNA damage (Wang et al. 2019). CRWN1 and CRWN4 play roles in the repair of DSBs, and crwn1 crwn4 double mutant showed the high sensitivity to methyl methanesulfonate (MMS) compared to the wild type (WT) (Hirakawa and Matsunaga 2019). Recent studies have demonstrated that CRWN complex and condensin II work synergistically to determine the distribution of centromeres during cell division, which is critical for maintaining genome integrity (Sakamoto et al. 2022).
Here we investigate how CRWN proteins are associated with DDR. Our data suggested that CRWN1 and CRWN2 were partially overlapped with γ-H2AX foci in response to DNA damage. In addition, we explored the physical and genetic interactions between CRWN1 and CRWN2 with RAD51D and SNI1. We further revealed that CRWN1 and CRWN2 undergo LLPS to nucleate the DNA repair machinery including RAD51D and SNI1. Our results shed light on the molecular and cellular mechanisms of Arabidopsis lamin-like proteins in DNA damage repair.
Results
DDR is activated in the crwn1 crwn2 double mutant
To study the mechanistic roles of CRWNs in DDR, we tested the effects of multiple DNA damaging agents, including cisplatin, methyl viologen (MV), and MMS (Wang et al. 2019; Hirakawa and Matsunaga 2019), on the growth of Col-0 (WT), crwn1, crwn2, and crwn1 crwn2 plants. Compared to WT, crwn1 crwn2 seedlings showed obviously increased sensitivity to these DSB-inducing agents (Supplemental Fig. S1, A to C). The transcript levels of DNA damage-responsive genes such as CYCLINB1;1 (CYCB1;1), POLY (ADP-RIBOSE) POLYMERASE 2 (PARP2), and RAD51 were upregulated in the crwn1-4 single mutants and crwn1 crwn3, crwn2 crwn3, and crwn2 crwn4 double mutants after the MMS treatment compared to WT plants (Wang et al. 2019). This prompted us to investigate how CRWN1 and CRWN2 are involved in DDRs. To this end, we first analyzed RNA-seq data of 10-d-old seedlings of crwn1 crwn2 mutant with WT as a control (Guo et al. 2017). Gene Ontology (GO) enrichment analysis indicated that the upregulated genes in crwn1 crwn2 are significantly enriched in the cell cycle process, DNA replication, DNA damage repair, and a wide range of stress responses (Supplemental Fig. S2).
Given that impaired DNA results in abnormal functions of root cells and even death (Li et al. 2016), we examined the root growth of crwn1 crwn2. We found that the length of primary roots in crwn1 crwn2 was dramatically decreased (1.45 cm for crwn1 crwn2 versus 2.32 cm for Col-0) compared to those in WT (Supplemental Fig. S1, D and E). We further examined the growth defects of the roots of 5-d-old seedlings with the propidium iodide (PI), which stains the cell walls of living cells but penetrates dead cells. Compared to Col-0, we found that the roots in crwn1 crwn2 seedlings had more dead cells which obviously increased when seedlings were transferred to MMS-containing medium (Supplemental Fig. S1F), consistent with the hypersusceptibility of crwn1 crwn2 to DNA damage agents.
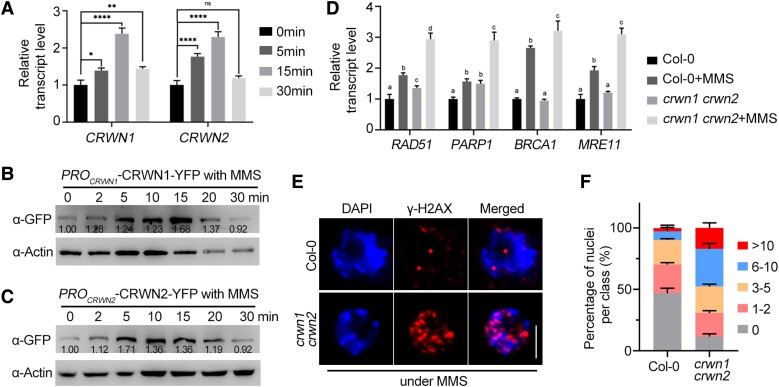
Next, we analyzed the changes of CRWN1 and CRWN2 transcript levels, protein levels, and localizations of CRWN1 and CRWN2 upon MMS treatment. The results showed that the transcript levels of CRWN1 and CRWN2 in root tips were significantly upregulated after MMS treatment and reached a maximum at about 15 min (Fig. 1A). The protein levels in the root tips of PROCRWN1-CRWN1-YFP and PROCRWN2-CRWN2-YFP seedlings were measured by immunoblots. The results showed that CRWN1 and CRWN2 levels were substantially elevated with the maximum accumulation in about 15 min after MMS treatment (Fig. 1, B and C). In MMS-treated or MS-treated root tips of crwn1 crwn2 and WT, we examined the transcript levels of genes associated with major DDR pathways in Arabidopsis (Song et al. 2015). Reverse transcription quantitative PCR (RT-qPCR) indicated that the transcription of RAD51 and POLY (ADP-RIBOSE) POLYMERASE 1 (PARP1) was upregulated in crwn1 crwn2 mutants compared to WT without MMS treatment (Fig. 1D). In addition, the transcription levels of RAD51, BREAST CANCER SUSCEPTIBILITY 1 (BRCA1), PARP1, and MEIOTIC RECOMBINATION 11 (MRE11) were significantly higher in crwn1 crwn2 plants than in Col-0 plants after MMS treatment (Fig. 1D).
Figure 1.
CRWN1 and CRWN2 are involved in DDR. A) The transcript levels of CRWN1 and CRWN2 after MMS treatment detected by RT-qPCR. The 7-d-old root tips were treated with MMS and collected at designated time points for RNA extraction. ACTIN2 was used as an internal control and the transcript level of CRWN1 or CRWN2 without MMS was set to 1. The data are mean ± Sd from 3 biological replicates. Statistical significance was analyzed by 1-way ANOVA followed by Tukey's multiple comparisons test. *P < 0.05, **P < 0.01, and ****P < 0.0001. B, C) Immunoblot analysis of CRWN1 B) and CRWN2 C) protein levels in 10-d-old root tips treated with MMS for designated time. Actin was used as loading control, the protein signals were quantified using ImageJ software, and the protein levels of CRWN1 B) and CRWN2 C) in untreated plants (0 h) were set as 1.00. D) RT-qPCR assays of DNA damage response factors in 7-d-old crwn1 crwn2 root tips with or without MMS treatment compared to Col-0. The data are mean ± Sd from 3 biological replicates. Statistical significance was analyzed by 1-way ANOVA followed by Tukey's multiple comparisons test. Means with different letters are significantly different from each other. E) Immunofluorescence analysis of nuclear γ-H2AX foci in root tip of Col-0 and crwn1 crwn2 mutant following 100 μg/mL MMS treatment. Phosphorylated H2AX (γ-H2AX) and DAPI were counterstained. Scale bar = 5 μm. F) Quantitative analysis of γ-H2AX immunostaining in E). For counting the number of γ-H2AX foci per nucleus, at least 100 nuclei per experiment were divided into 5 categories: nuclei without γ-H2AX foci, nuclei containing 0, 1 to 2, 3 to 5, 6 to 10, or more than 10 γ-H2AX foci, respectively. Three independent experiments were analyzed. Error bars indicate the Sd.
Phosphorylated H2AX (γ-H2AX) acts as a marker of DNA damage (Friesner et al. 2005). A recent study found that the number of γ-H2AX foci increases significantly in crwn1 crwn4 double mutant than that in WT and CRWN1 and CRWN4 function in stabilizing centromere distribution during interphase and protecting the genome stability (Sakamoto et al. 2022). Immunostaining using an antibody against γ-H2AX indicated that the nuclei of crwn1 crwn2 double mutant had more foci than those in WT upon treatment with 100 μg/mL MMS (over 48.5% of nuclei showed more than 5 foci per nucleus in crwn1 crwn2; in contrast, only 9.7% of nuclei have more than 5 foci per nucleus in Col-0) (Fig. 1, E and F). Together, these data suggested that DDR is activated in the crwn1 crwn2 double mutant.
CRWN1 and CRWN2 colocalize with DSBs upon DNA damage
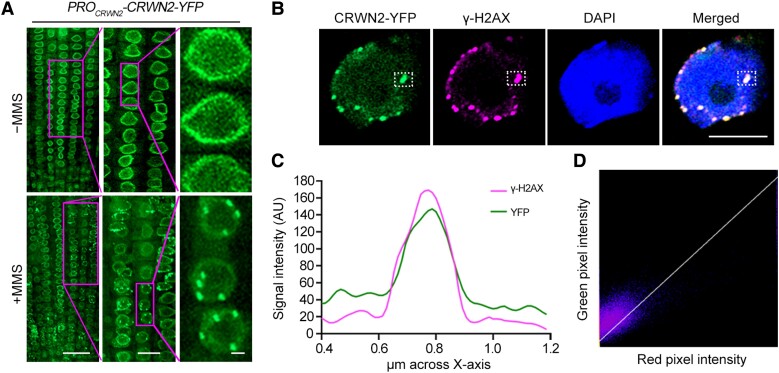
To study the role of CRWN1 and CRWN2 in DDR, we monitored the subnuclear localization of CRWN1 and CRWN2 upon MMS treatment. In root cells of PROCRWN1-CRWN1-YFP and PROCRWN2-CRWN2-YFP transgenic plants, CRWN1 and CRWN2 were found to have a dispersed and homogeneous distribution at the nuclear periphery as previously reported (Figs. 2A and S3A) (Sakamoto et al. 2020; Masuda et al. 2021). However, upon MMS treatment, both CRWN1-YFP and CRWN2-YFP nucleated in foci (Figs. 2A and S3A). Immunofluorescence labeling using anti-γ-H2AX antibody indicated that both CRWN1-YFP and CRWN2-YFP partially colocalized with γ-H2AX foci (Figs. 2B and S3B). We then scanned the overlapping regions in an optical section and found that the signal intensities of CRWN1/2 and γ-H2AX were highly associated (Figs. 2C and S3C).
Figure 2.
CRWN2 localizes at DSB sites upon DNA damage. A) Localization of yellow fluorescent protein-fused CRWN2 (CRWN2-YFP) in the root tips of PROCRWN2-CRWN2-YFP transgenic plants with or without MMS treatment. The 7-d-old seedlings were treated with (down) or without (up) 100 μg/mL MMS, and YFP signals were acquired by the confocal microscopy. The middle and right panels showed the enlarged views of the area in the magenta rectangle. Scale bars from left to right panels were 20, 10, and 1 μm, respectively. B) Representative localization of CRWN2-YFP and γ-H2AX by immunostaining analysis of root cells in PROCRWN2-CRWN2-YFP plants after MMS treatment, and DNA was counterstained with DAPI. The white dotted-line rectangle marks the colocalized foci. Scale bar = 5 μm. C) Fluorescence intensity of the overlapping region in 1 framed optical section in B) measured using ImageJ software for YFP and γ-H2AX channels. D) Coloc 2 in Fiji software was used to analyze the colocation of green and magenta fluorescence in B). Pearson's coefficient: 0.83 and Manders’ coefficients M1 = 1.000 (fraction of γ-H2AX overlapping CRWN2) and M2 = 0.779 (fraction of CRWN2 overlapping γ-H2AX).
Further analyses for these overlapped signals through different colocalization algorithms demonstrated the strong correlation between CRWN1/2 and γ-H2AX signals. For CRWN1, Pearson's R = 0.85, Manders’ M1 = 0.976 (fraction of γ-H2AX overlapping with CRWN1), and M2 = 0.940 (fraction of CRWN1 overlapping with γ-H2AX). For CRWN2, Pearson's R = 0.83, Manders’ M1 = 1.000 (fraction of γ-H2AX overlapping with CRWN2), and M2 = 0.779 (fraction of CRWN2 overlapping with γ-H2AX) (Figs. 2D and S3D). Together, these results demonstrated that both CRWN1 and CRWN2 colocalize with DSBs upon DNA damage.
CRWN1 and CRWN2 undergo LLPS in response to DNA damage
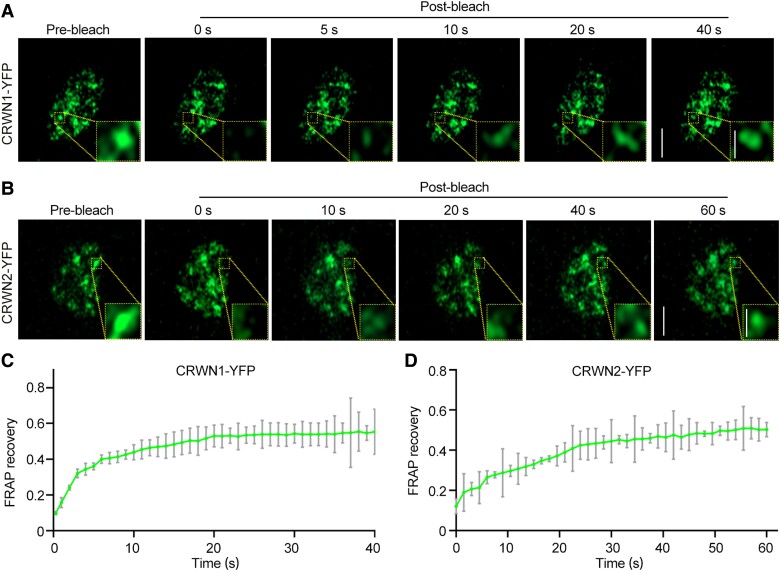
To characterize the protein dynamics in MMS-induced CRWN1 and CRWN2 puncta, we performed fluorescence recovery after photobleaching (FRAP) experiments on the root cells of PROCRWN1-CRWN1-YFP and PROCRWN2-CRWN2-YFP plants. After bleaching the droplets with a laser beam, the CRWN1-YFP and CRWN2-YFP fluorescence intensities in the bleached area were decreased to 9% and 12.1% of the original intensity, respectively. However, they recovered to 56% and 50.5% of the original fluorescence intensity in less than 50 s, indicating that CRWN1-YFP and CRWN2-YFP in these MMS-induced puncta are dynamic (Fig. 3).
Figure 3.
CRWN1 and CRWN2 form dynamic granule-like structures in vivo in response to DNA damage. A, B) Confocal micrographs of fluorescence intensity recovery of CRWN1-YFP A) and CRWN2-YFP B)-containing granule after photobleaching, respectively. The yellow dotted-line rectangle indicates the photobleached regions. Scale bars: 5 and 1 μm (A and B zoom in). C, D) Quantification of fluorescence intensity recovery was performed for the ROI in the FRAP experiment conducted in A) and B). The data are mean ± Sd from 3 biological replicates, with 10 nuclei included in the statistics for each replicate.
We then investigated how the puncta observed above were formed in the nuclei of PROCRWN1-CRWN1-YFP and PROCRWN2-CRWN2-YFP plants. To this end, we evaluated if LLPS promotes the formation of CRWN1 and CRWN2 foci upon DNA damage. First, we analyzed the phase separation potentials of CRWN1 and CRWN2 amino acid (aa) sequence. The DeePhase prediction (Saar et al. 2021) showed a score of 0.77 for CRWN1 (Supplemental Fig. S4B) and 0.81 for CRWN2 (Supplemental Fig. S4D), implying that CRWN1 and CRWN2 might have phase separation potential.
The intrinsically disordered region (IDR) plays a central role in mediating protein phase separation (Lin et al. 2015; Boeynaems et al. 2018; Alberti et al. 2019; Rawat et al. 2021). We analyzed the IDRs in CRWN1 and CRWN2 proteins by IUPred3 (Erdős et al. 2021) and the protein domains by SMART (Letunic et al. 2021). The results showed that the C-terminal domains of both CRWN1 and CRWN2 contain a predicted IDR (Supplemental Fig. S4, C and D) and multiple low-complexity domains (LCD) (Supplemental Fig. S4, A and B) usually required for phase separation (Molliex et al. 2015; Xie et al. 2021).
Next, we asked whether CRWN1 and CRWN2 undergo LLPS in vitro. Since the full-length proteins of CRWN1 and CRWN2 are large and were not successfully expressed in Escherichia coli, we purified the recombinant CRWN1-CC1 (the first coiled-coil, aa 73–366), CRWN1-CC2 (the second coiled-coil, aa 421–714), CRWN1-IDR (aa 799–1131), CRWN2-CC1 (the first coiled-coil, aa 71–365), CRWN2-CC2 (the second and third coiled-coil, aa 391–730), and CRWN2-IDR (aa 810–1127). Polyethylene glycol (PEG), Dextran, or Ficoll has been shown to produce a crowding effect that triggers protein phase separation (Wegmann et al. 2018; Andre and Spruijt 2020). When 10% (w/v) PEG 8000, 15% (w/v) Dextran 70, or 15% (w/v) Ficoll was added into CRWN1-IDR or CRWN2-IDR protein solution, spherical droplets could be observed under microscopy (Figs. 4A and S4E), and some of these droplets could fuse together within 5 s (Fig. 4B). In contrast, recombinant CRWN1-CC1 (aa 73–366), CRWN1-CC2 (aa 421–714), CRWN2-CC1 (aa 71–365), and CRWN2-CC2 (aa 391–730) lacking the IDR were unable to form droplets under LLPS conditions (Supplemental Fig. S5, A and B), demonstrating that IDR is necessary for phase separation of both CRWN1 and CRWN2.
Figure 4.
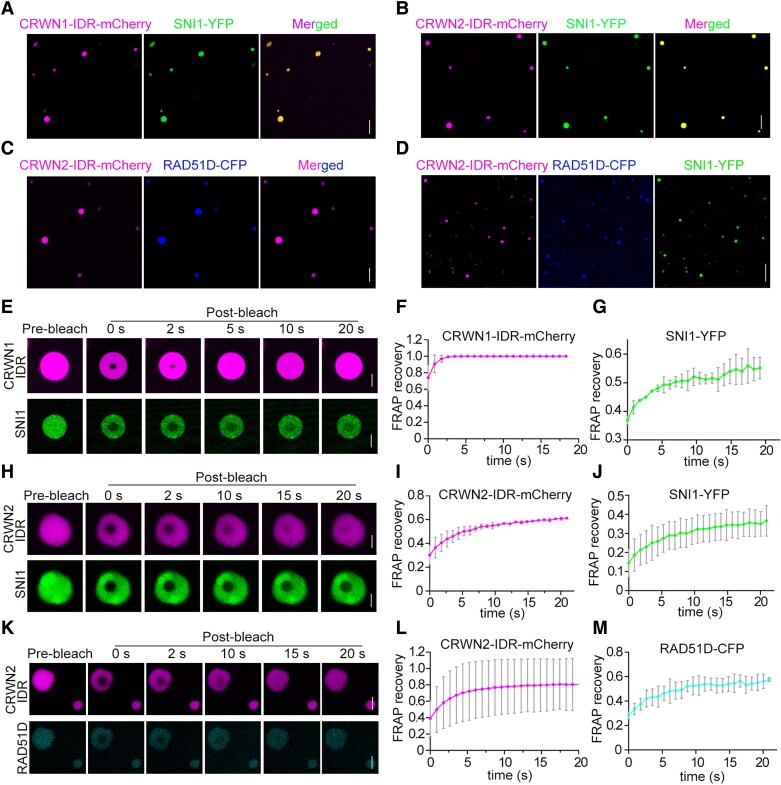
LLPS of CRWN1 and CRWN2 protein in vitro. A) Droplets were formed in CRWN1 or CRWN2 protein solution in vitro with liquid-like property. Representative differential interference contrast (DIC) and confocal images of purified CRWN1-IDR and CRWN2-IDR. IDR, intrinsically disordered region. LLPS condition: 2 mg/mL protein, 100 mM NaCl, pH 7.5, and 10% PEG 8000. Scale bar = 20 μm. B) Fusion of the phase-separated CRWN1-IDR-mCherry and CRWN2-IDR-mCherry droplets. Scale bar = 5 μm. C, D) The phase-separated CRWN1-IDR-mCherry C) and CRWN2-IDR-mCherry D) droplets were subjected to FRAP analysis. The white circles in C) and D) indicated the photobleached regions for fluorescence intensity recovery analysis. Scale bar = 5 μm. E, F) Quantification of fluorescence intensity recovery of a CRWN1-IDR-mCherry C) or CRWN2-IDR-mCherry D) droplets was analyzed from 3 independent experiments, with 15 droplets included in the statistics for each experiment; data are shown as mean ± Sd. The intensity of prebleach was set to 1. G) Co-LLPS of CRWN1-IDR and CRWN2-IDR in vitro. LLPS was performed with 2 mg/mL CRWN1-IDR or/and 2 mg/mL CRWN2-IDR, 100 mM NaCl, pH 7.5, and 10% PEG 8000. Scale bar = 20 μm.
To further probe the mobility of these in vitro CRWN1 and CRWN2 droplets, we performed FRAP experiments. The results showed that the fluorescence in CRWN1-IDR-mCherry and CRWN2-IDR-mCherry droplets recovered partially after laser bleaching (Fig. 4, C to F) in less than 30 s. Notably, the recombinant CRWN1-IDR-YFP and CRWN2-IDR-mCherry could cophase separate when combined in vitro (Fig. 4G).
Given that the IDRs in CRWN1 and CRWN2 are required for LLPS in vitro, we then tested whether IDR is necessary for LLPS of CRWN1 and CRWN2 in vivo. To this end, we generated CRWN1N (aa 1–769)-YFP, CRWN1C (aa 799–1132)-YFP, CRWN2N (aa 1–730)-YFP, and CRWN2C (aa 736–1128)-YFP transgenic plants. We found that CRWN1C-YFP and CRWN2C-YFP formed puncta whereas CRWN1N-YFP and CRWN2N-YFP cannot form obvious puncta under MMS treatment (Supplemental Fig. S5, C to F). Together, these results indicated that both CRWN1 and CRWN2 could undergo phase separation and IDR of CRWNs is important for their LLPS in vivo and in vitro.
CRWN2 interacts with SNI1 and RAD51D
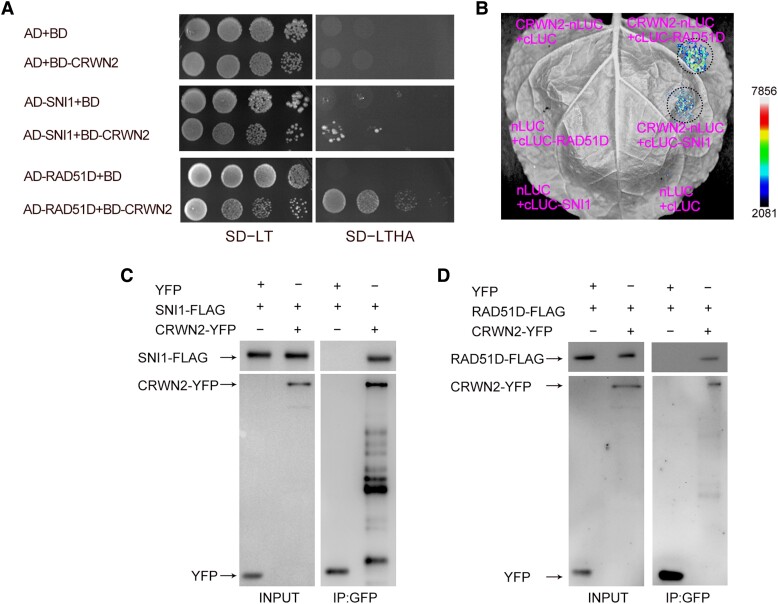
It was known that CRWN1 interacts with SNI1 (Guo et al. 2017), a subunit of the SMC5/6 complex. We tested if CRWN2 interacts with the subunits of the SMC5/6 complex including STRUCTURAL MAINTENANCE OF CHROMOSOME5 (SMC5), SMC6, METHYL METHANESULFONATE SENSITIVITY 21 (AtMMS21), NON-SMC ELEMENT 1 (NES1), NES3, NSE4, and ARABIDOPSIS SNI1-ASSOCIATED PROTEIN 1 (ASAP1), as well as known repressors of sni1 including RAD51D (SSN1), SWS1 (SSN2), BRCA2A (SSN3), RAD17 (SSN4), and RAD51. Interactions between CRWN2 with SNI1 and RAD51D were found in the yeast 2-hybrid (Y2H) and luciferase (LUC) complementation imaging assays (Fig. 5, A and B). We further performed coimmunoprecipitation (Co-IP) experiments in Nicotiana benthamiana and detected RAD51D and SNI1 in the immunoprecipitates of CRWN2 (Fig. 5, C and D).
Figure 5.
CRWN2 interacts with SNI1 and RAD51D. A) Y2H between CRWN2 with SNI1 and RAD51D. Relevant yeast colonies after continuous dilution were spotted onto SD medium (-Leu-Trp or -Leu-Trp-Ade-His). The pGBKT7 (empty bait vector; BD) and pGADT7 (empty prey vector; AD) were used as negative controls. Similar results came from 3 independent experiments. B) The interactions between CRWN2 with SNI1 and RAD51D detected by LUC complementation imaging assays. The empty vector nLUC and cLUC were used as negative controls. Similar results came from 3 independent experiments. The interaction strength is depicted by the color scale. C, D) Co-IP assays detected the interactions between CRWN2 and SNI1 C) or RAD51D D). Similar results came from 3 independent experiments. YFP acted as the negative control. Proteins were detected by immunoblots using anti-GFP and anti-FLAG antibodies.
We then used Y2H, pull-down, and Co-IP assays to map the interaction domains in CRWN2. We found that the N-terminal region of CRWN2 specifically interacts with SNI1 (Supplemental Fig. S6, A and B), and the C-terminal domain (aa 736–1128) of CRWN2, which contains IDR, is critical for its interaction with RAD51D (Supplemental Fig. S6, A, C, and D). Collectively, these results indicated that CRWN2 interacts with SNI1 and RAD51D.
CRWN1 and CRWN2 drive RAD51D and SNI1 to form the droplet-like structures
We then asked whether RAD51D and SNI1 could also form droplets like CRWN1 and CRWN2. The disordered regions of RAD51D and SNI1 were analyzed using IUPred3. The results showed that both RAD51D and SNI1 do not contain any IDRs (Supplemental Fig. S7, A and B). DeePhase prediction showed that RAD51D and SNI1 scores are less than 0.5, suggesting that RAD51D and SNI1 might not undergo LLPS. Recombinant full-length RAD51D and SNI1 were purified from E. coil, but no obvious spherical droplets were observed when 10% PEG 8000 was added into the RAD51D and SNI1 solution (Supplemental Fig. S7, C and D).
Recent studies have shown that the molecular interactions are the driving forces for phase separation of IDRs and play a vital role in maintaining droplet specificity (Borcherds et al. 2021; Liu et al. 2022). The interaction between CRWN1/2 and RAD51D/SNI1 prompted us to test if CRWN1/2 proteins could affect the phase separation of RAD51D and SNI1. When premixed protein solutions of CRWN1-IDR-mCherry with SNI1-YFP, CRWN2-IDR-mCherry with SNI1-YFP, CRWN2-IDR-mCherry with RAD51D-CFP, and CRWN2-IDR-mCherry with RAD51D-CFP/SNI1-YFP were added into 10% PEG 8000, SNI1 and RAD51D were observed to be incorporated into the CRWN-containing droplets (Fig. 6, A to D). The mobility of droplets formed by CRWN1/2 together with RAD51D/SNI1 was investigated by FRAP experiments.
Figure 6.
CRWN1 and CRWN2 drive the phase separation of RAD51D and SNI1. A) Representative confocal micrographs showing the incorporation of SNI1-YFP into droplets formed by CRWN1-IDR-mCherry. IDR, intrinsically disordered region. Scale bar = 20 μm. B) Representative images showing the incorporation of SNI1-YFP into droplets formed by CRWN2-IDR-mCherry. Scale bar = 20 μm. C) Representative images showing the incorporation of RAD51D-CFP into droplets formed by CRWN2-IDR-mCherry. Scale bar = 20 μm. D) Representative images showing the incorporation of RAD51D-CFP and SNI1-YFP into droplets formed by CRWN2-IDR-mCherry. Scale bar = 50 μm. E) The phase-separated CRWN1-IDR-mCherry and SNI1-YFP droplets were subjected to FRAP experiments. Scale bar = 5 μm. F, G) CRWN1-IDR-mCherry F) and SNI1-YFP G) fluorescence intensity statistics of the ROI region in E) were determined. The data are mean ± Sd from 3 biological replicates, and 10 droplets were counted for each replicate. The magenta curve corresponds to CRWN1-IDR-mCherry, and the green curve corresponds to SNI1-YFP. H) The phase-separated CRWN2-IDR-mCherry and SNI1-YFP droplets were subjected to FRAP experiments. Scale bar = 5 μm. I, J) CRWN2-IDR-mCherry I) and SNI1-YFP J) fluorescence intensity statistics of the ROI region in H) were determined. The data are mean ± Sd from 3 biological replicates, and 10 droplets were counted for each replicate. The magenta curve corresponds to CRWN2-IDR-mCherry, and the green curve corresponds to SNI1-YFP. K) The phase-separated CRWN2-IDR-mCherry and RAD51D-CFP droplets were subjected to FRAP experiments. Scale bar = 5 μm. L, M) CRWN2-IDR-mCherry L) and RAD51D-CFP M) fluorescence intensity statistics of the ROI region in K) were determined. The data are mean ± Sd from 3 biological replicates, and 10 droplets were counted for each replicate. The magenta curve corresponds to CRWN2-IDR-mCherry, and the cyan curve corresponds to RAD51D-CFP.
For CRWN1-IDR-mCherry with SNI1-YFP, after the droplets were bleached by a laser beam, the CRWN1-IDR-mCherry fluorescence intensity in the bleached region recovered from 74% to 99% of the original fluorescent intensity in 5 s. The SNI1-YFP fluorescence intensity in the bleached region recovered from 36% to 55% in 25 s (Fig. 6, E to G). For CRWN2-IDR-mCherry with SNI1-YFP, after the droplets were bleached, the mCherry fluorescence intensity in the bleached region recovered from 30% to 61% in 10 s. The YFP fluorescence intensity in the bleached region recovered to from 14% to 36% in 20 s (Fig. 6, H to J). For CRWN2-IDR-mCherry with RAD51D-CFP, after the droplets were bleached, the mCherry fluorescence intensity in the bleached region recovered from 39% to 81% in 15 s. The CFP fluorescence intensity in the bleached region recovered to from 28% to 57% in 15 s (Fig. 6, K to M). These results indicated that CRWN1/2 could drive the phase separation of RAD51D and SNI1.
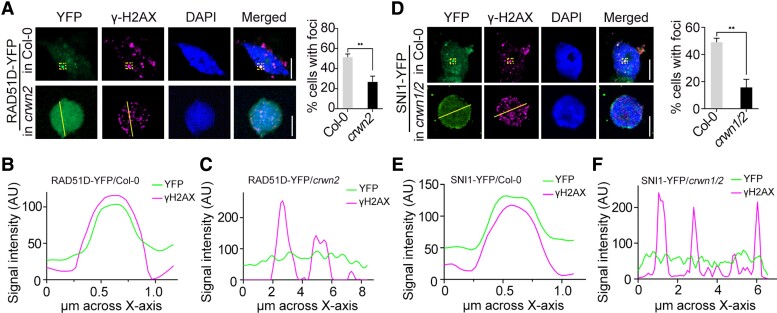
We then tested whether RAD51D or SNI1 also localizes at DSBs in response to DNA damage in the transgenic plants expressing RAD51D-YFP or SNI1-YFP. When DNA damage was induced by MMS, RAD51D-YFP (Fig. 7, A to C) and SNI1-YFP (Fig. 7, D to F) partially colocalized with γ-H2AX foci in the nucleus. To study the effect of CRWN1/2 on the localization of RAD51D/SNI1 to γ-H2AX foci in response to DNA damage, we generated RAD51D-YFP/crwn2 and SNI1-YFP/crwn1/2 transgenic plants. Under the MMS treatment, we found that RAD51D-YFP formed foci in RAD51D-YFP/Col-0 plants in about 51% of cells. In contrast, RAD51D-YFP formed foci in RAD51D-YFP/crwn2 plants in only about 26% of cells (Fig. 7, A to C). SNI1-YFP formed foci in SNI1-YFP/Col-0 plants in about 49% of cells. In contrast, SNI1-YFP formed foci in SNI1-YFP/crwn1 crwn2 plants in only about 15% of cells (Fig. 7, D to F). In addition, when CRWN2 and RAD51D or SNI1 were transiently coexpressed in N. benthamiana, they colocalize a small number of puncta-like structures in the nucleus without MMS treatment. However, these puncta-like structures were significantly increased with MMS treatment (Supplemental Fig. S8, A and B). The results suggested that CRWN proteins could facilitate RAD51D and SNI1 to nucleate at DSB sites.
Figure 7.
CRWNs promote the recruitment of RAD51D and SNI1 to DSB sites in response to DNA damage. A) Representative localization of yellow fluorescent protein-fused RAD51D (RAD51D-YFP) and γ-H2AX by immunostaining analysis of RAD51D-YFP/Col-0 or RAD51D-YFP/crwn2 plants after MMS treatment. Histogram data for the percentage of cells with DSB foci. More than 30 cells were scored in each replicate. The data are mean ± Sd from 3 independent experiments. Statistical significance was analyzed using 2-tailed Student's t test. **P < 0.01. Scale bar = 5 μm. B, C) Representative localization of RAD51D-YFP and γ-H2AX by immunostaining analysis of RAD51D-YFP/Col-0 B) or RAD51D-YFP/crwn2C) plants after MMS treatment. Fluorescence intensity of the overlapping region in yellow dotted-line rectangles framed optical section or on the yellow line in A) measured using ImageJ software for YFP and γ-H2AX channels. D) Representative localization of SNI1 and γ-H2AX by immunostaining analysis of SNI1-YFP/Col-0 or SNI1-YFP/crwn1 crwn2 plants after MMS treatment. Histogram data for the percentage of cells with DSB foci. More than 30 cells were scored in each replicate. The data are mean ± Sd from 3 independent experiments. Statistical significance was analyzed using 2-tailed Student's t test. **P < 0.01. Scale bar = 5 μm. E, F) Representative localization of SNI1-YFP and γ-H2AX by immunostaining analysis of SNI1-YFP/Col-0 E) or SNI1-YFP/crwn1 crwn2F) plants after MMS treatment. Fluorescence intensity of the overlapping region in yellow dotted-line rectangles framed optical section or on the yellow line in D) measured using ImageJ software for YFP and γ-H2AX channels.
CRWN1 and CRWN2 functionally associate with RAD51D and SNI1 in response to DNA damage
To test the genetic relationships among CRWN1, CRWN2, SNI1, and RAD51D, we generated multiple mutants by crossing sni1 rad51d with crwn1 crwn2 mutants. We could not obtain fertile seeds of homozygous crwn1 crwn2 sni1 triple mutant and crwn1 crwn2 rad51d sni1 quadruple mutant due to sterility. Therefore, we focused on the analysis of crwn1 sni1 and crwn2 sni1 double mutants and crwn1 crwn2 rad51d and crwn2 rad51d sni1 triple mutants in subsequent studies.
First, we characterized the genetic relationship between CRWN1 and CRWN2 and SNI1 or RAD51D in genomic DNA repair by comparing the seed germination rates of Col-0, rad51d, sni1, crwn1, crwn2, crwn1 crwn2, crwn1 sni1, crwn2 sni1, and crwn1 crwn2 rad51d mutants with or without MMS treatment. We found that the germination rates of WT, rad51d, sni1, crwn1, crwn2, crwn1 crwn2, crwn1 sni1, crwn2 sni1, and crwn1 crwn2 rad51d seeds showed no obvious difference without MMS treatment (Supplemental Fig. S9, A and B). However, upon treatment with 100 μg/mL MMS, crwn1 crwn2, rad51d, and crwn1 crwn2 rad51d seedlings showed increased MMS sensitivity compared to Col-0 (Supplemental Fig. S9, A and B) as indicated by the lowest germination rate for crwn1 crwn2 rad51d mutant (Supplemental Fig. S9, A and B). In addition, crwn1 sni1 and crwn2 sni1 double mutants showed lower germination rates than Col-0 or their single mutants upon MMS treatment (Supplemental Fig. S9, C and D). Overall, these results suggested that CRWN1 and CRWN2 act synergistically with RAD51D or SNI1 in the pathway of DDR.
Next, we analyzed the genetic relationship between CRWN2 with RAD51D and SNI1 by comparing the phenotypes of Col-0, crwn2, sni1, rad51d sni1, and crwn2 rad51d sni1 mutants. As CRWN2 interacts directly with both SNI1 and RAD51D, we examined the homozygous crwn2 rad51d sni1 triple mutant. The sni1 mutant exhibits pleiotropic phenotypes, such as short roots, reduced fertility, and narrow leaves (Li et al. 1999; Durrant Wendy et al. 2007); the rad51d single mutant displays normal vegetative and reproductive growth (Bleuyard et al. 2005; Durrant Wendy et al. 2007); and the rad51d mutation could largely restore WT morphology in the sni1 mutant (Durrant Wendy et al. 2007). The pleiotropic phenotypes of sni1 may be attributed to the insufficiency of DNA repair (Yan et al. 2013). In the rad51d sni1 double mutant, the HR pathway is blocked, allowing DNA damage to be repaired through alternative repair pathways (Song et al. 2011; Yan et al. 2013; Wang, Chen, et al. 2018). Similar to the sni1 single mutant, the crwn2 rad51d sni1 triple mutant plants are smaller and have narrower leaves (Figs. 8, A and B, and S10C) and more dead root meristematic cells than WT or rad51d sni1 (Fig. 8C), suggesting that the phenotypic recovery effect of rad51d on sni1 in the rad51d sni1 double mutant is CRWN2-dependent without MMS treatment.
Figure 8.
Phenotypic recovery in the rad51d sni1 double mutant was CRWN2-dependent. A) Visual phenotypes of Col-0, crwn2, rad51d, sni1, rad51d sni1, and crwn2 rad51d sni1 plants grown vertically on 1/2 MS medium without (upper panel) or with (lower panel) 100 μg/mL MMS at 8 d after germination. Scale bar = 10 mm. B) The statistical measurements of root lengths in A) without (upper panel) or with (lower panel) MMS treatment. The data in B) are mean ± Sd from 3 independent experiments (n = 20 seedlings per replicate). Statistical significance was analyzed by 1-way ANOVA followed by Tukey's multiple comparisons test. ****P < 0.0001. ns, nonsignificant. C) The root meristem structures of 5-d-old seedlings of Col-0, crwn2, rad51d, sni1, rad51d sni1, and crwn2 rad51d sni1 mutants stained with PI. Similar results were obtained from 3 independent experiments, and the representative results are shown. Scale bar = 20 μm.
To test if the role of CRWN2 in the phenotypic restoration effect of rad51d on sni1 is responsive to DNA damage, we compared the seed germination rates and primary root lengths of Col-0, crwn2, sni1, rad51d sni1, and crwn2 rad51d sni1 mutant plants with or without MMS treatment. The germination rates of Col-0, crwn2, sni1, rad51d sni1, and crwn2 rad51d sni1 showed no obvious differences without MMS treatment (Supplemental Fig. S10, A and B). Upon MMS treatment, all of these mutants showed increased MMS sensitivity compared to Col-0 as shown by the reduced germination rates and primary root growths (Supplemental Figs. S10, A and B, and 8, A and B). The sni1 single mutant had a germination rate of 74.3%, which was partly relieved by introducing rad51d mutation to the sni1 mutant (89.6% for rad51d sni1) under MMS treatment. In addition, the phenotypic restoration effect of rad51d mutation on sni1 mutant was partly impaired in crwn2 rad51d sni1 triple mutant (81.6%) (Supplemental Fig. S10, A and B). Similar results were also observed by comparing the primary root growths of sni1, rad51d sni1, and crwn2 rad51d sni1 mutants (Fig. 8, A and B). Together, these results indicated that CRWN2 associates functionally with RAD51D and SNI1 in response to DNA damage.
Discussion
Unlike animals, plants are immobile and rely on sunlight for photosynthesis, making them vulnerable to various kinds of endogenous and exogenous DNA-damaging factors throughout their lifetime. Thus, it is essential for them to develop efficient mechanisms to maintain genome stability. Given the diverse functions of CRWNs and the DDR-related phenotypes of crwn mutants, in-depth functional studies of CRWNs are crucial for understanding their molecular mechanisms in DDR.
CRWN1 and CRWN2 play direct roles in DDR
CRWNs, which are the candidates for the plant nuclear lamina proteins in Arabidopsis, regulate chromatin organization, nuclear size, and gene expression (Meier et al. 2017; Groves et al. 2018; Sakamoto 2020). Studies have reported that crwn mutants are hypersensitive to ABA treatment (Zhao et al. 2016) and multiple stresses, such as copper (Sakamoto et al. 2020) and drought (Yang et al. 2020). Interestingly, the crwn mutants displayed strong DNA DSB-induced phenotypes (Hirakawa and Matsunaga 2019; Wang et al. 2019; Sakamoto et al. 2022) (Supplemental Fig. S1, A to C). However, how CRWNs mediate DNA damage repair is not immediately clear.
Depletion of CRWN1 and CRWN2 may result in elevated DNA damage, as crwn1 crwn2 plants displayed short roots with more dead cells even without MMS treatment than WT (Supplemental Fig. S1, D to F). The crwn1 crwn2 and crwn1 crwn4 double mutants showed the accumulation of SA and activated defense responses in Arabidopsis seedlings (Guo et al. 2017; Choi et al. 2019). The production of SA, which is largely dependent on NONEXPRESSER OF PR GENES 1 (NPR1), restrains seedling development even under normal conditions. As a result, mutation of NPR1 in crwn1 crwn2 partly restored the primary root growth (Guo et al. 2017). The activation of DDR is an intrinsic mechanism during immune responses in plants (Yan et al. 2013). Some DNA damage repair proteins can be recruited to repair DNA damage sites or to the promoters of defense genes to promote gene expression (Yan et al. 2013). Thus, studying the potential role of CRWNs in the crosstalk between immune response and DNA damage repair in plants is significant.
γ-H2AX is a solid marker for DSBs in plant and animal cells (Friesner et al. 2005). The frequency of γ-H2AX foci colocalized with DAPI-stained chromosomes significantly increased in crwn1 crwn4 mutants under zeocin treatment (Sakamoto et al. 2022). Upon MMS treatment, γ-H2AX was induced in the crwn1 crwn2 mutant. In response to DNA damage, DNA repair foci, which are formed by DNA repairing factors, accumulate at damaged sites to promote DSB repair (Rothkamm et al. 2015; Horvath et al. 2017). In order to understand DSB repair mechanism, it is important to determine whether and how CRWN proteins are recruited to the DNA damage sites.
In this study, we revealed that CRWN1 and CRWN2 relocalized to DSB sites after DNA damage and at least partially overlap with γ-H2AX foci, which implicated a direct role of CRWN1 and CRWN2 in DDR. Nuclear membrane-associated proteins have an impact on the efficiency of DNA repair in eukaryotes (Dona and Mittelsten Scheid 2015). In Drosophila (Drosophila melanogaster), nuclear periphery as a specialized site for completing heterochromatic DSB repair and defects in anchoring DSBs at the nuclear periphery reduce the tolerance to γ-irradiation (Ryu et al. 2015). The plant-specific inner nuclear membrane proteins CRWN1 and CRWN2 may tether the DSBs to the nuclear periphery to mediate DNA repair. Further studies are required to address how CRWNs recognize and move to DNA damage sites.
CRWN1 and CRWN2 nucleate repair bodies through LLPS at the DNA damage sites in DDR
Biomolecular condensates have become a key player in cellular processes and stress responses (Alberti and Hyman 2021), and recent studies have shown that biomolecular condensates could be produced by LLPS, which plays important roles in diverse biological functions in different organisms, such as regulating gene expression, cell division, and cellular stress resistance (Wang, Conicella, et al. 2018; Woodruff 2018; Pessina et al. 2019; Zamudio et al. 2019; Jung et al. 2020; Claeys Bouuaert et al. 2021; Huang et al. 2021; Zhu et al. 2022). It was discussed that CRWN1 and PLANT NUCLEAR ENVELOPE TRANSMEMBRANE 2 (PNET2), components of the plant nuclear lamina, may undergo LLPS and contribute to the establishment of the separate phase/environment of plant nuclear lamina (Huang et al. 2021; Tang et al. 2021).
In Arabidopsis seedling and flower tissues, 985 proteins with phase separation potential, including CRWN1 and CRWN4, were identified from cell lysates in a recent study (Zhang, Peng, et al. 2022). We found that the IDR domains of CRWN1 and CRWN2 are critical for their LLPS in vitro (Figs. 4 and S5). CRWN1 and CRWN2 exhibit capacities for phase separation under MMS-treated conditions in vivo (Fig. 3). FRAP experiments showed that CRWN1 and CRWN2 form dynamic droplet-like structures. Interestingly, CRWN1 and CRWN2 protein levels are briefly accumulated and subsequently degraded following MMS induction in plant cells (Fig. 1, B and C).
Molecular interactions are the driving force for LLPS which allows relevant proteins to enter the droplet to maintaining droplet specificity. RAD51D and SNI1 cannot form droplets by themself, but can be facilitated by CRWN1/2 through LLPS due to the interactions between CRWN1 and CRWN2 with RAD51D and SNI1 (Fig. 6). Previous studies showed that SNI1 is a subunit of the SMC5/6 complex, which can be recruited to DNA damage sites to promote HR (Jianbin Lai 2018; Jiang et al. 2019). RAD51D has an essential function in DNA damage repair (Durrant Wendy et al. 2007; Wang et al. 2014). By studying the relationship between CRWN1 and CRWN2 with RAD51D and SNI1 proteins in DNA damage repair in vivo, we revealed that CRWN1 and CRWN2 can significantly promote the localization of RAD51D and SNI1 to DSB sites in response to DNA damage (Figs. 7 and S8). We therefore hypothesize that CRWN1and CRWN2 may drive RAD51D and SNI1 into the “DNA repair body” for efficient DNA repair.
SNI1 could form a complex with RAD51D and the rad51d mutation restores WT morphology to sni1 mutant in the rad51d sni1 double mutant. SNI1 and RAD51D may influence each other’s activity through physical interaction (Martín et al. 2006; Song et al. 2011; Yan et al. 2013). In this study, we determined that CRWN2 interacts with RAD51D and SNI1 (Fig. 5). The phenotypic recovery in the rad51d sni1 double mutant was CRWN2-dependent, and the crwn2 rad51d sni1 triple mutant is more sensitive to DSB-inducing agents than the rad51d sni1 double mutant (Figs. 8 and S10, A and B), indicating functional associations among CRWNs, RAD51D, and SNI1 in the DDR pathway. CRWN1 interacts with CRWN2 to form heterooligomers for building the meshwork structure (Sakamoto et al. 2020). CRWNs, SNI1, and RAD51D might form a complex to play dual roles in regulating both HR and the expression of defense-related genes (Durrant Wendy et al. 2007). Physical interactions between CRWNs and SNI1 and RAD51D might contribute to establishing a phase separation environment to form repair bodies at DNA damage sites.
In this study, we provided direct evidence to show how CRWNs function in DNA repair in plants. When DNA damage occurs, CRWN1 and CRWN2 interact with SNI1 and RAD51D to form the “DNA repair body” at γ-H2AX foci by LLPS for effective repair of the damaged DNA (Fig. 9). As CRWNs and their homologous proteins are widespread in diverse organisms, the current findings will shed light on DNA repair in other species.
Figure 9.
A working model for the role of CRWN1 and CRWN2 proteins in the regulation of DNA damage repair in Arabidopsis. When DNA damage occurs, CRWN1 and CRWN2 proteins are induced and undergo LLPS to localize at DNA DSB sites. In addition, CRWN1 and CRWN2 interact with SNI1 and RAD51D to nucleate “DNA repair body” at DSB sites for effective repair of the damaged DNA.
Materials and methods
Plant materials, growth conditions, and chemical treatments
Arabidopsis (A. thaliana) (accession Col-0), crwn1 crwn2 (Dittmer et al. 2007; Guo et al. 2017), sni1, rad51d, and rad51d sni1 (Durrant Wendy et al. 2007) (kind gifts from Dr. Shunping Yan) mutants were used in this study. The crwn1 crwn2 sni1 and crwn2 rad51d sni1 triple mutants were generated by crossing crwn1 crwn2 with sni1 rad51d. All mutants were verified by PCRs. The Arabidopsis seeds were surface-sterilized in 75% (v/v) ethyl alcohol for 15 min and then sown on half-strength MS medium (1/2 MS). Col-0 and N. benthamiana plants were grown in a growth room at 22 °C under a 16-h light/8-h dark light cycle, light emitting diode (LED) light intensity of 200 μmol/m2/s, the spectral range of 400 to 700 nm, and 70% humidity conditions. All transgenic plants were generated by the floral dip method (Zhang et al. 2006).
For chemical treatments, Col-0 and mutants were treated with MMS (Sigma M4016) (50/100 μg/mL) and the crosslinking agents cisplatin (Sigma P4394) (50 μM) and MV (Sigma 856177) (1 μM).
Constructs and transgenic plants
The coding sequence (CDS) of CRWN2 including its upstream regulatory sequence was subcloned into pCAMBIA 131-N1-YFP (Guo et al. 2017) to generate PROCRWN2-CRWN2-YFP. The coding sequences of CRWN1N (aa 1–769), CRWN1C (aa 799–1132), CRWN2N (aa 1–730), CRWN2C (aa 736–1128), RAD51D, and SNI1 were subcloned into vector pCAMBIA 131-35S-N-YFP (Guo et al. 2017) to generate PRO35S-CRWN1N-YFP, PRO35S-CRWN1C-YFP, PRO35S-CRWN2N-YFP, PRO35S-CRWN2C-YFP, PRO35S-RAD51D-YFP, and PRO35S-SNI1-YFP, respectively. Arabidopsis WT plants were transformed with the PRO35S-CRWN1N-YFP, PRO35S-CRWN1C-YFP, PRO35S-CRWN2N-YFP, PRO35S-CRWN2C-YFP, PRO35S-RAD51D-YFP, and PRO35S-SNI1-YFP individually to generate the corresponding overexpression lines. The PRO35S-RAD51D-YFP and PRO35S-SNI1-YFP transgenic lines were then crossed with crwn1 crwn2 to generate PRO35S-RAD51D-YFP/crwn2 and PRO35S-SNI1-YFP/crwn1 crwn2.
Transient expression
Agrobacterium-mediated transient protein expression was performed as described (Gu et al. 2016; Yan et al. 2017). Agrobacterium tumefaciens strain GV3101 harboring fused constructs was infiltrated into 3-wk-old N. benthamiana leaves with a 1-mL needleless syringe. For coexpression, N. benthamiana leaves were coinfiltrated with GV3101 carrying transgene constructs or an empty vector (the negative control) mixed in a 1:1 ratio to OD600 = 0.5. Samples were collected for microscopic analysis or immunoblotting analysis 48 h after infiltration. For MMS treatment, the N. benthamiana leaves were incubated with 50 μg/mL MMS for 6 h before imaging as described (Wang et al. 2022).
Y2H assays
Y2H was performed according to the Yeastmaker Yeast Transformation System 2 User Manual (Clontech). The full-length CDSs or truncated versions of interested genes were cloned into bait vector pGBKT7 or prey pGADT7, respectively, and then transformed into S. cerevisiae strain AH109. The transformed yeast cells were cultured on synthetic defined (SD) mediums, including double (SD-Leu-Trp) and quadruple (SD-Leu-Trp-His-Ala), at 30 °C for 3 to 5 d.
Co-IP
Fluorescence or epitope tag-conjugated proteins were transiently coexpressed in N. benthamiana leaves. Total proteins were extracted in protein extraction buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 1% [v/v] Triton X-100, 10% [v/v] glycerol, 1 mM PMSF, and 1×Cocktail), then incubated with anti-GFP mAb-Magnetic Beads (MBL, D153-11) overnight at 4 °C with gentle rotation. The beads were washed 5 times with washing buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM EDTA, and 1 × Cocktail). The immunoprecipitated proteins in 5 × SDS loading buffer were eluted by boiling at 100 °C for 10 min and loaded into SDS–PAGE, then subjected to immunoblot assays using anti-GFP (Abcam, ab290, dilution: 1:3,000) and anti-FLAG (Abcam, ab1170, dilution: 1:3,000) antibodies. The following secondary antibodies were used for immunoblotting: antirabbit horseradish peroxidase (HRP)-linked IgG antibody (7074, Cell Signaling, dilution: 1:8,000) and antimouse HRP-linked IgG antibody (7076, Cell Signaling, dilution: 1:8,000).
LUC complementation imaging assay
The firefly LUC complementation imaging assay was carried out as described (Chen et al. 2008; Guo et al. 2017). Full-length CDSs of CRWN1, CRWN2 genes, and SNI1 or RAD51D were fused with the N- or C-terminal fragments of LUC (NLUC or CLUC). The fused constructs were introduced into Agrobacterium strain GV3101 and then coinfiltrated into fully expanded 3-wk-old N. benthamiana leaves. After infiltration for 48 h, the leaves were infiltrated with 100 mM luciferin (Sangon, dissolved in water) and kept in the dark for 10 min for luminescence quenching. LUC activity was examined with a Chemiluminescence Imaging Analysis System (Tanon 5500). Each experiment was performed on 6 leaves of 3 individual plants and repeated at least 3 times.
In vitro pull-down assay
The pull-down assays were performed as described previously (Wang, Zhan, et al. 2021). Full length of SNI1 was cloned into vector pGEX4T (Beyotime, D2916) with a CFP tag, and CRWN2N1 (aa 1–365) was subcloned into pET28a (Yeasen, 11905ES03) with a mCherry tag. Recombinant proteins were expressed in E. coli BL21(DE3) and induced with 0.5 mM isopropyl β-D-thiogalactoside (IPTG) at 25 °C for 8 h. Cells were harvested and lysed by sonication. The His-CRWN2N1 was purified using His-tag Purification Resin according to the protocol. GST and GST-SNI1-CFP were purified using Glutathione Sepharose according to the protocol (GE Healthcare) and incubated with the purified His-CRWN2N1-mCherry protein in the phosphate buffer saline (PBS) buffer at 4 °C for 3 h, then the beads were washed 5 times with wash buffer (10 mM PBS [pH 7.4] and 10% glycerol). The beads were boiled in 1 × SDS loading buffer, followed by immunoblot analysis using anti-GST (abcam; ab111947, dilution: 1:3,000) and anti-mCherry (Engibody; AT0037, dilution: 1:3,000) antibodies.
RT-qPCR assays
For RT-qPCR, total RNA was extracted from root tips of 7-d-old seedlings using a TaKaRa MiniBEST Universal RNA Extraction Kit, then subjected to reverse transcription using ReverTra Ace qPCR RT Master Mix with gRNA Remover (TOYOBO) according to the manufacturer's instruction. The cDNA templates were used for RT-qPCRs using SYBR Premix Ex Taq (Takara) in a Bio-Rad CFX96 Real-Time system. All primers used for RT-qPCR are listed in Supplemental Table S1. Values were from 3 biological replicates.
Immunofluorescence assays
Immunofluorescence experiments were performed as previously described (Zhao et al. 2019) with slight modifications. Briefly, 10-d-old seedlings were treated with or without MMS. Seedlings were fixed with 4% (w/v) paraformaldehyde for ∼30 min using a vacuum system, washed with PBS 3 times (5 min each) at room temperature. Subsequently, extracted nuclei were spread onto a poly-lysine slide, incubated with the primary antibody (Zhou et al. 2016) (anti-γ-H2AX, kind gift from Dr. Zhu Yan) (1:100 dilution) at 4 °C overnight. The second antibody (the Alexa Fluor 555-coupled goat antirabbit, bioss; catalog no. bs-0295G-AF555) was used at 1:200 at 37 °C for 90 min. Finally, the slide was washed in PBS with 0.05% (v/v) Tween-20 (PBST) and counterstained in Fluoromount mounting media with DAPI (YEASEN 36308ES11). Immunofluorescence signals were detected under a microscopy (Zeiss ZEN 3.4) with the following excitation/emission wavelengths: YFP was excited at 514 nm and detected at 513 to 542 nm; DAPI was excited at 405 nm and detected at 410 to 488 nm; and Alexa Fluor 555 was excited at 555 nm and detected at 543 to 697 nm. The pixel intensity of a region of interest (ROI) was analyzed using the Plot Profile module of ImageJ and the fluorescence colocalization analysis was done with the ImageJ plugins Coloc2 (Dunn et al. 2011; Wang et al. 2017).
For root tips, immunofluorescence experiments were performed as described previously (Friesner et al. 2005; Hirakawa et al. 2017; Hirakawa and Matsunaga 2019). The root tips of 5-d-old seedlings treated with or without MMS were fixed with 4% (w/v) paraformaldehyde for ∼30 min, washed with PBS 3 times (5 min each) at room temperature. Tips were digested for 2 min at 37 °C in digestion buffer containing 0.3% (w/v) driselase (Sigma-Aldrich), 0.3% (w/v) cellulose R-10 (Wako), and 0.3% (w/v) pectolyase Y23 (Wako) in water. Samples were then washed 3 times for 5 min with PBS. Root tips were placed on a poly-lysine slide and squashed using a glass. The slides were immediately placed in liquid N2 and the cover glass was removed. The slides were dried and then immersed in 4% (w/v) paraformaldehyde for ∼30 min and then washed 3 times in PBS for 5 min each. The next steps were performed as for nuclear immunofluorescence.
Quantification of γ-H2AX foci in the nuclei
Foci of γ-H2AX were subjected to quantitative analysis using the Spot Pattern module of Imaris software (Bitplane AG). At least 100 nuclei per plant line and per experiment were counted, and they were divided into 4 categories according to the number of γ-H2AX foci in each nucleus: nuclei without γ-H2AX foci, nuclei with 0, 1 to 2, 3 to 5, 6 to 10, or more than 10 γ-H2AX foci.
Protein expression in E. coli and purification
All recombinant proteins were expressed in the E. coli Rosetta (DE3) strain. CRWN1/2-CCs (lacking the IDR) and CRWN1/2-IDR were cloned into pGEX4T (Beyotime, D2916) with a mCherry or YFP tag. Full lengths of RAD51D and SNI1 CDS were individually cloned into pGEX4T with a CFP tag or YFP tag. All proteins were induced with 0.5 mM IPTG at 25 °C for 8 h, collected and resuspended in lysis buffer (10 mM PBS [pH 7.4], 10% glycerol, 0.1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride). The cells were then lysed and centrifuged. The supernatant was added to Glutathione Sepharose beads at 4 °C for 3 h. Then wash beads 5 times in washing buffer (10 mM PBS [pH 7.4] and 10% glycerol) for 5 min each. The purified proteins were stored in stock buffer (50 mM Tris [pH 7.4], 500 mM NaCl, and 1 mM dithiothreitol) at −80 °C after being flash-frozen in liquid nitrogen.
In vitro phase separation assay
The GST tags of purified GST-CRWN1-IDR-YFP, GST-CRWN1-IDR-mCherry, GST-CRWN1-CCs-mCherry, GST-CRWN2-IDR-mCherry, GST-CRWN2-CCs-mCherry, GST-RAD51D-CFP, GST-SNI1-CFP, and GST-SNI1-YFP were cleaved with PreScission Protease overnight at 4 °C. PreScission Protease cleavage efficiency was verified by SDS–PAGE analysis. In vitro phase separation assay was performed in solution (50 mM Tris-HCl, pH 7.4, and 100 mM NaCl) containing the protein samples and different concentrations of PEG 8000 (Sigma, 89510), Ficoll 400 (Sangon Biotech), or Dextran-70 (Sangon Biotech). After incubation at room temperature for 1 h, droplets were observed by confocal microscopy. We used the following settings for excitation/emission of fluorescence: YFP was excited at 514 nm and detected at 513 to 542 nm; CFP was excited at 456 nm and detected at 460 to 488 nm; and mCherry was excited at 594 nm and detected at 600 to 650 nm.
FRAP
In vivo FRAP of CRWN1-YFP and CRWN2-YFP in Arabidopsis root tip cells was performed using a Zeiss LM980 confocal laser microscope equipped with a 60× oil immersion objective. A region of a CRWN1-YFP or CRWN2-YFP condensate was bleached using a 514-nm laser pulse with 50% intensity. Recovery was recorded every second for a total of 60 s after bleaching. In vitro FRAP experiments were performed with samples in slide using a Zeiss LM980 confocal laser microscope equipped with a 60× oil immersion objective. A small area of droplet was bleached with a 594-nm laser for mCherry, a 514-nm laser for YFP, and a 456-nm laser for CFP with 80% intensity. Fluorescence recovery was recorded every second for a total of 60 s after bleaching. For all the FRAP experiments, the fluorescence intensities of bleached, reference, and background regions were analyzed with Zeiss ZEN image software and Prism (GraphPad).
Accession numbers
Sequence data from this article can be found at https://www.arabidopsis.org/ (Arabidopsis Genome initiative) under the following accession numbers: CRWN1 (AT1G67230), CRWN2 (AT1G13220), SNI1 (AT4G18470), RAD51D (AT1G07745), BRCA1 (AT4G21070), RAD17 (AT5G66130), PARP1 (AT2G31320), MRE11 (AT5G54260), RAD51 (AT5G20850), and ACTIN2 (AT3G18780).
Supplementary Material
Acknowledgments
We thank Dr. Shunping Yan from Huazhong Agricultural University for sharing the rad51d sni1 mutant; Dr. Yan Zhu from Fudan University for sharing the γ-H2A.X; and Dr. Xiaoyu Tu from Shanghai Jiao Tong University for critical reading of the manuscript.
Contributor Information
Chunmei Yin, Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China.
Aiqing Sun, Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China.
Tongtong Guo, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai 200234, China.
Xuegao Mao, National key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200032, China.
Yuda Fang, Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China.
Author contributions
Y.F. and C.Y. designed the study; C.Y., A.S., T.G., and X.M. performed the experiments; C.Y. and Y.F. wrote the paper. All authors discussed the results and made comments on the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The crwn1 crwn2 double mutant seedlings show hypersensitive phenotypes to cisplatin, MMS, and MV.
Supplemental Figure S2 . Depletion of CRWN1 and CRWN2 results in the activation of DDR and cell cycle arrest.
Supplemental Figure S3 . CRWN1 localizes at DSBs upon DNA damage.
Supplemental Figure S4 . LLPS and sequence analyses of CRWN1 and CRWN2.
Supplemental Figure S5 . IDR of CRWN1 and CRWN2 is necessary to drive LLPS in vitro and in vivo.
Supplemental Figure S6 . The interactions between the different domains of CRWN2 and RAD51D or SNI1.
Supplemental Figure S7 . RAD51D and SNI1 have no phase separation potential.
Supplemental Figure S8 . RAD51D and SNI1 foci colocalize with CRWN2.
Supplemental Figure S9. CRWN1 and CRWN2 regulate synergistically the DNA damage repair with RAD51D and SNI1.
Supplemental Figure S10 . Loss of function of CRWN2 partially suppresses the phenotypic recovery of rad51d sni1 double mutant.
Supplemental Figure S11. Original images of the immunoblotting data.
Supplemental Table S1. List of primers used in this study.
Supplemental Date Set S1. The t test and ANOVA results in this study.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31871230 and 32170585 to Y.F. and 32100260 to A.S.).
References
- Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019:176(3):419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021:22(3):196–213. 10.1038/s41580-020-00326-6 [DOI] [PubMed] [Google Scholar]
- Amiard S, Gallego ME, White CI. Signaling of double strand breaks and deprotected telomeres in Arabidopsis. Front Plant Sci. 2013:4:405. 10.3389/fpls.2013.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre AAM, Spruijt E. Liquid-liquid phase separation in crowded environments. Int J Mol Sci. 2020:21(16):5908. 10.3390/ijms21165908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005:41(4):533–545. 10.1111/j.1365-313X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018:28(6):420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021:67:41–50. 10.1016/j.sbi.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Li M, Shao S, Li C, Ai S, Xue B, Hou Y, Zhang Y, Li R, Fan X, et al. Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell. 2022:13(4):258–280. 10.1007/s13238-020-00794-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):323–324. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Richards EJ. The role of CRWN nuclear proteins in chromatin-based regulation of stress response genes. Plant Signal Behav. 2020:15(1):1694224. 10.1080/15592324.2019.1694224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Strickler SR, Richards EJ. Loss of CRWN nuclear proteins induces cell death and salicylic acid defense signaling. Plant Physiol. 2019:179(4):1315–1329. 10.1104/pp.18.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M, Masuda K, Moreno Díaz de la Espina S. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J Exp Bot. 2013:64(6):1553–1564. 10.1093/jxb/ert020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys Bouuaert C, Pu S, Wang J, Oger C, Daccache D, Xie W, Patel DJ, Keeney S. DNA-driven condensation assembles the meiotic DNA break machinery. Nature. 2021:592(7852):144–149. 10.1038/s41586-021-03374-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw R, Gruenbaum Y, Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2018:28(1):34–45. 10.1016/j.tcb.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013:47(1):433–455. 10.1146/annurev-genet-110711-155540 [DOI] [PubMed] [Google Scholar]
- Diaz M, Pecinka A. Scaffolding for repair: understanding molecular functions of the SMC5/6 complex. Genes (Basel) 2018:9(1):36. 10.3390/genes9010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011:12(5):222. 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007:19(9):2793–2803. 10.1105/tpc.107.053231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donà M, Mittelsten Scheid O. DNA damage repair in the context of plant chromatin. Plant Physiol. 2015:168(4):1206–1218. 10.1104/pp.15.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011:300(4):C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant Wendy E, Wang S, Dong X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA. 2007:104(10):4223–4227. 10.1073/pnas.0609357104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdős G, Pajkos M, Dosztányi Z. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021:49(W1):W297–W303. 10.1093/nar/gkab408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation–dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and rad3-related. Molecular Biology of the Cell. 2005:16(5):2566–2576. 10.1091/mbc.e04-10-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric N, Genschik P, Noir S. Connections between the cell cycle and the DNA damage response in plants. Int J Mol Sci. 2021:22(17):9558. 10.3390/ijms22179558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs-Seymour I, Markiewicz E, Bekker-Jensen S, Mailand N, Hutchison CJ. Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell. 2015:14(2):162–169. 10.1111/acel.12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano S, Coll-Bonfill N, Teodoro-Castro B, Kuppa S, Jackson J, Shashkova E, Mahajan U, Vindigni A, Antony E, Gonzalo S. Lamin A/C recruits ssDNA protective proteins RPA and RAD51 to stalled replication forks to maintain fork stability. J Biol Chem. 2021:297(5):101301. 10.1016/j.jbc.2021.101301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves NR, Biel AM, Newman-Griffis AH, Meier I. Dynamic changes in plant nuclear organization in response to environmental and developmental signals. Plant Physiol. 2018:176(1):230–241. 10.1104/pp.17.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang B-H, Dong X. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell. 2016:166(6):1526–1538.e1511. 10.1016/j.cell.2016.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Hong B, Kong M, Zhao J. Application of ant colony algorithm in plant leaves classification based on infrared spectroscopy. AIP Conf Proc. 2014:1592(1):378–385. 10.1063/1.4872130 [DOI] [Google Scholar]
- Guo T, Mao X, Zhang H, Zhang Y, Fu M, Sun Z, Kuai P, Lou Y, Fang Y. Lamin-like proteins negatively regulate plant immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol Plant. 2017:10(10):1334–1348. 10.1016/j.molp.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Hernandez Sanchez-Rebato M, Bouatta AM, Gallego ME, White CI, Da Ines O. RAD54 is essential for RAD51-mediated repair of meiotic DSB in Arabidopsis. PLoS Genet. 2021:17(5):e1008919. 10.1371/journal.pgen.1008919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa T, Hasegawa J, White CI, Matsunaga S. RAD54 forms DNA repair foci in response to DNA damage in living plant cells. Plant J. 2017:90(2):372–382. 10.1111/tpj.13499 [DOI] [PubMed] [Google Scholar]
- Hirakawa T, Matsunaga S. Characterization of DNA repair foci in root cells of Arabidopsis in response to DNA damage. Front Plant Sci. 2019:10:990. 10.3389/fpls.2019.00990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, Ahmad Z, Sanchez-Perez GF, Perilli S, Blilou I, et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017:36(9):1261–1278. 10.15252/embj.201694561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang N, Bi X, Karaaslan ES, Weber AL, Zhu W, Berendzen KW, Liu C. Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 2019:20(1):87. 10.1186/s13059-019-1694-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhu S, Kumar P, MacMicking JD. A phase-separated nuclear GBPL circuit controls immunity in plants. Nature. 2021:594(7863):424–429. 10.1038/s41586-021-03572-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009:461(7267):1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jianbin Lai JJ, Wu Q, Mao N, Han D, Hu H, Yang C. The transcriptional coactivator ADA2b recruits a structural maintenance protein to double-strand breaks during DNA repair in plant. Plant Physiol. 2018:176(4):2613–2622. 10.1104/pp.18.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Mao N, Hu H, Tang J, Han D, Liu S, Wu Q, Liu Y, Peng C, Lai J, et al. A SWI/SNF subunit regulates chromosomal dissociation of structural maintenance complex 5 during DNA repair in plant cells. Proc Natl Acad Sci U S A. 2019:116(30):15288–15296. 10.1073/pnas.1900308116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020:585(7824):256–260. 10.1038/s41586-020-2644-7 [DOI] [PubMed] [Google Scholar]
- Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019:38(16):e101379. 10.15252/embj.2018101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Kuroda C, Masuda K. Differential nuclear envelope assembly at the end of mitosis in suspension-cultured Apium graveolens cells. Chromosoma. 2010:119(2):195–204. 10.1007/s00412-009-0248-y [DOI] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021:49(D1):D458–d460. 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhao Y, Yue M, Xue Y, Bao S. The protein arginine methylase 5 (PRMT5/SKB1) gene is required for the maintenance of root stem cells in response to DNA damage. J Genet Genomics. 2016:43(4):187–197. 10.1016/j.jgg.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999:98(3):329–339. 10.1016/S0092-8674(00)81962-5 [DOI] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015:60(2):208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu P, Ji T, Zheng L, Shen C, Ran S, Liu J, Zhao Y, Niu Y, Wang T, et al. Histone methyltransferase SUVR2 promotes the DSB repair via chromatin remodeling and liquid–liquid phase separation. Mol Plant. 2022:15(7):1157–1175. 10.1016/j.molp.2022.05.007 [DOI] [PubMed] [Google Scholar]
- Mahapatra K, Roy S. An insight into the mechanism of DNA damage response in plants—role of SUPPRESSOR OF GAMMA RESPONSE 1: an overview. Mutat Res. 2020:819–820:111689. 10.1016/j.mrfmmm.2020.111689 [DOI] [PubMed] [Google Scholar]
- Martín V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. Embo J. 2006:25(11):2564–2574. 10.1038/sj.emboj.7601141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Hikida R, Fujino K. The plant nuclear lamina proteins NMCP1 and NMCP2 form a filamentous network with lateral filament associations. J Exp Bot. 2021:72(18):6190–6204. 10.1093/jxb/erab243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Takahashi S, Nomura K, Arimoto M, Inoue M. Residual structure and constituent proteins of the peripheral framework of the cell nucleus in somatic embryos from Daucus carota L. Planta 1993:191(4):532–540. 10.1007/BF00195755 [DOI] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res. 1997:232(1):173–181. 10.1006/excr.1997.3531 [DOI] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014:6(9):a016428–a016428. 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Richards EJ, Evans DE. Cell biology of the plant nucleus. Annu Rev Plant Biol. 2017:68(1):139–172. 10.1146/annurev-arplant-042916-041115 [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj Anderson P, Kim Hong J, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015:163(1):123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Carr AM. Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol. 2008:9(2):177–182. 10.1038/nrm2309 [DOI] [PubMed] [Google Scholar]
- Nisa MU, Huang Y, Benhamed M, Raynaud C. The plant DNA damage response: signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front Plant Sci. 2019:10:653. 10.3389/fpls.2019.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek JJ. SMC5/6: multifunctional player in replication. Genes (Basel). 2018:10(1):7. 10.3390/genes10010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza-Garcia JA, Xiang Y, De Veylder L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. 2022:109(3):490–507. 10.1111/tpj.15567 [DOI] [PubMed] [Google Scholar]
- Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019:21(10):1286–1299. 10.1038/s41556-019-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat P, Boehning M, Hummel B, Aprile-Garcia F, Pandit AS, Eisenhardt N, Khavaran A, Niskanen E, Vos SM, Palvimo JJ, et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol Cell. 2021:81(5):1013–1026.e11. 10.1016/j.molcel.2021.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood AB, Perkins SM, Vanderwaal RP, Feng Z, Biehl KJ, Gonzalez-Suarez I, Morgado-Palacin L, Shi W, Sage J, Roti-Roti JL, et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011:10(15):2549–2560. 10.4161/cc.10.15.16531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Barnard S, Moquet J, Ellender M, Rana Z, Burdak-Rothkamm S. DNA damage foci: meaning and significance. Environ Mol Mutagen. 2015:56(6):491–504. 10.1002/em.21944 [DOI] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015:17(11):1401–1411. 10.1038/ncb3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu TH, Go YS, Choi SH, Kim JI, Chung BY, Kim JH. SOG1-dependent NAC103 modulates the DNA damage response as a transcriptional regulator in Arabidopsis. Plant J. 2019:98(1):83–96. 10.1111/tpj.14201 [DOI] [PubMed] [Google Scholar]
- Saar KL, Morgunov AS, Qi R, Arter WE, Krainer G, Lee AA, Knowles TPJ. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc Natl Acad Sci U S A. 2021:118(15). 10.1073/pnas.2019053118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Sakamoto Y, Grob S, Slane D, Yamashita T, Ito N, Oko Y, Sugiyama T, Higaki T, Hasezawa S, et al. Two-step regulation of centromere distribution by condensin II and the nuclear envelope proteins. Nat Plants. 2022:8(8):940–953. 10.1038/s41477-022-01200-3 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Sato M, Sato Y, Harada A, Suzuki T, Goto C, Tamura K, Toyooka K, Kimura H, Ohkawa Y, et al. Subnuclear gene positioning through lamina association affects copper tolerance. Nat Commun. 2020:11(1):5914. 10.1038/s41467-020-19621-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y. Nuclear lamina CRWN proteins regulate chromatin organization, gene expression, and nuclear body formation in plants. J Plant Res. 2020:133(4):457–462. 10.1007/s10265-020-01184-1 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM: expanding roles as a chief guardian of genome stability. Exp Cell Res. 2014:329(1):154–161. 10.1016/j.yexcr.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Song J, Durrant WE, Wang S, Yan S, Tan EH, Dong X. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe. 2011:9(2):115–124. 10.1016/j.chom.2011.01.011 [DOI] [PubMed] [Google Scholar]
- Song J, Keppler BD, Wise RR, Bent AF. PARP2 is the predominant poly(ADP-ribose) polymerase in Arabidopsis DNA damage and immune responses. PLoS Genet. 2015:11(5):e1005200. 10.1371/journal.pgen.1005200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampinato CP. Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell Mol Life Sci. 2017:74(9):1693–1709. 10.1007/s00018-016-2436-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998:122(1–2):42–66. 10.1006/jsbi.1998.3987 [DOI] [PubMed] [Google Scholar]
- Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016:51(3):195–212. 10.3109/10409238.2016.1172552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Dong Q, Wang T, Gong L, Gu Y. PNET2 is a component of the plant nuclear lamina and is required for proper genome organization and activity. Dev Cell. 2021:57(1):19–31. 10.1016/j.devcel.2021.11.002 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017:169(5):780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018:37(5). 10.15252/embj.201797452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang H, Huai J, Peng F, Wu J, Lin R, Fang X. Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat Chem Biol. 2022:18(12):1361–1369. 10.1038/s41589-022-01196-z [DOI] [PubMed] [Google Scholar]
- Wang L, Chen H, Wang C, Hu Z, Yan S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc Natl Acad Sci U S A. 2018:115(16):E3837–E3845. 10.1073/pnas.172009411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhan L, Zhao Y, Huang Y, Wu C, Pan T, Qin Q, Xu Y, Deng Z, Li J, et al. The ATR-WEE1 kinase module inhibits the MAC complex to regulate replication stress response. Nucleic Acids Res. 2021:49(3):1411–1425. 10.1093/nar/gkaa1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, La Y, Xia H, Zhou S, Zhai Z, La H. Roles of MEM1 in safeguarding Arabidopsis genome against DNA damage, inhibiting ATM/SOG1-mediated DNA damage response, and antagonizing global DNA hypermethylation. J Integr Plant Biol. 2021:64(1):87–104. 10.1111/jipb.13200 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu S, Lu C, La Y, Dai J, Ma H, Zhou S, Tan F, Wang X, Wu Y, et al. Roles of CRWN-family proteins in protecting genomic DNA against oxidative damage. J Plant Physiol. 2019:233:20–30. 10.1016/j.jplph.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu Q, Hu M, Liu B, Chai Z, Huang R, Wang Y, Xu H, Zhou L, Zheng L, et al. Ligand- and voltage-gated Ca (2+) channels differentially regulate the mode of vesicular neuropeptide release in mammalian sensory neurons. Sci Signal. 2017:10(484). 10.1126/scisignal.aal1683 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Wang H, Cheng Z, Li W, Zhu G, Wang Y, Ma H. The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol. 2014:201(1):292–304. 10.1111/nph.12498 [DOI] [PubMed] [Google Scholar]
- Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018:37(7). 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Demulder M, David P, Eekhout T, Yoshiyama KO, Nguyen L, Vercauteren I, Eeckhout D, Galle M, De Jaeger G, et al. Arabidopsis casein kinase 2 triggers stem cell exhaustion under Al toxicity and phosphate deficiency through activating the DNA damage response pathway. Plant Cell 2021:33(4):1361–1380. 10.1093/plcell/koab005 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, et al. The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 2016:35(19):2068–2086. 10.15252/embj.201593083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB. Assembly of mitotic structures through phase separation. J Mol Biol. 2018:430(23):4762–4772. 10.1016/j.jmb.2018.04.041 [DOI] [PubMed] [Google Scholar]
- Xie D, Chen M, Niu J, Wang L, Li Y, Fang X, Li P, Qi Y. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat Cell Biol. 2021:23(1):32–39. 10.1038/s41556-020-00606-5 [DOI] [PubMed] [Google Scholar]
- Yan Q, Xia X, Sun Z, Fang Y. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 2017:13(3):e1006663. 10.1371/journal.pgen.1006663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wang W, Marques J, Mohan R, Saleh A, Durrant WE, Song J, Dong X. Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol Cell. 2013:52(4):602–610. 10.1016/j.molcel.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chang Y, Qin Y, Chen D, Zhu T, Peng K, Wang H, Tang N, Li X, Wang Y, et al. A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. New Phytol. 2020:227(1):65–83. 10.1111/nph.16518 [DOI] [PubMed] [Google Scholar]
- Yoshiyama KO. SOG1: a master regulator of the DNA damage response in plants. Genes Genet Syst. 2016:90(4):209–216. 10.1266/ggs.15-00011 [DOI] [PubMed] [Google Scholar]
- Zamudio AV, Dall'Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, et al. Mediator condensates localize signaling factors to key cell identity genes. Mol Cell. 2019:76(5):753–766.e756. 10.1016/j.molcel.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Peng F, He C, Liu Y, Deng H, Fang X. Large-scale identification of potential phase-separation proteins from plants using a cell-free system. Mol Plant. 2022:16(2): 310–313. 10.1016/j.molp.2022.11.013 [DOI] [PubMed] [Google Scholar]
- Zhang L, Geng X, Wang F, Tang J, Ichida Y, Sharma A, Jin S, Chen M, Tang M, Pozo FM, et al. 53BP1 regulates heterochromatin through liquid phase separation. Nat Commun. 2022:13(1):360. 10.1038/s41467-022-28019-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006:1(2):641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhao S, Cheng L, Gao Y, Zhang B, Zheng X, Wang L, Li P, Sun Q, Li H. Plant HP1 protein ADCP1 links multivalent H3K9 methylation readout to heterochromatin formation. Cell Res. 2019:29(1):54–66. 10.1038/s41422-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Guan C, Feng J, Liang Y, Zhan N, Zuo J, Ren B. The /Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. J Integr Plant Biol. 2016:58(7):669–678. 10.1111/jipb.12448 [DOI] [PubMed] [Google Scholar]
- Zhou W, Gao J, Ma J, Cao L, Zhang C, Zhu Y, Dong A, Shen WH. Distinct roles of the histone chaperones NAP1 and NRP and the chromatin-remodeling factor INO80 in somatic homologous recombination in Arabidopsis thaliana. Plant J. 2016:88(3):397–410. 10.1111/tpj.13256 [DOI] [PubMed] [Google Scholar]
- Zhu L, Fernandez-Jimenez N, Szymanska-Lejman M, Pele A, Underwood CJ, Serra H, Lambing C, Dluzewska J, Bieluszewski T, Pradillo M, et al. Natural variation identifies SNI1, the SMC5/6 component, as a modifier of meiotic crossover in Arabidopsis. Proc Natl Acad Sci U S A. 2021:118(33). 10.1073/pnas.2021970118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Gu J, Yao J, Li Y, Zhang Z, Xia W, Wang Z, Gui X, Li L, Li D, et al. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev Cell. 2022:57(5):583–597.e6. 10.1016/j.devcel.2022.02.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.